Significance
Low levels of ATIP3 in breast tumors are associated with increased response to neoadjuvant chemotherapy, and ATIP3 silencing in breast cancer cells potentiates the effects of paclitaxel, highlighting the importance of this predictive biomarker to select breast cancer patients who are sensitive to taxane-based chemotherapy. ATIP3 depletion promotes mitotic abnormalities, including centrosome amplification and multipolar spindle formation, which is a source of chromosome segregation errors and aneuploidy. Excessive aneuploidy in ATIP3-deficient cells treated with low doses of paclitaxel results in massive cell death.
Keywords: MTUS1, ATIP3, taxanes, predictive biomarker, multipolar spindle
Abstract
Predictive biomarkers for tumor response to neoadjuvant chemotherapy are needed in breast cancer. This study investigates the predictive value of 280 genes encoding proteins that regulate microtubule assembly and function. By analyzing 3 independent multicenter randomized cohorts of breast cancer patients, we identified 17 genes that are differentially regulated in tumors achieving pathological complete response (pCR) to neoadjuvant chemotherapy. We focused on the MTUS1 gene, whose major product, ATIP3, is a microtubule-associated protein down-regulated in aggressive breast tumors. We show here that low levels of ATIP3 are associated with an increased pCR rate, pointing to ATIP3 as a predictive biomarker of breast tumor chemosensitivity. Using preclinical models of patient-derived xenografts and 3-dimensional models of breast cancer cell lines, we show that low ATIP3 levels sensitize tumors to the effects of taxanes but not DNA-damaging agents. ATIP3 silencing improves the proapoptotic effects of paclitaxel and induces mitotic abnormalities, including centrosome amplification and multipolar spindle formation, which results in chromosome missegregation leading to aneuploidy. As shown by time-lapse video microscopy, ATIP3 depletion exacerbates cytokinesis failure and mitotic death induced by low doses of paclitaxel. Our results favor a mechanism by which the combination of ATIP3 deficiency and paclitaxel treatment induces excessive aneuploidy, which in turn results in elevated cell death. Together, these studies highlight ATIP3 as an important regulator of mitotic integrity and a useful predictive biomarker for a population of chemoresistant breast cancer patients.
Breast cancer is a leading cause of cancer death among women worldwide. Neoadjuvant chemotherapy, administered before surgery, represents an option for a number of breast cancer patients (1). Preoperative chemotherapy decreases primary tumor burden, thus facilitating breast conservation (2, 3), and administration of chemotherapy on naïve tumors prior to surgery also provides the opportunity to rapidly measure tumor response and identify breast cancer patients who may gain an advantage from the treatment. The achievement of pathological complete response (pCR), characterized by complete eradication of all invasive cancer cells from the breast and axillary lymph nodes, is often considered a surrogate end point for cancer-free survival after neoadjuvant setting, especially in aggressive triple-negative breast tumors (4, 5). Clinical parameters, such as estrogen receptor-negative status, high histological grade, and high proliferative status, have been associated with greater sensitivity to chemotherapy (5, 6). However, the proportion of patients who achieve a pCR following preoperative chemotherapy remains low, reaching 15 to 20% in the whole population and 30 to 40% in ER-negative tumors (7, 8). Considering the rapidly growing area of personalized medicine, the identification of efficient molecular markers that can predict sensitivity to chemotherapy is crucial to select patients who may benefit from therapy, thereby avoiding unnecessary treatment and associated toxicities for those who remain resistant (9).
The most used regimens in the neoadjuvant setting of breast cancer patients include taxanes and anthracyclines, whose combination is associated with improved outcome compared to anthracyclines alone (3). Taxanes (paclitaxel and docetaxel) are microtubule-targeting agents that bind and stabilize microtubules (MT), inducing mitotic arrest and apoptosis (10, 11). At clinically relevant concentrations in the nanomolar range, these drugs suppress MT dynamic instability (11–13) and behave as mitotic poisons that target the mitotic spindle during mitosis, inducing multipolar spindles and centrosomal abnormalities (13). The assembly and dynamics of the mitotic spindle are tightly regulated by a number of MT-associated proteins (MAPs) and mitotic kinases (14, 15), suggesting that alterations of MAP expression and/or function in breast tumors may regulate their sensitivity to taxane-based chemotherapy. Gene expression studies indeed identified the MAP tau protein as a predictive biomarker whose down-regulation is associated with increased pCR rate in breast cancer patients (16–21), underlining the interest of studying MT-regulating proteins as predictors of chemotherapy efficacy.
In the present study, we analyzed a panel of 280 genes encoding MT-regulating proteins to evaluate their predictive value as biomarkers of neoadjuvant taxane-based chemotherapy in breast cancer patients. Seventeen genes were identified as being differentially expressed in tumors from patients achieving pCR from 3 independent multicenter randomized breast cancer clinical trials. We focused our interest on candidate tumor suppressor gene MTUS1 (22, 23) that encodes the MT-stabilizing protein ATIP3, previously reported as a prognostic biomarker of breast cancer patient survival (24, 25). We show here that low ATIP3 expression in breast tumors is associated with a higher pCR rate. Unexpectedly, ATIP3 deficiency, which is known to increase MT instability (25), improves rather than impairs cancer cell sensitivity to taxanes. Our results favor a model in which ATIP3 depletion sensitizes cancer cells to paclitaxel by increasing centrosome amplification and mitotic abnormalities, leading to massive aneuploidy and cell death.
Results
Gene Expression Studies Identify MTUS1 Gene as a Predictor of Breast Tumor Response to Neoadjuvant Chemotherapy.
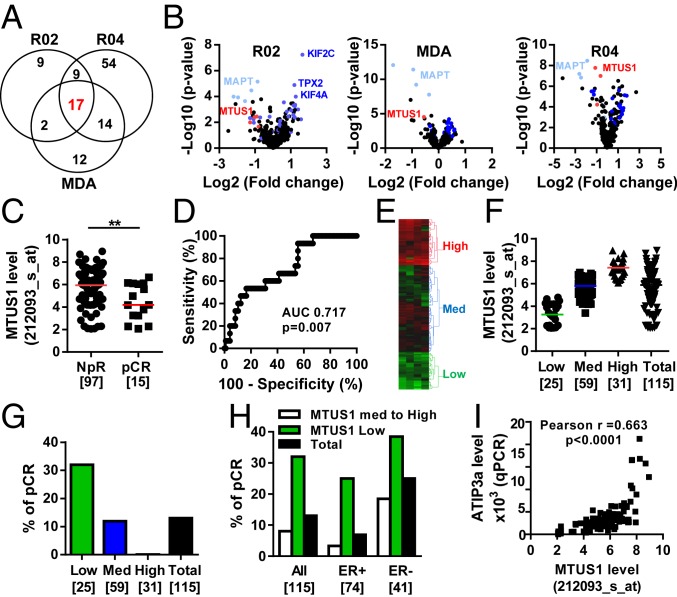
To identify predictive biomarkers of sensitivity to neoadjuvant chemotherapy in breast cancer, we analyzed a panel of 280 genes encoding MT regulatory proteins, including MAPs and mitotic kinases. We compared gene expression profiles with clinical data in 3 independent cohorts (R02, MDA, and R04) of 115, 133, and 142 breast cancer patients, respectively (SI Appendix, Table S1). We identified a total of 118 genes that were significantly differentially regulated (P < 0.01) in patients who achieved pCR compared to those (NpR) who did not. Among them, 17 were common to all 3 cohorts (Fig. 1 A and B, Table 1, and SI Appendix, Table S2). These genes encode structural MAPs that regulate MT stability (MAPT, MTUS1, STMN1), MT end-binding proteins (ASPM, GTSE1, RACGAP1), protein kinases and their regulators (AURKB, MAST4, TPX2), and molecular motors (a total of 8 kinesins) that control mitosis, cytokinesis, or intracellular transport. Of note, the MT-stabilizing protein tau encoded by MAPT was previously described as a potent predictor of taxane-based chemotherapy in breast cancer (16–21), therefore validating our gene profiling approach.
Fig. 1.
Low levels of MTUS1 predict the response to neoadjuvant chemotherapy. (A) Venn diagram of the number of differentially expressed genes between sensitive (pCR) and resistant (NpR) tumors in the REMAGUS02 (R02), REMAGUS04 (R04), and MD Anderson (MDA) cohorts, and common genes among them. (B) Volcano plots showing differentially expressed genes between pCR and NpR tumors from patients of the R02 (Left), MDA (Middle), and R04 (Right) cohorts. Each dot represents the fold change and the P value obtained for a single gene probe set. Genes common to all 3 cohorts are plotted in blue. MAPT is in light blue, and MTUS1 is in red. Names of the best candidates are indicated. (C) Scattered dot plot of the MTUS1 probe set (212093_s_at) intensity in tumors from patients of the R02 cohort with NpR or pCR after neoadjuvant chemotherapy. Numbers of samples are in brackets.**P < 0.01. (D) ROC curve evaluating the performance of MTUS1 expression for predicting complete response to neoadjuvant chemotherapy. (E) Heat map and hierarchical clustering of 115 breast tumor samples based on the intensities of 4 MTUS1 probe sets (212096_s_at, 212093_s_at, 212095_s_at, 239576_at). The heat map illustrates relative expression profiles of MTUS1 (column) for each tumor sample (line) in a continuous color scale from low (green) to high (red) expression. A dendrogram of the 3 selected tumor groups is shown on the right. (F) Scattered dot plot of MTUS1 expression in each of the 3 selected clusters based on the dendrogram shown in E. Numbers of samples are in brackets. (G) Proportion of patients with pCR according to the MTUS1 level in each selected cluster. Numbers of tumors in each group are indicated in brackets. (H) Proportion of patients with pCR according to the MTUS1 level in all tumors (All) and among ER+ and ER- tumors. Numbers of tumors in each group are indicated in brackets. (I) Correlation between MTUS1 (212093_s_at) probe set intensities and ATIP3 mRNA levels measured by real-time RT-PCR (qPCR) using oligonucleotides designed in 5′ exons that are specific to ATIP3 transcripts in 106 breast tumor samples of the R02 cohort.
Table 1.
MT-regulating genes differentially expressed in chemosensitive breast tumors
| Gene symbol | Gene name | Process |
| ASPM | Abnormal spindle microtubule assembly | MT minus-end binding, spindle organization |
| AURKB | Aurora kinase B | Ser/Thr protein kinase, spindle organization |
| GTSE1 | G2 and S phase-expressed 1 | MT plus-end binding, spindle organization, cell migration |
| KIF11 | Kinesin family member 11 | MT molecular motor, spindle organization |
| KIF14 | Kinesin family member 14 | MT molecular motor |
| KIF15 | Kinesin family member 15 | MT molecular motor |
| KIF18B | Kinesin family member 18B | MT depolymerizing, spindle organization |
| KIF20A | Kinesin family member 20A | MT molecular motor |
| KIF2C | Kinesin family member 2C | MT depolymerizing, spindle organization |
| KIF4A | Kinesin family member 4A | MT molecular motor, chromokinesin |
| KIFC1 | Kinesin family member C1 | MT molecular motor, spindle assembly |
| RACGAP1 | Rac GTPase-activating protein 1 | MT plus-end binding, spindle midzone assembly, cytokinesis |
| STMN1 | Stathmin 1 | MT-destabilizing, spindle organization |
| TPX2 | Targeting protein for Xklp2 | Aurora kinase A regulator, spindle organization |
| MAPT | Microtubule-associated protein tau | MT stabilizing, EB1 binding |
| MAST4 | Microtubule associated serine/threonine kinase family member 4 | Ser/Thr protein kinase |
| MTUS1 | Microtubule-associated tumor suppressor 1 | MT stabilizing, EB1 binding, tumor suppressor effects |
Properties of the 17 differentially regulated genes common to the 3 cohorts. Genes up-regulated in pCR tumors are in bold; those down-regulated are in italics.
Besides MAPT, the MTUS1 gene consistently reached higher fold change and better P value in all 3 cohorts (Fig. 1B). We thus focused our attention on MTUS1, whose gene product ATIP3 has been identified as a prognostic biomarker of patient survival with potent tumor suppressor effects in breast cancer (24, 25). In each cohort of breast cancer patients examined, MTUS1 Affymetrix probe set intensities were significantly lower in cases with pCR than in those with NpR (Fig. 1C and SI Appendix, Figs. S1–S3). Receiver operating curves (ROCs) revealed area under the curve (AUC) values of 0.717 (P = 0.007), 0.699 (P = 0.0005), and 0.769 (P < 0.0001) in the R02, MDA and R04 cohorts, respectively, indicating that MTUS1 levels predict the response to chemotherapy with good accuracy (Fig. 1D and SI Appendix, Figs. S1–S3).
Tumors were then classified into 3 groups expressing high, medium, and low MTUS1 levels according to heat map hierarchical clustering (Fig. 1 E and F). In the R02 cohort, the pCR rate was 32% (8/25) in tumors expressing low MTUS1 levels compared to 11.9% (7/59) and 0% (0/31) in those expressing medium and high levels of MTUS1, respectively (Fig. 1G), indicating that the subpopulation of low-MTUS1-expressing tumors is more likely to achieve complete response. Of note, 100% of tumors with high MTUS1 levels failed to achieve pCR, suggesting that high MTUS1 levels may identify patients unlikely to respond to chemotherapeutic treatment (Fig. 1G). Similar results were obtained using the 2 other cohorts of patients (SI Appendix, Figs. S2D and S3D).
Univariate logistic regression analysis including age, hormone receptors, HER2, tumor grade, stage, nodal status, and MTUS1 level identified hormonal receptors ER (odds ratio [OR]: 4.46; CI: 1.4 to 14.2) and PR (OR: 6.5; CI: 1.39 to 30.36), as well as MTUS1 (OR: 5.37; CI: 1.71 to 16.84), as predictive factors associated with pCR after neoadjuvant chemotherapy (Table 2 and SI Appendix, Table S3). In multivariate analysis, ER (OR: 4.66; CI: 1.46 to 14.81) and MTUS1 (OR: 12.16; CI: 3.88 to 38.07) were identified as independent predictors of pCR (Table 2 and SI Appendix, Table S3). These results indicate that low MTUS1 status may be used to identify patients with high response rates. Notably, 32% of low-MTUS1 tumors were associated with pCR, compared to 25% for ER-negative tumors. Combining ER status and low MTUS1 levels further increased the pCR rate from 7 to 25% among ER-positive tumors and 25 to 39% among ER-negative tumors of the R02 cohort (Fig. 1H). Similar results were obtained with the 2 other series of patients (SI Appendix, Figs. S2E and S3E). Real-time RT-PCR analysis performed in a panel of 106 breast tumors of the R02 cohort using 3 different pairs of oligonucleotides showed significant correlation between ATIP3 messenger RNA (mRNA) levels and MTUS1 Affymetrix probe set intensities (Fig. 1I and SI Appendix, Fig. S1 D–F). Together, these results indicate that low ATIP3 mRNA levels predict breast tumor response to chemotherapy.
Table 2.
Predictive factors of pCR in the R02 study
| Variable | pCR (%) | Univariate analysis | Multivariate analysis | ||
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| Age | |||||
| <50 | 8 (10.3) | 0.44 (0.145–1.33) | 0.147 | ||
| >50 | 7 (20.6) | ||||
| ER | |||||
| Negative | 10 (25) | 4.46 (1.4–14.2) | 0.011 | 4.66 (1.46–14.81) | 0.0076 |
| Positive | 5 (6.9) | ||||
| PR | |||||
| Negative | 13 (21.3) | 6.5 (1.39–30.36) | 0.017 | 1.88 (0.40–8.78) | 0.281 |
| Positive | 2 (4) | ||||
| HER2 | |||||
| Negative | 9 (12.2) | ||||
| Positive | 6 (15.8) | 1.35 (0.11–4.13) | 0.597 | ||
| Grade | |||||
| III | 10 (16.1) | 2.01 (0.59–6.89) | 0.262 | ||
| I/II | 4 (8.7) | ||||
| Tumor stage | |||||
| T2 | 10 (17.8) | 2.21 (0.70–6.96) | 0.172 | ||
| T3–4 | 5 (8.9) | ||||
| Nodal status | |||||
| N0 | 8 (19.1) | 2.05 (0.684–6.14) | 0.199 | ||
| N+ | 7 (10.3) | ||||
| MTUS1 level | |||||
| Low | 8 (32) | 5.37 (1.71–16.84) | 0.0039 | 12.16 (3.88–38.07) | <0.0001 |
| Medium/high (n = 7) | 7 (8.1) | ||||
| Continuous* | 1.78 (1.19–2.76) | 0.005 | |||
Entered as continuous variable (probe set 212096_s_at values). Significant P values are indicated in bold.
Low ATIP3 Expression in Breast Tumors and Cancer Cells Increases Sensitivity to Taxanes but Not DNA-Targeting Agents.
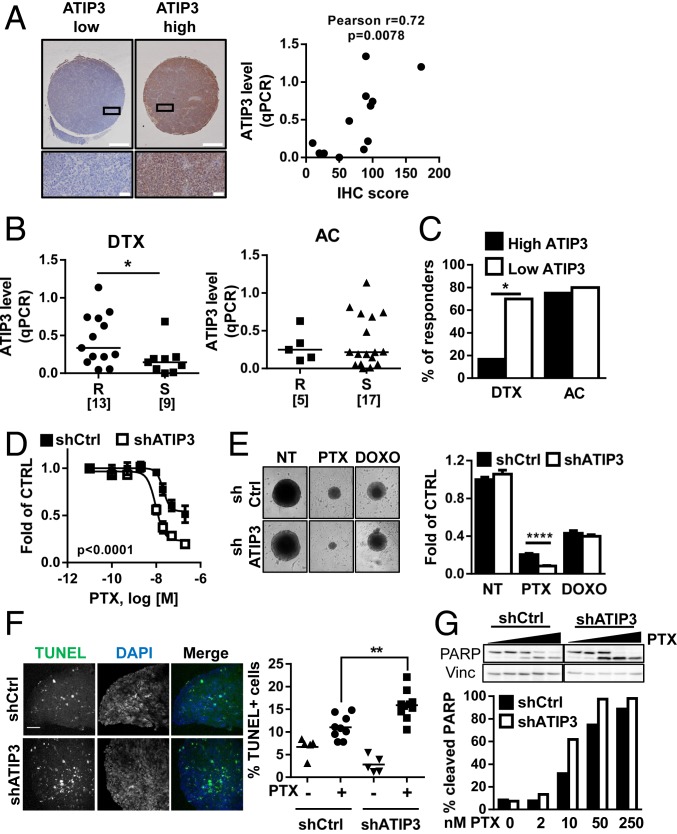
Preclinical studies were undertaken using patient-derived xenografts (26) to confirm and extend our results obtained on breast cancer patients. Twenty-two models of human breast cancer xenografts grown in mice (HBCx) were exposed to either docetaxel (DTX) or anthracycline plus cyclophosphamide (AC). ATIP3 expression levels in HBCx were evaluated by real-time RT-PCR and validated by immunohistochemistry (IHC) (Fig. 2A and SI Appendix, Table S4). ATIP3 mRNA levels were then plotted according to sensitivity or resistance of tumors to DTX and AC treatment. As shown in Fig. 2B, DTX-sensitive xenografts displayed significantly lower ATIP3 levels than DTX-resistant ones (median expression value of 0.14 versus 0.33). No significant difference in ATIP3 level was observed between AC-sensitive and AC-resistant HBCx (Fig. 2B), indicating that ATIP3 levels are associated with the response to the microtubule-targeting agent DTX rather than to DNA-targeting drugs. Accordingly, the response rate to DTX was significantly higher in low-ATIP3 compared to high-ATIP3 expressing HBCx (70 vs. 16.7%), whereas the response rate to AC remained similar in both groups of tumors (80 vs. 75%) (Fig. 2C).
Fig. 2.
Low ATIP3 levels are associated with sensitivity to taxanes in patient-derived xenografts and breast cancer MCSs. (A, Left) Immunohistochemistry performed on human breast cancer xenograft sections of a tissue microrarray using anti-MTUS1 monoclonal antibody. Shown are representative photographs of tumors expressing low (Left) or high (Right) levels of ATIP3. (Scale bar, 500 µm.) Close-ups are shown in the bottom. (Scale bar, 50 µm.) (A, Right) Correlation between ATIP3a mRNA expression level (by qPCR) and IHC score. (B) Scattered dot plot of ATIP3 (qPCR) mRNA expression level in HBCx treated with DTX (Left) or with (Right). Tumors are classified according to their response to drug treatment. R indicates resistance, and S indicates sensitivity. Numbers of samples are in brackets. *P < 0.05. (C) Percentage of responsive HBCx according to ATIP3 level. Tumors were subdivided into groups expressing high ATIP3 versus low ATIP3 levels based on the median value of ATIP3 measured by real-time RT-PCR. *P < 0.05. (D) Dose–response curves of SUM52PE spheroids (MCSs) expressing (shCtrl) or not expressing (shATIP3) ATIP3 and treated with increasing concentrations of PTX. (E) SUM52PE MCSs as in D were treated for 6 d with 50 nM PTX or 100 nM doxorubicin (DOXO) and photographed. The picture represents one MCS of the quadruplicate. Measures of spheroid area are plotted in the histogram on the Right. ****P < 0.0001. (F) Representative photographs of SUM52PE MCSs as in D treated with 50 nM of PTX for 72 h prior to staining with TUNEL reagent (green) and DAPI (blue). Quantification of apoptosis, measured as percent of TUNEL-positive cells, is shown on the Right. (Magnification, 20×.) (Scale bar, 100 µm.) **P < 0.01. (G) Western blot analysis of PARP cleavage in SUM52PE MCSs as in D treated for 72 h with increasing concentrations of PTX. Vinculin (Vinc) is used as the internal loading control. Quantification is shown below.
We then investigated the consequence of ATIP3 depletion on breast cancer cell viability upon exposure to chemotherapy. Breast cancer cells were grown in 3 dimensions as multicellular spheroids (MCSs) to mimic the main features and tissue architecture of solid tumors (27) and were treated with increasing doses of chemotherapeutic agents. ATIP3 silencing in SUM52-PE cells (SI Appendix, Fig. S4A) markedly improved the cytotoxic effects of both DTX and paclitaxel (PTX) (Fig. 2D and SI Appendix, Fig. S4 B–D) but had no effect on the cellular response to doxorubicin (Fig. 2E). Similar results were obtained in HCC1143 and MDA-MB-231 breast cancer cells treated with DTX and PTX, respectively (SI Appendix, Fig. S4 E and F), confirming that ATIP3 deficiency sensitizes breast cancer cells to taxanes but not to DNA-targeting drugs.
Taxanes are mitotic poisons that arrest cells in mitosis and trigger apoptosis. MCSs treated with clinically relevant doses of PTX (13) were arrested in mitosis, a phenotype that was further increased in ATIP3-deficient cells (SI Appendix, Fig. S5A). ATIP3 deficiency also increased the percentage of cells undergoing apoptosis following treatment with low doses of PTX (Fig. 2F and SI Appendix, Fig. S5B). Accordingly, molecular markers of apoptosis, such as cleavage of PARP (Fig. 2G) and of caspase-3 (SI Appendix, Fig. S5C) as well as decline in antiapoptotic protein Mcl-1 levels (SI Appendix, Fig. S5D), were elevated upon PTX treatment in ATIP3-deficient compared to ATIP3-proficient spheroids. Thus, ATIP3 deficiency improves the mitotic and proapoptotic effects of taxanes.
PTX-Induced Mitotic Defects Are Increased in ATIP3-Deficient Cells.
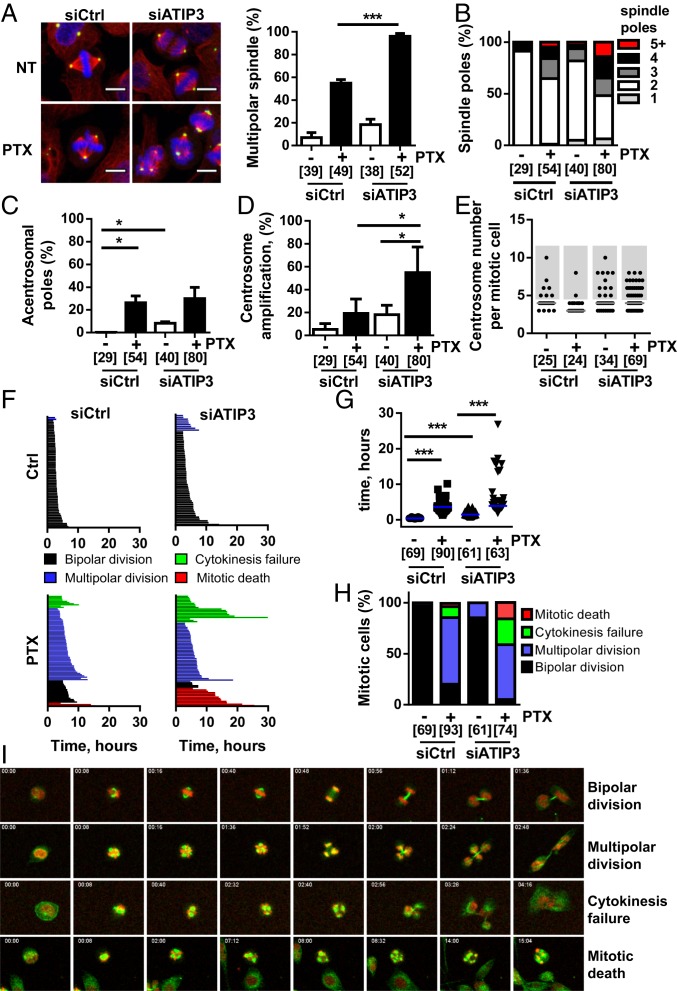
To get insight into the mechanism by which ATIP3 depletion sensitizes cancer cells to the effects of PTX, we examined the consequences of ATIP3 silencing on mitosis. HeLa cells were used as a reference model in these experiments because they express endogenous ATIP3 and are more suitable than SUM52 cells for cell imaging. As shown in Fig. 3A, ATIP3 depletion induced the formation of multipolar spindles (18%) and raised the percentage of multipolar cells from 55 to almost 100% in the presence of low doses of PTX. ATIP3 depletion also markedly increased the number of spindle poles formed upon PTX treatment, with a significant fraction of spindles showing more than 5 poles (Fig. 3B), some of them being acentrosomal (Fig. 3C), indicating excessive mitotic abnormalities. The increased number of spindle poles was mainly due to centrosome amplification, which was markedly increased when combining ATIP3 silencing and PTX treatment (Fig. 3D). Accordingly, ATIP3 silencing was associated with supernumerary centrosomes, and the number of centrosomes per mitotic cell was further elevated in ATIP3-deficient cells upon PTX treatment (Fig. 3E and SI Appendix, Fig. S6A). More than half of supernumerary centrosomes contained either one or no centriole (SI Appendix, Fig. S6B), underlining major centrosomal defects. Together, these results indicate that ATIP3 silencing induces centrosome amplification leading to multipolar spindles, a phenotype that is amplified upon PTX treatment.
Fig. 3.
Low ATIP3 levels increase PTX-induced mitotic defects. (A) Immunofluorescence photographs of HeLa cells expressing ATIP3 (siCtrl) or not (siATIP3) and treated or not (NT) with 5 nM of PTX prior to staining with antibodies against α-tubulin (red) and pericentrin (green). Nuclei are stained with DAPI (blue). Quantification of abnormal mitoses is shown on the Right. (B) Proportion of mitotic HeLa cells with an abnormal number of spindle poles upon 48 h ATIP3 silencing and 18 h PTX treatment (2 nM). (C) Percentage of mitotic HeLa cells containing acentrosomal poles following treatment as in B. (D) Percentage of mitotic HeLa cells containing more than 2 centrosomes. Cells were treated as in B. (E) Scattered dot plot of the number of centrosomes per mitotic HeLa cells treated as in B. (F) Cell fate profiles of control (Left) and ATIP3-silenced (Right) HeLa cells in the absence (Top) or in the presence of 2 nM PTX (Bottom). (G) Scattered dot plot of mitotic length measured from chromosome condensation to initiation of cytokinesis in HeLa cells silenced or not for ATIP3 and treated or not with 2 nM PTX. (H) Proportion of cell fate profiles measured in F. (I) Images from the time-lapse experiment performed in F, showing representative cell fates. Microtubules are stained in green; DNA is in red. Time, in hours:minutes, is indicated in top left of the picture. (Magnification, 20×.) (A–H) Number of mitotic cells is in brackets. *P < 0.05, ***P < 0.001.
We then examined the consequences of ATIP3 silencing and PTX treatment on cell fate at the single cell level using time-lapse video microscopy (Fig. 3F and Movies S1–S4). ATIP3 silencing increased the time in mitosis (Fig. 3G) and induced the formation of multipolar cells that were able to divide, giving rise to 2 or 3 viable daughter cells containing several nuclei (Fig. 3 H and I and Movie S2). PTX at low dose induced a majority (79%) of cells with multipolar spindles, among which 14% were unable to divide. These cells either died in mitosis or underwent cytokinesis failure, giving rise to groups of multinucleated cells that ultimately died during the following division (Fig. 3 H and I and Movie S3). ATIP3 silencing combined with a low dose of PTX induced massive (95%) formation of multipolar cells, 41% of which died during the first division from cytokinesis failure or mitotic death (Figs. 3 H and I and Movie S4). Thus, ATIP3 silencing exacerbates mitotic abnormalities and subsequent cell death induced by PTX treatment.
ATIP3 Depletion Is Associated with Increased Aneuploidy.
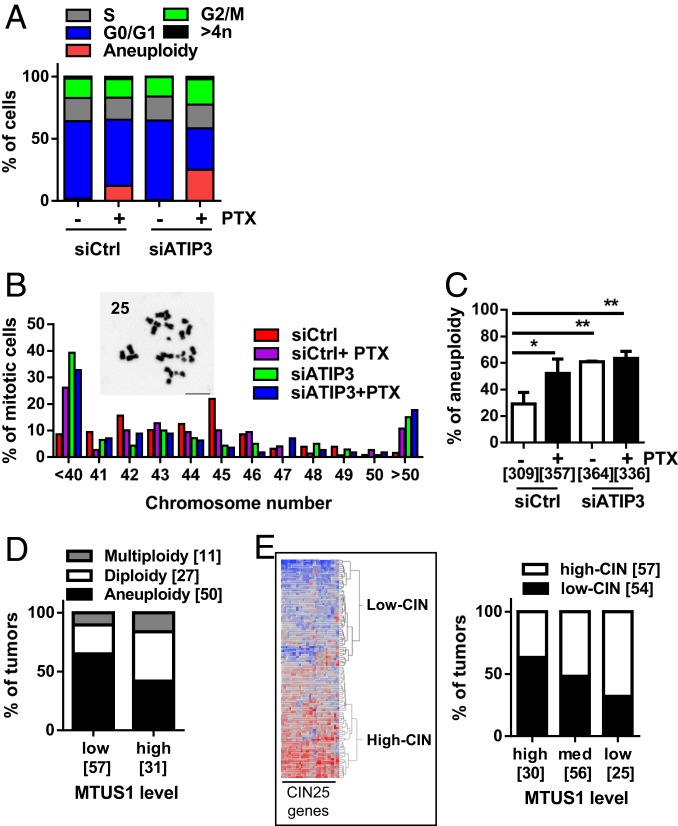
The formation of multipolar spindles is a source of chromosome missegregation and aneuploidy, suggesting that ATIP3 silencing may promote aneuploidy. To test this hypothesis, we analyzed cellular DNA content by flow cytometry to assess DNA ploidy in HeLa cells treated or not with low doses of PTX. In line with previous studies (11, 13), treatment with PTX at the nanomolar range resulted in a hypodiploid (<2N) population of cells (Fig. 4A and SI Appendix, Fig. S7A). This population of aneuploid cells disappeared at higher concentrations of PTX (100 nM) when cells were arrested in G2/M (SI Appendix, Fig. S7B) and was negative for annexin V labeling (SI Appendix, Fig. S7C), excluding the possibility that these cells may be apoptotic. In ATIP3-depleted cells, hypodiploidy could not be detected by flow cytometry procedures in the absence of treatment (Fig. 4A). However, following PTX exposure, the population of hypodiploid cells was raised from 12% in control cells to 25% in ATIP3-deficient cells, indicating that ATIP3 silencing increases PTX-induced aneuploidy.
Fig. 4.
Low ATIP3 level increases aneuploidy. (A) FACS analysis of DNA content of HeLa cells transfected with scramble (siCtrl) or siATIP3 prior treatment with 5 nM PTX for 18 h. Shown is the proportion of cells in each cell cycle fraction. (B) Proportion of HCT116 metaphase spreads containing abnormal numbers of chromosomes. Inset shows a representative image of metaphase spread. The corresponding number of chromosomes is indicated in the top left corner. (C) Percentage of aneuploid cells among HCT116 cells silenced (siATIP3) or not (siCtrl) for ATIP3 and treated or not with 5 nM PTX for 18 h. The number of metaphase spreads analyzed in 3 independent experiments is shown in brackets. *P < 0.05, **P < 0.01. (D) Proportion of tumors from the Curie cohort showing altered ploidy according to ATIP3 level. The number of tumors analyzed is in brackets. (E) Heat map and hierarchical clustering of 115 breast tumor samples of the R02 study based on the intensities of the 25 genes from the CIN signature (29). The proportion of tumors expressing a high level (high-CIN) or low level (low-CIN) of CIN signature according to ATIP3 levels is shown on the Right. The number of tumors is indicated in brackets.
Aneuploidy was also evaluated by counting the chromosome number per cell in metaphase chromosome spreads of HCT116 cells, used as a reference cellular model in these studies because they are nearly diploid and chromosomally stable. PTX treatment significantly increased the incidence of both hypodiploid (<40 chromosomes) and hyperdiploid (>50 chromosomes) cells, and ATIP3 silencing further increased aneuploidy (Fig. 4 B and C).
To assess the clinical relevance of our findings, we analyzed a series of 88 breast cancer patients in whom ploidy had been evaluated. Tumors were grouped according to low and high MTUS1 levels using heat map classification as reported (24, 28) and compared with ploidy status (SI Appendix, Table S5). As shown in Fig. 4D, 65% of low-MTUS1 tumors were found to be aneuploid compared to 42% of high-MTUS1 tumors. Since aneuploidy often results from chromosomal instability (CIN), we investigated whether MTUS1 levels may also be associated with CIN. A “CIN25 signature,” comprising a panel of 25 differentially regulated genes, was previously reported in breast cancer (29). We therefore classified breast tumors as high CIN or low CIN based on the CIN25 signature and compared the signature with MTUS1 levels. As shown in Fig. 4E and SI Appendix, Fig. S8, 40% of breast tumors with high MTUS1 levels were classified as high CIN compared to 60% of low-MTUS1 tumors. Together, these results support the notion that breast tumors expressing low levels of ATIP3 are more prone to chromosomal instability and aneuploidy.
Discussion
Based on transcriptional profiling of 3 independent cohorts of breast cancer patients treated with taxane-based neoadjuvant chemotherapy, we show here that microtubule-associated protein ATIP3 is an independent predictive biomarker of the response to treatment. Low levels of ATIP3 were significantly more frequent in breast tumors that achieved pCR compared with those that did not respond to treatment, suggesting that low ATIP3 expression may be used as a marker to identify breast cancers that are highly sensitive to taxane-containing chemotherapy. Importantly, in all 3 cohorts analyzed, 98 to 100% of tumors expressing high levels of ATIP3 failed to achieve pCR, indicating that ATIP3 may also be a useful biomarker to select patients unlikely to respond to conventional chemotherapy, which is of clinical importance to limit toxicity and side effects of ineffective treatments. Low ATIP3 levels predict the response to neoadjuvant chemotherapy even better than ER-negative status and can further identify responders among ER-negative tumors. This finding is of particular interest for triple-negative breast tumors for which chemotherapy remains the unique therapeutic option (30). Further validation in adjuvant trials is warranted to firmly establish the value of ATIP3 as a predictive biomarker in clinical practice in breast cancer. It will be interesting to broaden our study to other types of malignancies, such as prostate, lung, and ovarian cancer, where taxanes are frequently used.
Preclinical studies performed on breast cancer patient–derived xenografts and in 3-dimensional models of multicellular spheroids further allowed us to investigate the predictive value of ATIP3 in the response to taxanes compared to anthracyclines and showed that low levels of ATIP3 are associated with high sensitivity to docetaxel, with no impact on the response to DNA-targeting agents. Accordingly, ATIP3 silencing sensitizes cancer cells to low doses of paclitaxel and potentiates the well-known effects of the drug on mitotic arrest and apoptosis. Although consistent with data from breast cancer patients, this was an unexpected result given that ATIP3 silencing increases MT dynamics (25), which is opposite to the MT-stabilizing effects of taxanes.
Results presented here indicate that silencing of the MT-stabilizing protein ATIP3 induces multiple mitotic abnormalities that mimic those induced by PTX. We propose a mechanism in which ATIP3 silencing, by causing centrosome amplification and multipolar spindle formation, amplifies the effects of taxanes and thereby exacerbates mitotic abnormalities, chromosome segregation errors, CIN features, and aneuploidy, ultimately leading to cell death in response to treatment.
It has been widely shown that centrosome amplification, leading to spindle multipolarity and subsequent chromosome missegregation and aneuploidy, promotes tumor initiation and progression (31–33). Centrosome amplification is also associated with worse clinical outcome in breast cancer (34, 35). In this context, our findings that ATIP3 deficiency induces centrosome amplification are consistent with previous observations that low ATIP3 levels in breast tumors are associated with poor patient prognosis (25). Strikingly, our data also indicate that combining ATIP3 deficiency and PTX treatment causes excessive centrosome amplification and aneuploidy, which in turn triggers massive cell death in mitosis. This is in line with previous observations that increasing chromosome missegregation and aneuploidy beyond a critical threshold leads to cancer cell death and tumor suppression (36, 37) and supports our clinical results showing higher pCR for ATIP3-deficient breast cancer patients treated with taxane-based chemotherapy.
In conclusion, while the consequences of centrosome amplification and CIN for therapeutic responses in cancer patients still remain a matter of debate (38), our data emphasize the link between centrosome amplification and increased pCR rates for breast tumors. Our results highlight ATIP3 as a predictive biomarker to select a population of breast cancer patients who are likely to benefit from taxane-based chemotherapy and open the way to therapeutic strategies based on increasing centrosomal alterations to achieve chemosensitivity.
Materials and Methods
Studies using the R02 (6) and R04 cohorts of patients were reviewed by the French ethical committee of Paris-Bicêtre in compliance with the Helsinki Declaration. Studies on the MDA cohort (39) were approved by the institutional review boards of MD Anderson Cancer Center and Instituto Nacional de Enfermedades Neoplásicas and those on the Curie cohort (24) and patient-derived xenografts (26) were approved by the institutional review boards of Institut Curie (Paris, France). All patients signed an informed consent for voluntary participation in the trial. Details on patients and samples, clinical data, and gene profiling are provided in SI Appendix, Materials and Methods. These materials and methods also describe RNA extraction and real-time RT-PCR analysis, cells used, multicellular spheroids, Terminal deoxynucleotidyl transferase dUTP Nick End Labeling assay and Fluorescence Activated Cell Sorting analysis of apoptosis, DNA content analysis and chromosome spread, confocal and time-lapse imaging, analysis of mitotic defects, immunohistochemistry, and statistical analyses.
Supplementary Material
Acknowledgments
We thank the members of the REMAGUS02 and REMAGUS04 groups for their contribution to this work and K. Tran-Perennou for excellent technical assistance. We are grateful to Dr. Sophie Hamy-Petit (Institut Curie, Paris) for helpful discussion. This work has benefited from the facilities and expertise of the Imaging and Cytometry Platforms (thanks to the engineers Frederic De Leeuw, Yann Lecluse), Unité Mixte de Service 3655/US23, of the Gustave Roussy Cancer Campus, Villejuif, France. This work was funded by Gustave Roussy Research Center, the Agence Nationale pour la Recherche (ANR) Grant MMO ANR-10-IBHU-0001, the Taxe d’Apprentissage TA2018 (University Paris Saclay, France), the Comité Ile-de-France of the Ligue Nationale Contre le Cancer, the Ligue Contre le Cancer 94/Val-de-Marne, the GEFLUC Ile-de-France, the Fondation Association pour la Recherche contre le Cancer, CNRS, Institut national de la santé et de la recherche médicale, the Fondation Janssen Horizon, the Fonds de Dotation Agnès b., and association Odyssea and Prolific.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910824116/-/DCSupplemental.
References
- 1.Teshome M., Hunt K. K., Neoadjuvant therapy in the treatment of breast cancer. Surg. Oncol. Clin. N. Am. 23, 505–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mauri D., Pavlidis N., Ioannidis J. P. A., Neoadjuvant versus adjuvant systemic treatment in breast cancer: A meta-analysis. J. Natl. Cancer Inst. 97, 188–194 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Van de Wiel M., et al. , Neoadjuvant systemic therapy in breast cancer: Challenges and uncertainties. Eur. J. Obstet. Gynecol. Reprod. Biol. 210, 144–156 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Gralow J. R., et al. , Preoperative therapy in invasive breast cancer: Pathologic assessment and systemic therapy issues in operable disease. J. Clin. Oncol. 26, 814–819 (2008). [DOI] [PubMed] [Google Scholar]
- 5.von Minckwitz G., et al. , Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 30, 1796–1804 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Valet F., et al. , Challenging single- and multi-probesets gene expression signatures of pathological complete response to neoadjuvant chemotherapy in breast cancer: Experience of the REMAGUS 02 phase II trial. Breast 22, 1052–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wang-Lopez Q., et al. , Can pathologic complete response (pCR) be used as a surrogate marker of survival after neoadjuvant therapy for breast cancer? Crit. Rev. Oncol. Hematol. 95, 88–104 (2015). [DOI] [PubMed] [Google Scholar]
- 8.von Minckwitz G., Fontanella C., Comprehensive review on the surrogate endpoints of efficacy proposed or hypothesized in the scientific community today. J. Natl. Cancer Inst. Monogr. 2015, 29–31 (2015). [DOI] [PubMed] [Google Scholar]
- 9.de Gramont A., et al. , Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat. Rev. Clin. Oncol. 12, 197–212 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Chen J.-G., Horwitz S. B., Differential mitotic responses to microtubule-stabilizing and -destabilizing drugs. Cancer Res. 62, 1935–1938 (2002). [PubMed] [Google Scholar]
- 11.Weaver B. A., How Taxol/paclitaxel kills cancer cells. Mol. Biol. Cell 25, 2677–2681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White D., Honoré S., Hubert F., Exploring the effect of end-binding proteins and microtubule targeting chemotherapy drugs on microtubule dynamic instability. J. Theor. Biol. 429, 18–34 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Zasadil L. M., et al. , Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci. Transl. Med. 6, 229ra43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petry S., Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 85, 659–683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prosser S. L., Pelletier L., Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187–201 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Rouzier R., et al. , Microtubule-associated protein tau: A marker of paclitaxel sensitivity in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 8315–8320 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andre F., et al. , Microtubule-associated protein-tau is a bifunctional predictor of endocrine sensitivity and chemotherapy resistance in estrogen receptor-positive breast cancer. Clin. Cancer Res. 13, 2061–2067 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Wang K., et al. , Tau expression correlated with breast cancer sensitivity to taxanes-based neoadjuvant chemotherapy. Tumour Biol. 34, 33–38 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Zhou J., et al. , Predictive value of microtubule-associated protein tau in patients with recurrent and metastatic breast cancer treated with taxane-containing palliative chemotherapy. Tumour Biol. 36, 3941–3947 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Baquero M. T., et al. , Evaluation of prognostic and predictive value of microtubule associated protein tau in two independent cohorts. Breast Cancer Res. 13, R85–R102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonneau C., Gurard-Levin Z. A., Andre F., Pusztai L., Rouzier R., Predictive and prognostic value of the TauProtein in breast cancer. Anticancer Res. 35, 5179–5184 (2015). [PubMed] [Google Scholar]
- 22.Di Benedetto M., et al. , Structural organization and expression of human MTUS1, a candidate 8p22 tumor suppressor gene encoding a family of angiotensin II AT2 receptor-interacting proteins, ATIP. Gene 380, 127–136 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues-Ferreira S., Nahmias C., An ATIPical family of angiotensin II AT2 receptor-interacting proteins. Trends Endocrinol. Metab. 21, 684–690 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Rodrigues-Ferreira S., et al. , 8p22 MTUS1 gene product ATIP3 is a novel anti-mitotic protein underexpressed in invasive breast carcinoma of poor prognosis. PLoS One 4, e7239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molina A., et al. , ATIP3, a novel prognostic marker of breast cancer patient survival, limits cancer cell migration and slows metastatic progression by regulating microtubule dynamics. Cancer Res. 73, 2905–2915 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Marangoni E., et al. , A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 13, 3989–3998 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Antoni D., Burckel H., Josset E., Noel G., Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 16, 5517–5527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues-Ferreira S., et al. , Combinatorial expression of microtubule-associated EB1 and ATIP3 biomarkers improves breast cancer prognosis. Breast Cancer Res. Treat. 173, 573–583 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Carter S. L., Eklund A. C., Kohane I. S., Harris L. N., Szallasi Z., A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Bianchini G., Balko J. M., Mayer I. A., Sanders M. E., Gianni L., Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basto R., et al. , Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032–1042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganem N. J., Godinho S. A., Pellman D., A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine M. S., et al. , Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell 40, 313–322.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannu V., et al. , Rampant centrosome amplification underlies more aggressive disease course of triple negative breast cancers. Oncotarget 6, 10487–10497 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denu R. A., et al. , Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer 16, 47–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen A., Kops G. J., Medema R. H., Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. U.S.A. 106, 19108–19113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silk A. D., et al. , Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. U.S.A. 110, E4134–E4141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vargas-Rondón N., Villegas V. E., Rondón-Lagos M., The role of chromosomal instability in cancer and therapeutic responses. Cancers (Basel) 10, e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hess K. R., et al. , Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 24, 4236–4244 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.