Abstract
The resolution of the gar genome affords an opportunity to examine the diversification and functional specialization of immune effector molecules at a distant and potentially informative point in phylogenetic development. Although innate immunity is affected by a particularly large number of different families of molecules, the focus here is to provide detailed characterization of several families of innate receptors that are encoded in large multigene families, for which orthologous forms can be identified in other species of bony fish but not in other vertebrate groups as well as those for which orthologs are present in other vertebrate species. The results indicate that whereas teleost fish and the gar, as a holostean reference species, share gene families thought previously to be restricted to the teleost fish, the manner in which the members of the multigene families of innate immune receptors have undergone diversification is different in these two major phylogenetic radiations. It appears that both the total genome duplication and different patterns of genetic selection have influenced the derivation and stabilization of innate immune genes in a substantial manner during the course of vertebrate evolution.
Keywords: Toll-like receptors, novel immune-type receptors, diverse immunoglobulin domain-containing proteins, Actinopterygii, Holostei, teleost
INTRODUCTION
It has been suggested that the high level of diversity between innate immune receptor genes from different species reflects that evolutionary history of pathogen exposures for that species’ lineage (Jack, 2015). In addition, the co-evolution with commensal bacteria has likely further refined the host’s innate immune system (Thaiss et al., 2016). As the genomes of more diverse animals are reported, it has become clear that many species possess immune system strategies that differ dramatically from mammalian species (Buonocore and Gerdol, 2016). The merits of novel immune strategies include strong selection pressures which can influence rates of speciation and animal diversification (Malmstrøm et al., 2016). Ray-finned fish (Actinopterygii) comprise the largest group of vertebrate species with over 30,000 members (Eschmeyer et al., 2016) and nearly all of these (>99.8%) are teleost fishes (Nelson et al., 2016) which include model organisms, such as zebrafish (Danio rerio) and Japanese medaka (Oryzias latipes) and agriculturally important species (e.g. carps, catfish and salmonids). Genomic analyses have revealed variation in the sequence and numbers of innate immune receptors in diverse teleost species with much of this diversity arising from recent gene birth and death events within multiple multigene families of innate immune receptors (Malmstrøm et al., 2016; Wcisel and Yoder, 2016).
The diversification of immune-related genes can be accelerated through gene duplication (gene birth) events followed by species-specific adaptations. Whole genome duplications (WGD) provide a massive number of genes the freedom to evolve. The fate of duplicated genes may be neofunctionalization, (the acquisition of new functions), subfunctionalization, (all functions of the original gene are maintained, but divided between the gene copies) or nonfunctionalization (accumulation of deleterious mutations resulting in gene death) (Kuraku and Meyer, 2010). Two rounds of early vertebrate genome duplication (VGD1 and VGD2) (Dehal and Boore, 2005) and a subsequent teleost genome duplication (TGD) (Amores et al., 1998; Taylor et al., 2003; Glasauer and Neuhauss, 2014) has likely contributed to the great diversity of innate immune receptors present in teleosts (Malmstrøm et al., 2016; Wcisel and Yoder, 2016). In addition to the TGD, certain teleost lineages (e.g. salmonids, carps and suckers) have experienced additional, more recent WGD events (Pasquier et al., 2016).
Recently, the genome of spotted gar (Lepisosteus oculatus) (hereafter gar), a ray-finned fish but a holostean outgroup to the TGD, was analyzed and reported (Braasch et al., 2016). It was demonstrated that gar has the ability to bridge the gap between teleost and mammalian genomes, facilitating cross-species comparisons and illuminating hidden orthologies. The gar genome, when compared to teleosts, is considered slowly evolving thus, it has preserved a more ancient genome structure. This permits a unique perspective into the evolution of vertebrate immunity and the impact TGD may have had on immune-related loci.
Here, we offer a detailed description of multiple families of innate immune receptors encoded by gar including toll-like receptors (TLRs) and families of fish-specific immunoglobulin (Ig) domain containing innate immune receptors (IIIRs) (Fig. 1). We demonstrate that while many gar TLR genes share more sequence similarities with TLRs encoded by teleost fish (eg, zebrafish), their genomic organization often resembles that of human TLRs. We characterize gar sequences that encode inhibitory and activating forms of novel immune-type receptors (NITRs), which are predicted to function as natural killer cell receptors (Yoder and Litman, 2011). We identify gar sequences that encode diverse immunoglobulin domain containing proteins (DICPs), and demonstrate their linkage to major histocompatibility complex (MHC) sequences in cyprinids (zebrafish), Lepisosteiformes (gar), and Latimeria (coelacanth, a lobe-finned fish that branched from ray-finned fishes 450 mya and shares a common ancestor more recently with mammals than teleosts). The failure to identify orthologs of specific tetrapod innate immune receptors, indicates that, although gar is an outgroup to the TGD, its innate immune receptor repertoire more closely resembles that of teleosts than tetrapods. We have integrated these observations into an updated view of the evolutionary origins of these families of innate immune receptors.
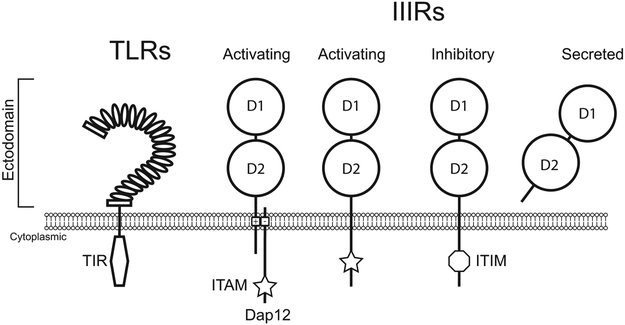
Figure 1. Typical protein structures of TLRs and immunoglobulin-domain contain innate immune receptors (IIIRs).
Receptors recognize ligands via ectodomains. TLR ectodomains consist of numerous LRR (ovals) giving a “horseshoe” conformation, which are flanked by LLRNT and LRRCT domains (rectangles). TLR signaling occurs in the cytoplasm via a highly conserved TIR domain. Members of IIIRs include NITRs, DICPs, PIGRLs, LITRs and NILTs, among many others (Yoder et al., 2004; Stet et al., 2005; Haire et al., 2012; Kortum et al., 2014). IIIR ectodomains possess at least one Ig domain (circles). NITRs and DICPs typically contain two Ig domains, coined D1 and D2, although proteins with a single Ig or more than two Ig domains are also observed. Activating IIIRs signal via ITAMs (star) which can be encoded directly on the receptor’s cytoplasmic tail or on an adaptor protein such as Dap12 which interacts with the IIIR through oppositely charged residues in their transmembrane domains. Inhibitory IIIRs signal through ITIMs (octagon) in their cytoplasmic tails. Secreted and ambiguous forms, which do not contain known signaling motifs, are also observed.
METHODS
Genes initially were identified by searching Ensembl’s database (release 79) of zebrafish and human TLRs, NITRs, and DICPs genes for gar orthologs identified by the Ensembl’s annotation and Gene Orthology/Paralogy pipeline (Vilella et al., 2009; Aken et al., 2016). For NITRs and DICPs, the surrounding genomic loci of the gar genome (LepOcu1) also was searched with tBLASTn using both zebrafish and gar NITR/DICP protein sequences for the presence of genes missed by prediction models (Altschul et al., 1990; Camacho et al., 2009). Syntenic comparisons were performed using Genomicus v84.01 (Louis et al., 2015). Multiple sequence alignments were performed with Clustal Omega (Sievers et al., 2011). Phylogenetic analyses were performed in MEGA7 (Kumar et al., 2016). Neighbor-joining trees were constructed using JTT matrix-based method to compute evolution distance, with pairwise deletion of ambiguous positions and 2000 bootstrap replicates (Saitou and Nei, 1987; Jones et al., 1992). Protein domains were identified using SMART (Letunic et al., 2015).
RESULTS AND DISCUSSION
Gar TLRs share evolutionary histories of both humans and zebrafish
The family of toll-like receptors (TLRs), which are present in vertebrates and invertebrates, recognize a variety of pathogen associated molecular patterns (PAMPs) providing one of the initial immune responses to infection (Aderem and Ulevitch, 2000). The TLR family includes ten receptors in humans (TLR1-TLR10) and twelve in mice (TLR1-TLR13, lacks TLR10). Teleost genomes can encode more than twenty TLR genes (Quiniou et al., 2013) which may reflect neofunctionalizaton after the TGD. For example, zebrafish and channel catfish (Ictalurus punctatus) contain duplicate copies of genes encoding TLR4, TLR5 and TLR8 and at least six other “non-mammalian” TLR genes (Jault et al., 2004; Palti, 2011; Quiniou et al., 2013; Kanwal et al., 2014). Vertebrate TLRs can be organized into six subfamilies based on amino acid sequence similarities: TLR1, TLR3, TLR4, TLR5, TLR7 and TLR11, each of which can contain multiple members (e.g., TLR7, TLR8 and TLR9 are all part of the TLR7 subfamily) (Roach et al., 2005).
TLRs recognize a variety of PAMPs through an N-terminal ectodomain consisting of numerous leucine-rich repeats (LRR) (Fig. 1). The number of LRRs varies between TLR members and assigning TLR gene homology across species based on LRR sequence alone can be difficult, due to the variable length of extensions within LRR motifs (Matsushima et al., 2005, 2007). Usually, cysteine clusters cap the N-terminal side of the LRRs and flank the C-terminal side and are termed LRRNT and LRRCT, respectively and are thought to stabilize the hydrophobic LRRs (Gay and Gangloff, 2007).
TLRs are type I transmembrane proteins and are expressed either on the cell surface or on endosomes. The cytoplasmic C-terminal region contains a TOLL/IL1 receptor (TIR) domain (Fig. 1). Upon ligand recognition, TLRs recruit cytoplasmic adapter molecules such as MyD88, MAL, TRIF, TRAM and SARM in order to initiate an immune response (reviewed in (Kawai and Akira, 2006). While the N-terminal LRR-containing regions are highly variable between species, the TIR domain is highly conserved and amenable to phylogenetic analyses (Beutler and Rehli, 2002; Jault et al., 2004; Quiniou et al., 2013; Boudinot et al., 2014a).
Ensembl’s annotation and Gene Orthology/Paralogy pipeline (Vilella et al., 2009) predicts seventeen TLR-related genes within the gar genome (Table 1). Thirteen of these seventeen gar sequences were annotated using zebrafish TLRs as reference, while four gar TLR sequences remain uncharacterized [tlr(a), tlr(b), tlr(c), tlr(d)]. Based on phylogenetic analysis of TIR domains (Fig. 2), we confirmed all but one of the annotations and account for the remaining uncharacterized TLRs utilizing West Indian Ocean coelacanth (Latimeria chalumnae) and channel catfish reference sequences. We find that representative members of all six TLR subfamilies are encoded in the gar genome. Here, we illustrate the genomic context of gar TLRs, infer syntenic relationships between human and zebrafish using gar as a reference, and make predictions of the evolutionary histories of these gene families.
Table 1.
Gar toll-like receptors (TLRs)
| TLR Subfamily |
Protein Name in Fig. 2 |
Recommended Gene Name |
Ensembl Identifier | Location | Syntenic Support1 |
Protein Domains2 |
|---|---|---|---|---|---|---|
| TLR 1 | Tlr1 | tlr1 | ENSLOCG00000012910 | LG4:62,158,233-62,160,668:1 | ZF | Sp/ED/Ct/TM/TIR |
| Tlr2 | tlr2 | ENSLOCG00000018220 | LG4:21,778,623-21,781,022:−1 | Hu | Sp/ED/Ct/TM/TIR | |
| Tlr18 | tlr18 | ENSLOCG00000007992 | LG24:6,943,613-6,955,636:1 | - | Sp/ED/Ct/TM/TIR | |
| Novel [Tlr(a)] | tlr25a | ENSLOCG00000016891 | LG8:45,599,630-45,602,032:−1 | - | Sp/ED/Ct/TM/TIR | |
| Novel [Tlr(b)] | tlr25b | ENSLOCG00000017911 | LG8:25,066,396-25,068,894:−1 | - | Sp/ED/Ct/TIR | |
| Novel [Tlr(c)] | tlr27 | ENSLOCG00000017732 | LG10:1,032,739-1,035,096:−1 | - | Sp/ED/TM/TIR | |
| Novel [Tlr(d)] | Pseudogene | ENSLOCG00000017625 | LG19:1,189,904-1,191,199:−1 | - | TIR/TM/TM | |
| TLR 3 | Tlr3 | tlr3 | ENSLOCG00000013826 | LG4:70,374,961-70,381,739:1 | Hu/ZF | Sp/ED/Ct/TIR |
| TLR 4 | Tlr4ba | tlr4 | ENSLOCG00000003751 | LG21:5,778,346-5,781,017:1 | Hu | ED/TM |
| TLR 5 | Tlr5a | tlr5 | ENSLOCG00000018000 | LG1:32,406,982-32,409,585:1 | Hu/ZF | ED/Ct/TM/TIR |
| TLR 7 | Tlr7 | tlr7 | ENSLOCG00000009977 | LG14:21,059,390-21,065,834:1 | Hu/ZF | SP/Nt/ED/Ct/TIR |
| Tlr8a | tlr8a | ENSLOCG00000009990 | LG14:21,075,973-21,079,392:1 | Hu/ZF | SP/Nt/ED/Ct/TM/TIR | |
| Tlr8b | tlr8b | ENSLOCG00000009982 | LG14:21,068,100-21,073,657:1 | Hu/ZF | SP/ED/Ct/TM/TIR | |
| Tlr9 [Tlr9(a)] | tlr9a | ENSLOCG00000014202 | LG5:48,972,573-48,981,170:−1 | Hu/ZF | SP/ED/Ct/TM/TIR | |
| Tlr9 [Tlr9(b)] | tlr9b | ENSLOCG00000014430 | LG5:50,478,835-50,482,640:1 | - | SP/ED/TIR | |
| TLR 11 | Tlr19 | Novel | ENSLOCG00000015687 | LG1: 13,198,517-13,200,058 | - | ED/Ct/TM/TIR |
| Tlr21/22 | tlr22 | ENSLOCG00000018316 | LG6:38,022,636-38,025,533:1 | - | Sp/ED/Ct/TM/TIR |
Evidence for conserved synteny presented in Supplementary Material Figure S1 and Table S1. ZF = zebrafish; Hu = human
Protein domains: Sp = signal peptide, Nt = N terminal LRR cap, ED = ectodomain, Ct = C terminal LRR cap, TM = transmembrane, TIR = TOLL/IL1 receptor domain
Figure 2. Phylogenetic relationships between gar, teleost, mammalian, avian, reptilian and coelacanth TLR TIR domains.
Four letter codes are used for each species: spotted gar (Lepisosteus oculatus, Leoc), coelacanth (Latimeria chalumnae, Lach), zebrafish (Danio rerio, Dare), human (Homo sapiens, Hosa), mouse (Mus musculus, Mumu), anole lizard (Anolis carolinensis, Anca), chicken (Gallus gallus, Gaga), and channel catfish (Ictalurus punctatus, Icpu). Coelacanth TLRs were described by Boudinot et al., (2014b). Coelacanth TLR paralogs ENSLACG00000006376 and ENSLACG00000004773, were included in this analysis although they may represent pseudogenes: ENSLACG00000006376 possesses a limited number of LRRs and ENSLACG00000004773 encodes a partial TIR domain. Channel catfish TLRs were annotated by Quiniou et al (Quiniou et al., 2013). Gar Tlr4 lacks a TIR domain and is excluded from this analysis. Bootstrap values are indicated next to the branches; values less than 50 are not shown. Sequences are provided in Supplementary Material Table S2.
TLR1 subfamily
Members of TLR1 subfamily bind a range of ligands which often originate from bacteria (reviewed in Gay and Gangloff, 2007). For example, mammalian TLR2 is able to heterodimerize with TLR1 in order to recognize triacyl lipopeptides originating from mycobacteria whereas TLR2/TLR6 heterodimers recognize diacylated lipopeptides (Takeuchi et al., 2002). Similar to vertebrates, teleost TLR1 and TLR1 subfamily members often lack the LRRNT flanking sequence (Zhang et al., 2014). This absence of LRRNT is thought to be an important feature allowing mammalian TLR1 and TLR2 molecules to dimerize and suggests that TLR1/TLR2 heterodimers might also occur in fish (Quiniou et al., 2013).
The TLR1 subfamily has been described as containing more species-specific adaptations than any other TLR subfamily (Roach et al., 2005). Seven gar genes were determined to belong to the TLR1 family based on the phylogenetic analysis of their TIR domains (Table 1, Figure 2). Gar tlr1 displays some synteny to zebrafish tlr1, however there is no evidence for conserved synteny with humans nor any other non-fish species (Table 1). The four gar TLRs which were not given an Ensembl annotation are likely due to gar- or fish-specific adaptations as comparisons with zebrafish and humans revealed only weak synteny (Supplementary Material Fig. S1 and Table S1). Although gar tlr(d) (ENSLOCG000000017625) groups out with a coelacanth TLR gene, it is unlikely either function as receptors given the arrangement of detected protein domains (TIR/TM/TM, Table 1). Through phylogenetic analyses of the TIR domains (Fig. 2), two additional un-annotated gar TLRs [tlr(a)/ENSLOCG00000016891 and tlr(b)/ENSLOCG00000017911] appear related to TLR25, which was first identified in catfish but determined to also be present in adrianichthyids, cichlids, cyprinids and plecoglossids suggesting its presence early in fish evolution (Quiniou et al., 2013). Gar tlr(b) shares significant synteny with tilapia TLR25 (not shown). Interestingly, the tilapia TLR25, ENSONIG00000014149, which is encoded on an unplaced scaffold in the latest Orenil1.1 genome assembly (Brawand et al., 2014), is considered synonymous with the gene name TLR1. TLR25 sequences from tilapia, catfish and gar [tlr(a) and tlr(b)] lack the LRRNT domain which may allow for the dimerization with TLR2, increasing the repertoire of ligand specificities. Together, this suggests that TLR25 resulted from a gene duplication event of TLR1 loci. Its presence in gar, a much more basal species, suggests that a TLR1 duplication and subsequent divergence may have predated the TGD, which may explain why so many TLR1 subfamily members are observed in teleosts (Fig. 2). Gar also encodes tlr(c) (ENSLOCG00000017732) that clusters very tightly with both coelacanth and elephant shark (Callorhinchus milii) TLR27 (Boudinot et al., 2014b; Wang et al., 2015). TLR10 was estimated to have diverged from its TLR1-like precursor ~300 million years ago (mya), nearly 150 million years after the divergence of ray-finned fishes (Beutler and Rehli, 2002). In agreement with this, gar and teleosts genomes do not encode orthologs of TLR10. Likewise, TLR6 is thought to have emerged 130 mya and thus is also absent from fish genomes.
TLR3 subfamily
It has been demonstrated that both mammalian and teleost TLR3 proteins are able to recognize and respond to dsRNA, the genetic material of many viruses, to induce the expression of type I interferons (Roach et al., 2005; Huang et al., 2011; Samanta et al., 2013). Phylogenetic analyses places gar tlr3 (ENSLOCG00000013826) in a group with teleosts (Fig. 2), however it shares an almost equivalent amount of synteny with human TLR3 loci (Table 1, Supplementary Material Fig. S1). Although it is tempting to predict that gar Tlr3 is also able to recognize and respond to dsRNA, caution should be used when generalizing the function of TLRs across species. For example, sea cucumber TLR3 (Apostichopus japonicus) is transcriptionally upregulated following exposure to bacterial PAMPs, suggesting a dual or alternative role of TLR3 (Lu et al., 2013). Many differences in TLR response to PAMPs are observed across species, and is well illustrated in the case of TLR4.
TLR4 subfamily
Mammalian TLR4, in conjunction with myeloid differentiation protein 2 (MD-2), initiates an immune response against lipopolysaccharide (LPS), the major cell wall component of Gram-negative bacteria (Beutler, 2000). Additionally, LPS-binding protein (LBP) and CD14 aid in separating LPS aggregates and delivering LPS to the TLR4/MD-2 complex (Wright et al., 1990; Beutler, 2000; Su et al., 2000; Kim et al., 2005). Prolonged activation of TLR4 can lead to endotoxic shock, a situation which can be fatal to the host and is characterized by the overproduction of cytokines such as tumor necrosis factor-a (TNFa) and interleukin-6 (IL-6) (Morrison and Ryan, 1987). Fish and amphibians display notable resistance to LPS-induced endotoxic shock when compared to mammals (Berczi and Bereznai, 1966). Beutler and Rehli (2002) suggested that LPS sensing by TLR4 may be a unique adaptation to mammals as TLR4 was not found in the available fish genomes. Following the completion of a quality draft of the zebrafish genome, Jault et al (2004) determined zebrafish contains 19 putative TLR variants including two copies of TLR4 (tlr4a and tlr4b), although accessory proteins LBP, MD-2 and CD14 remain absent. Although zebrafish Tlr4a and Tlr4b do not recognize LPS, over-expression of zebrafish Tlr4b TIR domain negatively regulates NF-kB activation (Sepulcre et al., 2009; Sullivan et al., 2009). As morpholino knockdowns of tlr4a, tlr4b and myd88 in zebrafish was not sufficient to disrupt the response to LPS, Sepulcre et al (2009) concluded that LPS sensing in zebrafish occurs independently of TLR4/MYD88 pathway.
Gar encodes a TLR4 ortholog (ENSLOCG00000003751), however it shares minimal synteny with humans and none with zebrafish (Table 1, Supplementary Material Fig. S1 and Table S1). The predicted protein sequence of gar Tlr4 lacks a TIR domain while the ectodomain displays a high degree of sequence divergence from other TLR4 genes (less than 31% identical to humans). Accessory proteins MD-2 and CD14 are not readily discernible in gar although LBP was identified (ENSLOCG00000006358). Therefore, we predict that LPS sensing by gar Tlr4 is unable to activate an immune response through either the MyD88-dependent pathway nor the TRIF-related adapter molecule (TRAM) signaling pathway as observed in mammals (Miggin and Neill). TLR4 likely diverged from a TLR2/TLR4 precursor around the time of the emergence of vertebrate, approximately 500 mya (Beutler and Rehli, 2002). The absence of TLR4 from coelacanth, and lack of Tlr4 TIR domain and accessory proteins in gar suggest that TLR4 function was dispensable in fish or that LPS sensing by TLR4 may be a mammalian innovation (Boudinot et al., 2014a).
TLR5 subfamily
Both mammalian and teleost TLR5 recognize bacterial flagellum (Muñoz et al., 2013). In some teleosts, such as rainbow trout (Oncorhynchus mykiss) and gilthead seabream (Sparus aurata), two TLR5 genes have been described: one encodes a membrane-bound TLR5M and the other encodes a soluble TLR5S which lacks the TIR domain necessary for classical TLR signaling. Even in the absence of a TIR domain, TLR5S is believed to enhance the immune response in rainbow trout (Tsujita et al., 2004). However, in gilthead seabream, it was observed that the binding of TLR5S to flagellum blocks TLR5M activation and therefore down-regulates the immune response (Muñoz et al., 2013). Coelacanth also are found to encode TLR5S and TLR5M (Boudinot et al., 2014b); however, since TLR5S lacks a TIR domain, orthology to teleost TLR5S was not examined. Nonetheless, the presence of TLR5S in coelacanth indicates an ancient origin for this sequence.
In other teleost species, such as Atlantic cod (Gadus morhua), TLR5 genes have been lost altogether. Atlantic cod has many immune system abnormalities including the apparent loss of MHC class II and large expansion of MHC class I molecules (Star et al., 2011; Buonocore and Gerdol, 2016). The single copy of gar tlr5 encodes a transmembrane domain and TIR domain and is therefore most similar to TLR5M. TLR5S was not detected in gar, and may be restricted to certain teleost species. Adjacent genomic regions of TLR5 remain highly conserved among human, gar, and zebrafish (Supplementary Material Fig. S1 and Table S1).
TLR7 subfamily
Mammalian members of the TLR7 subfamily recognize nucleic acid motifs, however it is not known if the ligands are conserved in fish (Rauta et al., 2014). The TLR7 subfamily, which includes TLR7, TLR8 and TLR9, may have arisen prior to the rise of vertebrates, some 500 million years ago and is therefore found throughout vertebrate taxa (Beutler and Rehli, 2002). Gar tlr7, tlr8a and tlr8b are found in tandem on LG14 in the same basic organization as observed in humans (Supplementary Material Fig. S1). While zebrafish tlr7 and tlr8 are encoded in tandem on chromosome 9, zebrafish tlr8b is encoded on chromosome 10. This may be reflective of a single gene transposition as syntenic genes surrounding zebrafish tlr8b are not observed (Supplementary Material Table S1). Duplicated copies of TLR9 are found on distinct loci on gar LG5 (Table 1). Gar TLR9 genes have diverged significantly and are only approximately 40% identical to each other. An expansion of TLR9 genes has been observed in Atlantic cod, which may be a compensatory adaptation in response to the loss of TLR5 and MHC class II sequences (Star and Jentoft, 2012). While gar does encode TLR5 and MHC class II sequences, an expansion of TLR9 may be advantageous for some fish lineages.
TLR11 subfamily
The TLR11 subfamily includes TLR11, TLR12, TLR13 and many others that may be species-specific. Humans lack a functional TLR11 ortholog whereas other vertebrate species have many TLR11 members. For example, TLR21 is common to birds, amphibians and fish but absent from humans. The ligands for most TLR11 subfamily members remain unknown; however, it has been determined that mouse TLR11 and TLR12 recognize ligands originating from protozoan parasites while TLR21 from chicken and zebrafish recognizes CpG DNA common to bacteria (Keestra et al., 2010; Yeh et al., 2013a; Zhang et al., 2014) and TLR22 in pufferfish (Takifugu rubripes) recognizes dsRNA (Matsuo et al., 2008). There are a number of fish-specific TLRs which belong to the TLR11 subfamily (e.g., catfish TLR26). The gar genome encodes two TLR11 subfamily members: tlr19 (ENSLOCG00000015687) and tlr21 (ENSLOCG00000018318), neither of which share synteny with zebrafish nor humans (Table 1). Gar tlr19 is highly diverged and is supported by low (34%) bootstrap values (Fig. 2). Therefore, this gene may represent a gar-specific adaptation and we recommend it remain classified as novel (Table 1).
As mentioned above, zebrafish Tlr21 is functionally similar to mammalian TLR9 in that it recognizes CpG DNA, although the specific CpG sequences recognized differ significantly between the two types of receptors (Yeh et al., 2013b; Zhang et al., 2014). Interestingly, gar tlr21 appears to share an evolutionary history with the TLR11 subfamily, instead of the TLR7 subfamily that might be expected based on shared functions with TLR9 (Fig. 2). While the functions of gar TLR genes have not yet been determined, it is an interesting observation that gar possess two divergent copies of TLR9 as well as a TLR21. This may indicate that gar has a heightened response to CpG and is able to recognize different CpG DNA motifs.
“Teleost-Specific” Innate Immune Receptors Are Present In Gar
Although TLRs are ubiquitous throughout the vertebrate kingdom and represent an ancient mechanism of innate immunity (Beutler and Rehli, 2002; Roach et al., 2005; Rauta et al., 2014), natural killer receptor (NKR) gene families have been described as recently and rapidly evolving (Manser et al., 2015; Carrillo-Bustamante et al., 2016). The recent evolution of NKRs has led to many species-specific adaptations. For example, in humans the killer cell immunoglobulin like receptor (KIR) gene family encodes activating and inhibitory NKRs that bind MHC class I or MHC class I-like molecules to differentiate between normal cells (“self”) and infected or transformed cells (“non-self”) (Jamil and Khakoo, 2011; Guethlein et al., 2015). Engagement of activating KIRs can result in the direct killing of target (‘non-self”) cells. In contrast, mice utilize the C-type lectin family of Ly49 NKRs to differentiate between “self” and “non-self” (Rahim and Makrigiannis, 2015). While fish species do not encode genetic orthologs of either the KIR or Ly49 families, it has been hypothesized that the NITR family, which belongs to the larger IIIR group of receptors and has been well described in teleosts, may function as NKRs in fish (Yoder, 2009; Yoder and Litman, 2011). Teleost NITRs are similar in structure to mammalian KIRs and when recombinant activating or inhibitory NITRs are expressed on human cells they can initiate NKR signaling pathways (Yoder et al., 2001; Wei et al., 2007). In addition to NITRs, teleost genomes encode other families of IIIRs that are similar in structure to NITRs such as: DICPs; polymeric immunoglobulin receptor-like proteins (PIGRLs); leukocyte immune-type receptors (LITRs); and novel immunoglobulin-like transcripts (NILTs) (Wcisel and Yoder, 2016).
Teleost IIIR gene families encode membrane bound and secreted proteins which possess one or more extracellular Ig-domains that are predicted to bind undefined ligands (Fig 1). Membrane bound IIIRs typically possess activating or inhibitory signaling motifs. Activating IIIRs may possess a cytoplasmic immunoreceptor tyrosine-based activation motif (ITAM: YxxI/Lx(6–12)YxxI/L) or a charged residue within the transmembrane domain which permits the association with an adaptor protein, such as DAP12 that possesses a cytoplasmic ITAM. Inhibitory IIIRs possess one or more immunoreceptor tyrosine-based inhibition motifs (ITIM: S/I/V/LxYxxI/V/L). ITIM signaling opposes ITAM signaling and the balance between these signals can determine the activation state of an immune cell (Barrow and Trowsdale, 2006).
Although genomic DICP sequences have been reported from the coelacanth (Boudinot et al., 2014a), NITRs, PIGRLs, LITRs and NILTs have previously been reported only in select teleost genomes (Wcisel and Yoder, 2016). Here, we investigated the gar genome for homologs of these gene families. We predict that gar encodes multiple inhibitory NITRs and a single activating NITR gene. We describe several DICP sequences from gar and substantiate the claim that DICPs are present in coelacanth. Lastly, we report that the putative single copy pigr gene in gar (ENSLOCG00000012073) did not undergo an expansion of PIGR-like genes as reported in certain teleost species (Kortum et al., 2014). We are currently unable to identify conclusive homologues of teleost NILTs or LITRs in gar (see below).
We also investigated the gar genome for homologs of a number of mammalian IIIR gene families. Human CD300s, orthologus to CMRFs in model species such mice, are a IIIR family comprised of 8–10 members, although many more CD300-like genes exist (reviewed in (Borrego, 2013). Ensembl identifies three potential orthologs to human CD300 on gar LG3. CD300s share many sequence similarities to teleost NILTs, which have only been described in species which did not have genomic resources at the time (Stet et al., 2005; Kock and Fischer, 2008; Østergaard et al., 2009; Ostergaard et al., 2010). Synteny was not observed between gar and any mammalian or teleost model in which NILTs have been described. Therefore, it is not clear at this time if these IIIR-like genes on gar LG3 are CD300s or NILTs. FcR-like molecules, which are found in mammals and teleost fish, are readily identifiable in gar (e.g. ENSLOCG00000009857, ENSLOCG00000009867, ENSLOCG00000009848, ENSLOCG00000000344, ENSLOCG00000000699, ENSLOCG00000000629) and dispersed throughout the gar genome (LG3, LG5 and one unplaced scaffold). As mentioned above, it is unclear if definitive orthologs of the teleost LITR gene family, a subset of which are markers for cytotoxic T cells (Taylor et al., 2016), are present in gar. Transcripts encoding inhibitory and activating LITRs have been described extensively in catfish (Stafford et al., 2006, 2007) and LITR Ig domains can be identified in the zebrafish genome (Rodríguez-Nunez et al., 2014). BLAST searches of the gar genome with catfish LITR sequences as queries identify multiple sequences annotated as FcR-like. The sequence similarity between LITR Ig domains and those of FcRs (as well as FcR-like, KIR, LILR and NKp46) has been reported (Stafford et al., 2006) and confounds the ability to definitively identify LITRs in gar. Further, the limited information on the genomic organization and syntenic relationships of teleost LITR gene cluster(s) currently prohibits the characterization of these genes in gar. Gar orthologs to human KIRs, Ly49s, NKp44, NKp46, and LILRs were not detected in our analyses.
Novel Immune Type Receptors (NITRs)
Teleost NITRs are expressed by lymphocytes (Yoder et al., 2010), function in allorecognition (Cannon et al., 2008) and, when expressed as recombinant proteins on mammalian cells, can influence NK cell function (Yoder et al., 2001; Wei et al., 2007). These features have led to their description as putative “functional orthologs” of mammalian NK receptors (Yoder and Litman, 2011). NITRs commonly possess two extracellular Ig-domains: one of the variable (V) type and one of the intermediate (I) type (Yoder, 2009). NITR V domains are highly similar to V domains encoded by immunoglobulin and T-cell receptor genes. A defining feature of NITR I domains is the presence of six cysteines which may influence the folding of this domain via intramolecular disulfide bonds. NITRs with a single Ig domain (V or I) have been reported. We previously identified 15 NITR sequences from gar that are encoded at two distinct genomic regions (Braasch et al., 2016). Here we provide an in-depth analyses of these sequences and identify additional gar NITR sequences. In total, we have identified seventeen gar ENSEMBL sequences that possess V and/or I domains that can be classified as NITRs (Figure 3). Thirteen sequences possess V and I domains, one lacks a V domain and three lack an I domain: these observations suggest that the presence of a V and an I domain is an ancient feature of NITRs. The original description of teleost NITRs noted the presence of sequences similar to joining (J) segments (FGxGTxLxV/L) that contribute to immunoglobulin (antibody) and T-cell receptor diversity via V(D)J recombination (Strong et al., 1999). Teleost NITR J segments are germline joined within the same exon as the V and I domains resulting in V-J or I-J sequences with no evidence for recombination. Most gar NITRs possess V-J and I-J sequences suggesting that this sequence organization precedes the divergence of teleosts from other ray-finned fish (Fig. 3). In addition, gar NITR genes encode V-J and I-J domains in adjacent exons which reflects the exon organization in zebrafish. This suggests that the organization of V-J-I-J in a single exon, which is observed in southern pufferfish (Sphoeroides nephelus) NITRs and in some Japanese medaka and European sea bass (Dicentrarchus labrax) NITRs, is a derived feature in these species (Strong et al., 1999; Desai et al., 2008; Ferraresso et al., 2009).
Figure 3. Conserved sequence features of gar NITRs.
Protein sequence alignment of predicted gar (Leoc) NITR proteins with prototypic zebrafish (Dare) inhibitory Nitr1a (Yoder et al., 2004). The longest predicted protein is included for each gar gene. Identical residues are highlighted in black and functionally similar residues highlighted in grey. The relative positions of sequences corresponding to peptide leader (L), variable Ig domain (V), intermediate Ig domain (I), J-related sequences (J), transmembrane domains (TM), and immunoreceptor tyrosine-based inhibition motifs (ITIM) sequences are indicated above the alignment. Not all sequences possess a L or TM which is indicated by a dashed line. Cysteines representative of Ig domains are indicated with single-headed arrow below the alignment. Cysteines representative of NITR I domains are indicated with double-headed arrows (Yoder, 2009; Wcisel and Yoder, 2016). Certain sequences possess additional cytoplasmic tyrosines that may mediate signaling and are indicated by asterisks below the alignment.
Most teleost NITRs encode a transmembrane domain although some are secreted. Membrane bound NITRs can be classified as activating, inhibitory or functionally ambiguous (Fig. 1; Yoder, 2009). Inhibitory NITRs encode ITIMs in their cytoplasmic tail. Activating NITRs possess an intramembranous charged residue which promotes the association with an activating adapter protein (Wei et al., 2007). Based on these features, fourteen of the gar NITR sequences are predicted to encode membrane proteins and, of these, ten are predicted to function as inhibitory NITRs, one is predicted to function as an activating NITR, and three lack these signaling motifs and are classified as functionally ambiguous (Fig. 3). Based on studies with an activating teleost NITR (Wei et al., 2007), we predict that the putative activating gar NITR with a positively charged arginine in the transmembrane domain (ENSLOCG00000008529), will associate with and signal via the adaptor protein, DAP12 (ENSLOCG00000003818). Three gar NITR sequences do not possess a transmembrane domain and are predicted to encode secreted proteins, although it is possible that there are errors in the ENSEMBL predictions.
In order to investigate possible conserved synteny between the gar NITR loci and the human and zebrafish genomes, gar NITRs were placed in genomic context (Fig. 4). Twelve NITRs are clustered on LG14 and five are present on unplaced scaffold JH591499.1. Only NITR sequences were identified on scaffold JH591499.1 providing no strategy for investigating conserved synteny. In contrast, gar sequences positioned within or adjacent to NITR clusters on LG14 can be identified, and include a single copy gene that encodes a secreted Ig domain and multiple G-coupled protein receptors including olfactory receptors. The secreted Ig domain encoded by this single copy gene (ENSLOCG00000011074) is more similar to members of the B7 family of proteins than it is to teleost NITRs. BLASTp searches of the human and zebrafish protein databases using this sequence as a query, identifies B7-H4 [encoded by the V-set domain-containing T-cell activation inhibitor 1-like (VTCN1) gene] as the most similar protein in both human and zebrafish; however, the gar vtcn1 gene has been placed on LG17 suggesting that this single copy gene encodes a gar-specific B7 protein. Orthologs of the olfactory receptors adjacent to the gar NITRs map to a region of human chromosome 12 which lacks sequences with identifiable similarity to NITRs. Although a number of olfactory receptor genes are linked to the T-cell receptor (TCR) alpha/delta locus, which undergoes V(D)J recombination, in human and mouse (Lane et al., 2002), the gar TCR alpha/delta locus is on LG24 (Braasch et al., 2016). Additional efforts to identify conserved synteny between NITRs and the human genome were unsuccessful as Genomicus and Ensembl indicate that NITRs have no mammalian homolog. Additionally, the genomic regions of gar that encode NITRs were compared to the genomic regions of teleosts that encode NITRs and no evidence for conserved synteny was apparent (not shown). Despite the lack of identifiable conserved synteny between NITR loci of gar and teleosts, their sequence conservation and presence of key NITR features provide compelling evidence that these regions of the gar genome encode homologs of teleost NITRs.
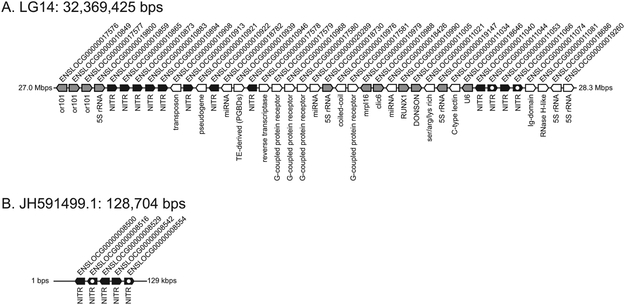
Figure 4. Genomic organization of NITR genes in spotted gar.
Genes and locations, not drawn to scale, are represented by pentagons, indicating transcriptional orientation. Black pentagons represent NITR genes as predicted by Ensembl gene annotation (Braasch et al., 2016). Black pentagons with white circles represent NITR genes identified in this study, but not considered paralogous by Ensembl. White pentagons are unannotated genes, with the PFAM domain summary listed below. Gray pentagons represent genes with Ensembl annotations, listed below, which are not NITRs. (A) NITR gene cluster on gar LG14. Several additional NITR genes may be downstream of this region on LG14 which can be identified by tBLASTn searches of this genomic region (using gar NITRs as queries), but are not currently defined by Ensembl gene models. (B) NITRs are also present on unplaced genomic scaffold JH591499.1. This scaffold encodes five genes which are all NITRs.
In order to examine the diversity of gar NITRs and compare them to teleost NITRs, one-to-one sequence comparisons were completed using 16 gar NITRs, 37 zebrafish NITRs, 44 Japanese medaka NITRs and 24 southern pufferfish NITRs and represented as a heatmap (Fig 5). This analysis utilized NITR V domains as they are highly variable and amenable to phylogenetic analyses. NITRs within each teleost species have been classified into subgroups, with the V domains with each subgroup sharing >70% sequence identity. For example, the 39 described zebrafish NITR genes are grouped into 14 subfamilies (named NITR1 through NITR14) (Yoder et al., 2008). The observation that any single NITR gene within a select species is more likely to be more similar to any other NITR within the same species, than to any NITR from a different species, demonstrates that there are no one-to-one NITR orthologs between teleosts and suggests lineage-specific gene birth and death events (Desai et al., 2008; Ferraresso et al., 2009; Yoder, 2009). In addition, the presence of NITR subgroups likely reflects recent gene duplications that differ between species. In contrast, gar NITR genes appear to be evolving more slowly. The 16 identified gar NITR V domains are all less than 70% identical to each other, however, the identity between a few pairs of gar NITRs approach 70% (Fig. 5). Although it is clear that the gar NITR genes have not undergone a large degree of diversifying selection, it could be stated that gar encodes three two-member subfamilies and eleven single copy NITRs. However, we propose defining gar NITRs as single copy genes based on the observation that the putative NITR gene duplication events (generating possible subfamilies) in gar are more ancient than those observed in teleost NITR subfamilies (Braasch et al., 2016).
Figure 5. NITR sequence diversity between and within species.
Pairwise sequence comparison of NITR V domains from gar, zebrafish, Japanese medaka and Southern pufferfish V-type Ig domains (Strong et al., 1999; Yoder et al., 2004, 2008; Desai et al., 2008). The darkest red indicates 70–100% sequence identity and the darkest blue indicates 0% sequence identity. Subfamilies of NITRs, represented by alternating black and gray rectangles (below), are classified based on at least ~70% identity of V-type Ig domains. One predicted gar NITR lacks a V domain (ENSLOCG00000008516) and was excluded from this analysis.
Diverse Immunoglobulin Domain-containing Proteins (DICPs)
Definitive DICPs have been identified in cyprinid fishes: zebrafish, grass carp (Ctenopharyngodon idella) and common carp (Cyprinus carpio) (Rodriguez-Nunez et al., 2016). A number of DICPs share many similarities to members of the mammalian CD300 family including the same basic protein structure (Fig. 1) and the ability to bind lipids (Haire et al., 2012). Each DICP possesses one or more extracellular Ig domains and two types of of Ig domains have been described, D1 and D2. A defining feature of both Ig domains is the presence of four cysteine residues that likely influence their folding via intramolecular disulfide bonds (Wcisel and Yoder, 2016). Secreted and membrane bound DICPs have been described from zebrafish. As described for NITRs (above), membrane bound DICPs include putative activating (identified by an intramembranous charged residue), inhibitory (identified by cytoplasmic ITIM), and functionally ambiguous forms (Fig. 1) (Wcisel and Yoder, 2016). Although DICP-like sequences have been identified in Atlantic salmon (Salmo salar), pufferfish (Takifugu rubripes and Tetraodon nigroviridis), and Nile tilapia (Oreochromis niloticus), which belong to the Salmonidae, Tetraodontidae, and Cichlidae families (Haire et al., 2012), it is unknown if they share a common evolutionary origin. DICP-like sequences recently were reported from coelacanth (Boudinot et al., 2014a).
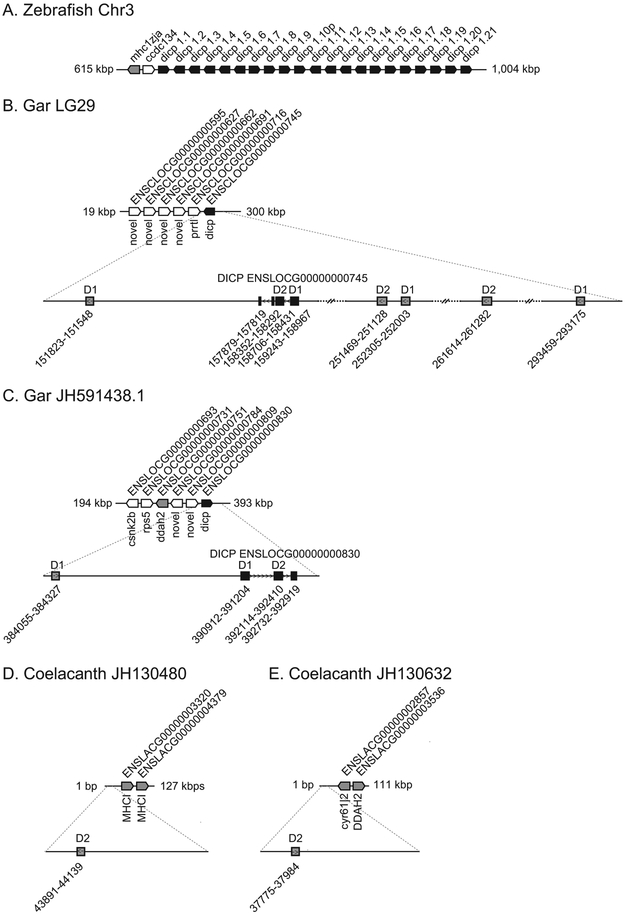
Here we identify two gar homologs of teleost DICPs on LG29 (ENSLOCG00000000745) and on unplaced scaffold JH591438 (ENSLOCG00000000830). Both of these DICP sequences are predicted to encode two Ig domains (D1 and D2) with no additional identifiable features. Using the predicted protein sequences from these gene models as tBLASTn queries against the spotted gar genome, we identified eight additional Ig domains on LG29 and scaffold JH591438 (Fig 6). These gar DICP Ig domains include six D1 domains and four D2 domains (Fig. 7). An alignment of gar DICP Ig domains with sequences from zebrafish, carps, salmon, pufferfish, tilapia and coelacanth reveals that each species, except coelacanth, possesses at least one DICP Ig domain with all conserved cysteines (Fig. 6). It will be of interest to determine if these gar and coelacanth genomic sequences encode inhibitory and activating DICPs as described in zebrafish.
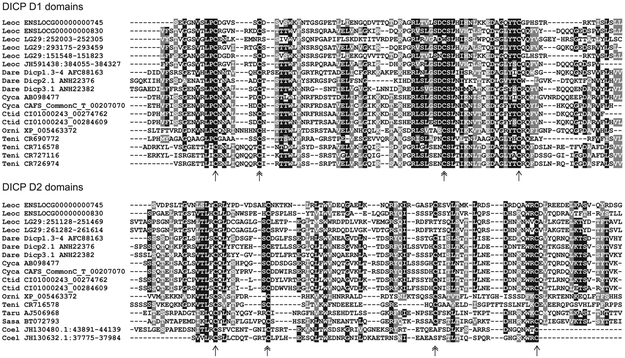
Figure 6. Conserved sequence features of gar DICP D1 and D2 domains.
Gar (Lepisosteus oculatus, Leoc) DICP D1 (top) and D2 (bottom) domains were aligned with previously described DICP domains from zebrafish (Danio rerio, Dare), carp (Cyprinus carpio, Cyca and Ctenopharyngodon idella, Ctid), pufferfish (Takifugu rubripes, Teni), salmon (Salmo salar, Sasa) and coelacanth (Latimeria chalumnae, Lach) (Haire et al., 2012; Boudinot et al., 2014a; Rodriguez-Nunez et al., 2016). Cysteines representative of Ig domains are indicated with single-headed arrow below the alignment. Cysteines representative of DICP Ig domains are indicated with double-headed arrows (Haire et al., 2012; Wcisel and Yoder, 2016).
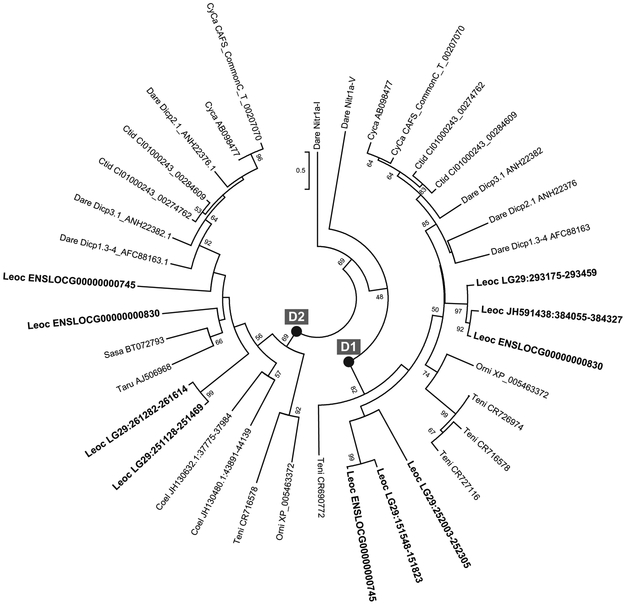
Figure 7. Evolutionary relations of DICP D1 and D2 domains.
The classification of gar DICP sequences as D1 or D2 domains is supported by phylogenetic analysis of Ig domains shown in Figure 6. Sequence nomenclature is as in Figure 6 with the addition of zebrafish Nitr1a Ig domains (V and I) as outgroups. Bootstrap values are indicated next to the branches; values less than 50 are not shown.
The identification of DICP sequences in gar and coelacanth indicates that DICPs are more ancient than NITRs. Two partial DICP D2 domain sequences (Fig. 6) were identified within the same unplaced scaffold (JH130480.1) of the coelacanth genome (Boudinot et al., 2014a). Although the coelacanth D2 domains lack one of the four conserved cysteines, we identify evidence for conserved synteny which suggests that coelacanth, gar, and zebrafish DICP sequences share a genetic ancestry (Fig. 8). The zebrafish DICP gene cluster present on chromosome 3 is adjacent to MHC class I genes (Haire et al., 2012; Rodriguez-Nunez et al., 2016). Similarly, one of the coelacanth DICP sequences also is adjacent to MHC class I sequences (Fig. 8). Although neither of the gar DICP clusters is found to be linked to MHC class I sequences, this may be due to the incomplete assembly of the gar genome. For example, a gar MHC class I sequence was identified in the NCBI genomic Sequence Read Archive (SRA: SRX090800) that was omitted from the final genomic assembly (Grimholt et al., 2015). Indeed, Braasch et al (2016) reported several gar MHC class I transcripts of the MHC class I Z and P lineages that did not map to the reference genome. These MHC class I lineages were previously thought to be restricted to teleost genomes (Dirscherl and Yoder, 2014; Dirscherl et al., 2014). Although DICP sequences are most similar to mammalian CD300 sequences, our analyses of the genomic region containing gar DICP sequences revealed no synteny with the human CD300 gene cluster on chromosome 17. Nevertheless, there is evidence linking gar DICPs to coelacanth DICPs - the DICP sequences on gar scaffold JH591438.1 and one of the coelacanth DICP sequences are both linked to the ddah2 (dimethylarginine dimethylaminohydrolase 2) gene (Fig. 8). Interestingly, DDAH2 is encoded within the human MHC locus on chromosome 6 (Flajnik and Kasahara, 2001). These observations support the model that zebrafish, gar and coelacanth DICP sequences share an evolutionary origin within the proto-MHC.
Figure 8. DICP genes of zebrafish, spotted gar and coelacanth are linked to genes associated with the MHC.
Syntenic relationships of DICP-containing loci are displayed for (A) zebrafish, (B, C) spotted gar and (D, E) coelacanth. Gene positions are intended only to give a sense of order and orientation and are not drawn to scale. DICP genes are shaded black. Genes with orthologs to the four paralogous human MHC loci (Flajnik and Kasahara, 2001) are shaded in gray. Novel genes or genes not known to be linked with MHC loci are white. (A) The linkage of zebrafish DICPs to MHC class I genes is well documented (Haire et al., 2012). (B-E) Genomic regions encoding gar and coelacanth DICP sequences are shown above with expanded views below. Exons from Ensembl-predicted DICP genes are shaded black, while additional DICP Ig domains are shaded grey, and drawn to scale. Dashed horizontal lines with a double slash represent gaps in genomic sequences introduced to reduce figure size. (D) Coelacanth MHC class I genes were previously described (Saha et al., 2014).
CONCLUSIONS
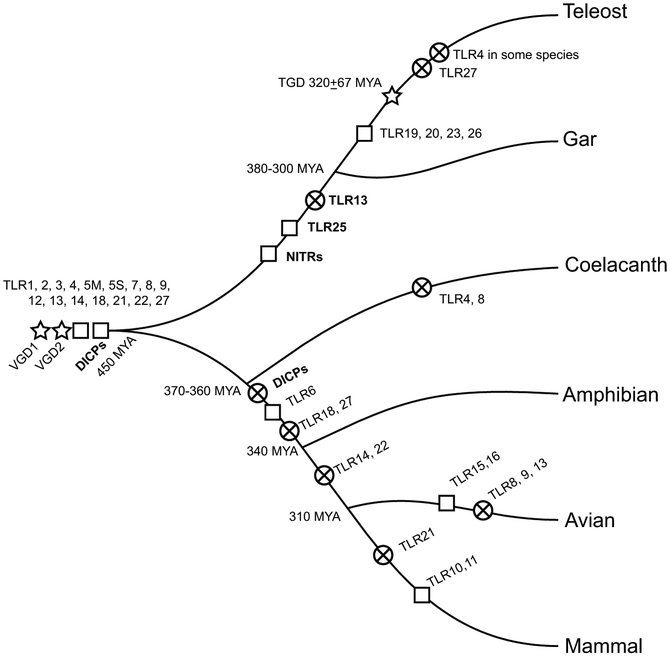
Multiple teleost species are being employed as model species for human diseases (Schartl, 2014); however, as seen here, the genomes of these species can be quite diverse from each other as well as from human. It is likely that the TGD combined with more recent lineage-specific expansions and diversification of a range of innate immune receptor gene families are responsible, at least in part, for these differences, especially in the molecular mediators of the immune system. Here, we have shown that although gar did not undergo the TGD, gar nonetheless possess a heightened repertoire of TLRs similar to teleosts. The gar genome also encodes NITRs and DICPs - two multigene families previously believed restricted to teleost species. This suggests that both NITRs and DICPs are characteristics of innate immunity that existed prior to the TGD. The evolutionary context of these gene families with respect to vertebrate evolution are summarized in Figure 9. As NITRs are absent in coelacanth, but present in gar and teleosts, we have placed their birth after the split of ray-finned fishes from lobe-finned fishes. As DICPs are present in teleost, gar and coelacanth, they have been placed in a more basal position on Figure 9. The “arms race” between host and pathogen as well as co-evolution between commensal microorganisms and host has likely shaped the genomic repertoire of immune response genes. The environment inhabited by fish ensures intimate interactions with microorganisms and may explain the expansions of some of these immune gene families. IIIR families often have few activating receptors but many inhibitory receptors; the ectodomains are often highly similar, but amino acid sequence differences may have evolved to tolerate the presence of certain ligands (e.g. commensal bacteria). The sequencing of additional fish genomes will likely reshape our view on the evolutionary history of some of these genes. For instance, TLR27 has been identified in coelacanth, gar, and elephant shark (Wang et al., 2015), but not yet described in teleost: therefore, we have TLR27 lost in teleost, but it may be identified in select teleost lineages at a later date. It should be kept in mind that sequence similarity does not always reflect functional similarity. For example, TLRs in Drosophila melanogaster and Caenorhabditis elegans play critical roles in embryonic development as well as immune function (Leulier and Lemaitre, 2008). Therefore, functional assignment of orthologous genes may require further experimental investigations.
Figure 9. Evolutionary context of TLRs, NITRs and DICPs among selected vertebrate lineages.
Stars represent whole genome duplications. Squares represent the earliest known occurrence of a gene based on its presence in at least one species of a vertebrate lineage while crossed circles represent the apparent loss of a gene or gene family in the remaining radiations. These gene representations are placed arbitrarily and the order of gains or losses are not considered. The approximate age of vertebrate lineages are described by Beutler and Rehli (Beutler and Rehli, 2002; Amores et al., 2011). This figure represents our current view on the histories of TLRs, NITRs and DICPs, based on information described in the main text (bold) and summarized by Hansen et al (Hansen et al., 2011; Boudinot et al., 2014b).
The gar genome has afforded a unique look into what may be an ancestral precursor of several immune gene families. Gar DICP and NITR genes have not undergone the large expansion and diversification observed in teleosts. In the case of immune genes, different gene families may substitute another gene family if they can function similarly. Perhaps the most famous example are mouse Ly49 and human KIRs. Therefore, it may be that novel mediators of immune function await discovery in gar. As genomic sequencing of novel genomes and functional annotation of these genes becomes available, we anticipate exciting revelations regarding the evolutionary processes driving gene gain/loss, gene expansion, and gene diversification. Importantly, understanding the similarities and differences between species is pinnacle to furthering our understanding of health and disease.
Supplementary Material
Supplementary Material Figure S1. Evidence for TLR conserved synteny. Genes of the same color outlined in black are homologs. Genes outlined in gray are paralogs of the genes in the same color. Light gray genes represent genes not found in the reference sequence. All TLR and TLR-like genes are white in color. The center-most gene, 16th gene, in white, is the TLR gene of interest or each panel. The top spotted gar linkage group is always the reference sequence for which gene names are given, when possible. Figures were generated using Genomicus 84.01 (Louis et al., 2015).
ACKNOWLEDGEMENTS
The authors thank Ingo Braasch, John Postlethwait and all who contributed to sequencing the spotted gar genome. We thank Chris Amemiya, Ronda Litman, Nil Ratan Saha, and Michael Criscitiello for discussions of gar immune genes. The authors declare no conflict of interest.
Grant sponsor: National Institutes of Health; Grant number: R01 AI057559
Grant sponsor: National Evolutionary Synthesis Center, National Science Foundation; Grant number: EF-0905606
Grant sponsor: Triangle Center for Evolutionary Medicine
REFERENCES
- Aderem A, Ulevitch RJ. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782–787. [DOI] [PubMed] [Google Scholar]
- Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Juettemann T, Keenan S, Laird MR, Lavidas I, Maurel T, McLaren W, Moore B, Murphy DN, Nag R, Newman V, Nuhn M, Ong CK, Parker A, Patricio M, Riat HS, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Wilder SP, Zadissa A, Kostadima M, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Cunningham F, Yates A, Zerbino DR, Flicek P. 2016. Ensembl 2017. Nucleic Acids Res [Internet]. Available from: 10.1093/nar/gkw1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188:799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282:1711–1714. [DOI] [PubMed] [Google Scholar]
- Barrow AD, Trowsdale J. 2006. You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol 36:1646–1653. [DOI] [PubMed] [Google Scholar]
- Berczi I, Bereznai T. 1966. COMPARATIVE STUDIES ON THE TOXICITY OF ESCHERICHIA COLI LIPOPOLYSACCHARIDE ENDOTOXIN IN VARIOUS ANIMAL SPECIES. CANADIAN JOURNAL OF MICROBIOLOGY J Gen Canadian Journal of Microbiology 12. [DOI] [PubMed] [Google Scholar]
- Beutler B 2000. Tlr4: central component of the sole mammalian LPS sensor. Curr Opin Immunol 12:20–26. [DOI] [PubMed] [Google Scholar]
- Beutler B, Rehli M. 2002. Evolution of the TIR, Tolls and TLRs: Functional Inferences from Computational Biology. In: Springer; Berlin Heidelberg: p 1–21. [DOI] [PubMed] [Google Scholar]
- Borrego F 2013. The CD300 molecules: an emerging family of regulators of the immune system. Blood 121:1951–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudinot P, Zou J, Ota T, Buonocore F, Scapigliati G, Canapa A, Cannon J, Litman G, Hansen JD. 2014a. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J Exp Zool B Mol Dev Evol 322:415–437. [DOI] [PubMed] [Google Scholar]
- Boudinot P, Zou J, Ota T, Buonocore F, Scapigliati G, Canapa A, Cannon J, Litman G, Hansen JD. 2014b. A tetrapod-like repertoire of innate immune receptors and effectors for coelacanths. J Exp Zool B Mol Dev Evol 322:415–437. [DOI] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Cañestro C, Sydes J, Beaudry FEG, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SMJ, Aken B, Yandell M, Schneider I, Yoder JA, Volff J-N, Meyer A, Amemiya CT, Venkatesh B, Holland PWH, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alföldi J, Lindblad-Toh K, Postlethwait JH. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, Turner-Maier J, Johnson J, Alcazar R, Noh HJ, Russell P, Aken B, Alföldi J, Amemiya C, Azzouzi N, Baroiller J-F, Barloy-Hubler F, Berlin A, Bloomquist R, Carleton KL, Conte MA, D’Cotta H, Eshel O, Gaffney L, Galibert F, Gante HF, Gnerre S, Greuter L, Guyon R, Haddad NS, Haerty W, Harris RM, Hofmann HA, Hourlier T, Hulata G, Jaffe DB, Lara M, Lee AP, MacCallum I, Mwaiko S, Nikaido M, Nishihara H, Ozouf-Costaz C, Penman DJ, Przybylski D, Rakotomanga M, Renn SCP, Ribeiro FJ, Ron M, Salzburger W, Sanchez-Pulido L, Santos ME, Searle S, Sharpe T, Swofford R, Tan FJ, Williams L, Young S, Yin S, Okada N, Kocher TD, Miska EA, Lander ES, Venkatesh B, Fernald RD, Meyer A, Ponting CP, Streelman JT, Lindblad-Toh K, Seehausen O, Di Palma F. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore F, Gerdol M. 2016. Alternative adaptive immunity strategies: coelacanth, cod and shark immunity. Mol Immunol 69:157–169. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Hernandez Prada JA, Bailey KM, Jakoncic J, Litman GW, Ostrov DA. 2008. A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity 29:228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Bustamante P, Keşmir C, de Boer RJ. 2016. The evolution of natural killer cell receptors. Immunogenetics 68:3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S, Heffelfinger AK, Orcutt TM, Litman GW, Yoder JA. 2008. The medaka novel immune-type receptor (NITR) gene clusters reveal an extraordinary degree of divergence in variable domains. BMC Evol Biol 8:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirscherl H, McConnell SC, Yoder JA, de Jong JLO. 2014. The MHC class I genes of zebrafish. Dev Comp Immunol 46:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirscherl H, Yoder JA. 2014. Characterization of the Z lineage Major histocompatability complex class I genes in zebrafish. Immunogenetics 66:185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschmeyer WN, Fricke R, van der Laan R. 2016. Catalog of fishes on-line database: http://www.calacademy.org/scientists/projects/catalog-of-fishes. Available from: http://www.calacademy.org/scientists/projects/catalog-of-fishes
- Ferraresso S, Kuhl H, Milan M, Ritchie DW, Secombes CJ, Reinhardt R, Bargelloni L. 2009. Identification and characterisation of a novel immune-type receptor (NITR) gene cluster in the European sea bass, Dicentrarchus labrax, reveals recurrent gene expansion and diversification by positive selection. Immunogenetics 61:773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. 2001. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15:351–362. [DOI] [PubMed] [Google Scholar]
- Gay NJ, Gangloff M. 2007. Structure and Function of Toll Receptors and Their Ligands. http://dx.doi.org/101146/annurev.biochem76060305151318. [DOI] [PubMed]
- Glasauer SMK, Neuhauss SCF. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics 289:1045–1060. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Tsukamoto K, Azuma T, Leong J, Koop BF, Dijkstra JM. 2015. A comprehensive analysis of teleost MHC class I sequences. BMC Evol Biol 15:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein LA, Norman PJ, Hilton HHG, Parham P. 2015. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev 267:259–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire RN, Cannon JP, O’Driscoll ML, Ostrov DA, Mueller MG, Turner PM, Litman RT, Litman GW, Yoder JA. 2012. Genomic and functional characterization of the diverse immunoglobulin domain-containing protein (DICP) family. Genomics 99:282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JD, Vojtech LN, Laing KJ. 2011. Sensing disease and danger: A survey of vertebrate PRRs and their origins. Dev Comp Immunol 35:886–897. [DOI] [PubMed] [Google Scholar]
- Huang X-N, Wang Z-Y, Yao C-L. 2011. Characterization of Toll-like receptor 3 gene in large yellow croaker, Pseudosciaena crocea. Fish Shellfish Immunol 31:98–106. [DOI] [PubMed] [Google Scholar]
- Jack RS. 2015. Evolution of Immunity and Pathogens. Results Probl Cell Differ 57:1–20. [DOI] [PubMed] [Google Scholar]
- Jamil KM, Khakoo SI. 2011. KIR/HLA interactions and pathogen immunity. J Biomed Biotechnol 2011:298348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault C, Pichon L, Chluba J. 2004. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol Immunol 40:759–771. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- Kanwal Z, Wiegertjes GF, Veneman WJ, Meijer AH, Spaink HP. 2014. Comparative studies of Toll-like receptor signalling using zebrafish. Dev Comp Immunol 46:35–52. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. 2006. TLR signaling. Cell Death Differ 13:816–825. [DOI] [PubMed] [Google Scholar]
- Keestra AM, de Zoete MR, Bouwman LI, van Putten JPM. 2010. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol 185:460–467. [DOI] [PubMed] [Google Scholar]
- Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. 2005. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J Biol Chem [Internet] 280 Available from: 10.1074/jbc.M414607200 [DOI] [PubMed] [Google Scholar]
- Kock H, Fischer U. 2008. A novel immunoglobulin-like transcript from rainbow trout with two Ig-like domains and two isoforms. Mol Immunol 45:1612–1622. [DOI] [PubMed] [Google Scholar]
- Kortum AN, Rodriguez-Nunez I, Yang J, Shim J, Runft D, O’Driscoll ML, Haire RN, Cannon JP, Turner PM, Litman RT, Kim CH, Neely MN, Litman GW, Yoder JA. 2014. Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics 66:267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Meyer A. 2010. Whole Genome Duplications and the Radiation of Vertebrates In: Evolution after Gene Duplication. John Wiley & Sons, Inc; p 299–311. [Google Scholar]
- Lane RP, Roach JC, Lee IY, Boysen C, Smit A, Trask BJ, Hood L. 2002. Genomic analysis of the olfactory receptor region of the mouse and human T-cell receptor alpha/delta loci. Genome Res 12:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Lemaitre B. 2008. Toll-like receptors — taking an evolutionary approach. Nat Rev Genet 9:165–178. [DOI] [PubMed] [Google Scholar]
- Louis A, Nguyen NTT, Muffato M, Roest Crollius H. 2015. Genomicus update 2015: KaryoView and MatrixView provide a genome-wide perspective to multispecies comparative genomics. Nucleic Acids Res 43:D682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Li C, Zhang P, Shao Y, Su X, Li Y, Li T. 2013. Two adaptor molecules of MyD88 and TRAF6 in Apostichopus japonicus Toll signaling cascade: Molecular cloning and expression analysis. Developmental & Comparative Immunology [Internet]:498–504. Available from: 10.1016/j.dci.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Star B, Snipen LG, Hansen TF, Baalsrud HT, Nederbragt AJ, Hanel R, Salzburger W, Stenseth NC, Jakobsen KS, Jentoft S. 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet 48:1204–1210. [DOI] [PubMed] [Google Scholar]
- Manser AR, Weinhold S, Uhrberg M. 2015. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev 267:178–196. [DOI] [PubMed] [Google Scholar]
- Matsuo A, Oshiumi H, Tsujita T, Mitani H, Kasai H, Yoshimizu M, Matsumoto M, Seya T. 2008. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J Immunol 181:3474–3485. [DOI] [PubMed] [Google Scholar]
- Matsushima N, Tachi N, Kuroki Y, Enkhbayar P, Osaki M, Kamiya M, Kretsinger RH. 2005. Structural analysis of leucine-rich-repeat variants in proteins associated with human diseases. Cell Mol Life Sci 62:2771–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima N, Tanaka T, Enkhbayar P, Mikami T, Taga M, Yamada K, Kuroki Y. 2007. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics 8:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miggin SM, Neill LAJO’. New insights into the regulation of TLR signaling. Available from: 10.1189/jlb.1105672 [DOI] [PubMed]
- Morrison DC, Ryan JL. 1987. Mechanisms. Annu Rev Med 38:417–432. [DOI] [PubMed] [Google Scholar]
- Muñoz I, Sepulcre MP, Meseguer J, Mulero V. 2013. Molecular cloning, phylogenetic analysis and functional characterization of soluble Toll-like receptor 5 in gilthead seabream, Sparus aurata. Fish Shellfish Immunol 35:36–45. [DOI] [PubMed] [Google Scholar]
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the World. Wiley. [Google Scholar]
- Ostergaard AE, Lubieniecki KP, Martin SAM, Stet RJM, Davidson WS, Secombes CJ. 2010. Genomic organisation analysis of novel immunoglobulin-like transcripts in Atlantic salmon (Salmo salar) reveals a tightly clustered and multigene family. BMC Genomics 11:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Østergaard AE, Martin SAM, Wang T, Stet RJM, Secombes CJ. 2009. Rainbow trout (Oncorhynchus mykiss) possess multiple novel immunoglobulin-like transcripts containing either an ITAM or ITIMs. Dev Comp Immunol 33:525–532. [DOI] [PubMed] [Google Scholar]
- Palti Y 2011. Toll-like receptors in bony fish: from genomics to function. Dev Comp Immunol 35:1263–1272. [DOI] [PubMed] [Google Scholar]
- Pasquier J, Cabau C, Nguyen T, Jouanno E, Severac D, Braasch I, Journot L, Pontarotti P, Klopp C, Postlethwait JH, Guiguen Y, Bobe J. 2016. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics 17:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiniou SMA, Boudinot P, Bengtén E. 2013. Comprehensive survey and genomic characterization of Toll-like receptors (TLRs) in channel catfish, Ictalurus punctatus: identification of novel fish TLRs. Immunogenetics 65:511–530. [DOI] [PubMed] [Google Scholar]
- Rahim MMA, Makrigiannis AP. 2015. Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev 267:137–147. [DOI] [PubMed] [Google Scholar]
- Rauta PR, Samanta M, Dash HR, Nayak B, Das S. 2014. Toll-like receptors (TLRs) in aquatic animals: signaling pathways, expressions and immune responses. Immunol Lett 158:14–24. [DOI] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. 2005. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci U S A 102:9577–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Nunez I, Wcisel DJ, Litman GW, Yoder JA. 2014. Multigene families of immunoglobulin domain-containing innate immune receptors in zebrafish: deciphering the differences. Dev Comp Immunol 46:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Nunez I, Wcisel DJ, Litman RT, Litman GW, Yoder JA. 2016. The identification of additional zebrafish DICP genes reveals haplotype variation and linkage to MHC class I genes. Immunogenetics 68:295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, Buonocore F, Canapa A, Cheng J-F, Amemiya CT. 2014. Genome complexity in the coelacanth is reflected in its adaptive immune system. J Exp Zool B Mol Dev Evol 322:438–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- Samanta M, Basu M, Swain B, Panda P, Jayasankar P. 2013. Molecular cloning and characterization of toll-like receptor 3, and inductive expression analysis of type I IFN, Mx and pro-inflammatory cytokines in the Indian carp, rohu (Labeo rohita). Mol Biol Rep 40:225–235. [DOI] [PubMed] [Google Scholar]
- Schartl M 2014. Beyond the zebrafish: diverse fish species for modeling human disease. Dis Model Mech 7:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre MP, Alcaraz-Pérez F, López-Muñoz A, Roca FJ, Meseguer J, Cayuela ML, Mulero V. 2009. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol 182:1836–1845. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JL, Bengtén E, Du Pasquier L, McIntosh RD, Quiniou SM, Clem LW, Miller NW, Wilson M. 2006. A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics 58:758–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JL, Bengtén E, Du Pasquier L, Miller NW, Wilson M. 2007. Channel catfish leukocyte immune-type receptors contain a putative MHC class I binding site. Immunogenetics 59:77–91. [DOI] [PubMed] [Google Scholar]
- Star B, Jentoft S. 2012. Why does the immune system of Atlantic cod lack MHC II? Bioessays 34:648–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrøm M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, Wetten OF, Lanzén A, Winer R, Knight J, Vogel J-H, Aken B, Andersen O, Lagesen K, Tooming-Klunderud A, Edvardsen RB, Tina KG, Espelund M, Nepal C, Previti C, Karlsen BO, Moum T, Skage M, Berg PR, Gjøen T, Kuhl H, Thorsen J, Malde K, Reinhardt R, Du L, Johansen SD, Searle S, Lien S, Nilsen F, Jonassen I, Omholt SW, Stenseth NC, Jakobsen KS. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stet RJM, Hermsen T, Westphal AH, Jukes J, Engelsma M, Lidy Verburg-van Kemenade BM, Dortmans J, Aveiro J, Savelkoul HFJ. 2005. Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics 57:77–89. [DOI] [PubMed] [Google Scholar]
- Strong SJ, Mueller MG, Litman RT, Hawke NA, Haire RN, Miracle AL, Rast JP, Amemiya CT, Litman GW. 1999. A novel multigene family encodes diversified variable regions. Proc Natl Acad Sci U S A 96:15080–15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su GL, Klein RD, Aminlari A, Zhang HY, Steinstraesser L, Alarcon WH, Remick DG, Wang SC. 2000. Kupffer cell activation by lipopolysaccharide in rats: Role for lipopolysaccharide binding protein and toll-like receptor 4. Hepatology 31:932–936. [DOI] [PubMed] [Google Scholar]
- Sullivan C, Charette J, Catchen J, Lage CR, Giasson G, Postlethwait JH, Millard PJ, Kim CH. 2009. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol 183:5896–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. 2002. Cutting Edge: Role of Toll-Like Receptor 1 in Mediating Immune Response to Microbial Lipoproteins. The Journal of Immunology 169:10–14. [DOI] [PubMed] [Google Scholar]
- Taylor EB, Moulana M, Stuge TB, Quiniou SMA, Bengten E, Wilson M. 2016. A Leukocyte Immune-Type Receptor Subset Is a Marker of Antiviral Cytotoxic Cells in Channel Catfish, Ictalurus punctatus. J Immunol 196:2677–2689. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. 2003. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res 13:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. [DOI] [PubMed] [Google Scholar]
- Tsujita T, Tsukada H, Nakao M, Oshiumi H, Matsumoto M, Seya T. 2004. Sensing Bacterial Flagellin by Membrane and Soluble Orthologs of Toll-like Receptor 5 in Rainbow Trout (Onchorhynchus mikiss). J Biol Chem 279:48588–48597. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang Z, Liu J, Li F, Chang F, Fu H, Zhao J, Yin D. 2015. Structural characterization and evolutionary analysis of fish-specific TLR27. Fish Shellfish Immunol 45:940–945. [DOI] [PubMed] [Google Scholar]
- Wcisel DJ, Yoder JA. 2016. The confounding complexity of innate immune receptors within and between teleost species. Fish Shellfish Immunol 53:24–34. [DOI] [PubMed] [Google Scholar]
- Wei S, Zhou J-M, Chen X, Shah RN, Liu J, Orcutt TM, Traver D, Djeu JY, Litman GW, Yoder JA. 2007. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics 59:813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. 1990. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249:1431–1433. [DOI] [PubMed] [Google Scholar]
- Yeh D-W, Liu Y-L, Lo Y-C, Yuh C-H, Yu G-Y, Lo J-F, Luo Y, Xiang R, Chuang T-H. 2013a. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc Natl Acad Sci U S A 110:20711–20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh D-W, Liu Y-L, Lo Y-C, Yuh C-H, Yu G-Y, Lo J-F, Luo Y, Xiang R, Chuang T-H. 2013b. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc Natl Acad Sci U S A 110:20711–20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA. 2009. Form, function and phylogenetics of NITRs in bony fish. Dev Comp Immunol 33:135–144. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Cannon JP, Litman RT, Murphy C, Freeman JL, Litman GW. 2008. Evidence for a transposition event in a second NITR gene cluster in zebrafish. Immunogenetics 60:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Litman GW. 2011. The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics 63:123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Litman RT, Mueller MG, Desai S, Dobrinski KP, Montgomery JS, Buzzeo MP, Ota T, Amemiya CT, Trede NS, Wei S, Djeu JY, Humphray S, Jekosch K, Hernandez Prada JA, Ostrov DA, Litman GW. 2004. Resolution of the novel immune-type receptor gene cluster in zebrafish. Proc Natl Acad Sci U S A 101:15706–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Mueller MG, Wei S, Corliss BC, Prather DM, Willis T, Litman RT, Djeu JY, Litman GW. 2001. Immune-type receptor genes in zebrafish share genetic and functional properties with genes encoded by the mammalian leukocyte receptor cluster. Proc Natl Acad Sci U S A 98:6771–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Turner PM, Wright PD, Wittamer V, Bertrand JY, Traver D, Litman GW. 2010. Developmental and tissue-specific expression of NITRs. Immunogenetics 62:117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kong X, Zhou C, Li L, Nie G, Li X. 2014. Toll-like receptor recognition of bacteria in fish: Ligand specificity and signal pathways. Fish Shellfish Immunol 41:380–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Figure S1. Evidence for TLR conserved synteny. Genes of the same color outlined in black are homologs. Genes outlined in gray are paralogs of the genes in the same color. Light gray genes represent genes not found in the reference sequence. All TLR and TLR-like genes are white in color. The center-most gene, 16th gene, in white, is the TLR gene of interest or each panel. The top spotted gar linkage group is always the reference sequence for which gene names are given, when possible. Figures were generated using Genomicus 84.01 (Louis et al., 2015).