Significance
Endothelial cells (ECs), which line all vertebrate blood vessels, are highly heterogeneous across different tissues. The present study uses a genetic approach to specifically tag mRNAs within ECs of the mouse, thereby allowing recovery and sequence analysis to evaluate the EC-specific gene expression program directly from intact organs. Our findings demonstrate marked heterogeneity in EC gene expression across different vascular beds under both normal and disease conditions, with a more accurate picture than can be achieved using other methods. The data generated in these studies advance our understanding of EC function in different blood vessels and provide a valuable resource for future studies.
Keywords: endothelial cell, gene expression, RNA sequencing
Abstract
Endothelial cells (ECs) are highly specialized across vascular beds. However, given their interspersed anatomic distribution, comprehensive characterization of the molecular basis for this heterogeneity in vivo has been limited. By applying endothelial-specific translating ribosome affinity purification (EC-TRAP) combined with high-throughput RNA sequencing analysis, we identified pan EC-enriched genes and tissue-specific EC transcripts, which include both established markers and genes previously unappreciated for their presence in ECs. In addition, EC-TRAP limits changes in gene expression after EC isolation and in vitro expansion, as well as rapid vascular bed-specific shifts in EC gene expression profiles as a result of the enzymatic tissue dissociation required to generate single-cell suspensions for fluorescence-activated cell sorting or single-cell RNA sequencing analysis. Comparison of our EC-TRAP with published single-cell RNA sequencing data further demonstrates considerably greater sensitivity of EC-TRAP for the detection of low abundant transcripts. Application of EC-TRAP to examine the in vivo host response to lipopolysaccharide (LPS) revealed the induction of gene expression programs associated with a native defense response, with marked differences across vascular beds. Furthermore, comparative analysis of whole-tissue and TRAP-selected mRNAs identified LPS-induced differences that would not have been detected by whole-tissue analysis alone. Together, these data provide a resource for the analysis of EC-specific gene expression programs across heterogeneous vascular beds under both physiologic and pathologic conditions.
Endothelial cells (ECs) form the inner lining of all vertebrate blood vessels, providing a critical barrier between circulating blood and parenchymal cells. In addition to maintaining blood fluidity and regulating vascular tone, ECs control the transport of nutrients and metabolites to and from underlying tissues while also contributing to host defense and the control of inflammatory processes. Consistent with their diverse microenvironments and specialized functions, ECs have evolved to display remarkable morphologic and structural heterogeneity across organs (1–5). Initial studies of well-established EC markers including von Willebrand factor (Vwf), platelet endothelial cell adhesion molecule (Pecam), and Cd34 demonstrated heterogeneous expression of these genes across the vascular tree and in distinct vascular beds (6, 7). However, more comprehensive characterization of the EC transcriptome has been largely limited by the challenge of purifying ECs, given their highly interspersed anatomic distribution among parenchymal cells in various tissues.
Previous studies have focused on human umbilical vein endothelial cells expanded in cell culture, or on isolation and in vitro expansion of ECs from distinct vascular beds such as the brain or liver (8–10). Although ECs expanded in vitro may retain some tissue-specific characteristics, the loss of microenvironmental cues could result in shifts in the expression program that no longer faithfully reflect the in vivo EC transcriptome (11–14). Several in vivo phage display approaches have been used to identify vascular bed-specific markers or “vascular zip codes,” although generally resulting in the identification of only a limited number of EC binding peptides (15–18). In addition, fluorescence-activated cell sorting of cells endogenously marked with a fluorescent protein (19–23) or via intravital staining (24), and more recently single-cell RNA sequencing (RNASeq) analyses (25–30), have been performed to determine cell type-specific transcriptomes. However, these latter methods rely on the preparation of single-cell suspensions after enzymatic tissue dissociation, often followed by flow sorting under high shear stress, which may affect gene expression programs.
Mouse transgenic approaches have enabled transcriptional profiling directly in vivo via translating ribosome affinity purification (TRAP) (31–36). This method relies on the inducible expression of an epitope-tagged ribosomal protein in a cell type-specific pattern, thus facilitating isolation of ribosome-associated transcripts, also known as the translatome (37), for particular cellular subsets directly from a complex mixture of cells. Here we report the application of EC-specific TRAP (EC-TRAP) combined with high-throughput RNASeq analysis to characterize EC translatomes across multiple vascular beds. Our findings demonstrate a high degree of tissue-specific endothelial heterogeneity, as well as notable shifts in EC mRNA content during the process of enzymatic tissue dissociation. Using EC-TRAP, we also demonstrate vascular bed-specific variation in endothelial reactivity in response to lipopolysaccharide (LPS) exposure, which would not have been identified by whole-tissue RNASeq data analysis.
Results and Discussion
In Vitro Expansion of ECs Leads to Phenotypic Drift.
Microarray analysis was performed on ECs isolated from heart or kidney, either directly after isolation (day 0) or after additional expansion in culture for 3 d. As shown by principal component analysis (PCA; Fig. 1), in vitro expansion of primary ECs results in major shifts in gene expression profiles, with the expression profiles of heart and kidney more closely resembling each other after 3 d in culture, than compared with freshly isolated ECs at day 0. These data suggest that removal of the native environment results in phenotypic drift with regression toward a common EC transcription profile. This finding supports previous reports (11–14) demonstrating an important role for microenvironmental cues in maintaining the molecular heterogeneity of ECs.
Fig. 1.
PCA of expression profiles from primary EC of wild-type heart and kidney capillaries obtained immediately after isolation (day 0) identified 4,783 differentially expressed genes (FDR < 10%) between tissue origins. In vitro expansion for 3 d resulted in significant shifts in expression programs, with the number of differentially expressed genes being reduced to 2,397 between heart and kidney ECs, indicating phenotypic drift occurring in the absence of a native microenvironment; n = 3 biologic replicates per condition.
Rpl22 Isoform Analysis to Assess Tissue-Specific EC Content.
To probe EC heterogeneity directly in vivo, we applied TRAP by taking advantage of the RiboTag mouse, which carries a conditional Rpl22 allele (35). Although a number of Cre transgenes have been used to target the endothelium (38), we and others have previously shown effective pan-EC targeting using Cre-recombinase driven by the Tek promoter in evaluating the cellular origin of coagulation factor VIII (39, 40). As shown in Fig. 2 A–D, analysis of mice carrying a Tek-Cre transgene (Tek-Cre+/0) (41, 42), together with the RosamTmG reporter (43) confirms efficient targeting of ECs in vivo. The high degree of EC sensitivity and specificity observed for the Tek-Cre transgene suggests that the extent of genomic Rpl22 excision in organs from Rpl22fl/fl, Tek-Cre+/0 mice should accurately reflect the relative fraction of ECs within those tissues. Quantitative analysis of Rpl22 targeting by direct high-throughput genomic sequence analysis demonstrated the lowest fraction of ECs in brain (5.5 ± 0.9%), followed by liver (13.5 ± 1.7%), kidney (16.3 ± 2.6%), and heart (38.6 ± 3.8), with the highest fraction observed in the lung (48.4 ± 6.7%; Fig. 2E). Although we cannot exclude a minor contribution from non-ECs (transiently) expressing Tek by this approach (see Correction of TRAP-Enriched Genes for Hematopoietic Cell Content), the quantitative Rpl22 data are consistent with our microscopic images from RosamT/mG, Tek-Cre+/0 mice (Fig. 2 A–D) and comparable to previous estimates of EC content based on histologic examination and fluorescence-activated cell sorting analysis (20, 44).
Fig. 2.
In vivo targeting of Tek-positive cells to determine tissue-specific EC content. (A–D) Histologic analysis of RosamT/mG,Tek-Cre+/0 brain (A), heart (B), kidney (C), and liver (D) sections. Tek-positive cells are identified by the expression of membrane-bound green fluorescent protein, whereas Tek-negative cells express membrane-bound Tomato red fluorescent protein. (Scale bar, 25 µm.) (E) Percentage of Tek-positive cells as a proxy for EC content in each tissue, determined by high-throughput DNA sequencing of Rpl22 isoforms in tissues from Rpl22fl/fl, Tek-Cre+/0 animals; n = 5 biologic replicates (mean ± SD).
Evaluation of EC-Specific Translatomes by TRAP.
TRAP specifically captures actively translated mRNAs (translatome), which may differ quantitatively from the total cellular mRNA pool (transcriptome). To determine whether tissue transcriptomes obtained before EC-TRAP are comparable to tissue translatomes, or whether corrections are needed for potential disproportional mRNA capture because of differences in ribosome density across mRNAs, we performed TRAP and RNAseq analysis on tissues of 10-wk-old male mice expressing HA-tagged ribosomes in all cell types (Rpl22fl/fl, EIIa-Cre+/0). Before tissue isolation, animals were perfused with cycloheximide to block translation and stabilize tagged ribosomes on their cognate mRNAs to minimize potential shifts in ribosome distribution, thereby maintaining the cellular state of translation during tissue collection and immunoprecipitation (45, 46) (SI Appendix, Fig. S1). Expression profiles obtained before and after TRAP were highly reproducible between biologic replicates, with distinct patterns evident across organs. Only minor changes were observed between the total mRNA and actively translated mRNA within a given tissue (Fig. 3A), resulting in a limited number of transcripts that were significantly different between the 2 mRNA pools (SI Appendix, Fig. S2 and Table S1). These data demonstrate that signatures obtained after EC-TRAP should primarily reflect cell-specific gene expression programs, with little or no contribution from shifts in ribosome distribution, and that the transcriptome of a tissue lysate can serve as a proxy for the corresponding tissue translatome.
Fig. 3.
Evaluation of transcriptome vs. translatome data after (EC) TRAP. (A) PCA of transcriptome vs. translatome data from 10-wk-old Rpl22fl/fl, EIIa-Cre+/0 male mice shows only minor differences between tissue-specific mRNA pools from brain, heart, kidney, liver, and lung, whereas (B) EC translatomes obtained after TRAP from Rpl22fl/fl, Tek-Cre+/0 tissues are highly distinct from the unselected tissue transcriptomes, with the exception of lung. n = 3 biologic replicates per tissue.
We next analyzed TRAP-selected mRNA fractions collected from multiple tissues of 10-wk-old Rpl22fl/fl, Tek-Cre+/0 male mice after cycloheximide perfusion. To avoid contamination of TRAP-selected mRNAs with transcripts originating from parenchymal cells (35), we also evaluated the accompanying unselected tissue transcriptomes by RNAseq analysis and calculated relative enrichment scores. These scores identify those transcripts that are more abundant in Tek+ cells (enriched genes, log2 fold change >0), or more abundant in Tek− cells (depleted genes, log2 fold change <0), where the former should represent EC-specific genes and the latter parenchymal cell-specific genes (SI Appendix, Fig. S2). As expected, gene ontology (GO) analysis of TRAP-enriched transcripts showed highly significant associations with endothelial and hematopoietic biologic processes including “vasculature development,” “immune response,” and “hemopoiesis,” while depleted gene sets showed signals consistent with parenchymal, tissue-specific processes (SI Appendix, Fig. S3).
Correction of TRAP-Enriched Genes for Hematopoietic Cell Content.
Although the Tek-Cre transgene provides an efficient and specific marker of the EC compartment, the Tek gene is also transiently expressed in hematopoietic cells (47), which could result in the false identification of mRNAs as EC-specific as a result of blood contamination of tissues. For example, despite exsanguination and cardiac perfusion before organ harvest to remove circulating blood cells, the genes encoding hemoglobin α and β (Hba-a1, Hba-a2, Hbb-bs, and Hbb-bt) were among the most enriched transcripts across all tissues, and although α-hemoglobin gene expression has been reported in arterial ECs, β-hemoglobin has not directly been associated with the endothelium (48, 49). Thus, the similar degree of enrichment observed for both α- and β-hemoglobin in our EC-TRAP suggests a contribution of HA-tagged polysomes derived from circulating blood cells that were not completely removed by perfusion before tissue isolation. To eliminate transcripts originating from hematopoietic cells, we applied additional computational filters based on data obtained from TRAP performed directly on peripheral blood samples from Rpl22fl/fl, Tek-Cre+/0 mice, as well as EC-TRAP on tissues collected from an Rpl22fl/fl, Tek-Cre+/0 recipient after bone marrow transplant using an Rpl22fl/fl, Tek-Cre− donor (SI Appendix). In addition to removing the hemoglobin genes and other known lymphoid and myeloid cell markers, including a number of established EC markers known to be expressed by megakaryocytes/platelets (50), applying these filters provides the most specific set of EC-enriched genes. GO analysis of this more restricted EC gene set now showed enrichment limited to vascular-related biologic processes (Fig. 4A) with loss of the GO terms for marrow-derived cell populations identified in the broader analysis (SI Appendix, Fig. S3).
Fig. 4.
Identification of enriched transcripts after EC-TRAP. (A) GO analysis of the 500 top-ranked genes with the highest enrichment scores after EC-TRAP (FDR < 10%) shows overrepresentation of transcripts involved in vascular-related processes. (B) Unsupervised hierarchical clustering of EC-enriched genes (enrichment score >2 in at least 1 tissue) shows distinct, highly heterogeneous vascular bed-specific EC expression patterns. (C) Comparison of the top 500 most enriched genes per tissue identifies a group of pan-endothelial and subsets of tissue-specific EC-enriched genes. Data based on n = 3 biologic replicates per tissue.
TRAP Identifies EC Heterogeneity Across Vascular Beds.
PCA of EC translatomes demonstrates distinct, reproducible patterns for each organ (Fig. 3B). This observation is confirmed by hierarchical clustering of enrichment scores where, consistent with the high percentage of ECs in the lung and heart, the degree of enrichment for EC transcripts in these organs is generally lower than that observed in brain, kidney, and liver (Fig. 4B).
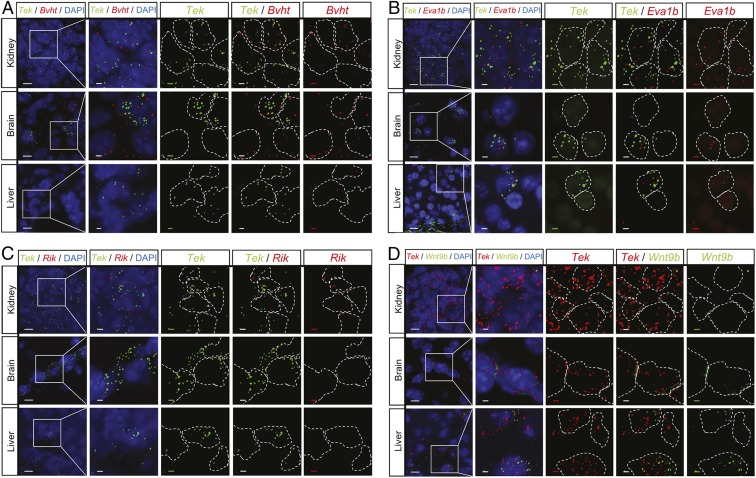
Overlap in expression across the different vascular beds for the 500 top-ranked genes based on enrichment scores for each tissue is shown in Fig. 4C, identifying 82 genes shared among all 5 vascular beds, including Tek, as expected, as well as other established pan-EC markers such as Cdh5, Nos3, Eng, and Robo4. This list also includes Ephb4, Dll4, and Flt4, specifically marking arterial, venous, and lymphatic ECs, respectively, demonstrating efficient targeting of all 3 EC subtypes (51, 52) (SI Appendix, Table S2). In addition to the pan-EC markers that have previously been associated with EC function and/or localization (23, 32, 53–55), our EC-TRAP analysis also identified potential novel pan-EC genes, including Gm20748 (also known as Bvht) and Eva1b, with in situ hybridization confirming colocalization of these transcripts with Tek transcripts in ECs of brain, kidney, and liver (Fig. 5 A and B).
Fig. 5.
Single-molecule in situ hybridization validates EC-enriched transcripts identified by TRAP. (A and B) Colocalization of Bvht and Eva1b, both identified as pan-EC markers by EC-TRAP, with Tek by in situ hybridization in kidney, brain, and liver sections. (C and D) 3110099E03Rik (Rik) and Wnt9b were identified as kidney- and liver-specific EC markers, respectively, and were only present in the corresponding tissues, where they also colocalized with Tek, confirming their EC origin. (Insets) Dotted lines outline the cell nuclei as indicated by DAPI counterstaining. (Scale bars, 10 µm in overviews [left column in each panel] and 2.5 µm in Insets [remaining columns]).
The EC-TRAP data identify marked heterogeneity of gene expression levels for EC-enriched genes across tissues, including genes restricted to a single or several vascular beds. An interesting subset of genes appears to be EC-enriched in 1 or more tissue or tissues, with expression levels more abundant in non-ECs in another tissue (SI Appendix, Fig. S4 and Dataset S1). For example, whereas several solute carriers play important roles in proximal tubules in the kidney, and are thus depleted after kidney EC-TRAP, a number of these genes are significantly enriched in brain ECs (Dataset S1). Focusing on EC-specific genes with expression restricted to a single vascular bed identified the largest number of unique markers for brain ECs, including several genes previously associated with the blood–brain barrier (e.g., Rad54b, Zic3, and Slco1c) (20, 25, 27, 56). Smaller subsets of tissue-specific EC markers were also identified for heart, kidney, liver, and lung (Table 1), with specific kidney EC expression of lincRNA 3110099E03Rik and liver EC expression of Wnt9b confirmed by in situ hybridization (Fig. 5 C and D).
Table 1.
Tissue-specific, EC restricted transcripts identified by EC-TRAP
| Tissue | Genes |
| Brain | 1700009J07Rik, 6030468B19Rik, Cldn13, Dyrk4, Edn3, Ermap, Fcnb, Foxl2, Foxq1, Gals3st4, Gm10384, Ndnf, Rad54b, Slc19a3, Slco1c1, Sox15, Tmem8c, Zic3 |
| Heart | A2m, Gm13154, Gm16565, Gm17234, Itgbl1 |
| Kidney | 3110099E03Rik, Calca, E130006D01Rik, Fam101a, Gata5, Krtdap, Lgr5, U90926 |
| Liver | 6030498E09Rik, Adamdec1, Gm13994, Il13ra2, Serpina3i, Smc1b, Sprr2b, Tmem132e, Ush1g, Wnt9b |
| Lung | 4930452B06Rik, Aard, Abca14, Ankrd63, B3gnt7, Ear1, Fignl2, Glp1r, Gm26878, Gm6116, Gm830, Hic2, Krt79, Mkrn3, Nr6a1, Pnpla5, Prdm8, Prr18, Prss12, Rnase1, Rprml, Rufy4, Speer4f, Tekt3, Tepp, Trgj1, Trim29 |
Transcripts significantly enriched (FDR < 10%) within a specific vascular bed while absent the transcriptomes of other tissues.
Comparison between EC-TRAP and Single-Cell RNASeq.
Recently developed single-cell RNASeq methods make it possible to distinguish cellular subtypes within complex mixtures of cells, including multiple subsets of ECs, as previously reported for the brain (23, 27). However, several studies raise the concern that the isolation procedures required to obtain individual cells for RNASeq analysis may perturb relative mRNA abundance (57–59). To evaluate the potential impact of such changes in EC expression programs across tissues, perfusion and enzymatic tissue dissociation were performed in the absence of cycloheximide to mimic the effects of single-cell isolation procedures (including continuing changes in mRNA transcription and translation), until the addition of cycloheximide-containing buffer before TRAP, ∼90 min after organ harvest.
Although some degree of vascular bed-specific gene expression patterns persist, substantial changes are observed when compared to the profiles obtained from cycloheximide-perfused animals (Fig. 6A). Whereas the kidney endothelium showed relatively small shifts in expression profiles between isolation methods, liver and brain ECs exhibit larger deviations as well as lower correlations between biologic replicates. Indeed, 48.2% of genes expressed in brain ECs (3,794/7,866) and 21.6% expressed in liver ECs (2,306/10,696) are differentially expressed when comparing the 2 preparation methods, with only a 7.4% difference observed for kidney EC genes (779/10,475). As a result, hierarchical clustering of EC translatomes identified liver and brain ECs as more similar based on sample preparation method than by tissue of origin, as shown in Fig. 6B.
Fig. 6.
Dissociation-induced changes in EC gene expression programs. (A) PCA analysis of brain, kidney, and liver EC translatomes obtained from TRAP samples after cycloheximide perfusion and mechanical dissociation vs. samples after enzymatic tissue dissociation without cycloheximide perfusion. (B) mRNA abundance based on log2(TPM) values, and presented as a yellow (low abundance) to blue (high abundance) gradient, shows clustering of brain and liver EC translatomes by preparation method rather than by tissue type. Data from n = 3 biologic replicates per tissue.
In addition to the potential for shifts in the EC expression program during the preparation of single cells, current single-cell RNASeq methods exhibit a relatively limited sensitivity to detect low abundance transcripts (60, 61). Consistent with these previous reports, of the 100 most abundant EC-enriched transcripts per tissue identified in our EC-TRAP dataset, many have also been identified in ECs on the basis of single-cell RNASeq data (median 50; range 28 to 77; SI Appendix, Table S3). In contrast, only a few of the 100 lowest abundant genes were detected by single-cell RNAseq (median 2; range 0 to 24) (25–27, 29).
Taken together, the EC-TRAP approach, combined with in vivo cycloheximide perfusion before mRNA preparation reported here, should result in a more accurate snapshot of native EC profiles across different vascular beds, including low abundant transcripts, than is possible with current single-cell isolation and RNASeq methods.
Endotoxemia Induces Tissue-Specific Changes in the EC Gene Expression Program.
The endothelium provides a key line of host defense in response to microbial pathogens. To examine the tissue-specific response in an animal model for systemic bacterial endotoxemia, 10-wk-old Rpl22fl/fl, Tek-Cre+/0 male mice received an i.p. injection with 1 mg/kg LPS. Expression profiles were evaluated 4 h after exposure, at which point significant gene expression changes are expected, while minimizing secondary downstream effects (62). Analysis of whole-organ transcriptomes (including both ECs and parenchymal cells) demonstrated marked changes in gene expression in response to LPS, with gene ontology analysis yielding highly significant values for terms associated with host defense and immune response (SI Appendix, Fig. S5A). The extent of gene changes was highly variable across tissues, with a markedly reduced number of LPS-responsive mRNAs evident for intact brain tissue (Fig. 7A). Analysis of the EC-specific response, however, showed similar numbers of genes affected by LPS treatment across all tissues, including brain (Fig. 7B). These data suggest a dramatic reduction in the effect of LPS on brain parenchymal cells compared with other tissues, consistent with a high degree of protection provided by the blood–brain barrier (63). In addition, this also illustrates a limitation of RNASeq of whole tissues, where cell type-specific responses can be masked by changes in more abundant cellular subsets. Interestingly, whereas most vascular bed-specific EC translatomes exhibited considerably more down-regulated than up-regulated transcripts in response to LPS, the opposite pattern was observed for brain ECs (Fig. 7B and SI Appendix, Fig. S5 B and C).
Fig. 7.
In vivo LPS-induced changes uncovered by EC-TRAP. (A and B) Volcano plots indicate genes significantly affected 4 h after LPS exposure (red), either for tissue transcriptomes (A) or in the EC translatome as identified by EC-TRAP (B). The horizontal dotted lines mark an FDR cutoff <10%, with the vertical dotted lines denoting log2 fold changes of >1 or <−1. Data averaged from n = 3 biologic replicates per tissue.
Consistent with previous reports (64–66), EC-enriched genes up-regulated by LPS included several adhesion molecules required for leukocyte recruitment and migration, while genes involved in maintenance of the EC barrier were down-regulated. Furthermore, a shift toward a procoagulant state was observed, with reduced expression of anticoagulant and profibrinolytic genes (SI Appendix, Table S4). Within this limited set of genes, expression of the protein C receptor (Procr) provides an example of vascular bed-specific variation in EC reactivity in response to LPS, with significant down-regulation of Procr in brain, kidney, and heart, whereas expression in liver and lung was increased. These data could explain the organ-specific susceptibility to increased vascular leakage after LPS challenge previously reported for mice with reduced Procr expression (67).
Conclusion
The interspersed anatomic distribution of ECs and marked dependence of their gene expression programs on microenvironmental cues have constituted major challenges to the study of EC function and responses to (patho)physiological stimuli in vivo. The EC-TRAP strategy described here provides a powerful tool for the analysis of the EC translatome across multiple vascular beds and in response to pathologic challenges in the whole animal. Our findings demonstrate that this method offers an accurate in vivo snapshot of tissue-specific EC expression profiles, resulting in better cellular resolution than whole-tissue RNASeq, while maintaining a high degree of sensitivity to detect low abundant transcripts. In addition, these data provide a database of tissue-specific EC gene expression that should be a useful reference for other studies, including the identification of tissue-specific targets for drug delivery. Future refinement of EC-TRAP taking advantage of intersectional genetic approaches based on combined expression of 2 or more EC markers (68) may lead to greater resolution of EC subsets within a tissue, particularly in combination with single-cell RNAseq methods. This would be particularly valuable as many of the widely used Cre models to target ECs, also target (subsets of) hematopoietic cells, including Tek-Cre as described here, as well as other commonly used EC-specific Cre transgenes (38, 69, 70). Finally, future applications of this approach should significantly advance our understanding of EC function in vivo under both physiologic and pathophysiologic conditions, potentially leading to the identification of new disease-specific biomarkers as well as potential novel therapeutic targets.
Materials and Methods
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Michigan. Detailed descriptions of the mice and procedures used in these studies, including sample preparations for TRAP and high-throughput RNA sequencing, data analysis and follow-up validation by single-molecule fluorescent in situ hybridization assays, are provided in the SI Appendix, Materials and Methods.
Data Availability.
All RNASeq data supporting these studies have been deposited in the Gene Expression Omnibus under accession number GSE138630.
Supplementary Material
Acknowledgments
We thank Lauren Janes, Kate Spokes, Peter Zwiers, Tamar Feinberg, and Nitin Kumar for their expert technical assistance with the in vitro experiments and Eliane Popa for the helpful discussions. This research was supported by grants from the NIH (R35HL135793 to D.G. and R01HL122684 to S.K.G.), and the American Heart Association (16POST27770093 to A.C.A.C.). D.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (accession no. GSE138630).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912409116/-/DCSupplemental.
References
- 1.Aird W. C., Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ. Res. 100, 174–190 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Potente M., Mäkinen T., Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 18, 477–494 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Ribatti D., Nico B., Vacca A., Roncali L., Dammacco F., Endothelial cell heterogeneity and organ specificity. J. Hematother. Stem Cell Res. 11, 81–90 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Atkins G. B., Jain M. K., Hamik A., Endothelial differentiation: Molecular mechanisms of specification and heterogeneity. Arterioscler. Thromb. Vasc. Biol. 31, 1476–1484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarron J. G., et al. , Heterogeneity and emergent behaviour in the vascular endothelium. Curr. Opin. Pharmacol. 45, 23–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aird W. C., et al. , Vascular bed-specific expression of an endothelial cell gene is programmed by the tissue microenvironment. J. Cell Biol. 138, 1117–1124 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pusztaszeri M. P., Seelentag W., Bosman F. T., Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J. Histochem. Cytochem. 54, 385–395 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Aman J., Weijers E. M., van Nieuw Amerongen G. P., Malik A. B., van Hinsbergh V. W., Using cultured endothelial cells to study endothelial barrier dysfunction: Challenges and opportunities. Am. J. Physiol. Lung Cell Mol. Physiol. 311, L453–L466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helms H. C., et al. , In vitro models of the blood-brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow Metab. 36, 862–890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer J., Gonelle-Gispert C., Morel P., Bühler L., Methods for isolation and purification of murine liver sinusoidal endothelial cells: A systematic review. PLoS One 11, e0151945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amatschek S., et al. , Blood and lymphatic endothelial cell-specific differentiation programs are stringently controlled by the tissue environment. Blood 109, 4777–4785 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Durr E., et al. , Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 22, 985–992 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Lacorre D. A., et al. , Plasticity of endothelial cells: Rapid dedifferentiation of freshly isolated high endothelial venule endothelial cells outside the lymphoid tissue microenvironment. Blood 103, 4164–4172 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Calabria A. R., Shusta E. V., A genomic comparison of in vivo and in vitro brain microvascular endothelial cells. J. Cereb. Blood Flow Metab. 28, 135–148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasqualini R., Ruoslahti E., Organ targeting in vivo using phage display peptide libraries. Nature 380, 364–366 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Rajotte D., et al. , Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J. Clin. Invest. 102, 430–437 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonson A. B., Schnitzer J. E., Vascular proteomic mapping in vivo. J. Thromb. Haemost. 5 (suppl. 1), 183–187 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Tang F. H. F., et al. , A ligand motif enables differential vascular targeting of endothelial junctions between brain and retina. Proc. Natl. Acad. Sci. U.S.A. 116, 2300–2305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunskill E. W., Potter S. S., Gene expression programs of mouse endothelial cells in kidney development and disease. PLoS One 5, e12034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daneman R., et al. , The mouse blood-brain barrier transcriptome: A new resource for understanding the development and function of brain endothelial cells. PLoS One 5, e13741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlereth K., et al. , The transcriptomic and epigenetic map of vascular quiescence in the continuous lung endothelium. eLife 7, e34423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabbagh M. F., et al. , Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. eLife 7, e36187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolan D. J., et al. , Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell 26, 204–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han X., et al. , Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Lother A., et al. , Cardiac endothelial cell transcriptome. Arterioscler. Thromb. Vasc. Biol. 38, 566–574 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Vanlandewijck M., et al. , A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 28.He L., et al. , Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data 5, 180160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anonymous; Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators , Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karaiskos N., et al. , A single-cell transcriptome atlas of the mouse glomerulus. J. Am. Soc. Nephrol. 29, 2060–2068 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heiman M., et al. , A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hupe M., Li M. X., Gertow Gillner K., Adams R. H., Stenman J. M., Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic Acids Res. 42, e14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., et al. , Cell-specific translational profiling in acute kidney injury. J. Clin. Invest. 124, 1242–1254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santhosh D., Huang Z., A Tie2-driven BAC-TRAP transgenic line for in vivo endothelial gene profiling. Genesis 54, 136–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanz E., et al. , Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U.S.A. 106, 13939–13944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P., et al. , Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc. Natl. Acad. Sci. U.S.A. 110, 15395–15400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King H. A., Gerber A. P., Translatome profiling: Methods for genome-scale analysis of mRNA translation. Brief. Funct. Genomics 15, 22–31 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Payne S., De Val S., Neal A., Endothelial-specific Cre mouse models. Arterioscler. Thromb. Vasc. Biol. 38, 2550–2561 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Everett L. A., Cleuren A. C., Khoriaty R. N., Ginsburg D., Murine coagulation factor VIII is synthesized in endothelial cells. Blood 123, 3697–3705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fahs S. A., Hille M. T., Shi Q., Weiler H., Montgomery R. R., A conditional knockout mouse model reveals endothelial cells as the principal and possibly exclusive source of plasma factor VIII. Blood 123, 3706–3713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Lange W. J., Halabi C. M., Beyer A. M., Sigmund C. D., Germ line activation of the Tie2 and SMMHC promoters causes noncell-specific deletion of floxed alleles. Physiol. Genomics 35, 1–4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koni P. A., et al. , Conditional vascular cell adhesion molecule 1 deletion in mice: Impaired lymphocyte migration to bone marrow. J. Exp. Med. 193, 741–754 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Hsieh P. C., Davis M. E., Lisowski L. K., Lee R. T., Endothelial-cardiomyocyte interactions in cardiac development and repair. Annu. Rev. Physiol. 68, 51–66 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingolia N. T., Lareau L. F., Weissman J. S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ingolia N. T., Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 470, 119–142 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Tang Y., Harrington A., Yang X., Friesel R. E., Liaw L., The contribution of the Tie2+ lineage to primitive and definitive hematopoietic cells. Genesis 48, 563–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lechauve C., et al. , Endothelial cell α-globin and its molecular chaperone α-hemoglobin-stabilizing protein regulate arteriolar contractility. J. Clin. Invest. 128, 5073–5082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Straub A. C., et al. , Endothelial cell expression of haemoglobin α regulates nitric oxide signalling. Nature 491, 473–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rowley J. W., et al. , Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood 118, e101–e111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aitsebaomo J., Portbury A. L., Schisler J. C., Patterson C., Brothers and sisters: Molecular insights into arterial-venous heterogeneity. Circ. Res. 103, 929–939 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podgrabinska S., et al. , Molecular characterization of lymphatic endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 99, 16069–16074 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhasin M., et al. , Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genomics 11, 342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallgard E., et al. , Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler. Thromb. Vasc. Biol. 28, 1469–1476 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Butler L. M., et al. , Analysis of body-wide unfractionated tissue data to identify a core human endothelial transcriptome. Cell Syst. 3, 287–301.e3 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Rosenberg A. B., et al. , Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adam M., Potter A. S., Potter S. S., Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development 144, 3625–3632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang S. S., Baker K. E., Wang X., Kocher J.-P., Fryer J. D., Translational profiling of microglia reveals artifacts of cell sorting. bioRxiv:10.1101/135566 (9 May 2017).
- 59.van den Brink S. C., et al. , Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Hicks S. C., Townes F. W., Teng M., Irizarry R. A., Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 19, 562–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuevas-Diaz Duran R., Wei H., Wu J. Q., Single-cell RNA-sequencing of the brain. Clin. Transl. Med. 6, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seemann S., Zohles F., Lupp A., Comprehensive comparison of three different animal models for systemic inflammation. J. Biomed. Sci. 24, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banks W. A., Robinson S. M., Minimal penetration of lipopolysaccharide across the murine blood-brain barrier. Brain Behav. Immun. 24, 102–109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aird W. C., The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood 101, 3765–3777 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Ince C., et al. ; ADQI XIV Workgroup , The endothelium IN sepsis. Shock 45, 259–270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komarova Y. A., Kruse K., Mehta D., Malik A. B., Protein interactions at endothelial junctions and signaling mechanisms regulating endothelial permeability. Circ. Res. 120, 179–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Drygalski A., Furlan-Freguia C., Ruf W., Griffin J. H., Mosnier L. O., Organ-specific protection against lipopolysaccharide-induced vascular leak is dependent on the endothelial protein C receptor. Arterioscler. Thromb. Vasc. Biol. 33, 769–776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pu W., et al. , Genetic targeting of organ-specific blood vessels. Circ. Res. 123, 86–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He Q., et al. , The Cdh5-CreERT2 transgene causes conditional Shb gene deletion in hematopoietic cells with consequences for immune cell responses to tumors. Sci. Rep. 9, 7548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monvoisin A., et al. , VE-cadherin-CreERT2 transgenic mouse: A model for inducible recombination in the endothelium. Dev. Dyn. 235, 3413–3422 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNASeq data supporting these studies have been deposited in the Gene Expression Omnibus under accession number GSE138630.