Significance
Myocardin-related transcription factor B (MRTFB) is a candidate tumor-suppressor gene identified in transposon mutagenesis screens of the intestine, liver, and pancreas. Using a combination of cell-based assays, in vivo tumor xenograft assays, and Mrtfb knockout mice, we demonstrate that MRTFB is a mouse and human colorectal cancer (CRC) tumor-suppressor gene that functions in part by inhibiting cell invasion and migration. Using whole transcriptome RNA sequencing in Mrtfb-knockdown cells, we also identify several MRTFB downstream genes, including a known tumor suppressor, MCAM, and a candidate tumor suppressor, SPDL1. Finally, we show that MCAM and SPDL1 are also human CRC tumor-suppressor genes that act downstream of MRTFB to regulate CRC growth and survival.
Keywords: MRTFB, colorectal cancer, tumor suppressor, RNA-seq, SPDL1
Abstract
Myocardin-related transcription factor B (MRTFB) is a candidate tumor-suppressor gene identified in transposon mutagenesis screens of the intestine, liver, and pancreas. Using a combination of cell-based assays, in vivo tumor xenograft assays, and Mrtfb knockout mice, we demonstrate here that MRTFB is a human and mouse colorectal cancer (CRC) tumor suppressor that functions in part by inhibiting cell invasion and migration. To identify possible MRTFB transcriptional targets, we performed whole transcriptome RNA sequencing in MRTFB siRNA knockdown primary human colon cells and identified 15 differentially expressed genes. Among the top candidate tumor-suppressor targets were melanoma cell adhesion molecule (MCAM), a known tumor suppressor, and spindle apparatus coiled-coil protein 1 (SPDL1), which has no confirmed role in cancer. To determine whether these genes play a role in CRC, we knocked down the expression of MCAM and SPDL1 in human CRC cells and showed significantly increased invasion and migration of tumor cells. We also showed that Spdl1 expression is significantly down-regulated in Mrtfb knockout mouse intestine, while lower SPDL1 expression levels are significantly associated with reduced survival in CRC patients. Finally, we show that depletion of MCAM and SPDL1 in human CRC cells significantly increases tumor development in xenograft assays, further confirming their tumor-suppressive roles in CRC. Collectively, our findings demonstrate the tumor-suppressive role of MRTFB in CRC and identify several genes, including 2 tumor suppressors, that act downstream of MRTFB to regulate tumor growth and survival in CRC patients.
Colorectal cancer (CRC) is the second-leading cause of cancer-related deaths worldwide, with 860,000 deaths and 1.8 million newly diagnosed cases each year (1). CRC imposes a large burden on the health care system, with approximately $14 billion spent annually to treat CRC in the US alone (2). Therefore, numerous studies have attempted to understand the etiology of CRC and apply the research findings to patient care and treatments.
In the last several years, cancer genomics has become a promising tool for furthering our understanding of CRC. Representative studies include The Cancer Genome Atlas molecular characterization of hundreds of CRCs to identify significantly mutated genes (3) and Sleeping Beauty (SB) transposon mutagenesis screens designed to uncover new CRC candidate driver genes in mouse models of CRC (4). A large number of candidate cancer driver genes have been identified by these studies, many of which have unknown roles in CRC, such as myocardin- related transcription factor B (MRTFB). Mrtfb also has been identified as a candidate cancer driver gene in other transposon mutagenesis screens in the gastrointestinal (GI) tract, including hepatocellular carcinoma (HCC) (5) and pancreatic ductal adenocarcinoma (PDAC) (6). These studies suggest a potentially important role for Mrtfb in GI tract cancers.
Mrtfb is an essential gene, as its whole-body knockout in mice leads to embryonic lethality at around embryonic day (E) 13.5, due mainly to cardiovascular defects (7, 8). Mrtfb has also been shown to regulate cell cycle progression (9) and HCC xenograft tumor growth (10). Functional validation using cell culture systems have also shown that reduced expression of MRTFB by RNA interference leads to increased CRC cell invasion (4), suggesting its important role in tumor progression.
Based on these results, we decided to conditionally delete Mrtfb in the mouse intestine to further explore its role in CRC. We found that tumor growth was significantly accelerated in the mouse intestine on Mrtfb knockout, which is critical for functional validation. We also showed that knockdown of MRTFB expression in human CRC cells resulted in accelerated xenograft tumor growth as well as increased invasion and migration of human CRC cells. We then performed whole transcriptome RNA sequencing using primary human colon cells with reduced MRTFB expression to identify MRTFB downstream genes, which led to the identification of a number of genes, including a known tumor suppressor, MCAM (melanoma adhesion molecule), and a candidate CRC tumor suppressor, SPDL1 (spindle apparatus coiled-coil protein 1). Subsequent follow-up studies showed that SPDL1 has tumor-suppressor activities in both cell-based and xenograft assays using human CRC cells, and that reduced SPDL1 expression levels are significantly associated with shorter overall survival in human CRC patients. Collectively, our studies have identified the human CRC tumor suppressor MRTFB, as well as several MRTFB downstream genes, including MCAM and SPDL1.
Results
Mrtfb Is a Candidate Driver Gene for GI Tract Cancers.
SB transposon mutagenesis screens performed in the intestines of mice carrying sensitizing mutations in genes that act at different stages of CRC development, including APC (Apcmin/+), KRAS (KrasG12D/+), SMAD4 (Smad4KO/+), and TP53 (p53R172H/+), identified 11 candidate tumor-suppressor genes that were mutated in all 4 cohorts (4), suggesting their critical roles in CRC. Seven of these are established cancer driver genes, including Abl1 (11), Ankrd11 (12), Arid1a (13), Ctnna1 (14), Gnb1 (15), Pik3r1 (16), and Zfp148 (17). Mrtfb was 1 of the 4 candidate tumor-suppressor genes identified, along with Dennd4c, Luc7l2, and Ppm1b, that have no clarified role in CRC or other types of cancer. However, a potentially important role for Mrtfb in GI tract cancer has been further suggested by other SB transposon mutagenesis screens, which identified Mrtfb as a candidate tumor-suppressor gene in HCC (5) and PDAC (6). Therefore, we focused on Mrtfb in the present study.
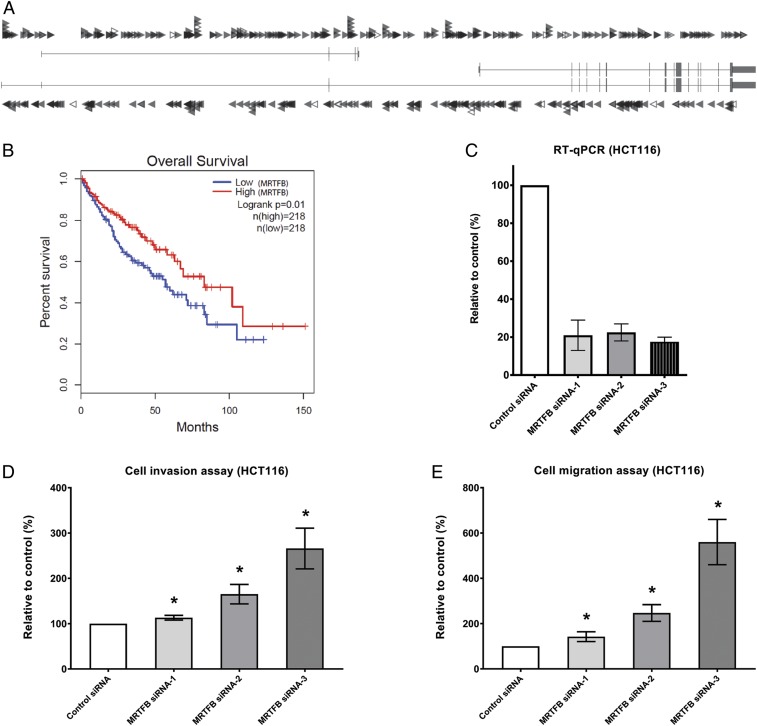
The insertion pattern of SB transposons in a given gene is indicative of whether it is functioning as an oncogene or a tumor-suppressor gene (18–20). As shown in Fig. 1A, transposon insertions in Mrtfb in mouse intestinal tumors are distributed almost evenly across the gene and in both orientations, which is indicative of a tumor-suppressor gene. Consistent with this, lower MRTFB expression levels are associated with shorter survival of patients with GI tract cancer (CRC and HCC) (Fig. 1B), further confirming MRTFB’s potential role as a GI tract tumor-suppressor gene. For patient survival, if only CRC is considered, low MRTFB expression is still correlated with shorter patient survival (SI Appendix, Fig. S1), although the results do not reach the level of statistical significance.
Fig. 1.
MRTFB depletion enhances cell migration and invasion in CRC cells. (A) Evenly distributed insertions of the SB transposon in both orientations indicate that Mrtfb is a tumor suppressor. Each arrowhead represents a specific insertion of an SB transposon, with right-pointing arrowheads denoting forward insertion events and left-pointing arrowheads denoting reverse insertion events. (B) Low expression levels of MRTFB are associated with poor patient survival in GI tract cancers. (C) RT-qPCR results demonstrated efficient knockdown of MRTFB expression in HCT116 CRC cells. (D) Knockdown of MRTFB led to increased cell invasion. *P < 0.05. (E) Knockdown of MRTFB led to increased cell migration. *P < 0.05.
Depletion of MRTFB in Human CRC Cells Increases Cell Motility and Accelerates Tumor Growth in Xenograft Assays.
We used small interference RNA (siRNA), a highly efficient and fast method, to knock down MRTFB expression in HCT116 cells, a human colon cancer cell line widely used in therapeutic research and drug screening. Depletion of MRTFB in HCT116 cells by each of 3 independent MRTFB siRNAs (Fig. 1C) led to increased cell invasion (Fig. 1D), consistent with previous findings (4).
We also tested whether MRTFB regulates CRC cell migration. As shown in Fig. 1E, MRTFB knockdown cells also displayed significantly increased cell migration. Similar results were also obtained from 2 other CRC cell lines, HT-29 and SW620, using the same siRNAs (SI Appendix, Fig. S2). These results indicate that MRTFB negatively regulates the motility of CRC cells. We also tested cell proliferation using the WST-1 assay but failed to observe significant changes with MRTFB knockdown, suggesting that MRTFB does not regulate CRC cell proliferation.
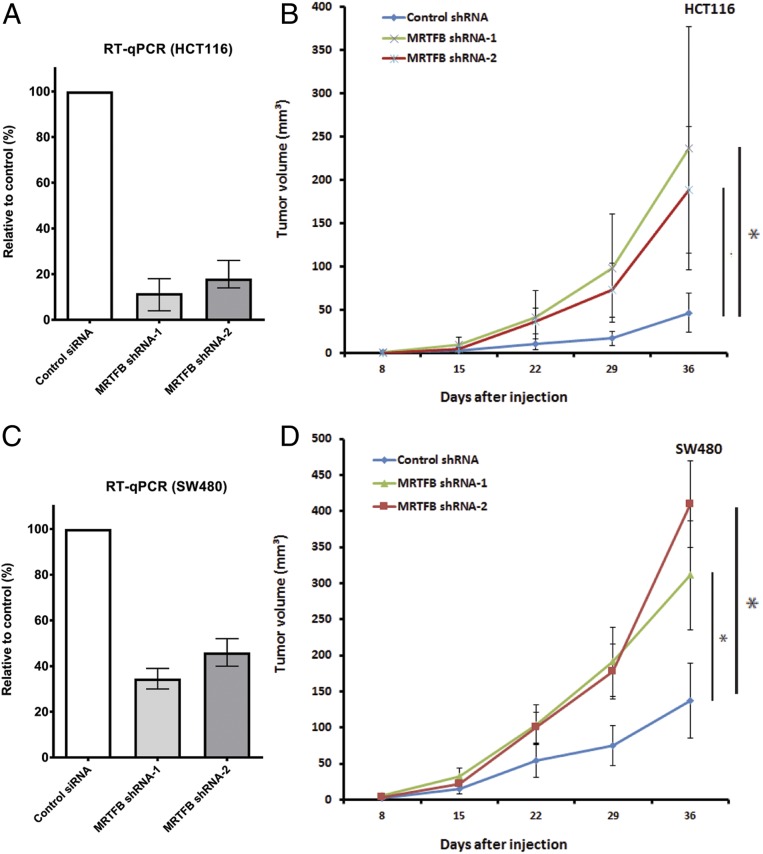
We next asked whether MRTFB depletion in HCT116 cells affects tumor growth in mouse xenograft assays. We used 2 different MRTFB short hairpin RNAs (shRNAs) to knockdown MRTFB expression in HCT116 cells (Fig. 2A). As shown in Fig. 2B, tumor growth was greatly accelerated in mouse xenograft assays when MRTFB expression was efficiently knocked down in HCT116 cells. To confirm these results, we performed the same experiment in a second commonly used CRC cell line, SW480. The MRTFB shRNA knockdown efficiency was slightly lower in SW480 cells (Fig. 2C); however, these knockdown cells also showed greatly accelerated tumor growth in mouse xenografts compared with control cells (Fig. 2D). These results further indicate that MRTFB is a CRC tumor- suppressor gene. In addition, we knocked down the expression of MRTFB in HuH7 HCC cells (SI Appendix, Fig. S1) and observed significant acceleration in xenograft tumor growth, which indicates that MRTFB’s suppressor role is not limited to CRC.
Fig. 2.
MRTFB depletion in CRC cells accelerates the growth of xenograft tumors. (A) RT-qPCR results demonstrated efficient knockdown of MRTFB expression in HCT116 CRC cells. (B) Knockdown of MRTFB in HCT116 CRC cells led to accelerated xenograft tumor growth. (C) RT-qPCR results demonstrated efficient knockdown of MRTFB expression in SW480 CRC cells. (D) Knockdown of MRTFB in SW480 CRC cells led to accelerated xenograft tumor growth.
Knockout of Mrtfb in Mouse Intestine Significantly Enhances Tumor Development in Apc Mutant Mice.
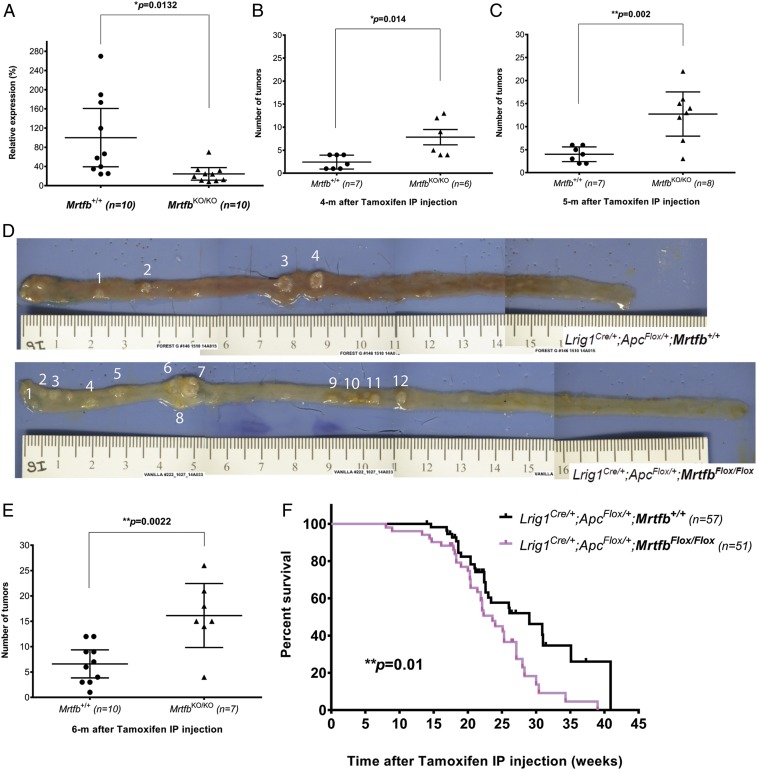
Since tumor development in knockout mice is critical for functional validation of a tumor-suppressor gene, we decided to knock out Mrtfb in the mouse intestine using a conditional MrtfbFlox allele (8). Lrig1Cre/+ was used as the Cre driver to specifically induce deletion of both copies of Mrtfb in the intestine, while ApcFlox/+ was used to sensitize these mice to intestinal tumor development (21). Lrig1Cre/+;ApcFlox/+;MrtfbFlox/Flox and Lrig1Cre/+;ApcFlox/+;Mrtfb+/+ mice were generated and treated with tamoxifen by intraperitoneal injection (IP) at 2 mg/d for 3 consecutive days, beginning at age 6 to 8 wk, to activate Cre expression. Knockout of Mrtfb expression in Lrig1Cre/+;ApcFlox/+;MrtfbFlox/Flox intestine was subsequently confirmed by quantitative reverse transcription polymerase chain reaction (RT-qPCR) (Fig. 3A), and the mice were monitored for tumor development at 4, 5, and 6 mo after tamoxifen injection. As shown in Fig. 3B, control animals developed an average of 2.4 tumors at the 4-mo time point, while Mrtfb knockout mice developed 7.8 tumors, a 3-fold increase. At the 5-mo time point, we observed an average of ∼13 tumors in each Mrtfb knockout mouse, compared with only ∼4 tumors in control animals (Fig. 3 C and D). A similar acceleration of tumor development was observed at the 6-mo time point (Fig. 3E), further confirming Mrtfb’s role as a CRC tumor suppressor.
Fig. 3.
Mrtfb knockout accelerates tumor development in mouse intestines. (A) RT-qPCR results showing loss of Mrtfb expression in intestinal tissues of Mrtfb KO mice. (B) Significantly increased (∼3-fold) tumor development at 4 mo after specific deletion of Mrtfb in intestines (small and large intestines combined). (C) Significantly increased (∼3-fold) tumor development at 5 mo after specific deletion of Mrtfb in intestines (small and large intestines combined). (D) Representative pictures of small intestines of control and Mrtfb KO mice at 5 mo show a significant difference in tumor development. (E) Significantly increased (∼2.5 fold) tumor development at 6 mo after specific deletion of Mrtfb in intestines (small and large intestines combined). (F) Specific deletion of Mrtfb in mouse intestines led to significantly shortened survival time.
We also collected tumor samples from control and Mrtfb knockout mice. Pathological analysis identified these tumors as adenomas (SI Appendix, Fig. S3). We did not observe adenocarcinomas in these animals, probably due to the intrinsic nature of the Lrig1Cre/+;ApcFlox/+ mouse model (21). The increased tumor burden may have contributed to the early death of the Mrtfb conditional knockout mice (Fig. 3F).
MRTFB Downstream Genes Were Identified by RNA-Seq in CRC Cells.
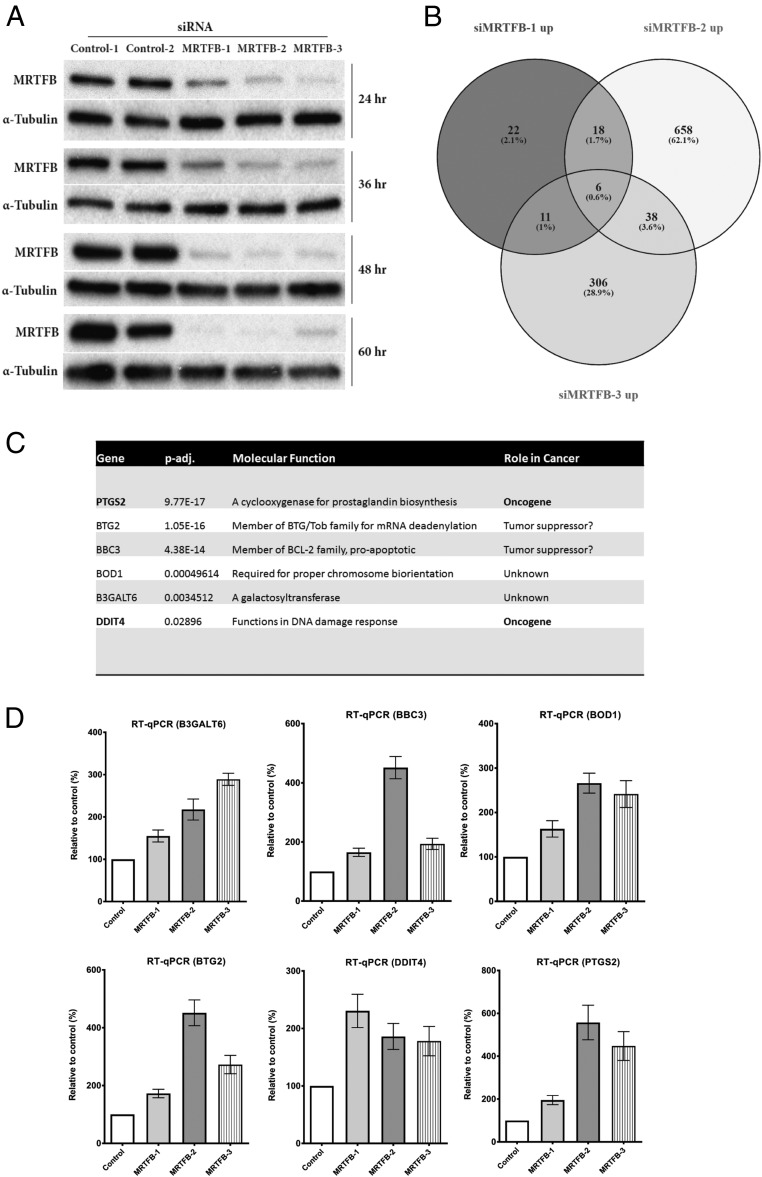
MRTFB is a transcriptional coactivator (22, 23); however, no CRC-specific genes that function downstream of MRTFB have been reported. To identify such genes, we performed whole transcriptome RNA sequencing using primary human colonic epithelial MRTFB siRNA knockdown cells (hCECs) (24). We first transfected control and MRTFB siRNAs into hCECs and collected the cells at different time points after transfection. As shown in Fig. 4A, >90% knockdown of MRTFB expression was achieved at the 48-h time point in all 3 siRNA samples. We selected the 72-h time point for RNA-seq with the consideration of providing extra time (24 h) for the cells to display the full effects of MRTFB depletion on transcription. Total RNA was then extracted from the transfected cells, and mRNA was enriched for library preparation and whole transcriptome sequencing.
Fig. 4.
RNA-seq of human epithelial colon cells with MRTFB depletion identified target genes of its transcriptional activity. (A) Western blot analysis results showed efficient knockdown of MRTFB expression in hCEC1 cells at different time points. (B) Venn diagram of comparison of significantly up-regulated genes among MRTFB siRNA samples. (C) Two known oncogenes were among the 6 common up-regulated genes that are potentially direct targets of MRTFB transcriptional activity. (D) PCR results validated the significant expression level changes of the 6 common up-regulated target genes of MRTFB identified by RNA-seq. All RT-qPCR analyses were performed 3 times (all P < 0.05).
A total of 5 samples were sequenced, including 2 control samples and 3 MRTFB knockdown samples. We obtained more than 106 million raw reads for each sample, >96% of which were clean reads. More than Over 92% of the clean reads could be mapped to the reference mouse genome, demonstrating a highly successful sequencing experiment.
Statistically significant differentially expressed genes from all 3 MRTFB siRNA samples were compared; a complete gene list is provided in Dataset S1. The expression levels of 6 genes—B3GALT6, BBC3, BOD1, BTG2, DDIT4, and PTGS2—were significantly up-regulated in all 3 MRTFB knockdown samples (Fig. 4 B and C). Subsequent validation of the RNA-seq results using RT-qPCR showed that the expression level changes of all 6 genes were consistent with the RNA-seq results (Fig. 4D).
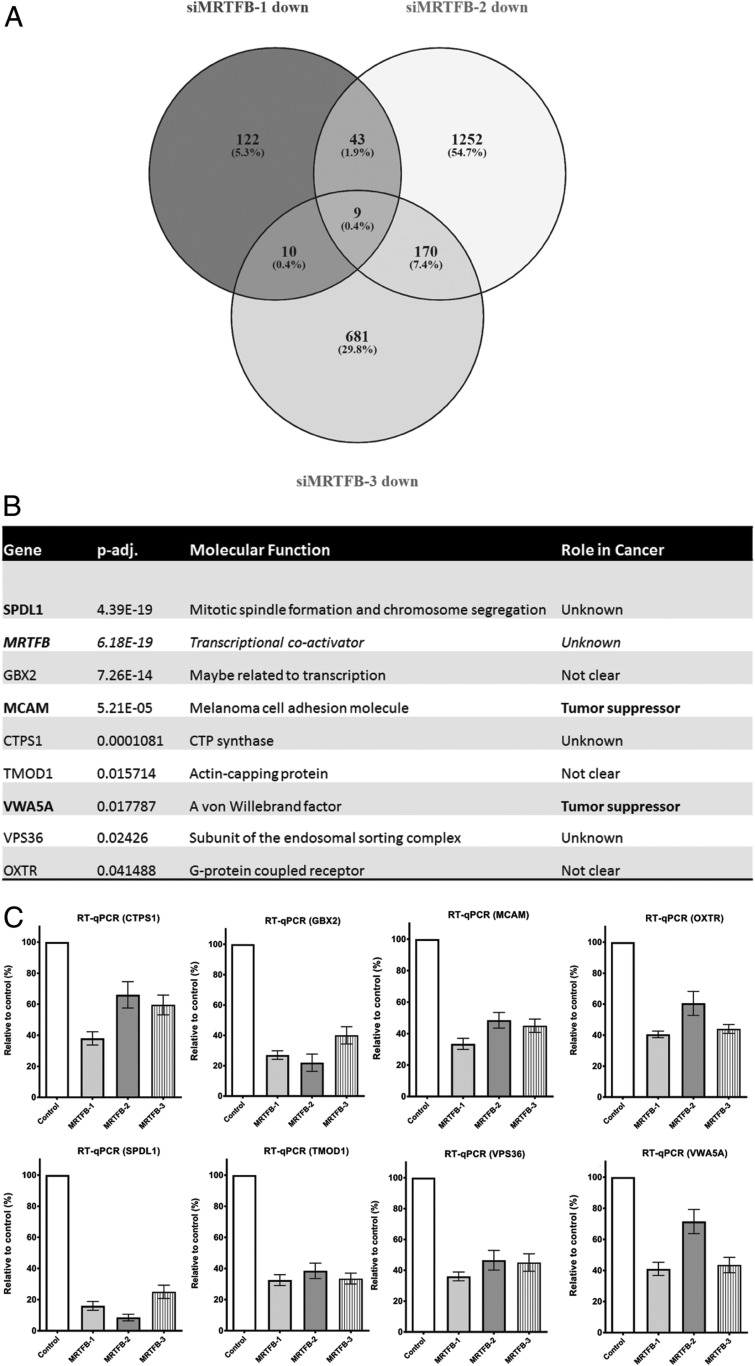
Nine genes displayed significantly reduced expression in all 3 MRTFB knockdown samples (Fig. 5A), including MRTFB, as expected, as well as SPDL1, GBX2, MCAM, CTPS1, TMOD1, VWA5A, VPS36, and OXTR (Fig. 5B). As shown in Fig. 5C, subsequent validation of the RNA-seq results using RT-qPCR showed that the expression level changes for all 8 genes were consistent with the RNA-seq results. There were 2 known tumor suppressor genes, MCAM (25–28) and VWA5A (29–31), among the 8 down-regulated genes.
Fig. 5.
The potential direct target genes of MRTFB include 2 known tumor suppressors. (A) Venn diagram of comparison of significantly down-regulated genes among MRTFB siRNA samples. (B) Two known tumor suppressors were among the 8 common down-regulated genes that are potentially direct targets of MRTFB transcriptional activity. (C) PCR results validated the significant expression level changes of the 8 common down-regulated target genes of MRTFB’s identified by RNA-seq.
MCAM has been associated with tumor progression in human malignant melanoma (32–34). Reduced MCAM expression has been shown to promote tumorigenesis and cancer stemness in CRC by activating Wnt/β-catenin signaling (26), while attenuation in cancer-associated fibroblast promotes pancreatic cancer progression (28). VWA5A is a tumor suppressor in melanoma (29) and is frequently deleted in breast cancer (30). VWA5A expression also has been shown to be dramatically up-regulated in CNE-2L2 human nasopharyngeal carcinoma cells with reduced malignancy, and thus VWA5A has been proposed as a candidate tumor suppressor gene for nasopharyngeal cancer (31).
MRTFB Targets SPDL1 and MCAM to Inhibit CRC Cell Motility and Tumor Growth.
Because MRTFB is a transcriptional coactivator, we decided to first focus on the functional validation of genes that were down-regulated in MRTFB knockdown cells (Fig. 5 B and C). In this study, we focused on SPDL1, since it was ranked first on our list of genes with reduced expression in MRTFB knockdown cells (Fig. 5B). SPDL1 transiently interacts with a protein complex on kinetochores to promote recruitment of dynein microtubule motor proteins for chromosome attachment of microtubules and formation of the metaphase plate (35). Its role in cancer is unknown. We used MCAM as a positive control since it is ranked third on our list of candidate genes with reduced expression in MRTFB knockdown cells (Fig. 5B) and is a known tumor suppressor (25–28).
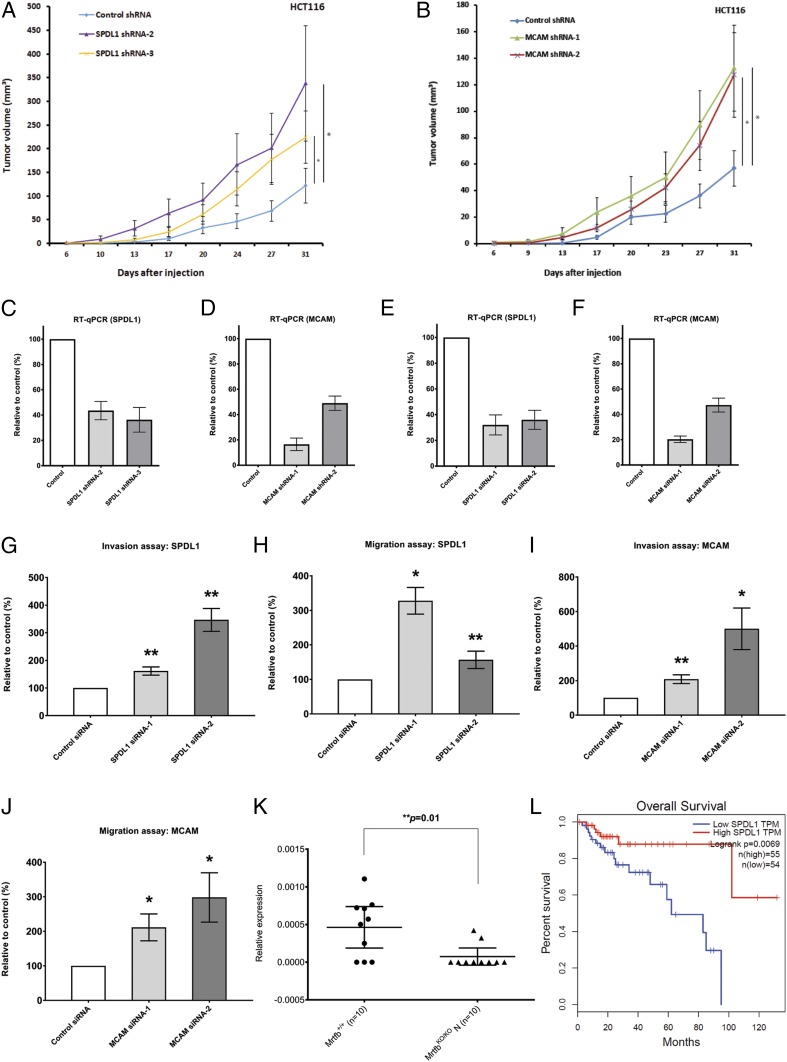
We first asked whether shRNA knockdown of SPDL1 and MCAM in HCT116 cells could accelerate tumor growth in mouse xenograft assays. As shown in Fig. 6 A–D, knockdown of SPDL1 or MCAM in HCT116 cells greatly accelerated tumor growth in mouse xenograft assays, further confirming that SPDL1 and MCAM are CRC tumor suppressor genes.
Fig. 6.
Depletion of MRTFB targets SPDL1 and MCAM promotes CRC tumor development and cell motility. (A) Knockdown of SPDL1 in HCT116 CRC cells led to accelerated xenograft tumor growth. *P < 0.05. (B) Knockdown of MCAM in HCT116 CRC cells led to accelerated xenograft tumor growth. *P < 0.05. (C) RT-qPCR results demonstrated efficient knockdown of SPDL1 expression by shRNAs in HCT116 CRC cells. (D) RT-qPCR results demonstrated efficient knockdown of MCAM expression by shRNAs in HCT116 CRC cells. (E) RT-qPCR results demonstrated efficient knockdown of SPDL1 expression by siRNAs in HCT116 CRC cells. (F) RT-qPCR results demonstrated efficient knockdown of MCAM expression by siRNAs in HCT116 CRC cells. (G) Knockdown of SPDL1 led to increased cell invasion. **P < 0.01. (H) Knockdown of SPDL1 led to increased cell migration. *P < 0.05; **P < 0.01. (I) Knockdown of MCAM led to increased cell invasion. *P < 0.05; **P < 0.01. (J) Knockdown of MCAM led to increased cell migration. *P < 0.05. (K) RT-qPCR results demonstrated significantly reduced expression of Spdl1 in mouse intestine tissues with Mrtfb knockout. (L) Lower expression levels of SPDL1 are associated with significantly shorter patient survival in CRC (http://gepia.cancer-pku.cn/).
Having shown that MRTFB negatively regulates cell invasion and migration, we decided to determine whether knockdown of SPDL1 and MCAM has similar effects on cell motility. Once again, we used small siRNAs to knockdown the expression of both genes in HCT116 CRC cells (Fig. 6 E and F). Similar to MRTFB knockdown cells, we found that depletion of SPDL1 in HCT116 cells led to increased cell invasion (Fig. 6G) and cell migration (Fig. 6H). Similarly, knockdown of MCAM in HCT116 cells also led to increased cell invasion (Fig. 6I) and migration (Fig. 6J). Similar consistent results were obtained from 2 other CRC cell lines, HT-29 and SW620, using the same siRNAs (SI Appendix, Fig. S4). These results show that SPDL1 and MCAM negatively regulate cell motility in a manner similar to MRTFB. We also tested cell proliferation using the WST-1 assay but failed to observe significant changes with either MCAM or SPDL1 knockdown, suggesting that MCAM and SPDL1 do not regulate CRC cell proliferation. Finally, we confirmed that Mrtfb positively regulates Spdl1 expression in the mouse intestine by RT-qPCR (Fig. 6K). In addition, we found that lower expression levels of SPDL1 are associated with reduced survival of CRC patients (Fig. 6L). Taken together, these results confirm that SPDL1 and MCAM are CRC tumor suppressors that function downstream of MRTFB in CRC.
Discussion
Thousands of human tumors have now been sequenced, with hundreds of candidate cancer genes identified (36–38). A critical and urgent challenge for the future is to functionally validate these genes in different types of cancers. The present study was designed to validate the role of MRTFB, a promising new tumor suppressor gene, in GI tract cancer. Several findings reported here confirm MRTFB’s role as a CRC tumor suppressor, including 1) SB transposon insertions in Mrtfb identified in mouse GI tract tumors have a typical tumor suppressor pattern, 2) knockdown of MRTFB expression in human CRC cells accelerates xenograft tumor growth in mouse xenograft assays, 3) knockout of Mrtfb in mouse intestines significantly accelerates tumor growth in Apc heterozygous knockout mice, and 4) reduced MRTFB expression in CRC patients is associated with reduced survival. Our study also suggests that MRTFB’s tumor suppressor role in CRC is exerted through its effect on cell invasion and migration, as MRTFB-depleted cells showed significantly increased invasion and migration compared with control cells.
The role of MRTFB in tumorigenesis is poorly understood. Prywes et al. (39) have shown that MRTFB and its homolog MRTFA mediate cancerous transformation in deleted in liver cancer 1 (DLC1)-deficient hepatocellular and mammary carcinoma cells. Interestingly, they also showed that depletion of MRTFA/B suppresses cell migration, cell proliferation, and anchorage-independent cell growth induced by DCL1 loss, which is somewhat different from what we observed for MRTFB in CRC. Recurrent chromosome fusions involving MRTFB have also been observed in some forms of cancer, including RREB1-MRTFB fusions in ectomesenchymal chondromyxoid tumors (40) and C11orf95-MRTFB fusions in chronic lipomas (41). MRTFA and MRTFB are key regulators of immediate early and muscle-specific gene expression and are required for skeletal myogenic differentiation. Both genes belong to a family of transcriptional coactivators that bind to serum response factor (SRF) and strongly activate transcription from promoters with SRF-binding sites (22, 23).
To help elucidate the role of MRTFB in CRC, we performed whole transcriptome RNA-seq on primary colonic epithelial MRTFB knockdown cells. While several previous studies have used microarray or RNA-seq analysis to identify MRTFB’s transcriptional targets in mouse bone marrow cells (42), mouse embryonic stem cells (8) and human breast cancer cell lines (43), no such studies have been done in intestinal cells. We sequenced 2 control samples and 3 MRTFB siRNA knockdown samples to eliminate off-target effects. From the RNA-seq analysis, we identified 15 genes that were differentially expressed in all 3 knockdown samples, 9 of which were down-regulated in MRTFB knockdown cells and 6 of which were up-regulated. Rt-qPCR for all 15 differentially expressed genes attested to the quality of our RNA-seq experiments. Interestingly, a recent paper has reported several potential target genes of MRTFB related to the Hippo/YAP/TEAD pathway (44); however, none of those genes are among the 14 genes identified in our present study.
Since MRTFB is a known coactivator of transcription, we focused on SPDL1, the gene most significantly down-regulated in MRTFB knockdown cells. We also included MCAM as a positive control, since it is a known tumor suppressor (25–28) and is ranked third in our list of genes down-regulated in MRTFB knockdown cells. Similar to what we observed for MRTFB, knockdown of SPDL1 or MCAM in CRC cells led to increased cell invasion, cell migration, and accelerated tumor growth in mouse xerograph assays. Spdl1 expression was also significantly down-regulated in Mrtfb knockout mouse intestine, another important indicator of the regulation of SPDL1 mRNA expression by MRTFB. Finally, we found worse survival in CRC patients with lower SPDL1 expression, similar to what we observed for MRTFB patient survival, further supporting a tumor suppressor role for SPDL1 in CRC.
SPDL1 (also referred to as Spindly/CCDC99) is required for efficient chromosome congression and mitotic checkpoint regulation (35), where it recruits a fraction of cytoplasmic dynein to kinetochores for poleward movement of chromosomes and control of mitotic checkpoint signaling. Human SPDL1 is also a cell cycle-regulated mitotic phosphoprotein that interacts with the Rod/ZW10/Zwilch complex and has been implicated in proinflammatory responses in prostate cancer patients (45). SPDL1 binds to the cell cortex and microtubule tips and colocalizes with dynein/dynactin at the leading edge of migrating human U2OS osteosarcoma cells (46). U2OS cells and primary fibroblasts that lack SPDL1 migrate slower in 2D cell culture than control cells, although centrosome polarization appears to occur normally in the absence of SPDL1. This is in contrast to CRC cells lacking SPDL1, which migrate faster. MCAM is a cell adhesion molecule that has been associated with tumor progression in human malignant melanoma. Importantly, reduced MCAM expression has been shown to promote tumorigenesis and cancer stemness in CRC by activating the Wnt/beta-catenin signaling pathway (26).
The expression of 6 genes—B3GALT6, BBC3, BOD1, BTG2, DDIT4, and PTGS2—was significantly up-regulated in all 3 MRTFB knockdown samples. PTGS2 (COX2), the top gene on our list, and DDIT4 are both thought to function as oncogenes. PTGS2 expression is associated with carcinogenesis and is overexpressed in many human malignancies (47). DDIT4 is a stress-related protein induced by adverse environmental conditions. DDIT4 protein is a transcriptional target of p53 induced following DNA damage (48) that has been shown to promote gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways (49). BBC3 (PUMA) has been proposed to be a direct mediator of p53-associated apoptosis (50). BTG2 encodes an antiproliferative protein (51) and, strikingly, is also a p53 target gene (52) and downstream effector of p53-dependent proliferation arrest (53).
At present, it is unclear whether any of the genes deregulated in MRTFB knockdown cells are direct targets of MRTFB. The promoter sequences of genes such as MCAM and SPDL1 were obtained by prediction (https://www.genecards.org/) and have not been experimentally verified, and chromatin immunoprecipitation assays examining whether MCAM and SPDL1 are direct targets of MRTFB have been inconclusive so far. CArG boxes are required for SRF to bind to DNA (8, 54, 55), but there are no CArG boxes in these predicted promoters of these genes. However, since these promoters have yet to be experimentally verified, it is difficult to judge the significance of this result. Imperfect CArG boxes that have no more than 1-bp deviation may still be functional for SRF-MRTFB activity (56), and we found 3 such imperfect CArG boxes in intron-1 of the SPDL1 gene and 1 imperfect CArG box in the last exon of the MCAM gene. Even though these imperfect CArG boxes are not in the predicted promoters, it is possible that such SRF-MRTFB binding sites in other regions of these genes may help activate the transcription of target genes. It is also possible, perhaps even likely, that MRTFB can bind to other transcription factors and regulate gene expression by as-yet unidentified means.
While it might seem difficult to conceive of a role for genes such as MRTFB, MCAM, and SPDL1 that effect cell motility in tumor growth, a recent report by Waclaw et al. (57) describing a spatial model for analyzing the forces that shape tumor growth predicts that cell motility is critical for tumor growth, even at the initial stage of tumor development. Their model shows that even small localized cellular movements are able to markedly reshape a tumor. Moreover, the model predicts that the rate of tumor growth can be substantially altered by a change in the dispersal rate of the cancer cells, even in the absence of any changes in doubling times or net growth rates of cells within the tumor.
In summary, here we report MRTFB’s tumor suppressor role in CRC and identify a number of genes whose expression is deregulated in MRTFB knockdown cells. We also show that 2 of these genes, SPDL1 and MCAM, are bona fide CRC tumor suppressor genes and provide a number of potential mechanisms for explaining MRTFB’s role in CRC.
Materials and Methods
Mouse Strains, Management, and Phenotypic Analysis.
The generation of conditional Mrtfb knockout mice has been described previously (8). The conditional knockout of Mrtfb in mouse intestines was obtained using the Lrig1-Cre strain (21). A floxed allele of Apc (58) was also used as a sensitizing mutation in this study, given that ∼80% human CRC patients carry mutations in Apc. For tumor development observations, we used Lrig1Cre/+;ApcFlox/+;Mrtfb+/+ mice as the control group and Lrig1Cre/+;ApcFlox/+;MrtfbFlox/Flox mice as the experimental group, with the only difference being the knockout of Mrtfb. All mice were in a mixed but mostly C57BL/6J background. Since genetic background is important for intestinal tumor development, we compared Mrtfb knockout mice with control animals from the same mating cages, if it became too difficult for all animals to come from the same litters because we used 3 mutant alleles (Lrig1Cre, ApcFlox, and MrtfbFlox) in the study. Furthermore, we used more than 50 animals in both the Mrtfb knockout and control groups, which further minimized the background effect.
Lrig1-Cre was activated by intraperitoneal injection of tamoxifen (21). Tamoxifen was dissolved in corn oil (10 mg/mL) at 65 °C for 1 h with occasional vortexing. Mice at age 6 to 8 wk were injected once daily with 2 mg tamoxifen for 3 consecutive days. Control group and experimental group animals were euthanized for tumor observation at 4, 5, and 6 mo after Cre activation (i.e., on tamoxifen injection) or when they became morbid. Histology analysis was done for extracted tumors and paired normal tissues.
The Houston Methodist Research Institute’s (HMRI) Institutional Animal Care and Use Committee (IACUC) approved all procedures.
Cell Lines and Reagents.
All cell lines used in this study were obtained from the American Type Culture Collection. HCT116 and HT-29 cells were cultured in McCoy’s 5a medium, and SW480 and SW620 cells were cultured in DMEM. The media were supplemented with 10% FBS and penicillin-streptomycin. All cell lines were confirmed to be free of pathogens and mycoplasma.
siRNA Transfection.
As described previously (59), the Silencer Select siRNA oligonucleotides targeting MRTFB were purchased from Thermo Fisher Scientific (assay IDs: s33157, s33158, and s33159). siRNA transfection was performed using the Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Knockdown efficiency was determined by RT-qPCR at 3 d after transfection.
Cell Invasion and Cell Migration Assays.
Cell invasion and migration assays were done as described previously (4).For cell invasion assays, cells were first transfected with siRNA oligos, then 3 d later, siRNA-transfected cells were separated by trypsinization and counted. Approximately 5 × 105 cells resuspended in FBS-free medium were seeded into the upper chamber of the QCM 24-well cell invasion assay kit (EMD Millipore; ECM 554), and the lower chamber was filled with normal medium containing 10% FBS as a chemoattractant. Approximately 22 h later, the invaded cells were detached from the bottom of the membrane and lysed, followed by quantification by fluorescence measurement with the CyQuant GR dye provided in the assay kit. The readings of the tested genes’ siRNAs were all normalized to those of the nontargeting negative control siRNA.
For cell migration assay, at 3 d after siRNA transfection, cells were separated by trypsinization and counted. Approximately 1 × 105 cells resuspended in FBS-free medium were seeded into the upper chamber of the 24-well cell migration assay kit (Trevigen; 3465–024-K), and the lower chamber was filled with normal medium containing 10% FBS as a chemoattractant. Approximately 24 h later, the migrated cells were detached from the bottom of the membrane. Detached cells were prelabeled with Calcein-AM and quantified by fluorescence measurement with a 485-nm excitation wavelength and 520-nm emission wavelength. The readings of the tested genes’ siRNAs were all normalized to those of the nontargeting negative control siRNA.
Lentiviral Transduction for MRTFB shRNA.
The MRTFB shRNA-1 (clone ID: V3LHS_318184) and MRTFB shRNA-2 (clone ID: V3LHS_402308) lentiviral particles were purchased from Dharmacon. As described previously (59), adding polybrene to the culture medium enhanced the transduction of lentiviral particles. Drug selection (puromycin for shRNAs) was started at 2 d after transduction. Knockdown efficiency was determined by RT-qPCR at 7 d after transduction.
Xenograft Assay.
The xenograft assay was conducted as described previously (59). Cell suspensions were injected subcutaneously into 6- to 8-wk-old female athymic nude mice obtained from Charles River Laboratories. All procedures were approved by the HMRI IACUC. Each xenograft assay was performed in 5 mice (10 flanks) of each group. One million HCT116 or SW480 cells were used for injection into each flank for MRTFB shRNA knockdown experiments. Tumor growth was measured twice weekly using a vernier caliper. Two perpendicular diameters of tumors, length (L) and width (W), were determined, with L defined as the larger of the 2 measurements. Tumor volume was calculated as W × (W × (L/2)).
RNA Extraction and RT-PCR.
Total RNA was extracted from cultured cells or mouse tissues by using the Qiagen RNeasy Plus Mini Kit. Reverse transcription was done with 1 μg of total RNA using the Promega Reverse Transcription System. The QuantStudio 12K Flex Real-Time PCR System (Life Technologies) was used for RT-qPCR analysis with TaqMan probes (Life Technologies) of MRTFB, SPDL1, MCAM, PTGS2, BTG2, BBC3, BOD1, B3GALT6, DDIT4, CTPS1, GBX2, TMOD1, VWA5A, VPS36, and OXTR, with ACTB and Actb (Life Technologies) used as internal controls for RT-qPCR for human cells and mouse tissues, respectively.
Whole Transcriptome Sequencing.
Total RNA was extracted using the above-described method. The sequencing was done as described previously (59). Two siRNA control samples (WT_1 and WT_2) and 3 MRTFB siRNA samples (MRTFB_1, MRTFB_2, and MRTFB_3) were sequenced. To construct sequencing libraries, mRNAs were enriched from the total RNA using poly-T oligo-attached beads. Enriched mRNAs were then fragmented and first-strand cDNAs were synthesized using random hexamer primer and M-MuLV reverse transcriptase. Second-strand cDNAs were synthesized using DNA polymerase I and RNase H. Double-stranded cDNAs were purified using AMPure XP beads (Beckman Coulter). After adenylation of 3′ ends, NEBNext adaptor (New England BioLabs) was ligated for hybridization, then 150- to 200-bp cDNA fragments were selected using the AMPure XP system. The final library was obtained after PCR amplification and PCR product purification using AMPure XP beads.
After passing quality assessment, the library was diluted to 1.5 ng/μL for sequencing using the Illumina HiSeq-4000 system. Raw data processing and quality assessment were done based on error rate distribution, GC content, and removal of low-quality reads or reads with adaptors. Each sample obtained 90 to 110 million clean reads, 92% to 93% of which were mapped to the human reference transcriptome (hg10).
RNA-seq read counts were proportional to gene expression level, gene length, and sequencing depth. FPKM (fragments per kilobase of transcript sequence per million base pairs sequenced) was used for gene expression level analysis.
Western Blot Analysis.
For Western blotting, whole cell lysates were prepared with RIPA buffer containing RNase I, and protein concentration was measured with the Bradford method. Protein lysates were loaded into precast SDS/PAGE gels (Bio-Rad) for separation, and separated proteins were transferred onto PVDF membrane using the quick semidry method (Bio-Rad). Membrane was incubated with anti-MRTFB primary antibody (Bethyl Laboratories; A302-768A, 1:1,000 dilution) for overnight at 4 °C or for 2 h at room temperature. Blotting with anti–α-tubulin antibody (Sigma-Aldrich; T6199) was used as loading control.
Correlation of Patient Survival with Gene Expression.
The correlation analysis between patient survival and gene expression levels was done using the published public web server GEPIA (gene expression profiling and interactive analyses) (http://gepia.cancer-pku.cn/) (60). All patients included in the analysis had a primary colorectal tumor without metastasis to either lymph nodes or other organs. The expression levels of both MRTFB and SPDL1 were not significantly different among different CRC stages; therefore, tumor stage was not included in the correlation analyses for MRTFB/SPDL1 expression and CRC patient survival.
Statistics.
All data are presented as mean ± SD unless indicated otherwise. Statistical analyses were performed using the Student t test or 1-way analysis of variance in GraphPad Prism. Differences with P < 0.05 were considered statistically significant.
Data Availability Statement.
All data will be available on request from the corresponding authors. No public depository exists.
Supplementary Material
Acknowledgments
We thank other research groups at Houston Methodist Cancer Center for equipment and technical support; the mouse facility at the Houston Methodist Research Institute for high-quality animal care; and Dr. Robert J. Coffey (Vanderbilt University) for the Lrig1-Cre mice. This work was supported by the faculty startup fund from Houston Methodist (Z.W.), the Japan Agency for Medical Research and Development (JP19cm0106437, to T.K.)., and the Cancer Prevention Research Institute of Texas (N.G.C. and N.A.J.).
Footnotes
The authors declare no competing interests.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910413116/-/DCSupplemental.
References
- 1.Bray F., et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Mariotto A. B., Yabroff K. R., Shao Y., Feuer E. J., Brown M. L., Projections of the cost of cancer care in the United States: 2010-2020. J. Natl. Cancer Inst. 103, 117–128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda H., et al. , Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat. Genet. 47, 142–150 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Bard-Chapeau E. A., et al. , Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat. Genet. 46, 24–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann K. M., et al. ; Australian Pancreatic Cancer Genome Initiative , Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 109, 5934–5941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh J., Richardson J. A., Olson E. N., Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc. Natl. Acad. Sci. U.S.A. 102, 15122–15127 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J., et al. , Myocardin-like protein 2 regulates TGFβ signaling in embryonic stem cells and the developing vasculature. Development 139, 3531–3542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaposhnikov D., Kuffer C., Storchova Z., Posern G., Myocardin-related transcription factors are required for coordinated cell cycle progression. Cell Cycle 12, 1762–1772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampl V., et al. , Depletion of the transcriptional coactivators megakaryoblastic leukaemia 1 and 2 abolishes hepatocellular carcinoma xenograft growth by inducing oncogene-induced senescence. EMBO Mol. Med. 5, 1367–1382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greuber E. K., Smith-Pearson P., Wang J., Pendergast A. M., Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 13, 559–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim S. P., et al. , Specific-site methylation of tumour suppressor ANKRD11 in breast cancer. Eur. J. Cancer 48, 3300–3309 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Wu R. C., Wang T. L., Shih IeM., The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 15, 655–664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi S. H., Estarás C., Moresco J. J., Yates J. R. 3rd, Jones K. A., α-Catenin interacts with APC to regulate β-catenin proteolysis and transcriptional repression of Wnt target genes. Genes Dev. 27, 2473–2488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoda A., et al. , Mutations in G protein β subunits promote transformation and kinase inhibitor resistance. Nat. Med. 21, 71–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philp A. J., et al. , The phosphatidylinositol 3′-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 61, 7426–7429 (2001). [PubMed] [Google Scholar]
- 17.Law D. J., Labut E. M., Merchant J. L., Intestinal overexpression of ZNF148 suppresses ApcMin/+ neoplasia. Mamm. Genome 17, 999–1004 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Moriarity B. S., Largaespada D. A., Sleeping Beauty transposon insertional mutagenesis-based mouse models for cancer gene discovery. Curr. Opin. Genet. Dev. 30, 66–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copeland N. G., Jenkins N. A., Harnessing transposons for cancer gene discovery. Nat. Rev. Cancer 10, 696–706 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Newberg J. Y., Mann K. M., Mann M. B., Jenkins N. A., Copeland N. G., SBCDDB: Sleeping Beauty Cancer Driver Database for gene discovery in mouse models of human cancers. Nucleic Acids Res. 46, D1011–D1017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell A. E., et al. , The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cen B., Selvaraj A., Prywes R., Myocardin/MKL family of SRF coactivators: Key regulators of immediate early and muscle-specific gene expression. J. Cell. Biochem. 93, 74–82 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Selvaraj A., Prywes R., Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J. Biol. Chem. 278, 41977–41987 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Chen H. J., et al. , A recellularized human colon model identifies cancer driver genes. Nat. Biotechnol. 34, 845–851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G. J., Zeng G. F., METCAM/MUC18 is a novel tumor and metastasis suppressor for the human ovarian cancer SKOV3 cells. BMC Cancer 16, 136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D., et al. , Reduced CD146 expression promotes tumorigenesis and cancer stemness in colorectal cancer through activating Wnt/β-catenin signaling. Oncotarget 7, 40704–40718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai Q., et al. , Decreased expression of mucin 18 is associated with unfavorable postoperative prognosis in patients with clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 8, 11005–11014 (2015). [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng B., et al. , CD146 attenuation in cancer-associated fibroblasts promotes pancreatic cancer progression. Mol. Carcinog. 55, 1560–1572 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Anghel S. I., et al. , Breast cancer suppressor candidate-1 (BCSC-1) is a melanoma tumor suppressor that down-regulates MITF. Pigment Cell Melanoma Res. 25, 482–487 (2012). Correction: 25, 834. [DOI] [PubMed] [Google Scholar]
- 30.Sinha S., et al. , Frequent alterations of LOH11CR2A, PIG8 and CHEK1 genes at chromosomal 11q24.1-24.2 region in breast carcinoma: Clinical and prognostic implications. Mol. Oncol. 5, 454–464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhou Y. Q., et al. , Tumor suppressor function of BCSC-1 in nasopharyngeal carcinoma. Cancer Sci. 100, 1817–1822 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu G. J., Ectopic expression of MCAM/MUC18 increases in vitro motility and invasiveness, but decreases in vivo tumorigenesis and metastasis of a mouse melanoma K1735-9 subline in a syngeneic mouse model. Clin. Exp. Metastasis 33, 817–828 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Johnson J. P., Cell adhesion molecules in the development and progression of malignant melanoma. Cancer Metastasis Rev. 18, 345–357 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Xie S., et al. , Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 57, 2295–2303 (1997). [PubMed] [Google Scholar]
- 35.Barisic M., et al. , Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell 21, 1968–1981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chin L., Hahn W. C., Getz G., Meyerson M., Making sense of cancer genomic data. Genes Dev. 25, 534–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blum A., Wang P., Zenklusen J. C., SnapShot: TCGA-analyzed tumors. Cell 173, 530 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Hutter C., Zenklusen J. C., The Cancer Genome Atlas: Creating lasting value beyond its data. Cell 173, 283–285 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Muehlich S., et al. , The transcriptional coactivators megakaryoblastic leukemia 1/2 mediate the effects of loss of the tumor suppressor deleted in liver cancer 1. Oncogene 31, 3913–3923 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Dickson B. C., et al. , Ectomesenchymal chondromyxoid tumor: A neoplasm characterized by recurrent RREB1-MKL2 fusions. Am. J. Surg. Pathol. 42, 1297–1305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flucke U., et al. , Presence of C11orf95-MKL2 fusion is a consistent finding in chondroid lipomas: A study of eight cases. Histopathology 62, 925–930 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Smith E. C., et al. , MKL1 and MKL2 play redundant and crucial roles in megakaryocyte maturation and platelet formation. Blood 120, 2317–2329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R., Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11, 257–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katschnig A. M., et al. , EWS-FLI1 perturbs MRTFB/YAP-1/TEAD target gene regulation inhibiting cytoskeletal autoregulatory feedback in Ewing sarcoma. Oncogene 36, 5995–6005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Saghire H., et al. , Intensity modulated radiotherapy induces pro-inflammatory and pro-survival responses in prostate cancer patients. Int. J. Oncol. 44, 1073–1083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conte C., Baird M. A., Davidson M. W., Griffis E. R., Spindly is required for rapid migration of human cells. Biol. Open 7, bio033233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmenkivi K., Haglund C., Ristimäki A., Arola J., Heikkilä P., Increased expression of cyclooxygenase-2 in malignant pheochromocytomas. J. Clin. Endocrinol. Metab. 86, 5615–5619 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Ellisen L. W., et al. , REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10, 995–1005 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Du F., et al. , DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. Cancer Commun. (Lond) 38, 45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B., PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Duriez C., et al. , The human BTG2/TIS21/PC3 gene: Genomic structure, transcriptional regulation and evaluation as a candidate tumor suppressor gene. Gene 282, 207–214 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Rouault J. P., et al. , Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 14, 482–486 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Boiko A. D., et al. , A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 20, 236–252 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohun T. J., Chambers A. E., Towers N., Taylor M. V., Expression of genes encoding the transcription factor SRF during early development of Xenopus laevis: Identification of a CArG box-binding activity as SRF. EMBO J. 10, 933–940 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sartorelli V., Webster K. A., Kedes L., Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 4, 1811–1822 (1990). [DOI] [PubMed] [Google Scholar]
- 56.Sun Q., et al. , Defining the mammalian CArGome. Genome Res. 16, 197–207 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waclaw B., et al. , A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuraguchi M., et al. , Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2, e146 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long Y., Marian T. A., Wei Z., ZFR promotes cell proliferation and tumor development in colorectal and liver cancers. Biochem. Biophys. Res. Commun. 513, 1027–1034 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Tang Z., et al. , GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data will be available on request from the corresponding authors. No public depository exists.