Abstract
Noroviruses are a very diverse group of viruses that infect different mammalian species. In humans, norovirus is a major cause of acute gastroenteritis. Multiple norovirus infections can occur in a lifetime as the result of limited duration of acquired immunity and cross-protection among different strains. A combination of advances in sequencing methods and improvements on surveillance has provided new insights into norovirus diversification and emergence. The generation of diverse norovirus strains has been associated with (1) point mutations on two different genes: ORF1, encoding the non-structural proteins, and ORF2, encoding the major capsid protein (VP1); and (2) recombination events that create chimeric viruses. While both mechanisms are exploited by all norovirus strains, individual genotypes utilize each mechanism differently to emerge and persist in the human population. GII.4 noroviruses (the most prevalent genotype in humans) present an accumulation of amino acid mutations on VP1 resulting in the chronological emergence of new variants. In contrast, non-GII.4 noroviruses present co-circulation of different variants over long periods with limited changes on their VP1. Notably, genetic diversity of non-GII.4 noroviruses is mostly related to the high number of recombinant strains detected in humans. While it is difficult to determine the precise mechanism of emergence of epidemic noroviruses, observations point to multiple factors that include host-virus interactions and changes on two regions of the genome (ORF1 and ORF2). Larger datasets of viral genomes are needed to facilitate comparison of epidemic strains and those circulating at low levels in the population. This will provide a better understanding of the mechanism of norovirus emergence and persistence.
Keywords: caliciviruses, norovirus, gastroenteritis, evolution, recombination
1. Introduction
Human noroviruses are a major cause of acute gastroenteritis worldwide. Symptoms include vomiting, diarrhea, abdominal pain, and cramps that develop within 12–48 h after contact with contaminated food or infected persons. Norovirus illness is self-limiting in healthy individuals; however, it can be life-threatening in vulnerable populations like young or malnourished children, the elderly, or immunocompromised individuals (Trivedi et al. 2013; Green 2014). It is estimated that norovirus is responsible for approximately 200,000 deaths worldwide, with most of those cases occurring in children from developing countries (Lanata et al. 2013; Lopman et al. 2016).
Norovirus is highly transmissible, with most of the outbreaks occurring in semi-enclosed settings such as child care facilities, schools, universities, nursing homes, and cruise ships. Because norovirus outbreaks peak during the coldest months of the year in temperate regions, norovirus infections are also known as ‘winter vomiting disease’. This seasonality is less evident in tropical regions (Rohayem 2009; Green 2013).
Noroviruses present a large genetic diversity, with over thirty different genotypes infecting humans. While multiple different viruses can be found co-circulating, usually a single virus causes large epidemics and spreads to different countries. In this article, I will summarize the evidence for how changes on two viral proteins (the capsid protein and the polymerase) have played a role in the emergence and predominance of new noroviruses in humans. I will also discuss the importance of studies that include full-genome analyses and the role of the other viral proteins for our comprehensive understanding of norovirus transmission, immunity, and emergence.
2. The norovirus genome and structure
The norovirus genome is a single-stranded, positive-sense, polyadenylated, RNA molecule of ∼7.5 kb in size that is organized into three or four open reading frames (ORFs). The ORFs are flanked by 5′- and 3′-end untranslated regions. ORF1 encodes six non-structural (NS) proteins involved in viral replication, which are: N-Term (NS1/2), NTPase (NS3), 3A-like (NS4), VPg (NS5), Protease (NS6), and RNA-dependent RNA polymerase (RdRp; NS7). ORF2 and ORF3 encode for the major (VP1) and minor (VP2) capsid proteins, respectively (Fig. 1A) (Green 2013). Murine noroviruses present a fourth ORF that encodes for VF1, a protein involved in antagonism of the innate response (McFadden et al. 2011).
The norovirus virion is composed of 180 copies of VP1 (90 dimers) arranged in a T = 3 icosahedral symmetry (Prasad et al. 1994, 1999) (Fig. 1B). During natural infections most of the immune responses are elicited against VP1; therefore, this protein has been the major target for vaccine development (Atmar et al. 2016; Kim et al. 2018). Expression of VP1 results in self-assembly of virus-like particles (VLPs) that antigenically resemble the native virion (Jiang et al. 1992). Structural analyses have shown that norovirus VP1 is divided into two domains, shell and protruding. The shell domain forms the scaffold for the icosahedral capsid, while the protruding domain projects from the shell domain to the outermost part of the capsid (Prasad et al. 1999). The major antigenic sites and the site of interaction with cellular factors, namely histo-blood group antigens (HBGA), have been mapped on the protruding domain (Cao et al. 2007; Choi et al. 2008; Debbink et al. 2012b; Shanker et al. 2016; Tohma et al. 2019) (Fig. 1B). Differential display of HBGA in epithelial cells has been identified as a genetic correlate of protection against certain norovirus strains (Ramani, Estes, and Atmar 2016). Moreover, the presence of antibodies that block the interaction of norovirus VLPs with HBGA has been shown to correlate with disease protection in human volunteers challenged with norovirus (Reeck et al. 2010; Atmar et al. 2015). In the absence of a traditional cell culture system to grow noroviruses, the blocking of HBGA carbohydrates by norovirus-specific serum has been considered a surrogate of norovirus neutralization in vaccine design (Atmar et al. 2016; Ramani, Estes, and Atmar 2016; Kim et al. 2018). Recently, using the stem cell-derived enteroids that support replication of human norovirus, Alvarado and colleagues have shown that human monoclonal antibodies with HBGA blocking activity are capable of neutralizing human norovirus (Ettayebi et al. 2016; Alvarado et al. 2018).
Figure 1.
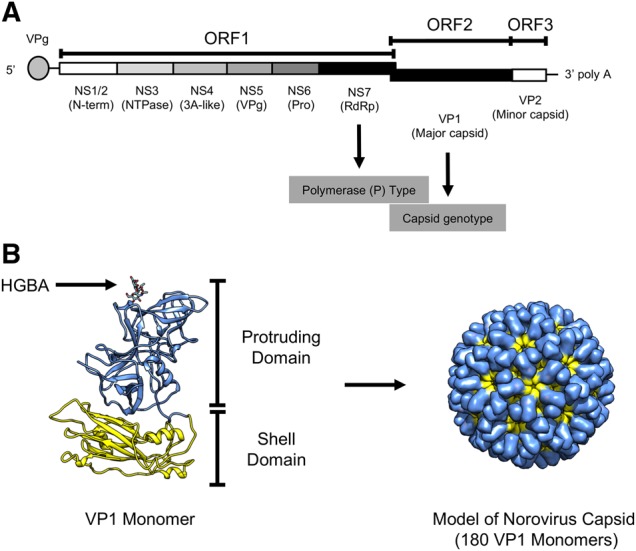
Norovirus structure and genome organization. (A) The ORFs and their encoded proteins are shown. ORF1 encodes six NS proteins involved in viral replication, ORF2 and ORF3 encode for the major (VP1) and minor (VP2) capsid proteins, respectively. The 5′-end of the genome is capped with the VPg (virion protein genome-linked) protein, while the 3′-end consists of an untranslated region and a poly-A tail. Genome regions utilized for norovirus characterization and typing include the RdRp and the major capsid protein (VP1). (B) Structural model of norovirus VP1 showing the protruding and shell domains. A model of the capsid (T:3) is shown at the right-side of the VP1. The molecular model of the VP1 was visualized using an X-ray solved structure (Protein Data Bank record: 1IHM) and rendered in Chimera (Pettersen et al. 2004).
3. Norovirus genotypes present host specificity
Norovirus characterization (typing) has been traditionally done based on sequence diversity within the capsid protein (Fig. 1). Thus, noroviruses can be classified into at least ten genogroups (GI-GX) and more than forty different genotypes (Fig. 2). Although the classification is done using phylogenetic distances (Chhabra et al. 2019), in general genogroups differ by about 40–60 percent of their amino acid sequence and genotypes by about 20–40 percent. Genotypes can be further divided into variants (Parra et al. 2017). Recently, the typing system for noroviruses has been revised, and the use of the RdRp-encoding region for dual typing of norovirus was updated (Chhabra et al. 2019). Thus, strains are designated by their genotype and P type, for example GI.1[P1].
Figure 2.
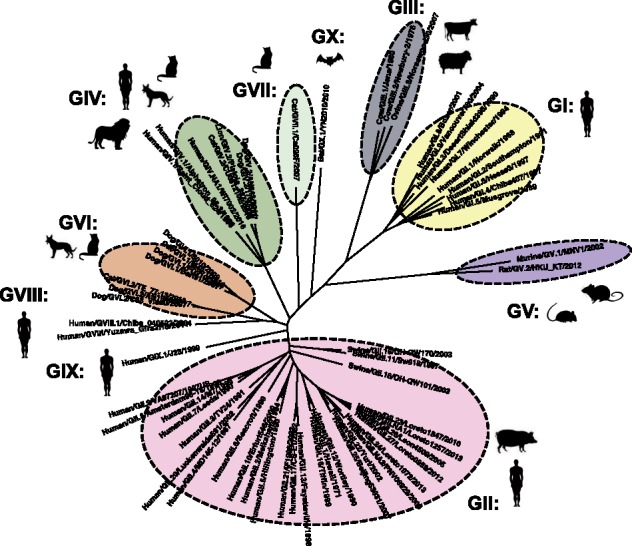
Classification of noroviruses based on the phylogeny of the major capsid protein (VP1). Genogroups are based on phylogenetic clustering and amino acid differences. Genogroups can be further divided into genotypes. Each genogroup is indicated with different colors and associated with infection of specific species (indicated by shadow figures). Viruses from genotypes GII.11, GII.18, and GII.19 infect porcine species, and viruses from GIV.1 and GIV.NA1 infect humans. Abbreviation for species: Bo, bovine; Ca, canine; Fe, feline; Hu, human; Mu, murine; Ov, ovine; Sw, swine. The phylogenetic tree was constructed using representative strains from each genotype and/or genogroup and the neighbor-joining method as implemented in MEGA (Kumar, Stecher, and Tamura 2016).
Despite that humans can be infected by multiple different noroviruses (>30 genotypes), usually a single virus causes large epidemics and is spread worldwide. The origin of these constantly changing pandemic noroviruses is unknown. Because noroviruses have been detected in different mammalian species (Chhabra et al. 2019), one hypothesis is that animals could be the source of these new viruses. However, while some evidence suggests possible inter-species transmission (Summa, von Bonsdorff, and Maunula 2012; Caddy et al. 2014, 2015), the different noroviruses exhibit marked host specificity. GI and GII viruses have been mostly associated with human infections (Green 2013), GIII viruses with bovine and ovine species (Oliver et al. 2004; Wolf et al. 2009), GV viruses with murine species (mice and rats) (Karst et al. 2003; Smith et al. 2012), and GIV, GVI, and GVII viruses with different species of carnivores (canine and felines) (Martella et al. 2008; Di Martino et al. 2016; Ford-Siltz et al. 2019). Two new genogroups (GVIII and GIX) have been described in humans, and one genogroup (GX) in bats (Chhabra et al. 2019) (Fig. 2). Some exceptions to the genogroup species specificity have been documented: Three GII genotypes (GII.11, GII.18, and GII.19) that infect pigs (Wang et al. 2005) have not been found in humans, and two GIV genotypes (GIV.1 and GIV.NA1) have only been found in human samples, and not in felines or canines. Although some studies have reported the presence of viral RNA from human strains in various animals (van Der Poel et al. 2000; Mattison et al. 2007; Nakamura et al. 2010; Summa, von Bonsdorff, and Maunula 2012; Farkas 2016), human strains present limited replication in animals (Souza et al. 2007, 2008; Bok et al. 2011; Jung et al. 2012). This marked species specificity has hampered the development of a robust animal model for human noroviruses. While the basis of norovirus species specificity is not completely understood and might be multifactorial, Haga and colleagues recently showed that expression of the murine norovirus receptor (molecules from the CD300 family) makes non-murine mammalian cells susceptible to murine norovirus (GV) infection (Haga et al. 2016), suggesting that the VP1–receptor interaction plays a major role in host specificity. In that regard, large differences have been mapped to the P2 subdomain of the VP1, which interacts with cellular factors, when comparing norovirus strains infecting different species (Singh et al. 2015; Ford-Siltz et al. 2019). Thus, based on the lack of conclusive data supporting inter-species transmission and the species-specific characteristics of VP1 (Fig. 2), the continuous emergence of new viruses to the human population through zoonotic events seems unlikely.
4. The role of immunocompromised individuals in norovirus evolution and transmission
In immunocompetent individuals, norovirus can be shed in stool weeks after symptoms have been resolved (Atmar et al. 2008, 2015; Green 2013; Parra and Green 2014; Saito et al. 2014). On the contrary, immunocompromised individuals can be chronically infected with norovirus for months or years, resulting in a high burden on this vulnerable population. Notably, during this chronic phase norovirus presents an extreme diversity that is not seen during the shedding phase in immunocompetent individuals (Bull et al. 2012; Green 2014; de Graaf, van Beek, and Koopmans 2016; van Beek et al. 2017). Thus, it was hypothesized that immunocompromised individuals could act as reservoirs for novel norovirus strains (Vega et al. 2014b; Karst and Baric 2015). While some experimental data have shown that new antigenic variants could emerge in these individuals (Debbink et al. 2014; Vega et al. 2014b), there is no direct evidence that immunocompromised individuals could act as sources of infection to healthy individuals. Rather, immunocompromised individuals seem to acquire new viruses upon contact with healthy individuals infected with viruses circulating in the population (Bok et al. 2016; Brown et al. 2019). Notably, Eden and colleagues recently showed, using noroviruses as a model, that immunocompromised individuals might play a minimal role in the global emergence of new viruses (Eden et al. 2017).
While the source and reservoir of novel norovirus strains is yet to be identified, norovirus diversity could be originated at inter- and intra-host levels in healthy populations. Thus, mutations could arise during transmission events in outbreak settings and/or during the shedding phase in healthy individuals (Bull et al. 2012; Cotten et al. 2014; Johnson et al. 2017; Parra et al. 2017), and probably persist in the population. Larger-studies in that regard could provide information on the emergence of new noroviruses.
5. Norovirus genotypes exhibit different evolutionary patterns on their capsid proteins
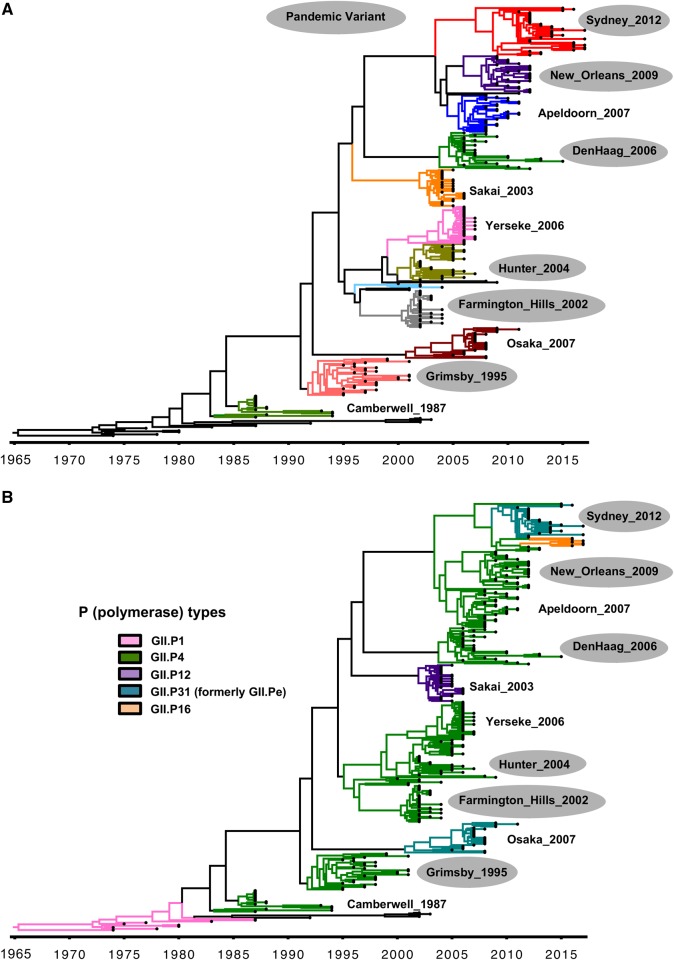
As the result of limited cross-protection among the different genotypes (Parra and Green 2014; Parra et al. 2017), individuals can undergo multiple norovirus infections throughout life (Saito et al. 2014). Notably, epidemiological observations coupled with phylogenetic analyses of all human norovirus genotypes suggest restrictions in which genotypes that cluster together could have some degree of cross-protection, thus reducing the number of types that individuals could be infected during a lifetime (Parra et al. 2017). While most genotypes circulate with variable incidences, a single genotype (GII.4) is the most prevalent worldwide (van Beek et al. 2018). The mechanisms for the persistence and dominance of the GII.4 viruses are not fully understood. The current working model is based on the epochal evolution of influenza A viruses (Koelle et al. 2006). Under this hypothesis, new GII.4 viruses chronologically emerge escaping from the immune responses developed against previous variants (Siebenga et al. 2007; Lindesmith et al. 2008; White 2014), and persist for years—without major phenotypic changes—until replaced by the next variant (Tohma et al. 2019). Thus, since the mid-1990s, six GII.4 variants (Grimsby_1996, Farmington_Hills_2002, Hunter_2004, Den_Haag_2006, New_Orleans_2009, and Sydney_2012) have emerged and caused global epidemics (Vinje, Altena, and Koopmans 1997; Siebenga et al. 2009; van Beek et al. 2013, 2018; Eden et al. 2014; Vega et al. 2014a) (Fig. 3A). Even though the shift of the predominant variants can occur rapidly (2–3 years), the Grimsby_1996 and Sydney_2012 variants each predominated for over 5 years in the human population (Parra et al. 2017; van Beek et al. 2018). Notably, contrary to influenza H3N2 viruses, the topology of the phylogenetic tree of VP1 suggests that each norovirus GII.4 variant presents a different origin (Tohma et al. 2019).
Figure 3.
Evolution of GII.4 noroviruses. (A) Maximum clade credibility (MCC) tree showing the circulation of different variants over time. Variants are identified by different colors and names. (B) Same phylogenetic tree indicating the RdRp (P) types associated with each variant. Variants that caused large epidemics and spread worldwide are indicated with gray shadows. The MCC tree was constructed using the BEAST package (Drummond et al. 2012) and visualized in FigTree v1.4.3. For tree reconstruction, thirty strains were randomly selected per variant, except for the Sydney_2012 variant in which forty strains were used (Tohma et al. 2019).
The major differences among these GII.4 variants have been mapped onto antigenic sites (A–G) within the Protruding domain (Tohma et al. 2019). Mouse- and human-derived antibodies against these sites have been shown to be involved in blocking the interaction of HBGA and GII.4 VLPs, supporting the role of immune pressure as a driver of GII.4 selection and evolution (Allen et al. 2009; Debbink et al. 2012a,b; Lindesmith et al. 2012a,b, 2013; Parra et al. 2012; Tohma et al. 2019). Recently, amino acids mapping to three of these antigenic sites (A, C, and G) have been linked to the emergence of new GII.4 variants (Tohma et al. 2019). This, together with the fact that variants have been shown to circulate prior to becoming the predominant strain, opens the opportunity to monitor and predict the emergence of new pandemic viruses (Eden et al. 2014; Tohma et al. 2019). Improvements on surveillance systems that include the sequencing of a large number of strains is needed to facilitate these efforts as pre-pandemic viruses might be circulating at low prevalence (Tohma et al. 2019).
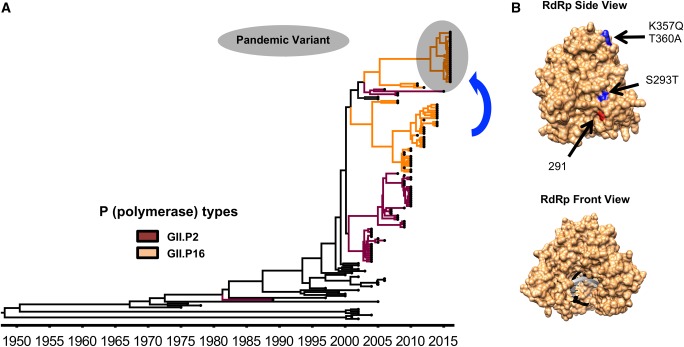
In contrast to GII.4 viruses, which can accumulate mutations and periodically replace antigenic variants, all other norovirus genotypes have a limited number of variants that can persist for decades with minimal modification in their VP1 (Parra et al. 2017). Although this evolutionary pattern restricts their overall prevalence, non-GII.4 viruses can also cause large outbreaks and—transiently—prevail over GII.4 viruses. This is evidenced by the recent emergence and increased incidence of two noroviruses, GII.17 and GII.2, in different countries (China, Japan, Hong Kong, Germany, Italy) (Chan et al. 2015b; Matsushima et al. 2015; Parra and Green 2015; Kwok et al. 2017; Lu et al. 2017; Niendorf et al. 2017; Medici et al. 2018; Nagasawa et al. 2018). Notably, these two viruses are antigenically distinct to GII.4 (Dai et al. 2017), and the emergence of epidemic GII.2 strains appears to be associated with changes in the RdRp (Matsushima et al. 2015; Tohma et al. 2017) (Fig. 4), highlighting the role of NS proteins in the evolution and emergence of predominant viruses.
Figure 4.
Evolution of GII.2 noroviruses. (A) MCC tree showing the circulation of different GII.2 noroviruses over time (Tohma et al. 2017). Branches with strains associated with the RdRp types GII.P2 and GII.P16 are indicated with dark red and orange, respectively. GII.2P[16] strains that caused large epidemics and spread worldwide are indicated with a gray shadow. The MCC tree was constructed using the BEAST package (Drummond et al. 2012) and visualized in FigTree v1.4.3. (B) A structural model of the norovirus RdRp is shown indicating the substitutions (blue) presented by the GII.2[P16] strains that predominated during 2016–17 in different countries (Kwok et al. 2017; Lu et al. 2017; Tohma et al. 2017). Substitutions on residue 291 (red) were shown to alter the mutation rate of GII.4 polymerases (Bull et al. 2010). The molecular model of the RdRp was visualized using an X-ray solved structure of norovirus RdRp (Protein Data Bank record: 4QPX) and was rendered in Chimera (Pettersen et al. 2004).
6. Non-structural proteins as drivers of emergence of new noroviruses
Similar to the VP1 encoding region (ORF2), ORF1 presents a genetic diversity that results in over sixty different types (Chhabra et al. 2019). Moreover, because noroviruses are prone to recombine at the boundary of ORF1/ORF2 (Bull et al. 2005; Bull, Tanaka, and White 2007), chimeric noroviruses, with different VP1 genotype and RdRp types, can be found (Ludwig-Begall, Mauroy, and Thiry 2018). Almost all human norovirus genotypes present recombinant viruses, with GII.2 and GII.3 noroviruses displaying the larger diversity of recombinant forms described up to date (Mahar et al. 2013; Tohma et al. 2017; Ludwig-Begall, Mauroy, and Thiry 2018).
The evolution and predominance of GII.4 strains constitute one of the best examples to illustrate the major role that changes in the NS proteins, by means of recombination, has played on the evolution and diversification of noroviruses. Analysis of archival samples has shown that GII.4 strains circulating in the 1970s and 1980s presented a GII.P1 RdRp type; however, in the late-1980s the GII.P1 RdRp was replaced by a GII.P4 RdRp (Bok et al. 2009; Mori et al. 2017; Parra et al. 2017) (Fig. 3B). Since then, GII.4[P4] viruses have been associated with five out of the six large outbreaks reported worldwide (van Beek et al. 2018) (Fig. 3A). It has been suggested that one of the reasons for the predominance of GII.4[P4] viruses is the higher mutation rate presented by GII.P4 RdRp over other norovirus polymerases (Bull et al. 2010; White 2014). While these differences on the mutational rate could provide a higher diversity at a given time, it seems to have limited impact on the overall evolutionary rate as all genotypes present similar evolutionary rates despite differences in their polymerases (P types) (Parra et al. 2017; Tohma et al. 2017, 2018). The last pandemic variant (GII.4_Sydney_2012) emerged in 2012 and was associated with the GII.P31 (formerly GII.Pe) type (Eden et al. 2013), and recently a new recombinant virus (GII.4_Sydney_2012[P16]) emerged, causing large number of gastroenteritis cases in different countries (Bidalot et al. 2017; Cannon et al. 2017; Ruis et al. 2017; Medici et al. 2018) (Fig. 3B). The emergence of the GII.4_Sydney_2012[P16] strain coincided with the predominance of GII.2[P16] viruses in different Asian and European countries (Cannon et al. 2017; Kwok et al. 2017; Niendorf et al. 2017; Medici et al. 2018; Nagasawa et al. 2018) (Fig. 4). These GII.2[P16] viruses evolved, with no specific changes in their VP1, from GII.2[P16] viruses that circulated during 2011–12. Only four conservative substitutions in the RdRp were found among the pre- and post-epidemic GII.2 strains (Tohma et al. 2017), which could have provided different characteristics to the epidemic viruses, again illustrating the role of NS proteins in norovirus emergence and predominance. Since GII.2[P16] strains pre-dated the GII.4_Sydney_2012[P16], it is most likely that the latter is a recombinant virus that originated from the parental pre-2016 Sydney_2012 viruses (van Beek et al. 2018).
Another example that highlights the role of recombination and/or NS proteins on norovirus disease is the emergence and sudden predominance of GII.17 strains in different Asian countries during 2013–15 (Chan et al. 2015b; Matsushima et al. 2015; Lu et al. 2016; Pan et al. 2016). Few GII.17 viruses circulating before 2013 have been detected, and those have been represented by two variants (GII.17_A and GII.17_B) that cluster into different VP1 lineages and were associated with different RdRp types: GII.P31 (formerly GII.Pe), GII.P4, GII.P16, and GII.P13 (Chan et al. 2015a; Matsushima et al. 2015; Parra and Green 2015; Tohma et al. 2018). The predominance of the GII.17 viruses during 2013–15 was associated with the emergence of a novel VP1 lineage and a new RdRp type, GII.P17 (Matsushima et al. 2015; Parra and Green 2015). Interestingly, using archival samples, Mori et al. showed that viruses similar to those emerging in 2013 (GII.17[P.17]) have been circulating since the 1970s in the human population (Mori et al. 2017). In addition to mutations in VP1, the emerging GII.17[P17] viruses also had a mutation (T293A) within the RdRp that resembles the one (S293T) presented by the GII.P16 strains that emerged and predominated in 2016 (Fig. 4). Thus, slow accumulation of mutations could have resulted in GII.17 viruses with novel biological properties that facilitated infection and transmission. Although the role of these mutations on norovirus biology needs to be explored further, different groups have shown that single-point mutations can affect the biological properties of the norovirus RdRp, which varied from increasing transmissibility of the virus to changing biochemical characteristics that affect replication (Bull et al. 2010; Arias et al. 2016). Thus, although some noroviruses explore a large number of capsid/polymerase combinations (e.g. GII.2 and GII.3), subtle changes might be sufficient to increase the fitness and epidemic potential of norovirus.
One important aspect of the evolution and epidemiology of noroviruses that merits further attention is that while some GII.4 and GII.17 variants predominated and spread worldwide, other variants sharing the same RdRp types only circulated at low levels or have been associated with geographically restricted epidemics (Fig. 3). Two notable examples are: (1) GII.4 variants that presented a GII.P4 type (GII.4_Sakai_2003, GII.4_Yerseke_2006, or GII.4_Apeldorn_2007), but did not became pandemic, and (2) variants (GII.4_Osaka_2007 and GII.17_A) that presented the GII.P31 (formerly GII.Pe) type but never reached the predominance showed by GII.4_Sydney_2012 strains. Thus, a better understanding of the lack of success of these strains to spread worldwide could provide critical information on the control of pandemic noroviruses. Studies designed to understand the role of NS proteins on immunity and transmissibility are warranted as little has been explored in that regards for human noroviruses (Ajami et al. 2012; White 2014; Hesse et al. 2016; Tohma et al. 2017; Ao et al. 2018).
7. Conclusions and final remarks
With the successful implementation of rotavirus vaccines in developed countries, noroviruses have become the most important cause of viral gastroenteritis in infants and children. These viruses exhibit a plasticity on their genome that could result in multiple variants co-circulating at the same time in the human population. Although it is difficult to determine the precise mechanism of emergence of a novel norovirus, all observations seem to point to a multifactorial event that involves host-virus interactions and changes on, at least, two different regions of the genome (ORF1 and ORF2).
Several works have utilized large datasets of full-genome sequences to study transmission and predominance of human noroviruses (Motomura et al. 2008; Kundu et al. 2013; Cotten et al. 2014; Brown et al. 2017a,b; Mizukoshi et al. 2017; Ruis et al. 2017; van Beek et al. 2017; Wang et al. 2018; Brown et al. 2019); however, very little effort has been made to determine the role of the other NS (NS1-6) on their emergence and epidemic significance (Ao et al. 2018). Improved surveillance systems (Cannon et al. 2017; van Beek et al. 2018) and novel cost-effective platforms recently developed for (nearly) full-length genome sequencing of noroviruses (Cotten et al. 2014; Vega et al. 2014b; Brown et al. 2016; Cotten and Koopmans 2016; Parra et al. 2017) would facilitate studies at the population and intra-host level. A better understanding of the mechanism of norovirus emergence and persistence could inform on the design of control strategies for norovirus.
Acknowledgements
The author would like to thank Kentaro Tohma, Lauren Ford-Siltz, and Robin Levis (DVP, FDA) for helping with figures and critical reading of the manuscript. Thank you to the Journal Editor for editorial assistance.
Funding
Financial support for this work was provided by the Food and Drug Administration intramural funds [Program Number Z01 BK 04012-01 LHV to G.I.P.].
Conflict of interest: None declared.
References
- Ajami N. J. et al. (2012) ‘Antibody Responses to Norovirus Genogroup GI.1 and GII.4 Proteases in Volunteers Administered Norwalk Virus’, Clinical and Vaccine Immunology, 19: 1980–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D. J. et al. (2009) ‘Characterisation of a GII-4 Norovirus Variant-specific Surface-exposed Site Involved in Antibody Binding’, Virology Journal, 6: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado G. et al. (2018) ‘Human Monoclonal Antibodies That Neutralize Pandemic GII.4 Noroviruses’, Gastroenterology, 155: 1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Y. et al. (2018) ‘Genetic Analysis of Reemerging GII.P16-GII.2 Noroviruses in 2016–2017 in China’, The Journal of Infectious Diseases, 218: 133–43. [DOI] [PubMed] [Google Scholar]
- Arias A. et al. (2016) ‘Polymerase Fidelity Contributes to Viral Transmission In Vivo’, mSphere, 1: e00279–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R. L. et al. (2008) ‘Norwalk Virus Shedding after Experimental Human Infection’, Emerging Infectious Diseases, 14: 1553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R. L. et al. (2015) ‘Serological Correlates of Protection against a GII.4 Norovirus’, Clinical and Vaccine Immunology, 22: 923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R. L. et al. (2016) ‘Rapid Responses to 2 Virus-like Particle Norovirus Vaccine Candidate Formulations in Healthy Adults: A Randomized Controlled Trial’, TheJournal of Infectious Diseases, 214: 845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidalot M. et al. (2017) ‘Emergence of New Recombinant Noroviruses GII.p16-GII.4 and GII.p16-GII.2, France, Winter 2016 to 2017’, Euro Surveill, 22: 30508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok K. et al. (2009) ‘Evolutionary Dynamics of GII.4 Noroviruses over a 34-year Period’, Journal of Virology, 83: 11890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok K. et al. (2011) ‘Chimpanzees as an Animal Model for Human Norovirus Infection and Vaccine Development’, Proceedings of the National Academy of Sciences of the United States of America, 108: 325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok K. et al. (2016) ‘Epidemiology of Norovirus Infection among Immunocompromised Patients at a Tertiary Care Research Hospital, 2010–2013’, Open Forum Infectious Diseases, 3: ofw169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R. et al. (2016) ‘Norovirus Whole-genome Sequencing by SureSelect Target Enrichment: A Robust and Sensitive Method’, Journal of Clinical Microbiology, 54: 2530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R. et al. (2017b) ‘Super-infections and Relapses Occur in Chronic Norovirus Infections’, Journal of Clinical Virology, 96: 44–8. [DOI] [PubMed] [Google Scholar]
- Brown L. K. et al. (2017a) ‘Norovirus Infection in Primary Immune Deficiency’, Reviews in Medical Virology, 27: e1926. [DOI] [PubMed] [Google Scholar]
- Brown J. R. et al. (2019) ‘Norovirus Transmission Dynamics in a Pediatric Hospital Using Full Genome Sequences’, Clinical Infectious Diseases, 68: 222–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R. A. et al. (2005) ‘Norovirus Recombination in ORF1/ORF2 Overlap’, Emerging Infectious Diseases, 11: 1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R. A., Tanaka M. M., White P. A. (2007) ‘Norovirus Recombination’, The Journal of General Virology, 88: 3347–59. [DOI] [PubMed] [Google Scholar]
- Bull R. A. et al. (2010) ‘Rapid Evolution of Pandemic Noroviruses of the GII.4 Lineage’, PLoS Pathogens, 6: e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R. A. et al. (2012) ‘Contribution of Intra- and Interhost Dynamics to Norovirus Evolution’, Journal of Virology, 86: 3219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddy S. et al. (2014) ‘Genogroup IV and VI Canine Noroviruses Interact with Histo-blood Group Antigens’, Journal of Virology, 88: 10377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddy S. L. et al. (2015) ‘Evidence for Human Norovirus Infection of Dogs in the United Kingdom’, Journal of Clinical Microbiology, 53: 1873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J. L. et al. (2017) ‘Genetic and Epidemiologic Trends of Norovirus Outbreaks in the United States from 2013 to 2016 Demonstrated Emergence of Novel GII.4 Recombinant Viruses’, Journal of Clinical Microbiology, 55: 2208–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S. et al. (2007) ‘Structural Basis for the Recognition of Blood Group Trisaccharides by Norovirus’, Journal of Virology, 81: 5949–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. C. et al. (2015a) ‘Complete Genome Sequence of a Novel Recombinant GII.Pe_GII.17 Norovirus Strain from Hong Kong in 2015’, Genome Announcements, 3: e01338–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M. C. et al. (2015b) ‘Rapid Emergence and Predominance of a Broadly Recognizing and Fast-evolving Norovirus GII.17 Variant in Late 2014’, Nature Communications, 6: 10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra P. et al. (2019) ‘Updated Classification of Norovirus Genogroups and Genotypes’, Journal of General Virology, 100: 1393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. M. et al. (2008) ‘Atomic Resolution Structural Characterization of Recognition of Histo-blood Group Antigens by Norwalk Virus’, Proceedings of the National Academy of Sciences of the United States of America, 105: 9175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M. et al. (2014) ‘Deep Sequencing of Norovirus Genomes Defines Evolutionary Patterns in an Urban Tropical Setting’, Journal of Virology, 88: 11056–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Koopmans M. (2016) ‘Next-generation Sequencing and Norovirus’, Future Virology, 11: 719–22. [DOI] [PubMed] [Google Scholar]
- Dai Y. C. et al. (2017) ‘Characterization of Antigenic Relatedness between GII.4 and GII.17 Noroviruses by Use of Serum Samples from Norovirus-infected Patients’, Journal of Clinical Microbiology, 55: 3366–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K. et al. (2012a) ‘Genetic Mapping of a Highly Variable Norovirus GII.4 Blockade Epitope: Potential Role in Escape from Human Herd Immunity’, Journal of Virology, 86: 1214–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K. et al. (2012b) ‘Norovirus Immunity and the Great Escape’, PLoS Pathogens, 8: e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K. et al. (2014) ‘Within-host Evolution Results in Antigenically Distinct GII.4 Noroviruses’, Journal of Virology, 88: 7244–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M., van Beek J., Koopmans M. P. (2016) ‘Human Norovirus Transmission and Evolution in a Changing World’, Nature Reviews Microbiology, 14: 421–33. [DOI] [PubMed] [Google Scholar]
- Di Martino B. et al. (2016) ‘A Novel Feline Norovirus in Diarrheic Cats’, Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 38: 132–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. et al. (2012) ‘Bayesian Phylogenetics with BEAUti and the BEAST 1.7’, Molecular Biology and Evolution, 29: 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J. S. et al. (2013) ‘Recombination within the Pandemic Norovirus GII.4 Lineage’, Journal of Virology, 87: 6270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J. S. et al. (2014) ‘The Emergence and Evolution of the Novel Epidemic Norovirus GII.4 Variant Sydney 2012’, Virology, 450–451: 106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J. S. et al. (2017) ‘Persistent Infections in Immunocompromised Hosts are Rarely Sources of New Pathogen Variants’, Virus Evolution, 3: vex018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettayebi K. et al. (2016) ‘Replication of Human Noroviruses in Stem Cell-derived Human Enteroids’, Science, 353: 1387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T. (2016) ‘Natural Norovirus Infections in Rhesus Macaques’, Emerging Infectious Diseases, 22: 1272–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford-Siltz L. A. et al. (2019) ‘Genomics Analyses of GIV and GVI Noroviruses Reveal the Distinct Clustering of Human and Animal Viruses’, Viruses, 11: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. Y. (2013) ‘Caliciviridae: The Noroviruses’, in Knipe D. M., Howley P. M. (eds.) Fields Virology, Philadelphia, PA: Lippincott, Williams & Wilkins. [Google Scholar]
- Green K. Y. (2014) ‘Norovirus Infection in Immunocompromised Hosts’, Clinical Microbiology and Infection, 20: 717–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K. et al. (2016) ‘Functional Receptor Molecules CD300lf and CD300ld within the CD300 Family Enable Murine Noroviruses to Infect Cells’, Proceedings of the National Academy of Sciences of the United States of America, 113: E6248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse S. et al. (2016) ‘Serological Responses to a Norovirus Nonstructural Fusion Protein after Vaccination and Infection’, Clinical and Vaccine Immunology, 23: 181–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. et al. (1992) ‘Expression, Self-assembly, and Antigenicity of the Norwalk Virus Capsid Protein’, Journal of Virology, 66: 6527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. A. et al. (2017) ‘A Large Outbreak of Acute Gastroenteritis in Shippensburg, Pennsylvania, 1972 Revisited: Evidence for Common Source Exposure to a Recombinant GII.Pg/GII.3 Norovirus’, Epidemiology and Infection, 145: 1591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. et al. (2012) ‘The Effects of Simvastatin or Interferon-alpha on Infectivity of Human Norovirus Using a Gnotobiotic Pig Model for the Study of Antivirals’, PLoS One, 7: e41619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S. M. et al. (2003) ‘STAT1-dependent Innate Immunity to a Norwalk-like Virus’, Science (New York, N.Y.), 299: 1575–8. [DOI] [PubMed] [Google Scholar]
- Karst S. M., Baric R. S. (2015) ‘What is the Reservoir of Emergent Human Norovirus Strains?’, Journal of Virology, 89: 5756–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L. et al. (2018) ‘Safety and Immunogenicity of an Oral Tablet Norovirus Vaccine, a Phase I Randomized, Placebo-controlled Trial’, JCI Insight, 3: 121077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle K. et al. (2006) ‘Epochal Evolution Shapes the Phylodynamics of Interpandemic Influenza A (H3N2) in Humans’, Science, 314: 1898–903. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016) ‘MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets’, Molecular Biology and Evolution, 33: 1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S. et al. (2013) ‘Next-generation Whole Genome Sequencing Identifies the Direction of Norovirus Transmission in Linked Patients’, Clinical Infectious Diseases, 57: 407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok K. et al. (2017) ‘Increased Detection of Emergent Recombinant Norovirus GII.P16-GII.2 Strains in Young Adults, Hong Kong, China, 2016–2017’, Emerging Infectious Diseases, 23: 1852–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanata C. F. et al. (2013) ‘Global Causes of Diarrheal Disease Mortality in Children <5 Years of Age: A Systematic Review’, PLoS One, 8: e72788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L. C. et al. (2008) ‘Mechanisms of GII.4 Norovirus Persistence in Human Populations’, PLoS Medicine, 5: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L. C. et al. (2012a) ‘Immunogenetic Mechanisms Driving Norovirus GII.4 Antigenic Variation’, PLoS Pathogens, 8: e1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L. C. et al. (2012b) ‘Monoclonal Antibody-based Antigenic Mapping of Norovirus GII.4-2002’, Journal of Virology, 86: 873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L. C. et al. (2013) ‘Emergence of a Norovirus GII.4 Strain Correlates with Changes in Evolving Blockade Epitopes’, Journal of Virology, 87: 2803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman B. A. et al. (2016) ‘The Vast and Varied Global Burden of Norovirus: Prospects for Prevention and Control’, PLoS Medicine, 13: e1001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2016) ‘The Evolution and Transmission of Epidemic GII.17 Noroviruses’, The Journal of Infectious Diseases, 214: 556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2017) ‘Association of GII.P16-GII.2 Recombinant Norovirus Strain with Increased Norovirus Outbreaks, Guangdong, China, 2016’, Emerging Infectious Diseases, 23: 1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Begall L. F., Mauroy A., Thiry E. (2018) ‘Norovirus Recombinants: Recurrent in the Field, Recalcitrant in the Lab—A Scoping Review of Recombination and Recombinant Types of Noroviruses’, Journal of General Virology, 99: 970–88. [DOI] [PubMed] [Google Scholar]
- Mahar J. E. et al. (2013) ‘The Importance of Intergenic Recombination in Norovirus GII.3 Evolution’, Journal of Virology, 87: 3687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V. et al. (2008) ‘Detection and Molecular Characterization of a Canine Norovirus’, Emerging Infectious Diseases, 14: 1306–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y. et al. (2015) ‘Genetic Analyses of GII.17 Norovirus Strains in Diarrheal Disease Outbreaks from December 2014 to March 2015 in Japan Reveal a Novel Polymerase Sequence and Amino Acid Substitutions in the Capsid Region’, Euro Surveill, 20: 21173. [DOI] [PubMed] [Google Scholar]
- Mattison K. et al. (2007) ‘Human Noroviruses in Swine and Cattle’, Emerging Infectious Diseases, 13: 1184–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden N. et al. (2011) ‘Norovirus Regulation of the Innate Immune Response and Apoptosis Occurs via the Product of the Alternative Open Reading Frame 4’, PLoS Pathogens, 7: e1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici M. C. et al. (2018) ‘Emergence of Novel Recombinant GII.P16_GII.2 and GII. P16_GII.4 Sydney 2012 Norovirus Strains in Italy, Winter 2016/2017’, New Microbiologica, 41: 71–2. [PubMed] [Google Scholar]
- Mizukoshi F. et al. (2017) ‘Molecular Evolution of the RNA-dependent RNA Polymerase and Capsid Genes of Human Norovirus Genotype GII.2 in Japan during 2004–2015’, Frontiers in Microbiology, 8: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. et al. (2017) ‘Comparison of Genetic Characteristics in the Evolution of Norovirus GII.4 and GII.17’, Journal of Medical Virology, 89: 1480–4. [DOI] [PubMed] [Google Scholar]
- Motomura K. et al. (2008) ‘Identification of Monomorphic and Divergent Haplotypes in the 2006–2007 Norovirus GII/4 Epidemic Population by Genomewide Tracing of Evolutionary History’, Journal of Virology, 82: 11247–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa K. et al. (2018) ‘Phylogeny and Immunoreactivity of Norovirus GII.P16-GII.2, Japan, Winter 2016–17’, Emerging Infectious Diseases, 24: 144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K. et al. (2010) ‘Frequent Detection of Noroviruses and Sapoviruses in Swine and High Genetic Diversity of Porcine Sapovirus in Japan during Fiscal Year 2008’, Journal of Clinical Microbiology, 48: 1215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niendorf S. et al. (2017) ‘Steep Rise in Norovirus Cases and Emergence of a New Recombinant Strain GII.P16-GII.2, Germany, Winter 2016’, Euro Surveill, 22: 30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. L. et al. (2004) ‘A Chimeric Bovine Enteric Calicivirus: Evidence for Genomic Recombination in Genogroup III of the Norovirus Genus of the Caliciviridae’, Virology, 326: 231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L. et al. (2016) ‘The Novel Norovirus Genotype GII.17 is the Predominant Strain in Diarrheal Patients in Shanghai, China’, Gut Pathogens, 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. I. et al. (2012) ‘Multiple Antigenic Sites are Involved in Blocking the Interaction of GII.4 Norovirus Capsid with ABH Histo-blood Group Antigens’, Journal of Virology, 86: 7414–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. I., Green K. Y. (2014) ‘Sequential Gastroenteritis Episodes Caused by 2 Norovirus Genotypes’, Emerging Infectious Diseases, 20: 1016–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. I., Green K. Y. (2015) ‘Genome of Emerging Norovirus GII.17, United States, 2014’, Emerging Infectious Diseases, 21: 1477–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G. I. et al. (2017) ‘Static and Evolving Norovirus Genotypes: Implications for Epidemiology and Immunity’, PLoS Pathogens, 13: e1006136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F. et al. (2004) ‘UCSF Chimera—A Visualization System for Exploratory Research and Analysis’, Journal of Computational Chemistry, 25: 1605–12. [DOI] [PubMed] [Google Scholar]
- Prasad B. V. et al. (1994) ‘Three-dimensional Structure of Baculovirus-expressed Norwalk Virus Capsids’, Journal of Virology, 68: 5117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B. V. et al. (1999) ‘X-ray Crystallographic Structure of the Norwalk Virus Capsid’, Science, 286: 287–90. [DOI] [PubMed] [Google Scholar]
- Ramani S., Estes M. K., Atmar R. L. (2016) ‘Correlates of Protection against Norovirus Infection and Disease—Where are We Now, Where Do We Go?’, PLoS Pathogens, 12: e1005334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeck A. et al. (2010) ‘Serological Correlate of Protection against Norovirus-induced Gastroenteritis’, The Journal of Infectious Diseases, 202: 1212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohayem J. (2009) ‘Norovirus Seasonality and the Potential Impact of Climate Change’, Clinical Microbiology and Infection, 15: 524–7. [DOI] [PubMed] [Google Scholar]
- Ruis C. et al. (2017) ‘The Emerging GII.P16-GII.4 Sydney 2012 Norovirus Lineage is Circulating Worldwide, Arose by Late-2014 and Contains Polymerase Changes That May Increase Virus Transmission’, PLoS One, 12: e0179572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M. et al. (2014) ‘Multiple Norovirus Infections in a Birth Cohort in a Peruvian Periurban Community’, Clinical Infectious Diseases, 58: 483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker S. et al. (2016) ‘Structural Basis for Norovirus Neutralization by an HBGA Blocking Human IgA Antibody’, Proceedings of the National Academy of Sciences of the United States of America, 113: E5830–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenga J. J. et al. (2007) ‘Epochal Evolution of GGII.4 Norovirus Capsid Proteins from 1995 to 2006’, Journal of Virology, 81: 9932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenga J. J. et al. (2009) ‘Norovirus Illness is a Global Problem: Emergence and Spread of Norovirus GII.4 Variants, 2001–2007’, The Journal of Infectious Diseases, 200: 802–12. [DOI] [PubMed] [Google Scholar]
- Singh B. K. et al. (2015) ‘Structural Analysis of a Feline Norovirus Protruding Domain’, Virology, 474: 181–5. [DOI] [PubMed] [Google Scholar]
- Smith D. B. et al. (2012) ‘Diversity of Murine Norovirus in Wild-rodent Populations: Species-specific Associations Suggest an Ancient Divergence’, Journal of General Virology, 93: 259–66. [DOI] [PubMed] [Google Scholar]
- Souza M. et al. (2007) ‘Cytokine and Antibody Responses in Gnotobiotic Pigs after Infection with Human Norovirus Genogroup II.4 (HS66 Strain)’, Journal of Virology, 81: 9183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza M. et al. (2008) ‘Pathogenesis and Immune Responses in Gnotobiotic Calves after Infection with the Genogroup II.4-HS66 Strain of Human Norovirus’, Journal of Virology, 82: 1777–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa M., von Bonsdorff C. H., Maunula L. (2012) ‘Pet Dogs—A Transmission Route for Human Noroviruses?’, Journal of Clinical Virology, 53: 244–7. [DOI] [PubMed] [Google Scholar]
- Tohma K. et al. (2017) ‘Phylogenetic Analyses Suggest That Factors Other than the Capsid Protein Play a Role in the Epidemic Potential of GII.2 Norovirus’, mSphere, 2: e00187–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohma K. et al. (2018) ‘Evolutionary Dynamics of Non-GII Genotype 4 (GII.4) Noroviruses Reveal Limited and Independent Diversification of Variants’, Journal of General Virology, 99: 1027–35. [DOI] [PubMed] [Google Scholar]
- Tohma K. et al. (2019) ‘Population Genomics of GII.4 Noroviruses Reveal Complex Diversification and New Antigenic Sites Involved in the Emergence of Pandemic Strains’, mBio, 10: e02202–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi T. K. et al. (2013) ‘Clinical Characteristics of Norovirus-associated Deaths: A Systematic Literature Review’, American Journal of Infection Control, 41: 654–7. [DOI] [PubMed] [Google Scholar]
- van Beek J. et al. (2013) ‘Indications for Worldwide Increased Norovirus Activity Associated with Emergence of a New Variant of Genotype II.4, Late 2012’, Euro Surveill, 18: 8–9. [PubMed] [Google Scholar]
- van Beek J. et al. (2017) ‘Whole-genome Next-generation Sequencing to Study within-host Evolution of Norovirus (NoV) among Immunocompromised Patients with Chronic NoV Infection’, The Journal of Infectious Diseases, 216: 1513–24. [DOI] [PubMed] [Google Scholar]
- van Beek J. et al. (2018) ‘Molecular Surveillance of Norovirus, 2005–16: An Epidemiological Analysis of Data Collected from the NoroNet Network’, The Lancet Infectious Diseases, 18: 545–53. [DOI] [PubMed] [Google Scholar]
- van Der Poel W. H. et al. (2000) ‘Norwalk-like Calicivirus Genes in Farm Animals’, Emerging Infectious Diseases, 6: 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega E. et al. (2014a) ‘Genotypic and Epidemiologic Trends of Norovirus Outbreaks in the United States, 2009 to 2013’, Journal of Clinical Microbiology, 52: 147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega E. et al. (2014b) ‘RNA Populations in Immunocompromised Patients as Reservoirs for Novel Norovirus Variants’, Journal of Virology, 88: 14184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinje J., Altena S. A., Koopmans M. P. (1997) ‘The Incidence and Genetic Variability of Small Round-structured Viruses in Outbreaks of Gastroenteritis in the Netherlands’, The Journal of Infectious Diseases, 176: 1374–8. [DOI] [PubMed] [Google Scholar]
- Wang C. et al. (2018) ‘Complete Genome Sequence of a Novel Recombinant GII.P16-GII.1 Norovirus Associated with a Gastroenteritis Outbreak in Shandong Province, China, in 2017’, Genome Announcements, 6: e01483-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. H. et al. (2005) ‘Porcine Noroviruses Related to Human Noroviruses’, Emerging Infectious Diseases, 11: 1874–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P. A. (2014) ‘Evolution of Norovirus’, Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases, 20: 741–5. [DOI] [PubMed] [Google Scholar]
- Wolf S. et al. (2009) ‘Molecular Detection of Norovirus in Sheep and Pigs in New Zealand Farms’, Veterinary Microbiology, 133: 184–9. [DOI] [PubMed] [Google Scholar]