Significance
Recent work implicates tumor neoantigens as the primary target of the antitumor immune response based on the correlates of tumor mutational burden and tumoral CD8+ T cell density. We provide evidence that, despite ubiquitous sharing of clonal and subclonal neoantigens among tumors, the CD8+ response is limited to a small (<10%) subset of antigens and is typically mono-/oligoclonal in nature. Vaccination promotes an increase in neoantigen breath and T cell clonotype diversity, suggesting ineffective neoantigen cross-presentation as the primary mechanism for immunological ignorance. Our findings support the notion that therapeutic vaccination is necessary as an adjunct to checkpoint inhibitor treatment, since most T cell clones specific for melanoma neoantigens are naïve and often below the limit of detection.
Keywords: neoantigen, melanoma, dendritic cells, cancer vaccine, CD8+ T cells
Abstract
The impact of intratumoral heterogeneity (ITH) and the resultant neoantigen landscape on T cell immunity are poorly understood. ITH is a widely recognized feature of solid tumors and poses distinct challenges related to the development of effective therapeutic strategies, including cancer neoantigen vaccines. Here, we performed deep targeted DNA sequencing of multiple metastases from melanoma patients and observed ubiquitous sharing of clonal and subclonal single nucleotide variants (SNVs) encoding putative HLA class I-restricted neoantigen epitopes. However, spontaneous antitumor CD8+ T cell immunity in peripheral blood and tumors was restricted to a few clonal neoantigens featuring an oligo-/monoclonal T cell-receptor (TCR) repertoire. Moreover, in various tumors of the 4 patients examined, no neoantigen-specific TCR clonotypes were identified despite clonal neoantigen expression. Mature dendritic cell (mDC) vaccination with tumor-encoded amino acid-substituted (AAS) peptides revealed diverse neoantigen-specific CD8+ T responses, each composed of multiple TCR clonotypes. Isolation of T cell clones by limiting dilution from tumor-infiltrating lymphocytes (TILs) permitted functional validation regarding neoantigen specificity. Gene transfer of TCRαβ heterodimers specific for clonal neoantigens confirmed correct TCR clonotype assignments based on high-throughput TCRBV CDR3 sequencing. Our findings implicate immunological ignorance of clonal neoantigens as the basis for ineffective T cell immunity to melanoma and support the concept that therapeutic vaccination, as an adjunct to checkpoint inhibitor treatment, is required to increase the breadth and diversity of neoantigen-specific CD8+ T cells.
There is strong evidence that tumor-derived missense mutations create amino acid substituted (AAS) peptides encoding neoantigens (1, 2). Even in high mutational burden (>10 mutations per megabase) malignancies, spontaneous antitumor immunity (in the absence of immune stimulation) in peripheral blood or TILs is often limited to a small fraction of neoantigens (3). Consistent with this observation, studies using experimental cancer models implicate ignorance (rather than anergy or clonal deletion) as the primary mechanism for the absence of spontaneous tumor antigen-specific immunity directed against most malignancies (4–6). Recent developments in high-throughput TCR sequencing have provided new insights into the clonal repertoire directed against tumor neoantigens, particularly in patients with high mutational burden cancers (7–9). Our prior experience suggested that melanoma patients treated with a personalized mature DC (mDC) vaccine developed an increased breadth and diversity of neoantigen-specific T cells (10). The recruitment of new TCR clonotypes resulted in a large population of neoantigen-specific CD8+ T cells that could be detected by p-MHC multimer staining ex vivo or after a 10-d culture period with neoantigen. However, the inability to detect low-frequency TCR clonotypes specific for tumor-encoded neoantigens remains a critical challenge for investigators and hampers the development of personalized immunotherapies such as cancer vaccines.
Intratumoral heterogeneity (ITH) is a widely recognized feature of solid tumors and poses distinct challenges related to the development of effective therapeutic strategies. In the context of cancer immunotherapy, rare tumor subclones that fail to express relevant target antigens are often responsible for immune escape, leading to disease recurrence after systemic treatment (11). The most notable example is the development of CD19-negative leukemia, which accompanies relapse in ∼20% of ALL patients treated with CAR T cell therapy (12). This phenomenon of immune editing with the outgrowth of antigen-loss variants after CAR T cell treatment has also been described in other malignancies such as NHL (13) and GBM (14). Similarly, NSCLC patients with prominent ITH as determined by whole exome sequencing, who typically respond poorly to anti–PD-1 treatment, are indicative of inadequate T cell immunity to low-frequency subclonal neoantigens (15). Here, we sought to investigate the issue of neoantigen heterogeneity within individual patients by predicting neoantigens from exome sequencing data of multiple resected metastases followed by treatment with a personalized tumor peptide mDC vaccine. Subsequent to vaccine administration, we evaluated T cell clonotype diversity changes compared to a baseline prevaccine sample. Downstream evaluation of TCR clonotypes with known specificity for ubiquitous clonal neoantigens was performed by high-coverage targeted DNA sequencing to establish the clonality of individual neoantigen-producing variants in each metastatic sample. We report that, despite ubiquitous expression of melanoma neoantigens, most tumors are bare of TCR clonotypes reactive to these antigens. Our data suggest a state of immunological ignorance in melanoma.
Results
Neoantigen Identification in a High Mutational Burden Melanoma, MEL66.
Patient MEL66, with no prior history of a primary cutaneous melanoma, presented to medical attention with several upper extremity subcutaneous (SQ) lesions. Excisional biopsy of 3 independent SQ lesions (SQ1 to 3) confirmed BRAF V600E-mutated metastatic melanoma, and a fourth independent SQ lesion (SQ4) was collected 3 mo later. Body imaging confirmed stage IV melanoma with interval development of several pulmonary metastases. Ipilimumab (3 mg/kg) was administered, and, 1 wk after the second dose, the patient developed severe (grade 3) colitis and was treated with corticosteroids. After resolution of the colitis, further imaging confirmed disease progression, and single-agent BRAF inhibitor for 2 mo followed by combination BRAF/MEK inhibitors were administered, with transient response for 6 mo. Upon disease progression, the patient underwent surgical resection of an enlarging solitary 2-cm pulmonary metastasis (PM) and a 3-cm soft-tissue retroperitoneal metastasis (RM). Both lesions were histologically confirmed as metastatic melanoma, and tumor-infiltrating lymphocytes (TILs) were isolated for analysis. The patient was rendered disease-free by surgery. Postoperative imaging 2 mo later confirmed no evidence of disease (NED). Repeat CT imaging 2 mo later confirmed NED, and the patient provided written consent to enroll in the clinical trial (NCT00683670) with mature DC vaccination against mutated neoantigens. In total, 6 tumors (all completely resected; SI Appendix, Fig. S1A) were available for combined genomic and TCR repertoire analysis.
Two index tumors (SQ1 and SQ4) were subjected to next-generation whole-exome and transcriptome sequencing. A PBMC-derived DNA extract was also subjected to whole-exome sequencing. The SQ1 sample revealed 3,442 single nucleotide variants (SNVs) with 2,182 missense mutations (MMs), and the SQ4 sample revealed 2,872 SNVs with 1,857 MMs, including a shared variant of uncertain significance (A184V substitution) in DNA polymerase ε (POLE). Taking into consideration the patient’s HLA class I alleles, HLA-A*02:01 and -B*08:01, putative transcriptionally expressed neoantigens were identified using pVACseq (pVACtools 1.0.7), a computational neoantigen prediction pipeline (16). Two hundred twenty and 204 HLA-A*02:01–restricted neoantigens and 92 and 77 HLA-B*08:01–restricted neoantigens were identified in SQ1 and SQ4, respectively (Dataset S1). In total, genomic analysis of these 2 index tumors identified 375 unique missense mutations in expressed genes that translate into 9-mer or 10-mer amino acid-substituted (AAS) peptides encoding putative neoantigens restricted to either HLA-A*02:01 or HLA-B*08:01 (SI Appendix, Fig. S1B).
Clonal and Subclonal Neoantigens Are Shared among Metachronous MEL66 Tumors.
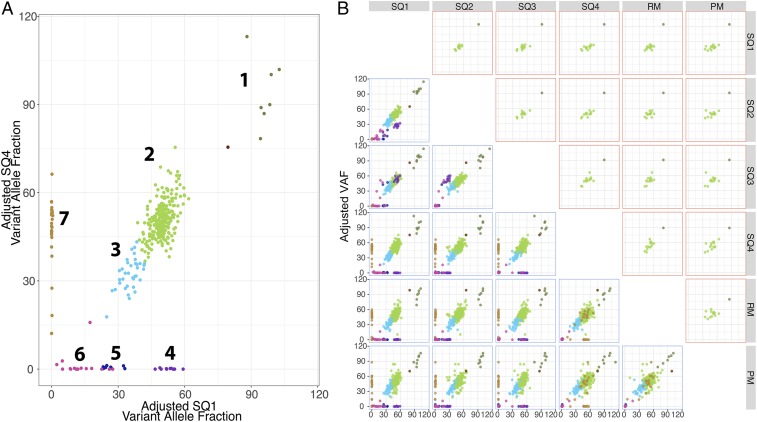
Next, we examined the intra/intertumoral heterogeneity of these 375 MMs in the 6 tumors using custom PCR primers designed to amplify each mutated locus followed by deep targeted DNA sequencing with the orthogonal Ion Torrent platform. Three hundred fifty primer pairs yielded amplicons with adequate coverage for the targeted sites, yielding an average sequencing depth between 1,504-fold (313 MMs in SQ4) and 2,047-fold coverage (295 MMs in PM; Dataset S2). Inference of the tumor neoantigen landscape, obtained from MM variant allele fraction (VAF) clustering using the Sciclone algorithm (17), detected 7 distinct neoantigen-encoding variant clusters as shown by the 2-dimensional analysis between SQ1 and SQ4 (Fig. 1A).
Fig. 1.
Neoantigen landscape of patient MEL66 tumors. Clonal neoantigen architecture of patient MEL66 tumors. Primers were designed to 375 MMs encoding putative neoantigens, and sequencing was performed using the Ion Torrent platform. A total of 350 of 375 targets yield sequencing results with an average depth of 1,776×. (A) Derived variant allelic fraction (VAF) values were used to infer clonal architecture of tumors using the SciClone algorithm, which revealed 7 neoantigen clusters as represented in the 2-dimensional analysis of SQ1 vs. SQ4. Each cluster is identified by a distinct color. (B, Bottom) Two-dimensional analysis representing distribution of neoantigen clusters across the 6 tumors obtained from patient MEL66. (B, Top) Two-dimensional analysis representing cluster distribution of 15 vaccine- and TIL-reactive neoantigens across MEL66 tumors. Identities of these neoantigens are detailed in SI Appendix, Fig. S1D.
Analysis of all 6 tumors from patient MEL66 indicated that neoantigens encoded by MMs in clusters 1 to 3 are shared by all tumors (Fig. 1 B, Bottom) and represented 76.57% (268 of 350) of the putative neoantigens (SI Appendix, Fig. S1C). The high VAF of neoantigens in cluster 1, several of which mapped to chromosome 7, is consistent with copy number amplification (CAN) or loss of heterozygosity (LOH) of the BRAF locus and surrounding regions. Neoantigens present in cluster 2 are encoded by clonal mutations not in copy number-altered loci and are present in all tumor cells in the 6 metastases, while those in cluster 3 are present in a subset of cells of all 6 tumors and hence represent shared subclonal mutations. In contrast, MM in clusters 4 to 7 encompassing 23.42% (82 of 350; Fig. 1) of putative neoantigens are not shared between the lesions and are considered private clonal/subclonal neoantigens. Noteworthy is the absence of cluster 4 neoantigens in RM and PM samples (Fig. 1 B, Bottom), both collected after the patient was treated with ipilimumab followed by targeted BRAF+MEK kinase inhibitors. This observation suggests possible therapeutic selection of cluster 4-carrying tumor cells due to systemic therapy. In summary, the genomic analysis revealed a landscape consisting of a majority of clonal and subclonal neoantigens expressed in all 6 tumors plus a small fraction of private clonal and subclonal neoantigens expressed in a subset of tumors.
Melanoma Neoantigen-Specific T Cells Remain Ignorant until Elicited by Vaccination.
To characterize neoantigen-reactive CD8+ T cells, we examined T cells in prevaccine peripheral blood and TILs using a panel of 70 AAS peptides with experimentally confirmed high binding affinity for HLA-A*02:01 (Dataset S3). These peptides represented clonal and subclonal neoantigens shared among all 6 tumors. In parallel, 10 of these mutated peptides (SI Appendix, Fig. S1D and Dataset S4), along with 2 control gp100 peptides, were formulated into a mature DC vaccine to evaluate immunogenicity. Details of the vaccine protocol have been previously described (10).
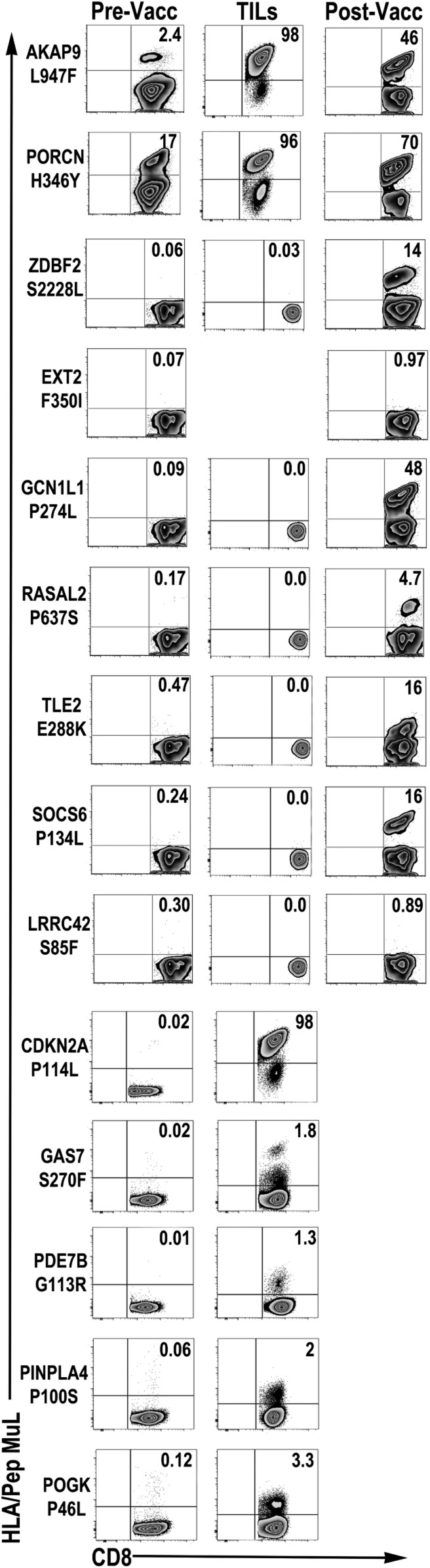
In prevaccine PBMCs, no T cell reactivity to any of the 70 candidate neoantigens was detectable by direct ex vivo staining of CD8+ T cells using custom neoantigen peptide-HLA (p-HLA) multimers. However, short-term (10-d) culture of prevaccine PBMCs with individual neoantigen peptides revealed T cell reactivity to 2 of the 70 neoantigen specificities, AKAP9 L947F and PORCN H346Y (Fig. 2, Left). In prevaccine fresh TILs derived from PM and RM tumors, AKAP9 L947F-specific T cells were readily detected (1.27 to 3.98%) by direct ex vivo p-HLA multimer staining, while no T cell reactivity to remaining vaccine neoantigen candidates was observed (SI Appendix, Fig. S2). However, short-term (10-d) culture of TILs with neoantigen peptides confirmed reactivity to AKAP9 L947F and revealed reactivity to 6 additional neoantigens—PORCN H346Y, CDKN2A P114L, GAS7 S270F, PDE7B G113R, PNPLA4 P100S, and POGK P46L—as detected by p-HLA multimer staining (Fig. 2, Middle). In total, spontaneous (prior to vaccination) T cell reactivities to 2 of 70 neoantigens (2.85%) were observed in peripheral blood, and 7 of 70 (10%) were observed in TILs. This result confirms previous observations that suggest higher frequencies of neoantigen-specific CD8+ T cells in TIL populations compared to the peripheral blood (9, 18).
Fig. 2.
Neoantigen-specific T cell responses in patient MEL66. Neoantigen T cell reactivities in prevaccine PBMCs (Pre-Vacc) and TILs and postvaccine PBMCs (Post-Vacc). Pre- and postvaccine PBMCs were cultured in vitro in the presence of peptide and IL-2 for 10 d, followed by p-HLA multimer staining. Prevaccine TILs were cultured in vitro in the presence of mDC/peptide and IL-2 for 10 d, followed by restimulation with artificial APCs pulsed with peptide for 10 d and staining with p-HLA multimers. Numbers in dot plot represent percentage of neoantigen-specific T cells in lymph/CD8+ gated cells. TIL reactivity to neoantigen EXT2 F350I could not be assessed due to insufficient cells.
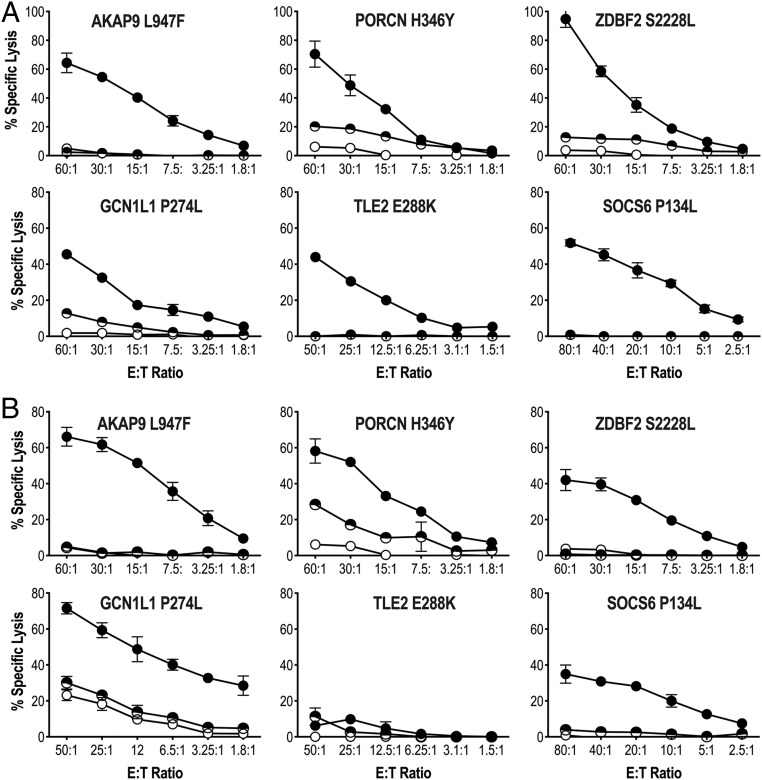
The immunogenicity of 10 selected candidate neoantigen peptides (Dataset S4) was characterized using postvaccine PBMC. Short-term PBMC cultures revealed vaccine-elicited responses to 5 additional antigens (ZDBF S2228L, GCN1L1 P274L, RASAL2 P637S, TLE2 E288K, and SOCS6 P134L) as well as increased responses to AKAP9 L947F and PORCN H346Y (Fig. 2, Right). The neoantigen specificity of each vaccine-elicited CD8+ T cell response was further confirmed by in vitro functional assays using short-term CD8+ T cell lines (Fig. 3). Each T cell line showed specificity for the corresponding AAS-peptide, but not the wild type (WT)-peptide sequence, as determined in standard 4-h 51Cr release assays using peptide-pulsed target cells (Fig. 3A). Processing and presentation of neoantigen was assessed using monoallelic cell lines expressing either HLA-A*02:01 or HLA-B*08:01 in conjunction with AAS- or WT-tandem minigene constructs (TMCs). Five (AKAP9 L947F, PORCN H346Y, ZDBF2 S2228L, GCN1L1 P275L, RASAL2 P637S, and SOCS6 P134L) of the 6 T cell lines tested recognized AAS-TMC but not WT-TMC (Fig. 3B). Only TLE2 E288K-specific T cells did not recognize AAS-TMC, suggesting that this epitope is cryptic and presumably destroyed by the proteasome or ER-resident peptidase. Importantly, as a confirmation, both WT- and AAS-AKAP9 and PORCN epitopes were identified as constituents of the MEL66 HLA-A*02:01 peptidome by LC-MS/MS (SI Appendix, Fig. S3). These functional data and biochemical analysis strongly support the notion that these mutated peptides represent bona fide neoantigens that are processed and presented by HLA-A*02:01. Vaccination had no effect on peripheral blood responses to the following neoantigens not included in the vaccine formulation: CDKN2A P114L, GAS7 S270F, PDE7B G113R, PNPLA4 P100S, and POGK P46L. These responses were exclusively observed in prevaccine TIL populations (Fig. 2). The observation that 5 neoantigen reactivities (ZDBF2 S2228L, GCN1L1 P275L, RASAL2 P637S, TLE2 E288K, SOCS6 P134L) are only found in postvaccination PBMCs and not detected in prevaccine peripheral blood (or TILs) supports the hypothesis that a precursor pool of naïve neoantigen-specific T cells is available and can be elicited upon mDC vaccination in the absence of additional adjunctive therapies (such as checkpoint inhibitors). In sum, spontaneous T cell immunity was confirmed for 7 of 70 (10%) neoantigen peptides using (prevaccine) TILs and 2 of 70 (2.85%) neoantigen peptides using (prevaccine) PBMCs. After mDC vaccination, T cell immunity could be detected for 7 of 10 (70% immunogenicity rate) of the neoantigen peptides tested using (postvaccine) PBMCs. At 60 wk postvaccination, CD8+ T cell responses to 4 (AKAP9 L947F, PORCN H346Y, ZDBF2 S2228L, GCN1L1 P275L) of the 5 HLA-A*02:01–restricted neoantigens incorporated in vaccine is detectable in peripheral blood upon cell culturing, suggesting long-term T cell memory. No response is observed to the 1 “ignored” HLA-A*02:01–restricted antigen, EXT2 F350I (SI Appendix, Fig. S4). Since the patient had NED at the time of vaccination, no postvaccine TILs were available for analysis; the patient remains with NED >42 mo after completion of vaccine protocol with no additional therapy required.
Fig. 3.
Antigen specificity of vaccine-induced T cell responses in patient MEL66. (A) Vaccine-induced CD8+ T cell recognition of AAS- (solid circle) and WT- (semicircle) peptide-pulsed HLA-A*02:01-expressing K562 line in a 4-h 51Cr-release assay. (B) Vaccine-induced CD8+ T cell recognition of AAS- (solid circle) and WT- (semicircle) TMC in HLA-A*02:01-expressing K562 line in a 4-h 51Cr-release assay. Open circle represents lysis obtained with parental K562 cell line. Percent specific lysis of triplicates (mean ± SD) is shown for each data point; spontaneous lysis <5%. Representative experiment of 2 to 3 independent evaluations is shown.
Intratumoral Neoantigen-Specific TCR Repertoires Are Clonally and Spatially Restricted.
To characterize intratumoral TCR repertoires, we performed TCR profiling on the 6 tumors obtained from patient MEL66. We obtained sequences from an average of 28,092 T cells per sample (ranging from 8,794 to 50,747) and observed an average of 9,852 unique TCRVB rearrangements per sample (ranging from 3,002 to 26,244).
Next, neoantigen-specific T cell receptor-β variable segment (TCRVβ) complementarity-determining region 3 (CDR3) sequence (TCRVB_CDR3) reference libraries (Dataset S5) were generated from antigen-expanded CD8+ T cells and used to probe the intratumoral TCR repertoires as well as unmanipulated peripheral CD8+ T cells (Fig. 4A and Dataset S6). We have previously described the use of TCRVB-CDR3 libraries as a platform to assess neoantigen-specific TCR repertoires in unmanipulated peripheral blood (10).
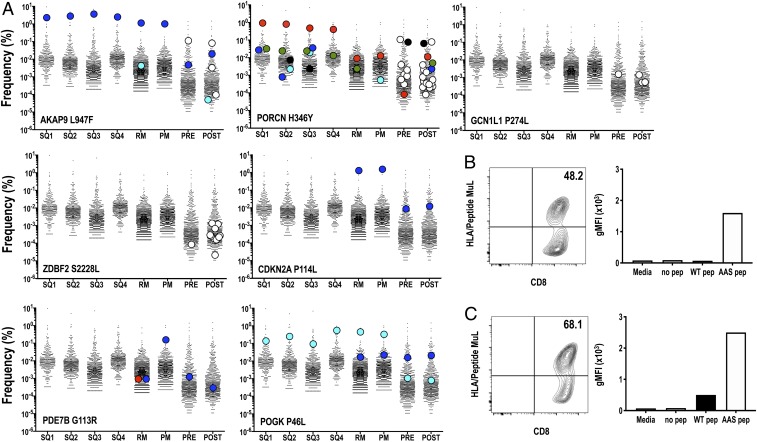
Fig. 4.
MEL66 neoantigen TCRVB CDR3 repertoires in blood and tumors. (A) Frequency of individual neoantigen-specific TCRVB clonotypes identified in tumors (SQ1–4, RM, and PM) and pre- and postvaccine peripheral blood CD8+ T cells as determined using neoantigen-specific TCRVB_CDR3 reference libraries. Each colored dot represents a unique neoantigen TCRVB clonotype. White dots represent neoantigen-specific TCRVB clonotypes found only in blood samples. For clarity, only TCR clonotypes with tumor frequencies >0.00001 are shown (Dataset S6 provides further details). TCRα/β sequencing of T cell clones isolated from (B) AKAP9 L947F- and (C) PORCN H346Y-specific stimulated TILs (SI Appendix, Fig. S4) yielded uniquely expressed TCRVB/TCRVA pairs (SI Appendix, Fig. S5). Lentiviral vector expression of these TCRα/β pairs into Jurkat 76 T cells revealed reactivity with HLA-A*02:01/AKAP9 L947F and HLA-A*02:01/PORCN H346Y multimers, respectively. Functionality of TCR was assessed upon stimulation of TCR+ Jurkat 76 T cells with K562 HLA-A2+ cells pulsed with 10 µg/mL WT or AAS peptide for 16 h. Geometric mean fluorescent intensity of NFAT-eGFP reporter is shown for a representative experiment.
The results reveal an absence or limited number of TCRVB clonotypes specific for most neoantigens among the metastases (SQ1-4, RM, PM) collected. For example, in neoantigen AKAP9 L947F, a single dominant clonotype (CASTPLSNQPQHF) is detected, at very high frequency (>1%), in all 6 metastases (Fig. 4A). This clonotype is also detected in prevaccine blood, along with a second clonotype (CASSIIGQRVEAFF) that curiously is not detected in any of the 6 resected metastases. Upon vaccination, 3 additional clonotypes are detected in peripheral blood. One of these clonotypes (CASSYFDSGKFAGELFF) is also found in RM tumor, resected after systemic therapy but prior to vaccination, at relatively low frequency (∼0.005%; Fig. 4A). TCR α/β sequencing of T cell clones isolated from AKAP9 L947F-stimulated TILs isolated from RM/PM tumors (SI Appendix, Fig. S5 A, Top) yielded a unique expressed TCRVβ CASTPLSNQPQHF (BV6-6/BJ1-5) chain in conjunction with a single rearranged TCRVα CAYGTGTASKLTF (AV38/AJ44) chain (SI Appendix, Fig. S5B, Top). Lentiviral vector expression of this TCRαβ in Jurkat (J76TPR) cells revealed dextramer binding and reactivity with the mutant AKAP9 L947F peptide, supporting the use of the TCRVB_CDR3 library platform for assessment of TCR repertoires in unmanipulated samples (Fig. 4B). For the PORCN H346Y neoantigen, 5 TCRVB clonotypes identified are found dispersed among the 6 tumors. A dominant TCRVB clonotype (CATSTRDTGNEQFF) is found at high frequency (∼1%) in 4 of 6 tumors (SQ1-4), and, after systemic therapy, at lower frequency (∼0.01%) in the RM and PM metastases (Fig. 4A). Interestingly, among the 9 PORCN H346Y-specific clonotypes identified in the prevaccine blood, this clonotype (CATSTRDTGNEQFF) represents the lowest frequency. Ten additional TCR clonotypes were revealed by vaccination (Fig. 4A). Of the 19 total PORCN H346Y TCRVB clonotypes identified, just 1 clonotype (CATSTRDTGNEQFF) is present in blood and all 6 tumors before vaccination. Again, PORCN H346Y-specific TILs isolated from RM/PM tumors (SI Appendix, Fig. S4 A, Bottom) yielded a unique expressed TCRVβ CATSTRDTGNEQFF (BV24-1/BJ2-1) in conjunction with 2 α chains, AV17/J58 and AV25/J39 (SI Appendix, Fig. S5 B, Bottom). Further experiments by gene transfer of paired TCRα/β heterodimers in J76TPR cells confirmed AV17/J58 as the correct partner conferring dextramer binding along with mutant PORCN H346Y peptide recognition; in addition, minor cross-reactivity to the nonmutated PORCN peptide is noted (Fig. 4C).
For vaccine-elicited neoantigens GCN1L1 P274L and ZDBF2 S228, no TCRVB clonotypes were detected in any of the metastases (Fig. 3A), and a single clonotype was detected in the prevaccine blood. However, upon vaccination, multiple new TCRVB clonotypes were revealed in the blood for both of these neoantigens (Fig. 4A). For the shared clonal neoantigens CDKN2A P114L and PDE7B G113R, no clonotypes are identified in the 4 tumor samples obtained prior to systemic therapy (Fig. 4A). However, a single dominant TCRVB clonotype specific for CDKN2A P114L and a single dominant clonotype specific for PDE7B G113R are found in both RM and PM tumors. A dominant CDKN2A P114L-specific clonotype is present at high frequency (∼1%) in 2 of 6 tumors and also is found in pre- and postvaccine blood; the PDE7B G113R neoantigen analysis is similar, with the presence of a dominant VB clonotype detectable only in blood and the PM/RM lesions but none of the SQ1 to 4 lesions. Finally, a single POGK P46L TCRVB clonotype (CAWTRLGIREQYF) is detected in all 6 metastases, with an additional clonotype detected in RM and PM tumors (Fig. 4A). Both of these clonotypes are also detected in pre- and postvaccine blood. Altogether, this sensitive molecular method allows the detection of neoantigen-specific TCR clonotypes based on the frequency (read count) and identity of the TCRVB CDR3 sequence.
In summary, these results demonstrate that spontaneous neoantigen-specific T cell immunity appears to be oligo-/monoclonal, randomly dispersed among both synchronous and metachronous tumors, and, in some instances, absent despite the presence of shared clonal neoantigens (Fig. 1 and SI Appendix, Fig. S1D). These findings suggest that a majority of neoantigen-specific CD8+ T cell clonotypes remain ignorant of the tumor despite homogeneous clonal expression of the cognate target antigen.
Immunological Ignorance Is a Common Feature of Neoantigen and Shared Tumor Antigens in Melanoma.
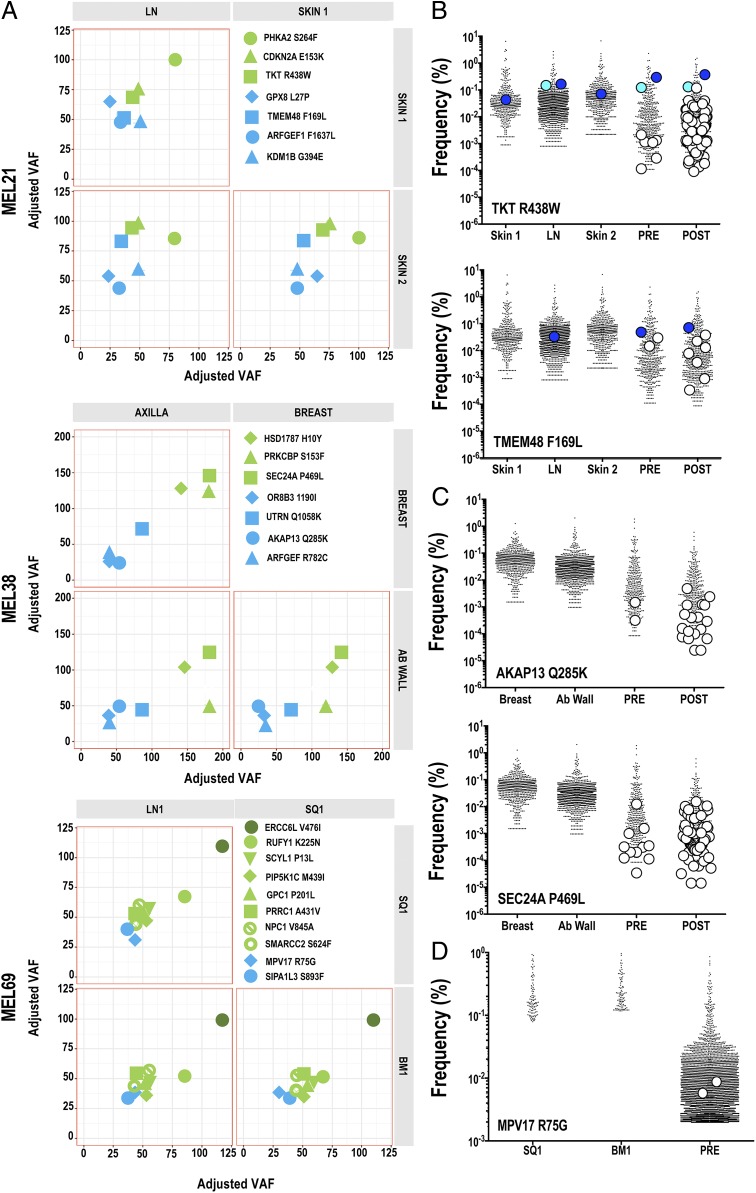
To extend our observations, we assessed the tumor antigenic landscape and intertumor antigen-specific TCR repertoires in 3 additional patients who received a personalized neoantigen mDC vaccine. All tumors were collected prior to vaccination. Genomic analysis for neoantigen identification has been previously reported for tumors of patients MEL21 and MEL38 (10), and analysis for patient MEL69 is reported here (SI Appendix, Fig. S6 and Dataset S7). For all patients’ tumors, inference of the candidate vaccine neoantigen landscape was obtained from MM variant allele fractions (VAFs) using the SciClone algorithm (17). All vaccine candidates were detected in clusters assigned as clonal or subclonal by SciClone and were shared among each patient’s tumors as shown in the 2-dimensional analysis shown in Fig. 5A. Of note, neoantigens to which preexisting immunity was observed were encoded by either clonal (MEL21: TKT R438W, TMEM48 F169L and MEL38: SEC24A P469L) or copy number-altered (MEL69: ERCC6L V476I) SNVs. Thus, we observed a ubiquitous sharing of clonal and subclonal HLA class I-restricted neoantigens in metachronous and synchronous tumors derived from additional melanoma patients.
Fig. 5.
Immunological ignorance is a feature in melanoma. (A) Using variant allelic fraction (VAF) values for SNV encoding neoantigens and the SciClone algorithm, the tumor architecture of these antigens was inferred in MEL21, MEL38, and MEL69 tumors. Two-dimensional analyses are shown, with each cluster defined by a distinct color. ID of mutated genes encoding neoantigens are indicated in figures. (B–D) Distribution of neoantigen-specific TCR clonotypes in tumors and blood. Tumors and peripheral blood pre- and postvaccine CD8+ T cells from patients (B) MEL21, (C) MEL38, and (D) MEL69 were characterized for neoantigen TCRVB repertoires using TCRVB_CDR3 reference libraries. Each colored dot represents a unique neoantigen TCR clonotype identified in tumors. White dots represent unique neoantigen TCR clonotypes found only in blood-derived samples. Identities of neoantigen-specific TCR clonotypes are listed in Dataset S10. For clarity, only TCR clonotypes with tumor frequencies >0.00001 are shown.
Vaccine results have been previously reported for patients MEL21 and MEL38 (10); both of these patients received ipilimumab as systemic therapy, without objective response, prior to vaccine administration. Patient MEL69, reported here, was vaccinated with autologous mDC plus 10 AAS peptides encoding putative neoantigens (SI Appendix, Fig. S6 and Dataset S8). Preexisting CD8+ T cell immunity to 2 neoantigens (MPV17 R75G, ERCC6L V476I) was observed prior to vaccination; T cell reactivity to 3 additional neoantigens (RUFY1 K225N, SIPA1L3 S893F, SMARCC2 S624F; SI Appendix, Fig. S6B) was revealed upon vaccination. To characterize neoantigen-specific TCR clonotypes in prevaccine tumors and peripheral blood CD8+ T cells, TCRVB-CDR3 reference libraries were generated from expanded/p-HLA multimer-sorted CD8+ T cells. For MEL21, we investigated the presence of TKT R438W- andTMEM48 F169L-specific VB clonotypes among 3 tumors (Skin1, LN, and Skin2; Fig. 5B and Dataset S9). Two dominant VB clonotypes for TKT R438W were present in the prevaccine blood. One or both clonotypes were detected in the 3 tumors at moderate frequency (∼0.1%), while none of the remaining 6 low-frequency TCRVB clonotypes detected in blood could be found in any tumor. For TMEM F169L, we detected 3 TCRVB clonotypes in prevaccine blood; however, just 1 clonotype (CASSRTGITDTQYF) was detected in 1 of 3 tumors at modest frequency (∼0.04%; Fig. 5B and Dataset S9). For MEL38, the presence of AKAP13 Q285K- and SEC24A P469L-specific VB clonotypes in 2 tumors (breast and ab wall) was investigated (Fig. 5C and Dataset S9). Two low-frequency TCRVB clonotypes specific for AKAP13 Q285K were detected in prevaccine blood, and yet neither clonotype could be detected in any tumor. One dominant SEC24A P469L-specific TCRVB (CASSVSNQPQHF) clonotype plus 10 lower-frequency clonotypes were found in prevaccine blood; however, all clonotypes were absent in both tumors (Fig. 5C and Dataset S9). As reported previously, mDC vaccination results in diverse TCR repertoires directed at each neoantigen (Fig. 5 B and C) (10). For MEL69, we investigated the presence of MPV17 R75G in 2 tumors (SQ1, BM1). Four low-frequency TCRVB clonotypes specific for MPV17 R75G were detected in prevaccine blood, but none of the clonotypes were detected in any tumor (Fig. 5D and Dataset S9).
Finally, in tumors derived from patients MEL66 (SI Appendix, Fig. S7A and Dataset S10) and MEL21 (SI Appendix, Fig. S7B and Dataset S10), we extended our analysis of intratumoral TCR repertoires to the shared gp100 antigen nonmutated epitopes (G209 and G280) restricted to HLA-A*02:01. Again, an absence or limited presence of TCRVB clonotypes reactive against these antigens is observed in most tumors despite gp100 expression. Thus, despite ubiquitously expressed neoantigens and shared antigens among tumors, a paucity of intratumoral T cells reactive against these antigens was observed. In sum, previously undetectable TCR clonotypes reactive against multiple tumor-encoded neoantigens can be readily elicited by mDC vaccination, consistent with a nonanergized, naive phenotype at low precursor frequencies.
Discussion
ITH is a well-recognized feature of cancer and poses a formidable challenge to therapeutic efficacy, particularly in the context of cancer immunotherapy (19). We investigated the distribution of putative neoantigens among multiple melanoma metastases prior to the administration of a personalized mDC vaccine. Among the patients studied, >75% of putative neoantigens are clonal and, as expected, could be detected in all resected metastases. By constructing TCRVB CDR3 reference libraries to identify and enrich for antigen-specific T cells, neoantigen-specific CD8+ T cells (defined by TCRVB CDR3seq) can be found in the tumor, sometimes at high frequency (∼1%); however, the diversity is limited and most often oligoclonal. In some instances, as exemplified by PORCN, AKAP9, and POGK neoantigens, a dominant neoantigen-specific TCR clonotype is present at high frequency in all metastases as well as peripheral blood. However, in each case examined, the majority of neoantigen-specific (T cell) clonotypes ignore the tumor and are not detectable in blood unless an mDC vaccine is administered. Given the low precursor frequencies of neoantigen-specific T cells in patients, new quantitative approaches are urgently needed to accurately assess tumor-reactive T cells in the peripheral blood, tumor, and various tissue compartments in a direct and facile manner.
Our findings agree with published studies related to the limited diversity of neoantigen-reactive T cells in cancer metastases. Rosenberg and colleagues have employed several strategies to characterize the mutant peptide specificity of both CD8+ and CD4+ T cells and have clearly demonstrated the oligoclonal nature of the response in patients with various malignancies (3). For example, TILs from a melanoma patient with >4,000 nonsynonymous mutations (720 highly expressed SNVs) exhibited a T cell response to 10 mutated tumor antigens; interestingly, all tumor antigens elicited a monoclonal T cell response except SRPX P55L, which elicited 5 unique TCR clonotypes (20). Similarly, in patients with GI malignancies (colon, bile duct, pancreas) (21), HPV-related cervical carcinoma (22), and breast carcinoma (23), neoantigen-reactive TILs were limited to 1 or 2 clonotypes per neoantigen; in some instances, these expanded TIL populations could mediate tumor regression when administered to patients with metastatic disease. A recent report characterizing the intratumoral TCR repertoire of TILs concluded that, in 2 of 4 tumors examined, no tumor-reactive T cells could be detected, while 5 tumor-reactive clonotypes were present in a case of colon cancer and just 1 tumor-reactive clonotype was found in an ovarian carcinoma (24); however, the target antigens in each case were not defined. Finally, a comprehensive genomic and proteomic analysis of melanoma patients provided several notable examples of validated (missense mutated) neoantigen epitopes identified using TIL from patient 12T. Whole-exome sequencing of tumor followed by proteomic analysis of peptides eluted from HLA class I molecules from 12T melanoma cells yielded 3 candidate neoantigens, including 2 mutant peptides (NCAPH2 S3Y and MED15 P4S) that were recognized by TILs (25). The 2 TCR clonotypes specific for NCAPH2 S3Y comprised 58% and 0.01% of the entire TIL population, while the 2 clonotypes specific for MED15 P4S comprised 5.95% and 4.02% of the entire TIL population. This finding agrees with published results from experimental tumor models that indicate that the host immune response is restricted to a small fraction of candidate neoantigens (26, 27).
Adoptive cell therapy studies provide evidence of clinical activity for solid tumors as a monotherapy, suggesting that targeting a single tumor antigen with a monoclonal (or oligoclonal) population may be sufficient. In 12 patients with metastatic synovial sarcoma, NY-ESO-1–specific TCR-transduced T cells provided compelling evidence of clinical activity, with an ORR of 50% and a median duration of response of 31 wk (28). Similarly, single-patient studies employing TILs recognizing only 1 (or several) unique tumor neoantigens mediated dramatic cancer regression in patients with melanoma or other solid tumors (3). Collectively, these data provide important clues about the requirements for antigen targeting when designing new combination strategies. An essential point to consider is the requirement for ubiquitous (clonal) expression of any candidate neoantigen or shared antigen that is selected for immune targeting. Genomic analysis of the clonal architecture of cutaneous melanoma demonstrates limited ITH compared to many other solid tumors, and, indeed, this feature may account for the relative success of checkpoint inhibitors and adoptive cell therapy in treating advanced melanoma (19).
Our work broadens the central tenet of immunological ignorance in cancer (5, 29) and reaffirms the view that most neoantigen-specific T cells remain naïve (4, 6, 30, 31). Therapeutic vaccination and related interventions that facilitate antigen cross-presentation in secondary lymphoid organs will likely be required to increase the breadth, diversity, and frequency of neoantigen-specific CD8+ T cells as an adjunct to checkpoint inhibitor treatment for cancer (32). In support of this proposal, personalized cancer vaccine studies from several investigative teams report the induction of mutant peptide-specific CD8+ and CD4+ T cells directed against ubiquitous clonal neoantigens for melanoma (10, 33, 34), glioblastoma (35, 36), and ovarian carcinoma (37). However, confirmed clinical responses are infrequent. Several patients who received anti-PD1 following neoantigen vaccine subsequently went on to exhibit a confirmed clinical response, suggesting that a combinatorial approach will be required for broader utilization. A test for investigators will be to better define the tumor neoantigen architecture particularly in malignancies with high ITH. The prospective identification of bona fide tumor rejection antigens would appear to represent a most significant challenge for scientists in the cancer vaccine field.
Materials and Methods
Human Subjects and Tumor Samples.
Patients were enrolled in a clinical trial (NCT00683670, BB-IND 13590) and signed informed consents that had been approved by the institutional review board of Washington University. All subjects were HLA-A*02:01+, had no evidence of autoimmune disorder, and were negative for HIV, HBV, and HCV. Leukapheresis was performed prior to treatment and after the third mature dendritic cell (mDC) vaccination at Barnes Jewish Hospital blood bank. Informed consent for genome sequencing was obtained for all patients on a protocol approved by the institutional review board of Washington University. Tumor samples were either flash-frozen or formalin-fixed and paraffin-embedded. Peripheral blood mononuclear cells (PBMCs) were cryopreserved as cell pellets. DNA samples were prepared using a QIAamp DNA Mini Kit (Qiagen), and RNA samples were prepared using a High Pure RNA Paraffin kit (Roche). DNA and RNA quality were determined by Nanodrop 2000 (Thermo Fisher Scientific) and quantitated by Qubite (Thermo Fisher Scientific).
Next-generation sequencing and neoantigen prediction.
For exome sequencing, tumor/PBMC (normal) matched genomic DNA samples were processed with 1 normal and 2 tumor libraries, each using 500 ng DNA input. Mean depth for coverage was targeted at 50× coverage for tumor and 20× coverage for normal germline (PBMC) genome. Exome and cDNA-capture sequencing were performed and analyzed as described in SI Appendix and previously reported (10). Raw data for exome and cDNA-capture data are available on the NCBI dbGAP database (accession no. phs001005) (38).
Ion Torrent Sequencing.
Using the Ion AmpliSeq Designer version 5.2 (https://ampliseq.com/), a BED file of MEL66 targets was submitted for primer design under the FFPE DNA workflow. Primers were delivered prepooled at 2× concentration. In conjunction with the AmpliSeq Library Kit 2.0, 10 ng of input DNA in 6 µL was combined with 10 µL of the 2× custom AmpliSeq primer pool and 4 µL of the 5× Ion AmpliSeq HiFi Mix. All reactions were cycled: 99 °C for 2 min followed by 24 cycles of 99 °C for 15 s and 60 °C for 4 min. After amplification, primer sequences were digested by adding 2 L of FuPa Reagent and cycling at 50 °C for 10 min, 55 °C for 10 min, and 60 °C for 20 min. Adapters were ligated onto the samples through the addition of 4 µL of Switch Solution, 2 µL of Ion Xpress Barcode adapter mix, and 2 µL of DNA Ligase. Samples were purified using 45 µL of Ampure XP beads (Agencourt/Beckman Coulter). The libraries were quantified through use of the KAPA Library Quantification Kit for Ion Torrent and diluted to 26 pM with Ion Torrent Low TE. Template preparation was carried out on the Ion OneTouch 2 instrument in conjunction with the Ion Personal Genome Machine (PGM) Template OT2 200 Kit according to revision 3.0 of the corresponding Ion PGM Template OT2 200 protocol (publication part number MAN0007221). The amplification reaction consisted of 25 µL of nuclease-free water, 500 µL of Ion PGM Template OT2 200 Reagent Mix, 300 µL of Ion PGM Template OT2 200 PCR Reagent B, 50 µL of Ion PGM Template OT2 200 Enzyme Mix, 25 µL of the library diluted to 26 pM, and 100 µL of Ion PGM Template OT2 200 Ion Sphere Particles. Following automated template prep on the OneTouch 2, enrichment of template-positive ISPs was done on the Ion OneTouch ES instrument. From each template prep, we loaded enriched ISPs onto loaded onto an Ion 318 Chip v2 sequenced samples using the Ion PGM Sequencing 200 Kit v2 on the Ion Torrent PGM.
Cluster Analysis.
Clusters were defined using called somatic variants as described earlier. Briefly, variants used in clustering were required to have a tumor coverage ≥50×. Variants for each individual were then clustered using the R library SciClone (v1.1.0) (17). Clustering was performed in 2 dimensions using the samples SQ1/SQ4 and SQ1/SQ2 for patients MEL66 and MEL69, respectively. These cluster assignments were then applied to variants from the remaining samples for each patient. Clusters were additionally defined for auxiliary patients MEL21 and MEL38 using vaccine candidates called in each sample for that patient. In these samples, the maximum number of clusters was limited to 5, and variants used must have ≥10× depth. Tumor purity correction for all samples was applied after clustering using the apex of a density estimation for the dominant clones as a correction factor.
DC Manufacturing.
Dendritic cell manufacturing and vaccine preparation was performed as detailed in SI Appendix and previously described (39).
Neoantigen T Cell Responses.
Kinetics and magnitude of T cell response to AAS-encoding and gp100-derived peptides were assessed using PBMCs collected weekly as described previously (39).
TCR Repertoire Analysis.
Short-term ex vivo-expanded neoantigen-specific T cells were purified to 97 to 99% purity by cell sorting in a Sony sy3200 BSC (Sony Biotechnology) fitted with a 100-µm nozzle, at 30 psi, using 561-nm (585/40) and 642-nm (665/30) lasers and cell pellets prepared. DNA isolation and TCRVB sequencing was performed by Adaptive Biotechnologies. Sequencing was performed at either survey (∼5 × 105 cells for neoantigen-specific T cell reference libraries, TILs, and tumor samples) or deep (∼106 cells for pre- and postvaccine unmanipulated CD8+ T cell populations) level. TCRB V-, D-, J-genes of each CDR3 regions were defined using IMGT (ImMunoGeneTics)/Junctional algorithms and data uploaded into the ImmunoSeq Analyzer (Adaptive Biotechnologies) for analysis. A complete amino acid identity between reference library and 1) tumor, 2) prevaccine, or 3) postvaccine unmanipulated CD8+ T cell samples was required for assigning a TCRB match. TCRB clonotypes with frequencies above 0.1% (>100-fold sequencing depth) in a reference library were set as a threshold for identification of neoantigen-specific TCRVB CDR3 sequences within tumor, TILs, and CD8+ T cell populations.
TCR Sequencing from Cell Lines.
RNA from cell pellets was extracted using the RNeasy Plus Micro Kit (Qiagen, cat. no. 74034). TCR Vβ and Vα chain rearrangement libraries were prepared with SMARTer Human TCR α/β Profiling Kit following the manufacturer’s directions (Takara Bio, cat. no. 635014). Libraries were loaded onto an Illumina MiSeq in the human immunology core facility at the University of Pennsylvania using 2 × 300-bp paired-end kits (Illumina MiSeq Reagent Kit v3, 600-cycle; Illumina, cat. no. MS-102-3003). Sequencing data were analyzed with MiXCR (version 2.1.12) and VDJtools (version 1.1.10) using the default settings (40, 41).
Supplementary Material
Acknowledgments
We thank Tim Ley, Matt Walter, and John Welch for advice and helpful discussion. This project was supported by NIH Grants R21CA205794 (G.P.L.), R01CA204261 (B.M.C.), and P30-CA016520 and the Parker Institute for Cancer Immunotherapy (B.M.C. and G.P.L.). M.G. was supported by the National Human Genome Research Institute (NHGRI) of the National Institutes of Health (NIH) under Award Number R00HG007940 and the V Foundation for Cancer Research under Award Number V2018-007. We also thank Nina Luning-Prak and Wenzhao Meng and the Human Immunology Core facility for performing the RNA-based TCR sequencing on the cell lines.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.C. is a guest editor invited by the Editorial Board.
Data deposition: Raw exome and transcriptome data have been deposited on the National Center for Biotechnology Information (NCBI) dbGaP Genotypes and Phenotypes database (accession no. phs001005).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906026116/-/DCSupplemental.
References
- 1.Coulie P. G., Van den Eynde B. J., van der Bruggen P., Boon T., Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat. Rev. Cancer 14, 135–146 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Schumacher T. N., Scheper W., Kvistborg P., Cancer neoantigens. Annu. Rev. Immunol. 37, 173–200 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Tran E., Robbins P. F., Rosenberg S. A., ‘Final common pathway’ of human cancer immunotherapy: Targeting random somatic mutations. Nat. Immunol. 18, 255–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochsenbein A. F., Immunological ignorance of solid tumors. Springer Semin. Immunopathol. 27, 19–35 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Chen L., Immunological ignorance of silent antigens as an explanation of tumor evasion. Immunol. Today 19, 27–30 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Melero I., et al. , Immunological ignorance of an E7-encoded cytolytic T-lymphocyte epitope in transgenic mice expressing the E7 and E6 oncogenes of human papillomavirus type 16. J. Virol. 71, 3998–4004 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reuben A., et al. , TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: An association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 7, 1088–1097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riaz N., et al. , (2017) Tumor and microenvironment evolution during immunotherapy with Nivolumab. Cell 171, 934–949.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasetto A., et al. , Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol. Res. 4, 734–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carreno B. M., et al. , Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozic I., Nowak M. A., Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 15964–15968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotillo E., et al. , Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 5, 1282–1295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shalabi H., et al. , Sequential loss of tumor surface antigens following chimeric antigen receptor T-cell therapies in diffuse large B-cell lymphoma. Haematologica 103, e215–e218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Rourke D. M., et al. , A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl. Med. 9, eaaa0984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGranahan N., et al. , Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hundal J., et al. , pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 8, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller C. A., et al. , SciClone: Inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput. Biol. 10, e1003665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen C. J., et al. , Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest. 125, 3981–3991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGranahan N., Swanton C., Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Prickett T. D., et al. , Durable complete response from metastatic melanoma after transfer of autologous T cells recognizing 10 mutated tumor antigens. Cancer Immunol. Res. 4, 669–678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran E., et al. , Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stevanović S., et al. , Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 356, 200–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zacharakis N., et al. , Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24, 724–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheper W., et al. , Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 25, 89–94 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Kalaora S., et al. , Combined analysis of antigen presentation and T-cell recognition reveals restricted immune responses in melanoma. Cancer Discov. 8, 1366–1375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav M., et al. , Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Gubin M. M., et al. , Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Angelo S. P., et al. , Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 c259T cells in synovial sarcoma. Cancer Discov. 8, 944–957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsenbein A. F., et al. , Immune surveillance against a solid tumor fails because of immunological ignorance. Proc. Natl. Acad. Sci. U.S.A. 96, 2233–2238 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochsenbein A. F., et al. , Roles of tumour localization, second signals and cross priming in cytotoxic T-cell induction. Nature 411, 1058–1064 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Spiotto M. T., et al. , Increasing tumor antigen expression overcomes “ignorance” to solid tumors via crosspresentation by bone marrow-derived stromal cells. Immunity 17, 737–747 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Chen D. S., Mellman I., Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Ott P. A., et al. , An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin U., et al. , Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Keskin D. B., et al. , Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilf N., et al. , Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Tanyi J. L., et al. , Personalized cancer vaccine effectively mobilizes antitumor T cell immunity in ovarian cancer. Sci. Transl. Med. 10, eaao5931 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Griffith M. et al., Search for MEL66 and MEL69 data . dbGaP Genotypes and Phenotypes database. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001005.v2.p1. Deposited 22 October 2019.
- 39.Carreno B. M., et al. , IL-12p70-producing patient DC vaccine elicits Tc1-polarized immunity. J. Clin. Invest. 123, 3383–3394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolotin D. A., et al. , MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Shugay M., et al. , VDJtools: Unifying post-analysis of T cell receptor repertoires. PLOS Comput. Biol. 11, e1004503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.