Significance
Telomere length homeostasis is an important mechanism for maintaining genomic stability. Telomere length is regulated by numerous events that include protein–DNA interactions, the length and structure of telomere DNA, and recruitment of telomerase. Here we used hydroxyl radical footprinting to identify environmental changes in the telomere end-binding heterodimer, POT1-TPP1, as a function of telomere length. Our data identified a specific residue (histidine 266) of the POT1 protein that reports differences in solvent accessibility as a function of telomere DNA length. We further show that the chronic lymphocytic leukemia-related H266L POT1 mutation disrupts the ability of POT1-TPP1 to negatively regulate telomerase activity in vitro and in cancer cells.
Keywords: hydroxyl radical, footprinting, shelterin, chronic lymphocytic leukemia
Abstract
Telomeres cap the ends of linear chromosomes and terminate in a single-stranded DNA (ssDNA) overhang recognized by POT1-TPP1 heterodimers to help regulate telomere length homeostasis. Here hydroxyl radical footprinting coupled with mass spectrometry was employed to probe protein–protein interactions and conformational changes involved in the assembly of telomere ssDNA substrates of differing lengths bound by POT1-TPP1 heterodimers. Our data identified environmental changes surrounding residue histidine 266 of POT1 that were dependent on telomere ssDNA substrate length. We further determined that the chronic lymphocytic leukemia-associated H266L substitution significantly reduced POT1-TPP1 binding to short ssDNA substrates; however, it only moderately impaired the heterodimer binding to long ssDNA substrates containing multiple protein binding sites. Additionally, we identified a telomerase inhibitory role when several native POT1-TPP1 proteins coat physiologically relevant lengths of telomere ssDNA. This POT1-TPP1 complex-mediated inhibition of telomerase is abrogated in the context of the POT1 H266L mutation, which leads to telomere overextension in a malignant cellular environment.
Telomeres are specialized nucleoprotein complexes that cap the ends of linear chromosomes (1). After thousands of base pairs of a repeating G-rich sequence, telomeres terminate in a short single-stranded DNA (ssDNA) overhang (2). In proliferating and transformed cells, telomerase, a reverse transcriptase ribonucleoprotein complex, base pairs with the telomere ssDNA overhang to synthesize new telomere ssDNA (1). Telomere DNA is protected by a set of specialized proteins called shelterin that help regulate telomerase activity and prevent telomeres from being misidentified as sites of DNA damage (3). POT1 and TPP1 are 2 shelterin proteins that interact with the ssDNA overhang to help maintain telomere integrity (3, 4). Whereas POT1 binds specifically to telomere ssDNA, TPP1 interacts with POT1 and other shelterin proteins to localize POT1 to telomeres. Recently, more than 300 single-nucleotide polymorphisms (SNPs) within the coding region of POT1 (cBioPortal) have been identified in patients with malignancies including chronic lymphocytic leukemia (CLL) (5), familial melanoma (6, 7), familial glioma (8), and cardiac angiosarcoma (9). Most cancer-associated SNPs result in mutations that localize to the 2 N-terminal oligosaccharide–oligonucleotide (OB) folds of the POT1 protein, with many residing near the ssDNA binding cleft. Meanwhile, TPP1 helps to regulate interactions with telomerase for its recruitment to telomeres in a cell cycle-dependent manner (10–14). Therefore, the POT1-TPP1 heterodimer plays diverse roles in protecting the telomere ssDNA from degradation and repair, while also facilitating access of telomere ssDNA for telomerase-mediated extension (3, 4, 15, 16).
In Saccharomyces cerevisiae, telomere length homeostasis is regulated by a protein-counting mechanism, whereby the length of the telomere DNA and the relative number of telomere proteins bound to it differentially influence telomerase-mediated extension (17, 18). The length of the telomere ssDNA overhang is also an important regulator of telomere homeostasis in most eukaryotes as its ability to adopt disparate secondary structures including G quadruplexes (19, 20) and T-loops (21) poses obstacles for telomere extension. The binding of telomere proteins alleviates secondary structures to promote telomerase accessibility and extension (22–25). In mammalian cells, the 50 to 100 copies of POT1 and TPP1 proteins per telomere is more than enough to fully coat the telomere ssDNA overhang (26, 27). Together, these studies indicate that telomere ssDNA length, binding of shelterin proteins, and telomere DNA structure collectively contribute to telomere homeostasis. Despite these findings, the molecular switch governing whether these regulatory elements contribute positively or negatively to telomere extension is not well understood.
Here we probed the functional influences of telomere ssDNA length and binding of multiple POT1-TPP1 complexes on nucleoprotein-mediated regulation of telomerase. Hydroxyl radical footprinting (HRF) coupled with mass spectrometry was used to identify key alterations in POT1-TPP1 complex structural environments as a function of telomere ssDNA length. Specifically, our data present a model in which short POT1-TPP1-ssDNA complexes enhance telomerase activity, while longer tracts of POT1-TPP1-ssDNA complexes negatively regulate telomerase-mediated extension. Furthermore, our biophysical and functional data highlight significant environmental changes surrounding histidine 266 of POT1. Indeed, the cancer-associated H266L POT1 mutant exhibits defects in telomere ssDNA binding and telomerase regulation that are dependent on the length of ssDNA and the number of POT1-TPP1 proteins bound to it. The observed defects of telomerase inhibition and telomere length regulation associated with the H266L mutant were further confirmed using CRISPR-Cas9 technology to introduce this substitution into malignant cells. Together, our data establish an elaborate model in which POT1-TPP1 binding to ssDNA differentially regulates telomerase in a manner that is dependent upon ssDNA length and degree of POT1-TPP1 saturation of that ssDNA. Additionally, our data identified the H266 residue of POT1 as playing a key role in transitioning the POT1-TPP1 complex from a positive to a negative processivity factor of telomerase.
Results
Hydroxyl Radical Footprinting of Monomeric Versus Multimeric POT1-N Binding to Different Lengths of ssDNA Highlights Different Structural Environments.
The physiological length of the telomere ssDNA overhang is maintained at a length of 50 to 200 nucleotides in mammalian cells (28) and can accommodate binding of several POT1 proteins or POT1-TPP1 complexes (27, 29). In order to explore the structural environment changes induced by multiple protein binding events on physiologically relevant lengths of ssDNA, we employed hydroxyl radical footprinting (HRF) to characterize single and multiple POT1 binding events to telomere ssDNA of differing lengths. Hydroxyl radicals generated from exposure to X-ray beams oxidatively modify solvent-accessible amino acid side chains, providing high-resolution information on protein–protein and protein–DNA interactions (30–32).
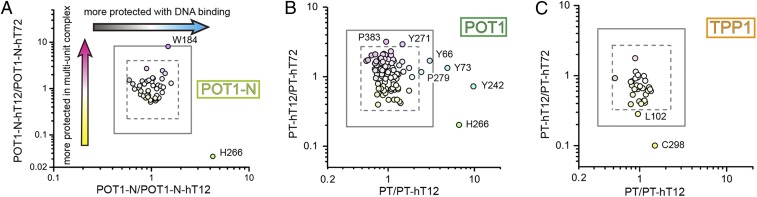
Initially, HRF experiments were performed with a splice variant of POT1 (POT1-N) that represents the DNA binding domain of full-length protein, and the availability of its structure solved by X-ray crystallography (33) provides a detailed guide for mapping and analyzing the individual residues identified in the footprinting studies. HRF experiments were performed to probe POT1-N protein alone (POT1-N), a single protein bound to the minimal 12-nt ssDNA substrate (POT1-N-hT12), and a complex assembled from 6 POT1-N proteins bound to a 72-nt, physiologically relevant length of telomere ssDNA (POT1-N-hT72) (SI Appendix, Fig. S1 A and B). The modification ratio compares solvent accessible reaction rates for an individual amino acid measured in different sample environments, as previously described (31, 34, 35). Thus, a normalized modification ratio equal to 1 indicates that the solvent accessibility of a residue is consistent across 2 different sample environments. Meanwhile, a normalized modification ratio value of less than 1 identifies residues that experience a gain in solvent accessibility in the state represented in the denominator of the ratio. Similarly, a normalized modification ratio that is greater than 1 highlights residues that are more protected from solvent in the state that is represented in the denominator of the ratio. For experimental data comparing POT1-N to POT1-N-hT12, the majority of modification ratios fall in the range of mean ± 2 SD, indicating that the solvent accessibility of POT1-N protein is not significantly altered upon hT12 binding (Fig. 1A and SI Appendix, Fig. S2A and Tables S1 and S2). However, residue H266 exhibited a modification ratio greater than the mean + 3 SD upon hT12 binding. This finding suggests that H266 is significantly more protected from solvent in the presence of hT12 DNA. This conclusion is supported by the X-ray crystal structure which describes intimate interactions between H266 and 2 separate nucleotides (T8 and G10) in the short ssDNA substrate (SI Appendix, Fig. S2C) (33).
Fig. 1.
HRF identifies POT1-N and POT1-TPP1 residues undergoing environmental changes upon binding of differing telomere ssDNA substrates. (A) Normalized modification ratio of POT1-N-hT12/POT1-N-hT72 versus normalized modification ratio of POT1-N/POT1-N-hT12 for all detectable POT1-N residues. Gray squares indicate boundaries of mean ± 3 SD (solid) and mean ± 2 SD (dashed). (B) POT1 and (C) TPP1 residue modification ratios of PT-hT12/PT-hT72 versus normalized modification ratio of PT/PT-hT12 in the context of POT1-TPP1 binding to ssDNA substrates. Data are plotted as described in A.
A comparison between POT1-N-hT12 and POT1-N-hT72 samples (Fig. 1A and SI Appendix, Fig. S2B) revealed W184 and H266 as the only residues with modification ratios outside the mean ± 2 SD range. Specifically, W184 became significantly more protected in complexes assembled from 6 POT1-N proteins coating the physiologically relevant ssDNA substrate when compared to a single POT1-N protein interacting with the short ssDNA substrate. Meanwhile, H266 displayed a modification rate that was dramatically elevated with the protein-coated hT72 ssDNA sample (25.1 s−1) compared to that of a single POT1 -N protein bound to the hT12 ssDNA (1.5 s−1) (SI Appendix, Fig. S3A). Together, these data highlight H266 as a residue in POT1 protein that becomes significantly more solvent accessible in samples containing protein-coated hT72 ssDNA versus the hT12-bound complex.
POT1 H266 Is Solvent Accessible in POT1-TPP1 Complexes Coating Physiologically Relevant ssDNA Substrates.
To further explore the structural changes of telomere protein–DNA interactions in an expanded physiological context, the POT1-TPP1 heterodimer was similarly assembled with differing lengths of telomere ssDNA (referred to as PT, PT-hT12, and PT-hT72; SI Appendix, Fig. S1C), and HRF experiments were conducted. Similar to POT1-N, POT1-TPP1 exhibited a significantly reduced modification rate (∼6- to 7-fold) for POT1 H266 upon binding to hT12 ssDNA substrates (Fig. 1B and SI Appendix, Fig. S4A and Tables S3 and S4). The modification rate ratios for residues Y73 and Y242 in the context of hT12 ssDNA binding were similarly determined to be beyond the mean ± 3 SD range for this analysis (Fig. 1B and SI Appendix, Fig. S4A). Both Y73 and Y242 are located at the interface between the 2 OB-fold domains of POT1, and a hydrogen bond is formed between the backbone carbonyl group of the Y73 and Y242 side chain hydroxyl group in the previously solved X-ray crystal structure (SI Appendix, Fig. S4C) (33). The enhanced protection of these 2 tyrosine residues is indicative of a conformational change, upon hT12 ssDNA binding, that brings the 2 OB folds of POT1 closer together to limit solvent accessibility. As these changes were not observed in the POT1-N experiments, it is likely that the inclusion of TPP1 helps to promote this conformational change of POT1, which could help explain the role of TPP1 in enhancing the binding affinity of POT1 for hT12 ssDNA by an order of magnitude (16).
A comparison of the modification rates between PT-hT12 and PT-hT72 identified few structural changes that are altered upon binding to telomere ssDNA of differing lengths as the majority of ratios were within the mean ± 2 SD range (Fig. 1B and SI Appendix, Fig. S4B). Consistent with POT1-N experiments, however, H266 exhibited a marked increase in modification rate when in complex with hT72 ssDNA and the lowest ratio of normalized modification rates when comparing PT bound to hT12 versus hT72 ssDNA (Fig. 1B). Once again, these data reveal that H266 is more protected upon POT1-TPP1 binding to hT12 ssDNA, but this protective capacity is lost upon binding to the longer, physiologically relevant hT72 ssDNA substrate. Compared to POT1, TPP1 is less involved in the POT1-TPP1–DNA and interprotein interactions that are inherent to multiple protein binding events. The modification rate ratios of TPP1 residues were primarily in the normalized range of mean ± 3 SD, with the exception of C298 which exhibits greater solvent accessibility when bound to hT72 versus hT12 ssDNA (Fig. 1C and SI Appendix, Fig. S4B). These data suggest that the interface encompassing C298 of TPP1 undergoes a DNA length-dependent rearrangement; however, delineation of the role of such a conformational change in POT1-TPP1 function will require further investigation.
Cancer Associated POT1 H266L Mutant Differentially Regulates POT1-TPP1 Binding to Telomere ssDNA Substrates in a Length-Dependent Manner.
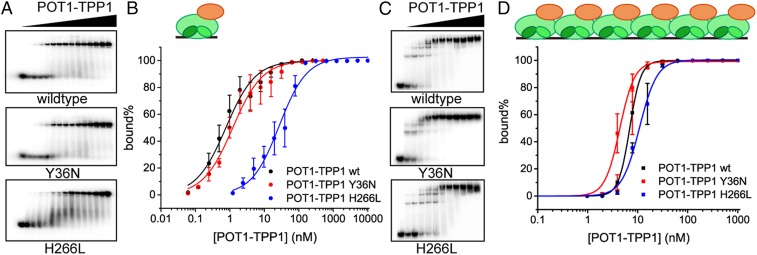
With the identification of the H266L POT1 mutation in patients diagnosed with CLL (5), we hypothesized that the pathology observed may be related to altered H266 function in coordinating interactions between POT1 and telomere ssDNA. Like H266, single nucleotide polymorphism resulting in Y36 mutations (Y36N) at the POT1 protein level has been identified in CLL patients (5). Although both H266 and Y36 interact directly with telomere ssDNA in the X-ray crystal structure (33), Y36 did not exhibit significant changes in solvent accessibility in the HRF experiments described above (SI Appendix, Fig. S3B). One potential explanation for this observation is localization of H266 and Y36 in the POT1 DNA-binding domain; Y36 resides in OB1, whereas H266 localizes to OB2. Since both OB folds cooperate in recognizing telomere ssDNA (25, 33), we first sought to determine whether CLL-associated mutations introduced at the Y36 or H266 position differentially alter ssDNA interactions. To do so, POT1-TPP1 heterodimeric protein with a H266L or a Y36N mutation introduced in the POT1 protein were expressed and purified. The ability for each construct to interact with telomere ssDNA of different lengths (hT12 and hT72), and identical to those used in HRF experiments, was quantitatively measured. Electrophoretic mobility shift assays (EMSAs) were performed to compare the equilibrium dissociation constant (KD) of wild-type and mutant POT1-TPP1 proteins for ssDNA substrates. Initial experiments for a single protein bound to the minimal hT12 ssDNA substrate yielded a calculated KD value for wild-type POT1-TPP1 to be 1.0 ± 0.5 nM, consistent with previous reports (16, 33, 36). Similar experiments revealed POT1 Y36N moderately decreased the affinity of POT1-TPP1 protein for hT12 ssDNA (KD = 2.2 ± 1.1 nM), whereas POT1 H266L mutation significantly reduced DNA binding affinity by ∼30-fold (KD = 29.1 ± 9.3 nM) (Fig. 2 A and B). Similar results were obtained using an alternative 12-nt telomere ssDNA substrate (TTAGGG)2 (SI Appendix, Fig. S5). In neither case was there evidence to suggest that human POT1-TPP1 proteins oligomerize on a minimal ssDNA substrate, as has been observed for Schizosaccharomyces pombe Pot1 protein (37).
Fig. 2.
POT1(H266L)-TPP1 mutant exhibits differential binding defects that are length-dependent of ssDNA substrate. (A) EMSA assays were performed under equilibrium binding conditions to determine the effects CLL-associated POT1 mutations had on POT1-TPP1 binding to short telomere ssDNA substrate (hT12). Protein concentration ranged from 0 to 500 nM for wild type and Y36N and from 0 to 10 µM for H266L. (B) Quantification of EMSA data for POT1-TPP1 and mutant protein binding to hT12. Error bars represent the mean ± SD (n = 3). (C) EMSAs were performed to determine the effects of CLL-associated POT1 mutations on multiple POT1-TPP1 complexes coating long telomere ssDNA substrates (hT72). (D) Quantification of EMSA data for POT1-TPP1 and mutant protein coating hT72. Error bars represent the mean ± SD (n = 3). Schematic models indicate POT1-TPP1 complexes bound to differing telomere ssDNA hT12 in B and hT72 in D. POT1 is shown as green ellipse with 2 OB domains. TPP1 is shown as orange ellipse, and telomere ssDNA is depicted as black lines.
EMSAs were then employed to characterize the ability of multiple POT1-TPP1 proteins to bind to the physiologically relevant hT72 ssDNA substrate. The apparent KD value (Kapp) was calculated to describe the efficiency of 6 POT1-TPP1 proteins to simultaneously bind to, thereby coating, the hT72 ssDNA substrate. In these experiments, the Kapp was determined to be 6.9 ± 0.6, 4.5 ± 0.4, and 11.1 ± 1.1 nM for wild-type, Y36N, and H266L constructs, respectively (Fig. 2 C and D). These data indicate that the CLL associated Y36N or H266L mutations only modestly (less than 2-fold) alter the apparent equilibrium dissociation constant of POT1-TPP1 to coat physiological lengths of telomeric ssDNA. This finding is in stark contrast to the 30-fold difference determined for H266L POT1-TPP1 protein binding to short, hT12 ssDNA substrate. We next investigated whether the H266L mutant alters the preference for 3′ end binding exhibited by native POT1-TPP1 protein (16). Accordingly, EMSAs were conducted with a3 and a5 ssDNA substrates that contain nucleotide substitutions to impact POT1-TPP1 binding at either the 5′ or the 3′ ends of an 18-nt telomere ssDNA substrate (16). Whereas wild-type protein exhibits a 10-fold higher affinity for binding to the 3′ recognition motif, this preference is reduced to only 2.5× higher for the H266L mutant (SI Appendix, Fig. S6). Altogether, these data recapitulate the HRF results described above identifying H266 as being critical for ssDNA substrate binding and dictating the differential roles for POT1-TPP1 binding to short hT12 ssDNA versus the coating of multiple heterodimers on long hT72 ssDNA substrates.
H266L POT1 Mutant Abrogates Inhibitory Role of Multiple POT1-TPP1 Binding Events on Telomerase Activity.
In addition to telomere ssDNA protection, the POT1-TPP1 heterodimer plays a critical role in regulating telomere length homeostasis (11, 36, 38). Although the POT1-TPP1 complex is known to increase both activity and processivity of human telomerase (16, 36), it remains unclear what regulatory elements prevent telomerase from perpetually extending telomere ssDNA. To address the role of POT1-TPP1 in this context, we employed an in vitro direct telomerase incorporation and extension assay to determine the role of POT1-TPP1 in regulating telomerase activity and processivity with ssDNA substrates of differing lengths, specifically hT12 and hT72 (Fig. 3 and SI Appendix, Fig. S7). Similar experiments using the CLL-associated POT1 mutants, Y36N and H266L, were carried out to determine if these mutations disrupt the ability of POT1-TPP1 complexes to properly regulate telomerase activity.
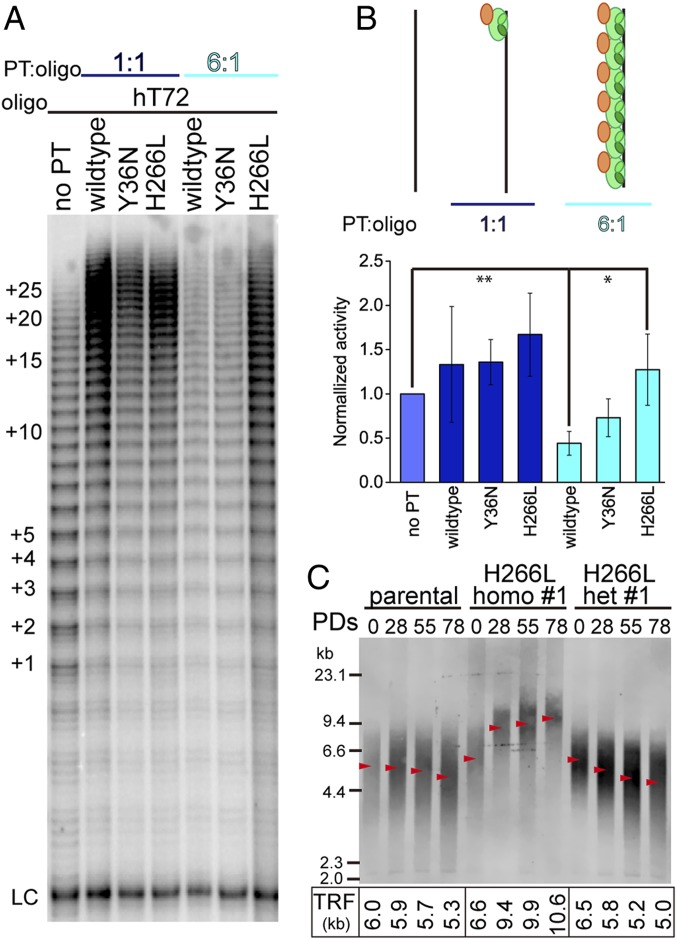
Fig. 3.
H266L POT1 mutant abrogates POT1-TPP1 complex-mediated inhibition of telomerase in a substrate length-dependent manner promoting telomere extension. (A) Direct in vitro telomerase assay performed on hT72 ssDNA. Lane 1, no POT1-TPP1 added. Lanes 2 to 4, stoichiometric concentration of POT1-TPP1 wild type, Y36N, and H266L were added, respectively. Lanes 5 to 7, POT1-TPP1 wild type, Y36N, and H266L were added to saturate all binding sites on hT72. LC, loading control. The number of telomere repeats being added is indicated at left. (B) Quantification of lanes in A displaying normalized telomerase activity. Schematic models indicate different binding states of POT1-TPP1 complexes bound to hT72. Error bars represent the mean ± SD (n = 3). **P < 0.005; 0.005 < *P < 0.05. (C) TRF analysis of genomic DNA from parental and CRISPR-Cas9 edited HCT 116 cell lines containing either homozygous H266L (homo #1) or heterozygous H266L (het #1) mutations at the indicated population doublings (PDs) in culture. Quantification of TRF length is indicated by red arrows with corresponding values shown below the gel.
The introduction of either Y36N or H266L POT1 mutations did not significantly alter telomerase activity or processivity provided by POT1-TPP1 on the hT12 ssDNA (SI Appendix, Fig. S7 A and B). Because longer G-rich ssDNA substrates form complex secondary structures that affect telomerase activity (22, 24, 39), we reasoned that telomere length alone may be a determining feature in preventing telomerase-mediated overextension of telomere ssDNA. Alternatively, longer ssDNA substrates sheathed in multiple POT1-TPP1 complexes may potentially impair the accessibility of ssDNA by telomerase, thereby limiting telomere extension. To test this, we conducted in vitro telomerase extension assays as described above but with hT72 ssDNA substrate (Fig. 3A). These data revealed that in the absence of POT1-TPP1 complexes, telomerase extends hT72 ssDNA substrates to a similar extent as hT12 (Fig. 3A and SI Appendix, Fig. S7A). When stoichiometric concentrations of POT1-TPP1 complexes were included (such that 1 protein binds to 1 ssDNA), telomerase activity was enhanced slightly while processivity was elevated by about 3-fold (SI Appendix, Fig. S7C). Under these conditions, it would be expected that a single POT1-TPP1 protein preferentially occupies the 3′ position of each hT72 substrate. In similar experiments, the inclusion of either Y36N or H266L mutated POT1-TPP1 proteins regulated telomerase in a manner that was indistinguishable from that of wild-type protein. Finally, the impact of multiple POT1-TPP1 binding events on the regulation of telomerase activity and processivity was assessed in the presence of hT72 ssDNA substrates completely sheathed in protein complexes. In this scenario, coating of hT72 ssDNA substrate with wild-type POT1-TPP1 complexes impaired telomerase activity, while no significant change in processivity was observed (Fig. 3B and SI Appendix, Fig. S7C). POT1-TPP1 complexes with the Y36N mutation exhibited a similar affect to wild-type protein on telomerase regulation. However, the introduction of the H266L mutation supported telomerase activity, thereby abrogating the inhibitory role of wild-type POT1-TPP1 complexes on telomerase extension of physiologically relevant lengths of ssDNA.
Since native POT1-TPP1 binding destabilizes G-quadruplexes (25, 40), we asked whether the H266L mutation impairs this ability to contribute to unregulated telomere elongation via telomerase. To do so, we used circular dichroism (CD) spectroscopy, which illustrates a positive band at 295 nm and a negative band at 265 nm, both of which are signatures associated with G-quadruplex structures (41, 42) (SI Appendix, Fig. S8A). POT1-TPP1 protein was mixed with hT72 ssDNA at a concentration sufficient to saturate all binding sites using a stopped-flow device. The gradual decrease in the circular dichroism spectra at 295 nm is consistent with the G-quadruplex structures being resolved (41, 43) upon addition of POT1-TPP1 protein (SI Appendix, Fig. S8B). These data indicate that the H266L mutant POT1-TPP1 protein resolves G-quadruplex structures but at rates (kobs1 = 0.27 ± 0.06 s−1, kobs2 = 0.019 ± 0.002 s−1) that are slower than those of wild-type protein (kobs1 = 0.63 ± 0.27 s−1, kobs2 = 0.030 ± 0.004 s−1).
To further explore the biologic role of POT1 aberrations associated with the H266L mutation, the SNP coding for the appropriate amino acid change was introduced into the genome of HCT116 colon adenocarcinoma cancer cells using CRISPR-Cas9. Two homozygous and 2 heterozygous cell lines with the successful introduction of the H266L substitution at the protein level were recovered and validated using next-generation and Sanger sequencing (SI Appendix, Fig. S9A). Each of these 4 cell lines, as well as parental HCT116 cells, were passaged for 78 d, and changes in telomere length were determined using telomere restriction fragment (TRF) analysis (Fig. 3C and SI Appendix, Fig. S9B). Parental cells displayed a slight shortening in telomere length with increasing population doublings. In contrast, the homozygous H266L mutant cells demonstrated robust telomere lengthening over the 78 d of cell growth. Heterozygous H266L mutant cells demonstrated either minimal telomere shortening (het #1) or maintained a relatively consistent telomere length (het #2) over the 78-d time course. While our results help to define an important role of POT1 H266 in properly regulating telomerase-mediated extension, they are unlikely to fully explain the pathology of the H266L mutation as patients with CLL are heterozygous for the mutation (5). POT1 mutations that are associated with CLL, and including H266L, also correlate with a higher frequency of sister chromatid fusions and stalled replication forks at telomeres (5). Nonetheless, consistent with the in vitro direct telomerase assay, our cellular data further demonstrate that the H266 residue of POT1 plays a critical role in regulating telomere maintenance and that mutation of this residue results in enhanced telomere elongation.
Discussion
The ability of telomerase to maintain telomere DNA at a constant length is a fundamental, yet complex, process of physiology that involves many regulatory elements (4). In this report, we show that the length of telomere ssDNA and the number of POT1-TPP1 heterodimers bound to it are key contributing factors that differentially influence telomerase-mediated extension. In the context of binding short hT12 ssDNA, the added protection of H266 demonstrated in our HRF experiments can be attributed to direct interactions that exist between H266 of POT1 and T8 and G10 of bound telomere ssDNA (33). Some amino acid side-chains exhibit lower intrinsic reactivity to hydroxyl radicals (44), which might explain why more POT1 residues were not identified upon DNA binding in the HRF experiments. However, DMS methylation and pyrrolidine cleavage similarly highlighted only G10 in the DNA to be protected when bound to POT1 protein, even though other residues interact with ssDNA in the crystal structure of the protein–DNA complex (33). These independent results suggest that the presence of ssDNA does not significantly alter solvent accessibility and/or chemical reactivity for many of the protein–DNA interactions that were identified in the X-ray crystal structure, at least for 10- to 12-nt substrates.
Highlighting a dynamic nature between POT1 and telomere ssDNA, our findings elucidate a critical and opposing role of residue H266 in POT1 protein for binding telomere ssDNA of different lengths. In our HRF experiments using POT1-TPP1 protein, the modification rate of POT1 H266 decreased by ∼6 times when bound to hT12 ssDNA as compared to free POT1-TPP1 protein (SI Appendix, Table S2). However, a similar comparison of modification rates for hT12- versus hT72-bound protein indicates an elevation in modification rate of POT1 H266 by 5 times. As the modification rate for the hT72-bound complex represents the average rate of modification for the 6 proteins coating the hT72 ssDNA, it is plausible that only 1 of the 6 proteins is more protected by ssDNA binding while the others are solvent accessible. The enhanced binding interaction for POT1-TPP1 interacting with the 3′ hydroxyl of ssDNA (33, 45), as compared to the 5 proteins binding an internal ssDNA sequence on the hT72 substrate, could account for the difference in rates observed. Alternatively, the arrangement of protein on the longer, more structured ssDNA might provide a modest protection against solvent accessibility to result in a subtly reduced modification rate as compared to free protein. In any event, the modification rate of H266 in the context of 6 wild-type proteins bound to hT72 ssDNA more closely resembles that of free protein than the hT12-bound complex.
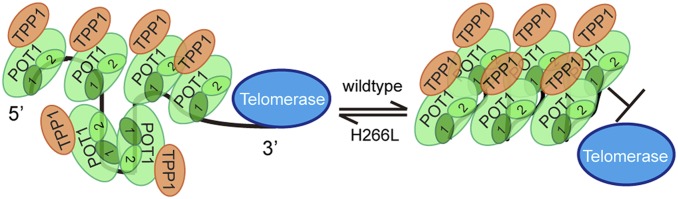
Another possible explanation for the discrepancy in POT1-TPP1 interactions with long versus short telomere ssDNA may be related to different secondary structures adopted by longer ssDNA substrates. The G-rich sequence makes telomere ssDNA prone to forming complex secondary structures such as T-loops and G-quadruplexes, which are dependent on the length of ssDNA (19–21). The binding of POT1 to telomere ssDNA normally helps to relieve such structures to properly regulate telomere maintenance (24, 25, 40). Therefore, it is conceivable that the first OB-fold domain of POT1 recognizes telomere ssDNA regardless of its secondary structure, while the second OB fold of POT1 partially remains solvent accessible allowing for dynamic interactions with telomere ssDNA with potential to form secondary structures. Our data demonstrate that the introduction of the H266L mutation slows the rate at which G-quadruplex structures are relieved by POT1-TPP1 protein. As G-quadruplex structures generally inhibit telomerase-mediated extension of telomere ssDNA (24, 39) and the POT1 H266L mutant impairs this ability, it would be expected that the POT1 H266L mutation would be associated with shorter telomeres. Therefore, it is more likely that the weaker affinity and reduced preference for 3′ binding that is associated with the H266L POT1 mutation negate some of the protective properties of the protein, thereby allowing telomerase to more readily access the 3′ end of telomeres to promote overextension in the context of the H266L mutation (Fig. 4).
Fig. 4.
The proposed mechanism of POT1-TPP1 complex-mediated regulation of telomerase. Telomere ssDNA is capable of forming higher ordered structures. POT1-TPP1 wild-type complexes bind to telomere ssDNA thereby forming a compact structure and protecting the 3′ end of the telomere ssDNA overhang from telomerase accessibility (Right). However, POT1-TPP1 complexes harboring the H266L POT1 mutant impair POT1-TPP1 complex function, leaving the 3′ end of the telomere ssDNA overhang accessible for telomerase extension (Left).
Building upon studies demonstrating that the POT1-TPP1 heterodimer acts as a telomerase processivity factor (16, 36), our investigation further defines this role as being conditional for telomere ssDNA length and the number of POT1-TPP1 proteins bound to it. Specifically, our data describe an opposing role of the telomere-binding heterodimer by which longer ssDNA products are coated with multiple proteins to inhibit telomerase activity. Taken together, these data present a scenario where POT1-TPP1 binds short telomere ssDNA substrates to promote telomerase recruitment and activity (16, 36). However, once the ssDNA overhang reaches a specific threshold length sheathed by multiple POT1-TPP1 complexes, telomerase binding and subsequent extension is prevented. This inhibitory regulation of telomerase in the presence of long ssDNA overhangs is lost in the case of H266L POT1 mutant protein indicating the H266 residue is critical for telomerase regulation. In this regard, and potentially separate from its role in resolving DNA secondary structure, the mutant POT1 is unable to sufficiently protect the extreme 3′ end of telomere ssDNA promoting telomerase accessibility and unbridled telomere extension. While the average length of the ssDNA overhang of telomeres is usually in the range of 50 to 200 nt, this length is not always fixed and is subject to being regulated (46). Our results provide a potential regulatory event by which telomerase extends the ssDNA of telomeres in S-phase following telomere replication (47). After this initial extension event, our data indicate that the longer G-rich products are coated with native POT1-TPP1 protein to prevent additional telomerase-mediated extension events. This balance would insure that telomerase adds approximately the same number of nucleotides that are lost due to the end-replication problem for each cell cycle and would contribute to maintaining a relatively constant telomere length in telomerase-positive cells. In the case of the POT1 H266L mutation, the ability to depress telomerase-mediated extension is lost, resulting in more telomerase-mediated extension events and subsequently longer telomeres at each round of replication.
In summary, our results support a model in which POT1-TPP1 regulates telomere length homeostasis by coating long ssDNA substrates to render them inaccessible for telomerase extension. The presence of the CLL-associated H266L POT1 mutant impairs this ability of POT1-TPP1 to destabilize long telomere ssDNA, thereby leaving the telomere in a state that is accessible for telomerase-mediated extension, regardless of telomere ssDNA substrate length.
Materials and Methods
Protein expression and purification, HRF analysis, EMSA assays, telomere restriction fragment assay, and direct telomerase assay were performed using published protocols (24, 31, 35, 48). CRISPR-Cas9–mediated introduction of SNP was performed following published protocols (49). See SI Appendix, SI Materials and Methods, for full details.
Data Availability.
All data generated and analyzed over the course of the current study are included within the manuscript or SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Magdalena Małgowska and Sukanya Srinivasan for comments on the manuscript and for helpful discussion regarding experimental design. We thank Dr. Wei Huang for computational assistance related to the footprinting data and structural models and Sayan Gupta and Jennifer Bohon for assistance with sample irradiation. We acknowledge the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO, for services rendered by the Genome Engineering and Induced Pluripotent Stem Cell Center. These services included genome engineering to introduce the described H266L mutation into the POT1 gene of HCT116 cells using CRISPR-Cas9. Work in the D.J.T. laboratory is funded by grants from the NIH (R01 GM133841 and R01 CA240993). The Siteman Cancer Center is supported in part by a National Cancer Institute Cancer Center Support Grant (P30 CA091842). The Advanced Light Source synchrotron is supported in part by grants from the NIH (R01 GM126218) and Department of Energy (DE-AC02-05CH11231). The Brookhaven National Laboratory synchrotron at the National Synchrotron Light Source is supported in part by an NIH grant (P30 EB009998).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.L.O. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905381116/-/DCSupplemental.
References
- 1.Blackburn E. H., Telomeres and telomerase: Their mechanisms of action and the effects of altering their functions. FEBS Lett. 579, 859–862 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Moyzis R. K., et al. , A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U.S.A. 85, 6622–6626 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange T., Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Nandakumar J., Cech T. R., Finding the end: Recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 14, 69–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsay A. J., et al. , POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 45, 526–530 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Shi J., et al. ; NCI DCEG Cancer Sequencing Working Group; NCI DCEG Cancer Genomics Research Laboratory; French Familial Melanoma Study Group , Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 46, 482–486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trigueros-Motos L., Mutations in POT1 predispose to familial cutaneous malignant melanoma. Clin. Genet. 86, 217–218 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Bainbridge M. N., et al. ; Gliogene Consortium , Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl. Cancer Inst. 107, 384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvete O., et al. , A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat. Commun. 6, 8383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandakumar J., et al. , The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abreu E., et al. , TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 30, 2971–2982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sexton A. N., Youmans D. T., Collins K., Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 287, 34455–34464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong F. L., et al. , TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt J. C., Zaug A. J., Cech T. R., Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumann P., Cech T. R., Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Wang F., et al. , The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Marcand S., Gilson E., Shore D., A protein-counting mechanism for telomere length regulation in yeast. Science 275, 986–990 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Teixeira M. T., Arneric M., Sperisen P., Lingner J., Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117, 323–335 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Williamson J. R., Raghuraman M. K., Cech T. R., Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell 59, 871–880 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Sundquist W. I., Klug A., Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 342, 825–829 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Griffith J. D., et al. , Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Zahler A. M., Williamson J. R., Cech T. R., Prescott D. M., Inhibition of telomerase by G-quartet DNA structures. Nature 350, 718–720 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Wang Q., et al. , G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 39, 6229–6237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullins M. R., et al. , POT1-TPP1 binding and unfolding of telomere DNA discriminates against structural polymorphism. J. Mol. Biol. 428, 2695–2708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang H., Buncher N., Opresko P. L., Myong S., POT1-TPP1 regulates telomeric overhang structural dynamics. Structure 20, 1872–1880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takai K. K., Hooper S., Blackwood S., Gandhi R., de Lange T., In vivo stoichiometry of shelterin components. J. Biol. Chem. 285, 1457–1467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor D. J., Podell E. R., Taatjes D. J., Cech T. R., Multiple POT1-TPP1 proteins coat and compact long telomeric single-stranded DNA. J. Mol. Biol. 410, 10–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarov V. L., Hirose Y., Langmore J. P., Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88, 657–666 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Corriveau M., Mullins M. R., Baus D., Harris M. E., Taylor D. J., Coordinated interactions of multiple POT1-TPP1 proteins with telomere DNA. J. Biol. Chem. 288, 16361–16370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain S. S., Tullius T. D., Footprinting protein-DNA complexes using the hydroxyl radical. Nat. Protoc. 3, 1092–1100 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Kiselar J., Chance M. R., High-resolution hydroxyl radical protein footprinting: Biophysics tool for drug discovery. Annu. Rev. Biophys., 10.1146/annurev-biophys-070317-033123 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Chance M. R., Protein footprinting comes of age: Mass spectrometry for biophysical structure assessment. Mol. Cell. Proteomics 16, 706–716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei M., Podell E. R., Cech T. R., Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 11, 1223–1229 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Deperalta G., et al. , Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. MAbs 5, 86–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takamoto K., Chance M. R., Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Xin H., et al. , TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445, 559–562 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Nandakumar J., Cech T. R., DNA-induced dimerization of the single-stranded DNA binding telomeric protein Pot1 from Schizosaccharomyces pombe. Nucleic Acids Res. 40, 235–244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaug A. J., Podell E. R., Nandakumar J., Cech T. R., Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24, 613–622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaug A. J., Podell E. R., Cech T. R., Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U.S.A. 102, 10864–10869 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray S., Bandaria J. N., Qureshi M. H., Yildiz A., Balci H., G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc. Natl. Acad. Sci. U.S.A. 111, 2990–2995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dapić V., et al. , Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 31, 2097–2107 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambrus A., et al. , Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray D. M., et al. , Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality 20, 431–440 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Huang W., Ravikumar K. M., Chance M. R., Yang S., Quantitative mapping of protein structure by hydroxyl radical footprinting-mediated structural mass spectrometry: A protection factor analysis. Biophys. J. 108, 107–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loayza D., Parsons H., Donigian J., Hoke K., de Lange T., DNA binding features of human POT1: A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279, 13241–13248 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Huffman K. E., Levene S. D., Tesmer V. M., Shay J. W., Wright W. E., Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem. 275, 19719–19722 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y., et al. , Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajavel M., et al. , Dynamic peptides of human TPP1 fulfill diverse functions in telomere maintenance. Nucleic Acids Res. 44, 10467–10479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sentmanat M. F., Peters S. T., Florian C. P., Connelly J. P., Pruett-Miller S. M., A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8, 888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed over the course of the current study are included within the manuscript or SI Appendix.