Significance
Damaged mitochondria are eliminated by lysosomes, a process called mitophagy. However, the brain tissue specific roles and mechanisms of mitophagy remain unknown. We report that Rhes, a protein highly enriched in the striatum, eliminates damaged mitochondria via mitophagy, a function that may regulate the optimum number and function of mitochondria in the striatum. However, when mitochondria are irreversibly damaged, as in the presence of 3-NP (an inhibitor of complex II, SDH), Rhes exacerbates mitophagy in association with Nix, a mitophagy receptor and promotes cell death. Intriguingly, Rhes can travel via membranous protrusions from a healthy cell to the neighboring cell and interacts with the damaged mitochondria via Nix. Collectively, Rhes acts as “mitochondrial surveillant,” and excessive mitophagy may account for striatal vulnerability in brain.
Keywords: striatal neuronal vulnerability, tunneling nanotubes, mitophagy ligand, SUMO-E3 ligase, mitophagosomes
Abstract
Elimination of dysfunctional mitochondria via mitophagy is essential for cell survival and neuronal functions. But, how impaired mitophagy participates in tissue-specific vulnerability in the brain remains unclear. Here, we find that striatal-enriched protein, Rhes, is a critical regulator of mitophagy and striatal vulnerability in brain. In vivo interactome and density fractionation reveal that Rhes coimmunoprecipitates and cosediments with mitochondrial and lysosomal proteins. Live-cell imaging of cultured striatal neuronal cell line shows Rhes surrounds globular mitochondria, recruits lysosomes, and ultimately degrades mitochondria. In the presence of 3-nitropropionic acid (3-NP), an inhibitor of succinate dehydrogenase, Rhes disrupts mitochondrial membrane potential (ΔΨm) and promotes excessive mitophagy and cell death. Ultrastructural analysis reveals that systemic injection of 3-NP in mice promotes globular mitochondria, accumulation of mitophagosomes, and striatal lesion only in the wild-type (WT), but not in the Rhes knockout (KO), striatum, suggesting that Rhes is critical for mitophagy and neuronal death in vivo. Mechanistically, Rhes requires Nix (BNIP3L), a known receptor of mitophagy, to disrupt ΔΨm and promote mitophagy and cell death. Rhes interacts with Nix via SUMO E3-ligase domain, and Nix depletion totally abrogates Rhes-mediated mitophagy and cell death in the cultured striatal neuronal cell line. Finally, we find that Rhes, which travels from cell to cell via tunneling nanotube (TNT)-like cellular protrusions, interacts with dysfunctional mitochondria in the neighboring cell in a Nix-dependent manner. Collectively, Rhes is a major regulator of mitophagy via Nix, which may determine striatal vulnerability in the brain.
Understanding mitophagy mechanisms and its dysregulation leading to pathological abnormalities in human diseases is a major challenge in modern biology. Reduced mitochondrial functions are linked to aging and many neurodegenerative disorders. Parkinson disease (PD) is the best example linked to mitochondrial dysfunction, because the most vulnerable neurons of PD, the substantia nigra pars compacta, show mitochondrial abnormalities. Familial PD genes, such as PTEN-induced kinase-1 (Pink1), parkin, and leucine-rich repeat kinase 2, are implicated in mitochondrial dysfunction (1–3). Decline in mitochondrial enzyme activity is reported more in Alzheimer disease and amyotrophic lateral sclerosis patients compared to control subjects (4). Impaired mitochondrial function is observed in Huntington disease (HD) as patients continue to drop weight, even though they have a high caloric intake (5). Oligomers of amyloid, mutant superoxide dismutase 1, and mutant huntingtin can affect mitochondrial membrane potentials and impair mitochondria trafficking and function (6–12). Despite these important studies, how mitochondrial dysfunction leads to selective neuronal vulnerability remains unknown. For example, although the familial PINK1 and parkin mutants are ubiquitously expressed (13), the mechanisms by which they elicit lesion in the substantia nigra remains largely unclear. Similarly, how mitochondrial toxins induce tissue-specific lesion in the brain is also unclear. It is well known that 3-nitropropionic acid (3-NP) promotes lesion in the striatum but not in the cortex or cerebellar neurons, resulting in HD-like motor deficits (14, 15). MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine) or rotenone promotes substantia nigra pars compacta neurodegeneration, sparing the cortex or striatum, resulting in PD-like symptoms (16–18). The major difference between these toxins that promote brain region-specific lesion are that 3-NP blocks complex II, succinate dehydrogenase (SDH) (19), whereas rotenone and MPTP block complex I (NADH [nicotinamide adenine dinucleotide, reduced] dehydrogenase) (20, 21). Thus, what determines the substantia nigra neuron to MPTP-induced lesion and striatal neuron to 3-NP−induced lesion remains unclear. The differences in their brain penetrability may not account for the tissue-specific lesion, as these toxins block mitochondrial function throughout the brain and peripheral tissue. Therefore, although mitochondrial dysfunction is ubiquitous, it is insufficient to elicit selective neuronal death, suggesting that there are additional mechanisms that may play a role in the brain (22, 23).
Rhes belongs to a small GTPase (guanosine triphosphate hydrolase) family of proteins highly enriched in the brain’s striatum, which controls psychiatric, cognitive, and motor functions. Rhes is induced by thyroid hormones and can inhibit the cAMP/PKA (cyclic adenosine monophosphate/protein kinase A) pathway, dopaminergic signaling, and N-type Ca2+ channels (Cav 2.2) (24–27). Over the years, we have found several roles for Rhes in the striatum. Rhes can regulate the mammalian target of rapamycin complex 1 (mTORC1), SUMOylation, and HD toxicity in cell and mouse models (28–32). Independent studies show a link for Rhes in striatal toxicity in various models of HD (33–38). Rhes knockout (KO) mice are also resistant to 3-NP−induced striatal lesion (39). Yet the mechanisms by which Rhes promotes neuronal vulnerability in the brain remains unclear.
Here we report that Rhes is a critical regulator of mitophagy in the striatum. Using ultrastructure, biochemical, and cell and molecular biology tools, we demonstrate that Rhes up-regulates mitophagy via Nix receptor, leading to striatal cell death. Our study reveals mitophagy mechanisms by which Rhes might promote striatal vulnerability in the brain.
Results
Rhes Associates with Mitochondria and Lysosomes In Vivo and In Vitro.
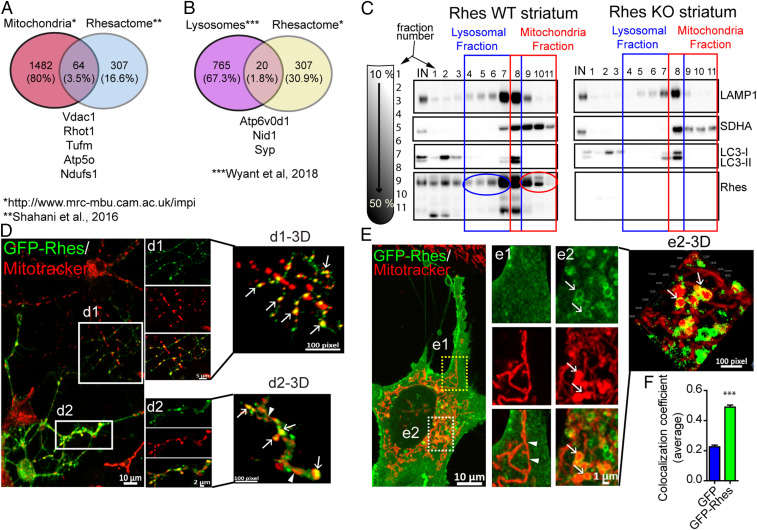
We took an unbiased approach by comparing our previously identified in vivo striatal interactome of Rhes (40) with a publicly available database of mitochondrial (www.mrc-mbu.cam.ac.uk/impi) and lysosomal proteins (41). We found 64 (3.5%) and 20 (1.8%) out of 307 high-confidence striatal interactors of Rhes with potential association to mitochondria (for example, Vdac1, Rhot1) and lysosomes (Atp6v0d1, Nid1), respectively (Fig. 1 A and B). Next, using sucrose gradient density, we separated organelles from the striatal brain tissue homogenate from wild-type (WT) and Rhes KO and found that endogenous Rhes cosedimented in mitochondrial (as detected by SDH subunit A [SDHA]) and lysosomal (Microtubule associated protein light chain 3 [LC3] and lysosome associated membrane protein 1) fractions in WT striatum, and, as expected, no Rhes signal was detected in KO (Fig. 1C). In order to further understand the Rhes’s mitochondrial role, we employed striatal neuronal cell lines (also known as STHdhQ7/Q7) isolated from the striatum of knock-in mice, which contain a targeted insertion of a chimeric mouse with human Htt exon 1 with 7 polyglutamine repeats (control) (42). These cells do not express endogenous Rhes (30, 43), so we think this is a suitable model for studying the function of exogenously added Rhes, as it is the only source of Rhes, and it would not compete with endogenous function of Rhes. Similar to endogenous Rhes in the striatum, the cultured striatal neuronal cell line expressing GFP-Rhes, not GFP alone (control), also cosedimented in the mitochondrial and lysosomal fractions (SI Appendix, Fig. S1A). Next, we carried live-cell time-lapse fluorescence confocal microscopy to determine Rhes interaction and function with mitochondria and lysosomes. We found that 1) Rhes interacted preferentially with globular mitochondria in primary striatal neurons processes (Fig. 1 D, Insets d1, d2, and 3D rendered images, d1-3D, d2-3D, arrows) and striatal neuronal cell line cytoplasm (Fig. 1 E, Insets e1, e2, and e2-3D) but not with the elongated mitochondria (Fig. 1 D and E, arrowheads); 2) Rhes formed a circular structure around the globular mitochondria (Fig. 1 E, e2-3D, arrow), and strongly colocalizes with it (Fig. 1F); 3) Rhes colocalizes with lysosomes in primary striatal neuronal processes (SI Appendix, Fig. S1 B, Insets b1 and b1-3D, arrow) as well as striatal neuronal cell lines (SI Appendix, Fig. S1 C, Inset c1-3D, arrow and Fig. S1D); and 4) triple staining reveals Rhes localizes with both mitochondria and lysosomes in striatal neuronal cell (SI Appendix, Fig. S1 E, Insets e1 and e1-3D). GFP alone did not show colocalization with mitotracker or lysosome in striatal neuronal cells or primary neurons (SI Appendix, Fig. S2). Next, we performed live-cell imaging of striatal neuronal cells transfected with GFP-Rhes (green) and costained with mitotracker (red) and lysotracker (blue) (Movie S1 with Insets). We found that GFP-Rhes was enriched with globular mitochondria, which becomes positive for lysosome in a time-dependent fashion (Movie S1, arrows). Intriguingly, enhanced GFP-Rhes signal intensity was associated with loss of mitotracker intensity, suggestive of mitochondrial degradation (Movie S1, arrow in GFP-Rhes). Collectively, these data indicated that Rhes associates with mitochondrial as well as lysosomal components in the intact striatum, primary neurons, and the striatal neuronal cell line, where we found stronger association with globular mitochondria.
Fig. 1.
Rhes associates with mitochondria and lysosome in vitro and in vivo. (A and B) Venn diagram showing mitochondrial and lysosomal proteins found in Rhes interactome in vivo, “Rhesactome,” compared with available database of mitochondrial and lysosomal proteins (40, 41). (C) Sucrose density gradient and Western blotting of striatal tissue from WT or Rhes KO mice indicating lysosomal (blue) and mitochondrial (red) proteins across different fractions. (D and E) Representative confocal images (their respective Insets or 3D rendered images) of (D) primary striatal neuron and (E) striatal neuronal cells transfected with GFP-Rhes and costained with mitotracker orange (red). Arrow and arrowhead depict globular and elongated mitochondria, respectively, in neuronal processes. (F) Bar graph depicting average Pearson’s coefficient of colocalization (n = 35 to 40 striatal neuronal cells per group, Student’s t test; ***P < 0.001; data are mean ± SEM).
Rhes Affects Basal Mitophagy but Not Mitochondrial Functions.
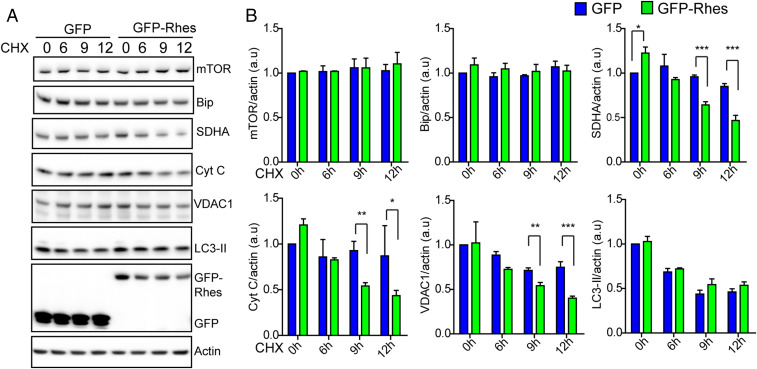
Lysosomal localization of Rhes-positive globular mitochondria (Movie S1 and SI Appendix, Fig. S1E) leads us to investigate the role of Rhes in mitochondrial degradation or mitophagy. Using cycloheximide (CHX) chase experiments, we tested whether Rhes affects basal mitophagy. As expected, CHX treatment resulted in the degradation of SDHA (inner mitochondrial protein), VDAC1 (outer mitochondrial protein), and cytochrome C (intermembrane mitochondrial protein), whose levels were further diminished by GFP-Rhes compared to GFP-alone overexpression (Fig. 2A). Note that levels of mTOR or Bip were not altered by CHX between the groups in these cells, and down-regulation of LC3-II was similar between GFP-expressing and GFP-Rhes−expressing cells (Fig. 2B). To further confirm the role of Rhes in regulation of basal mitophagy, we assessed the mitochondrial mass by CHX chase experiment in striatal neuronal cells infected with validated adenovirus empty (Ad-null) or adenovirus-Rhes (Ad-Rhes) (32), using mitotracker green, a molecular probe that binds to mitochondria regardless of its membrane potential (44). We found that Rhes-positive cells stained with mitotracker green showed less fluorescent intensity in CHX treatment compared to control, indicating rapid mitochondrial degradation, suggesting Rhes regulates basal mitophagy (SI Appendix, Fig. S3 A and B). Next, we wondered, whether Rhes can affect overall mitochondrial functions. To test this, we used seahorse assay, which measures the mitochondrial respiratory functions. We infected striatal neuronal cell lines with Ad-null or Ad-Rhes. As shown in SI Appendix, Fig. S3 C and D, Rhes expression did not affect the mitochondrial functions basally or in the absence or the presence of oligomycin (an inhibitor of ATP [adenosine triphosphate] synthase), Carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, uncouples oxidative phosphorylation), or rotenone (complex I inhibitor). Similarly, Rhes expression did not affect FCCP-induced mitophagy (SI Appendix, Fig. S3 E and F). Thus, Rhes enhances basal mitophagy in CHX conditions, but may not affect the overall mitochondrial functions basally or in the presence of certain pharmacological modulators of mitochondria.
Fig. 2.
Rhes regulates basal mitophagy. (A) Western blots from GFP-transfected or GFP-Rhes−transfected striatal neuronal cells treated with vehicle or cycloheximide (CHX, 100 µM) at indicated time point. Blots were probed with indicated antibodies. (B) Bar graph shows quantification of band intensities of indicated proteins in A. Protein levels were normalized to actin (n = 3, Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001; data are mean ± SEM).
Rhes Diminishes Mitochondrial Functions and Up-Regulates Mitophagy in the Presence of 3-NP.
To further assess the role of Rhes in mitochondrial functions, we investigated the effect of Rhes in the presence of 3-NP in seahorse assay. The rationale for testing 3-NP is 2-fold: 1) 3-NP is known to promote lesion selectively in the striatum, and 2) previously, we found that Rhes KO mice are totally prevented from 3-NP−induced striatal lesion compared to WT control (39). In sea horse assay, as in SI Appendix, Fig. S3C, Rhes alone did not affect mitochondrial function in vehicle conditions. But, in the presence of 3-NP, which diminished both the basal and maximal mitochondrial respiration, Rhes further potentiated the effect (SI Appendix, Fig. S4). This indicated that Rhes selectively worsens the 3-NP−induced mitochondrial dysfunctions, but not oligomycin, FCCP, or rotenone.
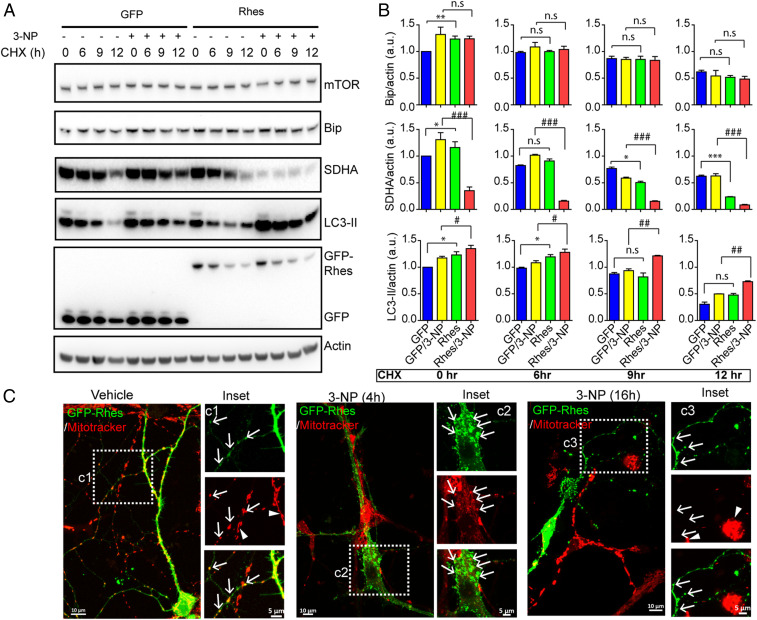
To investigate the mechanisms, we assessed the effect of Rhes on mitophagy in striatal neuronal cell line or primary striatal neurons using biochemical and confocal imaging approaches. We used cycloheximide chase experiment to assess the mitophagy, and Rhes markedly enhances mitophagy. As shown in Fig. 3 A and B, in GFP alone, there was a gradual loss of SDHA, in both vehicle and 3-NP conditions at 6, 9, and 12 h after CHX treatment. In the presence of GFP-Rhes, mTOR and Bip protein levels are not affected, but loss of SDHA is more rapid in vehicle and almost all (>90%) of SDHA was eliminated in 3-NP, which is also accompanied by enhanced LC3-II production (Fig. 3 A and B). Next, we transiently expressed GFP, GFP-Rhes WT, and GFP-Rhes C263S (mutant defective in membrane binding) in striatal neuronal cells by staining them with mitotracker and exposing them to vehicle or 3-NP (SI Appendix, Fig. S5). As expected, in vehicle-treated neuronal cells, we found that GFP-Rhes WT interacted with mitochondria (SI Appendix, Fig. S5, Inset a4-2.5D, arrow) and that mitotracker staining was observed throughout the GFP-Rhes WT−transfected (yellow arrowhead) and untransfected (blue arrowhead) neuronal cells (SI Appendix, Fig. S5, Inset a3). Contrary to this observation, in 3-NP−treated neuronal cells, the dispersed mitotracker staining was completely disrupted in the GFP-Rhes−transfected cells (SI Appendix, Fig.S5, Inset a9, yellow arrowhead) but not in untransfected neuronal cells (SI Appendix, Fig. S5, Inset a9, blue arrowhead). Also, in the presence of 3-NP, numerous globular mitochondria that were positive for Rhes were observed in GFP-Rhes−transfected cells (SI Appendix, Fig. S5, Inset a10-2.5D, arrow). Consistent with our previous work (28, 30, 45), we found that mutation of Rhes C263, which is a farnesylation consensus site, leads to its mislocalization into the nucleus (SI Appendix, Fig. S5, Insets a5 and a11). Note that, in GFP-alone− or GFP-Rhes C263S-transfected cells, the mitotracker signal appears undisrupted, and no obvious association of GFP or GFP-Rhes C263S with mitochondria was observed in either vehicle (SI Appendix, Fig. S5, Insets a2-2.5D and a6-2.5D, arrowhead) or 3-NP−treated cells (SI Appendix, Fig. S5, Insets a8-2.5D and a12-2.5D, arrowhead). We further analyzed the mitotracker intensity by flow cytometry in striatal neuronal cells, infected with either Ad-null or Ad-Rhes, treated with vehicle or 3-NP. We found that mitotracker intensity was reduced by 30% in Ad-Rhes−infected cells compared to Ad-null infection after 3-NP treatment (SI Appendix, Fig. S6), indicating Rhes robustly diminishes mitochondrial intensity upon 3-NP treatment. Analogous to striatal cell lines, in primary striatal neurons, we also found that GFP-Rhes interacted with mitochondria (Fig. 3 C, Inset c1, arrow) and that mitotracker staining was observed throughout the GFP-Rhes WT-transfected and untransfected primary neuronal processes (Fig. 3 C, Inset c1, arrowhead). However, in 3-NP−treated primary neurons at 4 h, the mitotracker staining was reduced and globular mitochondria were surrounded by GFP-Rhes (Fig. 3 C, Inset c2, arrows). At a later time point of 3-NP treatment (16 h), mitotracker staining was almost completely abolished in the GFP-Rhes−transfected cells (Fig. 3 C, Inset c3, arrow) but not in untransfected neuronal cells (Fig. 3 C, Inset c3, arrowhead), indicating that Rhes diminishes mitochondrial functions in a time-dependent manner in the presence of 3-NP. Note that GFP-alone (control)−transfected primary neuron does not show reduction in mitotracker staining after 3-NP treatment (SI Appendix, Fig. S7). Together, these data indicate that Rhes exacerbates mitochondrial dysfunction and promotes mitophagy in the presence of SDHA inhibitor 3-NP.
Fig. 3.
Rhes promotes mitophagy in the presence of SDH inhibitor 3-NP. (A) Western blots of striatal neuronal cells transfected with GFP or GFP-Rhes, treated with vehicle or 3-NP (10 mM for 1 h) or with CHX (100 µM). (B) Bar graph shows the quantification of indicated proteins in A (*P < 0.05, **P < 0.01, ***P < 0.001, between GFP/vehicle and GFP-Rhes/Vehicle; #P < 0.05, ##P < 0.01, ###P < 0.001 between GFP/3-NP and GFP-Rhes/3-NP; 1-way ANOVA test followed by Tukey post hoc test; n = 3; data are mean ± SEM; n.s, not significant). (C) Representative confocal image and corresponding Insets of GFP-Rhes−transfected primary striatal neuron treated with vehicle or 3-NP (10 mM) at indicated time points. Cells were stained with mitotracker orange (red). Arrow indicates GFP-Rhes−positive neuronal processes colocalized with mitochondria in vehicle, and 4-h 3-NP–treated neuron but not in 16-h 3-NP–treated neuron. Arrowheads indicate that the mitotracker intensity is not affected in untransfected neighboring cell in the same field in either vehicle or 3-NP treatment.
Rhes Promotes Mitophagy Flux in the Presence of 3-NP.
Next, we biochemically estimated whether Rhes alters the mitophagy flux in striatal neuronal cell lines in gradient fractionation (SI Appendix, Fig. S8A) and in total lysate (SI Appendix, Fig. S8 B–E). In Ad-null condition (control), as expected, we found an accumulation of SDHA in the mitochondrial fraction in the vehicle-treated groups (SI Appendix, Fig. S8A). Addition of 3-NP resulted in a robust accumulation of SDHA in the lysosomal fractions (compare green circles). Blocking autophagy with chloroquine (CQ), however, resulted in a diminished accumulation of SDHA and enhanced LC3-II levels in control, indicating that 3-NP might induce new mitochondrial production, in control condition. In Ad-Rhes, however, higher levels of SDHA were found in vehicle-treated conditions. Treatment of 3-NP in Ad-Rhes condition resulted in a reduction of SDHA levels in the lysosomal fractions (compare red circles). Cotreatment with CQ showed increased SDHA levels in lysosomal fraction, which is also accompanied by a rapid up-regulation of LC3 conversion (SI Appendix, Fig. S8A).
Apart from mitophagy, we also checked mitobiogenesis by investigating expression level of genes such as Nrf1, Nrf2, Pgc1α, and Tfam involved in the mitochondrial biogenesis in GFP-containing and GFP-Rhes−containing cells treated with 3-NP alone or in combination with CQ. We found that 3-NP treatment up-regulated the expression of mitobiogenesis regulatory genes, but the combination of 3-NP and CQ diminished their expression in both control and Rhes-containing cells (SI Appendix, Fig. S9). This indicates that the lysosomal activity is necessary for biogenesis in 3-NP−treated cells. Mitochondrial degradation and biosynthesis are tightly regulated processes (46). Thus, the reduced SDHA levels in 3-NP + Rhes are presumably due to higher degradation of mitochondria compared to biogenesis. This notion is consistent with enhanced abundance of SDHA in GFP-Rhes + 3-NP + CQ, compared to GFP + 3-NP + CQ (SI Appendix, Fig. S8A). Together, biochemical fractionation data suggest that Rhes up-regulates mitophagy flux in 3-NP condition.
In total lysate, in both vehicle-treated GFP-expressing and GFP-Rhes−expressing cells, we found there was no major difference in SDHA levels (SI Appendix, Fig. S8 B and C). Upon 3-NP treatment, however, there were increased SDHA levels in GFP-expressing cells, but, in GFP-Rhes−expressing cells, the SDHA levels were diminished, which is also accompanied by a rapid up-regulation of autophagy, as measured by increased LC3 conversion (SI Appendix, Fig. S8 B and C). Addition of CQ has further increased the SDHA and LC3-II levels in 3-NP condition (SI Appendix, Fig. S8 D and E). Together, these data indicate that Rhes increases autophagosome and lysosome accumulation around mitochondria and mediates mitophagy flux in the presence of 3-NP. Similarly, in immunocytochemistry, SDHA staining is also diminished in GFP-Rhes−transfected striatal neuronal cells, treated with 3-NP compared to vehicle treatment (SI Appendix, Fig. S10). Next, we investigated whether Rhes recruits lysosomes to globular mitochondria using lysotracker and GFP-LC3, by confocal microscopy. As predicted, we observed colocalization of lysotracker with GFP-Rhes−positive globular mitochondria (stained with mitotracker) in vehicle-treated cells, and this colocalization was further enhanced in the presence of 3-NP (SI Appendix, Fig. S11 A and B, arrow). The 3-NP−treated, GFP-alone−transfected cells also showed increase in colocalization between mitochondria and lysosomes, but only GFP-Rhes−positive cells show disrupted mitotracker staining (SI Appendix, Fig. S11, arrow). Similarly, there was an enhanced number of GFP-LC3 puncta (SI Appendix, Fig. S12 A and B, arrow) and increased colocalization between GFP-LC3 and mCherry TOMM20, a mitochondrial marker, in 3-NP–treated Myc-Rhes–expressing cells (SI Appendix, Fig. S12C). Thus, Rhes associates with mitochondria and lysosomes and activates mitophagy, upon 3-NP treatment.
Rhes Promotes the Formation of Mitophagosomes In Vivo.
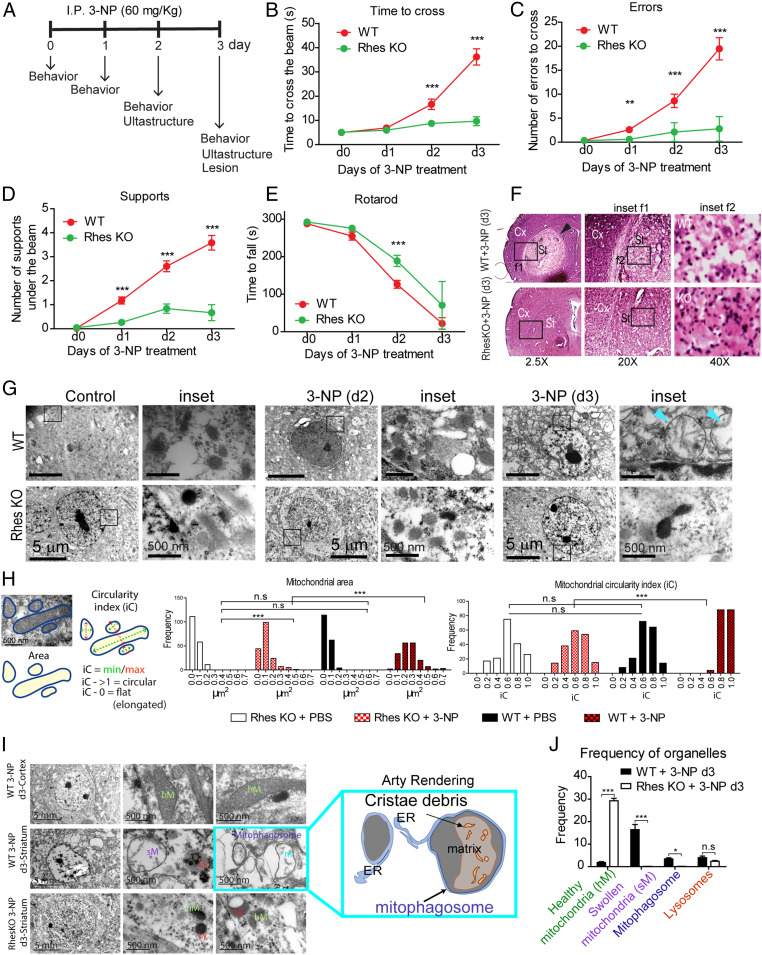
Since Rhes interacted with globular mitochondria and lysosomes (Fig. 1 and SI Appendix, Fig. S1) and modulates basal mitophagy which is further up-regulated in 3-NP (Figs. 2 and 3), we posited that Rhes may activate mitophagy in vivo. To test this thoroughly in vivo, we carried out behavioral, pathological, and ultrastructural analysis (using transmission electron microscopy) of the striatum in WT and Rhes KO mice after systemic injection of 3-NP (see experimental scheme, Fig. 4A). Consistent with our earlier report (39), 3-NP administration to WT mice promoted motor abnormalities that were diminished in Rhes KO mice (Fig. 4 B–E). The 3-NP also induced striatal-specific lesion in WT striatum, which was completely prevented in the Rhes KO striatum (Fig. 4F, arrowhead) (39). Microscopic analysis of the 3-NP−induced striatal-specific lesions found pyknotic nuclei and a diminished neuronal staining in WT mice, while KO striatum displayed healthy nuclei and normal neural staining (Fig. 4F, Insets f1 and f2, compare WT and KO) (39). Thus, 3-NP promotes lesions selectively in the striatum of the WT mice but not in the Rhes KO striatum, a highly reproducible phenotype (39). To address why Rhes KO are resistant to 3-NP−induced lesion, we hypothesized that ultrastructural analysis might provide a clue. We carried out time-lapse ultrastructural analysis using serial corticostriatal section electron microscopy of the second and third days after systemic administration of 3-NP. On the second day (d2), we found no gross changes in the overall cell morphology or mitochondria ultrastructure between WT and KO, which were similar to control (Fig. 4 G, Insets). On the third day (d3), in WT, we found neuronal shrinkage and abnormal, swollen mitochondria with broken inner membrane and matrix in the striatum (Fig. 4 G, Insets, arrowhead). And, those abnormal striatal neuronal changes were completely absent in the Rhes KO (Fig. 4G). Mitochondrial circularity index (length/breadth) and area analysis showed that the frequency of circular mitochondria is high in Rhes WT compared to Rhes KO neuron after 3-NP treatment (Fig. 4H). Cortex from either genotype showed no lesion (SI Appendix, Fig. S13, arrowhead) or mitochondrial abnormalities (SI Appendix, Fig. S13, Insets a1 and b1), indicating that 3-NP−induced lesion (arrow) and mitochondrial abnormalities (SI Appendix, Fig. S13, Inset b2) are selective to the striatum, and require Rhes. Further examination of ultrastructure revealed numerous mitophagosomes, where the mitochondria with cristae debris were engulfed within the double-layer membranes resembling autophagosomes (Fig. 4I, and arty rendering), and there was a diminished frequency of the number of healthy looking mitochondria, but enhanced swollen mitochondria and mitophagosomes in Rhes WT striatum, compared to Rhes KO (Fig. 4J). Thus, Rhes deletion prevents the formation of neuronal shrinkage, mitochondrial swelling, and induction of mitophagosomes by 3-NP treatment. All together, these data indicated that Rhes physiologically regulates 3-NP−induced mitochondrial dysfunction and promotes the formation of globular mitochondria and mitophagosomes, which coincides with worsening motor phenotype in vivo.
Fig. 4.
Rhes promotes mitophagy in vivo. (A) Experimental design for 3-NP treatment in mice (I.P.: intraperitoneal injection). (B–D) Beam walk analysis during 3-NP treatment for WT (red circles) and Rhes-KO (green circles) groups. (E) Changes in the latency to fall for the rotarod test during 3-NP treatment. **P < 0.01, ***P < 0.001; 2-way ANOVA followed by Bonferroni’s post hoc test. (F) Representative hematoxylin/eosin-stained sections for WT and Rhes KO mice after 3 d of 3-NP treatment; Cx, cortex; St, striatum. Insets f1 and f2 show the magnified region from the selected area (arrowhead shows the 3-NP striatal lesion). (G) Representative electron micrographs from striatum of WT and Rhes KO mice at day 2 (d2) and day 3 (d3) of 3-NP treatment; Insets are high magnification (blue arrowheads show mitochondria with broken inner membrane). (H) Ultrastructural changes analysis using frequency histograms of the mitochondrial area and circularity index (iC) for the WT and Rhes KO groups treated with vehicle (PBS) or 3-NP. One-way ANOVA (Bartlett’s test), Bonferroni’s post hoc test. ***P < 0.001; n.s, not significant. (I) Representative electron micrographs from striatum and cortex of indicated groups after 3 d of 3-NP treatment. Arty Rendering shows the swollen mitochondria in mitophagosome. (J) Quantification of the healthy and swollen mitochondria, lysosomes, and mitophagosomes frequency in the striatum of WT and Rhes KO mice at day 3 of 3-NP treatment. *P < 0.05, ***P < 0.001, n.s: not significant, determined by 2-way ANOVA, Bonferroni’s post hoc test.
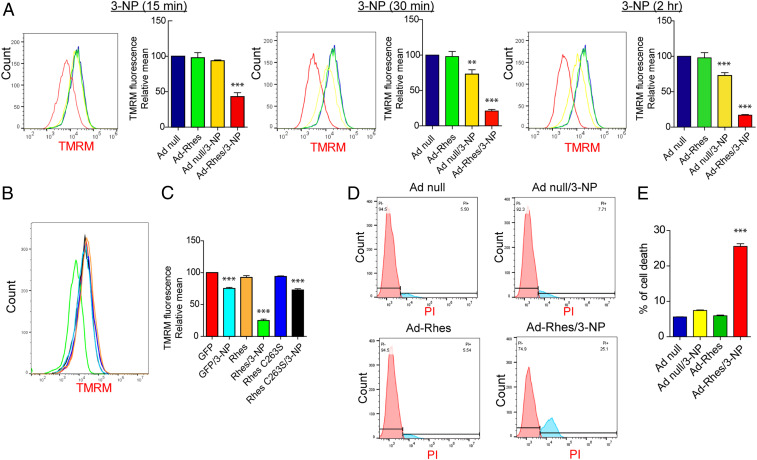
Rhes Diminishes Mitochondrial Potential (ΔΨm) and Promotes Cell Death in the Presence of 3-NP.
To understand the mechanisms, and because Rhes can interact with VDAC1 (Fig. 1A), which is a component of membrane permeability pore, we rationalized that Rhes may affect mitochondrial potential (ΔΨm). To investigate this, we tested whether Rhes alters ΔΨm using tetramethyl rhodamine (TMRM), a widely used cell-permeable fluorescent dye that binds mitochondria with intact ΔΨm. We infected striatal neuronal cell lines with Ad-null or Ad-Rhes for ∼32 h and added TMRM. We quantified TMRM signal intensity using FACS (fluorescence-activated cell sorting) in vehicle or 3-NP treatment at 15 min, 30 min, and 2 h (Fig. 5A). Notably, TMRM (red) signals were not altered in vehicle condition in both Ad-null− and Ad-Rhes−expressing cells. But, in the presence of 3-NP, the TMRM signal intensity is rapidly diminished only in Ad-Rhes−expressing cells but not Ad-null in a time-dependent manner, indicating a rapid loss of ΔΨm by Rhes (Fig. 5A). This effect is specific to Rhes WT, as GFP-Rhes C263S mutant failed to alter TMRM signal, which is like GFP control (Fig. 5 B and C). Next, as Rhes is required for 3-NP induced striatal lesion in vivo (ref. 39 and Fig. 4), we investigated whether Rhes modulates striatal cell death in vitro, using propidium iodide (PI) staining, a fluorescent dye that enters cells only when they are compromised (47), and FAC sorting. As shown in Fig. 5D, we found that Ad-null control expressing cells did not show changes in PI+ cells in either vehicle or 3-NP condition (∼5 to 7%). Similarly, Ad-Rhes−expressing cells in vehicle did not show changes in PI+ cells (∼5%). However, in Ad-Rhes and 3-NP condition, there was a robust increase in PI+ cells (∼25%; Fig. 5E). To further establish that cell death effect is specific to Rhes WT, we FAC-sorted GFP alone, GFP-Rhes WT, and GFP-Rhes C263S cells and added vehicle or 3-NP (see scheme in SI Appendix, Fig. S14A). As shown in SI Appendix, Fig. S14 B and C, Rhes WT showed ∼27% cell death as measured by PI staining in the presence of 3-NP, whereas GFP or GFP-Rhes C263S showed ∼5 to 10% cell death. Collectively, these data indicate Rhes diminishes ΔΨm and promotes cell death in the presence of 3-NP.
Fig. 5.
Rhes diminishes mitochondrial membrane potential (ΔΨm) and promotes cell death in the presence of 3-NP. (A) Representative flow cytometry plot of striatal neuronal cells infected with Ad-null or Ad-Rhes in the presence of vehicle or 3-NP (10 mM, for 15 min, 30 min, or 2 h). Cells were stained for TMRM (ΔΨm indicator) and analyzed in flow cytometry. Bar graph shows the relative TMRM mean intensity. **P < 0.01, ***P < 0.001 vs. Ad-null and vehicle (n = 3 per group; data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test). (B) Flow cytometry plot of striatal neuronal cells transfected with GFP or GFP-Rhes or GFP-Rhes C263S treated with vehicle or 3-NP (10 mM and for 2 h) and stained for TMRM. (C) Bar graph shows the relative TMRM mean intensity for B. ***P < 0.001 vs. GFP/Vehicle sample (n = 3 per group; data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test). (D) Flow cytometry plot of striatal neuronal cells infected with Ad-null or Ad-Rhes treated with vehicle or 3-NP (10 mM for 8 h), stained with PI. (E) Bar graph shows the percent of cell death in indicated groups. ***P < 0.001 vs. Ad-null/Vehicle (n = 3 per group; data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test).
Rhes Promotes Mitophagy, Disrupts ΔΨm, and Promotes Cell Death via Nix.
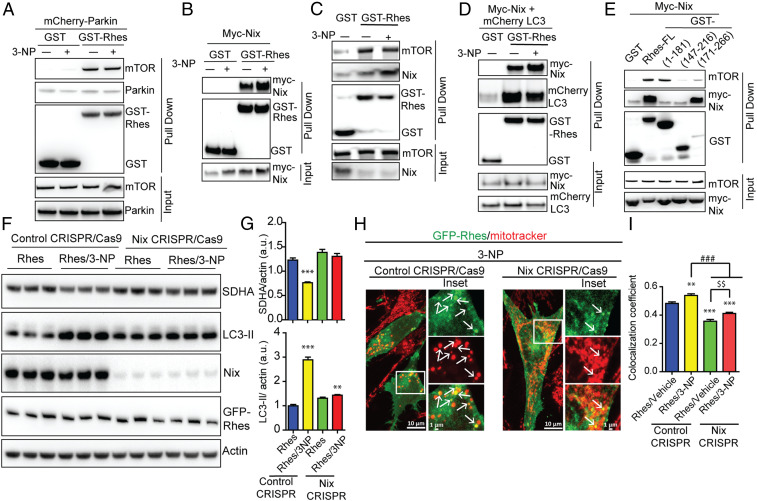
To investigate the mechanisms by which Rhes may activate mitophagy, we considered whether Rhes interacts with well-known mitophagy modulators, such as Parkin, PINK1, DRP1 (Dynamin related protein 1), or Nix (48). In an affinity purification experiment, GST-Rhes did not interact with Parkin but readily interacted with mTOR (Fig. 6A), as shown in our previous report (28). We did not observe interaction of Rhes with either DRP1 or Pink1 (SI Appendix, Fig. S15 A and B). However, GST-Rhes readily interacted with Nix, a known mitophagy receptor (49) (Fig. 6 B and C). Rhes interaction with overexpressed Nix (Fig. 6B) or endogenous Nix (Fig. 6C), increases with 3-NP treatment; however, Rhes interaction with mTOR remains unaltered (Fig. 6C). GST-Rhes is also affinity-purified with LC3 without or with 3-NP and showed a consistent and enhanced interaction with Nix in the presence of 3-NP (Fig. 6D). Next, we found that the C-terminal SUMO E3 ligase domain (171 to 266 aa), but not the N-terminal GTPase (1 to 181 aa), or the fragment (147 to 216 aa) interacted strongly with Nix (Fig. 6E). Additionally, we found that membrane-binding defective mutant Rhes C263S, but not GTP-binding mutant (Rhes S33N), fails to interact with Nix (SI Appendix, Fig. S15C). Interaction of Rhes with mTOR, on the other hand, seems to be more toward the N-terminal side of Rhes (Fig. 6E). Thus, Rhes associates strongly with Nix in the presence of 3-NP, and the binding requires intact membrane binding domain.
Fig. 6.
Rhes interacts with Nix and promotes mitophagy. (A–E) Representative Western blot of indicated proteins after glutathione affinity purification of HEK 293 cells transfected with GST or GST-Rhes (full length [FL] or fragments), mCherry-Parkin, myc-Nix, or mCherry-LC3 plasmids that are exposed to vehicle or 3-NP (10 mM, 2 h) wherever indicated. Input is 5% of total lysate. (F) Representative Western blot of control or CRISPR/Cas9-mediated Nix-depleted striatal neuronal cells that were transfected with GFP or GFP-Rhes and treated with vehicle or 3-NP (10 mM, 2 h). (G) Bar graph shows the quantification for normalized SDHA and LC3-II proteins. **P < 0.01, ***P < 0.001 vs. Rhes control CRISPR/vehicle (n = 4, data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test). (H) Representative confocal images and their Insets of control CRISPR/Cas9 or Nix CRISPR/Cas9 striatal neuronal cells transfected with GFP-Rhes and costained for mitotracker (red) that were treated with 3-NP (10 mM for 2 h). Arrows represents the globular mitochondria, positive for GFP-Rhes. (I) Bar graph shows the average of Pearson’s coefficient of colocalization between GFP-Rhes and mitochondria in indicated groups (n = 33 to 37 cells per group; **P < 0.01, ***P < 0.001 vs. Rhes/vehicle control CRISPR; ###P < 0.001, $$P < 0.01 between indicated groups; data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test).
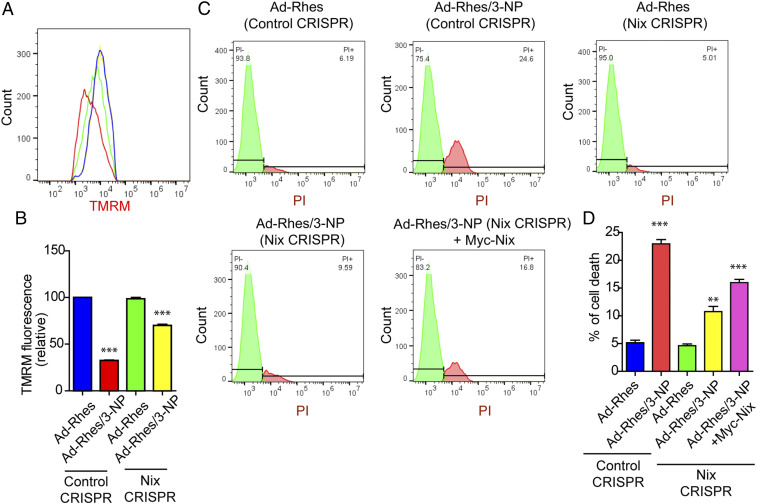
Because Rhes interacted strongly with Nix, we hypothesized that Rhes may promote mitophagy via Nix. To test this hypothesis, we depleted endogenous Nix in the striatal cells using CRISPR/Cas9 tools. While GFP-Rhes promoted a significant loss of SDHA and increased LC3-II productions in CRISPR/Cas9 control cells, it completely failed to do so in CRISPR/Cas9 Nix-depleted cells (Fig. 6 F and G), which resulted in ∼90% depletion of Nix (SI Appendix, Fig. S19A). This finding indicated that Nix is essential for Rhes-induced mitophagy. Furthermore, when we reconstituted Nix in Nix-depleted cells, Rhes was able to restore mitophagy and increase LC-3 conversion (SI Appendix, Fig. S16 A and B), suggesting that Nix is critical for Rhes-mediated mitophagy. Consistent with this, in confocal data, we found that, in CRISPR/Cas9 control cells, upon 3-NP treatment, Rhes promoted rapid loss of mitotracker intensity, and formation of numerous globular mitochondria colocalized with GFP-Rhes (Fig. 6H, arrow). However, in Nix-depleted GFP-Rhes expressing cells, most of the mitotracker signal intensity remained unaltered by 3-NP treatment and colocalization between GFP-Rhes and mitotracker was significantly reduced (Fig. 6 H, arrows and I). In vehicle treatment, Rhes was unable to interact with globular mitochondria in Nix-depleted cells compared to control cells (SI Appendix, Fig. S16C, arrowhead). Together, these data suggest that Rhes interacts with globular mitochondria and promotes mitophagy via Nix.
Next, as Rhes decreases ΔΨm in the presence of 3-NP, we wondered whether this process requires Nix. As shown in Fig. 7 A and B, while Rhes diminishes TMRM signals in control cells, this effect is not observed in Nix-depleted cells. Similarly, the observed Rhes-induced cell death in the presence of 3-NP in control cells was also markedly diminished in Nix-depleted cells (Fig. 7 C and D). Moreover, reconstitution of Nix significantly elevated Rhes-mediated cell death in 3-NP–treated cells (Fig. 7D). Nix is localized to endoplasmic reticulum (ER) as well as mitochondria (50). Using ER-targeted Nix, where transmembrane (TM) domain of Nix was replaced by CytoB, and mitochondria-targeted Nix, where TM domain was replaced with monoamine oxidase located in the outer mitochondrial membrane (50), we found Rhes affinity with both ER- and mitochondrial-targeted Nix (SI Appendix, Fig. S17 A and B). However, in a reconstitution FACS/TMRM experiment, we found that replenishing only the mitochondria-targeted Nix, but not ER-targeted Nix, restores the loss of TMRM signal by Rhes (SI Appendix, Fig. S17 C and D). Collectively, these data indicate that Rhes interacts with both ER- and mitochondrial-targeted Nix and disrupts ΔΨm mostly via the latter.
Fig. 7.
Rhes promotes cell death and disrupts mitochondrial potential (ΔΨm) via Nix. (A) Flow cytometry plots of control CRISPR/Cas9 and or Nix CRISPR/Cas9 striatal neuronal cells, infected with either Ad-null or Ad-Rhes viral particles. (B) Bar graph shows the relative TMRM fluorescence intensity for A. ***P < 0.001 (n = 3 per group; data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test). (C) Flow cytometry plot of control CRISPR/Cas9 and or Nix CRISPR/Cas9 striatal neuronal cells infected with Ad-null or Ad-Rhes treated with vehicle or 3-NP (10 mM for 8 h) that are stained with PI. Nix CRISPR/Cas9 striatal neuronal cells reconstituted with Nix by transfecting myc-Nix cDNA. (D) Bar graph shows the percent of cell death in indicated groups (**P < 0.01, ***P < 0.001; n = 3 per group; data are mean ± SEM; 1-way ANOVA followed by Tukey post hoc test).
Rhes Travels from Cell to Cell and Interacts with the Globular Mitochondria in the Neighboring Cells via Nix.
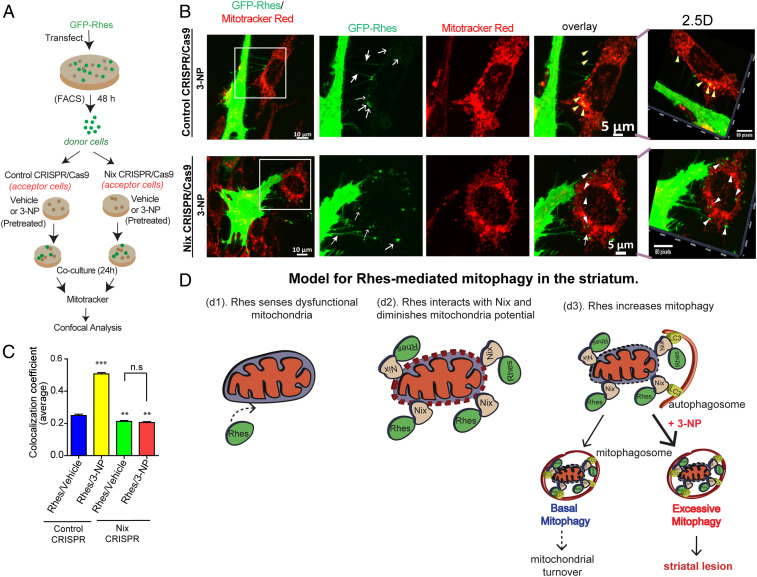
We recently reported that Rhes induces the biogenesis of tunneling nanotube (TNT)-like cellular protrusion, “Rhes tunnel,” through which Rhes travels from cell to cell (45). It is unknown whether intercellular signaling can regulate mitochondrial turnover. So, we considered whether Rhes can interact with damaged mitochondria in the neighboring cells. We designed an experiment in which we cocultured FACS-sorted GFP-Rhes−expressing cells (donor cells) with the acceptor cells that were exposed to 3-NP or vehicle (acceptor cells), followed by confocal imaging with mitotracker (see scheme in Fig. 8A). As expected, we found GFP-Rhes−positive TNT-like protrusions (closed arrow) and numerous GFP-Rhes (open arrow) in the neighboring cell in both vehicle-treated and 3-NP−treated cells (Fig. 8B and SI Appendix, Fig. S18). Interestingly, we found numerous GFP-Rhes puncta that were colocalized with globular mitochondria in cells that are exposed to 3-NP (yellow arrowhead) in control acceptor cell. But, when GFP-Rhes donor cells are cocultured with Nix-depleted acceptor cells, we do observe TNT-like protrusions (closed arrow) and numerous puncta (open arrow), but we failed to observe GFP puncta (white arrowhead) localization with globular mitochondria in the Nix-depleted acceptor cell treated with 3-NP (Fig. 8B). Pearson’s colocalization coefficient further confirmed a marked reduction in Rhes localization with mitochondria in Nix-depleted cells (Fig. 8C). This indicates that Rhes can travel from cell to cell and interact with damaged mitochondria via Nix. Thus, these data raise a possibility that Rhes can act as an intercellular mitochondrial surveillance factor.
Fig. 8.
Rhes travels Intercellularly and interacts with damaged mitochondria via Nix. (A) Experimental design for B. (B) Representative confocal image of GFP-Rhes (FAC-sorted) striatal neuronal cells (donor cells) cocultured with vehicle-treated or 3-NP−treated control or Nix-depleted (Nix CRISPR) cells (acceptor cells). Insets show the magnified region from selected area. Corresponding Insets were processed for 2.5-dimensional rendering. Closed arrow indicates Rhes-induced TNT-like protrusions. Open arrow represents GFP-Rhes puncta in acceptor cell. Yellow and white arrowheads indicate presence or lack, respectively, of colocalization of GFP-Rhes with mitotracker. (C) Bar graph shows average of Pearson’s coefficient of colocalization between GFP puncta and mitotracker in neighboring acceptor cells where Rhes is transported from donor cell; n = 72 to 90 GFP-Rhes puncta per group were counted from 13 to 18 cells per group. **P < 0.01, ***P < 0.001; 1-way ANOVA followed by Tukey post hoc test; n.s, not significant. (D) Model for Rhes-mediated mitophagy in the striatum. Rhes senses dysfunctional mitochondria (d1). Rhes interacts with Nix and diminishes membrane potential (d2). Rhes-Nix interaction recruits autophagosomes via LC3 and formation of mitophagosomes to promote mitochondrial degradation (d3). Basal mitophagy may lead to mitochondrial turnover; however, excessive mitophagy in the presence of mitochondrial toxin, 3-NP, may promote striatal lesion.
Discussion
To date, the brain tissue-specific regulator(s) of mitophagy or its role in neuronal vulnerability remains poorly understood. By using neuronal and mouse model combined with electron and live-cell confocal microscopy, we have systematically investigated the role of Rhes in mitophagy. We found striatal-enriched protein Rhes promotes mitophagy via Nix, a known mitophagy receptor (49). Despite abundant and comparable expression of Nix in the striatum of both WT and Rhes KO mice (SI Appendix, Fig. S19B), the removal of damaged mitochondria via mitophagy requires Rhes demonstrating its “mitophagy ligand-like” capabilities in vivo. In addition, this study demonstrate the role of Rhes in intercellular surveillance of mitochondria through the cellular protrusion that we recently discovered (45) and lays a foundation for understanding the unprecedented complexity by which Rhes may signal mitophagy within and outside the striatum.
Half-lives of mitochondria in rat whole brain were estimated to be ∼24 d (51). Striatum is considered to be one of the metabolically active regions of the brain (52); the exact half-life of mitochondria remains unknown. Although Rhes appears to regulate basal mitophagy, the functional relevance of this has yet to be determined. Rhes KO mice are hyperactive to dopaminergic drugs, but whether the lack of mitophagy may contribute to such hyperactive phenotype remains largely unknown. It has long been known that 3-NP elicits striatal-specific lesion (53, 54), and mechanisms such as oxidative stress and excitotoxicity were implicated (55, 56). Oxidative stress or excitotoxicity can also occur in the cortex; however, 3-NP does not elicit lesion in the cortex (SI Appendix, Fig. S13). Therefore, the molecular details that contribute to 3-NP−induced striatal lesion has remained enigmatic. Now this study provides a clear molecular route for striatal lesion by 3-NP. Our data indicate that Rhes’s role is to protect neurons by removing damaged mitochondria via mitophagy. However, upon exposure to mitochondrial toxin such as 3-NP, which irreversibly damages the mitochondria, the Rhes-mediated mitophagy processes are exacerbated that lead to depletion of the mitochondria and neuronal death (Fig. 8D). Because Rhes selectively mediates 3-NP−induced toxicity, it raises the question of whether Rhes contains special mechanisms to sense SDHA dysfunction in the brain.
Based on the data presented in this report, our current model predicts that Rhes may sense the dysfunctional mitochondria and binds to Nix to disrupt the ΔΨm and initiate mitophagy. Rhes and Nix together may alter the membrane permeability pore, leading to the diminishment of ΔΨm. Nix alone cannot disrupt ΔΨm, because there was no difference in TMRM signal intensity between Nix WT and Nix KO cells with or without 3-NP (Fig. 7). Moreover, as mentioned above, Nix alone cannot elicit mitophagy in vivo because the levels of Nix between WT and Rhes KO striatum were similar (SI Appendix, Fig. S19B), and yet Rhes KO shows diminished mitophagy compared to WT (Fig. 4). Mechanistically, as loss of ΔΨm is a major initial step in the mitophagy up-regulation, we hypothesize that altering the ΔΨm by Rhes is the likely step to initiate mitophagy. Rhes’s effect on mitophagy appears to be specific to 3-NP but not FCCP (Fig. 2 and SI Appendix, Fig. S3 E and F). How is this specificity accomplished? One possibility would be that Rhes, via yet unknown mechanisms, may particularly react to dysfunctional complex II of mitochondria. Analogous to this, some factors in the substantia nigra may sense MPTP- or rotenone- induced mitochondrial dysfunction related to complex I. Such tissue-specific regulator may set the stage for eliciting tissue-specific vulnerability through mitophagy. But the identity of such regulators in the substantia nigra remains unknown.
An earlier study showed that mitochondria from the axonal protrusions could be degraded in astrocytes via mitophagy, supporting the existence of intercellular mitophagy mechanisms in the brain (57). The implication that Rhes can travel from cell to cell and interacts with damaged mitochondria thus raises the intriguing possibility that Rhes may act like “mitochondrial surveillant” in the striatum. We imagine that Rhes “scans” for damaged mitochondria in the surrounding cells and travels there via TNT-like membranous tubes to eliminates them. Conceptually, such a process in brain sounds like “science fiction.” Considering the complexity of brain, the surveillance ability of Rhes to monitor defective mitochondria may be needed for an effective coordination of billions of densely packed neurons in the brain. Collectively, this study reveals Rhes as a mediator of mitophagy and its impact on the regulation of striatal vulnerability. Development of therapeutic approaches that modulates mitophagy in the striatum may offer benefits for dysfunction related to the striatal region of the brain.
Materials and Methods
Cell Culture and Chemicals.
Mouse normal striatal neuronal cells (STHdhQ7/Q7) (42) were cultured in growth medium containing Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) with 10% fetal bovine serum, 1% penicillin−streptomycin, as described in our previous work (30, 40, 58).
Generation of Nix KO Cell Line.
Nix KO cell line was generated using Nix CRISPR/Cas9 plasmids from Santa Cruz Biotechnologies. First, we transfected the striatal neuronal cells with Nix CRISPR/Cas9 plasmid (SC-419357) or CRISPR/Cas9 control plasmid (SC-418922) in a 10-cm dish. After 48 h, we sorted the cells based on GFP fluorescence and recultured them. We passaged them 2 to 3 times and prepared lysate to confirm the Nix protein depletion by Western blotting using Nix antibody.
Mice.
For in vivo experiments, we used Rhes KO mice, and we used C57BL/6J mice as a control group. Rhes KO mice were obtained from A. Usiello (25) and were backcrossed with C57BL/6J mice at least 8 generations; homozygous Rhes KO were used for all of the experiments. WT mice (C57BL/6) were obtained from Jackson Laboratory and maintained in our animal facility according to Institutional Animal Care and Use Committee at The Scripps Research Institute. The 3-NP injections were based on previous studies (14, 39). Briefly, 3-NP was dissolved in sterile phosphate-buffered saline (PBS; 0.1 M, 10 mg/mL), and pH was adjusted to pH 7.4 using sodium hydroxide. Mice received intraperitoneal injections twice a day (60 mg/kg doses), with 2 h between injections for 3 consecutive days. In this study, we used female and male mice; all of them were between 16 wk and 20 wk of age.
Supplementary Material
Acknowledgments
We thank Dr. Long Yan of Max Plank Institute of Neuroscience, Jupiter, FL, for imaging help; and Alta Johnson and Bivian Torres of Flow Cytometry Core, Scripps Research, Jupiter, FL, for help in cell sorting and data analysis. We thank Melissa Benilous for administrative support, and Sumitha Rajendra Rao and Stephen Zorc for technical assistance. We thank Rodolfo Paredes for electron microscopic technical assistance. This part of the work was supported by Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (Project IN206719). This research was partially supported by a training grant in Alzheimer’s Drug Discovery from the Lottie French Lewis Fund of the Community Foundation for Palm Beach and Martin Counties. This research was supported by funding from NIH/National Institute of Neurological Disorders and Stroke grant R01-NS087019-01A1, NIH/National Institute of Neurological Disorders and Stroke grant R01-NS094577-01A1, and grants from Cure Huntington Disease Initiative (CHDI) Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912868116/-/DCSupplemental.
Change History
October 22, 2020: This article has been updated to correct an author name.
References
- 1.Barodia S. K., Creed R. B., Goldberg M. S., Parkin and PINK1 functions in oxidative stress and neurodegeneration. Brain Res. Bull. 133, 51–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X., et al. , LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet. 21, 1931–1944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vives-Bauza C., et al. , PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., et al. , Altered brain energetics induces mitochondrial fission arrest in Alzheimer’s disease. Sci. Rep. 6, 18725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins B. G., Koroshetz W. J., Beal M. F., Rosen B. R., Evidence for impairment of energy metabolism in vivo in Huntington’s disease using localized 1H NMR spectroscopy. Neurology 43, 2689–2695 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Panov A. V., et al. , Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat. Neurosci. 5, 731–736 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Browne S. E., et al. , Oxidative damage and metabolic dysfunction in Huntington’s disease: Selective vulnerability of the basal ganglia. Ann. Neurol. 41, 646–653 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Zhang L., et al. , Differential effect of amyloid beta peptides on mitochondrial axonal trafficking depends on their state of aggregation and binding to the plasma membrane. Neurobiol. Dis. 114, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan S. D., et al. , An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer’s disease. Nature 389, 689–695 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Mattiazzi M., et al. , Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 277, 29626–29633 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Oliveira J. M., Nature and cause of mitochondrial dysfunction in Huntington’s disease: Focusing on huntingtin and the striatum. J. Neurochem. 114, 1–12 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Shi P., Gal J., Kwinter D. M., Liu X., Zhu H., Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1802, 45–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truban D., Hou X., Caulfield T. R., Fiesel F. C., Springer W., PINK1, parkin, and mitochondrial quality control: What can we learn about Parkinson’s disease pathobiology? J. Parkinsons Dis. 7, 13–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouillet E., Jacquard C., Bizat N., Blum D., 3-Nitropropionic acid: A mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington’s disease. J. Neurochem. 95, 1521–1540 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Brouillet E., et al. , Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. Proc. Natl. Acad. Sci. U.S.A. 92, 7105–7109 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker W. D. Jr, Boyson S. J., Parks J. K., Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann. Neurol. 26, 719–723 (1989). [DOI] [PubMed] [Google Scholar]
- 17.Cannon J. R., et al. , A highly reproducible rotenone model of Parkinson’s disease. Neurobiol. Dis. 34, 279–290 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langston J. W., Ballard P., Tetrud J. W., Irwin I., Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science 219, 979–980 (1983). [DOI] [PubMed] [Google Scholar]
- 19.Huang L. S., et al. , 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 281, 5965–5972 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramsay R. R., et al. , Interaction of 1-methyl-4-phenylpyridinium ion (MPP+) and its analogs with the rotenone/piericidin binding site of NADH dehydrogenase. J. Neurochem. 56, 1184–1190 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Nicklas W. J., Vyas I., Heikkila R. E., Inhibition of NADH-linked oxidation in brain mitochondria by 1-methyl-4-phenyl-pyridine, a metabolite of the neurotoxin, 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. Life Sci. 36, 2503–2508 (1985). [DOI] [PubMed] [Google Scholar]
- 22.Banerjee R., Starkov A. A., Beal M. F., Thomas B., Mitochondrial dysfunction in the limelight of Parkinson’s disease pathogenesis. Biochim. Biophys. Acta 1792, 651–663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas B., Mohanakumar K. P., Melatonin protects against oxidative stress caused by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine in the mouse nigrostriatum. J. Pineal Res. 36, 25–32 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Vargiu P., et al. , The small GTP-binding protein, Rhes, regulates signal transduction from G protein-coupled receptors. Oncogene 23, 559–568 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Errico F., et al. , The GTP-binding protein Rhes modulates dopamine signalling in striatal medium spiny neurons. Mol. Cell. Neurosci. 37, 335–345 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Ghiglieri V., et al. , Rhes influences striatal cAMP/PKA-dependent signaling and synaptic plasticity in a gender-sensitive fashion. Sci. Rep. 5, 10933 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison L. M., He Y., Rhes and AGS1/Dexras1 affect signaling by dopamine D1 receptors through adenylyl cyclase. J. Neurosci. Res. 89, 874–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramaniam S., et al. , Rhes, a striatal-enriched small G protein, mediates mTOR signaling and L-DOPA-induced dyskinesia. Nat. Neurosci. 15, 191–193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramaniam S., et al. , Rhes, a physiologic regulator of sumoylation, enhances cross-sumoylation between the basic sumoylation enzymes E1 and Ubc9. J. Biol. Chem. 285, 20428–20432 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramaniam S., Sixt K. M., Barrow R., Snyder S. H., Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science 324, 1327–1330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramaniam S., Snyder S. H., Huntington’s disease is a disorder of the corpus striatum: Focus on Rhes (Ras homologue enriched in the striatum). Neuropharmacology 60, 1187–1192 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Swarnkar S., et al. , Ectopic expression of the striatal-enriched GTPase Rhes elicits cerebellar degeneration and an ataxia phenotype in Huntington’s disease. Neurobiol. Dis. 82, 66–77 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Okamoto S., et al. , Balance between synaptic versus extrasynaptic NMDA receptor activity influences inclusions and neurotoxicity of mutant huntingtin. Nat. Med. 15, 1407–1413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seredenina T., Gokce O., Luthi-Carter R., Decreased striatal RGS2 expression is neuroprotective in Huntington’s disease (HD) and exemplifies a compensatory aspect of HD-induced gene regulation. PLoS One 6, e22231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baiamonte B. A., Lee F. A., Brewer S. T., Spano D., LaHoste G. J., Attenuation of Rhes activity significantly delays the appearance of behavioral symptoms in a mouse model of Huntington’s disease. PLoS One 8, e53606 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sbodio J. I., Paul B. D., Machamer C. E., Snyder S. H., Golgi protein ACBD3 mediates neurotoxicity associated with Huntington’s disease. Cell Rep. 4, 890–897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu B., Palacino J., A novel human embryonic stem cell-derived Huntington’s disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 27, 1820–1829 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Argenti M., “The role of mitochondrial dysfunction in Huntington’s disease pathogenesis and its relation with striatal rhes protein,” PhD thesis, Università degli Studi di Padova, Padova, Italy (2014).
- 39.Mealer R. G., Subramaniam S., Snyder S. H., Rhes deletion is neuroprotective in the 3-nitropropionic acid model of Huntington’s disease. J. Neurosci. 33, 4206–4210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahani N., et al. , RasGRP1 promotes amphetamine-induced motor behavior through a Rhes interaction network (“Rhesactome”) in the striatum. Sci. Signal. 9, ra111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyant G. A., et al. , NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trettel F., et al. , Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 9, 2799–2809 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Eshraghi M., et al. , Global ribosome profiling reveals that mutant huntingtin stalls ribosomes and represses protein synthesis independent of fragile X mental retardation protein. bioRxiv:10.1101.629667 (7 May 2019).
- 44.Doherty E., Perl A., Measurement of mitochondrial mass by flow cytometry during oxidative stress. React. Oxyg. Species (Apex) 4, 275–283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma M., Subramaniam S., Rhes travels from cell to cell and transports Huntington disease protein via TNT-like protrusion. J. Cell Biol. 218, 1972–1993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ploumi C., Daskalaki I., Tavernarakis N., Mitochondrial biogenesis and clearance: A balancing act. FEBS J. 284, 183–195 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Subramaniam S., et al. , ERK activation promotes neuronal degeneration predominantly through plasma membrane damage and independently of caspase-3. J. Cell Biol. 165, 357–369 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin S. M., Youle R. J., PINK1- and Parkin-mediated mitophagy at a glance. J. Cell Sci. 125, 795–799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novak I., et al. , Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mughal W., et al. , Myocardin regulates mitochondrial calcium homeostasis and prevents permeability transition. Cell Death Differ. 25, 1732–1748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menzies R. A., Gold P. H., The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 246, 2425–2429 (1971). [PubMed] [Google Scholar]
- 52.Brown L. L., et al. , Differential metabolic activity in the striosome and matrix compartments of the rat striatum during natural behaviors. J. Neurosci. 22, 305–314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beal M. F., et al. , Neurochemical and histologic characterization of striatal excitotoxic lesions produced by the mitochondrial toxin 3-nitropropionic acid. J. Neurosci. 13, 4181–4192 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cirillo G., Cirillo M., Panetsos F., Virtuoso A., Papa M., Selective vulnerability of basal ganglia: Insights into the mechanisms of bilateral striatal necrosis. J. Neuropathol. Exp. Neurol. 78, 123–129 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Albin R. L., Greenamyre J. T., Alternative excitotoxic hypotheses. Neurology 42, 733–738 (1992). [DOI] [PubMed] [Google Scholar]
- 56.Beal M. F., Energetics in the pathogenesis of neurodegenerative diseases. Trends Neurosci. 23, 298–304 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Davis C. H., et al. , Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 111, 9633–9638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pryor W. M., et al. , Huntingtin promotes mTORC1 signaling in the pathogenesis of Huntington’s disease. Sci. Signal. 7, ra103 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.