Significance
The effects of diverse environmental factors on the crop circadian clock have not been systemically studied. We devised a reliable and cost-effective discovery pipeline and demonstrated its feasibility by mapping the abiotic stress “inputome” of the soybean clock. We found that the interfaces between the soybean clock and abiotic stress signals were quite different from those in Arabidopsis. Furthermore, alkaline stress was identified as a circadian clock modulator. These findings highlight the complexity of the soybean clock, which cannot be simply extrapolated from knowledge obtained from Arabidopsis. Our discovery pipeline thus offers a broadly applicable and affordable tool for similar large-scale circadian clock studies in diverse species since it does not rely on transgenic circadian reporters or expensive imaging systems.
Keywords: soybean circadian clock, abiotic stress, molecular timetable, RASL-seq, comprehensive map
Abstract
The plant circadian clock evolved to increase fitness by synchronizing physiological processes with environmental oscillations. Crop fitness was artificially selected through domestication and breeding, and the circadian clock was identified by both natural and artificial selections as a key to improved fitness. Despite progress in Arabidopsis, our understanding of the crop circadian clock is still limited, impeding its rational improvement for enhanced fitness. To unveil the interactions between the crop circadian clock and various environmental cues, we comprehensively mapped abiotic stress inputs to the soybean circadian clock using a 2-module discovery pipeline. Using the “molecular timetable” method, we computationally surveyed publicly available abiotic stress-related soybean transcriptomes to identify stresses that have strong impacts on the global rhythm. These findings were then experimentally confirmed using a multiplexed RNA sequencing technology. Specific clock components modulated by each stress were further identified. This comprehensive mapping uncovered inputs to the plant circadian clock such as alkaline stress. Moreover, short-term iron deficiency targeted different clock components in soybean and Arabidopsis and thus had opposite effects on the clocks of these 2 species. Comparing soybean varieties with different iron uptake efficiencies suggests that phase modulation might be a mechanism to alleviate iron deficiency symptoms in soybean. These unique responses in soybean demonstrate the need to directly study crop circadian clocks. Our discovery pipeline may serve as a broadly applicable tool to facilitate these explorations.
Plants, like many other living organisms, have an internal timekeeper called the circadian clock, which synchronizes physiological processes to allow the anticipation of recurring daily and seasonal environmental changes to coordinate metabolic activities. The current understanding of the molecular architecture of the plant circadian clock derives primarily from studies performed in the model organism Arabidopsis thaliana. In Arabidopsis, the clock system is composed of 3 parts: a central oscillator driven by interlocked transcription–translation feedback loops that generate the endogenous circadian rhythms; input pathways that integrate environmental cues to the oscillator function; and output pathways that control diverse physiological processes, including growth, flowering time, and stress responses (1, 2). While light is considered the dominant input into the plant circadian clock, accumulating studies have demonstrated that a wide spectrum of stress signals also provide feedback regulation of the circadian clock in Arabidopsis (1). These emerging discoveries suggest that the plant circadian clock may function as a central signaling hub that integrates diverse stress signals to balance the energy needs for stress tolerance and growth.
In contrast to Arabidopsis, for most crop plants, although circadian clock genes have been repeatedly identified to be associated with key agronomic traits, little is known about the impacts of environmental stresses on the circadian clock (3–5). Artificial selection during domestication has even resulted in the deceleration of the circadian clock in cultivated tomatoes to allow better adaptation to the longer summer days that they encountered as they were moved further away from the equator (6). Our limited understanding of the environmental factors that influence crop circadian clocks represents a key knowledge gap that impedes further germplasm improvement through more targeted selection for specific circadian traits. To overcome these obstacles, we developed a reliable and cost-effective 2-module discovery pipeline in soybean to comprehensively map abiotic stress inputs to the circadian clock.
Soybean (Glycine max L. Merrill) is the primary source of the world’s supply of vegetable protein and oil. Demand for soybean has grown rapidly due to soybean’s wide range of applications in food, feed, and industrial products. However, soybean production is threatened by several abiotic stresses, such as drought (7), soil salinity (8, 9), and nutrient deficiency (10, 11). The effects of these environmental stresses on the soybean circadian clock and their interface with specific components of the central oscillator remain largely unknown, even though the clock has already been suggested to influence soybean yield. Modulation of central clock components by overexpressing a B-box protein involved in light signaling has led to soybean grain yield increases year after year in multiple transgenic events in multilocation field trials (5). Natural variation in soybean ELF3, a key component of the central oscillator network, has been shown to improve soybean’s adaptation to the tropics (12). Therefore, deciphering the interacting network between environmental stress signals and the soybean circadian clock may help enhance the abiotic stress tolerance of soybean.
Our 2-module discovery pipeline consists of a molecular timetable module involving computational heuristics and a module based on RASL-seq (RNA-mediated oligonucleotide annealing, selection, and ligation with next-generation sequencing), a large-scale targeted sequencing technology. The molecular timetable method is a method for genome-scale global rhythm analysis first developed for mice (13) and later adapted for the analysis of Arabidopsis (14) and tomato (15). It relies on the average expression profiles of time-indicating genes to assess the global rhythm based on a single-time-point genome-wide expression profile rather than a time-course experiment. Therefore, it can be readily applied to existing genome-wide expression profiles. Using these computational heuristics, we surveyed publicly available soybean transcriptomes related to abiotic stresses, including 19 datasets with 306 microarrays and RNA-seq samples (7–11, 16–28), and identified specific abiotic stresses that dramatically perturbed the global rhythm. To enable the affordable experimental validation of these computational predictions, RASL-seq was adapted for soybean. RASL-seq is a probe-based targeted sequencing method that can accommodate the profiling of hundreds of genes in hundreds of samples in a single HiSeq. 2000/2500 sequencing lane (29), making large-scale expression profiling cost-effective. By applying RASL-seq to over 300 time-course experiment samples from various abiotic stress treatments, we confirmed our computational predictions and further identified specific circadian clock components targeted by each abiotic stress. By integrating the molecular timetable method and the RASL-seq technology, we discovered that the soybean circadian clock responds to short-term iron deficiency in a significantly different manner from that in Arabidopsis. We also discovered that alkaline stress has a widespread impact on the circadian clock components in plants.
Results
Identification of Time-Indicating Genes.
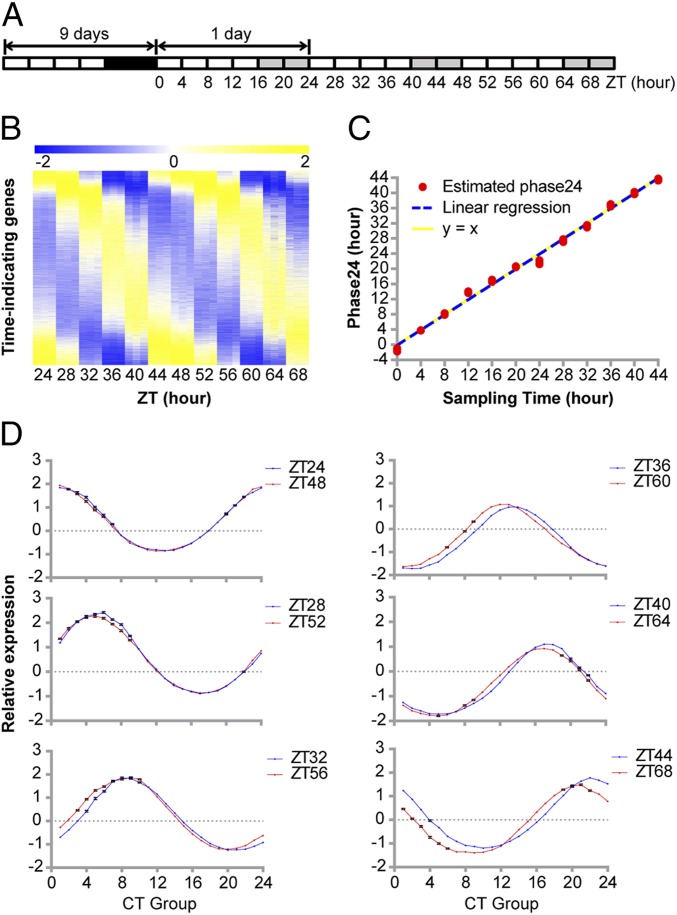
Central to the molecular timetable method is the availability of time-indicating genes derived from genome-wide circadian time-course expression profiles. To identify time-indicating genes in soybean, the transcriptome of soybean cultivar Williams 82 was analyzed via RNA-seq in samples subjected to constant light conditions during a 2-d time-course experiment (Fig. 1A). Briefly, soybean seedlings were grown in soil under diurnal conditions (16 h light/8 h dark) for 9 d. At the end of the ninth day, they were exposed to constant light conditions and unifoliolate leaves were collected starting at ZT (Zeitgeber time) 24 after 1 d of acclimation in constant light. The samples were harvested every 4 h for 2 d and subjected to 100-base pair paired-end Illumina sequencing. There was an average of 57.6 million clean reads per sample and a maximum of 65.3 million clean reads, demonstrating a high-coverage expression profile. Sampling was performed under the constant light conditions to eliminate the effect of photoperiod and reveal the function of the endogenous circadian clock.
Fig. 1.
Identification of time-indicating genes and validation of the molecular timetable method in soybean. (A) Sampling scheme for circadian time-course RNA-seq of soybean unifoliolate leaves. The numbers mark the sampling time. White boxes: day; black boxes: night; gray boxes: subjective night under the constant light conditions. (B) Heat map of the standardized expression levels of 3,695 time-indicating genes. Three biological replicates are shown as adjacent columns within each sampling time. Genes were sorted based on their peak expression times and are organized into rows. (C) The molecular timetable method can estimate actual sampling time with high precision. The estimated Phase24 was plotted against actual sampling time. Linear regression was performed, and the y = x line was plotted as a reference. (D) Normalized expression levels of time-indicating genes binned into 24 CT groups based on their phases. Samples from the same time of a day from days 1 and 2 are plotted in blue and red, respectively. Mean and SE are plotted. SEs of some data points are smaller than the size of the data symbol, so they may appear invisible.
To identify the time-indicating genes, the expression profiles of each gene from triplicated samples combined into a single profile were fit against 1,440 cosine curves (1 curve per min) to assess their rhythmicity. The coefficient of variation (CV) of each gene’s expression profile during the time course was used to select for oscillatory genes with high relative amplitude; this strategy ensured robustness against noise (see Materials and Methods for details). In total, 3,695 genes were determined to be time-indicating genes (Fig. 1B); their phases therefore reflect the time of the day (Dataset S1). Based on the calculated phase, these 3,695 genes were further assigned to 24 phase groups with peak expression between CT (circadian time) 0 and CT23. The 24 time-indicating groups are hereafter termed CT groups.
To assess the performance of these time-indicating genes, we first applied the molecular timetable method to our time-course RNA-seq dataset itself. Through nonlinear regression, we derived Phase24, a period-corrected estimation of circadian sampling time, for each RNA-seq sample and fit it to the actual sampling time (Fig. 1C). The estimated linear relationship was statistically indistinguishable from a diagonal line, clearly showing that the molecular timetable method using these time-indicating genes can provide a good reflection of the actual sampling time. Interestingly, while the global rhythm of samples collected at ZT24 and ZT48 appeared visually indistinguishable, asynchrony started to emerge over time as the global rhythm of samples collected at ZT44 and ZT68 had significant differences (Fig. 1D). This indicates that Phase24 does not simply equal the sampling time but rather reflects the actual endogenous time of the sample, which is set by the circadian clock (13).
To further validate the performance of these time-indicating genes, we tested them in a well-designed time-course profiling experiment related to mild drought stress (Fig. 2 A and B) originally performed by Rodrigues et al. (7). Although the experimental conditions, including cultivar, photoperiod, light intensity, temperature, and humidity, were quite different from our circadian time-course RNA-seq experiment (Dataset S2), the estimated Phase24 still closely matched the sampling time of the control samples (Fig. 2B). Therefore, the time-indicating genes that we identified can be used reliably in the molecular timetable method to accurately reflect the global rhythm of each individual biological sample based on its genome-wide expression profile.
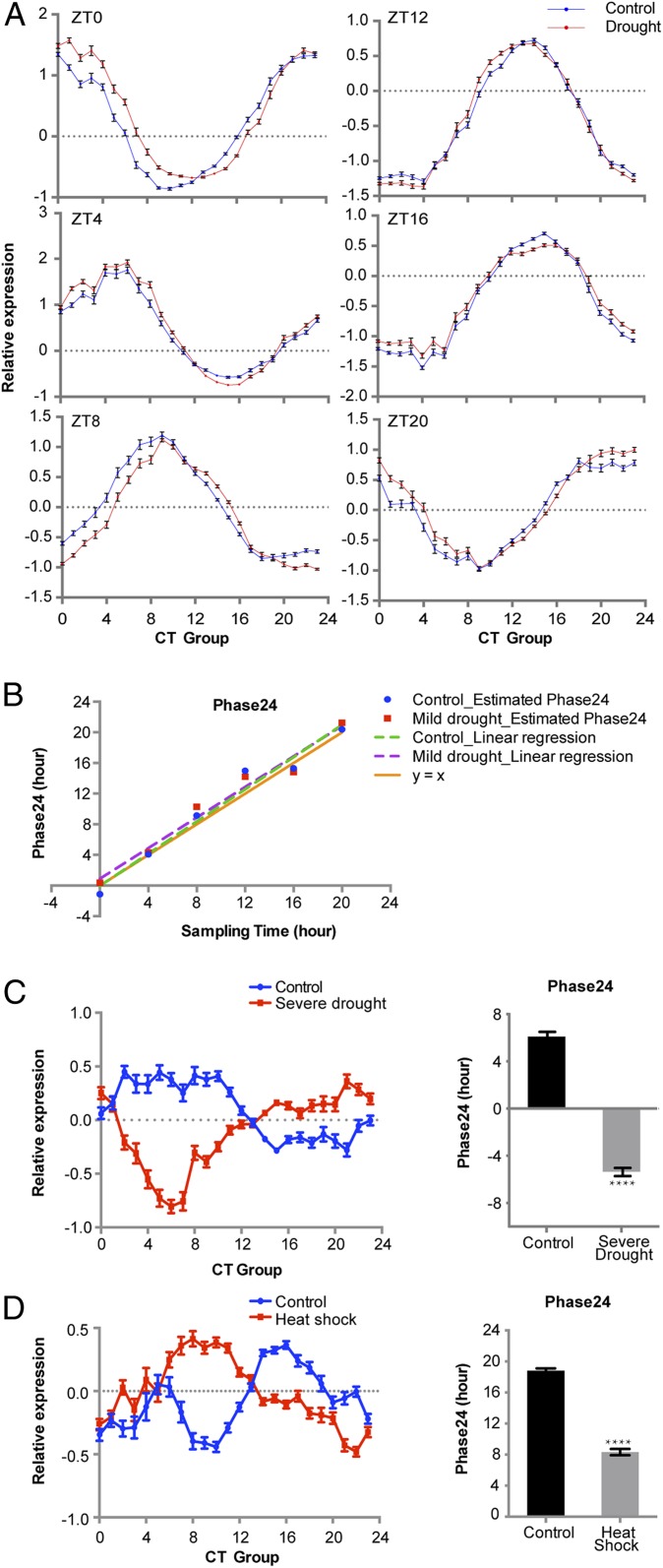
Fig. 2.
Drought severity correlates with perturbations of global rhythm, while heat shock has dramatic impact on soil-grown soybean seedlings. (A) Normalized expression levels of time-indicating genes in control and mild drought-treated samples. Treatments were applied at ZT0, and samples were harvested at the indicated times. Mean and SE are plotted. SEs of some data points are smaller than the size of the data symbol, so they may appear invisible. (B) Estimated Phase24 has a good linear relationship with reported sampling time. Linear regression was performed using estimated Phase24 and reported sampling time. Adjusted R2 = 0.9999 for both control and drought-treated samples. y = x is plotted as a reference. (C) Severe drought stress perturbs global rhythm and causes a dramatic phase shift. (D) Heat shock stress (30-min heat treatment at 42 °C) perturbs global rhythm dramatically. Mean and SE are plotted. ****P < 0.0001 (Student’s t test). A and B are derived from GSE69469. C is derived from GSE40627. D is derived from GSE26198.
Application of the Molecular Timetable Method.
To identify specific abiotic stresses that may perturb the global rhythm in soybean, we first assembled the publicly available soybean transcriptomes related to abiotic stresses. These included 19 datasets with 306 RNA-seq and microarray samples generated from different platforms (Dataset S2). To enhance comparability, whenever available the raw data were resummarized and standardized (30). Then the expression matrices of the time-indicating genes from these datasets (Dataset S3) were subjected to nonlinear regression to estimate Phase24, the key oscillation parameter (Dataset S4).
Due to the large degree of freedom used in the statistical test, the molecular timetable method can detect subtle changes, but here we focused on large perturbations. First, we investigated the datasets involving soil-grown plants, the most similar growth conditions to our circadian time-course profiling experiment. Mild drought (without irrigation for 3 d) caused negligible changes to the global rhythm (Fig. 2 A and B) (7). However, more-severe drought (without irrigation for 6 d) induced a drastic phase shift (Fig. 2C) (17). In nature, drought stress can be accompanied by heat stress. However, unlike mild drought, even a 30-min heat shock was strong enough to cause a phase advancement of more than 8 h (Fig. 2D) (22). Surprisingly, prolonged heat stress resulted in a milder perturbation of the global rhythm (SI Appendix, Fig. S1) (23). This difference may be due to the capacity of the soybean circadian clock to compensate for temperature after adaptation to prolonged heat stress. These results suggest that in soybean the responses of the global rhythm to stresses like drought and heat are duration-dependent.
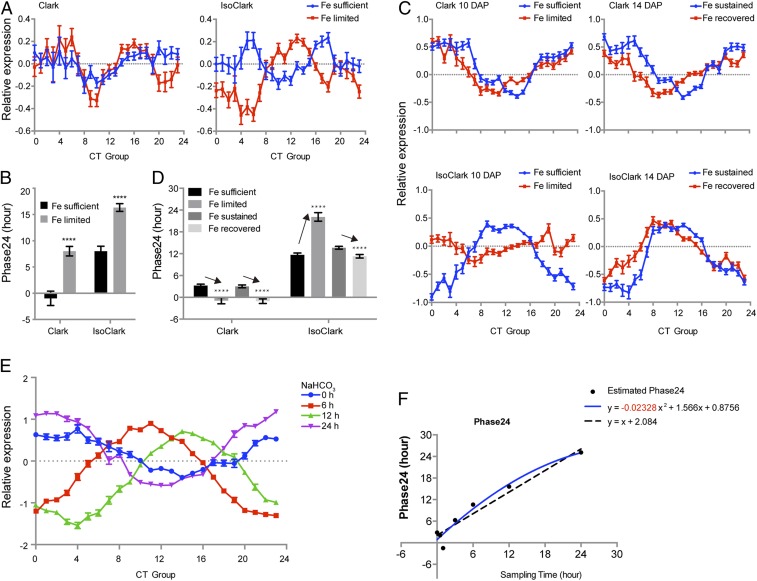
Next, we applied the molecular timetable to datasets with hydroponically grown plants from which both leaf and root transcriptomes had been obtained. Prolonged exposure to iron deficiency caused a phase delay of the circadian clock in leaves in the 2 near-isogenic soybean lines, Clark (iron-efficient) and IsoClark (iron-inefficient) (Fig. 3 A and B) (11). However, in roots, iron deficiency resulted in opposite phase changes in Clark and IsoClark: The phase advanced in Clark and but was delayed in IsoClark. When iron was replenished, these perturbations largely disappeared in IsoClark but not in Clark (Fig. 3 C and D) (10). These results imply that the responses to iron deficiency may be organ-specific, probably due to their different functions: Roots are responsible for iron assimilation, while leaves are where most of the iron is stored and utilized. In addition to the organ-specific responses in Clark, it is also important to note that in IsoClark iron deficiency caused a phase delay in both the leaves and roots. These differences in the iron utilization efficiency of these 2 near-isogenic lines suggest a strong connection between the phase change and iron utilization efficiency.
Fig. 3.
Circadian phases are altered in hydroponically grown soybean seedlings under long-term iron deficiency and alkaline stress. (A and B) Long-term iron deficiency causes different rhythm changes in soybean leaves from cultivars with different iron utilization efficiencies. Normalized expression levels of time-indicating genes are plotted in A. Estimated Phase24 is shown in B. Clark: iron-efficient; IsoClark: iron-inefficient. Fe sufficient: seedlings grown with 100 μM Fe (NO3)3; Fe limited: seedlings grown with 50 μM Fe (NO3)3. ****P < 0.0001 (Student’s t test with Holm–Sidak multiple comparison correction). (C and D) Long-term iron deficiency causes different rhythm changes in soybean roots from cultivars with different iron utilization efficiencies. Normalized expression levels of time-indicating genes are plotted in C. Estimated Phase24 is shown in D. DAP: days after planting. Fe sufficient: seedlings grown with 100 μM Fe (NO3)3; Fe limited: seedlings grown with 50 μM Fe (NO3)3; Fe sustained: seedlings grown with sufficient iron for 14 d; Fe recovered: seedlings grown with limited iron for 12 d and then with sufficient iron for another 2 d. ****P < 0.0001 (Student’s t test with Holm–Sidak multiple comparison correction). (E and F) Alkaline stress causes a phase advance in leaves. Normalized expression levels of time-indicating genes are shown in E. Estimated Phase24 is plotted against reported sampling time in F. A quadratic curve (solid blue line) fits the data better than a straight line (dashed black line), P < 0.0001 (exact F test). The coefficient for the quadratic is significantly smaller than 0, suggesting phase advancement induced by alkaline stress. P < 0.0001 (exact F test). Mean and SE are plotted. A and B are derived from GSE10730. C and D are derived from GSE22227. E and F are derived from GSE20323.
In addition to iron deficiency, alkaline stress also causes organ-specific changes in the wild soybean Glycine soja (Fig. 3 E and F and SI Appendix, Fig. S2) (8, 9). Unlike the iron deficiency samples, the alkaline-treated leaf and root samples did not include corresponding control samples at each time point. Therefore, a direct comparison to identify potential changes in phase was not possible. To solve this problem, we performed regression analysis and used model selection to look for phase changes. If no phase shift was induced by alkaline treatment, the estimated Phase24 should have shown a linear relationship with the actual sampling time. However, an F test indicated that Phase24 for the leaf sample had a quadratic relationship with the sampling time (P < 0.0001), and the coefficient of this quadratic was significantly lower than zero (P < 0.0001) (Fig. 3 E and F) (9). This result means alkaline treatment causes statistically significant phase advances of the global rhythm in leaves. In roots, however, a quadratic curve fit no better than a line (SI Appendix, Fig. S2) (8). Therefore, alkalinity does not cause a significant phase shift in roots.
Finally, we extended the molecular timetable method to all of the abiotic stress-related soybean transcriptomes and summarized the corresponding Phase24 in Dataset S4. Except for a few experiments without appropriate controls from which we could not draw conclusions, most abiotic stresses perturbed the phase to some extent. This result suggests that the phase of the soybean circadian clock is sensitive to environmental cues, probably allowing rapid responses to the ever-changing environment by adjusting the gene expression accordingly.
Validation of Global Rhythm Changes and Identification of Specific Targets.
The molecular timetable method offers an efficient and sensitive way to detect phase perturbations triggered by diverse stress signals. Such phase shifts may reflect a true phenotype if the corresponding control and treated samples were taken at a similar time of day. However, sampling in some published experiments did not take this into consideration. Obvious phase shifts were detected even among biological replicates in some datasets. Therefore, it was imperative to validate these computational predictions experimentally.
We performed 2-d circadian time-course experiments with 4 stresses, mild drought, heat shock, short-term iron deficiency, and short-term alkaline stress. For soil-based experiments, we sampled unifoliolate leaves to minimize the effects of developmental stages on transcript abundance. For hydroponically grown plants, we collected the first trifoliolate leaf and root tissues for the iron-deficiency experiment and only the first trifoliolate leaf for alkaline stress testing. With 3 biological replicates and shared control samples, these experiments generated 315 samples (Dataset S5). Since we were only interested in examining the effects of these stresses on the circadian clock components and clock output genes, it was not cost-effective to perform genome-wide expression profiling of this large number of samples using microarrays or RNA-seq. Instead, we adapted RASL-seq, a multiplexed targeted sequencing method (31).
Using bioinformatics analysis, we first identified 49 soybean orthologs of 16 key Arabidopsis circadian clock components. Also included in the RASL-seq analysis were 6 soybean orthologs of 4 Arabidopsis circadian clock output genes, Chlorophyll A/B-Binding Protein 2 (CAB2), Catalase 2 (CAT2), Catalase 3 (CAT3), and Cold, Circadian Rhythm, and RNA Binding 2 (CCR2) (SI Appendix, Table S1). For each of these genes, 3 pairs of gene-specific primers were tested during the preliminary analysis, and the most efficient pair was chosen for RASL-seq (Dataset S6) (29).
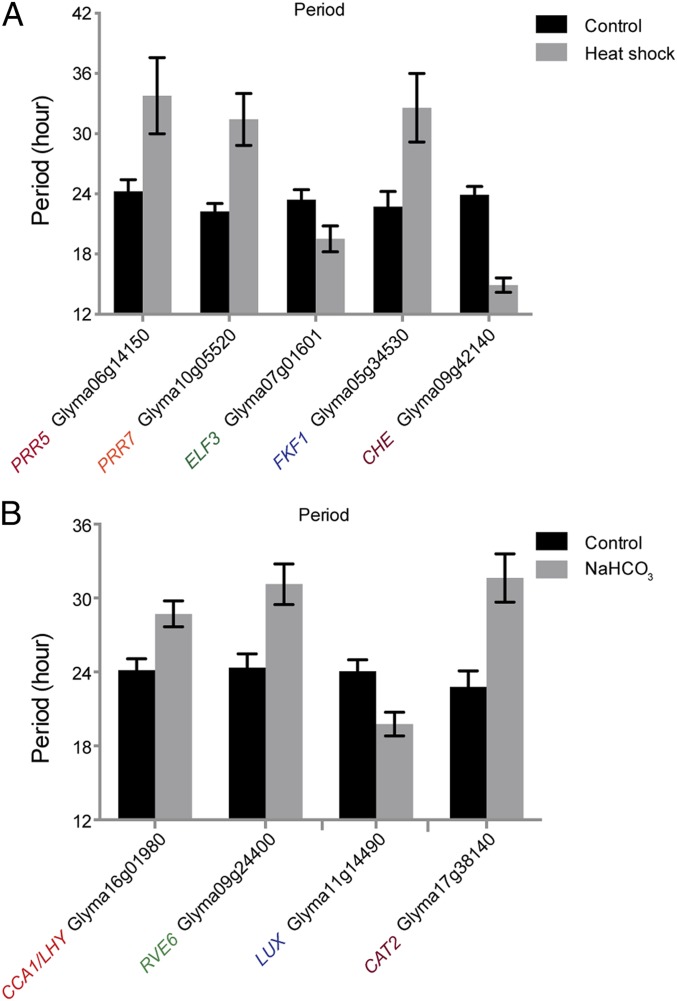
Due to the large number of soybean clock gene orthologs surveyed, we applied a very high stringency level to the robustness of the circadian oscillation (P < 10−10) to focus on those genes with robust circadian oscillation and statistically significant changes caused by stress treatments (false discovery rate [FDR] < 0.05) (Figs. 4 and 5). To better present the data, only those with periods between 22 and 26 h without a treatment and phase shifts over 4 h with a treatment are shown in the main figures. Consistent with the prediction from the molecular timetable analysis, short-term mild drought did not cause any statistically significant phase changes (SI Appendix, Fig. S3). Heat shock caused both phase advances and period changes of one FKF1 ortholog, one CHE ortholog, and one PRR7 ortholog (Figs. 4A and 5A). After heat-shock treatment, the phases of 3 PRR genes, a PRR5 ortholog, a PRR7 ortholog, and a PRR9 ortholog, advanced more than 5 h. These observed phase changes agreed with the observations derived from the molecular timetable analysis (Fig. 2D).
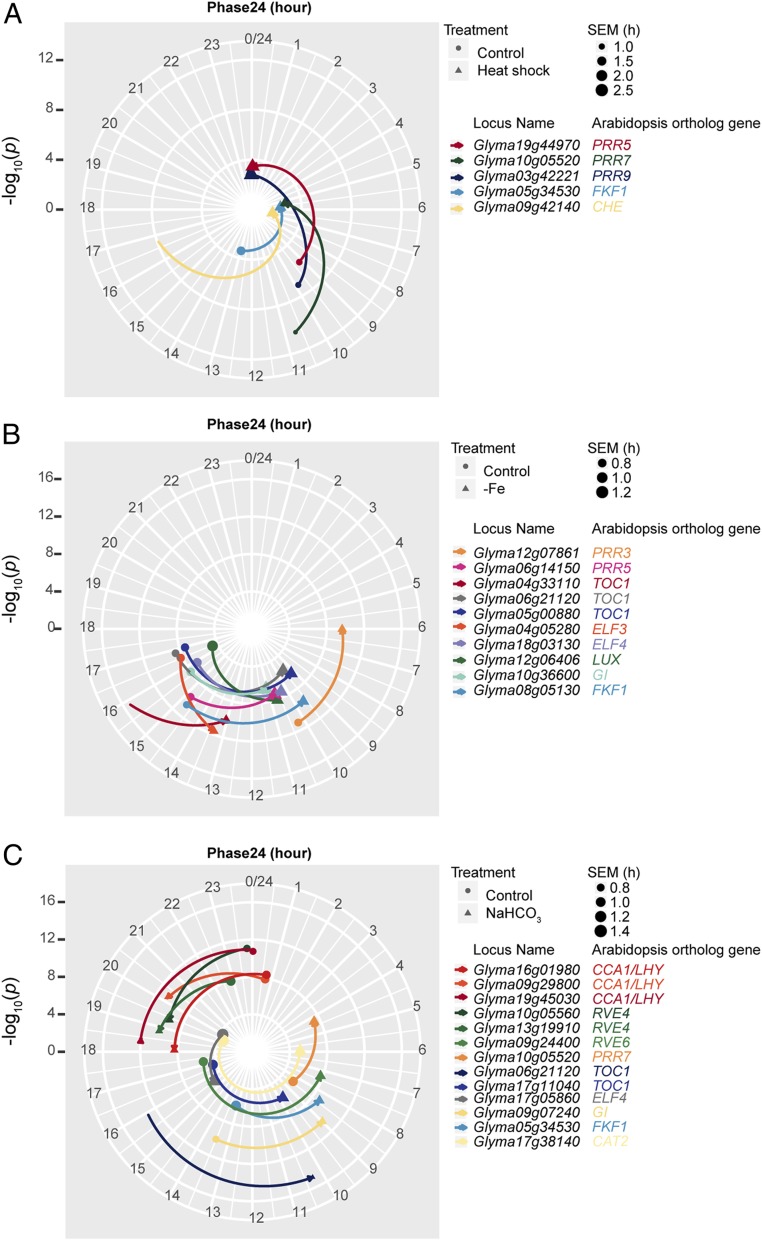
Fig. 4.
Heat shock (A), short-term iron deficiency (B), and short-term alkaline stress (C) induce phase advances in specific circadian components in soybean leaves. The angular coordinates represent Phase24. The −log10-transformed oscillation P values represent the robustness of the oscillation and are plotted as radial coordinates. Circles represent control samples, and the triangles represent treated samples. The size of the symbols is proportional to the SE (SEM) of Phase24, as illustrated in the key. The arrows pointing from control to treated samples represent the direction of the phase shift. Student’s t tests with the Benjamini–Hochberg multiple comparison correction were used to compare control and treated samples and derive FDR. To highlight statistically significant changes and apply a high stringency level to oscillation robustness, only genes with oscillation P values < 10−10 and FDRs < 0.05 were plotted.
Fig. 5.
Heat shock (A) and short-term alkaline stress (B) change the period of specific circadian components in soybean leaves. Student’s t test with the Benjamini–Hochberg multiple comparison correction was used to compare control and treated samples and derive the FDR. To highlight statistically significant changes and apply a high stringency level to oscillation robustness, only genes with oscillation P values < 10−10 and FDRs < 0.05 were plotted. Mean and SE are plotted.
In the iron-inefficient line IsoClark, short-term iron deficiency mainly caused phase changes in leaves rather than roots and resulted in phase advances of evening genes, including 8 orthologs of 6 key evening components (Fig. 4B). Short-term iron deficiency only caused a phase advance in one CCA1/LHY ortholog in roots, but under the low-stringency condition, the periods of 4 morning genes were altered (SI Appendix, Fig. S3). Two CCA1/LHY orthologs and one RVE6 ortholog had periods of less than 18 h under the control condition in the iron-inefficient IsoClark. This suggests that iron inefficiency shortened the circadian period in soybean and that morning genes may play a role in regulating iron assimilation in roots. This organ-specific response was unlikely to be the less synchronized circadian clock in roots. Under low stringency, only 10 out of the 49 clock genes lacked robust circadian oscillation in roots, while 6 failed the oscillation test in leaves (SI Appendix, Fig. S3). Therefore, consistent with the implications of the molecular timetable analysis, the soybean circadian clock’s response to iron deficiency is organ-specific.
Different treatment durations also elicited different circadian clock responses. Compared with long-term iron deficiency (Fig. 3 A–D, IsoClark), short-term treatment induced more perturbations in the leaf circadian clock than in the root clock. Furthermore, the changes were opposite: Long-term iron deficiency caused phase delays in the leaf circadian clock, while short-term stress led to phase advances.
Surprisingly, short-term alkaline stresses in hydroponically grown soybean had a profound impact on the circadian clock in leaves. Alkaline stress triggered a phase shift in 8 out of 16 different key clock components under the high-stringency condition (Fig. 4C) and in 25 out of all of the 55 soybean clock gene and output gene orthologs tested under the low-stringency condition (SI Appendix, Fig. S3). Interestingly, consistent with the molecular timetable analysis, all these phase changes were phase advances, suggesting a similar perturbation to the entire circadian clock network. Despite this strong influence on the phase, alkaline stress also disturbed the period, lengthening the period of 2 morning genes, including one CCA1/LHY ortholog and one RVE6 ortholog, and one ortholog of the CAT2 under the high-stringency condition and shortening the period of one LUX ortholog (Fig. 5B).
To enable a more holistic view of the whole RASL-seq analysis, we used a lower-stringency condition (oscillation P < 0.05, FDR < 0.05) and summarized all of the phase and period changes together in a heat map (SI Appendix, Fig. S3). Except drought, which did not cause any changes in the circadian phase, all stresses advanced the phases of different genes to different degrees. The periods were not altered as dramatically as the phases. Among the 4 stresses, heat shock had the strongest impact on period: It caused arrhythmicity in more than half of the soybean circadian clock orthologs tested, suggesting that the soybean circadian clock is highly sensitive to heat shock. Different orthologs of the same clock components had similar responses to the same stress. For example, short-term alkaline stress and iron deficiency advanced the phases of 2 TOC1 orthologs, Glyma04g33110 and Glyma06g21120. Heat shock and short-term alkaline stress lengthened the periods of 2 ELF3 orthologs, Glyma07g01601 and Glyma08g21115. Nevertheless, the periods of different CCA1/LHY orthologs in roots responded differently to iron deficiency. The heat map also allowed us to identify additional functional orthologs (SI Appendix, Fig. S3). Among the 6 CCA1/LHY orthologs, 5, including the 2 with lower sequence similarities, responded to short-term alkaline stress.
Assessment of the Effects of Abiotic Stresses on Circadian Leaf Movement.
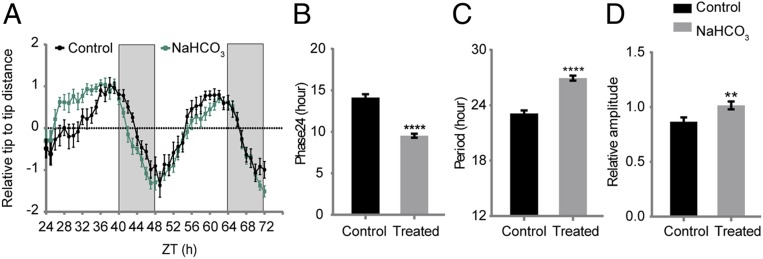
To test whether the stress-induced transcript abundance changes in the soybean clock genes could influence clock-controlled physiological outputs, we chose to measure circadian leaf movement, as it has been widely used as a proxy to study the plant circadian clock. Using the imaging setup, the specific soybean cultivars or species and plant growth conditions provided in Materials and Methods and in Dataset S5, we performed 2-d time-course experiments using alkaline stress, mild drought, heat shock, and short-term iron deficiency. The circadian parameters including Phase24 and period were derived via nonlinear regression. The wild soybean G. soja showed very robust circadian leaf movement, and alkaline stress caused a phase advancement (Fig. 6), which is consistent with the results of the molecular timetable analysis (Fig. 3F) and RASL-seq analysis (Fig. 4C). However, it seems there was a phase delay on day 2 with alkaline treatment (Fig. 6A), which could be explained by the period lengthening in response to alkalization (Fig. 6C). Interestingly, while mild drought caused negligible perturbations to the clock gene expression in the molecular timetable (Fig. 2B) and RASL-seq analyses (SI Appendix, Fig. S3), it resulted in significant changes in Phase24 and the period of leaf movement (SI Appendix, Fig. S4A). By contrast, heat shock and short-term iron deficiency did not cause significant changes in leaf movement (SI Appendix, Fig. S4 B and C) despite their impacts on the clock gene expression (SI Appendix, Fig. S3). These data are consistent with the idea that different physiological processes in plants are controlled by distinct subsets of clock components (1). Perturbations in individual or subsets of clock components may not alter every clock-mediated behavior (32).
Fig. 6.
Wild soybean has robust circadian leaf movements, and alkaline stress changes this global rhythm dramatically. (A) Leaf movements of soybean under alkaline stress. Seven-day-old seedlings of G. soja were transferred from germination paper to a hydroponic unit system and grown for 10 d under the ambient light condition in the greenhouse. The seedlings were kept in constant light on the ninth day. On the 10th day, half of the seedlings were treated with 50 mmol/L NaHCO3 (pH 8.5) at ZT 24, and then the movement of the first unifoliolate leaf was recorded hourly. Leaf movement data represent mean ± SE. (B) Alkaline stress induced phase advances in circadian leaf movement. (C) The period of circadian leaf movement was lengthened after alkaline stress. (D) Alkaline stress increased the relative amplitude of circadian leaf movement. Circadian rhythm parameters, including Phase24, period, and relative amplitude, were derived via nonlinear regression. The white and gray regions in the trace plot indicate subjective light and dark periods, respectively. **P < 0.01; ****P < 0.0001 (Student’s t test).
Discussion
Our 2-module discovery pipeline to assess rhythmicity changes requires sensitivity and accuracy. To that end, we tested different algorithms with different design principles, assumptions, and complexities. An online rhythmicity analysis system called BioDare (33) was developed with 6 period analysis algorithms: fast Fourier transform–nonlinear least squares (FFT-NLLS) (34), mFourfit (35), maximum entropy spectral analysis (36), the Enright periodogram (37, 38), the Lomb–Scargle periodogram (LS) (39), and spectrum resampling (40). In this study, we also devised a nonlinear regression strategy based on a cosine function combined with a linear trend (COS) (see Materials and Methods for details). To compare the performance of these 7 methods, we used the time-course RNA-seq data of 55 clock candidate genes as the test dataset (Dataset S7). The smallest variation in estimated periods was achieved by LS, followed by COS and then FFT-NLLS (SI Appendix, Fig. S5A). However, the LS algorithm had a high false negative rate. The expression profiles of genes with clear oscillations (e.g., Glyma03g42221 and Glyma04g33110) were considered as arrhythmic by LS (SI Appendix, Fig. S5B and Dataset S7). FFT-NLLS also suffered from the same problem with genes like Glyma03g42221, Glyma08g05130, and Glyma11g14490 (SI Appendix, Fig. S5B and Dataset S7). In contrast, the COS method developed in our study achieved a sound assessment of rhythmicity without the high false negative rate. Therefore, COS was used in our analysis pipeline to examine the influences that abiotic stresses have on the soybean circadian clock.
Our previous knowledge of the effects of abiotic stress on the soybean circadian clock is limited. Marcolino-Gomes et al. (41) observed that mild and severe drought stresses perturbed the amplitudes of representative clock genes in a drought-sensitive soybean cultivar under diurnal conditions. However, these observations were confounded by both the endogenous circadian clock and changes in the exogenous periodic light conditions, making it hard to distinguish whether the clock or the light was the major contributor to this phenotype. Our RASL-seq analysis suggests that mild drought had a negligible effect on the phase and period of circadian clock gene expression in soybean (SI Appendix, Fig. S3). Therefore, it is likely that the effects recorded in Marcolino-Gomes et al. (41) were mainly due to the exogenous periodic light changes instead of the endogenous circadian clock.
The effect of iron deficiency on the circadian clock has been investigated extensively in Arabidopsis (42–45). Short-term iron deficiency causes period lengthening under circadian conditions (43) and phase delays under diurnal conditions (44). Despite these molecular details, the biological significance of these phenotypes remains unclear (44). The observed soybean circadian clock responses in our analysis of soybean have provided some clues. Iron is a key element for photosynthesis and redox regulation. Thus, iron homeostasis is tightly controlled through the competition between the iron uptake and iron storage pathways. The circadian clock regulates many genes involved in iron homeostasis. The iron uptake pathway may be induced during the day to facilitate photosynthetic processes (46). To maintain homeostasis, and avoid iron toxicity, excess iron is stored in ferritin proteins. At night, iron uptake is repressed and stored iron is released (46). RASL-seq analysis suggested that short-term iron deficiency mainly targeted evening-phased circadian components for phase advances (Fig. 4B). Long-term iron deficiency caused phase delay in both the iron-efficient wild-type cultivar, Clark, and the inefficient Clark mutant, IsoClark, in leaves (Fig. 3C). These phase delays may prolong the iron uptake period to compensate for the iron deficiency. Although IsoClark utilized the same strategy as Clark, under short-term iron deficiency several genes displayed lengthened periods, including 1 LUX ortholog in leaves and 3 morning clock gene orthologs in roots; these differences were probably due to the iron inefficiency for IsoClark (SI Appendix, Fig. S3). This behavior is similar to that in Arabidopsis mutants defective in iron homeostasis, which also have longer periods, even when grown under iron-sufficient conditions (42). Future experiments comparing Clark and IsoClark will provide more mechanistic insights into the biological function of the circadian clock in the regulation of the short-term and long-term iron deficiency.
Alkaline stress is also a major abiotic stress that limits crop productivity. To our knowledge, its impact on the circadian clock has never been explored in plants. However, this connection was captured by our computational analysis (Fig. 3 E and F) and the pervasive modulations of the phase of clock components by alkaline stress was experimentally revealed (Figs. 4C, 5B, and 6 and SI Appendix, Fig. S3). Interestingly, salinity stress, which usually cooccurs with alkaline stress, also causes very strong perturbations in phase (SI Appendix, Fig. S6). Since alkaline stress reduces the solubility of many minerals, including iron (46), its pervasive impacts on the phase echoes the effect of iron deficiency. Future studies will determine whether alkaline stress and iron deficiency share similar signaling components to modulate the circadian clock and will uncover the biological significance of these modulations.
One interesting general observation from our study is that only some soybean circadian clock components were perturbed by different abiotic stresses (SI Appendix, Fig. S3). This observation is consistent with results from other studies. In Arabidopsis, comparisons of the genome-wide expression profiles of samples under thermocycles and circadian conditions also found that the phases of only a subset of clock-related genes were shifted by thermocycles (47). In rice, while temperature and solar radiation had strong effects on the expression of certain clock-related genes, the internal time of the plants was not affected by weather, photocycle, or developmental age in the field (32). In our study, the period changes varied and were observed in a limited number of genes with variations. For example, alkaline stress lengthened the period of CCA1/LHY (Glyma16g01980), RVE6 (Glyma09g24400), and CAT2 (Glyma17g38140), while shortening the period of LUX (Glyma11g14490) under the same stress condition. These observations raised an interesting question: Why do perturbations in a subset of clock-related genes not transmit to other clock genes or even the circadian clock system as a whole? One possible explanation is that for a limited number of clock components the interlocked multiloop structure and nonlinearity of the circadian clock system have enhanced the robustness against the endogenous and exogenous perturbations (48). Another possibility is that because the time course lasted only 48 h, the clock was undergoing a transient and shifting perturbation. What we observed might have been the initiation of clock changing. Based on our bioinformatics analysis, the soybean genome has many more copies of clock-related genes than does the Arabidopsis genome. Thus, the regulatory network of the soybean circadian clock system may be more complex and potentially more robust than the Arabidopsis circadian clock. In-depth mechanistic and systems studies are required to better understand the soybean circadian clock.
Similar levels of complexity were observed for the effects of abiotic stresses on the clock outputs. Using leaf movement as a proxy for clock behavior, we found a phase advancement in leaf movement that was consistent with our gene expression analysis for the response to alkaline stress, but not with that for mild drought, heat shock, or short-term iron deficiency. We hypothesize that the differences between the rhythmic leaf movement and clock gene expression in response to abiotic stresses are probably due to the distinct molecular mechanisms underlying the rhythmic leaf movement of Arabidopsis and soybean. Although the molecular details of the control of Arabidopsis leaf movement have not been fully resolved, circadian leaf movement is conferred by the differential growth of the adaxial and abaxial sides of the petiole (49, 50). However, leguminous plants like soybean have a specialized leaf motor organ, the pulvinus, to regulate reversible changes in leaf position. Opposing volume changes resulting from ion and water fluxes in the opposite parts of the pulvinus drive reversible leaf movement without requiring growth (51). Isolated bean pulvinus protoplasts could maintain their rhythmic swelling under continuous light for more than 8 d (52). Therefore, while the circadian regulation of growth is likely to be the main contributor to the rhythmic leaf movement of Arabidopsis, the circadian regulation of pumps, carriers, ion, and water channels in the pulvinus is probably the key driving force in soybean (51, 52). Alkaline stress is likely to affect the ion flux through its direct impact on pH and mild drought may significantly influence the water flux, thus perturbing the rhythmic swelling of the pulvinus. The current literature is unclear about whether heat shock or short-term iron deficiency affects the swelling of the pulvinus. Clock components involved in the response to heat shock and short-term iron deficiency may not regulate pulvinus swelling. An alternative explanation for the differences may be the tissue specificity of the circadian clock (53). Since different tissues were studied for the leaf movement assay (pulvini of the first trifoliolate leaf) and RASL-seq analysis (blades of unifoliolate or trifoliolate leaves), the circadian clocks in these tissues may not have been tightly coupled.
Taken together, our discovery pipeline has helped reveal insights into the environment–circadian clock interplay in soybean that could not have been extrapolated from what has been learned in Arabidopsis. The comprehensive high-resolution datasets generated are also valuable assets for quantitative modeling of the crop circadian clock system, which is paramount to the systems-level understanding of this complex network. However, it cannot fully replace the circadian time-course transcriptomic analysis. Rather, the discoveries generated by our pipeline may guide the investment in the more laborious and expensive full-scale time-course transcriptomic experiments on selected environmental stimuli. Importantly, this discovery pipeline is not limited to applications in soybean; it can be readily applied to any species with genome information and publicly available transcriptome datasets.
Materials and Methods
Plant Materials, Growth Conditions, and Treatments.
Seeds of soybean cultivars Williams 82, Clark, and IsoClark and of Glycine soja were used in this study. For the RNA-seq analysis, Williams 82 seedlings were grown in soil (Sun Gro Horticulture) under long-day conditions (16 h light/8 h dark) for 9 d in a growth chamber under the following conditions: 28 °C, 50% relative humidity, and a light intensity of 100 μmol⋅m−2⋅s−1. On the tenth day, the light was switched to constant conditions. Seedlings were sampled every 4 h for 44 h starting at ZT24 on the 11th day. For each time point, 3 biological replicates were obtained, and each sample comprised pooled material from the unifoliolate leaves of 3 individual seedlings. The plant materials, growth conditions, and treatments related to the time-course experiment for RASL-seq are summarized in Dataset S5.
RNA-Seq and Data Analysis.
Total RNA was extracted using RNeasy Plant Mini Kits (Qiagen). RNA quality was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). Transcriptome sequencing was performed using the Illumina HiSeq platform (Illumina, Inc.) and conducted at BGI Americas (https://www.bgi.com/us/). The data processing was performed mainly according to Li et al. (54). Briefly, for each sample, RNA-seq data were collected from 3 lanes, and the raw reads were trimmed and cleaned separately. The cleaned reads were combined and aligned to the soybean reference genome Glycine_max_Wm2.a1.v1.0 using the spliced alignment identification tool TopHat (V2.1.0). Glycine_max_Wm2.a1.v1.0 was used for better comparison since most of the transcriptome data used for the molecular timetable analysis were originally mapped to this version of the reference genome. The aligned reads were counted using HTseq-count (version 0.6.0). Only genes with nonzero FPKM (fragments per kilobase per million mapped fragments) values in all 36 samples were included for further analysis. The RNA-seq data generated in this study have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (accession no. GSE94228).
Identification of Time-Indicating Genes.
To identify time-indicating genes that had robust circadian oscillation with high relative amplitude, the FPKM values of each gene were analyzed via 2 filters. First, the expression profile of each gene was fit against 1,440 test cosine curves. The curves all had unit amplitudes and 24-h periods, but they had different phases (0 to 24 h) measured at 1-min increments. The correlation coefficient (R) and phase of the best-fitting curve were assigned to each gene. Genes with R ≥ 0.9 were selected for further filtering. Second, the CV (coefficient of variation) was used as a proxy for relative amplitude and calculated for each gene using FPKM values. A total of 3,695 genes with CV ≥ 0.3 and R ≥ 0.9 were selected as time-indicating genes. The phases of these time-indicating genes were rounded to integers from 0 to 23. The time-indicating genes with the same rounded phase were considered as one CT group, resulting in 24 CT groups.
Assembly of Soybean Abiotic Stress-Related Transcriptomes and Normalization.
Abiotic stress-related soybean transcriptome datasets were assembled from diverse online sources including NCBI’s Gene Expression Omnibus, EMBL-EBI’s ArrayExpress, and PLEXdb. Whenever available, raw data from single-color microarray datasets were renormalized using the RMA algorithm with baselines of each array adjusted to the median. Both channels from 2-color microarray datasets were individually normalized using a quantile normalization method and then median-adjusted. RPKM (reads per kilobase per million mapped reads) and FPKM values were generated and used for single-end and paired-end RNA-seq datasets, respectively. Whenever the raw datasets were not available in the submitted files, processed expression matrices supplied by the original authors were used directly. The original experimental designs of these transcriptomes are listed in Dataset S2, the normalized expression matrices used for molecular timetable analysis are provided on GitHub (30), and a list of time-indication genes is provided in Dataset S3.
Application of the Molecular Timetable Method.
The expression profiles of time-indicating genes were extracted from the renormalized transcriptome datasets. Standardization was first applied to each time-indicating gene across different samples within one experiment so that each time-indicating gene had the same average expression level and SD. Standardization was then applied to each sample within one experiment so that each sample had the same average expression level and SD. Three sufficient statistics, including number of genes and the mean and SD of the standardized expression levels of time-indicating genes, were calculated for each of the 24 CT groups and used to estimate the oscillation parameters for each sample. To estimate the circadian rhythm parameters, the 3 sufficient statistics for the 24 CT groups of each sample were fit to the following equation in GraphPad Prism 6:
Period was constrained to be more than 12 h but less than 36 h. Samples with nonconverging fitting were considered arrhythmic. The resulting best-fitted value, SE, and degree of freedom for each parameter were used for statistical tests. To improve comparability and reduce the confounding effects of period differences on the comparisons of phases (peak expression time in a day), the phase was scaled to a 24-h period, yielding Phase24.
Identification of Soybean Circadian Clock and Output Genes.
To identify orthologs of the Arabidopsis circadian clock and output genes in the soybean genome, the amino acid sequences (The Arabidopsis Information Resource, https://www.arabidopsis.org/) of the corresponding Arabidopsis proteins were used as queries in BLAST searches (tBLASTn) against the G. max genome v_1.0 using the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Gmax). The most similar representative sequences were selected on the basis of whether they had alignment e-values equal to 0 or less than the cutoff E-value of 10−12.
RASL-Seq.
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. The RASL-seq procedure was performed as described (29). The primers are listed in Dataset S6. The sequencing data were demultiplexed and aligned with the gene-specific primers. The resulting count matrix was first blanked using the reads from the average reads of the blank wells and then standardized. The standardized expression matrix is provided in Dataset S8. Nonlinear regression was applied to derive Phase24 and period. The best fit value, SE, and degree of freedom of Phase24 and the period were used for statistical tests with multiple comparison corrections to obtain the FDR. The predicted values from the nonlinear regression were fit against the standardized expression profile of each gene in each experimental condition through linear regression. The corresponding P values from associated ANOVA tests on these linear regressions were used as measurements of the robustness of the circadian oscillation.
Leaf Movement Assay.
Leaf movement was measured as described previously with minor modifications (55). Images under constant light were recorded hourly for 2 d using a 4K Ultra HD Camera (GoPro Hero 5). Leaf movement was assessed by measuring the distance from tip to tip of the symmetrical leaf pair of the first trifoliolate using Fiji/ImageJ (NIH). Data normalized by Z-score transformation were used in the analysis of circadian rhythm. The results represent the mean ± SE. Four biological replicates were obtained. The plant materials, growth conditions, and treatments were used the same as those used for the RASL-seq experiments.
Data Availability.
All data discussed in the paper are available in Datasets S1–S8, and on GitHub (30).
Supplementary Material
Acknowledgments
We thank G. Tylka and S. Whitham for sharing soybean seeds and P. Benfey for enlightening discussions about the project. We thank E. Pasoreck for editing and improving the manuscript. We thank BGI for processed sequencing data. This work was supported by startup funds from State Key Laboratory for Protein and Plant Gene Research, Peking University, startup funds from Peking-Tsinghua Center for Life Sciences, USDA National Institute of Food and Agriculture, Hatch project 3808 (to W.W.), grant 31871648 from the National Natural Science Foundation of China and startup funds (2809911) from Guangzhou University to M.L., grants from the NIH (R35GM118036-04) and the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (GBMF3032, to X.D.), and funds from US Department of Agriculture Agricultural Research Service project 3625-21220-005-00D (to J.A.O.).
Footnotes
The authors declare no competing interest.
Data deposition: The RNA-sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE94228). Expression profiles of soybean transcriptome datasets under abiotic stress treatments are available on GitHub (https://github.com/wanglab-PKU/PNAS-Dataset-S3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708508116/-/DCSupplemental.
References
- 1.Greenham K., McClung C. R., Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hsu P. Y., Harmer S. L., Wheels within wheels: The plant circadian system. Trends Plant Sci. 19, 240–249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendix C., Marshall C. M., Harmon F. G., Circadian clock genes universally control key agricultural traits. Mol. Plant 8, 1135–1152 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Kim M. Y., Shin J. H., Kang Y. J., Shim S. R., Lee S. H., Divergence of flowering genes in soybean. J. Biosci. 37, 857–870 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Preuss S. B., et al. , Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS One 7, e30717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller N. A., et al. , Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48, 89–93 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues F. A., et al. , Daytime soybean transcriptome fluctuations during water deficit stress. BMC Genomics 16, 505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge Y., et al. , Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol. 10, 153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge Y., et al. , Alkaline-stress response in Glycine soja leaf identifies specific transcription factors and ABA-mediated signaling factors. Funct. Integr. Genomics 11, 369–379 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Severin A. J., et al. , An integrative approach to genomic introgression mapping. Plant Physiol. 154, 3–12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke J. A., et al. , Integrating microarray analysis and the soybean genome to understand the soybeans iron deficiency response. BMC Genomics 10, 376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu S., et al. , Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Ueda H. R., et al. , Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 101, 11227–11232 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerwin R. E., et al. , Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23, 471–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashi T., et al. , Detection of diurnal variation of tomato transcriptome through the molecular timetable method in a sunlight-type plant factory. Front. Plant Sci. 7, 87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi P., Rabara R. C., Shen Q. J., Rushton P. J., Transcriptomics analyses of soybean leaf and root samples during water-deficit. Genom. Data 5, 164–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le D. T., et al. , Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS One 7, e49522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha C. V., et al. , Comparative analysis of root transcriptomes from two contrasting drought-responsive Williams 82 and DT2008 soybean cultivars under normal and dehydration conditions. Front. Plant Sci. 6, 551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho H. H., et al. , The molecular chaperone binding protein BiP prevents leaf dehydration-induced cellular homeostasis disruption. PLoS One 9, e86661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prince S. J., et al. , Comparative analysis of the drought-responsive transcriptome in soybean lines contrasting for canopy wilting. Plant Sci. 240, 65–78 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Tamang B. G., Magliozzi J. O., Maroof M. A. S., Fukao T., Physiological and transcriptomic characterization of submergence and reoxygenation responses in soybean seedlings. Plant Cell Environ. 37, 2350–2365 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Weston D. J., et al. , Comparative physiology and transcriptional networks underlying the heat shock response in Populus trichocarpa, Arabidopsis thaliana and Glycine max. Plant Cell Environ. 34, 1488–1506 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Kidokoro S., et al. , Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 81, 505–518 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Belamkar V., et al. , Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics 15, 950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei W., et al. , Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duressa D., Soliman K. M., Chen D., Mechanisms of magnesium amelioration of aluminum toxicity in soybean at the gene expression level. Genome 53, 787–797 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Duressa D., Soliman K., Chen D., Identification of aluminum responsive genes in Al-tolerant soybean line PI 416937. Int. J. Plant Genomics 2010, 164862 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whaley A., et al. , RNA-seq analysis reveals genetic response and tolerance mechanisms to ozone exposure in soybean. BMC Genomics 16, 426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Qiu J., Fu X. D., RASL-seq for massively parallel and quantitative analysis of gene expression. Curr. Protoc. Mol. Biol. 98, 4.13.1–4.13.9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M., et al. , PNAS-Dataset-S3. GitHub. https://github.com/wanglab-PKU/PNAS-Dataset-S3. Deposited 12 July 2019.
- 31.Wang W., et al. , Timing of plant immune responses by a central circadian regulator. Nature 470, 110–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuzaki J., Kawahara Y., Izawa T., Punctual transcriptional regulation by the rice circadian clock under fluctuating field conditions. Plant Cell 27, 633–648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zielinski T., Moore A. M., Troup E., Halliday K. J., Millar A. J., Strengths and limitations of period estimation methods for circadian data. PLoS One 9, e96462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plautz J. D., et al. , Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12, 204–217 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Edwards K. D., et al. , Quantitative analysis of regulatory flexibility under changing environmental conditions. Mol. Syst. Biol. 6, 424 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burg J. P., The relationship between maximum entropy spectra and maximum likelihood spectra. Geophysics 37, 375–376 (1972). [Google Scholar]
- 37.Enright J. T., The search for rhythmicity in biological time-series. J. Theor. Biol. 8, 426–468 (1965). [DOI] [PubMed] [Google Scholar]
- 38.Sokolove P. G., Bushell W. N., The chi square periodogram: Its utility for analysis of circadian rhythms. J. Theor. Biol. 72, 131–160 (1978). [DOI] [PubMed] [Google Scholar]
- 39.Lomb N. R., Least-squares frequency-analysis of unequally spaced data. Astrophys. Space Sci. 39, 447–462 (1976). [Google Scholar]
- 40.Costa M. J., et al. , Inference on periodicity of circadian time series. Biostatistics 14, 792–806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcolino-Gomes J., et al. , Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS One 9, e86402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong S., Kim S. A., Guerinot M. L., McClung C. R., Reciprocal interaction of the circadian clock with the iron homeostasis network in Arabidopsis. Plant Physiol. 161, 893–903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y. Y., et al. , Iron is involved in the maintenance of circadian period length in Arabidopsis. Plant Physiol. 161, 1409–1420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salomé P. A., Oliva M., Weigel D., Krämer U., Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. EMBO J. 32, 511–523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner L., Schmal C., Staiger D., Danisman S., The plant leaf movement analyzer (PALMA): A simple tool for the analysis of periodic cotyledon and leaf movement in Arabidopsis thaliana. Plant Methods 13, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darbani B., et al. , Dissecting plant iron homeostasis under short and long-term iron fluctuations. Biotechnol. Adv. 31, 1292–1307 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Michael T. P., et al. , Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4, e14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saithong T., Painter K. J., Millar A. J., The contributions of interlocking loops and extensive nonlinearity to the properties of circadian clock models. PLoS One 5, e13867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polko J. K., et al. , Ethylene-induced differential petiole growth in Arabidopsis thaliana involves local microtubule reorientation and cell expansion. New Phytol. 193, 339–348 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Rauf M., et al. , NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 25, 4941–4955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Moran N., “Rhythmic leaf movements: Physiological and molecular aspects” in Rhythms in Plants: Dynamic Responses in a Dynamic Environment, Mancuso S., Shabala S., Eds. (Springer, 2007), pp. 57–95. [Google Scholar]
- 52.Mayer W. E., Fischer C., Protoplasts from Phaseolus coccineus L. pulvinar motor cells show circadian volume oscillations. Chronobiol. Int. 11, 156–164 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Endo M., Tissue-specific circadian clocks in plants. Curr. Opin. Plant Biol. 29, 44–49 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Li L., et al. , QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. U.S.A. 112, 14734–14739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J., Kim Y., Yeom M., Kim J. H., Nam H. G., FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20, 307–319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available in Datasets S1–S8, and on GitHub (30).