Abstract
The success of gene technologies hinges on our ability to engineer superior encapsulation and delivery vectors. Cubosomes are lipid-based nanoparticles where membranes, instead of enveloping into classic liposomes, intertwine into complex arrays of pores well-ordered in a cubic lattice. These complex nanoparticles encapsulate large contents of siRNA compared to a liposomal analogue. Importantly, the membranes that form cubosomes have intrinsic fusogenic properties that promote fast endosomal escape. Despite the great potential, traditional routes of forming cubosomes lead to particle sizes too large to fulfill the state-of-the art requirements of delivery vectors. To overcome this challenge, we utilize a microfluidic nanomanufacturing device to synthesize cubosomes and siRNA-loaded cubosomes, termed cuboplexes. Utilizing cryogenic TEM and small angle X-ray scattering we elucidate the time-resolved mechanisms in which microfluidic devices allow the production of small cubosomes and cuboplexes (75 nm) that outperform commercially available delivery vectors, as well as liposome-based systems.
Keywords: microfluidics, cubosomes, cuboplexes, siRNA, lipids, bicontinuous cubic
Graphical Abstract
Cubosomes, first observed during fat digestion1 and described by K. Larsson,2 are the colloidally stable form of bulk bicontinuous phases. Lipid bicontinuous cubic structures consist of bilayers with midplanes curved into gyroid, diamond, or primitive minimal surfaces that separate into two independent water channel domains. While structurally complex, these structures can be thermodynamically stabilized by a single molecular component (e.g., glycerol monooleate) with increasing amounts of water. Lipid cubic phases have an important role in nature3–5 including transformations in mitochondrial membranes upon starvation6 and viral infections.7 Cubosomes are continuously compartmentalized into hydrophilic and hydrophobic domains at high surface to volume ratio, making them attractive as drug carriers.8–13 Recently, we developed cubosomes encapsulating siRNA, cuboplexes,14 for applications in RNAi therapy. Cuboplexes enable a mechanism for endosomal escape that does not rely on the proton sponge effect or electrostatically driven membrane fusion. Instead, owing to their intrinsic membrane elasticity properties, nanoparticles made of membranes arranged in bicontinuous cubic structures have the ability to effectively puncture pores in endosomal membranes.15 With the recent success of nucleic acid therapies16,17 it is a timely opportunity to develop safe, biocompatible, and low-toxicity delivery systems. In this context, endosomal escape remains an enduring challenge in nucleic acid delivery systems.15,18–21 A common feature of the most varied nucleic acid delivery systems is their ability to puncture membrane pores.22 In addition to this “fusogenic” property, cubosomes and cuboplexes will also have to conform to the state-of-the-art requirements of nanomedicines including narrow size distribution and colloidal stability for successful in vivo applications.23
The traditional approach to fragment bicontinuous cubic phases into sub-micrometer particulates with colloidal stability is top-down homogenization in the presence of a nonlipidic component (e.g., a pluronic polymer),24–31 which is not amenable to the coassembly of many biological molecules such as nucleic acids. In addition, particles prepared by random shear stress (homogenization or sonication) yield large size and polydispersity.32–34 Bottom-up approaches utilize volatile solvents to solubilize lipids that are then added dropwise into bulk water. Owing to the low solubility of lipids in water, nanoprecipitates are formed and their size can be controlled by volatile solvent/water mixing rate. Upon volatile solvent extraction, the nanoprecipitates develop as cubosomes suspended in water. - In general, cubosomes formed using this bottom-up approach are smaller and have better physical stability.38 However, because of bulk-scale mixing of ethanol and water, this method also results in high polydispersity of particle sizes.36,37
We have shown that colloidal stability of cuboplexes can be attained when siRNA is coassembled with a ternary lipid mixture composed of glycerol monooleate (GMO), residual amounts of cationic phospholipid DOTAP (1,2-dioleoyl-3-trimethylammonium propane), and GMO covalently linked to polyethylene glycol 2 kDa (GMOPEG). This ternary mixture emerged as a fruitful polymorphic system, and all three symmetry groups (Ia3d, Pn3m, and Im3m) of bicontinuous cubic phases could be stabilized in excess water.14,39 This is in contrast to pure GMO, where only the Pn3m is stable.40 Importantly, the formation of the cubosomes was successful simply by ultrachilled sonication, a method completely analogous to producing a liposome. However, the obtained particle size and size distribution were rather wide, restricting potential applications in nanomedicine. In this study, we show that microfluidic devices41–43 can be used to prepare colloidally stable cubosomes and cuboplexes with excellent control over size, size distributions, and composition. We employ cryogenic transmission electron microscopy (Cryo-EM) and small angle X-ray scattering (SAXS) to elucidate that microfluidic synthesis of cubosomes and gene silencing cubosomes (cuboplexes) is controlled by membrane fusion of ethanol-in-water nanoemulsions. The cubosomes and cuboplexes prepared by this microfluidics method exhibit significantly smaller size and narrower size distributions compared to conventional bottom-up or top-down large-scale approaches. Furthermore, Cryo-EM unveils how siRNA interacts with cubosome membranes and water nanochannels to form cuboplexes. Our study provides important insight on microfluidic synthesis of lipid-based nanoparticles with and without biological cargo that can be extended to a variety of nanomedicines development.
RESULTS AND DISCUSSION
Our study is focused on the preparation of cubosomes and cuboplexes having small average size and narrow size distributions, as well as colloidal stability, which are basic requirements for applications in nanomedicine. To accomplish this goal, we employ a bottom-up microfluidic approach to naonomanufacture a ternary lipid system previously shown to self-organize into well-ordered (but very polydisperse) spheroidal membranes with internal bicontinuous cubic structure.14 We utilize a ternary mixture where each lipid serves a specific function. The first component is GMO, which is a bicontinuous cubic phase forming lipid. The second lipid is DOTAP, which is positively charged and is commonly used to encapsulate negatively charged therapeutics via electrostatic pinning. In addition, it has a favorable interaction with negatively charged cell membranes. Finally, GMOPEG is utilized for colloidal stability. We hypothesize that microfluidic synthesis combined with lipid compositional tuning enables the stabilization of cubosomes and cuboplexes with sizes smaller than 100 nm and narrow size distribution. We fabricated and utilized a staggered herringbone mixer (SHM) that provides chaotic and complete mixing profiles in small volumes, which is crucial for size-controlled nanoparticle synthesis.41
Microfluidic Synthesis.
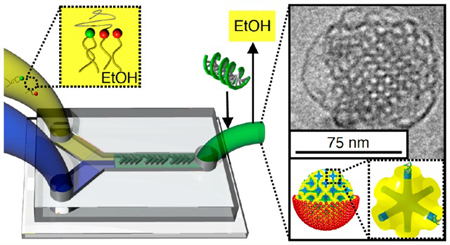
Figure 1 schematically represents the process of cubosome fabrication using the SHM device. The lipid feeding solution is injected in one inlet and consists of three different lipids (GMO, DOTAP, and GMOPEG) at compositions such that a bicontinuous cubic phase is the equilibrium structure in bulk and excess water. On the basis of our previous studies of this tricomponent lipid system, we fixed GMOPEG content to 1 mol %, which showed good colloidal stabilization properties and DOTAP contents to 2 mol %.14,39 At this regime, the stable space group of the bicontinuous cubic phase in excess water is the Im3m (see cartoon in Figure 2), but in general any type of cubic phase or other geometry can be attained and is determined by lipid composition. In a second inlet, an aqueous phase is concurrently loaded in the microfluidic device using a syringe pump. The lipid and water feeding solutions are delivered with a total flow rate and flow rate ratio (water/lipid solution) that are optimized for a particular lipid system. Supporting Note 1 describes the optimized conditions to produce cubosomes. The ethanol and aqueous solutions are then subjected to chaotic advection, which is induced by the herringbone structure of the microchannels,41 reaching intermediate Reynold’s numbers (2 < Re < 500 for 0.02 mL/min < total flow rate < 4 mL/min).42 Under these conditions an ethanol-in-water emulsion of nanosized droplets is produced with a lipid layer stabilizing the interface between the solvents. The emulsion is collected from the outlet (after chaotic mixing for under a second) and treated on a rotary evaporator to extract the volatile solvent (less than 20 min). The resultant solution consists of a dispersion of cubosomes of small size and narrow size distribution. The size and size distribution of the resultant cubosomes are completely coupled with the properties of the emulsion droplets. There are multiple variables that can affect emulsion sizes including flow conditions, lipid concentration, and lipid composition (see Supporting Note 1).
Figure 1.
Schematic illustration of cubosome microfluidic synthesis. Water and lipid/ethanol feeding solutions are simultaneously injected in two separated inlets. A herringbone pattern is designed to promote the chaotic mixture of water and lipid/ethanol solutions. The mixing step takes 0.6 s and leads to the formation of small and monodisperse ethanol-in-water emulsion droplets stabilized by a lipid layer. Subsequent removal of ethanol from the emulsions by rotary evaporation (at 57 °C and P = 75 mbar) produces monodisperse and small cubosomes in less than 15 min.
Figure 2.
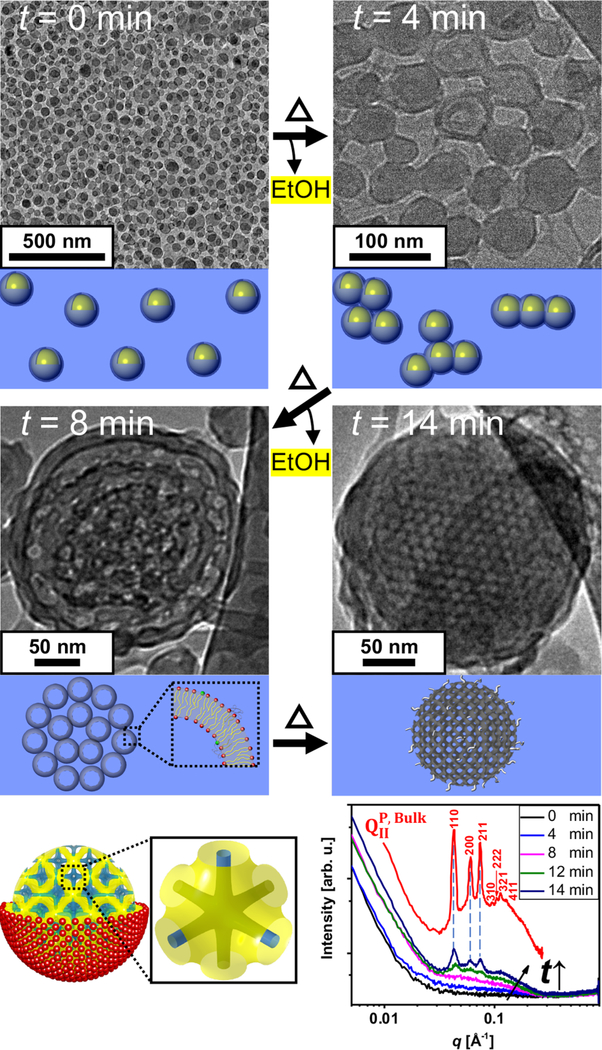
Mechanism of cubosome formation. Cryo-EM images obtained for microfluidic solutions of ethanol-in-water droplets stabilized by a ternary lipid mixture (GMO/DOTAP/GMOPEG, 97/2/1, mole ratio) at different time points (t) of ethanol evaporation. At t = 0 min monodisperse emulsions (droplet size 50 nm) are observed. At t = 4 min, emulsion droplets begin to fuse into string-like and cluster morphologies. After four more minutes of the treatment (t = 8 min) the membranes in the emulsion are fully fused and starting to display a periodic array. This is the stage where most of nanoparticles show a rim that is consistent with a lipid bilayer structure. At the final stage of the treatment (t = 14 min), lipid membranes now depleted from ethanol rearrange into a periodic and highly ordered primitive bicontinuous cubic structure. Schematics of a cubosome having a primitive bicontinuous cubic structure enclosed by a single lipid leaflet are shown where yellow represents the midplane of the lipid bilayer and blue the water channels. A single unit cell is also represented to the right of the schematic. Synchrotron SAXS scans of the ternary mixture (GMO/DOTAP/GMOPEG, 97/2/1, mole ratio) at different rotary evaporation time points are shown in the bottom right. SAXS scan of a bulk cubic phase with the same lipid composition is included in the plot (red line). The sample treated from 0 and 4 min shows weak scattering due to low lipid concentration. At t = 8 min, a bilayer form factor signal starts to be visible, and at t =12 min three clear Bragg peaks at q = 0.043, 0.061, and 0.075 Å −1 are observed exactly matching the q positions of the bulk phase. The three peaks correspond to the [110], [200], and [211] planes that index to a bicontinuous cubic structure with Im3m space group, QIIP. With two more minutes of treatment (t =14 min), more intense Bragg peaks are observed, indicating that more and highly ordered cubosomes are formed.
Mechanism of Cubosome Formation.
Upon extraction of the volatile solvent (in a rotary evaporator operating at P = 75 mbar and T = 57 °C), cubosomes are formed in an estimated time of 14 min. The effective removal of ethanol is a crucial step for the formation of well-ordered cubosomes, as for significant alcohol contents a discorded bicontinuous structure (referred to as a sponge phase) is favored instead.44 To understand how emulsions evolve into cubosomes in the rotary evaporator, we collected solutions at 0, 4, 8, 12, and 14 min time points (t). Figure 2 shows Cryo-EM images as well as SAXS profiles obtained for each collected solution at various t. Initially (t = 0 min) Cryo-EM images indicate that the solution consists of an emulsion of rather monodisperse and well-isolated droplets at an average diameter of 50 nm. Supporting Note 1 shows dynamic light scattering (DLS) results that corroborate the droplet sizes observed by Cryo-EM. At t = 4 min, emulsions start to fuse, forming “beads on a string” morphologies as well as sheet-like structures. At this point, the darker rim of the droplets is readily visible and indicates the presence of a lipid bilayer membrane. Further thermal treatment and evaporation (t =8 min) leads to a periodic arrangement of fused membranes confined within a spheroidal particle. These particles display a clear membranous rim, most likely consisting of lipid bilayers. After full thermal treatment (t = 14 min), we found that nanoparticles comprise membranous interiors highly ordered into a bicontinuous cubic lattice, a cubosome. The size of the cubosomes observed by Cryo-EM is around 200 nm. Dynamic light scattering (Supporting Note 2) confirms an average diameter of the microfluidics cubosomes of 201 nm and a polydispersity index (PDI) of 0.04. It is noteworthy that traditional large-scale techniques yield larger sizes (average diameter of 200–500 nm) and much higher polydispersity (PDI = 0.11).36,37
In order to clearly elucidate the structure of the nanoparticles in larger ensembles, we conducted SAXS of the collected solutions at different time points. The SAXS profile for the equilibrium bulk bicontinuous cubic phase in excess water is displayed as a control. This consists of several diffraction peaks located at q positions corresponding to a bulk primitive Im3m cubic phase (QIIP). These seven peaks are at the reciprocal lattice vectors corresponding to [110], [200], [211], [310], [222], [321], and [411] reflections, respectively. The peak indexes are satisfying the QIIP structure rules: (i) (h + k + l) = 2n, (ii) 0kl (k + l = 2n), (iii) hhl (l = 2n), (iv) h00 (h = 2n), (v) hkl (k, l = 2n) (with h, k, l permutable and n is an integer).45 The Im3m unit cell is schematically presented in Figure 2 with the midplane of the lipid bilayer represented in yellow and the water channels in blue. The calculated lattice spacing a = 20.7 nm is well matched with the measured spacing from Cryo-EM images (a = 19.8 nm). As time (t) evolves from 0 to 14 min the solutionscattering profiles develop three structure factor peaks with increased intensity and at the q positions compared to the concentrated bulk phase (red line). Already after t =12 min the SAXS profile displays prominent Bragg peaks indicating that the solution consists of dispersed and well-ordered cubosomes.
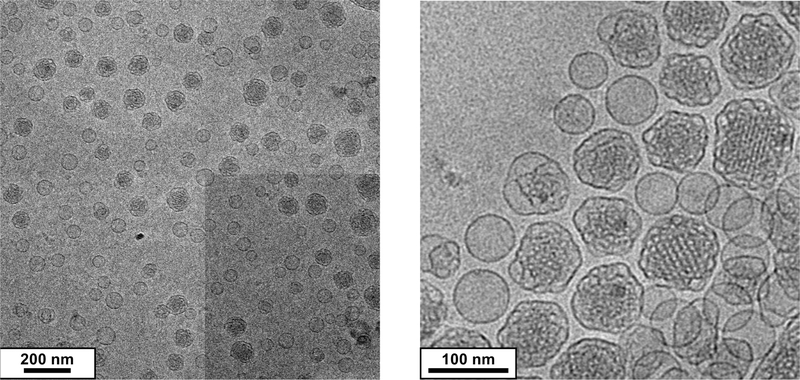
In addition to microfluidics flow conditions and lipid concentration, another handle in the ternary system is lipid composition. On the basis of the fact that cubosome formation is mediated by the fusion of droplet membranes, we hypothesize that increasing GMOPEG stabilizer content (while keeping within the bicontinuous cubic phase regime14,39) will cap particle growth by hindering fusion events. This is important because nanoparticles for therapeutics are now well understood to have an optimal size of 50–100 nm. Figure 3 shows Cryo-EM data for microfluidic cubosomes produced when the GMOPEG content is increased from 1 to 2 mol %. It is noteworthy that the cubosomes reach low diameters32–37 of aproximately 75 nm without compromising the internal ordering of the bicontinuous cubic membrane.
Figure 3.
Ultrasmall cubosome synthesis. Increased amount of GMOPEG (from 1 to 2 mol % gives rise to microfluidic cubosomes of about 75 nm in diameter without compromising ordering on the bicontinuous cubic membranes. Left and right show different magnification of the cubosomes.
siRNA Loading.
In order to produce cuboplexes, siRNA needs to be efficiently loaded into cubosomes without disrupting their structure, colloidal stability, or size distribution. We load siRNA into cubosomes under conditions of nominal charge neutrality (one charged DOTAP per nucleic acid base, ρ = nDOTAP/nNA = 1). Various methods were carried out to achieve successful incorporation of siRNAs within the cubosomes. First, an siRNA solution was used as the feeding aqueous solution instead of pure water during the microfluidic emulsification process. This approach has been successful for the loading of nucleic acids into regular liposomes (lipoplexes) produced by microfluidic devices.42,46 These previous studies employ the same methodology of ethanol-based emulsions but at higher flow rates to limit the size of emulsions. In addition, instead of using a rotary evaporator, the removal of the volatile solvent was conducted by dialysis where no fusion of emulsion droplets is observed. Hence, the size of lipoplexes is controlled solely by microfluidic flow conditions and lipid compositions reaching dimeters as low as 20 nm. In our case of cubosome manufacturing, a crucial step is emulsion fusion, and solvent removal by rotary evaporator is preferable. We found that addition of siRNA into the aqueous feeding line is detrimental because siRNA hinders the fusion events between emulsions. As a result, if siRNA is added directly into the aqueous feeding line, only a few cuboplexes were found and the majority of emulsions containing siRNA were transformed into regular lipoplexes. Traditional lipoplex nanoparticles consist of concentrically organized lipid bilayers intercalated with siRNA.21,47 In an alternative approach, siRNA solutions were mixed with premanufactured microfluidic cubosomes. After a day-long incubation, the solution is centrifuged and filtered to eliminate unbound siRNAs. The concentration of siRNA in the eluted solution was determined by UV absorption using the molar extinction coefficient at 260 nm for the firefly siRNA used in this work, ε260 = 367 392 L mol−1 cm−1. It was found that the siRNA encapsulation rate reaches more than 90%. Figure 4 shows Cryo-EM images of cuboplexes. The postincorporation method yields very well ordered cuboplexes seemingly unaffected by siRNA loading (Figure 4A). Figure 4B shows two-dimensional (2D) electron density maps of cuboplexes in the square region (70 nm × 70 nm) marked in Figure 4A. The relative electron density contrast achieved from Cryo-EM was converted into a color scale (higher to lower electron density goes from blue to red). A well-ordered lipid membrane (green color) is forming a square lattice aligned in the [100] direction, and the red color in the map represents the water channels (less electron dense region). While it is expected that siRNA is loaded within the water channels of the cubosomes,14 as schematically represented by the cartoons of Figure 4, the contrast obtained for the nucleic acids is not sufficient to resolve siRNA partition within the cuboplex. In order to map out the location of siRNA within cuboplexes with nanometer resolution, a strategy was devised where small gold nanoparticles (AuNPs, 1.8 nm diameter) were conjugated with siRNA molecules. We employed a 21 bp siRNA (basepair sequence designed for luciferase knockdown) modified with a 5′ thiol end-group, which is rather specific for AuNP pairing via a thiol—gold bond. After purification through gel electrophoresis, monovalent siRNA—AuNP conjugates were obtained and were incorporated to cubosomes after the microfluidic synthesis as described earlier. Cryo-EM images of cuboplexes loaded with siRNA—AuNP are shown in Figure 4C. AuNP conjugation does not seem to affect the structure of cuboplexes, nor their size. In Figure 4C yellow circles mark the regions where AuNPs can clearly be detected. Figure 4D shows the 2D electron density profiles expanded to the square section, as indicated in Figure 4C. In this case, cuboplexes loaded with siRNA—AuNP show blue and purple regions (high electron density), corresponding to the presence of siRNA—AuNPs. In Supporting Notes 3 and 4, additional 1D and 2D electron density maps clearly elucidate that siRNA—AuNP molecules are partitioned inside the water channels and in close proximity to the lipid membrane, presumably due to the Coulombic attraction between negative charges of siRNA and positively charged lipids. These results are important because they provide a direct imaging proof of siRNA incorporation within colloidal dispersions of lipid bicontinuous cubic membranes.
Figure 4.
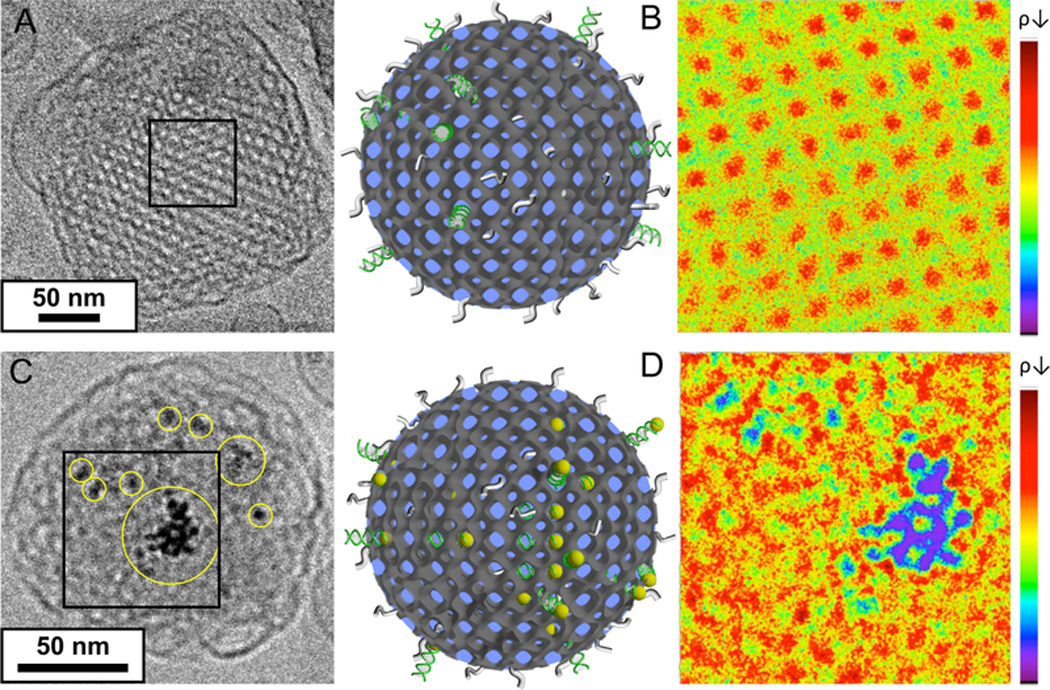
siRNA loading. (A, C) Cryo-EM images of GMO/DOTAP/GMOPEG (97/2/1, mole ratio) cubosomes having (A) siRNA and (C) siRNA-AuNP conjugates. siRNA-AuNP conjugates provide much higher contrast to locate siRNA molecules within the cubosomes. (B, D) 2D electron density maps of selected boxes (70 nm × 70 nm) shown in A and C. The relative electron density contrast achieved from Cryo-EM images was converted into a color scale shown on the right (red for low and blue for high electron density). In B, the well-ordered lipid membrane (green color) is forming a square lattice aligned in the [100] direction. The red color in the map represents less electron dense regions, the water channels. On the other hand, D shows additional blue and purple regions (high electron density) corresponding to AuNP locations and hence the approximate siRNA location. This is a direct proof of siRNA incorporation into a cubosome. Schematics of cuboplexes are also shown in the middle of the figure. The dark gray, green, gray, and yellow colors represent lipid membrane, siRNA, polyethylene glycol moieties, and gold nanoparticles, respectively.
Figure 5 displays additional Cryo-EM data of cuboplexes prepared by siRNA incubation with ultrasmall microfluidic cubosomes (diameter of 75 and 100 nm at 2 mol % and 1.5 mol % GMOPEG, respectively). Cuboplexes can be as small as the cubosomes (<100 nm), which is perfectly suitable for applications in nanomedicine.23 SAXS of cuboplexes (red line) in comparison to cubosomes (black line, from Figure 2C) is shown in Figure 5 as well. There are three sharp Bragg peaks found at q = 0.046, 0.065, and 0.079 A −1 for the cuboplex solution. The three peaks are located at the reciprocal lattice vectors corresponding to [110], [200], and [211] reflections, respectively. The peak indexes are satisfying the QIIP (Im3m space group) structure rules.45 Compared to the SAXS profile of cubosome solutions (black curve), there are two main important points. First, Bragg peak widths with and without siRNA are similar, indicating that incorporation of siRNAs within cubosome water channels does not perturb the ordering of bicontinuous cubic membranes. Second, siRNA inclusion retains the Im3m space group but shifts the Bragg peaks to larger q (wave vector) values. In real space, the unit cell dimension is decreased from a= 20.7 to 19.1 nm upon siRNA inclusion. Changes in unit cell dimensions are indirect proofs of siRNA incorporation in the cubosomes and has been observed for different cationic lipid—nucleic acid complex systems.14,15 By combining Cryo-EM and SAXS data we can qualitatively infer that siRNA molecules are located in the water channels but in tight contact with the cubosome membranes presumably due to electrostatic attraction to the charged DOTAP lipids.
Figure 5.

Small cuboplexes. Small cuboplexes (loaded with siRNA-AuNP) are prepared from small cubosomes that have higher stabilizer composition (top: GMOPEG 2 mol %; middle: GMOPEG 1.5 mol %). In both Cryo-EM images, siRNA-AuNP conjugate locations are distinguishable as black dots, representing higher electron density. Synchrotron SAXS scans (bottom figure) of cubosomes and cuboplexes fabricated by the SHM device. There are three Bragg peaks found at q = 0.046, 0.065, and 0.079 Å−1 for the cuboplex solution (red curve). The three peak ratios correspond to [110], [200], and [211] reflections of the primitive bicontinuous cubic lattice. siRNA inclusion shifts Bragg peaks into larger q, meaning that the unit cell dimension is decreased from 20.4 to 19.1 nm.
Gene Silencing Cuboplexes.
From our previous work14 we found that cuboplexes prepared by top-down approaches were able to efficiently deliver siRNA in cell culture and initiate specific knockdown effects. Traditional lipoplexes consist of a lamellar phase of lipid bilayers intercalated by siRNA21,47 (see Figure 6C) that is known to be less fusogenic with endosomal membranes compared to cubic phases, resulting in lower siRNA delivery efficiency. Importantly, when compared to standard transfecting agents or traditional lipoplexes, cuboplexes displayed a far superior performance at lower toxicity. This was attributed to the fact that cuboplexes have much lower content of toxic cationic lipids, and the escape from the endosomes occurs via favorable membrane topological transitions. However, a systematic investigation of gene knockdown efficiency requires that the ensemble has uniform physicochemical properties such as charge density, size, and shape, as all these factors contribute significantly to the success of a given nanoparticle to deliver their cargo.
Figure 6.
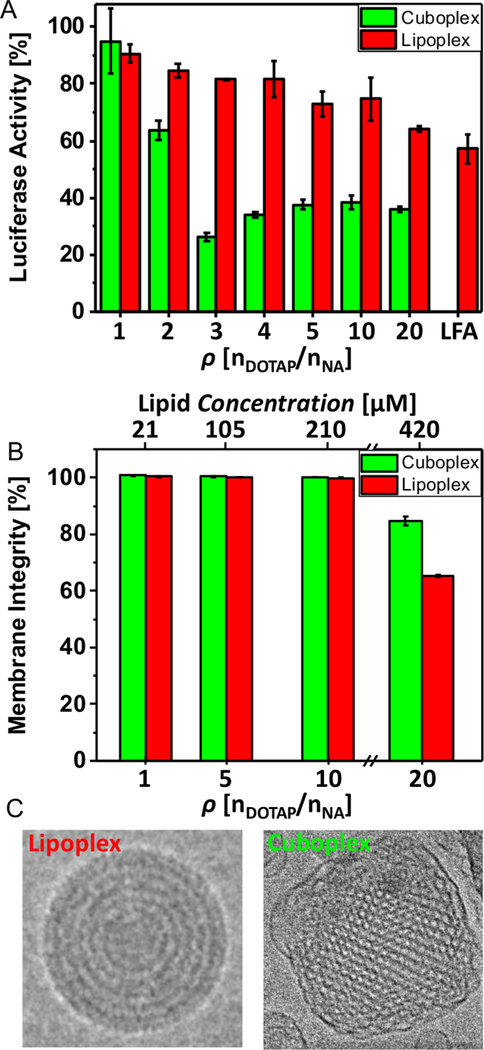
Gene silencing cuboplexes. (A) Luciferase gene knockdown in HeLa-Luc cells of GMO/DOTAP/GMOPEG (97/2/1, mol ratio)-siRNA cuboplexes in contrast to DOPC/DOTAP/DOPEPEG (97/2/1, mol ratio)-siRNA lipoplexes at different charge ratios HeLa-Luc cells were treated with either cuboplexes (green bar) or lipoplexes (red bar) containing two different siRNA molecules (50 nM): siGL2, which targets the firefly luciferase mRNA, and “scrambled” siRNA, which has a random gene sequence. Percentage of luciferase activity is calculated by normalizing the signal ratio of siGL2 to scrambled siRNA at each ρ. As a reference knockdown product, a commercially available liposomal formulation (Lipofectamine 2000, LFA) is also used. For cuboplexes, the maximum knockdown efficiency of 73.6% is attained at ρ = 3 versus 35.8% at ρ = 20 from lipoplexes (higher content of cationic lipid). Importantly, the knockdown efficiency of cuboplexes is much higher than what is obtained with commercially available LFA (45.8%). (B) Membrane integrity test of HeLa-Luc cells when incubated with cuboplexes and lipoplexes. Up to ρ = 10, no significant membrane disruption is observed. At ρ = 20 (0.42 mM of total lipid), however, cuboplexes and lipoplexes show 15.4% and 34.8% decrease of membrane integrity, indicating carriers cannot be used reliably at these high ρ values. (C) Representative Cryo-EM images of traditional DOPC/DOTAP-based lamellar-phase lipoplexes compared to GMO/DOTAP-based cubic-phase cuboplexes.
In this study we investigate the ability of small cuboplexes to deliver appropriate contents of siRNA and initiate gene knockdown in HeLa-Luc cells that are genetically engineered to stably express luciferase. We compare cuboplex performance to a lipoplex system that was designed analogously to cuboplexes. Lipoplexes are composed of DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), instead of the GMO neutral lipid, and a phospholipid-based “PEGylation” agent (DOPE-PEG, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]). The molar fractions of neutral, cationic, and “PEGylated” lipids were kept constant for both systems as well as the siRNA content and charge ratio Figure 6A shows the normalized luciferase activity measured in cells after luciferase-targeting siRNA was delivered to the cells using cuboplexes (green bars) or lipoplexes (red bars) as a function of ρ. As a control, the performance of the cuboplexes and lipoplexes is compared to that obtained by Lipofectamine, a commercially available transfecting agent (LFA). The maximum gene-knockdown efficiency at 73.6% is attained at ρ = 3 for cuboplexes compared to 35.8% at ρ = 20 for lipoplexes, with LFA displaying 45.8% efficiency. Both cuboplexes and lipoplexes showed increased knockdown activity as ρ increases. However, knockdown activity by cuboplexes is saturated at ρ = 3. The dramatic increase of knockdown activity from ρ = 1 to ρ = 3 is presumable due to the fact that Coulomb attraction between cuboplexes and cell membranes is optimized at ρ = 3. It is also plausible that at higher charge ratios siRNAs are not readily decoupled from the cuboplex, even after endosomal escape, due to strong Coulombic attraction to the cubosome membranes.19–21 It is noteworthy that cuboplexes outperform lipoplexes and LFA virtually at all charge ratios. This is important because cationic lipids are toxic and a system at low charge ratio is a requirement for nanomedicine design.
Figure 6B shows the results of cell toxicity, in particular plasma membrane integrity, for cuboplexes and lipoplexes as a function of p. Both systems display comparable toxicity in all workable charge ratios. Up to ρ = 10, both cuboplexes and lipoplexes do not cause significant damage to the integrity of the cell membranes, but at ρ = 20 (0.42 mM of total lipid concentration) cuboplexes and lipoplexes showed 15.4% and 34.8% decrease in membrane integrity, respectively. Figure 6C shows representative Cryo-EM images of traditional DOPC/DOTAP-based lamellar-phase lipoplexes compared to GMO/DOTAP-based cubic-phase cuboplexes.
CONCLUSION
In this study we utilize a microfluidic approach to synthesize monodisperse cubosomes and gene-loaded cubosomes (cuboplexes) with high control over size and size distribution. The cuboplexes significantly outperform commercially available products in their ability to deliver and elicit specific gene knockdown in cells. The microfluidic device enables chaotic mixing of small volumes of lipids dissolved in ethanol and aqueous solutions. As a result, an emulsion of 50 nm droplets of ethanol-in-water is formed and is stabilized with a lipid layer. Utilizing Cryo-EM and SAXS we elucidate the mechanism at which lipid layers of the emulsion droplets fuse as ethanol is evaporated from the sample. After full ethanol evaporation membranes self-organize into periodic bicontinuous cubic arrays to form cubosome particles of about 200 nm and very low polydispersity index (PDI = 0.04). The process relies on a judicious choice of lipids that are known to be stable in bulk bicontinuous cubic phases in excess water, and we use GMO/DOTAP/GMOPEG (97/2/1, molar fraction). We expected that cubosome polydispersity and size to be controllable by microfluidic flow conditions, and while this is true, we found a surprising effect on the amount of GMOPEG stabilizer. Because cubosome development is mediated by membrane fusion, increasing GMOPEG, which is a steric stabilizer, from 1 to 2 mol % caps cubosome growth at sizes as small as 75 nm without losing internal structure ordering. This is important because the use of nanoparticles for nanomedicine requires sizes in the range of 50–100 nm.
siRNA can be loaded to cubosomes to form cuboplexes without perturbing the size, structure, or polydispersity. By conjugating small (1.8 nm) gold nanoparticles to siRNA molecules (siRNA-AuNP) we were able to gain enough Cryo-EM contrast to resolve how siRNA is distributed within the cubosome structure. siRNA molecules are filling the water channels and attracted to the vicinity of lipid membranes through Coulombic interaction. This observation is supported by SAXS by a decrease in unit cell dimension of the cubic lattice upon siRNA loading.
This work highlights the importance of exploring microfluidic synthesis of soft nanoparticles as a route to gain control of size, polydispersity, composition, and nanostructure, which is crucial for the applications in cell delivery.
EXPERIMENTAL DETAILS
Materials.
Glycerol monooleate was purchased from Sigma-Aldrich (MO, USA) and PEGylated GMO was custom designed and ordered from NOF America (NY, USA). DOTAP was purchased from Avanti Polar Lipids (AL, USA). GMO, DOTAP, and GMOPEG are dissolved in fresh chloroform solvent (Sigma-Aldrich, MO, USA) at the desired composition and used without further purification. Citric acid capped 1.8 nm diameter gold nanoparticles were purchased from Nanopartz (CO, USA). Su-8 2025 photoresist was purchased from MicroChem (MA, USA). Tetrahydrooctyl-dimethylchlorosilane was purchased from Gelest (PA, USA). Sylgard 184 (polydimethylsiloxane, PDMS) was purchased from Dow Corning (MI, USA). Lipofectamine 2000 was purchased from Invitrogen (CA, USA).
Staggered Herringbone Mixer Fabrication.
The SHM is fabricated using conventional photolithography methods as described elsewhere.42 Briefly, Su-8 2025 negative photoresist was spin coated on a three-inch sized silicon wafer. Upon soft baking (75 °C for 3 min and 105 °C for 9 min), the bottom channel pattern was formed through UV exposure using a mask aligner (MJB3, SUSS MicroTec, Garching bei Munchen, Germany). The pattern was then baked postexposure at 75 °C for 2 min and 105 °C for 7 min. After the baking process, a single channel pattern is visible by eye. The second layer of Su-8 2025 was then spin coated on top of the first layer and soft baked at 105 °C for 6 min. The second layer was then UV exposed and baked postexposure at 75 °C for 1 min and 105 °C for 5 min. After confirming that the herringbone structure was on top of the single channel, the pattern was developed with an SU-8 developer and hard baked at 150 °C for 3 min. The feature dimensions were 200 X 79 μm for the channel, and the herringbone structure is 31 μm high and 50 μm thick. The fabricated features on the wafer were then cleaned with oxygen plasma (Harrick plasma cleaner, Harrick Plasma, NY, USA) for 1 min, and the surface was treated with tetrahydrooctyl-dimethylchlorosilane to easily detach the PDMS mold. The PDMS solution was made by mixing the base to curing agent at a 10 to 1 ratio. The PDMS solution was poured carefully onto the surfacetreated patterns and put in a vacuum to remove trapped bubbles. Once bubbles were removed, PDMS was cured at room temperature overnight and carefully detached from the pattern. The three holes (two inlets and one outlet) were made on PDMS using a biopsy puncture with 1 mm diameter. The PDMS was finally checked through optical microscopy for defects and bonded to the glass slide through oxygen plasma treatment. The fabricated device was then washed with ethanol more than three times and dried under vacuum before each use.
Cubosome and Cuboplex Synthesis with SHM Devices.
The lipid feeding solution was prepared in ethanol (10 mM total lipid concentration) at the desired composition. A syringe pump (NE-400, New Era Pump Systems, NY, USA) was used to regulate the flow conditions of lipid and water feeding solutions. For most of the samples, flow conditions and flow rate ratio were fixed at 0.05 mL/min and 4, respectively. Obtained emulsions were then collected in a round-bottom 50 mL glass flask. Typically, 1.5 mL of lipid feeding solution was used to fabricate one cubosome solution. Collected emulsion solutions (total lipid concentration: 10 mM) were transferred into a rotary evaporator (RV 10, IKA, NC, USA) operating at P = 75 mbar and T =57 °C for about 14 min. The solutions displayed a color change from clear transparent to opaque white, but there were no macroscale precipitates. Resultant cubosome solutions were filtered with a centrifugal filter (Nanosep 100 K MWCO, Sigma-Aldrich, MO, USA) to remove unreacted emulsions and used for DLS, SAXS, and Cryo-EM studies. Cuboplex solutions were prepared by mixing siRNA and cubosome solutions at the desired charge ratio. After a day of incubation, the solution was then purified three times with a centrifugal filter (Nanosep 3k MWCO, Sigma-Aldrich, MO, USA) to remove unbound siRNA.
Small-Angle X-ray Scattering
Cubosome and cuboplex solutions were analyzed by Synchrotron SAXS at beamline 12-ID-B, Advanced Photon Source at Argonne National Lab. The synchrotron source has an average photon energy of 14 keV with a beam size of 300 μm × 20 μm (H × V). 2D scattering data were radially averaged upon acquisition on a Pilatus 300 K 20 Hz hybrid pixel detector (Dectris, Switzerland) and integrated using FIT2D software (http://www.esrf.eu/computing/scientific/FIT2D) from European Synchrotron Radiation Facility.
CryogenicTransmission Electron Microscopy.
Lipid and lipid-siRNA samples for cryogenic transmission electron microscopy (JEOL 2100 Cryo-TEM at 200 kV) were prepared on a lacey carbon-coated copper grid (Structure Probe Incorporation, PA, USA) using the semiautomated Vitrobot (Vitrobot Mark II, FEI). Briefly, 3 μL of 0.1 M cubosome or cuboplex solution was casted on top of a carbon grid at 100% humidity and 25 °C. Rapid immersion of the grid into liquid ethane after two blotting sessions (1 s blotting time) effectively vitrifies the sample with thin (<500 nm) ice thickness. Note that the samples should be kept under −170 °C until successfully transferred into the Cryo-EM instrument in order to prevent ice crystallization. The prepared sample was transferred to the Cryo-EM with a CT-3500 cryo transfer holder (Gatan, CA, USA). The images are obtained at a defocus of ~ 4000 nm. Higher magnification images were obtained by having smaller apertures to minimize drift and electron beam damage on the sample.
Particle Sizing.
Particle size (Z-average) and polydispersity index of the dispersions were determined using a dynamic light scattering instrument (90Plus particle size analyzer, Brookhaven Instruments Corporation) at an angle of 90°. Measurements were performed at 25 °C, and the data presented correspond to the mean of three successive measurements. The dispersions were diluted with water to adjust the signal level if needed. The PDI is taken to be the width of the particle size distribution and calculated using the 90Plus particle sizing software.
Cell Culture and Gene Knockdown.
Stable overexpressing luciferase HeLa cells (HeLa-Luc, SL-0102 by Signosis) were cultured in Dulbecco’s modified Eagle’s medium (mixture of 10% fetal bovine serum and 1% penicillin-streptomycin) at 37 °C with 5% carbon dioxide (CO2). siRNA knockdown experiments with HeLa-Luc cells were conducted by the method provided by the manufacturer with slight modifications. Briefly, 10 000 cells are seeded per well in a 96-well plate on day 1. On day 2, HeLa-Luc cells were then incubated with cuboplexes, lipoplexes, and Lipofectamine 2000 (Invitrogen, USA) with 50 nM siRNA targeting firefly luciferase genes (siGL2, Dharmacon) or 50 nM “scrambled” siRNA (Allstars negative control siRNA, QIAGEN) per well at different charge ratios (ρ) for about 6 h. “No cells” and “cells-only” control groups were also used. Gene knockdown efficiency was then evaluated using the luciferase assay system (Promega, USA). Specifically, the produced light from each well of interest was measured by a plate reader (Victor 3 multilabel reader, PerkinElmer) and normalized by the mass of cells in each well. The relative light unit per mg of protein (RLU/mg protein) was expressed in percentage of luciferase activity as RLU/mg protein ratio for siGL2 with respect to scrambled siRNA at each p. Each reported result is the result of three independent cell culture experiments performed on different days. In addition, each well during one experiment is conducted in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health under grant no. 1DP2EB024377–01. This research used resources of the Advanced Photon Source, beamline 12-ID-B, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. This research was carried out in part at the Frederick Seitz Materials Research Laboratory, University of Illinois.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b03770.
Effect of microfluidic flow conditions on emulsion size, DLS data, and mapping of siRNA partition in Cryo-EM images by electron density in 1D and 2D (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Patton JS; Carey MC Watching Fat Digestion. Science 1979, 204, 145–148. [DOI] [PubMed] [Google Scholar]
- (2).Larsson K. Cubic Lipid-Water Phases: Structures and Biomembrane Aspects. J. Phys. Chem 1989, 93, 7304–7314. [Google Scholar]
- (3).Xiao Q; Wang Z; Williams D; Leowanawat P; Peterca M; Sherman SE; Zhang S; Hammer DA; Heiney PA; King SR; Markovitz DM; Andre S; Gabius H-J; Klein ML; Percec V. Why Do Membranes of Some Unhealthy Cells Adopt a Cubic Architecture? ACS Cent. Sci 2016, 2, 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Almsherqi ZA; Landh T; Kohlwein SD; Deng Y. Chapter 6 Cubic Membranes: The Missing Dimension of Cell Membrane Organization. Int. Rev. Cell Mol. Biol 2009, 274, 275–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Almsherqi ZA; Kohlwein SD; Deng Y. Cubic Membranes: a Legend Beyond the Flatland* of Cell Membrane Organization. J. Cell Biol 2006, 173, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Daniels EW; Breyer EP Starvation Effects on the Ultrastructure of Amoeba Mitochondria. Cell Tissue Res 1968, 91, 159–169. [DOI] [PubMed] [Google Scholar]
- (7).Deng Y; Almsherqi ZA; Ng MML; Kohlwein SD Do Viruses Subvert Cholesterol Homeostasis to Induce Host Cubic Membranes? Trends Cell Biol 2010, 20, 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yaghmur A; Glatter O. Characterization and Potential Applications of Nanostructured Aqueous Dispersions. Adv. Colloid Interface Sci 2009, 147–148, 333–342. [DOI] [PubMed] [Google Scholar]
- (9).Barauskas J; Johnsson M; Tiberg F. Self-Assembled Lipid Superstructures: Beyond Vesicles and Liposomes. Nano Lett 2005, 5, 1615–1619. [DOI] [PubMed] [Google Scholar]
- (10).Spicer PT Progress in Liquid Crystalline Dispersions: Cubosomes. Curr. Opin. Colloid Interface Sci 2005, 10, 274–279. [Google Scholar]
- (11).Farokhzad OC; Langer R. Impact of Nanotechnology on Drug Delivery. ACS Nano 2009, 3, 16–20. [DOI] [PubMed] [Google Scholar]
- (12).Malmsten M. Soft Drug Delivery Systems. Soft Matter 2006, 2, 760–769. [DOI] [PubMed] [Google Scholar]
- (13).Pan X; Han K; Peng X; Yang Z; Qin L; Zhu C; Huang X; Shi X; Dian L; Lu M; Wu C. Nanostructured Cubosomes as Advanced Drug Delivery System. Curr. Pharm. Des 2013, 19, 6290–6297. [DOI] [PubMed] [Google Scholar]
- (14).Kim H; Leal C. Cuboplexes: Topologically Active siRNA Delivery. ACS Nano 2015, 9, 10214–10226. [DOI] [PubMed] [Google Scholar]
- (15).Leal C; Bouxsein NF; Ewert KK; Safinya CR Highly Efficient Gene Silencing Activity of siRNA Embedded in a Nanostructured Gyroid Cubic Lipid Matrix. J. Am. Chem. Soc 2010, 132, 16841–16847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).George LA; Sullivan SK; Giermasz A; Rasko JEJ; Samelson-Jones BJ; Ducore J; Cuker A; Sullivan LM; Majumdar S; Teitel J; McGuinn CE; Ragni MV; Luk AY; Hui D; Wright JF; Chen Y; Liu Y; Wachtel K; Winters A; Tiefenbacher S; et al. Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant. N. Engl. J. Med 2017, 377, 2215–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Thi EP; Mire CE; Lee ACH; Geisbert JB; Zhou JZ; Agans KN; Snead NM; Deer DJ; Barnard TR; Fenton KA; Fenton KA; MacLachlan I; Geisbert TW Lipid Nanoparticle siRNA Treatment of Ebola-Virus-Makona-Infected Nonhuman Primates. Nature 2015, 521, 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sahay G; Querbes W; Alabi C; Eltoukhy A; Sarkar S; Zurenko C; Karagiannis E; Love K; Chen D; Zoncu R; Buganim Y; Schroeder A; Langer R; Anderson DG Efficiency of siRNA Delivery by Lipid Nanoparticles is Limited by Endocytic Recycling. Nat. Biotechnol 2013, 31, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ahmad A; Evans HM; Ewert K; George CX; Samuel CE; Safinya CR New Multivalent Cationic Lipids Reveal Bell Curve for Transfection Efficiency versus Membrane Charge Density: LipidDNA Complexes for Gene Delivery. J. Gene Med 2005, 7, 739–748. [DOI] [PubMed] [Google Scholar]
- (20).Lin AJ; Slack NL; Ahmad A; George CX; Samuel CE; Safinya CR Three-Dimensional Imaging of Lipid Gene-Carriers: Membrane Charge Density Controls Universal Transfection Behavior in Lamellar Cationic Liposome-DNA Complexes. Biophys. J 2003, 84, 3307–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bouxsein NF; McAllister CS; Ewert KK; Samuel CE; Safinya CR Structure and Gene Silencing Activities of Monovalent and Pentavalent Cationic Lipid Vectors Complexed with siRNA. Biochemistry 2007, 46, 4785–4792. [DOI] [PubMed] [Google Scholar]
- (22).Chiappini C; De Rosa E; Martinez JO; Liu X; Steele J; Stevens MM; Tasciotti E. Biodegradable Silicon Nanoneedles Delivering Nucleic Acids Intracellularly Induce Localized in vivo Neovascularization. Nat. Mater 2015, 14, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Tang L; Yang X; Yin Q; Cai K; Wang H; Chaudhury I; Yao C; Zhou Q; Kwon M; Hartman JA; Dobrucki IT; Dobrucki LW; Borst LB; Lezmi S; Helferich WG; Ferguson AL; Fan TM; Cheng J. Investigating the Optimal Size of Anticancer Nanomedicine. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 15344–15349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Buchheim W; Larsson K. Cubic Lipid-Protein-Water Phases. J. Colloid Interface Sci 1987, 117, 582–583. [Google Scholar]
- (25).Chong JYT; Mulet X; Waddington LJ; Boyd BJ; Drummond CJ Steric Stabilisation of Self-Assembled Cubic Lyotropic Liquid Crystalline Nanoparticles: High Throughput Evaluation of Triblock Polyethylene Oxide-Polypropylene OxidePolyethylene Oxide Copolymers. Soft Matter 2011, 7, 4768–4777. [Google Scholar]
- (26).Zhai J; Waddington L; Wooster TJ; Aguilar M-I; Boyd BJ Revisiting ß-Casein as a Stabilizer for Lipid Liquid Crystalline Nanostructured Particles. Langmuir 2011, 27, 14757–14766. [DOI] [PubMed] [Google Scholar]
- (27).Dulle M; Glatter O. Internally Self-Assembled Submicrometer Emulsions Stabilized with a Charged Polymer or with Silica Particles. Langmuir 2012, 28, 1136–1141. [DOI] [PubMed] [Google Scholar]
- (28).Chong JYT; Mulet X; Waddington LJ; Boyd BJ; Drummond CJ High-Throughput Discovery of Novel Steric Stabilizers for Cubic Lyotropic Liquid Crystal Nanoparticle Dispersions. Langmuir 2012, 28, 9223–9232. [DOI] [PubMed] [Google Scholar]
- (29).Barauskas J; Misiunas A; Gunnarsson T; Tiberg F; Johnsson M. Sponge” Nanoparticle Dispersions in Aqueous Mixtures of Diglycerol Monooleate, Glycerol Dioleate, and Polysorbate 80. Langmuir 2006, 22, 6328–6334. [DOI] [PubMed] [Google Scholar]
- (30).Almgren M; Borne J; Feitosa E; Khan A; Lindman B. Dispersed Lipid Liquid Crystalline Phases Stabilized by a Hydro-phobically Modified Cellulose. Langmuir 2007, 23, 2768–2777. [DOI] [PubMed] [Google Scholar]
- (31).Chong J; Mulet X; Boyd B; Drummond C. Steric Stabilizers for Cubic Phase Lyotropic Liquid Crystal Nanodispersions (Cubosomes). Adv. Planar Lipid Bilayers Liposomes 2015, 21, 131–187. [Google Scholar]
- (32).Deshpande S; Singh N. Influence of Cubosome Surface Architecture on Its Cellular Uptake Mechanism. Langmuir 2017, 33, 3509–3516. [DOI] [PubMed] [Google Scholar]
- (33).Wang D; Luo L; Zheng S; Niu Y; Bo R; Huang Y; Xing J; Li Z; Liu Z. Cubosome Nanoparticles Potentiate Immune Properties of Immunostimulants. Int. J. NanoMed 2016, 11, 3571–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nazaruk E; Majkowska-Pilip A; Bilewicz R. Lipidic Cubic-Phase Nanoparticles-Cubosomes for Efficient Drug Delivery to Cancer Cells. ChemPlusChem 2017, 82, 570–575. [DOI] [PubMed] [Google Scholar]
- (35).Spicer PT; Hayden KL Novel Process for Producing Cubic Liquid Crystalline Nanoparticles (Cubosomes). Langmuir 2001, 17, 5748–5756. [Google Scholar]
- (36).Martiel I; Sagalowicz L; Handschin S; Mezzenga R. Facile Dispersion and Control of Internal Structure in Lyotropic Liquid Crystalline Particles by Auxiliary Solvent Evaporation. Langmuir 2014, 30, 14452–14459. [DOI] [PubMed] [Google Scholar]
- (37).Kim D-H; Lim S; Shim J; Song JE; Chang JS; Jin KS; Cho EC A Simple Evaporation Method for Large-Scale Production of Liquid Crystalline Lipid Nanoparticles with Various Internal Structures. ACS Appl Mater. Interfaces 2015, 7, 20438–20446. [DOI] [PubMed] [Google Scholar]
- (38).Garg G; Saraf S; Saraf S. Cubosomes: an Overview. Biol. Pharm. Bull 2007, 30, 350–353. [DOI] [PubMed] [Google Scholar]
- (39).Kim H; Song Z; Leal C. Super-Swelled Lyotropic Single Crystals. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 10834–10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Larsson K. Two Cubic Phases in Monoolein-Water System. Nature 1983, 304, 664–664. [Google Scholar]
- (41).Stroock AD; Dertinger SKW; Ajdari A; Mezi I; Stone HA; Whitesides GM Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. [DOI] [PubMed] [Google Scholar]
- (42).Belliveau NM; Huft J; Lin PJ; Chen S; Leung AK; Leaver TJ; Wild AW; Lee JB; Taylor RJ; Tam YK; Hansen CL; Cullis PR Microfluidic Synthesis of Highly Potent Limit-Size Lipid Nanoparticles for in vivo Delivery of siRNA. Mol. Ther.--Nucleic Acids 2012, 1, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Rungta RL; Choi HB; Lin PJ; Ko RW; Ashby D; Nair J; Manoharan M; Cullis PR; MacVicar BA Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain. Mol. Ther.-Nudeic Acids 2013, 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Oka T; Hojo H. Single Crystallization of an Inverse Bicontinuous Cubic Phase of a Lipid. Langmuir 2014, 30, 8253–8257. [DOI] [PubMed] [Google Scholar]
- (45).Aroyo MI; Arnold H; Bertaut EF; Burzlaff H; Chapuis G; Fischer W; Flack HD; Glazer AM; Grimmer H; Gruber B; Hahn TH; Klapper H; Koch E; Konstantinov P; Kopsky V; Litvin B; Looijenga-Vos A; Muller U; Momma K; Shmueli U; et al. International Tables for Crystallography Vol. A: Space-Group Symmetry In International Tables for Crystallography; International Union of Crystallography: Chester, England, 2016; Vol. A, pp 193–687. [Google Scholar]
- (46).Li Y; Lee RJ; Huang X; Li Y; Lv B; Wang T; Qi Y; Hao F; Lu J; Meng Q; Teng L; Zhou Y; Xie J; Teng L. SingleStep Microfluidic Synthesis of Transferrin-Conjugated Lipid Nanoparticles for siRNA Delivery. Nanomedicine 2017, 13, 371–381. [DOI] [PubMed] [Google Scholar]
- (47).Desigaux L; Sainlos M; Lambert O; Chevre R; Letrou-Bonneval E; Vigneron J-P; Lehn P; Lehn J-M; Pitard B. Self-Assembled Lamellar Complexes of siRNA with Lipidic Aminoglycoside Derivatives Promote Efficient siRNA Delivery and Interference. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16534–16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.