Significance
Vertebrate vision relies on a specialized sensory cilium, the outer segment, and its cellular lifeline, the connecting cilium (CC). This study applies sample preparation methods developed for rod cells to 2 nanoscale imaging modalities, cryoelectron tomography (cryo-ET) and single-molecule localization (STORM) fluorescence microscopy, to understand these structures and their disruption in blinding ciliopathies. Cryo-ET maps of the CC and mother centriole reveal new details: Y-link structures, axonemal microtubule doublet twisting, and a central terminal plate-like structure. STORM reveals the subciliary localization of key molecular component resolutions inaccessible by conventional imaging. Intraflagellar transport (IFT) trains and other interaction partners for Bardet–Biedl syndrome (BBS) proteins are imaged with improved resolution in normal CCs and in mouse models of BBS.
Keywords: primary cilia, photoreceptors, ciliopathies, superresolution, cryoelectron tomography
Abstract
Primary cilia carry out numerous signaling and sensory functions, and defects in them, “ciliopathies,” cause a range of symptoms, including blindness. Understanding of their nanometer-scale ciliary substructures and their disruptions in ciliopathies has been hindered by limitations of conventional microscopic techniques. We have combined cryoelectron tomography, enhanced by subtomogram averaging, with superresolution stochastic optical reconstruction microscopy (STORM) to define subdomains within the light-sensing rod sensory cilium of mouse retinas and reveal previously unknown substructures formed by resident proteins. Domains are demarcated by structural features such as the axoneme and its connections to the ciliary membrane, and are correlated with molecular markers of subcompartments, including the lumen and walls of the axoneme, the membrane glycocalyx, and the intervening cytoplasm. Within this framework, we report spatial distributions of key proteins in wild-type (WT) mice and the effects on them of genetic deficiencies in 3 models of Bardet–Biedl syndrome.
Primary cilia have been intensively studied in recent years because of their importance in sensory signaling, development, and physiological homeostasis. A host of inherited defects in cilium or basal body structure and function cause ciliopathies, a group of human diseases whose clinical symptoms include mental retardation, polydactyly, obesity, cognitive defects, cystic kidneys, and blinding retinal degeneration (1).
Mammalian primary cilia contain hundreds of different polypeptide components (2–4). In most cases, their precise locations and functions are not known. They include molecules necessary for precisely regulated transport of molecules into and out of cilia, such as the intraflagellar transport (IFT) proteins of the IFT complexes, as well as numerous receptors, ion channels, transporters, and structural proteins.
Photoreceptors in the vertebrate retina contain specialized sensory cilia known as outer segments (OSs) (5), cylindrical structures roughly 20 μm in length and 1.5 μm in diameter, packed with membrane disks containing proteins of the phototransduction cascade. They connect to the inner segment (IS) of the cell by a short (∼1.1-μm) connecting cilium (CC), which contains an axoneme extending from a basal body (BB) complex in the IS, and resembles the transition zone of other cilia (6). A pericentriolar region surrounds the BB and the membrane adjacent to primary cilia forms a ciliary pocket, which in photoreceptors takes the form of a periciliary membrane that extends along 1 side of the CC (7).
The microtubule doublets of the axoneme within the CC are surrounded by a 50-nm layer of cytoplasm and enclose a 150-nm central lumen for a total CC diameter of ∼300 nm (8). The requirement for daily renewal of ∼10% of the OS membrane mass through transport up the CC (9) and the high metabolic demands of phototransduction place stringent demands on this structure, as evidenced by the number of ciliopathies which lead to retinal degeneration involving progressive rod cell death and blindness (10). Understanding the structure and organization of the CC is important for understanding these blinding diseases. The CC serves as an informative model for mammalian primary cilia and is particularly accessible to studies of in vivo structure and function.
Two groups of supramolecular complexes important for photoreceptor function and health and in transport mechanisms conserved among eukaryotic cilia are the IFT complexes and the BBSome complex formed by proteins associated with Bardet–Biedl syndrome (BBS). BBS is a retinal ciliopathy caused by mutations to over a dozen BBS genes. The BBSome is a large protein complex composed of 8 BBS ciliary proteins: BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9, and BBS18 (11, 12), whose formation requires essential BBS chaperone proteins including BBS17/LZTFL1 (leucine-zipper transcription factor-like 1) (13). Bbs mutant mouse models display progressive retinal degeneration, with loss of most photoreceptors by 6 mo of age (14, 15). Rod cell death is preceded by defects in protein localization and aberrant subcellular morphology. In Bbs4−/− mice at early stages, 35-nm vesicles accumulate within the lumen and near the base of the CC (8). In Bbs1M390R and Lztf1l/Bbs17−/− mutants, aberrant membranes were observed in the OS near the distal end of the CC (13).
The IFT-A and IFT-B transport complexes form large “trains” that bind to microtubule motor proteins kinesin-II and dynein to traverse the microtubules of the axoneme and deliver cargo proteins to or retrieve them from primary cilia (16). The retinal degeneration observed in Ift88/Tg737 mutant mice reveals their importance in rod cilia (6). Live-cell imaging of primary cilia in cultured cells revealed the BBSome moving bidirectionally along the axoneme at the same rate as IFT proteins (12), suggesting BBSome–IFT interactions and shared trafficking mechanisms.
The functions of the BBSome and other ciliopathy-associated complexes in CCs depend on their precise locations. A major limitation in determining their subciliary locations has been the lack of an accurate map of the nanoscale molecular organization of the CC and surrounding regions. Confocal immunofluorescence microscopy has limited ability to reveal subciliary distributions because the entire 300-nm width of the CC is only slightly wider than the ∼250-nm FWHM of the narrowest point-spread function practically achievable with a confocal microscope (17). Conventional electron microscopy (EM) has provided a somewhat fuzzy picture of the ultrastructure of the CC (5), confounded by the ambiguity imposed by multiple contrast mechanisms in samples stained with heavy-metal salts and by structural distortions introduced by fixation, embedding, and ultrathin sectioning.
Two complementary nanoscopy techniques that overcome many of these limitations are cryoelectron tomography (cryo-ET) and superresolution single-molecule localization fluorescence. Cryoelectron tomography has provided insights into the rod cell and CC structure (8, 18), but previous studies have not made use of the dramatic improvements in signal-to-noise and resolution that can be obtained through the technique of subtomogram averaging (19). This technique has been applied with great success to eukaryotic flagella (20–22), which are motile cilia, and to centrioles (23) but is just beginning to be applied to mammalian primary cilia (5).
Superresolution fluorescence nanoscopy makes it possible to localize specific proteins at resolutions well below the diffraction limit by immunofluorescence. Stochastic optical reconstruction microscopy (STORM) uses antibodies conjugated to organic “photoswitching” fluorophores that cycle from a dark state to an active emitting state, and switch off again, repetitively. In STORM, many thousands of these random single-molecule photoswitching events are captured and each of their images is fit to a Gaussian profile whose center defines the molecule location with high precision in the x–y dimensions that are collectively used to generate reconstruction maps (24).
In this work, we have used cryoelectron tomography and subtomogram averaging to define the 3-dimensional (3D) architecture of the basal connecting cilium and mother centriole. We combined the results with STORM imaging to define the radial subcompartments of the CC via subdiffraction reconstruction of immunotargets. The molecular constituents of the CC were reconstructed into 4 distinct layers, and localization details were revealed within these layers, including the IFT trains and their partial colocalization of the BBSome as protruding complexes in the connecting cilium. Finally, STORM imaging was used to determine the subciliary effects of BBS mutations on the organization and localization of the rod cilium and its components.
Results
Structure of the Connecting Cilium by Cryo-ET and Subtomogram Averaging.
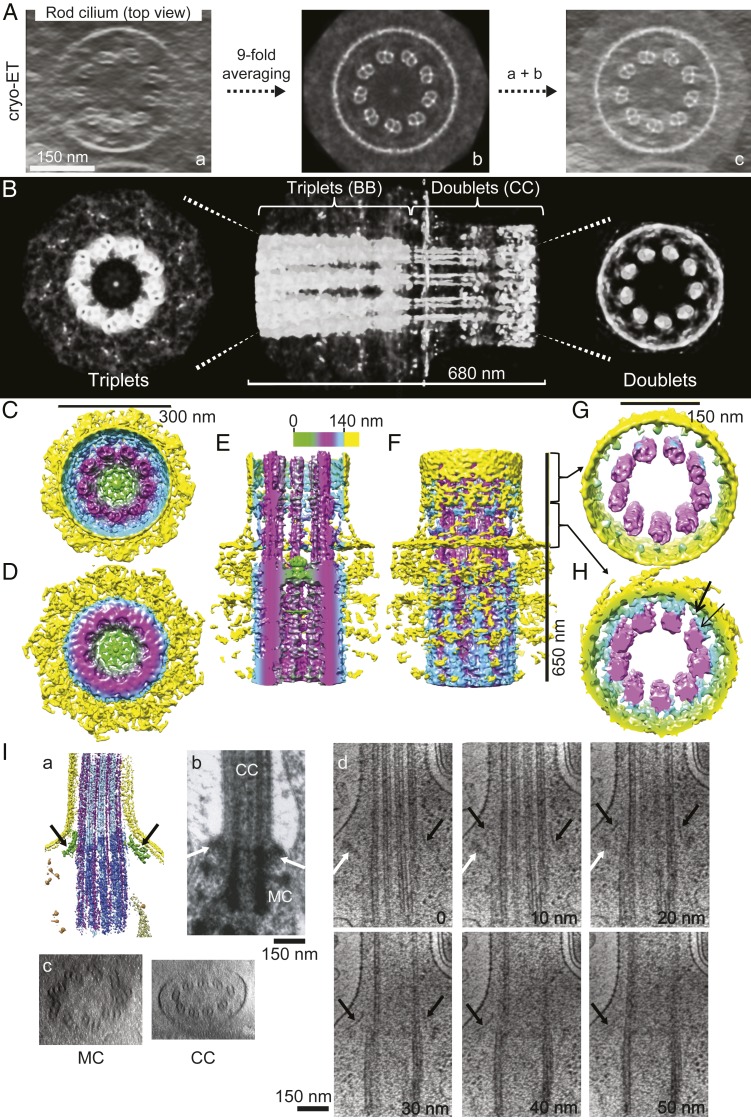
To determine the repeating structural features of the CC and define the subdomains’ geometries, we collected cryo-ET data on rods as described previously (5, 8, 25) and selected an unflattened region of the basal 1/3 of a single CC for subtomogram averaging, based on 9-fold symmetry, using an approach previously used to refine the structure of a mouse rod daughter centriole (5). The resulting map (26) (Fig. 1 A–H) showed good signal to noise and many clearly resolved nanometer-scale features from the base of the mother centriole through the entire length of the selected region of the axoneme. These include the microtubule triplets of the centriole, the microtubule doublets of the axoneme, the ciliary membrane, and connections between the microtubules and the membrane. One interesting observation is a structure in the axoneme lumen at the junction between triplets and doublets, which is reinforced by 9-fold averaging, and which resembles the terminal plate of motile cilia (Fig. 1 C–E, green). There are numerous filamentous and planar structures emanating from the mother centriole of the basal body, which are reinforced by 9-fold averaging; these bear little resemblance to the pinwheel-like or weather vane-like structures referred to as “distal appendages,” “transition fibers,” or “alar sheets” in conventional transmission electron microscopy (TEM) images of other basal bodies (e.g., Fig. 1 I, b), but likely correspond to transition fibers.
Fig. 1.
Structure of the connecting cilium and basal body by cryo-ET and subtomogram averaging. (A) Views down the CC axis of the tomogram used for 9-fold averaging. (A, a) Before averaging. (A, b) After 9-fold averaging. (A, c) a and b superimposed. (B) Volume-rendered representations of the final map after 9-fold averaging and 15-nm low-pass filtering. (C–F) Surface representations of the entire map (C, D, and F) or a portion sectioned along the ciliary axis (E) from the side. (C) Top view (CC end). (D) Bottom view (BB end). (E and F) Side view. Coloring is by radius as shown. The terminal plate is the green structure in the lumen at the boundary between the triplet microtubules of the mother centriole and the doublets of the CC axoneme (C–E). (G and H) Surface representations of the transition zone of the CC. In the more distal portion (G), no 9-fold symmetric connections are seen between the microtubules and the membrane. In the more basal portion (H), there are 2 distinct types of connections between the axoneme and plasma membrane at lower (thin arrow) and more distal (thick arrow) axial positions. (I) Views of the CC–BB junction without averaging. Black arrows show features likely corresponding to transition fibers or distal appendages. White arrows show features routinely observed extending in an arc away from the microtubule triplet–doublet junction. (I, a) A surface rendering of a segmented version of a typical tomogram, with these features highlighted in green. (I, b) A conventional TEM region of the same region for comparison. (I, c and d) Volume-rendered sections through another typical tomogram (different from those in A, H, and I, a). (I, c) End-on views up the axis of the mother centriole (MC) or down the axis of the CC axoneme (CC); both show flattening, which precluded subtomogram averaging. (I, d) Side views of successive 10-nm sections through the mother centriole and CC. (Scale bars: A, 150 nm; B, 680 nm; C and D, 300 nm; E and F, 650 nm; G and H, 150 nm; I, 150 nm.)
In maps not subjected to 9-fold averaging, there are structures associated with the mother centriole near the base of the axoneme (Fig. 1I). The strongest and most continuous feature begins near the axoneme base and slopes away from the CC and the centriole with a slight curve (Fig. 1I, black arrows). Nearby, there are arrangements of clumps of density that extend in a roughly triangular shape away from the basal body (Fig. 1I, white arrows). Although these are much less dense than strong features such as the microtubules or the curved structure, it is likely that these appear much darker in conventional TEM images, such as Fig. 1 I, b, due to disproportionate accumulation of stain relative to their mass, giving rise to the distal appendages.
There are also connections between the basal region of the axoneme and the ciliary membrane that display 9-fold symmetry, and likely correspond to the structures commonly referred to as “Y-shaped links” or “champagne glass structures” (Fig. 1H). Although these do have a Y-shaped appearance in certain projection views, they actually consist of 2 different types of connections, each type emerging from the axoneme at a different angle and appearing at different axial positions.
The symmetrized map (Fig. 1 A–H) reveals several subciliary structural domains. These include the mother centriole, containing triplet microtubules and a lumen, along with protruding appendages; the central structure at the boundary between the triplet microtubules of the centriole and the doublet microtubules of the CC; and the axoneme composed of microtubule doublets, the lumen of the axoneme, the volume between the axoneme and the ciliary membrane, the membrane itself, and the glycocalyx extending from it. This last feature is not visible in our map, likely due to its lack of 9-fold symmetry and relatively low electron density. These clearly demarcated structural domains provide a framework for interpreting immunolocalization results in the context of a 3D model.
A feature of this structure not readily apparent in conventional TEM images is the difference in geometries between the a and b microtubules of the centriole and those of the axoneme. The inner diameter of the ring formed by the a microtubules contracts from 176 nm at the base of the centriole to 136 nm in the axoneme, with a 5° inward rotation of the line connecting the centers of the a and b microtubules. The microtubules are not perfectly straight. As can be seen in Fig. 1F, there is a 16° right-hand twist of the a and b microtubules along the 650-nm length of the centriole and axoneme in the map.
Superresolution Fluorescence Nanoscopy of Rod Cilium Domain Markers.
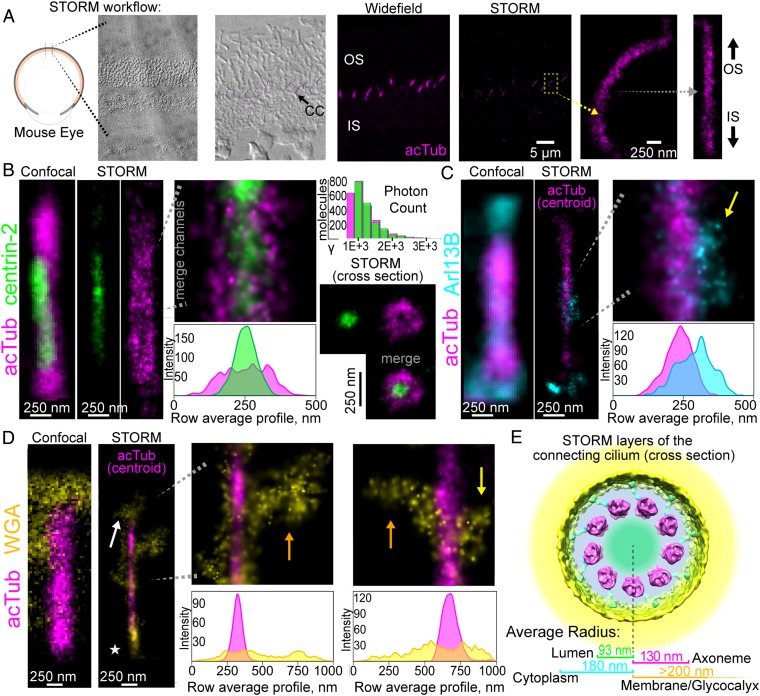
In order to define subdomains in molecular terms, we tried several approaches to acquiring STORM data from immunostained mouse rods, beginning with a preparation used previously to determine structural features of the rod cilium by cryoelectron tomography (8, 25), and including methods we used previously for immunolocalization in retinal whole mounts (8, 25), with a variety of fixation and membrane permeabilization methods. These all suffered from either poor photon yield or poorly preserved cellular morphology. We eventually developed a protocol adapted from one used previously to image retinal interneurons (27) in which retinas are labeled with organic dye-conjugated antibodies, followed by embedding and sectioning with an ultramicrotome, and etching with sodium ethoxide. This procedure yields observable photoswitching with reasonable photon yields and good preservation of cell morphology. Two different types of reconstructions were generated. Using stringent filtering parameters to exclude nearly all overlapping “blinking” events (2 molecules with simultaneously overlapping diffraction-limited images), we generated true STORM single-molecule maps of labeled antigens with nominal localization accuracies of ≤30 nm (Fig. 2B). For marking the central axis of the CC, we used antibodies specific for 2 antigens whose localization is well-characterized within the cilium: acetylated alpha-tubulin (acTub), the major component of the microtubule doublets of the axoneme, and centrin-2, a calcium-binding protein known to occupy the lumen of the axoneme (28) (Fig. 2B). Once the true distributions of our standard markers for the axoneme, acTub and centrin-2, were established, we generated reconstructions of these with reduced stringency that included both single- and double-molecule events and additional events above and below the plane of focus. These reconstructions provide very accurate representations of the central axis of the CC with higher sampling frequency than those from more stringent filtering (SI Appendix, Fig. S1C) and are labeled “centroid” in the figures. These were used for computational straightening and as reference axes for other antigens (see SI Appendix, Materials and Methods for further details). Although the stringent reconstructions yield precise locations of individual molecules, the resolution of their depiction of the cilium structure is limited by the size of antibody labeling complexes, flattening during sample preparation, and sampling frequencies of true single-molecule events.
Fig. 2.
STORM reconstruction of 4 distinct layers of the rod connecting cilium. (A) STORM workflow. A differential interference contrast image of the retina is superimposed on a STORM reconstruction of acetylated alpha tubulin to show the CC location. The STORM image is compared with a wide-field image. A single CC acTub reconstruction is isolated (yellow arrow) and straightened (gray arrow). All CCs are oriented with the top directed toward the outer segment. (B–D) Reconstructions of clusters for subdomain markers in 2-color STORM experiments: acTub (magenta) to mark the axoneme; centrin-2 (green; B) to mark the lumen; Arl13B (cyan; C) to mark the ciliary cytoplasm; and wheat germ agglutinin (yellow; D) to mark the ciliary membrane and glycocalyx. Confocal images of CCs immunostained with the same antibodies are shown adjacent to the STORM reconstructions. Centroid acTub reconstructions that accurately define the CC centers are superimposed onto single-molecule STORM reconstructions of Arl13B in C and WGA in D. In B, a histogram of photon counts per event is presented for the adjacent acTub (magenta) and centrin-2 (green) reconstructions. (B, Bottom Right) STORM reconstruction of a CC cross-section. Row average intensity profiles are below corresponding STORM images. (E) Diagram depicting STORM localization results in a cross-sectional view of the CC cryo-ET map, showing average radial locations from STORM images for each CC layer. Yellow arrows indicate STORM clusters in CCs. Orange arrows indicate wide WGA STORM clusters in CC. White arrow indicates a WGA STORM cluster in the OS. The star indicates a basal body/pericentriolar region.
Based on the inside diameter of the axoneme, determined by cryoelectron tomography to be 156 nm, and a nominal STORM localization error of ∼25 nm, we might expect the profile of the luminal centrin-2 staining to have a profile with a width of ∼181 nm. Indeed, the observed width (distance between profile positions with amplitudes of 1/e times the maximum) is 187 ± 15 nm (Fig. 2B and SI Appendix, Fig. S1B; n = 17; see SI Appendix, Table S1 for all experimental replicate information). This result leads to 2 striking conclusions: 1) Localization can be determined with resolutions of tens of nanometers, and 2) centrin-2 is confined to the central region of the axoneme and is not detectable beyond the microtubules. This narrow range suggests that centrin-2 may be a component of the proteinaceous particles previously observed within the central lumen of the axoneme by cryoelectron tomography (8). Not surprisingly, the acTub signal has a wider profile, consistent with it forming the microtubules surrounding the lumen. The average width is 260 ± 37 nm (from all experiments in Figs. 2–4 and SI Appendix, Figs. S1–S3; n = 73), consistent with a structure 200 nm wide, labeled with both a primary and secondary antibody that could extend beyond the surface of the microtubules by as much as 20 nm on either side and be subject to moderate flattening. In cross-sectional views (e.g., Fig. 2B, Bottom Right), the central location of centrin-2 within the acTub staining is apparent. The distributions of photon counts per single-molecule event are presented for the stringently filtered acTub and centrin-2 STORM reconstructions in Fig. 2B.
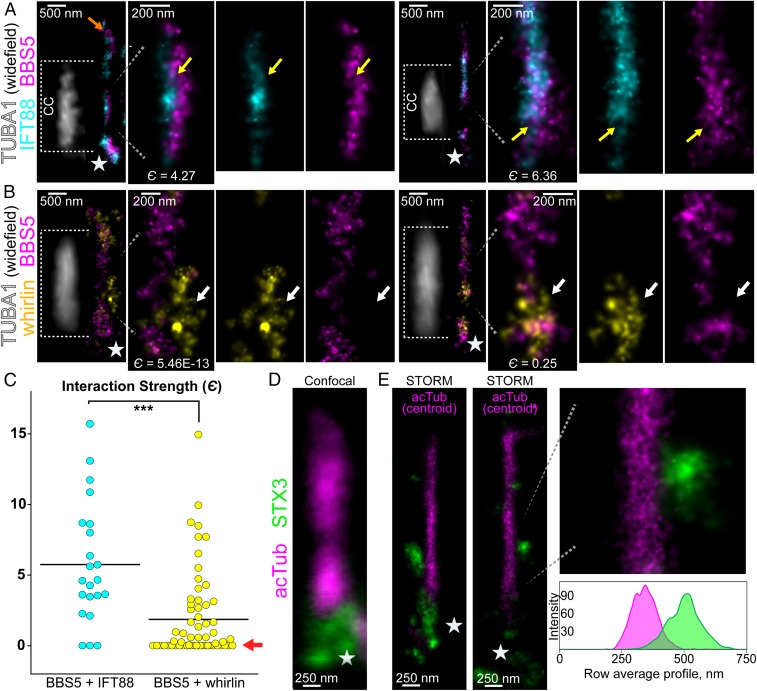
Fig. 4.
Two-color localization of BBS5 with IFT88 or whirlin, and localization of syntaxin-3 in CCs. (A) Isolated connecting cilia from 2-color STORM of IFT88 (cyan) and BBS5 (magenta), with adjacent wide-field fluorescence of α-tubulin 1 (TUBA1) staining (white) from the same region to mark the CC. See SI Appendix, Fig. S4 for a full-retina view of this staining. Dashed lines indicate the linear extent of CCs corresponding to each TUBA1 profile and magnified CC regions showing colocalized IFT88 and BBS5 clusters. The orange arrow indicates a STORM cluster in the OS. Yellow arrows indicate overlapping/colocalized BBS5 and IFT88 clusters. (B) Two examples of isolated CCs with 2-color STORM of whirlin (yellow) and BBS5 (magenta). TUBA1 image and dashed lines are as in A. The interaction strength is listed under the reconstructions. White arrows indicate nonoverlapping BBS5 and whirlin clusters. (C) Plot of the interaction strengths for all reconstructed CCs in BBS5 + IFT88 (cyan, n = 23) and BBS5 + whirlin (yellow, n = 60) experiments. ***P < 0.001, unpaired t test. Black bars indicate the mean є for each group. Fractions of clusters with “no interaction” (red arrow): BBS5 + IFT88 3/23 (13%); BBS5 + whirlin 28/63 (44.4%). (D) Confocal image of a CC immunostained for acetylated α-tubulin (magenta) and syntaxin-3 (green). (E) STORM reconstructions in the rod CC for STX3 (single molecule) and acTub (centroid), with a row average profile underneath. White stars indicate basal body/pericentriolar regions.
The small GTPase Arl13B, a cilium protein with a less well established subciliary localization within the photoreceptor connecting cilium (29), served as a marker for the ciliary membrane and the cytoplasmic domain outside the axoneme (Fig. 2C and SI Appendix, Fig. S1 D and E). The signal from this antigen was much less uniform, and extended well beyond the edge of the axoneme, consistent with association of this palmitoylated protein (30) with the ciliary membrane. Arl13B is not restricted to a subdomain of the CC but is found throughout its length. However, unlike the centrin-2 and acTub labeling, and in contrast to its appearance at confocal resolution (e.g., Fig. 2C, Left), the signal was not distributed uniformly along the length of the CC, and appeared primarily in clusters at irregular spacings along the long axis. A similar pattern for Arl13B was also observed via superresolution imaging in other primary cilia (31, 32). This observation of clustering has important implications for Arl13B function in the CC, suggesting its association with large sparse complexes along the ciliary axis. It has been reported to act as a guanine nucleotide exchange protein for a related small GTPase, Arl3 (33, 34), which in turn plays a critical role in releasing farnesylated proteins from complexes with PDE6δ/prenyl-binding protein during ciliary trafficking. In addition to the consistently strong labeling of the CC, Arl13B signal was also seen in the proximal outer segments, with a diffuse pattern not confined to the region near the axoneme (orange arrows in SI Appendix, Fig. S1 D and E).
To assess the signal for a marker that should be external to the ciliary membrane, we used the lectin wheat germ agglutinin (WGA) to label the glycocalyx of the cilium. In previous immuno-EM studies, WGA–gold conjugates strongly labeled an irregular glycoconjugate cloud that partially enveloped the CC of rods (35). The STORM reconstruction of WGA was more than 700 nm wide in some cases, consistent with a glycocalyx extending hundreds of nanometers beyond the ciliary membrane (Fig. 2D and SI Appendix, Fig. S1F).
Localization of IFT Complexes.
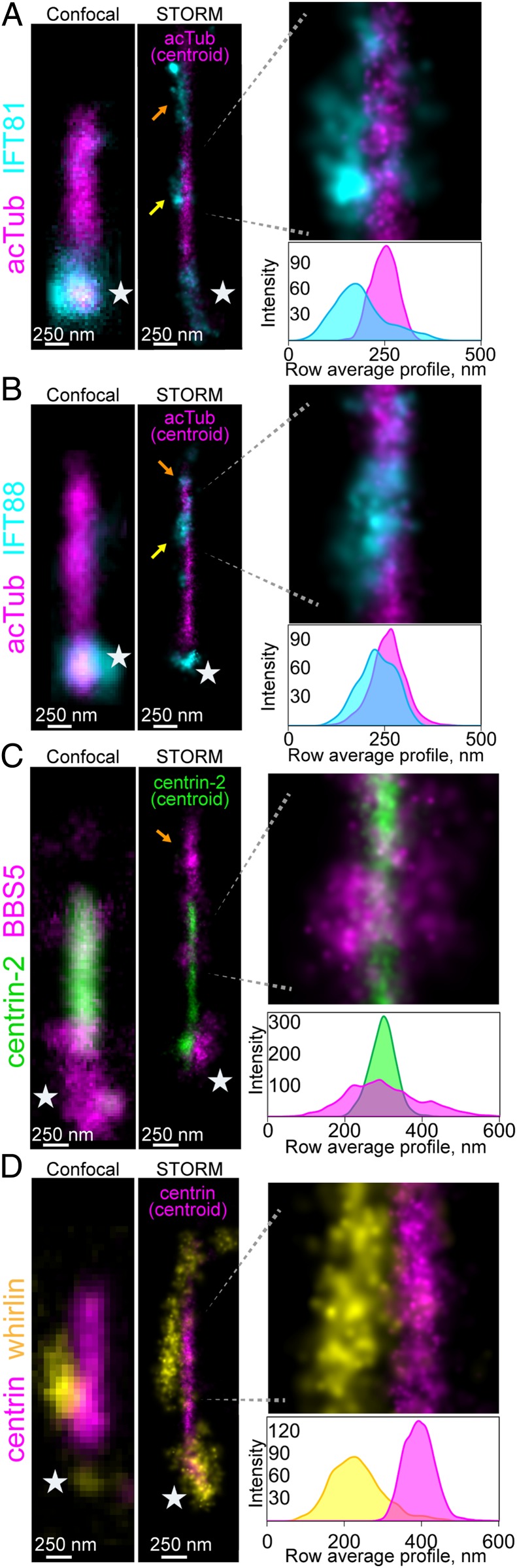
IFT81 and IFT88 proteins were targeted as subunits of the highly conserved core of the IFT-B primary cilium complex. With conventional fluorescence microscopy in rods, IFT protein signal was generally concentrated at either end of the CC with less clear localization along the CC axoneme (6), whereas IFT88 subcellular localization via immuno-EM was prominently along the outer face of the axoneme microtubules (36), consistent with IFT81 being a component of an IFT tubulin-binding module (37). Both kinesin-II, the anterograde motor protein for the IFT complex, and dynein, the retrograde motor, have been similarly immunolocalized to the CC (38, 39). We observed a very bright signal around the BB for IFT81 and IFT88, as well as along the axoneme in the OS. Multiple IFT clusters were observed along the CC (Fig. 3 A and B and SI Appendix, Fig. S2 B and C), and these STORM clusters were somewhat larger and less numerous than the clusters observed for Arl13B but were located within the same radial domain. Averaged along the length of the CC, the average radii were 150 ± 30 nm (IFT81, n = 9) and 154 ± 32 nm (IFT88, n = 26). These radial localizations are in the range of those observed with Arl13B (180 ± 51 nm, n = 23), consistent with a reported in vitro Arl13B–IFT-B complex interaction (40). The large size of the IFT81 and IFT88 STORM clusters suggests they likely represent IFT particles, or trains, and in some cases, these are very closely associated with the surface of the axoneme (Fig. 3B); in others, they fill the radial span between the axoneme surface and the ciliary membrane (Fig. 3A), consistent with immunoelectron microscopy results showing IFT staining of the axoneme and the ciliary membrane (36).
Fig. 3.
Superresolution localization of IFT proteins, BBS5, and whirlin. Example STORM reconstructions in the rod CC for (A) acTub (magenta) and IFT81 (cyan), (B) acTub (magenta) and IFT88 (cyan), (C) centrin-2 (green) and BBS5 (magenta), and (D) centrin (magenta) and whirlin (yellow). Centroid reconstructions for each antigen marker (acTub in A and B, centrin-2 in C, and centrin in D are superimposed to mark the CC centers). For each antigen reconstructed with STORM at the CC, a confocal image of an identically immunostained CC is shown at the same scale for comparison. Magnified regions are presented with corresponding row average profiles aligned directly beneath. Yellow arrows indicate STORM clusters in CCs. Orange arrows indicate STORM clusters in OSs. Stars indicate basal body/pericentriolar regions.
Localization of the BBSome and Periciliary Membrane Proteins.
To localize the BBSome, we relied on a monoclonal antibody specific for the BBSome subunit BBS5, whose localization within the CC is unresolved (41). Out of many antibodies for BBS subunits we tested, this was the only one whose staining indicated good specificity with consistently reliable results under our labeling and imaging conditions and which we were able to confirm with knockout retinas (SI Appendix, Fig. S8). STORM reconstructions with a BBS9 antibody also yielded specific patterns (which we have not validated with knockouts), but which matched and overlapped with the BBS5 localization pattern at the CC (SI Appendix, Fig. S2 E and F). To correlate BBS5 signal with the defined radial domains, we colabeled for centrin-2. As shown in Fig. 3C and SI Appendix, Fig. S2D, BBS5 in the wild-type (WT) retina is located in 3 subcellular regions at different levels, with very intense staining at the basal body complex (white stars), somewhat less intense staining along the axoneme in the outer segment (orange arrows), and less intense but consistently detectable clusters in the CC volume outside the axoneme. Somewhat surprisingly, the signal from these CC clusters extends in many cases considerably more than 200 nm from the center of the axoneme marked by “centroid” reconstructions of centrin-2 (mean BBS5 radius 231 ± 57 nm, n = 22), so that BBS5 signal extends beyond the nominal 300-nm width of the CC, even if the extra ∼40-nm width introduced by the antibodies and the localization uncertainty is taken into account. These appear to represent localized protrusions of the membrane into which the BBSome is inserted, although contributions from adjacent periciliary membrane cannot be ruled out (see below). Local membrane distortions would be consistent with the observation that the BBSome is able to form a coat on the surface of phospholipid bilayers (42). In contrast to the IFT proteins which are very closely associated with the BB, BBS5 staining near the BB is more diffuse, suggesting its association with the pericentriolar material.
Due to the radial extension of the BBSome and IFT proteins from the CC axoneme, we next considered the nearby localization of the periciliary pocket membrane (SI Appendix, Fig. S3A). This membrane extension of the IS, which envelopes a portion of the CC, is linked to the adjoining CC membrane by the periciliary adhesion membrane complex (PMC) composed of proteins associated with Usher syndrome (7). Many of the PMC proteins are also localized within the CC cytoplasm, including myosin VII, SANS, and whirlin. Together, these scaffold the transmembrane receptor proteins of the PMC, including VLGR1 (very large G protein-coupled receptor 1), which maintain the periciliary–CC membrane interface (43). With STORM, we localized the PMC in rod cells by staining for whirlin, which we reconstructed as large clusters that show very infrequent overlap with centrin (Fig. 3D and SI Appendix, Fig. S3B); these clusters had a mean radius of 276 ± 75 nm (n = 24), marking the extension of periciliary localized proteins. We confirmed this segregated pattern of localization for PMC by also staining for VLGR1, which was also localized to 1 side of the CC (SI Appendix, Fig. S3C).
Colocalization of the BBSome with IFT and Periciliary Proteins.
To determine the extent of colocalization of BBS5 with other proteins within the CC or with periciliary pocket membrane proteins, it was necessary to include a third label for alpha-tubulin 1 (TUBA1) to mark the CC and distinguish these proteins within the CC from their distinct populations in the inner and outer segments. As our experimental setup precluded simultaneous STORM imaging of the third label, we superimposed wide-field images of TUBA1 staining on 2-color superresolution reconstructions of BBS5 with either IFT88 (Fig. 4A and SI Appendix, Fig. S4 A–C) or whirlin (Fig. 4B and SI Appendix, Fig. S4D). The results for BBS5 + IFT88 reveal that there are membrane protrusions at the CC containing colocalized IFT88 and BBS5 clusters (Fig. 4A), and others containing only one or the other (SI Appendix, Fig. S4C). The results for BBS5 + whirlin reveal that in the majority of rod CCs analyzed, the BBS5 and whirlin molecule clusters are nonoverlapping and are not apparently contiguous (Fig. 4B), indicating that they are not colocalized. The extents of STORM colocalization in these images were quantified with a spatial interaction analysis performed using the estimated molecule coordinates from each imaging channel. The interaction potentials from these coordinate patterns generate an interaction strength parameter (є) to quantify the degree of interaction or colocalization. Supporting our STORM image observation, the average interaction strength is significantly greater for BBS5 + IFT88 compared with BBS5 + whirlin (Fig. 4C), and a larger percent of total cilia with no interaction strength (defined as a strength <0.01; red arrow in Fig. 4C) was observed for BBS5 + whirlin (28/63, 44.4%) cilia vs. BBS5 + IFT88 (3/23, 13.0%).
Thus, there are at least 4 distinct populations of BBS5 in rod cilia: One associated with the centrioles and pericentriolar material, a second associated with the axoneme in the outer segment, a third within the CC colocalized with and possibly participating in complexes with IFT particles containing IFT88, and a fourth present in the CC in regions not containing IFT88. The low interaction score of BBS5 with the PMC marker whirlin implies that BBS5, and by extension the BBSome, does not preferably localize to the periciliary pocket membrane. This diversity of locations and distributions supports proposed roles for the BBSome in dynamic trafficking of cargo throughout the CC in cooperation with IFT particles and is consistent with continual movement of BBSome subunits observed in live-cell imaging. When BBS5 clusters were adjacent to IFT88 clusters, they were consistently patterned at the edge of IFT88 clusters and extended away from the CC as if surrounding the IFT88 clusters. This pattern is consistent with a model for transport machinery at the CC in which BBS5/the BBSome interacts with the IFT complex as part of a larger transport complex with a molecular arrangement of IFT proteins near the axoneme microtubules via motor proteins that are surrounded by BBSome complexes which extend out into the cytoplasm and to the CC membrane.
Localization of Syntaxin-3.
Syntaxin-3 (STX3) is a tSNARE transmembrane protein whose retinal expression is predominantly in the IS of rod cells (SI Appendix, Fig. S4F), including the region surrounding the BB at the base of the connecting cilium in rodent rods (44). Furthermore, STX3 is proposed to be important for rhodopsin vesicle trafficking (44) and is severely mislocalized in Bbs mutant rods (13, 45) (see below; SI Appendix, Fig. S5B), consistent with a model for STX3 localization requiring a fully functional BBSome for retrograde trafficking in the CC.
In STORM images of the CC, clusters are observed for STX3 in 3 major locations (Fig. 4 D and E and SI Appendix, Fig. S3D): extensive signal at the BB (white stars) and pericentriolar region, large punctate clusters along the CC membrane, and staining occasionally observed near the axoneme in the OS (orange arrows in SI Appendix, Fig. S3). The STX3 clusters at the CC are more compact than those observed for IFT proteins and BBS5 and are variable in size; as measured along the longitudinal axis of the CC, the average length was 490 ± 94 nm (n = 10). The average radius of these STX3 clusters was 275 ± 76 nm (n = 28) from the CC center. Thus, along the CC, STX3 was localized as large and cohesive STORM clusters which were located in variable and likely transient positions at the exterior CC membrane STORM layer, possibly as membrane transport structures in transit between the IS and OS. Interestingly, we also found rare examples of CCs in which we observe a significantly larger gap between STX3 clusters and the acTub axoneme; these STX3s are localized radially ∼500 nm from the CC and potentially lie within the periciliary inner segment or an adjacent inner segment (SI Appendix, Fig. S3E).
Effects of Bbs Gene Deficiencies on Localization.
The 8 BBS proteins of the BBSome (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, BBS9, and BBS18) are expressed primarily as a full complex in rod sensory cilia as in ciliated mammalian cells; however, select BBSome subunits can form stable subcomplexes upon coexpression and during complex assembly (12), leading to the possibility of functional BBSome subcomplexes in rod cilia. We immunolabeled BBS5 for STORM reconstruction in rod cells from Bbs2−/−, Bbs4−/−, and Bbs7−/− mutant mouse models of BBS retinal degeneration to determine if BBS5 and the remaining incomplete BBSome are mislocalized in these mutant backgrounds.
For these STORM experiments in mouse mutants with degenerating retinas, ages between 5 and 8 wk were selected as time points when enough rod cells remain intact for sufficient sampling size yet there has been sufficient aging to visualize substantial protein accumulation or mislocalization defects. Bbs2−/− mutant mice have a rate of photoreceptor degeneration comparable to Bbs4−/− (14), in which apoptosis was not apparent until 6 wk, whereas apoptosis was not measured in Bbs7−/− mice (15). By 8 wk, all Bbs mutant retinas demonstrated evidence of photoreceptor degeneration through retinal thinning; however, all mutant retinas still maintained numerous intact rod cells in the photoreceptor cell layer (SI Appendix, Fig. S4E).
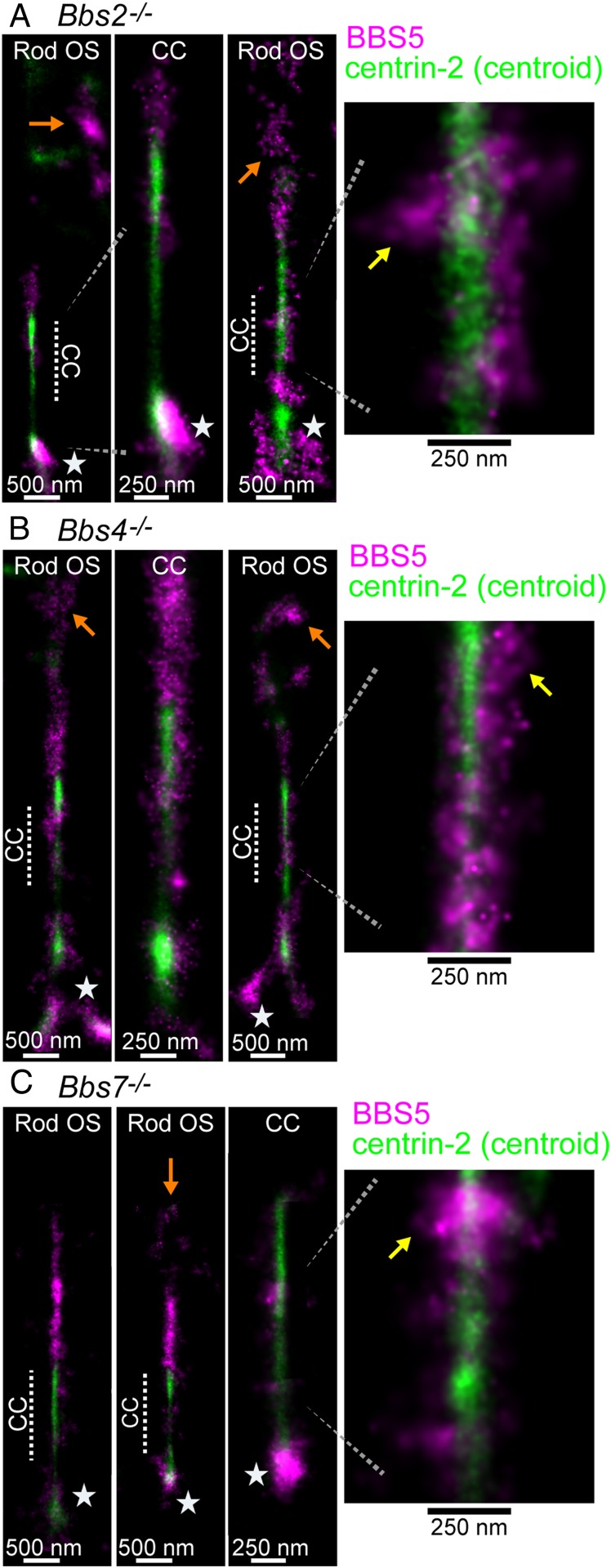
Surprisingly, the distribution of BBS5 among ISs, OSs, and CCs was not severely altered in any of the 3 Bbs mutant mouse strains compared with WT and heterozygous controls (compare Figs. 3C and 5 and SI Appendix, Figs. S2C and S6). Mislocalization within the OS was observed. In wide-field fluorescence images, small abnormal BBS5 puncta were observed in the OS layer of the mutant retinas that were not present in wild-type and heterozygous controls (SI Appendix, Fig. S5A) and, in STORM reconstructions, BBS5 misaccumulation away from the axoneme is evident in the OS (Fig. 5 and SI Appendix, Fig. S6) of the mutants. However, subciliary localization within the CC was unaffected in any of the BBS knockouts: BBS5 STORM localizations extended to the membrane STORM layer of the CC as in the WT and were found along the length of the CC. Surprisingly, BBS5 STORM clusters at the BB/IS were also not significantly affected in any of the Bbs mutant rods. These results indicate that BBS5 and BBSome subciliary localizations within the CC are maintained despite BBSome deficiencies that lead to rod cell death. Thus, incomplete BBSome complexes retain at least partial function to the extent of being properly localized, and perhaps contribute to moderating defects in CC trafficking.
Fig. 5.
BBS5 mislocalization in Bbs mutant mouse rods. Example STORM reconstructions for BBS5 (magenta) superimposed with centrin-2 (green) centroid reconstructions from (A) Bbs2−/− rods (age 8 wk), (B) Bbs4−/− rods (age 8 wk), and (C) Bbs7−/− rods (age 7 wk). See Fig. 3C for the WT reference control. For each mutant, rod outer segment STORM examples from a wider view are depicted to demonstrate BBS5 (orange arrows) mislocalization to the OS. In these reconstructions, the CC subregion is indicated with a dashed line. Yellow arrows indicate BBS5 STORM clusters along the CC. Stars indicate basal body/inner segment BBS5 clusters.
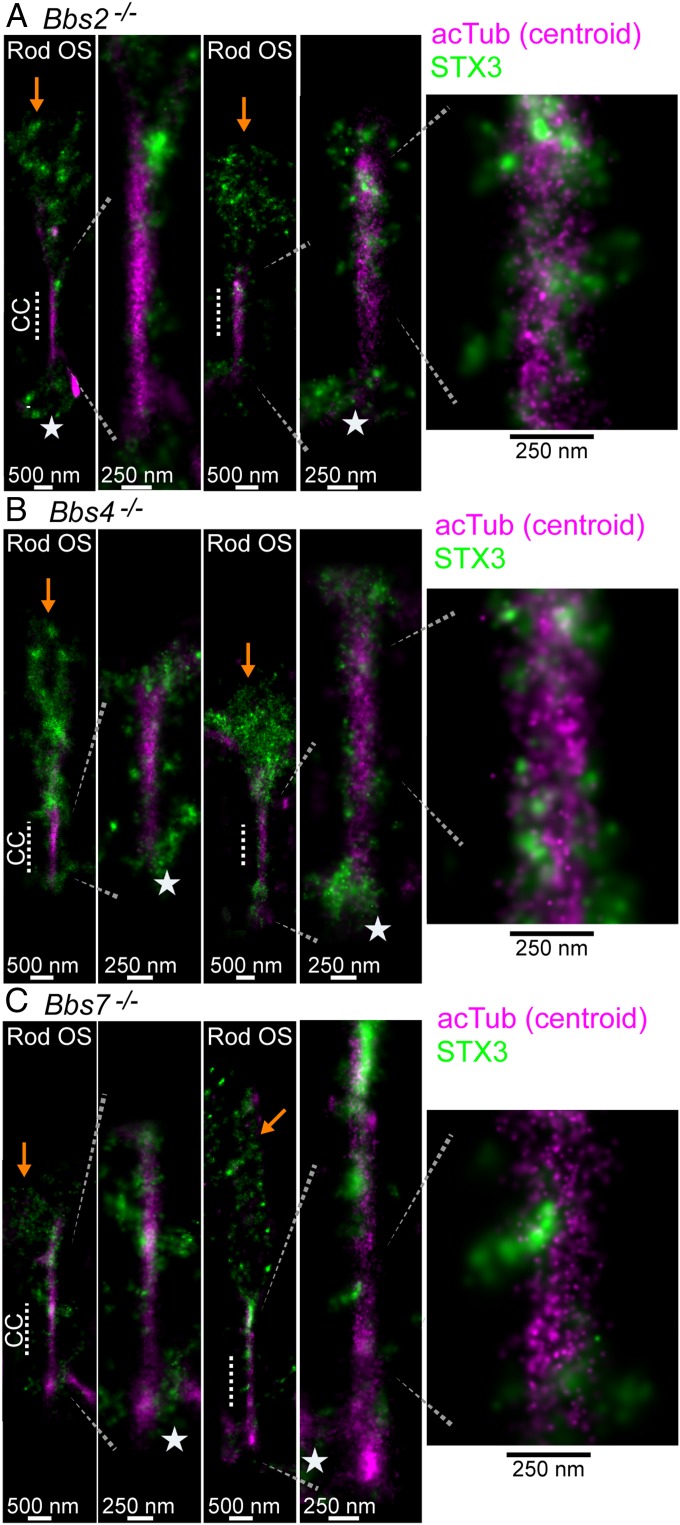
We also used STORM to localize STX3 in rods from Bbs2−/−, Bbs4−/−, and Bbs7−/− mutant mice to determine if any subciliary localization differences of STX3 in the CC are caused by disrupted BBSome composition and function. In STORM reconstructions of STX3 from each Bbs mutant line, STX3 was grossly mislocalized from the IS to the OS compared with WT or Bbs heterozygous controls (SI Appendix, Fig. S5B). This pattern of mislocalization is also apparent in STORM reconstructions of individual rod cells of all of the Bbs mutants, in which STX3 clusters in the OS were typically punctate and located along the edge of the OS membrane and sometimes diffusely localized throughout the entire OS of mutant rods (Fig. 6 and SI Appendix, Fig. S5C). Despite OS accumulation, STX3 was normally localized to the BB in each Bbs mutant. Along the CC, the overall radial distribution of STX3 localization to the exterior membrane layer of the CC was maintained; however, Bbs mutant STX3 clusters were either more diffuse, less structured, or smaller than the large WT STX3 clusters (compare Figs. 4E and 6). Therefore, based on these STORM results in 3 different Bbs mutant mouse lines, subciliary localization of STX3 is only partially attenuated in the CC membrane layer despite gross OS accumulation. Thus, anterograde and possibly partial retrograde trafficking evidently continue to some degree despite an incomplete BBSome composition and ciliopathy-related OS stress caused by protein accumulation and other mutant defects.
Fig. 6.
Mislocalization of syntaxin-3 in Bbs mutant mouse rods. Example STORM reconstructions for syntaxin-3 (green) superimposed with centroid acetylated α-tubulin (magenta) reconstructions from (A) Bbs2−/− rods (age 8 wk), (B) Bbs4−/− rods (age 5 wk), and (C) Bbs7−/− rods (age 8 wk). See Fig. 4E for the WT reference control. For each mutant, rod outer segment STORM examples from a wider view are depicted to demonstrate STX3 mislocalization to the OS (orange arrows). Stars indicate basal body/inner segment STX3 localization.
Discussion
Whereas primary cilia are important throughout the body, rod cell degeneration in ciliopathies likely reflects both general and photoreceptor-specific functions of cilium components (46). Combining cryo-ET and superresolution fluorescence nanoscopies have allowed us to map the repeating structures and localization of protein targets with nanoscale precision in the rod sensory cilium.
Our application of 9-fold subtomogram averaging of cryoelectron tomograms has identified features of CC structure (Fig. 1), including the 3D structure of Y-shaped links, the twisting of the axoneme microtubule doublets at a slight tilt with respect to the ciliary axis, and a central, luminal structure akin to the terminal plate of motile cilia (47).
The cryo-ET map provides a framework for the molecular map revealed by STORM, which provides localization information for individual components at sufficient resolution to assign them to CC subcompartments. For example, the axoneme lumen marker centrin-2 (28) was reconstructed with a diameter of 187± 16 nm, in good agreement with the cryo-ET map. Functionally, the lumen remains an uncharacterized subcompartment in rods, filled with heterogeneous structures and densities (48), now known to include centrin-2 in the CC. Despite some variability in the maximum widths between acTub STORM reconstructions, likely due to flattening, the average of 260 nm (±37 nm) is consistent with the ultrastructure of the microtubules of the axoneme revealed by electron tomography.
STX3 was found to form clusters along the axoneme of the CC, which extend to the ciliary membrane. There is considerable evidence for the occasional presence of similar membrane protrusions, or “buds,” on the surface of primary cilia (49, 50); these represent a possible reason for some membrane STX3 clusters appearing outside the apparent CC boundary. Another possible explanation is STX3 localization in the adjacent ciliary pocket membrane. We found some examples of rods in which STX3 was apparently localized in the adjacent periciliary inner segment (SI Appendix, Fig. S3E). However, STX3 clusters have shorter profiles along the longitudinal axis than either whirlin or VLGR1 (compare Fig. 4E and SI Appendix, Fig. S3 D and E with Fig. 3D and SI Appendix, Fig. S3 B and C), suggesting that STX3 is not a part of the PMC.
Perhaps the most striking difference between conventional confocal imaging and our STORM is labeling for Arl13B (Fig. 2C), often used as a marker for the primary cilium axoneme because of its apparent continuous localization along the axoneme at lower resolution. Our results reveal Arl13B localized in clusters along the axoneme, similar to our localizations of the IFT-B proteins, BBS5, and STX3. The clustered localization of the IFT-B proteins IFT81 and IFT88 (Fig. 3 A and B), tightly overlapped with the axoneme, supports their organization together with motor proteins in IFT trains, which have been visualized in Chlamydomonas flagella by cryo-ET and cryo-EM (51, 52) and have been proposed in rod cilia to correspond to large cytoplasmic electron densities in TEM micrographs (53). IFT88 was also recently localized in distinct clusters along the axoneme via superresolution imaging of primary cilia in cultured epithelial cells (54, 55). Our STORM reconstructions of these ciliary components in clusters suggest they are each organized along the axoneme in CCs within a series of large supramolecular complexes. Colocalization of large subsets of BBS5 and IFT88 clusters, quantified by interaction analysis, is consistent with their association in trains and, together with their colocalization at the BB, provides evidence for in vivo interactions between the BBSome and the IFT complex as active transporters in rod cilia, as previously hypothesized in other primary and motile cilium models (56, 57).
STORM analysis of CC protein distributions in the Bbs knockouts yielded surprising results. It had been reported that loss of individual BBS subunits, except for BBS4 (58), severely disrupts BBSome formation in vivo, yet the localization phenotypes of Bbs4−/− retinas were remarkably similar to those for Bbs2−/− and Bbs7−/−. Most surprising was that localization of ciliary proteins, including BBS5 within the CC, was very similar to that in the WT in all 3 mutant models. The most dramatic mislocalization in Bbs mutant rods was observed for STX3, which has been extensively used as a mislocalization marker for Bbs pathogenesis in the retina (13, 45, 59). In Bbs mutants, there is extensive aberrant STX3 accumulation in the outer segment, as expected (SI Appendix, Fig. S5B and diagram in SI Appendix, Fig. S8C), but, surprisingly, we found the STX3 distribution outside of the OS to be much less severely affected. However, the reconstructed STX3 clusters at the CC of Bbs mutant rods were smaller and less organized than in WT rods, indicating that in addition to preventing STX3 accumulation within the OS, a fully functional BBSome is necessary for the organization of STX3 within the CC. These observations held across all 3 Bbs knockout animals.
In addition to the value of our findings in their own right, they demonstrate the power of the methodology we present and the cellular/molecular map we have produced. Emerging technologies in both static and dynamic imaging (27, 31, 49, 60, 61) will likely continue to enhance both the resolution and the functional content of such maps.
Materials and Methods
Extended materials and methods are provided in SI Appendix.
Animals.
All wild-type mice used for this study were C57BL/6 between ages 3 wk and 3 mo. Examples of replicate STORM images are provided for all conditions in SI Appendix, and replicate information including the number of mouse replicates and number of cilia analyzed by STORM is described in SI Appendix, Table S1. Bbs mutant mouse lines are described in SI Appendix, Materials and Methods. All procedures adhered to the NIH Guide for the Care and Use of Laboratory Animals (62), and were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
Cryo-ET.
Rod cell fragments containing the CC were collected from WT mice by isoosmotic density-gradient centrifugation as described previously (8, 25). Fragments were applied to carbon-coated holey grids and plunge-frozen in liquid ethane. The frozen-hydrated specimens were imaged with a Polara G2 electron microscope (FEI) equipped with a field emission gun and a direct detection device (Gatan K2 Summit). Images of the CC were collected at a magnification of 9,400× at 300 kV as single-axis tilt series image stacks. MotionCorr (63) was used to correct image drift within each stack. Alignment and 3D reconstruction were performed using IMOD (64, 65). Good-quality CCs were used to search extensively for cilium and basal body-associated features, such as transition fibers, and to generate figures illustrating selected features. Subtomogram averaging was carried out as described previously (22) using Chimera (66) and EMAN2 (67) (additional details are in SI Appendix, Materials and Methods).
Antibodies and Labeling Reagents.
Primary antibody descriptions, including validation references, are available in SI Appendix, Table S2. The BBS5 antibody was validated via immunofluorescence in knockout mouse tissue (SI Appendix, Fig. S8 A and B). Wheat germ agglutinin (10 µg) conjugated to Alexa 647 (Molecular Probes) was used as in immunostaining procedures for glycoprotein labeling of mouse retinas.
STORM.
Mouse retinas were immunolabeled for STORM following a 2-step protocol. First, retinas were dissected in unfixed Ames’ media (Sigma) and blocked in 10% normal goat serum (Fitzgerald Industries) + 0.3% saponin (Sigma) + 1× Protease Inhibitor Mixture (GenDepot) diluted in 1× Ames’ media for 2 h at 4 °C. Primary antibodies (5 to 10 μg each) were added to the blocking buffer and incubated at 4 °C for 20 to 22 h. For antibody information, see SI Appendix, Table S2. The BBS5 antibody was validated via immunofluorescence in knockout mouse tissue (SI Appendix, Fig. S8 A and B). Retinas were washed and secondary antibody-stained at 4 °C for 2 h. Secondary antibodies used were (8 µg each) F(ab′)2-goat anti-mouse IgG Alexa 647 and F(ab′)2-goat anti-rabbit IgG Alexa 555 (Thermo Fisher). In WGA–Alexa 647 labeling experiments, F(ab′)2-goat anti-rabbit IgG Alexa 555 (Thermo Fisher) was used for dual labeling. Retinas were washed and fixed in 4% formaldehyde for 15 min at room temperature (RT). Next, retinas were reblocked in 10% normal goat serum + 0.2% Triton X-100 for 2 h at RT. Primary antibodies were readded to the blocking buffer and incubated for 2 d at 4 °C. Retinas were washed and probed again with the same secondary antibody overnight at 4 °C. Finally, retinas were washed and postfixed in 3% formaldehyde for 1 h at RT. Stained and fixed retinas were then embedded in Ultra Bed Epoxy Resin (SI Appendix, Materials and Methods) and cured on the top shelf of a 65 °C baking oven for 20 h; 500-nm to 1-μm sections were cut on a UCT or UC6 Leica ultramicrotome and dried directly onto glass-bottom dishes (MatTek 35-mm dishes, no. 1.5 coverslip).
Immediately prior to imaging, sections were etched in a mild sodium ethoxide solution (27). The following STORM imaging buffer was added on top of the sections: 45 mM Tris (pH 8.0), 9 mM NaCl; oxygen-scavenging system: 0.7 mg⋅mL−1 glucose oxidase (Amresco) + 42.5 µg⋅mL−1 catalase (Sigma), 10% (wt/vol) glucose + 100 mM mercaptoethylamine (i.e., l-cysteamine; Chem-Impex) + 10% VECTASHIELD (Vector Laboratories). Imaging was performed on the Nikon N-STORM System equipped with 200-mW, 561- and 647-nm solid-state lasers. For STORM image acquisition, sections were first scanned with low laser power to locate a region with multiple and sufficiently bright CCs. Both the 561- and 647-nm laser lines were then increased to maximum power to photobleach/quench the fluorescence and initiate photoswitching. Imaging frames were collected at ∼56 frames per s; 20,000 to 50,000 frames were collected for each imaging experiment, and 561- and 647-nm frames were collected sequentially.
Two-dimensional STORM analysis of STORM acquisition frames was performed using NIS-Elements AR analysis software. See SI Appendix, Materials and Methods for details. In Fiji/ImageJ, regions of interest (ROIs) of digitally straightened STORM reconstructions were measured using row average profiling, which plots the average intensity across the width of the ROI for each row of pixels along the length of the ROI. Pixels were converted to nanometers for accurate scaling. From these row average profiles, the edges of STORM clusters were set as 1/e times the maximum-intensity value for any given cluster profile. The MosaicIA ImageJ plugin (68) was used to perform the spatial interaction analysis in Fig. 4.
Data Availability.
All datasets used to generate the figures in the paper are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01-EY007981 and R01-EY026545 and the Welch Foundation (Q0035) (to T.G.W.), the Welch Foundation (AU-1714) (to J.L.), the National Center for Macromolecular Imaging (P41-GM103832) (to M.F.S.), NIH Fellowships F31-EY028025 (to V.L.P.) and F32-EY027171 (to M.A.R.), and the Baylor College of Medicine Vision Research Core (P30-EY002520). We thank Dr. Ivan Anastassov, Dr. Melina Agosto, Dr. Abigail Moye, and Mr. Christopher Hampton for useful insights and discussions. We also thank Dr. Clay Smith and Dr. Uwe Wolfrum for sharing resources and Mr. Matthew Mitschelen (Nikon) for technical advice and guidance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.V.N. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Electron Microscopy Data Bank (EMDB ID code EMD-8992).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902003116/-/DCSupplemental.
References
- 1.Ware S. M., Aygun M. G., Hildebrandt F., Spectrum of clinical diseases caused by disorders of primary cilia. Proc. Am. Thorac. Soc. 8, 444–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Q., et al. , The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 6, 1299–1317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao L., et al. , Integrative subcellular proteomic analysis allows accurate prediction of human disease-causing genes. Genome Res. 26, 660–669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiter J. F., Leroux M. R., Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18, 533–547 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wensel T. G., et al. , Structural and molecular bases of rod photoreceptor morphogenesis and disease. Prog. Retin. Eye Res. 55, 32–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pazour G. J., et al. , The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157, 103–113 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., et al. , Ablation of whirlin long isoform disrupts the USH2 protein complex and causes vision and hearing loss. PLoS Genet. 6, e1000955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilliam J. C., et al. , Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell 151, 1029–1041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young R. W., Droz B., The renewal of protein in retinal rods and cones. J. Cell Biol. 39, 169–184 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams N. A., Awadein A., Toma H. S., The retinal ciliopathies. Ophthalmic Genet. 28, 113–125 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Loktev A. V., et al. , A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev. Cell 15, 854–865 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Nachury M. V., et al. , A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Datta P., et al. , Accumulation of non-outer segment proteins in the outer segment underlies photoreceptor degeneration in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E4400–E4409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mykytyn K., et al. , Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. U.S.A. 101, 8664–8669 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q., et al. , BBS7 is required for BBSome formation and its absence in mice results in Bardet-Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J. Cell Sci. 126, 2372–2380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung C. H., Leroux M. R., The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol. 15, 1387–1397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox G., Sheppard C. J., Practical limits of resolution in confocal and non-linear microscopy. Microsc. Res. Tech. 63, 18–22 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Nickell S., Park P. S., Baumeister W., Palczewski K., Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J. Cell Biol. 177, 917–925 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid M. F., Single-particle electron cryotomography (cryoET). Adv. Protein Chem. Struct. Biol. 82, 37–65 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Bui K. H., Ishikawa T., 3D structural analysis of flagella/cilia by cryo-electron tomography. Methods Enzymol. 524, 305–323 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Nicastro D., et al. , Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. U.S.A. 108, E845–E853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyfman A. Y., et al. , Structure of Trypanosoma brucei flagellum accounts for its bihelical motion. Proc. Natl. Acad. Sci. U.S.A. 108, 11105–11108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S., Fernandez J. J., Marshall W. F., Agard D. A., Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 31, 552–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rust M. J., Bates M., Zhuang X., Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wensel T. G., Gilliam J. C., Three-dimensional architecture of murine rod cilium revealed by cryo-EM. Methods Mol. Biol. 1271, 267–292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Liu J., Schmid M. F., Wensel T. G., Cryo-electron tomogram of mouse rod connecting cilium and mother centriole with C9 sub-tomogram averaging. Electron Microscopy Data Bank. https://www.ebi.ac.uk/pdbe/entry/emdb/EMD-8992. Deposited 25 July 2018. [Google Scholar]
- 27.Sigal Y. M., Speer C. M., Babcock H. P., Zhuang X., Mapping synaptic input fields of neurons with super-resolution imaging. Cell 163, 493–505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trojan P., et al. , Centrins in retinal photoreceptor cells: Regulators in the connecting cilium. Prog. Retin. Eye Res. 27, 237–259 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Liem K. F., Jr, et al. , The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J. Cell Biol. 197, 789–800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cevik S., et al. , Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188, 953–969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao M., et al. , Expansion stimulated emission depletion microscopy (ExSTED). ACS Nano 12, 4178–4185 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Yang T. T., et al. , Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9, 2023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotthardt K., et al. , A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. eLife 4, e11859 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humbert M. C., et al. , ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. U.S.A. 109, 19691–19696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horst C. J., Forestner D. M., Besharse J. C., Cytoskeletal-membrane interactions: A stable interaction between cell surface glycoconjugates and doublet microtubules of the photoreceptor connecting cilium. J. Cell Biol. 105, 2973–2987 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sedmak T., Wolfrum U., Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J. Cell Biol. 189, 171–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhogaraju S., et al. , Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341, 1009–1012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beech P. L., et al. , Localization of kinesin superfamily proteins to the connecting cilium of fish photoreceptors. J. Cell Sci. 109, 889–897 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Mikami A., et al. , Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 115, 4801–4808 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Cevik S., et al. , Active transport and diffusion barriers restrict Joubert syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet. 9, e1003977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith T. S., et al. , Light-dependent phosphorylation of Bardet-Biedl syndrome 5 in photoreceptor cells modulates its interaction with arrestin1. Cell. Mol. Life Sci. 70, 4603–4616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin H., et al. , The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maerker T., et al. , A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 17, 71–86 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Chuang J. Z., Zhao Y., Sung C. H., SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell 130, 535–547 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu Y., et al. , BBSome function is required for both the morphogenesis and maintenance of the photoreceptor outer segment. PLoS Genet. 13, e1007057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dharmat R., et al. , SPATA7 maintains a novel photoreceptor-specific zone in the distal connecting cilium. J. Cell Biol. 217, 2851–2865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munn E. A., Fine structure of basal bodies (kinetosomes) and associated components of Tetrahymena. Tissue Cell 2, 499–512 (1970). [DOI] [PubMed] [Google Scholar]
- 48.Chuang J. Z., Hsu Y. C., Sung C. H., Ultrastructural visualization of trans-ciliary rhodopsin cargoes in mammalian rods. Cilia 4, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang F., et al. , Ultra-high resolution 3D imaging of whole cells. Cell 166, 1028–1040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon J., et al. , Revealing nanoscale morphology of the primary cilium using super-resolution fluorescence microscopy. Biophys. J. 116, 319–329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jordan M. A., Diener D. R., Stepanek L., Pigino G., The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20, 1250–1255 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Pigino G., et al. , Electron-tomographic analysis of intraflagellar transport particle trains in situ. J. Cell Biol. 187, 135–148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenbaum J. L., Cole D. G., Diener D. R., Intraflagellar transport: The eyes have it. J. Cell Biol. 144, 385–388 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi X., et al. , Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat. Cell Biol. 19, 1178–1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weng R. R., et al. , Super-resolution imaging reveals TCTN2 depletion-induced IFT88 lumen leakage and ciliary weakening. Biophys. J. 115, 263–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker S. A., Freeman K., Luby-Phelps K., Pazour G. J., Besharse J. C., IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J. Biol. Chem. 278, 34211–34218 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Lechtreck K. F., et al. , The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117–1132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q., Yu D., Seo S., Stone E. M., Sheffield V. C., Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet-Biedl syndrome protein complex, the BBSome. J. Biol. Chem. 287, 20625–20635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dilan T. L., et al. , Bardet-Biedl syndrome-8 (BBS8) protein is crucial for the development of outer segments in photoreceptor neurons. Hum. Mol. Genet. 27, 283–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson E., et al. , Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins. Sci. Rep. 5, 9583 (2015). Erratum in: Sci. Rep.6, 22681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D., et al. , Correlative stochastic optical reconstruction microscopy and electron microscopy. PLoS One 10, e0124581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 63.Li X., et al. , Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Shivanandan A., Radenovic A., Sbalzarini I. F., MosaicIA: An ImageJ/Fiji plugin for spatial pattern and interaction analysis. BMC Bioinformatics 14, 349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets used to generate the figures in the paper are available from the corresponding author upon reasonable request.