Abstract
Context-specific GEnome-scale metabolic Network REconstructions (GENREs) provide a means to understand cellular metabolism at a deeper level of physiological detail. Here, we use transcriptomics data from chemically-exposed rat hepatocytes to constrain a GENRE of rat hepatocyte metabolism and predict biomarkers of liver toxicity using the Transcriptionally Inferred Metabolic Biomarker Response algorithm. We profiled alterations in cellular hepatocyte metabolism following in vitro exposure to four toxicants (acetaminophen, carbon tetrachloride, 2,3,7,8-tetrachlorodibenzodioxin, and trichloroethylene) for six hour. TIMBR predictions were compared with paired fresh and spent media metabolomics data from the same exposure conditions. Agreement between computational model predictions and experimental data led to the identification of specific metabolites and thus metabolic pathways associated with toxicant exposure. Here, we identified changes in the TCA metabolites citrate and alpha-ketoglutarate along with changes in carbohydrate metabolism and interruptions in ATP production and the TCA Cycle. Where predictions and experimental data disagreed, we identified testable hypotheses to reconcile differences between the model predictions and experimental data. The presented pipeline for using paired transcriptomics and metabolomics data provides a framework for interrogating multiple omics datasets to generate mechanistic insight of metabolic changes associated with toxicological responses.
Keywords: metabolism, constraint-based modeling, toxicology, transcriptomics, metabolomics
Toxicity is an unintended effect of many compounds, resulting in significant health complications. The liver, kidney, and heart are often subject to adverse, potentially toxic effects because of their role in drug metabolism (Albini et al., 2010; Awdishu and Mehta, 2017; Chen et al., 2015). Hepatotoxicity is of particular concern (Church and Watkins, 2017; Rueda-Zárate et al., 2017; Zimmerman, 1999), highlighting the need to understand how liver metabolism is altered as a result of toxicity. Understanding the metabolic changes to the liver can facilitate understanding the mechanisms associated with toxicity, thereby guiding development of novel strategies to counterbalance any toxic effects. Furthermore, with such mechanistic interrogation of liver metabolism, we can identify potential biomarkers associated with toxicity and potential intervention points involved with toxicological processes.
Genome-scale metabolic network reconstructions (GENREs) have emerged as useful tools for the study of cellular metabolism (Gille et al., 2010; Karlstädt et al., 2012; Mardinoglu et al., 2013; Väremo et al., 2015). GENREs represent metabolic reactions in a stoichiometric matrix that accounts for the stoichiometric coefficients of chemical transformations and the associated metabolites. GENREs also account for gene-protein-reaction (GPR) rules that map relationships between genes, the proteins they encode, and the reactions they catalyze in the network. With the GPR mappings and stoichiometric matrix to account for associated metabolic reactions, GENREs can be used to predict gene essentiality, changes in metabolites secreted, and the ability of a cell to catabolize particular carbon substrates; because of these characteristics, GENREs are increasingly applied to tackle questions about cellular toxicological responses (Bartell et al., 2014; Brunk et al., 2018; Carbonell et al., 2017; Gatto et al., 2015; Pannala et al., 2018).
The incorporation of omics data into GENREs allows for cell-type-specific interrogation of metabolism. Transcriptomics and proteomics data are frequently integrated into GENREs to create cell-type-specific models. Several algorithms to integrate omics data into GENREs have been developed (Shlomi et al., 2008; Zur et al., 2010). Often with such methods, the integration of omics data constrains the GENRE by turning “on” and “off” genes and their associated reactions, reflecting gene expression in different conditions. These expression data integration algorithms help to contextualize these omics data and improve predictions of cellular metabolic functions.
Biomarkers are currently used in the diagnosis of cancer, cardiac function, and renal function (Jungbauer et al., 2016; Lotan et al., 2018; Pan et al., 2018; Shlipak et al., 2012) among other pathologies, often associating the presence or absence of a molecule with a specific diagnosis. For example, alanine aminotransferase is a protein that is used frequently as a biomarker of liver function (Dufour et al., 2000; Zimmerman, 1999); high levels of this protein indicate that the liver has been damaged. A recently developed computational method for predicting biomarkers called Transcriptionally Inferred Biomarker Response (TIMBR) (Blais et al., 2017) uses gene expression data contextualized in a GENRE to estimate relative changes in secreted metabolite levels. In a previous study (Blais et al., 2017), TIMBR predicted changes in extracellular metabolite levels based on gene expression data for cells exposed to various chemical compounds. Predictions of a limited number of metabolite biomarkers for one chemical were validated, but a global evaluation of how well the biomarker predictions matched experimental data was missing. In this study, predictions from TIMBR are compared with paired metabolomics data to observe the differences between computational predictions and experimental data. Agreement between predictions and experimental data can be illustrative of mechanism behind an observed biomarker; disagreements between the computational model and experimental data can facilitate the development of specific testable hypotheses.
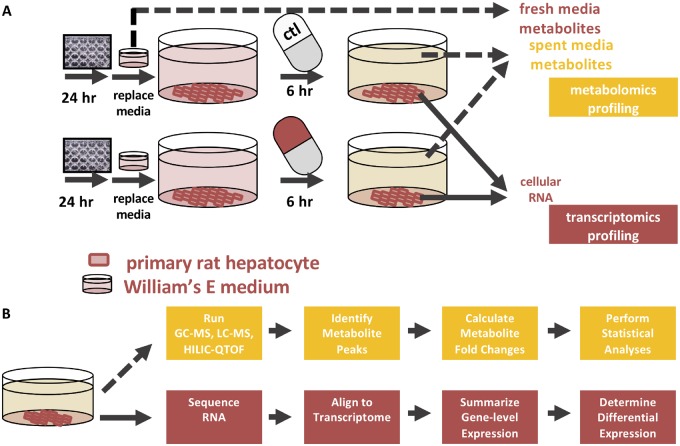
Here, we exposed primary rat hepatocytes to four chemical compounds and characterized their acute metabolic response (Figure 1). After exposure, transcriptomics and metabolomics data were collected from the same sample. We characterized the response of the hepatocytes to the compounds through changes in gene expression and metabolite levels, and evaluated similarities and differences between the cell’s responses across all conditions. The transcriptomics data were integrated into a GENRE of rat metabolism via iMAT (Zur et al., 2010) to create a hepatocyte-specific network model, then the TIMBR algorithm was used to predict changes in the secreted metabolite profile. We compared these predictions with the coupled metabolomics data. With this methodology, we present a comprehensive strategy to characterize the toxicological response of hepatocytes to compounds of interest, and provide a framework to identify further areas of study in hepatocyte drug and toxicity metabolism.
Figure 1.
Schematic of the experimental set up. A, Primary rat hepatocytes were plated in 12-well format and exposed to acetaminophen, carbon tetrachloride, 2,3,7,8-tetrachlorodibenzodioxin (TCDD), or trichloroethylene for six h. After compound exposure, supernatants were collected and sent for metabolomics analysis. Hepatocytes were lysed and RNA was collected for sequencing. B, Cellular RNA was isolated and sequenced by Genewiz. With the raw sequencing reads as an input, the program kallisto was used to align sequencing reads to a reference transcriptome. The R packages TxImport and DESeq2 were used to summarize transcript counts to the gene level and to perform differential gene analysis respectively. Spent media from the hepatocytes were collected and sent for GC-MS, LC-MS, and HILIC-QTOF metabolomics at West Coast Metabolomics. After receiving metabolite peak intensities, the data were processed in R to generate a list of differentially abundant metabolites in each condition.
MATERIALS AND METHODS
Hepatocyte growth conditions
Frozen, primary rat hepatocytes (male, Sprague-Dawley) were purchased from ThermoFisher Scientific and cultured according to the manufacturer’s directions. Briefly, cells were rapidly thawed in a water bath (37°C), resuspended in plating media (William’s E media base supplemented with FBS, dexamethasone, penicillin/streptomycin, insulin, GlutaMAX, and HEPES; Gibco #CM3000), pelleted (50 ×g, five min), and plated at approximately 85% confluence in 12-well tissue culture plates. After 24 h, plating media was replaced with maintenance media (William’s E media base supplemented with dexamethasone, penicillin/streptomycin, ITS+, GlutaMAX and HEPES; Gibco #CM4000) and cells were incubated at 37°C under 5% CO2 for the remainder of the experiment.
Hepatocyte exposure to compounds
Hepatocytes were exposed to a hepatotoxicant and general toxicants at subtoxic levels. Subtoxic levels were defined as concentrations that resulted in minimal cell death but observed phenotypic changes (e.g., decreases in albumin production, ATP levels, increases in cytochrome p450 activity) (Supplementary Figs 1-3). The compounds were acetaminophen (APAP) at 3 mM, carbon tetrachloride (CCl4) at 10 mM, trichloroethylene (TCE) at 1 mM, and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) at 1 nM. APAP and CCl4 are known hepatotoxicants, whereas TCDD and TCE are not considered primary hepatotoxicants typically, but do indeed induce hepatotoxicity. APAP, TCDD, and TCE conditions have four replicates, whereas CCl4 and the DMSO controls have three replicates. Solutions were made in WEM containing 0.1% DMSO with 0.1% DMSO as a control. Cells were exposed to the compounds for six hour. Concentrations and the six hour time point were selected based on literature evidence of comparable studies and conditions (Aly and Domènech, 2009; Cai et al., 2005; Dere et al., 2011; Forgacs et al., 2013; Kienhuis et al., 2009; Mitchell et al., 1985; Uehara et al., 2010; Xu et al., 2012).
RNA isolation, sequencing, and analysis
After supernatants were collected, cells from each condition were treated with TRIzol and then scraped and collected into tubes. Chloroform was added to each tube and after shaking, cells were poured into prespun phase-lock gel tubes (5PRIME). Tubes were then spun in a cold room, the upper phase was collected, and isopropanol and glycogen were added to each tube followed by gentle inversion. Supernatants were again spun in a cold room and the resulting pellet was washed twice with 75% ethanol. The pellet was semidried and then dissolved in nuclease-free H2O. RNA samples were treated with DNA-free DNA removal kit (Ambion/Invitrogen), according to the manufacturer’s instructions, to remove any remaining DNA. RNA was quantified using the Qubit RNA broad range kit and sample integrity assessed using Agilent. RNA samples were subjected to rRNA depletion prior to library construction and sequencing; all services were performed by GENEWIZ. Libraries were sequenced using the Illumina HiSeq2500 platform in a 2× 100 bp pair-end configuration in High Output mode (V4 chemistry). The Unix-based program Kallisto v. 0.43.0 (Bray et al., 2016) was used to process RNA sequence data in fastq format and quantify transcript abundances. Normalized transcript abundance values (transcripts per million) were calculated by Kallisto, using default settings, and imported to R for differential analysis. To quantify transcript abundances and aggregate toward the gene level, the package tximport in R was used (Soneson et al., 2015). Differential gene expression was then performed with the standard DESeq2 R package (Love et al., 2014) to obtain a list of differentially expressed genes (DEGs) with their log2-fold change values.
Metabolomics
After hepatocytes were exposed to the different compounds, supernatants were collected and stored at −20°C. Supernatants were then shipped to West Coast Metabolomics (http://metabolomics.ucdavis.edu/) at the University of California, Davis and untargeted analysis of primary metabolites, complex lipids, and biogenic amines was conducted on each sample, DMSO controls, and on blank media. An extraction solvent of 3:3:2 acetonitrile/isopropanol/water was prepared to use with the collected samples for Gas Chromatography-Mass Spectrometry (GC-MS) to analyze primary metabolites. External and internal standards for quality control were also prepared along with the samples. Raw results were reported as peak heights for quantification ion at the specific retention index. A full description of the protocol was outlined previously (Fiehn, 2016). Lipidomics analysis was performed by preparing samples with methanol, methyl tert-butyl ether, and water before running Liquid Chromatography-Mass Spectrometry (LC-MS). LipidBlast was used to identify and annotate lipids, and peak heights were reported according to the published protocol (Cajka and Fiehn, 2017).
Biogenic amine peak heights were quantified using Hydrophilic Interaction Chromatography Quadrupole Time of Flight (HILIC-QTOF) Mass Spectrometry, and peak heights were calculated following methods previously described (Meissen et al., 2015). Samples were processed and analyzed according to West Coast Metabolomics protocols. Proteins and small polar hydrophilic small molecules were separated from lipids according to the protocol published by Matyash et al. (2008). Data were acquired using the following chromatographic parameters. Ultrapure water with 10 mM ammonium formate and 0.125% formic acid (pH 3) for mobile phase A, and 95:5 (vol/vol) acetonitrile: ultrapure water with 10 mM ammonium formate with 0.125% formic acid (pH 3) for mobile phase B. A column temperature of 40°C, with the flow rate of 0.4 ml/min and injection volume of 3 µl for ESI (+) and temperature of 4°C was used. The ESI Capillary voltage was +4.5 kV for ESI (+), the scan range was m/z 60−1200 Da, and the mass resolution was 10 000 for ESI (+) on an Agilent 6530 QTOF MS. After raw peaks were obtained, they were processed by mzMine 2.0 software to find peaks in up to 300 chromatograms. Relative peak intensities of both identified and unidentified metabolites were generated and used for further analyses.
Data analysis
Before differential expression analysis, genes with no counts were removed from analysis to avoid skewing the results. A gene was considered significantly differentially expressed if the false discovery rate (FDR) corrected p-value was <.1. Standard Euclidean hierarchical clustering was performed on all the gene expression data and clustering was done by each individual gene. For the metabolomics dataset, primary metabolites, lipids, and biogenic amines were read in and combined into one data frame to analyze the data similarly. Experimental replicates were averaged together and fold changes were calculated from the cell samples and the fresh media samples. Significance of metabolite differences was determined with a p-value <.05 using the Mann-Whitney U test. All statistical analyses were performed using R version 3.4.0.
Gene enrichment analysis
To perform gene enrichment analysis, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resource was used with a list of DEGs for each compound (Huang et al., 2009a,b). The Functional Annotation Tool was used to determine which Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were overrepresented, or enriched. Entrez gene IDs were submitted to the DAVID Bioinformatics Resource website and the Rattus norvegicus species was selected. The category “KEGG pathways” and functional annotation clustering were selected. KEGG pathway terms were considered significantly enriched if the FDR corrected p-value was <.1.
Flux balance analysis and the creation of tissue-specific models
The stoichiometric matrix (S matrix) was analyzed using the COBRA toolbox v. 2.0.6 (Schellenberger et al., 2011). The iRno reconstruction of rat metabolism, which accounts for the function of 5620 metabolites, 2324 genes, and 8268 reactions, was used to make computational predictions (Blais et al., 2017). iRno has been curated to perform liver-specific metabolic tasks, making it appropriate as a base model of liver metabolism. Flux balance analysis was performed using the optimizeCBmodel function in the COBRA toolbox in MATLAB v. R2016b. Condition-specific models were then created using the iMAT algorithm in the COBRA toolbox. The createTissueSpecificModel function in the COBRA toolbox was used, with iMAT set as the method for expression data integration, using reactions associated with DEGs and exchange reactions as high confidence reactions to include in the model. Log-fold changes for DEGs were supplied as inputs along with a model with genes created for exchange reactions, whereas the hepatocyte-specific model was provided as an output.
TIMBR algorithm
The TIMBR algorithm combines the transcriptomics data with the iRno network reconstruction to determine production scores for each exchangeable metabolite relative to a control as previously described (Blais et al., 2017). The transcriptomics data was used to generate weights for a control case and a treatment case on each reaction in the reconstruction. Next, for each metabolite, the weighted flux through each reaction was minimized whereas maintaining positive flux through that metabolite’s exchange reaction for the control and treatment conditions. Production scores are normalized using the previously described formula (Blais et al., 2017) to determine whether a metabolite has increased or decreased production relative to the control and used for further downstream analysis. The scripts used to generate each of the datasets can be found on the github site (www.github.com/csbl) published with the TIMBR algorithm.
RESULTS
Transcriptomics Data Reveal Compound-Specific Responses of Hepatocytes
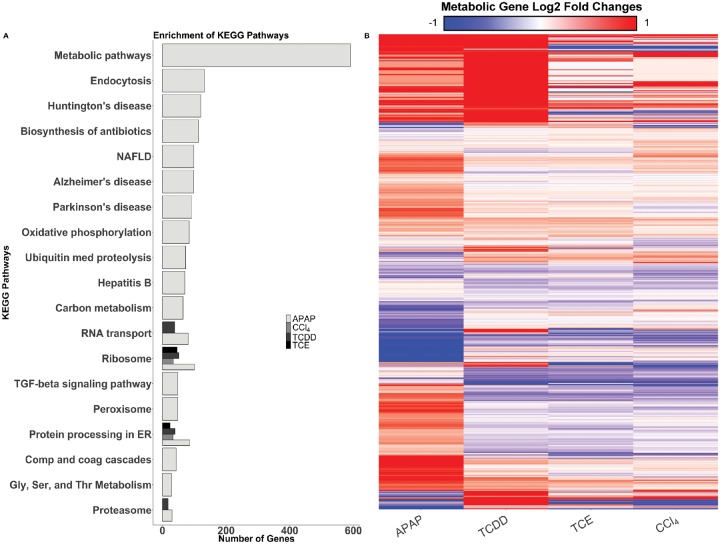
Hepatocytes were exposed to APAP, CCl4, TCDD, or TCE for six hours to characterize the differential toxicity-induced metabolic response. Figure 1 shows the experimental layout; after hepatocytes were exposed to each compound, supernatants were collected for metabolomics analyses and RNA was isolated for transcriptomics analysis. DMSO was used as a nondrug control. The number of DEGs for each condition and time point were determined (Table 1) and a list of genes from the differential gene analysis was produced (Supplementary Data 1). APAP induced the most DEGs in the hepatocytes, whereas TCE induced the least number of DEGs. To further analyze the genes that were differentially expressed, we used the DAVID Bioinformatics platform to identify enriched KEGG pathways for each compound. Figure 2A shows the enrichment results of the DEGs for APAP, CCl4, TCDD, and TCE (Complete enrichment results are shown in Supplementary Data 2). APAP at six hours showed an enrichment for metabolic pathways, whereas CCl4, TCDD, and TCE at six hours did not (Figure 2), suggesting that the hepatocyte’s metabolism was more altered globally in response to APAP compared with the other three compounds. As evidenced in the enrichment analysis, APAP exposure induced a wide variety of gene expression changes, whereas gene expression changes after CCl4, TCDD, and TCE exposure appeared focused toward RNA and protein processing. After investigating the broad effects of the compounds, we then focused on metabolic genes to evaluate how each compound perturbed hepatocyte metabolism.
Table 1.
Comparison of the Number of Differentially Expressed Genes in Response to Each Chemical Compound at the six hour Time Point, Compared With Their Respective Controls
| Chemical Compound | Number of Differentially Expressed Genes (FDR <0.1) | Number of Differentially Expressed Genes (FDR <0.1) in the iRno Model |
|---|---|---|
| APAP – 6 h | 7370 | 1009 |
| CCl4 – 6 h | 824 | 131 |
| TCDD – 6 h | 2493 | 304 |
| TCE – 6 h | 907 | 151 |
Figure 2.
Gene enrichment and metabolic gene expression data. A, DAVID enrichment of KEGG Pathways for six hours in APAP-, CCl4-, TCDD-, and TCE-induced toxicity conditions. The heat map above shows the log2 fold changes of the metabolic genes from sequencing (B). Each condition is listed on the x-axis, and the individual genes are listed on the y-axis. Genes that are upregulated are shown in red, whereas downregulated genes are shown in blue. Genes on the x-axis are clustered by Euclidean distance, using complete linkage.
Figure 2B shows a heat map of the log2-fold changes of all the metabolic DEGs with a Benjamini-Hochberg adjusted p-value of <.1 in at least one condition. This heatmap shows that CCl4 and TCE elicit similar changes in gene expression. We observed changes in expression for the Cyp450 family of genes, often associated with metabolizing drugs (Guengerich, 2008). We saw a decrease in Cyp3a4 in APAP (Supplementary Figure 3) but no changes in the other compounds, likely because other Cyp450 genes play a role in rat metabolism of compounds (Tran et al., 2001; Zuber et al., 2002). Specifically, Cyp2e1 is induced in hepatotoxicity (Jaeschke et al., 2002; McGill et al., 2012). We saw upregulation of Cyp2e1 in APAP-induced toxicity, but not for TCDD- and TCE-induced toxicity; however, there were other genes in the Cyp450 family that were differentially expressed in these other conditions. In APAP- and TCDD-induced toxicity, the Cyp450 gene Cyp2d4, also associated with the metabolism of drugs (Mizuno et al., 2003), was upregulated. TCDD-induced toxicity resulted in upregulation of most other Cyp450-related genes, whereas TCE-induced toxicity resulted in downregulation for many of the same genes. This result highlights that even though there are common pathways of toxicity associated with the liver, these compounds ultimately result in different specific effects on the hepatocytes. In an effort to identify potential biomarkers specific to each compound, we next interrogated the metabolomics data to identify differential effects of each compound (Table 2).
Table 2.
Comparison of the Number of Differentially Changed Metabolites for Each Chemical Compound at the 6-h Time Point, Compared With Their Respective Controls
| Chemical Compound | Statistically Significant Metabolites Changed | Subset of Statistically Changed Metabolites Identified |
|---|---|---|
| APAP – 6 h | 82 | 8 |
| CCl4 – 6 h | 11 | 3 |
| TCDD – 6 h | 102 | 6 |
| TCE – 6 h | 84 | 6 |
Metabolomic Data Discriminates the Response of the Primary Hepatocytes Specific to Each Treatment
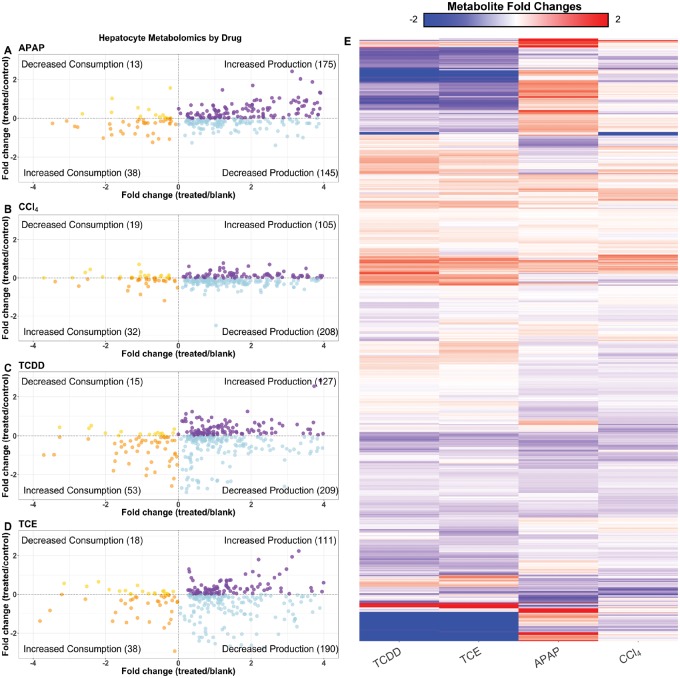
APAP produces the most distinct signature of the three compounds, whereas TCDD and TCE display a similar profile. The metabolomics data are illustrated in scatter plots for APAP (Figure 3A), CCl4 (Figure 3B), TCDD (Figure 3C), and TCE (Figure 3D) exposure conditions and fold changes with respect to the DMSO control is described in Supplementary Data 3. The scatter plots show each metabolite, with the fold change of average relative metabolite peak intensity compared with blank medium on the x-axis, and compared with the DMSO controls on the y-axis. With this arrangement, metabolites are classified as having increased or decreased production if the fold change relative to blank is positive, or increased or decreased consumption if the fold change relative to blank is negative. Metabolites are also color coordinated, to help distinguish metabolites that were increased or decreased in their production or consumption. Only metabolites that were significantly changed in either the treated versus control, or treated versus blank cases are displayed. From these data, we see that APAP induces the greatest number of metabolites with an increase in production, whereas the other compounds induced a decrease in production of most measured metabolites. There is a trend for metabolites to either be increased in production (upper right) or decreased in production (lower right). This trend is clear in each condition, as these were the two categories with the most metabolites, although many of these metabolites are not yet identified. In APAP-induced toxicity, there were several amino acids that decreased in production compared with the control case (Supplementary Figure 4A, bottom left and right). This result indicates that hepatocytes consumed more amino acids after being exposed to APAP. TCDD and TCE both caused hepatocytes to decrease production of fatty acids (Supplementary Figs. 4C and 4D, bottom right), whereas APAP triggered an increased production of fatty acids (Supplementary Figure 4A, top right). The results from the metabolomics data suggest a clear metabolic difference in the hepatocytes treated with different compounds, and that the mechanism of action or off target effects of the toxicants may be the likely cause of this shift.
Figure 3.
Overview of the metabolomics data. The scatter plots show the distribution of metabolites that are significantly (p<.05) changed when compared with either the control media or blank media, and colored according to their levels when compared with both sets of media. Metabolites in the top left corner have decreased overall consumption, metabolites in the bottom left corner have increased overall consumption, the bottom right corner indicates decreased overall production, whereas the top right corner shows increased overall production, all with respect to the control media. Plots are shown for APAP- (A), CCl4- (B), TCDD- (C), and TCE- (D) induced toxicity conditions at six hours. The heat map above shows the log2 fold changes for metabolites compared with their respective controls (E). Each condition is listed on the x-axis, and the metabolites are listed on the y-axis. Metabolites on the x-axis are clustered by Euclidean distance, using complete linkage.
We next decided to interrogate the total metabolic response of the hepatocytes to further discriminate treatment conditions. Figure 3E shows a heatmap of the individual metabolite levels, and whether or not the amount of the metabolites increased or decreased with respect to the control condition. Again, we noticed that TCE and TCDD showed a similar but distinct pattern of changes in metabolite levels. Valine and leucine were uniquely increased in TCE, whereas tryptophan, serine, and glutamate were uniquely decreased in TCDD. Between both compounds, nicotinate, glucose-1-phosphate, and aminomalonate all decreased. There were only seven metabolites that increased for both TCDD and TCE, 1,3-diheptadecanoyl-2-(10Z-heptadecenoyl)-glycerol d5 and six unidentified metabolites. There were 80 metabolites that decreased between both compounds including both identified and unidentified metabolites. In the heatmap in Figure 3E there are a few prominent clusters of metabolites. There was a small cluster of unidentified metabolites in the TCDD and TCE condition whose levels were decreased when compared with the control condition. APAP did not follow this trend, as a number of those same metabolites were increased. Within this large cluster the only identified metabolite was nicotinate. Of the 559 metabolites we were able to detect, only 115 could be identified. Of the identified metabolites, we then looked at the unique metabolites altered by each condition to compare and contrast each compound’s effect on the hepatocytes. Common metabolites that consistently decreased across all conditions were l-lactate, glycerate, and alpha-ketoglutarate (AKG), which have been shown to decrease in other toxicity studies (Kim and Moon, 2012). Other studies have shown decreases in citrate and AKG (Ishihara et al., 2006), which have been attributed to disruptions of the TCA cycle. Finally, there were increased lipid levels in TCE and TCDD compared with their controls, suggesting a strong alteration in lipid metabolism in response to these compounds.
TIMBR Predictions Suggest Unique Responses to Each Toxicant
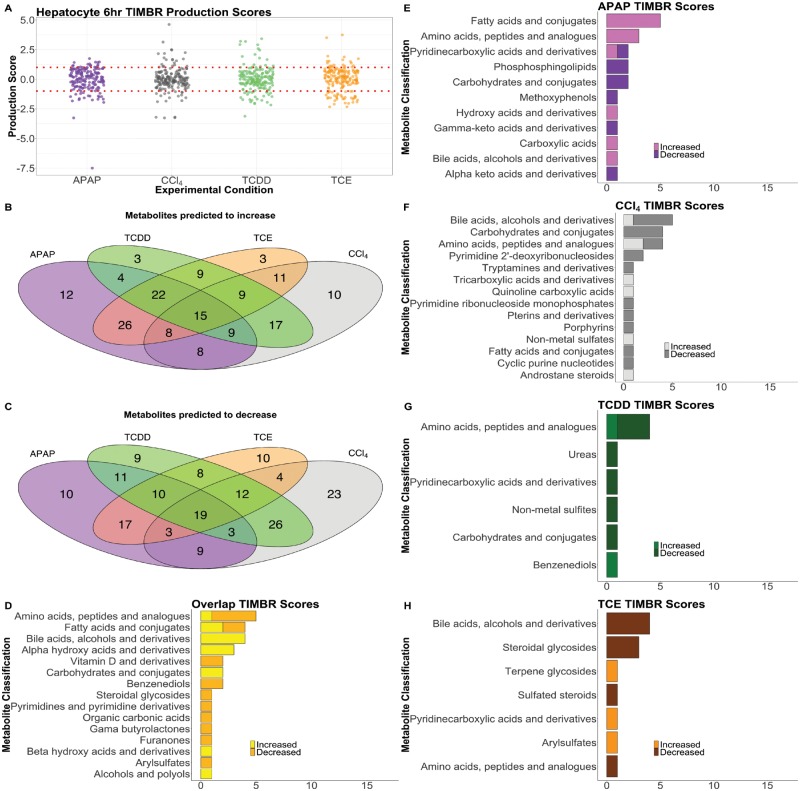
To make predictions on metabolite production levels relative to control from the gene expression data, we created a hepatocyte-specific metabolic model from the unconstrained iRno GENRE using iMAT (Zur et al., 2010) along with the gene expression data described earlier. The Transcriptionally Inferred Metabolic Biomarker Response (TIMBR) response algorithm (Blais et al., 2017) was used to create normalized production scores for each metabolite that could be secreted by the model and we compared these values with the fold changes calculated from the metabolomics data above (Supplementary Data 4). Figure 4A shows a distribution of normalized TIMBR production scores by compound, with the median indicated by the notches and black line and the mean represented by the white diamond in the middle of the box plot. In Figure 4A, we see that each group has its mean at about zero, however, the median for each group is different. The analysis of the APAP and TCE conditions show that more scores have positive TIMBR scores while CCl4 has slightly more negative TIMBR scores. This result suggests that hepatocytes are predicted to produce more metabolites in response to APAP and TCE exposure compared with other toxicant conditions.
Figure 4.
Summary and Distribution of TIMBR production scores. The distribution of TIMBR production scores are shown (A) indicating that the ranges are similar, but scores have a slight skew according to their condition. The APAP condition results in more negative production scores, whereas TCE results in more positive production scores. Lines mark y = 1 and y = −1. Venn diagrams compare all positive (B) and negative (C) production scores for each compound, and the overlap between the 3 conditions. TIMBR scores that are common across all conditions (D), and unique to APAP (E), CCl4 (F), TCDD (G), TCE (H) are illustrated. Here, metabolites are classified into categories taken from the subclass names from the Human Metabolome DataBase (HMDB) if available. Metabolite in a category that increases were given a light color, whereas metabolites in a category that decrease were given a darker color.
We then compared common metabolites that were predicted to increase or decrease after all treatments, which indicate common metabolic shifts in response to drug treatment. Figures 4B and 4C shows Venn diagrams of the metabolites that were predicted to commonly increase or decrease in production, or uniquely increase or decrease in response to APAP, CCl4, TCDD, or TCE at six hours, respectively. Similar to the trend noted in all of the TIMBR production scores (Figure 4A), CCl4 exposure was predicted to decrease a higher number of metabolite production scores (Figure 4C) that do not also decrease in other conditions. However, APAP exposure was predicted to result in the increase of more metabolite production scores that were not increased in other conditions (Figure 4B), which is consistent with the prediction of more positive production scores. We then classified metabolites according to their Human Metabolome Database (HMDB) subclassification (Supplementary Data 5) that were changing in each condition from the results shown in Figures 4B and 4C. The bar charts in Figures 4D−H indicate the number of metabolites uniquely predicted to increase or decrease production after toxicant exposure according to their subclassification. For shared metabolites across all conditions (Figure 4D), a small number of amino acids are predicted to decrease, whereas bile acids are predicted to increase. APAP exposure (Figure 4E) resulted in the highest number of fatty acids predicted to increase in production, followed by amino acids. The increase in bile acids and amino acids suggests alterations in these pathways in response to liver injury, and has been observed in literature (Kumar et al., 2012; Sun et al., 2013). In CCl4 exposure (Figure 4F), carbohydrate compounds are predicted to decrease in production, whereas these same metabolites were predicted to increase in the other three conditions. With TCDD exposure (Figure 4G), amino acids are predicted to decrease in production while with TCE exposure bile acids are predicted to decrease (Figure 4H) which is similar to CCl4. Overall the TIMBR predictions illustrate that the response of the hepatocytes to each compound is primarily due to carbohydrate and amino acid metabolism, which could represent a generic response toward toxic compounds. However, predictions from APAP exposure indicate a distinct response in fatty acid metabolism, with CCl4 and TCE eliciting more of a change in bile acid metabolism, suggesting that we can predict compound-specific effects on the hepatocytes.
Comparing TIMBR Predictions and Metabolomics Data
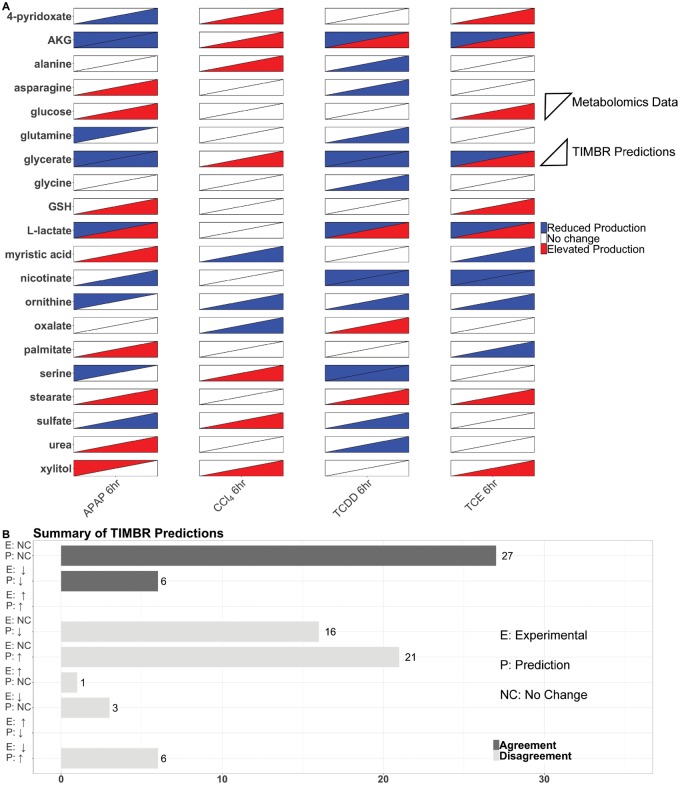
We next wanted to quantify the similarity and dissimilarity between the predictions and metabolomics data, to determine how indicative gene expression changes were in predicting metabolite levels. In addition to the fold changes calculated from the metabolomics data, the Mann-Whitney U test was used to determine statistical significance at the p < .05 level. A change in metabolite levels with p > .05 when compared with the control condition, was classified as “no change,” and represented with a fold change of zero. We then took the subset of secreted, identified metabolites and compared this list with our TIMBR predictions which resulted in 20 metabolites we could validate for each experimental condition. For the TIMBR predictions, metabolite production scores were ranked, and metabolites in the middle 50% of the list were classified as no change and given a value of zero for comparing with the metabolomics data. Figure 5A shows a heatmap of the metabolomics data and the production scores for metabolites on the y-axis with each condition on the x-axis. Of the 20 metabolites in each condition, we predicted five correctly in the APAP condition, 10 correctly in the CCl4 condition, 9 correctly in the TCDD condition, and 9 correctly in the TCE condition. From the list of successful predictions nicotinate, and glycine were correct in three of the conditions, whereas 9 metabolites were correct in two of the conditions. There were six amino acids in the set of 20 that we could make predictions for, and of those six, we correctly predicted 2 in the APAP condition, three in the CCl4 condition, whereas only one prediction was correct in the TCDD condition and five in the TCE condition. Although we were able to predict broad changes in carbohydrate and energy metabolism from the TIMBR predictions as described above, the data were too limited to draw the same conclusions from the subset of experimental data that we were able to validate.
Figure 5.
Validation of TIMBR production scores using metabolomics data. The heat map (A) shows the results from the metabolomics data, and the TIMBR production scores for each metabolite we were able to make a prediction for and validate. Each condition is listed on the x-axis, and the metabolites are listed on the y-axis. Metabolomics data are shown in the upper left triangle, and TIMBR production scores are shown in the bottom right triangle. The bar chart (B) shows the categories a prediction can fall into on the y-axis ranging from increase, decrease, or no change for both the experimental data and the TIMBR predictions. The x-axis contains the number of predictions that fall into the category on the y-axis. Predictions that agree with the experimental data have dark colored bars, whereas disagreement between the data show light colored bars.
Figure 5B quantifies our validation results, and shows exactly where predictions were right and where predictions were wrong. The bulk of the correct predictions came from identifying no change in both the experimental condition and the computational prediction. Overall, our accuracy for our predictions was 41%. We made no correct predictions on metabolites that were measured as increased or predicted to increase. Thirty-seven of the predictions were incorrect from detecting a change and predicting there was none, or vice versa. Our sensitivity for detecting no change was 42% and lower for predicting an increase (0%) or decrease (40%). Our specificity for no change or decrease was high at 75%, but was lower (65%) for the no change condition.
DISCUSSION
There is limited information on biomarkers of toxicity; therefore, novel approaches to elucidate and validate relevant biomarkers are needed. A promise of metabolomics as an approach for identifying biomarkers is its connection to cell phenotype as a change in metabolite levels may represent changes in the functional state of the cells. Here, we present the first use of paired transcriptomics and metabolomics with GENREs to study hepatocytes exposed to different compounds and to integrate these data with metabolic network models to provide insight into the changes that are occurring. At the concentrations and timepoint we selected for exposure, we used standard measures of hepatocyte function (albumin production, Cyp450 activity, etc.) to ensure we were not killing the cells (Supplementary Figs. 1–3). Although we observed minimal changes in traditional measurements of toxicity after six hour exposure (Supplementary Figs. 1 − 3), we observed changes in metabolism as indicated by the transcriptomics and metabolomics data. Additionally, connecting transcriptomic changes to secreted metabolites even at low-toxic compound concentration can be useful in clinical settings, as these secreted metabolites can be measured to gain an early indication of hepatic injury. Secreted metabolites can then be connected back to transcriptional changes using metabolic network models, which allows us to generate mechanistic insight into observed changes in transcript or metabolite levels.
From this study, we observed a number of transcriptional changes in metabolic genes of hepatocytes following exposure to compounds. Analyses of transcriptional changes highlighted that APAP produced the largest change in hepatocyte gene expression, as expected (Ben-Shachar et al., 2012; McGill and Jaeschke, 2013; Sjogren et al., 2014; Taguchi et al., 2015). There were 31 differentially expressed metabolic genes that were shared across all four compounds. There were five genes that were upregulated in each of the four treatment conditions. Among the group of upregulated genes includes two glutathione S-transferase genes, indicative of detoxification mechanisms given glutathione is used to conjugate toxic metabolites (Guengerich, 2008; Monks et al., 1990). Furthermore, glutathione S-transferase is responsible for the detoxification of NAPQI, a toxic metabolite that is generated from metabolizing APAP (Henderson et al., 2000). In the metabolomics data, glutathione production was decreased in APAP (albeit p = .34). For the TIMBR predictions in the APAP condition, we did predict decreased production of glutathione, which is attributed toward glutathione detoxifying NAPQI in the hepatocytes.
Twelve differentially expressed metabolic genes that were shared across all compounds were downregulated. Among this group was isocitrate dehydrogenase 3 (Idh3a), which is responsible for the NAD+ dependent conversion of isocitrate to AKG. In the metabolomics data, we observed decreased production of AKG in response to APAP, TCDD, and TCE compared with their respective controls. We also computationally predicted this decrease in AKG in the APAP condition (Figure 5). These examples suggest that there are some transcriptional changes that are indicative of downstream metabolite changes.
Glycolysis and the TCA cycle were disrupted as a result of compound exposure. In the metabolomics data, we observed that glycerate was decreased in response to exposure to APAP, TCDD and TCE, and glucose-1-phosphate was decreased after treatment with TCDD and TCE. Both glucose-1-phosphate and glycerate can feed into glycolysis and then progress to the TCA cycle. Decreases in these metabolites indicate that the hepatocytes are inefficiently producing ATP via the TCA cycle. This observation is also further supported by the measured decrease in AKG in most of the conditions as well as the decrease in citrate in response to APAP. Carbohydrates also feed into glycolysis, and a decrease in carbohydrates can also decrease TCA activity. In Figure 4, we observe that carbohydrates were predicted to decrease in production after exposure to APAP and CCl4, which are both hepatotoxicants. Although we were not able to correctly predict changes in glycerate production in every condition (Figure 5), we were able to predict this shift in metabolism via the carbohydrates, which is supported by the metabolomics data. Thus, TIMBR predictions can be useful for suggesting pathway level differences of a treatment that can be experimentally validated.
We compared our in vitro and computational results with other in vivo toxicity studies that have been done. Across the different studies, lipid metabolism, amino acid metabolism, and energy metabolism (TCA Cycle) were all affected by exposure to different compounds. One study that focused on TCDD-induced transcriptomic changes identified several genes associated with these pathways that were both upregulated and down regulated (Boverhof et al., 2006). From a metabolomics perspective, TCA cycle intermediates were down regulated in response to APAP (Sun et al., 2008), which agreed with our data. These same pathways came up in common with our TIMBR predictions (Figure 4) that are based on our measured transcriptional changes. One study noted that in response to APAP-induced toxicity, metabolite levels for glycerol and kynurenine were increased, whereas threonine, serine, ornithine, lysine, glycerate, and glutathione were reduced (Pannala et al., 2018). The authors also observed enrichment in the glycine, serine, and threonine metabolic pathway (Pannala et al., 2018). Although we did not observe the decrease in glutathione levels, we did note enrichment in the glycine, serine, and threonine pathway in the APAP condition (Figure 2A). The decrease in glutathione in APAP was shown to occur at later time points, due to increasing progression of liver injury as noted by the authors (Pannala et al., 2018). Lastly, CCl4 is known to cause hepatocytes to increase urinary bile acid levels (Yang et al., 2008). We observed that there were a few bile acids predicted to increase (Figure 4D), but unique to CCl4 was the observation that most of the bile acids were predicted to decrease (Figure 4F). Because in vitro conditions do not fully capture in vivo conditions due to differences in time-scales, actual exposure concentrations, among other variables, there is not complete agreement between the in vitro and in vivo results as expected. However, our in vitro experiment provides a means to study changes in hepatocyte metabolism without the variability of an in vivo experiment. Although results may not fully match, general trends in metabolic changes do agree, as indicated by the shift in fatty acid metabolism from TCA cycle and amino acid breakdown noted earlier, which highlights the utility of in vitro systems for interrogating toxicological responses.
One limitation of this study that affected the ability to make predictions was the lack of overlap between the metabolomics data and metabolites for which we were able to make TIMBR predictions. For the primary metabolites in the metabolomics dataset, only 115 out of 559 were identifiable. Of these 115, there were only 21 metabolites in the subset that were secreted and that were accounted for in our current network reconstruction, as shown in Figure 5. Although the number of correct predictions was limited, we were still able to make predictions on glycolysis, the TCA cycle, and amino acid metabolism that were supported by either the metabolomics data or literature from other toxicity studies (Beger et al., 2010; Kumar et al., 2012). There are opportunities for further curation of the network reconstruction to account for more metabolites and metabolic reactions, as well as further curation of the metabolomics data.
This study used transcriptomics data paired with metabolomics data to provide insight into the changes induced by these toxicants on hepatocytes. Protein fold changes could be used in place of gene expression data and ultimately could have been used for TIMBR predictions because we can map such data to the metabolic reactions accounted for in the metabolic network reconstruction. As large data sets are made accessible or easy to collect, the use of multi-omic datasets to predict and validate modeling results becomes critical in interrogating specific phenotypes of interest for a chosen system. Our paired experimental and computational approach is one step toward characterizing the cellular response to a compound and identifying potential biomarkers indicative of cell state.
DATA AVAILABILITY
The sequencing and processed data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) with the accession number GSE129814. All supplementary files are stored on the Dryad Digital Repository website (Rawls et al., 2019), accessible at https://doi.org/10.5061/dryad.04vk390. Source data needed to reproduce figures can be obtained via the code available at github.com/csbl/hepatocyte_omicsdata. All other data supporting the findings of this study are available within the article and its supplementary information files.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary Material
ACKNOWLEDGMENTS
Support for this project was provided by the United States Department of Defense (W81XWH-14-C-0054 to J.P.). The opinions and assertions contained herein are the private views of the authors and are not to be construed as the official or as reflecting the views of the U.S. Army, the U.S. Department of Defense, or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF). This manuscript has been approved for public release with unlimited distribution. The authors thank Salman Khetani for his help with starting the experiments. The authors also thank Laura J. Dunphy for assistance with figure design.
AUTHORS’ CONTRIBUTIONS
K.R., E.B., G.K., A.W., and J.P. conceived the study. K.R., G.K., and B.D. performed the experiments to collect transcriptomics and metabolomics data. K.R. wrote the initial draft of the manuscript. K.R., E.B., B.D., G.K., V.P., K.V., A.W., and J.P. edited and wrote the final manuscript.
REFERENCES
- Albini A., Pennesi G., Donatelli F., Cammarota R., De Flora S., Noonan D. M. (2010). Cardiotoxicity of anticancer drugs: The need for cardio-oncology and cardio-oncological prevention. J. Natl. Cancer Inst. 102, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly H. A. A., Domènech Ò. (2009). Cytotoxicity and mitochondrial dysfunction of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) in isolated rat hepatocytes. Toxicol. Lett. 191, 79–87. [DOI] [PubMed] [Google Scholar]
- Awdishu L., Mehta R. L. (2017). The 6R’s of drug induced nephrotoxicity. BMC Nephrol. 18, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartell J. A., Yen P., Varga J. J., Goldberg J. B., Papin J. A. (2014). Comparative metabolic systems analysis of pathogenic Burkholderia. J. Bacteriol. 196, 210–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beger R. D., Sun J., Schnackenberg L. K. (2010). Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicol. Appl. Pharmacol. 243, 154–166. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar R., Chen Y., Luo S., Hartman C., Reed M., Nijhout H. F. (2012). The biochemistry of acetaminophen hepatotoxicity and rescue: A mathematical model. Theor. Biol. Med. Model 9, 55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais E. M., Rawls K. D., Dougherty B. V., Li Z. I., Kolling G. L., Ye P., Wallqvist A., Papin J. A. (2017). Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat. Commun. 8, 14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof D. R., Burgoon L. D., Tashiro C., Sharratt B., Chittim B., Harkema J. R., Mendrick D. L., Zacharewski T. R. (2006). Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 Mice. Toxicol. Sci. 94, 398–416. [DOI] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brunk E., Sahoo S., Zielinski D. C., Altunkaya A., Dräger A., Mih N., Gatto F., Nilsson A., Preciat Gonzalez G. A., Aurich M. K. (2018). Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 36, 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Gong L., Qi X., Li X., Ren J. (2005). Apoptosis initiated by carbon tetrachloride in mitochondria of rat primary cultured hepatocytes. Acta Pharmacol. Sin. 26, 969–975. [DOI] [PubMed] [Google Scholar]
- Cajka T., Fiehn O. (2017). LC−MS-based lipidomics and automated identification of lipids using the LipidBlast in-silico MS/MS library. Methods Mol. Biol. 1609, 149–170. [DOI] [PubMed] [Google Scholar]
- Carbonell P., Lopez O., Amberg A., Pastor M., Sanz F. (2017). Hepatotoxicity prediction by systems biology modeling of disturbed metabolic pathways using gene expression data. ALTEX 34, 219–234. [DOI] [PubMed] [Google Scholar]
- Chen M., Suzuki A., Borlak J., Andrade R. J., Lucena M. I. (2015). Drug-induced liver injury: Interactions between drug properties and host factors. J. Hepatol. 63, 503–514. [DOI] [PubMed] [Google Scholar]
- Church R. J., Watkins P. B. (2017). The transformation in biomarker detection and management of drug-induced liver injury. Liver Int. 37, 1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Lee A. W., Burgoon L. D., Zacharewski T. R. (2011). Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells. BMC Genomics 12, 193 doi:10.1186/1471-2164-12-193. Accessed Jun 28, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D. R., Lott J. A., Nolte F. S., Gretch D. R., Koff R. S., Seeff L. B. (2000). Diagnosis and monitoring of hepatic injury. I. Performance characteristics of laboratory tests. Clin. Chem. 46, 2027–2049. [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. (2016). Metabolomics by gas chromatography-mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 114, 30.4.1–30.4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgacs A. L., Dere E., Angrish M. M., Zacharewski T. R. (2013). Comparative analysis of temporal and dose-dependent TCDD-elicited gene expression in human, mouse, and rat primary hepatocytes. Toxicol. Sci. 133, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto F., Miess H., Schulze A., Nielsen J. (2015). Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism. Sci. Rep. 5, 10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C., Bölling C., Hoppe A., Bulik S., Hoffmann S., Hübner K., Karlstädt A., Ganeshan R., König M., Rother K., et al. (2010). HepatoNet1: A comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol. Syst. Biol. 6, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich F. P. (2008). Cytochrome P450 and chemical toxicology. Chem. Res. Toxicol. 21, 70–83. [DOI] [PubMed] [Google Scholar]
- Henderson C. J., Wolf C. R., Kitteringham N., Powell H., Otto D., Park B. K. (2000). Increased resistance to acetaminophen hepatotoxicity in mice lacking glutathione S-transferase Pi. Proc. Natl. Acad. Sci. USA 97, 12741–12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K., Katsutani N., Aoki T. (2006). A metabonomics study of the hepatotoxicants galactosamine, methylene dianiline and clofibrate in rats. Basic Clin. Pharmacol. Toxicol. 99, 251–260. [DOI] [PubMed] [Google Scholar]
- Jaeschke H., Gores G. J., Cederbaum A. I., Hinson J. A., Pessayre D., Lemasters J. J. (2002). Mechanisms of hepatotoxicity. Toxicol. Sci. 65, 166–176. [DOI] [PubMed] [Google Scholar]
- Jungbauer C. G., Uecer E., Stadler S., Birner C., Buchner S., Maier L. S., Luchner A. (2016). N-acteyl-ß-d-glucosaminidase and kidney injury molecule-1: New predictors for long-term progression of chronic kidney disease in patients with heart failure. Nephrol. Carlton Vic. 21, 490–498. [DOI] [PubMed] [Google Scholar]
- Karlstädt A., Fliegner D., Kararigas G., Ruderisch H. S., Regitz-Zagrosek V., Holzhütter H.-G. (2012). CardioNet: A human metabolic network suited for the study of cardiomyocyte metabolism. BMC Syst. Biol. 6, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienhuis A. S., van de Poll M. C. G., Wortelboer H., van Herwijnen M., Gottschalk R., Dejong C. H. C., Boorsma A., Paules R. S., Kleinjans J. C. S., Stierum R. H., et al. (2009). Parallelogram approach using rat-human in vitro and rat in vivo toxicogenomics predicts acetaminophen-induced hepatotoxicity in humans. Toxicol. Sci. 107, 544–552. [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Moon A. (2012). Drug-induced nephrotoxicity and its biomarkers. Biomol. Ther. 20, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B. S., Chung B. C., Kwon O.-S., Jung B. H. (2012). Discovery of common urinary biomarkers for hepatotoxicity induced by carbon tetrachloride, acetaminophen and methotrexate by mass spectrometry-based metabolomics. J. Appl. Toxicol. 32, 505–520. [DOI] [PubMed] [Google Scholar]
- Lotan Y., Woldu S. L., Sanli O., Black P., Milowsky M. I. (2018). Modeling cost-effectiveness of a biomarker-based approach to neoadjuvant chemotherapy for muscle-invasive bladder cancer. BJU Int. 122, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A., Agren R., Kampf C., Asplund A., Nookaew I., Jacobson P., Walley A. J., Froguel P., Carlsson L. M., Uhlen M., et al. (2013). Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol. Syst. Biol. 9, 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D. (2008). Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. R., Jaeschke H. (2013). Metabolism and disposition of acetaminophen: Recent advances in relation to hepatotoxicity and diagnosis. Pharm. Res. 30, 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill M. R., Sharpe M. R., Williams C. D., Taha M., Curry S. C., Jaeschke H. (2012). The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Invest. 122, 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissen J. K., Hirahatake K. M., Adams S. H., Fiehn O. (2015). Temporal metabolomic responses of cultured HepG2 liver cells to high fructose and high glucose exposures. Metabolomics 11, 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. B., Acosta D., Bruckner J. V. (1985). Role of glutathione depletion in the cytotoxicity of acetaminophen in a primary culture system of rat hepatocytes. Toxicology 37, 127–146. [DOI] [PubMed] [Google Scholar]
- Mizuno D., Takahashi Y., Hiroi T., Imaoka S., Kamataki T., Funae Y. (2003). A novel transcriptional element which regulates expression of the CYP2D4 gene by Oct-1 and YY-1 binding. Biochim. Biophys. Acta 1627, 121–128. [DOI] [PubMed] [Google Scholar]
- Monks T. J., Anders M. W., Dekant W., Stevens J. L., Lau S. S., van Bladeren P. J. (1990). Glutathione conjugate mediated toxicities. Toxicol. Appl. Pharmacol. 106, 1–19. [DOI] [PubMed] [Google Scholar]
- Pan X., Quan J., Li Z., Zhao L., Zhou L., Jinling X., Weijie X., Guan X., Li H., Yang S., et al. (2018). miR-566 functions as an oncogene and a potential biomarker for prognosis in renal cell carcinoma. Biomed. Pharmacother. Biomed. Pharmacother. 102, 718–727. [DOI] [PubMed] [Google Scholar]
- Pannala V. R., Wall M. L., Estes S. K., Trenary I., O’Brien T. P., Printz R. L., Vinnakota K. C., Reifman J., Shiota M., Young J. D., et al. (2018). Metabolic network-based predictions of toxicant-induced metabolite changes in the laboratory rat. Sci. Rep. 8, 11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls K. D., Blais E. M., Dougherty B. V., Pannala V. R., Vinnakota K. C., Wallqvist A., Kolling G. L., Papin J. A. (2019). Data from: Genome-scale characterization of toxicitiy-induced metabolic alterations in primary hepatocytes. Dryad Digital Repository. doi.org/10.5061/dryad.04vk390. [DOI] [PMC free article] [PubMed]
- Rueda-Zárate H. A., Imaz-Rosshandler I., Cárdenas-Ovando R. A., Castillo-Fernández J. E., Noguez-Monroy J., Rangel-Escareño C. (2017). A computational toxicogenomics approach identifies a list of highly hepatotoxic compounds from a large microarray database. PLoS One 12, e0176284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J., Que R., Fleming R. M. T., Thiele I., Orth J. D., Feist A. M., Zielinski D. C., Bordbar A., Lewis N. E., Rahmanian S., et al. (2011). Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlipak M. G., Scherzer R., Abraham A., Tien P. C., Grunfeld C., Peralta C. A., Devarajan P., Bennett M., Butch A. W., Anastos K., et al. (2012). Urinary markers of kidney injury and kidney function decline in HIV-infected women. J. Acquir. Immune Defic. Syndr. 1999 61, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T., Cabili M. N., Herrgård M. J., Palsson B. Ø., Ruppin E. (2008). Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 26, 1003–1010. [DOI] [PubMed] [Google Scholar]
- Sjogren A.-K., Liljevald M., Glinghammar B., Sagemark J., Li X.-Q., Jonebring A., Cotgreave I., Brolén G., Andersson T. B. (2014). Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch. Toxicol. 88, 1427–1437. [DOI] [PubMed] [Google Scholar]
- Soneson C., Love M. I., Robinson M. D. (2015). Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ando Y., Ahlbory-Dieker D., Schnackenberg L. K., Yang X., Greenhaw J., Pence L., Qian F., Salminen W., Mendrick D. L., et al. (2013). Systems biology investigation to discover metabolic biomarkers of acetaminophen-induced hepatic injury using integrated transcriptomics and metabolomics. J. Mol. Biomark. Diagn S1:002. doi: 10.4172/2155-9929.S1-002. [Google Scholar]
- Sun J., Schnackenberg L. K., Holland R. D., Schmitt T. C., Cantor G. H., Dragan Y. P., Beger R. D. (2008). Metabonomics evaluation of urine from rats given acute and chronic doses of acetaminophen using NMR and UPLC/MS. J. Chromatogr. B 871, 328–340. [DOI] [PubMed] [Google Scholar]
- Taguchi K., Tokuno M., Yamasaki K., Kadowaki D., Seo H., Otagiri M. (2015). Establishment of a model of acetaminophen-induced hepatotoxicity in different weekly-aged ICR mice. Lab. Anim. 49, 294–301. [DOI] [PubMed] [Google Scholar]
- Tran A., Tréluyer J.-M., Rey E., Barbet J., Ferracci G., d'Athis P., Vincent J., Pons G. (2001). Protective effect of stiripentol on acetaminophen-induced hepatotoxicity in rat. Toxicol. Appl. Pharmacol. 170, 145–152. [DOI] [PubMed] [Google Scholar]
- Uehara T., Ono A., Maruyama T., Kato I., Yamada H., Ohno Y., Urushidani T. (2010). The Japanese toxicogenomics project: Application of toxicogenomics. Mol. Nutr. Food Res. 54, 218–227. [DOI] [PubMed] [Google Scholar]
- Väremo L., Scheele C., Broholm C., Mardinoglu A., Kampf C., Asplund A., Nookaew I., Uhlén M., Pedersen B. K., Nielsen J. (2015). Proteome- and transcriptome-driven reconstruction of the human myocyte metabolic network and its use for identification of markers for diabetes. Cell Rep. 11, 921–933. [DOI] [PubMed] [Google Scholar]
- Xu X., Liu Y., Lu L., Ke Y., Mao J., Mao K. (2012). Altered expression of hepatic metabolic enzyme and apoptosis-related gene transcripts in human hepatocytes treated with trichloroethylene. Hum. Exp. Toxicol. 31, 861–867. [DOI] [PubMed] [Google Scholar]
- Yang L., Xiong A., He Y., Zaiyong W., Wang C., Zhengtao W., Li W., Yang L., Hu Z. (2008). Bile acids metabonomic study on the CCl4 - and α-naphthylisothiocyanate-induced animal models: Quantitative analysis of 22 bile acids by ultraperformance liquid chromatography−mass spectrometry. Chem. Res. Toxicol. 21, 2280–2288. [DOI] [PubMed] [Google Scholar]
- Zimmerman H. J. (1999). Hepatotoxicity: The Adverse Effects of Drugs and Other Chemicals on the Liver, 2nd ed Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- Zuber R., Anzenbacherová E., Anzenbacher P. (2002). Cytochromes P450 and experimental models of drug metabolism. J. Cell Mol. Med. 6, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur H., Ruppin E., Shlomi T. (2010). iMAT: An integrative metabolic analysis tool. Bioinformatics 26, 3140–3142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing and processed data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) with the accession number GSE129814. All supplementary files are stored on the Dryad Digital Repository website (Rawls et al., 2019), accessible at https://doi.org/10.5061/dryad.04vk390. Source data needed to reproduce figures can be obtained via the code available at github.com/csbl/hepatocyte_omicsdata. All other data supporting the findings of this study are available within the article and its supplementary information files.