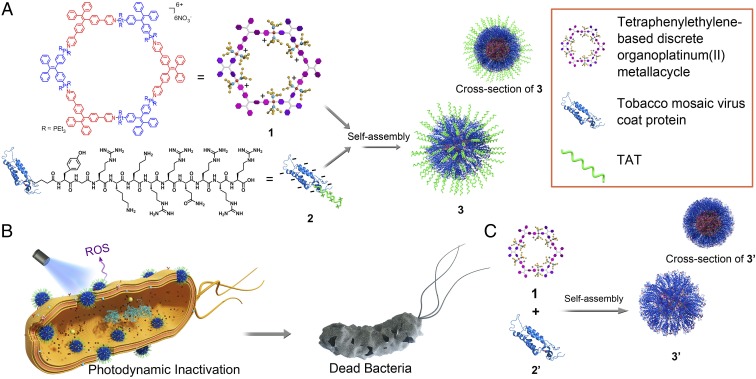
Significance
Photodynamic inactivation (PDI), which has led to little antibiotic resistance, plays a promising role in the control of bacterial infection. Its main mechanism is the damage of membrane components by reactive oxygen species (ROS). However, achieving bacterial membrane intercalation of the photosensitizers remains a challenge. Here, we report the self-assembly of an aggregation-induced emission active photosensitizer with a cell-penetrating peptide-decorated virus coat protein. This assembly exhibits both ROS generation and a strong membrane-intercalating capacity, resulting in significantly enhanced PDI efficiency against bacteria. Especially for Escherichia coli possessing outer membrane, this assembly decreases the survival rate to nearly zero upon light irradiation. This study has implications from the control of bacterial infection to the generation of multifunctional nanomaterials.
Keywords: photodynamic inactivation, bacteria, membrane-intercalating, virus coat protein, aggregation-induced emission
Abstract
Antibiotic resistance has become one of the major threats to global health. Photodynamic inactivation (PDI) develops little antibiotic resistance; thus, it becomes a promising strategy in the control of bacterial infection. During a PDI process, light-induced reactive oxygen species (ROS) damage the membrane components, leading to the membrane rupture and bacteria death. Due to the short half-life and reaction radius of ROS, achieving the cell-membrane intercalation of photosensitizers is a key challenge for PDI of bacteria. In this work, a tetraphenylethylene-based discrete organoplatinum(II) metallacycle (1) acts as a photosensitizer with aggregation-induced emission. It self-assembles with a transacting activator of transduction (TAT) peptide-decorated virus coat protein (2) through electrostatic interactions. This assembly (3) exhibits both ROS generation and strong membrane-intercalating ability, resulting in significantly enhanced PDI efficiency against bacteria. By intercalating in the bacterial cell membrane or entering the bacteria, assembly 3 decreases the survival rate of gram-negative Escherichia coli to nearly zero and that of gram-positive Staphylococcus aureus to ∼30% upon light irradiation. This study has wide implications from the generation of multifunctional nanomaterials to the control of bacterial infection, especially for gram-negative bacteria.
Bacterial infections pose a major threat to global health (1, 2) and have become increasingly serious with increasing antibiotic resistance (3, 4). Despite efforts to find new antibacterial agents, the development of new drugs still lags far behind the evolution of antibiotic resistance (5–8). The newly emerging photodynamic inactivation (PDI) strategy has attracted attention because it involves minimal invasiveness, exhibits specific spatiotemporal selectivity, and is subject to limited antibiotic resistance (9–12). In the presence of light and O2, photosensitizers can generate reactive oxygen species (ROS), such as singlet oxygen (1O2) and hydroxyl radical (•OH), that damage membrane lipids, DNA, or proteins and consequently cause irreversible bacterial death (13–16). The main concern with a traditional photosensitizer is its aggregation-caused quenching behavior, which results in self-quenched fluorescence and reduced ROS generation in the aggregated state. However, aggregation-induced emission (AIE) active photosensitizers can maintain their ROS production ability, even in the aggregated state, and are thus better PDI agents (17).
There are 2 factors that should be taken into consideration to achieve a high PDI efficiency: 1) The bacterial membrane can effectively protect bacteria from foreign substances, including photosensitizers (7); and 2) the half-life and reaction radius of ROS are relatively limited (18). To address these problems, scientists have conjugated a membrane-intercalating moiety to the photosensitizer to enhance its membrane-intercalating capacity. Tang and coworkers (19) modified AIE luminogens with amphiphilic molecules consisting of alkyl chains and a positive charge to facilitate bacterial membrane intercalation of the PDI. Bazan and coworkers (20) designed a membrane-intercalating conjugated oligoelectrolyte to achieve high PDI activity against gram-negative bacteria. In addition to chemical conjugation, self-assembly through physical interactions also provides a general strategy to prepare photosensitizers with multiple functions, which have already been widely adopted for tumor-targeting photodynamic therapy in cancer treatment (21–24).
Herein, we report a self-assembly strategy to obtain a membrane intercalation-enhanced PDI system for the treatment of bacterial infections. Specifically, a tetraphenylethylene-based discrete organoplatinum(II) metallacycle (1) was self-assembled with tobacco mosaic virus coat protein, which was decorated with a transacting activator of transduction (TAT) peptide (2). Metallacycle 1, with the AIE property (25–27), was used as the photosensitizer. The heavy atoms in this molecule, platinum, can promote intersystem crossing from a singlet state to a triplet state to enhance ROS generation (28–30). In 2, the negatively charged protein moiety tends to self-assemble with the positively charged 1 (31), and the TAT moiety provides a strong membrane-intercalating capacity (32, 33). The resulting assembly (3) shows both ROS generation and membrane-intercalating behavior and may be a good candidate for PDI in bacterial inhibition, especially for gram-negative bacteria possessing outer membranes.
Results and Discussion
Preparation and Self-Assembly of Metallacycle 1 and TAT-Decorated Protein 2.
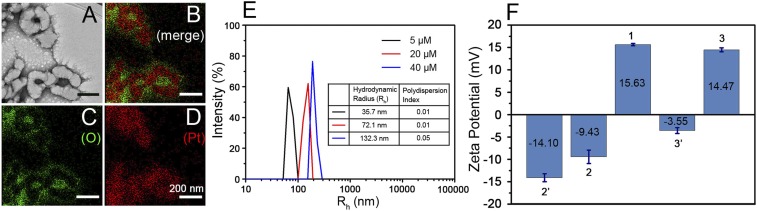
Metallacycle 1 was obtained according to a reported method. Its AIE and self-assembly properties were investigated in detail in previous studies (25–27). Protein 2 was prepared via copper (I)-catalyzed azide-alkyne cycloaddition between 5-azidopentanoic acid-YGRKKRRQRRR (TAT-N3) and the alkyne-modified tobacco mosaic virus coat protein (2′) at a specific site (SI Appendix, Fig. S1). Sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) analysis (SI Appendix, Fig. S2) revealed the emergence of a new protein band with higher molecular mass in the lane of protein 2. This new protein band was digested in-gel by trypsin and analyzed by matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry (SI Appendix, Fig. S3). During the trypsin-digestion process, the protein was cut into peptide segments at the carboxyl side of arginine (R). According to the amino acid sequence of protein 2′ and TAT (34), the theoretical molecular mass of peptide segment in which TAT conjugated (sequence shown in SI Appendix, Fig. S3D) was 1,401 Da, which is consistent with the mass spectra results. The SDS/PAGE and MALDI-TOF mass results proved that TAT was successfully decorated on protein 2′ (35). Based on our previous work on the self-assembly of the positively charged metallacycle 1 with negatively charged protein-based nanoparticles (31), protein 2 tends to self-assemble with 1 through electrostatic interactions (Fig. 1A). Transmission electron microscopy (TEM) images (Fig. 2A) and energy-dispersive X-ray spectroscopy (EDS) mapping (Fig. 2 B–D) showed the formation of assembly 3, in which metallacycle 1 (shown by Pt) was in the core and protected by a protein 2 shell (shown by O). More TEM and EDS mapping images are supplied in SI Appendix, Fig. S4. Free 1 or 2 showed no regular structure (SI Appendix, Fig. S5). Dynamic light scattering (DLS) analysis in 10 mM (pH 7.4) KH2PO4–K2HPO4 buffer showed a broad distribution for metallacycle 1 due to its aggregation status in aqueous solution (SI Appendix, Fig. S6), while assembly 3 with different proportions exhibited a narrow particle size distribution (Fig. 2E). At a protein 2 concentration of 15 μM, the size of assembly 3 increased with the concentration of 1, confirming the 1 in 2 structure.
Fig. 1.
(A) Schematic illustration of the self-assembly of metallacycle 1 and protein 2. (B) Antibacterial mechanism of assembly 3. (C) Schematic illustration of the self-assembly of metallacycle 1 and protein 2′.
Fig. 2.
(A) TEM image of negatively stained assembly 3. (B–D) Overlaid EDS element map (B) and individual EDS maps for O (C) and Pt (D) in A. (Scale bars for A–D: 200 nm.) (E) DLS data for assembly 3 prepared at a protein 2 concentration of 15 μM and at different concentrations (5, 20, and 40 μM) of metallacycle 1. (F) Zeta potential of 15 μM protein 2′, 15 μM protein 2, 40 μM metallacycle 1, assembly 3′ (prepared at 40 μM metallacycle 1 and 15 μM protein 2′), and assembly 3 (prepared at 40 μM metallacycle 1 and 15 μM protein 2) at 10 mM (pH 7.4) KH2PO4–K2HPO4 buffer. Data are shown as mean ± SD (n = 3). Error bars represent the SD of 3 replicates.
Zeta potential was examined to confirm the self-assembly mechanism and the composition of assembly 3. As shown in Fig. 2F, protein 2′ was negatively charged (−14.10 mV) in 10 mM (pH 7.4) KH2PO4–K2HPO4 buffer. Decoration with the positively charged peptide TAT slightly decreased its negative charge, but the entire protein 2 remained negatively charged (−9.43 mV). In the absence of TAT, the assembly of 1 and 2′ (denoted as 3′) showed a negatively charged surface, demonstrating that 1 was in the core and protein 2′ was in the shell (Fig. 1C). With TAT decoration, assembly 3 exhibited a positive charge (14.47 mV) at pH 7.4, demonstrating that the positively charged TAT peptide was exposed outside the surface of 3. This specific structure (shown in Fig. 1A) with the TAT peptide on the exterior surface will be beneficial to cell-membrane intercalation (shown in Fig. 1B).
ROS Generation of Assembly 3.
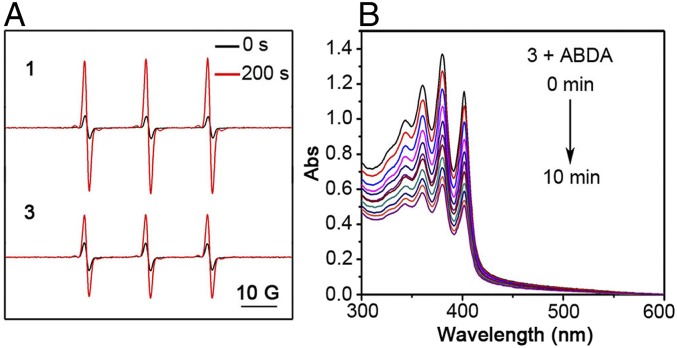
Electron spin-resonance spectrometry (ESR) with 2,2,6,6-tetramethylpiperidine (TEMP) as a spin trap is frequently used for the detection of 1O2, which is an important ROS generated from the type II photochemical reaction. For both metallacycle 1 and assembly 3, there was a strong increase in ESR intensity upon 405-nm irradiation (Fig. 3A), indicating significant 1O2 generation by energy transfer from the excited triplet state of metallacycle 1 to oxygen. The 9,10-anthracenediyl-bis(methylene)dimalonic acid (ABDA) is another commonly used probe for 1O2. With the generation of 1O2, the absorbance of ABDA at 360, 380, and 400 nm will gradually decrease. We first tested the absorption spectra of 1 and 3 in the absence of ABDA under light irradiation to eliminate interference from photoinstability. As shown in SI Appendix, Fig. S7 A and B, the absorption intensity at 400 nm did not show a significant decrease, and the absorption peak at 400 nm of ABDA was therefore applied to identify 1O2 in this work. SI Appendix, Fig. S7C and Fig. 3B show that the absorbance of ABDA at 400 nm significantly decreased upon light irradiation of 1 and 3, indicating 1O2 generation. By eliminating the interference of photoinstability and calculating the relative absorption intensity, we acquired the normalized absorption intensity of ABDA for comparison (SI Appendix, Fig. S7D). From both the ABDA test (SI Appendix, Fig. S7D) and the ESR test (Fig. 3A), assembly 3 did not generate as much 1O2 as free metallacycle 1. This may come from 2 factors: 1) Protein 2 may compete with the spin-trap TEMP or probe ABDA to react with 1O2 (36); and 2) because the photosensitizer is protected by the protein shell, the ROS generation from assembly 3 may be more sensitive to the consumption of ambient O2.
Fig. 3.
(A) With TEMP as a spin trap, ESR spectra of metallacycle 1 and assembly 3 stored in the dark or irradiated with a mercury lamp equipped with a 405-nm bandpass filter for 200 s at room temperature. ESR spectra were obtained after subtracting the background. g = 2.00642, αN = 17.12 G. (B) Ultraviolet-visible spectra of ABDA in the presence of assembly 3 under light irradiation (25 mW cm−2, 405 nm). Abs, absorbance. C1 = 10 μM; C2 = 7.0 μM. ESR and ABDA tests were taken in air.
Intracellular Distribution of Assembly 3 in Gram-Negative Bacteria.
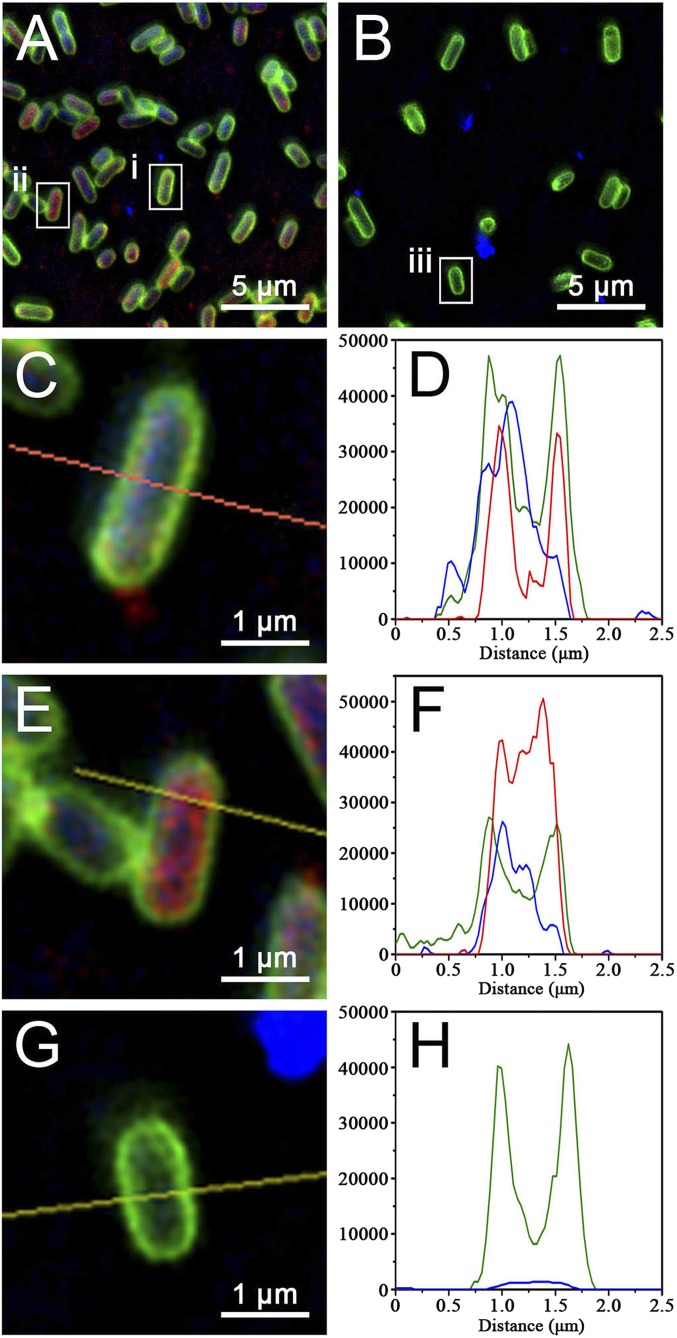
In a PDI process for bacterial inhibition, due to the short half-life and limited reaction radius of ROS, ROS generation and bacterial accumulation are both required for the photosensitizers. Here, we labeled protein 2 with rhodamine B (RB) (red) and the Escherichia coli (E. coli) cell membrane with FM 1-43FX membrane stain (green). The intracellular distribution of assembly 3 could then be monitored by using structured illumination fluorescence microscopy (SIM) (Fig. 4). The fluorescence intensity profiles for the enlarged images were further calculated to analyze the colocalization in detail. As shown in Fig. 4, assembly 3 intercalated in the cell membrane of E. coli, causing the green and red fluorescence in the intensity profiles to coincide closely (Fig. 4 A, C, and D). Some assembly 3 even entered the bacteria by disturbing the integrity of the membrane, as shown by a “red-in-green” fluorescence distribution in the intensity profiles (Fig. 4 A, E, and F). The high fluorescence intensity of blue in Fig. 4 D and F provided evidence of abundant uptake of photosensitizer. In contrast to assembly 3, metallacycle 1 without membrane-intercalating properties just accumulated only outside the bacteria (Fig. 4 B and G). There was little blue fluorescence in the E. coli cells (see the fluorescence intensity profiles in Fig. 4H). To better quantify the intracellular distribution, Pearson’s correlation for Fig. 4 and more SIM images were analyzed through Nikon software (SI Appendix, Fig. S8). The high values of Pearson’s correlation (0.69 for green and red; 0.60 for green and blue) indicated that, through self-assembly, assembly 3 achieved a significantly enhanced membrane-intercalating property (Fig. 1B), which is beneficial for the PDI process.
Fig. 4.
(A and B) SIM images of E. coli cells treated with assembly 3 (A) and metallacycle 1 (B) in 10 mM (pH 7.4) KH2PO4–K2HPO4 buffer. (C–H) Enlarged images (C, E, and G) and their corresponding intensity profiles (D, F, and H) from squares i (C and D), ii (E and F), and iii (G and H). Green fluorescence indicates the membrane of E. coli, blue fluorescence indicates 1, and red fluorescence indicates 2 labeled with RB. Y-axis in D, F, and H indicates the fluorescence intensity with the unit of a.u.
Antibacterial Activity on E. coli.
We investigated the antibacterial effect of assembly 3 on E. coli in the dark and upon 420-nm irradiation for 15 min. Metallacycle 1 and assembly 3′ were applied for comparison. A standard plate count that judged the bacterial proliferative capacity was employed to assess the antibacterial activity. In Fig. 5, assembly 3 and metallacycle 1, which were positively charged (Fig. 2F), created higher dark toxicity than assembly 3′ because of the negative charge on the bacterial membrane. Upon light irradiation, the survival rate of E. coli treated with assembly 3 decreased from ∼55% to nearly 0% at metallacycle 1 concentrations of 20 and 40 μM (Fig. 5A). However, E. coli incubated with free metallacycle 1 at the same concentrations showed little change in survival rate after 420-nm irradiation for 15 min compared to that in the dark (Fig. 5B). The PDI efficiency of assembly 3 was significantly higher than that of 1, which benefited from the enhanced bacterial accumulation capacity of 3. We then calculated the total PDI efficiency of protein 2 (15.3%) and metallacycle 1 (46.3%) at a concentration of 40 μM. This value (61.6%) was much lower than the PDI efficiency of assembly 3 (96.3%) at the same concentration, demonstrating a synergistic antibacterial effect of this system. To explore the importance of the membrane-intercalating property of protein 2 in this PDI system, we further assessed the PDI efficiency of assembly 3′ without TAT decoration for comparison. As shown in Fig. 5C, at a metallacycle 1 concentration of 40 μM, the survival rate of E. coli was reduced only from 74% (in the dark) to 63% upon light irradiation. This result confirmed the important contribution of the membrane-intercalating capacity to the PDI efficiency.
Fig. 5.
E. coli viability against assembly 3 (A), metallacycle 1 (B), and assembly 3′ (C) in the dark and in 25-mW/cm2, 420-nm light irradiation for 15 min. Assembly 3 was prepared with 15 μM protein 2 and different concentrations of metallacycle 1. Assembly 3′ was prepared with 15 μM protein 2′ and different concentrations of metallacycle 1. Data are shown as mean ± SD (n = 3). Error bars represent the SD of 3 replicates. *P < 0.05; **P < 0.01.
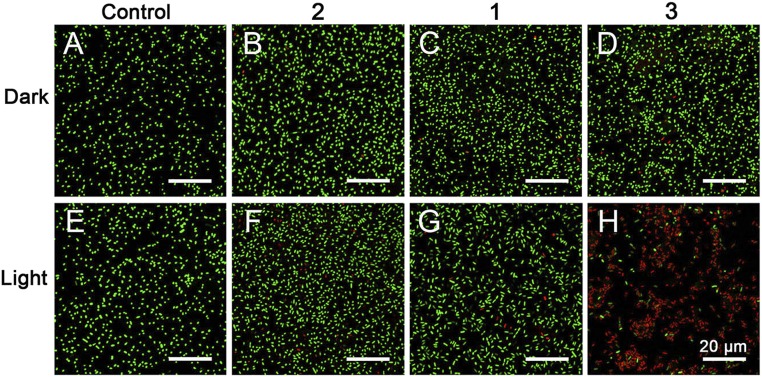
The antibacterial effect was further investigated by the Live/Dead assay. E. coli cells adsorbed on lysine-embedded coverslips were incubated with protein 2 (15 μM), metallacycle 1 (40 μM), or assembly 3 at the same concentration, then kept in the dark or irradiated with 420-nm light (25 mW/cm2) for 15 min. Live/Dead kit staining and confocal laser-scanning microscopy (CLSM) observation (Fig. 6) showed a similar trend in PDI efficiency to that obtained by the standard plate-count method (Fig. 5). Only assembly 3 together with light irradiation led to significant bacterial death (Fig. 6H). Only 16.0% of bacterial cells were alive (SI Appendix, Fig. S9). Both metallacycle 1 without cell-penetrating capacity and assembly 3 without light irradiation could induce only a small amount of death (Fig. 6 C, G, and D). More than 90.0% of bacterial cells were still alive (SI Appendix, Fig. S9). This result indicates that the photosensitizer, light, and membrane-intercalating moiety are all indispensable in this PDI system.
Fig. 6.
Live/Dead analysis of E. coli without any treatment (A and E) or incubated with protein 2 (B and F), metallacycle 1 (C and G), or assembly 3 (D and H) in the dark (A–D) or with 420-nm light (25 mW/cm2) irradiation for 15 min (E–H). Green (SYTO9 nucleic acid stain) indicates live E. coli, and red (propidium iodide) indicates dead E. coli. (Scale bars: 20 μm.)
Cytotoxicity of assembly 3 toward mammalian cells was assessed with L929 and HeLa cells (SI Appendix, Fig. S10). The cells were treated with assembly 3 for 2 h, irradiated or kept in the dark for 15 min, and then incubated for another 12 h. The cell viabilities of L929 and HeLa cells were above 60%, confirming that assembly 3 has good biocompatibility toward mammalian cells.
Antibacterial Mechanism.
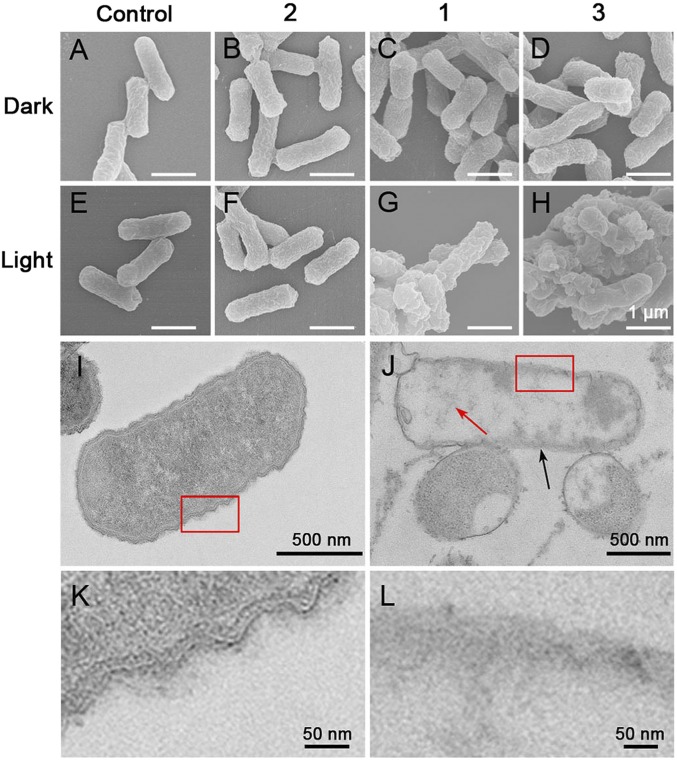
To explore the antibacterial mechanism of assembly 3, we observed the morphology of bacteria by scanning electron microscopy (SEM) and TEM. After incubation with assembly 3 under irradiation, significant rupture of the bacterial membrane of E. coli was observed due to the oxidation of membrane phospholipids, proteins, DNA, and other molecules in the PDI process (Fig. 7H). However, the surface of E. coli incubated with metallacycle 1 under irradiation only became rough, indicating slight damage, but not major destruction, of the cell membrane (Fig. 7G). E. coli cells in the dark or in the absence of photosensitizers still had a typical rod-like shape and intact morphology (Fig. 7 A–F). From TEM section observation, assembly 3 under light irradiation caused bacterial-membrane perturbation (black arrow in Fig. 7J), double-membrane structural damage (red squares in Fig. 7 I and J, which are enlarged as Fig. 7 K and L), and significant cell-content leakage from E. coli cells (red arrow in Fig. 7J). From these SEM and TEM images, membrane lysis by ROS and the subsequent cell-content leakage are the main antibacterial mechanism of assembly 3.
Fig. 7.
(A–H) Representative SEM images of E. coli cells without treatment (A and E) or incubated with protein 2 (B and F), metallacycle 1 (C and G), or assembly 3 (D and H) in the dark (A–D) or with 420-nm light (25 mW/cm2) irradiation for 15 min (E–H). (Scale bars for A–H: 1 μm.) (I and J) TEM section images of E. coli without any treatment in the dark (I) or incubated with assembly 3 with 420-nm light (25 mW/cm2) irradiation for 15 min (J). K and L are enlarged images of the red squares in I and J, respectively.
Broad-Spectrum Antibacterial Properties.
To test whether this system has a broad-spectrum antibacterial effect, we assessed the PDI efficiency of assembly 3 against gram-positive Staphylococcus aureus (S. aureus). As shown in Fig. 8, upon 420-nm light irradiation, the S. aureus survival rate decreased from 71 to 47% at a metallacycle 1 concentration of 20 μM and from 64 to 30% at a metallacycle 1 concentration of 40 μM. This result indicated a convincing PDI effect on gram-positive bacteria, but the PDI efficiency was not as high as that against gram-negative E. coli (cell viability reduced to nearly zero). The superiority of assembly 3 in gram-negative bacterial inhibition may come from a difference in cell-wall structure. The cell wall of gram-negative E. coli consists of the outer membrane, a thin peptidoglycan layer, and the inner membrane, while the cell wall of gram-positive S. aureus consists of a thick peptidoglycan layer and the cytoplasmic membrane. Due to the affinity between TAT peptide and the membrane structure, assembly 3 exhibited superior membrane-intercalating capacity in gram-negative bacteria, thus achieving higher PDI efficiency against E. coli.
Fig. 8.
S. aureus viability against assembly 3 in the dark and with 420-nm light irradiation for 15 min. Assembly 3 was prepared with 15 μM protein 2 and different concentrations of metallacycle 1. Data are shown as mean ± SD (n = 3). Error bars represent the SD of 3 replicates. *P < 0.05.
With the broad-spectrum antibacterial properties and low cytotoxicity against mammalian cells, assembly 3 has a great potential for the infection control of wounds at superficial locations, such as at skin, eye, and epithelium of the gastrointestinal tract. The PDI process could be realized by a pretreatment of assembly 3 solution on the infectious wounds and a subsequent blue-light irradiation. To broaden the clinical applications, we may exploit an AIE-active and near-infrared (NIR) light-excited photosensitizer (37) in this assembly in the future. This PDI system may be more favorable for infection control of deep tissues through NIR laser irradiation upon the infection sites.
Conclusion
In summary, we developed a membrane-intercalation-enhanced PDI system for bacterial inhibition. Metallacycle 1, an organoplatinum(II) metallacycle with AIE activity, was self-assembled with TAT-decorated protein 2 through electrostatic interactions. In assembly 3, metallacycle 1 in the core provided ROS-generation capacity, and the TAT peptide exposed on the surface provided membrane-intercalating capacity. Through the interaction between TAT peptide and the bacterial cell membrane, assembly 3 achieved greatly enhanced membrane-intercalating ability in bacteria. Upon light irradiation, assembly 3 significantly inhibited gram-negative E. coli, with a survival rate of nearly zero. SEM and TEM section images showed that lysis by ROS and subsequent cell-content leakage may be the main antibacterial mechanism. In addition, assembly 3 also exhibited PDI ability against gram-positive S. aureus. The PDI efficiency against gram-positive bacteria was not as high as that against gram-negative bacteria, which possess an outer membrane. Assembly 3 shows membrane-intercalation-enhanced PDI behavior and may become a promising candidate for PDI in bacterial elimination, especially for gram-negative bacteria.
Materials and Methods
Preparation of Assembly 3.
Assembly 3 was prepared by mixing metallacycle 1 with protein 2 in multiple proportions. Metallacycle 1 was prepared according to published procedures (25–27). Protein 2 was synthesized through click reaction between the azide group on 5-azidopentanoic acid-YGRKKRRQRRR (TAT-N3) and the alkyne group on alkyne-protein 2′ (38). Alkyne-protein 2′ was prepared according to the published procedures (39). Detailed methods are mentioned in SI Appendix.
Identification of ROS.
ROS was identified by ESR and chemical method. ESR spectra were recorded on a Bruker E-500 magnetic resonance instrument in air and at room temperature. TEMP was used as a spin trap for ESR. Chemical method was determined by absorption spectra in air, and ABDA was used as a trapping agent. Detailed methods are mentioned in SI Appendix.
Bacteria Culture.
E. coli (ATCC 25922) and S. aureus (ATCC 6538) were incubated in tryptic soy broth medium in a shaking incubator (200 rpm) at 37 °C overnight.
Antimicrobial Activity Measurement.
A 105 colony-forming units (CFU)/mL bacteria suspension was incubated with different samples at room temperature for 15 min in the dark. Then, it was irradiated with a light-emitting diode lamp (25 mW/cm2, 420 nm) or kept in the dark for 15 min. The obtained bacterial solutions were spread on sterile Mueller–Hinton agar plates and incubated upside at 37 °C. The number of CFUs was counted after 24 h. Bacterial solution without any treatment in the dark was used as a control. Bacterial viability was calculated by the following equation:
where Ctest and Ccontrol correspond to the CFU number for the test group and control group, respectively.
Characterizations.
The distribution of assembly 3 in E. coli was observed by Nikon SIM. A Live/Dead assay was performed on a Nikon CLSM. SEM images were recorded on a Hitachi S-4800. TEM images were taken on a JEOL-2100F or Hitachi HT-7700. Detailed methods are mentioned in SI Appendix.
Data Availability.
All data included in this study are available upon request by contacting with the corresponding authors.
Supplementary Material
Acknowledgments
This work was supported by National Key R&D Program of China Grant 2018YFC1105300; Beijing Natural Science Foundation Grant 7182110; National Natural Science Foundation of China Grants 21474123 and 51703230; Youth Innovation Promotion Association of the Chinese Academy of Sciences Grant 2017039; and the Presidential Foundation of Technical Institute of Physics and Chemistry. X.Y. was supported by National Natural Science Foundation of China Grant 21901161; the Program for Eastern Scholar of Shanghai; and start-up funds from Shanghai Jiao Tong University. P.J.S. was supported by NIH Grant R01 CA215157. We thank Na Tian for the help with the light source.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911869116/-/DCSupplemental.
References
- 1.Heymann D. L., Resistance to anti-infective drugs and the threat to public health. Cell 124, 671–675 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Molyneaux P. L., et al. , The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 190, 906–913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chait R., Craney A., Kishony R., Antibiotic interactions that select against resistance. Nature 446, 668–671 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Martínez J. L., Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Brown E. D., Wright G. D., Antibacterial drug discovery in the resistance era. Nature 529, 336–343 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Baym M., Stone L. K., Kishony R., Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakradhar S., Breaking through: How researchers are gaining entry into barricaded bacteria. Nat. Med. 23, 907–910 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Velema W. A., et al. , Optical control of antibacterial activity. Nat. Chem. 5, 924–928 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Celli J. P., et al. , Imaging and photodynamic therapy: Mechanisms, monitoring, and optimization. Chem. Rev. 110, 2795–2838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucky S. S., Soo K. C., Zhang Y., Nanoparticles in photodynamic therapy. Chem. Rev. 115, 1990–2042 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Li X., Lee S., Yoon J., Supramolecular photosensitizers rejuvenate photodynamic therapy. Chem. Soc. Rev. 47, 1174–1188 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Mao D., et al. , Metal-organic-framework-assisted in vivo bacterial metabolic labeling and precise antibacterial therapy. Adv. Mater. 30, e1706831 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Bäumler W., et al. , Photo-oxidative killing of human colonic cancer cells using indocyanine green and infrared light. Br. J. Cancer 80, 360–363 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravanat J.-L., Di Mascio P., Martinez G. R., Medeiros M. H., Cadet J., Singlet oxygen induces oxidation of cellular DNA. J. Biol. Chem. 275, 40601–40604 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Wright A., Bubb W. A., Hawkins C. L., Davies M. J., Singlet oxygen-mediated protein oxidation: Evidence for the formation of reactive side chain peroxides on tyrosine residues. Photochem. Photobiol. 76, 35–46 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y., et al. , Polycationic synergistic antibacterial agents with multiple functional components for efficient anti-infective therapy. Adv. Funct. Mater. 28, 1706709 (2018). [Google Scholar]
- 17.Hu F., Xu S., Liu B., Photosensitizers with aggregation-induced emission: Materials and biomedical applications. Adv. Mater. 30, e1801350 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Sharman W. M., Allen C. M., van Lier J. E., Role of activated oxygen species in photodynamic therapy. Methods Enzymol. 319, 376–400 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Zhao E., et al. , Light-enhanced bacterial killing and wash-free imaging based on AIE fluorogen. ACS Appl. Mater. Interfaces 7, 7180–7188 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Wang B., Wang M., Mikhailovsky A., Wang S., Bazan G. C., A membrane-intercalating conjugated oligoelectrolyte with high-efficiency photodynamic antimicrobial activity. Angew. Chem. Int. Ed. Engl. 56, 5031–5034 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Li M., et al. , One-step formulation of targeted aggregation-induced emission dots for image-guided photodynamic therapy of cholangiocarcinoma. ACS Nano 11, 3922–3932 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Abbas M., Zou Q., Li S., Yan X., Self-assembled peptide- and protein-based nanomaterials for antitumor photodynamic and photothermal therapy. Adv. Mater. 29, 1605021 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y., et al. , Design of an amphiphilic iRGD peptide and self-assembling nanovesicles for improving tumor accumulation and penetration and the photodynamic efficacy of the photosensitizer. ACS Appl. Mater. Interfaces 10, 31674–31685 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Yu G., et al. , A discrete organoplatinum(II) metallacage as a multimodality theranostic platform for cancer photochemotherapy. Nat. Commun. 9, 4335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan X., Cook T. R., Wang P., Huang F., Stang P. J., Highly emissive platinum(II) metallacages. Nat. Chem. 7, 342–348 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Yan X., et al. , Light-emitting superstructures with anion effect: Coordination-driven self-assembly of pure tetraphenylethylene metallacycles and metallacages. J. Am. Chem. Soc. 138, 4580–4588 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Yan X., et al. , Endo- and exo-functionalized tetraphenylethylene M12L24 nanospheres: Fluorescence emission inside a confined space. J. Am. Chem. Soc. 141, 9673–9679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z., et al. , Heterometallic Ru-Pt metallacycle for two-photon photodynamic therapy. Proc. Natl. Acad. Sci. U.S.A. 115, 5664–5669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Bai H., Xu J. F., Wang S., Zhang X., Supramolecular porphyrin photosensitizers: Controllable disguise and photoinduced activation of antibacterial behavior. ACS Appl. Mater. Interfaces 9, 13950–13957 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Lu K., He C., Lin W., Nanoscale metal-organic framework for highly effective photodynamic therapy of resistant head and neck cancer. J. Am. Chem. Soc. 136, 16712–16715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y., Yan X., Saha M. L., Niu Z., Stang P. J., Hierarchical self-assembly of responsive organoplatinum(II) metallacycle-TMV complexes with turn-on fluorescence. J. Am. Chem. Soc. 138, 12033–12036 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Rao Y., Kwok S. J., Lombardi J., Turro N. J., Eisenthal K. B., Label-free probe of HIV-1 TAT peptide binding to mimetic membranes. Proc. Natl. Acad. Sci. U.S.A. 111, 12684–12688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douat C., et al. , A cell-penetrating foldamer with a bioreducible linkage for intracellular delivery of DNA. Angew. Chem. Int. Ed. Engl. 54, 11133–11137 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Alonso J. M., Górzny M. L., Bittner A. M., The physics of tobacco mosaic virus and virus-based devices in biotechnology. Trends Biotechnol. 31, 530–538 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Tian Y., et al. , Integration of cell-penetrating peptides with rod-like bionanoparticles: Virus-inspired gene-silencing technology. Nano Lett. 18, 5453–5460 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Leshem G., et al. , Photoactive chlorin e6 is a multifunctional modulator of amyloid-β aggregation and toxicity via specific interactions with its histidine residues. Chem. Sci. 10, 208–217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trivedi E. R., et al. , Chiral porphyrazine near-IR optical imaging agent exhibiting preferential tumor accumulation. Proc. Natl. Acad. Sci. U.S.A. 107, 1284–1288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlick T. L., Ding Z., Kovacs E. W., Francis M. B., Dual-surface modification of the tobacco mosaic virus. J. Am. Chem. Soc. 127, 3718–3723 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Fraenkel-Conrat H., Degradation of tobacco mosaic virus with acetic acid. Virology 4, 1–4 (1957). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in this study are available upon request by contacting with the corresponding authors.