Abstract
Annexins are a superfamily of calcium-dependent phospholipid-binding proteins that are implicated in a wide range of biological processes. The annexin superfamily comprises 13 members in humans (ANXAs), the majority of which are frequently dysregulated in cancer. However, the expression patterns and prognostic values of ANXAs in liver cancer are currently largely unknown. The present study aimed to analyze the expression levels of ANXAs and survival data in patients with liver cancer from the Oncomine, GEPIA, Kaplan-Meier plotter and cBioPortal for Cancer Genomics databases. The results demonstrated that ANXA1, A2, A3, A4 and A5 were upregulated, whereas ANXA10 was downregulated in liver cancer compared with normal liver tissues. The expression of ANXA10 was associated with pathological stage. High expression levels of ANXA2 and A5 were significantly associated with poor overall survival (OS) rate whereas ANXA7 and A10 were associated with increased OS. The prognostic values of ANXAs in liver cancer were determined based on sex and clinical stage, which revealed that ANXA2, A5, A7 and A10 were associated with OS in male, but not in female patients. In addition, the potential biological functions of ANXAs were identified by Gene Ontology functional annotation and Kyoto Encyclopedia of Genes Genomes pathway analysis; the results demonstrated that ANXAs may serve a role in liver cancer through the neuroactive ligand-receptor interaction pathway. In conclusion, the results of the present study suggested that ANXA1, A2, A3, A4, A5 and A10 may be potential therapeutic targets for liver cancer treatment, and that ANXA2, A5, A7 and A10 may be potential prognostic biomarkers of liver cancer.
Keywords: liver cancer, annexins, prognosis, Oncomine, GEPIA, Kaplan-Meier plotter
Introduction
Liver cancer is the second leading cause of cancer-associated mortality worldwide, resulting in >500,000 deaths per year (1). Despite recent advances in surgical resection and liver transplantation, the 5-year survival rate of patients with liver cancer remains <17% (2). In the majority of cases, liver cancer is diagnosed at an advanced stage with limited therapeutic options. Only 10–20% of tumors are considered surgically resectable at the time of diagnosis (3), and the long-term survival of the patients remains unsatisfactory due to postsurgical recurrence. Thus, an improved understanding of the molecular mechanisms, as well as identification of prognostic biomarkers and potential therapeutic targets of liver cancer are desirable.
Annexins (ANX) are a superfamily of calcium-dependent phospholipid-binding proteins with a structural homology of 40–60% (4). In humans, annexins comprise 13 members (ANXA1-A11, A13 and A8L1; A12 is unassigned) (4), the nomenclature of which is summarized in Table I. Each ANXA is composed of two major domains; the amino-terminus allows interactions with cytoplasmic proteins, whereas the carboxyl-terminus contains the calcium- and membrane-binding sites (4). ANXAs are involved in a wide range of biological processes such as vesicle trafficking (5), anti-inflammation (6), anti-coagulation (7), calcium signaling (8), cell differentiation, apoptosis and proliferation (4). A number of studies have reported aberrant expression levels of ANXAs in various types of cancer, and that ANXAs may function as tumor suppressors or promoters depending on the cancer type. For example, ANXA1 serves a tumor-promoting role in colorectal and lung cancer (9,10), but functions as a tumor suppressor in esophageal and gastric cancer (11). The role of ANXA1 has also been investigated in hepatocellular carcinoma (HCC), but the results are controversial (12). In addition, other ANXAs, including ANXA2, A3, A4, A6, A7 and A10, have been reported to be dysregulated in liver cancer (13–18). However, the majority of these studies only reported the expression levels of ANXAs and lacked any prognosis data. The expression patterns, prognostic roles and functions of ANXAs in liver cancer remain unclear.
Table I.
Nomenclature and characterization of the ANXAs.
| ANXA | Symbol | Synonyms | Chromosomal location | Mass, Da (length, a.a.) |
|---|---|---|---|---|
| Annexin A1 | ANXA1 | ANX1, LPC1 | 9q21.13 | 38,714 (346) |
| Annexin A2 | ANXA2 | ANX2, ANX2L4, CAL1H, LPC2, LIP2 | 15q22.2 | 38,694 (339) |
| Annexin A3 | ANXA3 | ANX3 | 4q21.21 | 36,375 (323) |
| Annexin A4 | ANXA4 | ANX4 | 2p13.3 | 35,883 (319) |
| Annexin A5 | ANXA5 | ANX5, ENX2 | 4q27 | 35,937 (320) |
| Annexin A6 | ANXA6 | ANX6 | 5q33.1 | 75,873 (673) |
| Annexin A7 | ANXA7 | ANX7 | 10q22.2 | 52,739 (488) |
| Annexin A8 | ANXA8 | ANX8 | 10q11.22 | 30,693 (276) |
| Annexin A8 Like 1 | ANXA8L1 | ANXA8L2 | 10q11.22 | 36,879 (327) |
| Annexin A9 | ANXA9 | ANX31 | 1q21.3 | 38,364 (345) |
| Annexin A10 | ANXA10 | ANX14 | 4q32.3 | 37,278 (324) |
| Annexin A11 | ANXA11 | ANX11 | 10q22.3 | 54,390 (505) |
| Annexin A13 | ANXA13 | ANX13 | 8q24.13 | 35,463 (316) |
In the present study, the distinct expression patterns, prognostic values and potential functions of ANXAs in liver cancer were investigated by analyzing the gene expression, copy number variation and survival data published online.
Materials and methods
Ethics statement
The present study was approved by the Academic Committee of Zhengzhou University (Zhengzhou, Henan, China) and performed in accordance with the principles of the Declaration of Helsinki. Online databases were used to retrieve the datasets.
ANXA mRNA expression level evaluation
The Oncomine database (https://www.oncomine.org) was used to evaluate the mRNA expression levels of ANXAs in different types of cancer. The mRNA levels of ANXAs in cancer tissues and normal controls were compared using a Student's t-test, with P≤0.01 and fold change (FC)>2.
GEPIA (http://gepia.cancer-pku.cn) is an online database for analyzing tumor and normal sample RNA sequencing data from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) and the Genotype-Tissue Expression (GTEx) projects (http://commonfund.nih.gov/GTEx/). In the present study, the GEPIA database was used to analyze the mRNA expression levels of ANXAs in liver cancer and normal liver samples. Each ANXA was entered into the database separately and analyzed with the settings |log2(FC)|≥1 and P≤0.01. The method for differential gene expression analysis is one-way ANOVA.
Kaplan-Meier survival analysis
The Kaplan-Meier (KM) plotter (http://kmplot.com) database was used to analyze the associations between ANXA mRNA expression levels and OS of patients with liver cancer and the log-rank test was used to obtain the P-values. Briefly, ANXAs were entered into the database and analyzed using different settings of clinical parameters [e.g. sex and pathological stage (19)]. The cases were divided into high or low expression groups based on the median expression level of each gene. Differences in OS were tested by Cox proportional hazards regression. KM survival plots were obtained with the number-at-risk, hazard ratio (HR), 95% confidence intervals (CI) and P-value displayed on the webpage. P<0.05 was considered to indicate a statistically significant difference.
cBioPortal for cancer genomics
The cBioPortal for Cancer Genomics (www.cbioportal.org) provides visualization, analysis and downloads of large-scale cancer genomics datasets (20). The liver HCC (TCGA, provisional) dataset (21), which contained 442 patients with HCC, was selected for analyses of ANXAs. The alterations of ANXAs, as well as the network between ANXAs and the 50 most frequently altered neighboring genes were obtained according to the instructions on the cBioPortal.
Gene function and pathway enrichment analysis
Gene Ontology (GO; http://geneontology.org/) analysis was used to predict the enriched biological functions of ANXAs and genes associated with ANXA alterations (neighboring genes). A Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/) pathway analysis was used to determine the enriched biological pathways of ANXAs and their neighboring genes. The Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool (https://david.ncifcrf.gov/) was used to perform the GO functional annotation and KEGG pathway enrichment analysis.
Results
mRNA expression levels of ANXAs in patients with liver cancer
The mRNA expression levels of 13 ANXAs in liver cancer were compared with those in normal samples using the Oncomine database. As presented in (Fig. 1), ANXA1, A2, A3 and A4 were upregulated, whereas ANXA10 was downregulated in liver cancer compared with normal tissues. Among these, ANXA2 was upregulated in four datasets [Mas (22), Roessler2 (23), Roessler (23) and Wurmbach (24)] with fold-changes of 2.409, 3.506, 3.528 and 2.350, respectively (Table II). In addition, the Mas dataset exhibited higher mRNA levels of ANXA1, A3 and A4 in HCC compared with those in normal liver, with fold-changes of 5.649, 5.253 and 2.270, respectively. ANXA10 was the only downregulated ANXA gene in liver cancer, with fold-changes of −6.349, −5.586 and −10.711 in the Roessler, Roessler2 and Wurmbach datasets, respectively. However, for the transcription levels of ANXA5, A6, A7, A8, A8L1, A9, A11 and A13, no significant difference was identified between tumors and normal liver samples (data not shown).
Figure 1.
mRNA levels of ANXAs in different types of cancer, generated from the Oncomine database. The number of datasets with statistically significant (P<0.01) were indicated. Blue represents downregulated and red represents upregulated genes (Cancer vs. corresponding normal tissues). The numbers represent the number of datasets with differentially expressed genes.
Table II.
Significant changes of ANXA expression at the transcriptional level between liver cancer and normal liver tissues using the Oncomine database.
| ANXA | Comparison | Fold change | P-value | t-test | Database | (Refs.) |
|---|---|---|---|---|---|---|
| ANXA1 | HCC vs. Normal | 5.649 | 1.60×10−13 | 9.880 | Mas | (22) |
| ANXA2 | HCC vs. Normal | 3.506 | 1.10×10−65 | 20.365 | Roessler2 | (23) |
| HCC vs. Normal | 2.409 | 1.28×10−10 | 8.629 | Mas | (22) | |
| HCC vs. Normal | 3.528 | 2.69×10−7 | 6.044 | Roessler | (23) | |
| HCC vs. Normal | 2.350 | 2.78×10−4 | 4.128 | Wurmbach | (24) | |
| ANXA3 | HCC vs. Normal | 5.253 | 2.18×10−28 | 17.617 | Mas | (22) |
| ANXA4 | HCC vs. Normal | 2.270 | 1.03×10−11 | 8.673 | Mas | (22) |
| ANXA10 | HCC vs. Normal | −10.711 | 1.36×10−10 | −8.183 | Wurmbach | (24) |
| HCC vs. Normal | −5.586 | 2.50×10−69 | −22.733 | Roessler2 | (23) | |
| HCC vs. Normal | −6.439 | 8.55×10−8 | −7.275 | Roessler | (23) |
HCC, hepatocellular carcinoma.
ANXA mRNA levels are associated with clinicopathological parameters of patients with liver cancer
The mRNA expression levels of 13 ANXAs in liver cancer were accessed from the GEPIA database. As demonstrated in (Fig. 2), four dysregulated members were identified. The expression levels of ANXA2, A5 and A11 were significantly higher (P<0.01), whereas ANXA10 was significantly lower in liver cancer compared with those in normal liver tissues (P<0.01). In addition, the ANXA mRNA levels were compared between groups of patients with different pathological stages [classified according to American Joint Committee on Cancer TNM (19)] of liver cancer. The results revealed that the expression levels of ANXA3, A8, A8L1 and A10 were significantly different between patients with different pathological stages (Fig. 3). However, the results of ANXA3, A8 and 8L1 levels need to be interpreted with caution due to their very low expression levels.
Figure 2.
mRNA expressions of ANXAs in liver cancer and normal tissues as accessed by GEPIA database. *|Log2FC|>1 and P<0.01.
Figure 3.
One-way ANOVA was used to test the association between ANXAs expression and tumor stage in liver cancer (GEPIA database). The Pr (<F) corresponds to the P-value of the test.
Prognostic values of ANXAs in patients with liver cancer
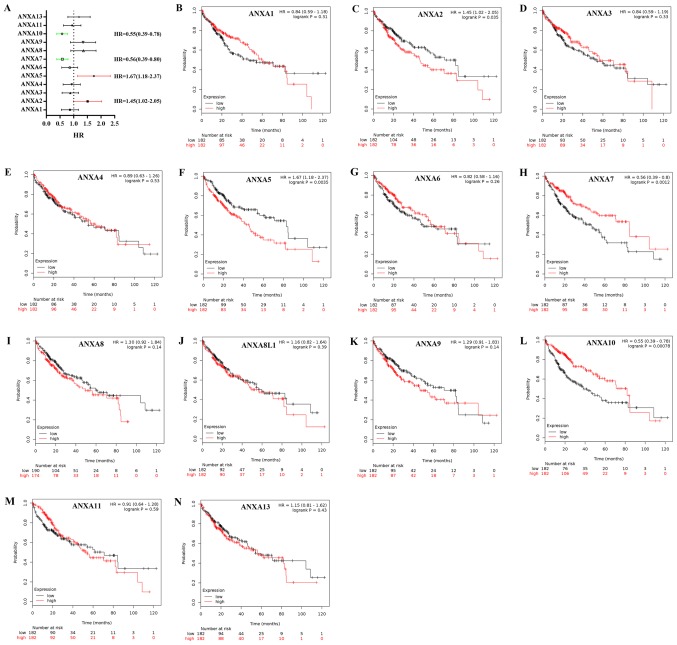
The prognostic values of 13 ANXAs in liver cancer were determined using the KM plotter database. The results demonstrated that four members were significantly associated with prognosis. As presented in (Fig. 4), the survival curves revealed that high mRNA expression levels of ANXA2 and A5 were significantly associated with poor prognosis (HR, 1.45; 95% CI, 1.02–2.05; P=0.035 and HR, 1.67; 95% CI, 1.18–2.37; P=0.0035, respectively), whereas high mRNA expression levels of ANXA7 and A10 were significantly associated with improved prognosis compared with the respective low expression groups (HR, 0.56; 95% CI, 0.39–0.80; P=0.0012 and HR, 0.55;95% CI,0.39–0.78; P=0.00078, respectively), although the high expression of ANXA10 resulted in decreased survival time. The other ANXAs, including ANXA1 (HR, 0.84; 95% CI, 0.59–1.18; P=0.31), A3 (HR, 0.84; 95% CI, 0.59–1.19; P=0.33), A4 (HR, 0.89; 95% CI, 0.63–1.26; P=0.53), A6 (HR, 0.82; 95% CI, 0.58–1.16; P=0.26), A8 (HR, 1.30; 95% CI, 0.92–1.84; P=0.14), A8L1 (HR, 1.16; 95% CI, 0.82–1.64; P=0.39), A9 (HR, 1.29; 95% CI, 0.91–1.83; P=0.14), A11 (HR, 0.91; 95% CI, 0.64–1.28; P=0.59) and A13 (HR, 1.15; 95% CI, 0.81–1.62; P=0.43) were not associated with OS.
Figure 4.
Prognostic value of ANXAs in Kaplan-Meier plotter. (A) Prognostic HRs and (B-N) survival curves of individual ANXAs plotted in liver cancer. HR, hazard ratio.
Prognostic values of ANXAs in patients with liver cancer according to clinicopathological features
The association between individual ANXAs and other clinicopathological features such as sex and clinical stages were further analyzed. As presented in Table III, in male patients with liver cancer, high mRNA expression levels of ANXA2 and A5 were significantly associated with poor OS (HR, 1.95; 95% CI, 1.23–3.18; P=0.036 and HR, 1.93;95% CI, 1.22–3.05; P=0.004, respectively), whereas high mRNA expression levels of ANXA7 and A10 were significantly associated with improved OS compared with the low expression group (HR, 0.53; 95% CI, 0.33–0.83; P=0.0051 and HR, 0.49; 95% CI, 0.31–0.79; P=0.0023, respectively), which was similar to that in all patients with liver cancer. None of the ANXAs were identified as associated with OS in female patients. High expression of ANXA5 was significantly associated with poor OS in patients with stage I (HR, 2.21; 95% CI, 1.17–4.18; P=0.012) and stage III (HR, 2.06; 95% CI, 1.10–3.84; P=0.021) liver cancer; high expression of ANXA7 (HR, 0.41; 95% CI, 0.22–0.78; P=0.0048) and A10 (HR, 0.37;95% CI, 0.20–0.70; P=0.0017) were significantly associated with improved OS in patients with stage III liver cancer (Table IV). The expression levels of other ANXAs were not significantly associated with OS in patients at different clinical stages, although the expression of ANXA2 (HR, 1.71; 95% CI, 0.92–3.18; P=0.088) was modestly associated with poor OS in patients with stage I liver cancer.
Table III.
Association between ANXAs and the sex of patients with liver cancer.
| ANXAs | Sex | Cases, n | HR (95% CI) | P-value |
|---|---|---|---|---|
| ANXA1 | Male | 246 | 0.72 (0.46–1.12) | 0.1390 |
| Female | 118 | 0.97 (0.56–1.68) | 0.8994 | |
| ANXA2 | Male | 246 | 1.95 (1.23–3.18) | 0.0036 |
| Female | 118 | 1.28 (0.74–2.24) | 0.3754 | |
| ANXA3 | Male | 246 | 0.81 (0.52–1.26) | 0.3489 |
| Female | 118 | 0.80 (0.45–1.41) | 0.4426 | |
| ANXA4 | Male | 246 | 1.02 (0.66–1.59) | 0.9253 |
| Female | 118 | 0.93 (0.53–1.63) | 0.7996 | |
| ANXA5 | Male | 246 | 1.93 (1.22–3.05) | 0.0040 |
| Female | 118 | 1.42 (0.81–2.47) | 0.2176 | |
| ANXA6 | Male | 246 | 0.72 (0.46–1.12) | 0.1458 |
| Female | 118 | 0.88 (0.50–1.53) | 0.6474 | |
| ANXA7 | Male | 246 | 0.53 (0.33–0.83) | 0.0051 |
| Female | 118 | 0.97 (0.55–1.69) | 0.9079 | |
| ANXA8 | Male | 246 | 1.28 (0.82–1.99) | 0.2792 |
| Female | 118 | 1.33 (0.76–2.33) | 0.3218 | |
| ANXA8L1 | Male | 246 | 1.30 (0.83–2.02) | 0.2463 |
| Female | 118 | 0.96(0.55–1.67) | 0.8717 | |
| ANXA9 | Male | 246 | 1.44 (0.92–2.26) | 0.1061 |
| Female | 118 | 1.15 (0.66–2.01) | 0.6123 | |
| ANXA10 | Male | 246 | 0.49 (0.31–0.79) | 0.0023 |
| Female | 118 | 0.61 (0.35–1.07) | 0.0817 | |
| ANXA11 | Male | 246 | 0.84 (0.54–1.31) | 0.4366 |
| Female | 118 | 0.88 (0.51–1.53) | 0.6490 | |
| ANXA13 | Male | 246 | 1.50 (0.96–2.35) | 0.0748 |
| Female | 118 | 0.83 (0.48–1.45) | 0.5123 |
P<0.05 was considered to indicate a statistically significant difference. HR, hazard ratio; CI, confidence interval.
Table IV.
Association between ANXAs and the clinical stage of patients with liver cancer.
| ANXAs | Clinical stagea | Cases, n | HR (95% CI) | P-value |
|---|---|---|---|---|
| ANXA1 | I | 170 | 0.82 (0.45–1.50) | 0.5175 |
| II | 83 | 0.87 (0.4–1.90) | 0.7345 | |
| III | 83 | 0.74 (0.41–1.33) | 0.3048 | |
| ANXA2 | I | 170 | 1.71 (0.92–3.18) | 0.0884 |
| II | 83 | 0.99 (0.45–2.14) | 0.9710 | |
| III | 83 | 1.46 (0.81–2.64) | 0.2051 | |
| ANXA3 | I | 170 | 0.93 (0.50–1.7) | 0.8034 |
| II | 83 | 0.70 (0.32–1.56) | 0.3842 | |
| III | 83 | 0.6 (0.33–1.11) | 0.1002 | |
| ANXA4 | I | 170 | 0.84 (0.46–1.54) | 0.5760 |
| II | 83 | 0.74 (0.34–1.6) | 0.4393 | |
| III | 83 | 1.43 (0.78–2.62) | 0.2393 | |
| ANXA5 | I | 170 | 2.21 (1.17–4.18) | 0.0122 |
| II | 83 | 0.81 (0.37–1.76) | 0.5909 | |
| III | 83 | 2.06 (1.1–3.84) | 0.0209 | |
| ANXA6 | I | 170 | 1.04 (0.57–1.91) | 0.8932 |
| II | 83 | 0.94 (0.43–2.06) | 0.8786 | |
| III | 83 | 0.58 (0.31–1.07) | 0.0761 | |
| ANXA7 | I | 170 | 0.60 (0.32–1.11) | 0.0988 |
| II | 83 | 0.70 (0.31–1.56) | 0.3820 | |
| III | 83 | 0.41 (0.22–0.78) | 0.0048 | |
| ANXA8 | I | 170 | 0.76 (0.41–1.41) | 0.3879 |
| II | 83 | 1.87 (0.84–4.17) | 0.1216 | |
| III | 83 | 1.10 (0.61–2.01) | 0.7468 | |
| ANXA8L1 | I | 170 | 0.98 (0.54–1.80) | 0.9551 |
| II | 83 | 1.65 (0.74–3.68) | 0.2183 | |
| III | 83 | 0.65 (0.35–1.19) | 0.1583 | |
| ANXA9 | I | 170 | 1.29 (0.70–2.39) | 0.4065 |
| II | 83 | 1.78 (0.80–3.96) | 0.1532 | |
| III | 83 | 1.47 (0.81–2.68) | 0.2039 | |
| ANXA10 | I | 170 | 0.60 (0.32–1.12) | 0.1019 |
| II | 83 | 0.76 (0.35–1.65) | 0.4800 | |
| III | 83 | 0.37 (0.20–0.70) | 0.0017 | |
| ANXA11 | I | 170 | 0.97 (0.53–1.77) | 0.9131 |
| II | 83 | 0.48 (0.21–1.08) | 0.0689 | |
| III | 83 | 1.01 (0.56–1.81) | 0.9758 | |
| ANXA13 | I | 170 | 1.17 (0.63–2.14) | 0.6189 |
| II | 83 | 1.30 (0.59–2.84) | 0.5122 | |
| III | 83 | 1.59 (0.87–2.89) | 0.1273 |
The data of stage IV was not shown as the sample number was too low for meaningful analysis. HR, hazard ratio; CI, confidence interval.
Predicted functions and pathways of ANXAs in liver cancer
Using the cBioPortal online tool, alterations (copy number variation, mutations and mRNA expression change of ANXAs) were identified in 205 out of 442 patients with liver cancer (46.4%) in the selected dataset (Fig. 5A). An interaction network for ANXAs and the 50 most frequently altered neighboring genes was constructed. The results demonstrated that signal transduction-associated genes, such as EGFR, PRKCA, HCRT, GNAI1 and GNBs, were associated with ANXA alterations (Fig. 5B).
Figure 5.
Analysis of the expression and mutation of ANXAs in liver cancer (cBioPortal). (A) The proportion and distribution of cases with ANXAs alterations. (B) The interaction network of ANXAs and their neighboring genes constructed by cBioPortal. The nodes with bold black outlines represent the seed genes while nodes with thin outlines represent neighbor genes. The color of the nodes represents the alteration frequency of the indicated gene. A darker red represent a higher alteration frequency. The brown line indicates that two genes belong to the same component while the blue line indicates that the two genes interact with each other.
In order to investigate the potential biological functions of ANXAs in liver cancer, GO functional annotation and KEGG pathway enrichment analyses were performed for ANXAs, and the 50 most frequently altered neighboring genes using the DAVID. The results of the GO analysis revealed that ANXAs and their neighboring genes were involved in several biological processes, such as ‘neuropeptide signaling pathway’, ‘phospholipase C-activating G-protein coupled receptor signaling pathway’, ‘positive regulation of cytosolic calcium ion concentration’, ‘G-protein coupled receptor signaling pathway’ and ‘signal transduction’ (Fig. 6A). The cellular component analysis revealed that ANXAs and their neighboring genes were primarily located in ‘plasma membrane’, ‘axon terminus’, ‘dendrite’ and ‘extracellular exosome’ (Fig. 6B). The molecular function analysis suggested that ANXAs and the neighboring genes were primarily enriched in ‘calcium-dependent phospholipid binding’, ‘calcium-dependent protein binding’, ‘calcium ion binding’, ‘G-protein coupled receptor binding’ and ‘neuropeptide receptor activity’ (Fig. 6C). In addition, the results of the KEGG pathway analysis demonstrated that ANXAs and the neighboring genes were primarily enriched in ‘neuroactive ligand-receptor interaction’, ‘GABAergic synapse’, ‘chemokine signaling pathway’, ‘calcium signaling pathway’ and ‘pathways in cancer’ (Fig. 6D).
Figure 6.
Top 10 enrichment function and pathway terms of ANXAs and their neighboring genes in liver cancer. (A-C) Gene Ontology analysis. (D) Kyoto Encyclopedia of Genes and Genomes pathway analysis.
Discussion
Aberrant expression of ANXAs is common in various types of cancer such as melanoma, pancreatic cancer, gastric cancer, lung cancer and breast cancer (8). ANXAs have been demonstrated to be involved in carcinogenesis and progression of liver cancer according to previous studies (12–18). However, the complex roles of ANXAs in the carcinogenesis, progression and prognosis of liver cancer remain to be elucidated. In the present study, the mRNA expression and prognostic values of different ANXAs in liver cancer were investigated by bioinformatics analysis.
ANXA1 is the first known member of ANXAs, which has been reported to be involved in a wide range of cell signaling pathways including inflammatory, cell differentiation, proliferation and apoptosis (25–27). ANXA1 enhances growth and migration in breast cancer by mediating alternative macrophage polarization in the tumor microenvironment (28). In addition, ANXA1 regulates TGF-β signaling and promotes metastasis of basal-like breast cancer cells (29). In liver cancer, the expression pattern of ANXA1 in previous studies is controversial. Suo et al (30) reported that ANXA1 was upregulated in liver cancer compared with nontumor tissues, and that high levels of ANXA1 expression were significantly associated with tumor grade. Lin et al (12) reported that high ANXA1 expression predicted poor prognosis and enhanced the malignant phenotype of tumor cells in liver cancer. However, Hongsrichan et al (31) identified no ANXA1 protein expression in liver cancer by immunohistochemistry, and Xue et al (32) reported decreased ANXA1 expression levels in liver cancer using tissue microarray analysis. In the present study, the expression of ANXA1 was increased in liver cancer compared with normal tissues, which was consistent with Suo's and Lin's studies. However, the present study did not observe a significant association between ANXA1 expression and OS of patients with liver cancer. The prognostic value of ANXA1 in liver cancer requires further investigation.
ANXA2 is one of the most abundant ANXAs, and is widely distributed in the nucleus, cytoplasm, endosomes and extracellular space. The potential role of ANXA2 in cancer has been widely investigated. Previous studies have reported that ANXA2 is upregulated in various types of cancer including lung (33), breast (34), gastric (35), pancreatic (36), colorectal (37) and liver (38) cancers. Increased expression of ANXA2 is associated with cancer development and poor prognosis (39,40). In addition, knockdown of ANXA2 effectively suppresses tumor progression in vitro and in vivo; studies that focused on the underlying molecular mechanisms have demonstrated that ANXA2 either promotes tumor cell invasion by forming a heterotetramer with S100A10 (41), contributing to heterotypic cell-cell interactions between tumor cells and microvascular endothelial cells, or facilitates tumor cell proliferation and chemoresistance by inhibiting p53 expression and activating the transcription factors STAT3 and NFκB (37,42,43). In liver cancer, protein and mRNA expression levels of ANXA2 were identified as upregulated and associated with poor prognosis (38,44). Knockdown of ANXA2 suppressed liver cancer cell migration and invasion by regulating the trafficking of CD147-harboring membrane microvesicles (45). In addition, the expression of ANXA2 was also elevated in the serum of patients with liver cancer compared with healthy controls (46). The results of the present study confirmed that ANXA2 was upregulated in liver cancer and that high expression of ANXA2 was significantly associated with poor OS.
ANXA3 has been demonstrated to function either as a tumor suppressor or promoter candidate in different types of cancer. Upregulation of ANXA3 enhances drug resistance and promotes tumor metastasis in breast cancer (47) and promotes tumor growth and predicts poor prognosis in gastric cancer (48), whereas in prostate cancer, ANXA3 protein expression is downregulated, which is associated with the tumor stage and Gleason score (49). In liver cancer, ANXA3 is upregulated and preferentially expressed in cancer stem cells. High levels of ANXA3 maintains cancer cell stemness via hypoxia-inducible factor α/Notch and JNK signaling pathways (50,51). ANXA3 promotes tumorigenesis and drug resistance in liver cancer, which makes it a potential therapeutic target (18). In addition, serum ANXA3 is also increased in liver cancer compared with normal liver tissues; therefore, ANXA3 is a promising biomarker for the diagnosis, prognosis and therapeutic response evaluation of liver cancer (52). In the present study, ANXA3 was upregulated in liver cancer compared with healthy tissues, but the mRNA level of ANXA3 was not associated with OS.
ANXA4 is primarily expressed in epithelial cells (53). Recent studies have demonstrated that ANXA4 is upregulated and acts as an oncogene in multiple types of cancer, including lung (54), colorectal (55), cervical (56) and gallbladder cancer (57). In liver cancer, ANXA4 has been reported as upregulated, particularly in patients with early recurrence or metastasis (58). High expression levels of ANXA4 predicted early recurrence or metastasis and poor OS of patients with liver cancer (58). In addition, inhibition of ANXA4 suppresses liver cancer cell proliferation, migration and invasion both in vivo and in vitro (15,58). Consistent with these results, the results of the present study demonstrated that the expression of ANXA4 was higher in liver cancer compared with normal tissues, but it was not associated with OS.
ANXA5 exhibits tumor promoter activity in the majority of different types of tumor, including liver cancer (59). Guo et al (60) analyzed protein expression in five pairs of matched primary tumor and tumor thrombus samples using two-dimensional gel electrophoresis, which revealed that ANXA5 was upregulated in tumor thrombus samples. Sun et al (61) reported that the expression of ANXA5 was positively associated with the progression and metastasis of liver cancer, and that ANXA5 promoted carcinogenesis via the integrin and mitogen-activated protein kinase kinase-extracellular signal-regulated kinase pathways. Consistent with these studies, the results of the present study demonstrated that ANXA5 expression was increased in liver cancer compared with normal liver tissues. Furthermore, to the best of our knowledge, the prognostic role of ANXA5 in liver cancer has not yet been reported. In the present study, it was revealed that high expression of ANXA5 was significantly associated with poor OS.
There are limited studies available that focus on ANXA6 in liver cancer. The protein level of ANXA6 is decreased, whereas the mRNA level is increased in liver cancer compared with non-tumorous tissues, suggesting post-transcriptional regulation of ANXA6 (16). To the best of our knowledge, there are currently no reports stating the prognostic value of ANXA6 in liver cancer. In the present study, no significant differences were observed in the mRNA level of ANXA6 between liver cancer and normal tissues, and ANXA6 expression was not associated with OS. In addition, the roles of ANXA8, A8L1, A9, A11 and A13 in liver cancer have rarely been reported. The present study demonstrated that these ANXAs were not associated with OS in patients with liver cancer.
ANXA7 has been reported to be upregulated in liver cancer and to promote tumor cell migration and invasion by interacting with galectin-3 or receptor of activated protein C kinase 1 (62–64), whereas the inhibition of ANXA7 decreases tumor cell invasion and migration (65). Consistent with these studies, the present study revealed modestly increased expression levels of ANXA7 in liver cancer compared with normal liver tissues. In contrast, high expression levels of ANXA7 were associated with improved OS. This contradictory result was also reported by Wang et al (66), who demonstrated that upregulation of ANXA7 increased liver cancer cell migration in vitro, but decreased lymph node metastasis in vivo. In addition, the aforementioned study reported a dynamic change of ANXA7 expression during liver cancer progression (66). Therefore, the exact role of ANXA7 in liver cancer remains unclear and requires further investigation.
ANXA10 is the latest ANXA member to be identified (67). In previous studies, aberrant expression of ANXA10 was associated with carcinogenesis and progression of various types of cancer (68–70), which suggested a possible tumor promoter or suppressor role, although its functional role remains to be clarified. In liver cancer, downregulation of ANXA10 is associated with the malignant phenotype of tumor cells and poor prognosis (14). Overexpression of ANXA10 inhibits proliferation and promotes apoptosis of HepG2 cells (71). Consistent with these studies, the results of the present study confirmed that ANXA10 was decreased and associated with clinical stage in patients with liver cancer. In addition, high expression of ANXA10 was associated with an improved prognosis compared with patients with low ANXA10 expression.
Previous studies have suggested that the risk of developing liver cancer in males is higher compared with that in females (72–75). Irrespective of the etiology, the morbidity of liver cancer in males is 2–4-fold higher compared with that in females (3). A number of studies have suggested that the androgen receptor (AR) may be responsible for the sex disparity observed in liver cancer (76,77). Of note, a study has reported that the transcription and splicing of ANXA7 is regulated by AR-signaling in prostate cancer (78). However, to the best of our knowledge, the role of ANXAs in the sex disparity of liver cancer has rarely been studied. In the present study, the mRNA expression levels of ANXA2, A5, A7 and A10 were significantly associated with OS in male, but not in female patients, which suggested a possible role for these ANXAs in sex disparity.
The potential biological functions of ANXAs in liver cancer were also investigated in the present study through GO functional annotation and KEGG pathway enrichment analyses. The GO analysis revealed that ANXAs and their neighboring genes primarily participated in ‘neuropeptide signaling pathway’, ‘G-protein coupled receptor signaling pathway’, ‘positive regulation of cytosolic calcium ion concentration’ and ‘signal transduction’, which were consistent with previous studies reporting that ANXAs serve important roles in calcium signaling (7,79). In addition, the KEGG pathway analysis demonstrated that ANXAs and their neighboring genes were primarily involved in ‘neuroactive ligand-receptor interaction’, ‘GABAergic synapse’, ‘chemokine signaling pathway’, ‘calcium signaling pathway’ and ‘pathways in cancer’. Among them, the most significantly enriched pathway was the ‘neuroactive ligand-receptor interaction’, suggesting that ANXAs may function in liver cancer through the neuroactive ligand-receptor interaction pathway.
In the present study, the expression levels, prognostic roles and potential biological functions of 13 ANXAs in liver cancer were systemically investigated. The results indicated that ANXA1, A2, A3, A4, A5 and A10 may be potential therapeutic targets for liver cancer treatment, whereas ANXA2, A5, A7 and A10 may be potential prognostic biomarkers of liver cancer. However, functional experiments are required in order to confirm the role of ANXAs in liver cancer progression as well as their specificity and sensitivity as biomarkers. The results of the present study introduced ANXA2/5/10 as good candidates for future experimental works.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81700514) and the Key Scientific Research Projects in Colleges and Universities of Henan Province (grant no. 19A320064).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in the (TCGA) and (Oncomine) repositories, (https://portal.gdc.cancer.gov/, http://www.oncomine.org/).
Authors' contributions
CZ and LM conceived and designed the study. CZ, PW, TS and LZ collected and analyzed the data. CZ and PW wrote the initial draft of the manuscript.. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Academic Committee of Zhengzhou University (Zhengzhou, Henan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Enrich C, Rentero C, Grewal T. Annexin A6 in the liver: From the endocytic compartment to cellular physiology. Biochim Biophys Acta Mol Cell Res. 2017;1864:933–946. doi: 10.1016/j.bbamcr.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Sugimoto MA, Vago JP, Teixeira MM, Sousa LP. Annexin A1 and the resolution of inflammation: Modulation of neutrophil recruitment, apoptosis, and clearance. J Immunol Res. 2016;2016:8239258. doi: 10.1155/2016/8239258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rand JH, Wu XX, Quinn AS, Taatjes DJ. Resistance to annexin A5 anticoagulant activity: A thrombogenic mechanism for the antiphospholipid syndrome. Lupus. 2008;17:922–930. doi: 10.1177/0961203308095029. [DOI] [PubMed] [Google Scholar]
- 7.Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 8.Mussunoor S, Murray GI. The role of annexins in tumour development and progression. J Pathol. 2008;216:131–140. doi: 10.1002/path.2400. [DOI] [PubMed] [Google Scholar]
- 9.Ydy LR, do Espirito Santo GF, de Menezes I, Martins MS, Ignotti E, Damazo AS. Study of the annexin A1 and its associations with carcinoembryonic antigen and mismatch repair proteins in colorectal cancer. J Gastrointest Cancer. 2016;47:61–68. doi: 10.1007/s12029-015-9791-3. [DOI] [PubMed] [Google Scholar]
- 10.Biaoxue R, Xiling J, Shuanying Y, Wei Z, Xiguang C, Jinsui W, Min Z. Upregulation of Hsp90-beta and annexin A1 correlates with poor survival and lymphatic metastasis in lung cancer patients. J Exp Clin Cancer Res. 2012;31:70. doi: 10.1186/1756-9966-31-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y, Chen Y, Xu D, Wang J, Yu G. Differential expression of ANXA1 in benign human gastrointestinal tissues and cancers. BMC Cancer. 2014;14:520. doi: 10.1186/1471-2407-14-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y, Lin G, Fang W, Zhu H, Chu K. Increased expression of annexin A1 predicts poor prognosis in human hepatocellular carcinoma and enhances cell malignant phenotype. Med Oncol. 2014;31:327. doi: 10.1007/s12032-014-0327-7. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim MM, Sun MZ, Huang Y, Jun M, Jin Y, Yue D, Jiasheng W, Zhang J, Qazi AS, Sagoe K, Tang J. Down-regulation of ANXA7 decreases metastatic potential of human hepatocellular carcinoma cells in vitro. Biomed Pharmacother. 2013;67:285–291. doi: 10.1016/j.biopha.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Liu SH, Lin CY, Peng SY, Jeng YM, Pan HW, Lai PL, Liu CL, Hsu HC. Down-regulation of annexin A10 in hepatocellular carcinoma is associated with vascular invasion, early recurrence, and poor prognosis in synergy with p53 mutation. Am J Pathol. 2002;160:1831–1837. doi: 10.1016/S0002-9440(10)61129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YY, Ge C, Tian H, Jiang JY, Zhao FY, Li H, Chen TY, Yao M, Li JJ. The transcription factor Ikaros inhibits cell proliferation by downregulating ANXA4 expression in hepatocellular carcinoma. Am J Cancer Res. 2017;7:1285–1297. [PMC free article] [PubMed] [Google Scholar]
- 16.Meier EM, Rein-Fischboeck L, Pohl R, Wanninger J, Hoy AJ, Grewal T, Eisinger K, Krautbauer S, Liebisch G, Weiss TS, Buechler C. Annexin A6 protein is downregulated in human hepatocellular carcinoma. Mol Cell Biochem. 2016;418:81–90. doi: 10.1007/s11010-016-2735-9. [DOI] [PubMed] [Google Scholar]
- 17.Mohammad HS, Kurokohchi K, Yoneyama H, Tokuda M, Morishita A, Jian G, Shi L, Murota M, Tani J, Kato K, et al. Annexin A2 expression and phosphorylation are up-regulated in hepatocellular carcinoma. Int J Oncol. 2008;33:1157–1163. [PubMed] [Google Scholar]
- 18.Pan QZ, Pan K, Weng DS, Zhao JJ, Zhang XF, Wang DD, Lv L, Jiang SS, Zheng HX, Xia JC. Annexin A3 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma. Mol Carcinog. 2015;54:598–607. doi: 10.1002/mc.22126. [DOI] [PubMed] [Google Scholar]
- 19.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more ‘personalized’ approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 20.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network. Electronic address, corp-author. wheeler@bcm.edu; Cancer Genome Atlas Research Network: Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 25.Sheikh MH, Solito E. Annexin A1: Uncovering the many talents of an old protein. Int J Mol Sci. 2018;19(pii):E1045. doi: 10.3390/ijms19041045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Acquisto F, Piras G, Rattazzi L. Pro-inflammatory and pathogenic properties of Annexin-A1: The whole is greater than the sum of its parts. Biochem Pharmacol. 2013;85:1213–1218. doi: 10.1016/j.bcp.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Locatelli I, Sutti S, Jindal A, Vacchiano M, Bozzola C, Reutelingsperger C, Kusters D, Bena S, Parola M, Paternostro C, et al. Endogenous annexin A1 is a novel protective determinant in nonalcoholic steatohepatitis in mice. Hepatology. 2014;60:531–544. doi: 10.1002/hep.27141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moraes LA, Kar S, Foo SL, Gu T, Toh YQ, Ampomah PB, Sachaphibulkij K, Yap G, Zharkova O, Lukman HM, et al. Annexin-A1 enhances breast cancer growth and migration by promoting alternative macrophage polarization in the tumour microenvironment. Sci Rep. 2017;7:17925. doi: 10.1038/s41598-017-17622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Graauw M, van Miltenburg MH, Schmidt MK, Pont C, Lalai R, Kartopawiro J, Pardali E, Le Dévédec SE, Smit VT, van der Wal A, et al. Annexin A1 regulates TGF-beta signaling and promotes metastasis formation of basal-like breast cancer cells. Proc Natl Acad Sci USA. 2010;107:6340–6345. doi: 10.1073/pnas.0913360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suo A, Zhang M, Yao Y, Zhang L, Huang C, Nan K, Zhang W. Proteome analysis of the effects of sorafenib on human hepatocellular carcinoma cell line HepG2. Med Oncol. 2012;29:1827–1836. doi: 10.1007/s12032-011-0013-y. [DOI] [PubMed] [Google Scholar]
- 31.Hongsrichan N, Rucksaken R, Chamgramol Y, Pinlaor P, Techasen A, Yongvanit P, Khuntikeo N, Pairojkul C, Pinlaor S. Annexin A1: A new immunohistological marker of cholangiocarcinoma. World J Gastroenterol. 2013;19:2456–2465. doi: 10.3748/wjg.v19.i16.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue LY, Teng LH, Zou SM, Ren LQ, Zheng S, Luo W, Bi R, Lü N. Expression of annexin I in different histological types of carcinomas. Zhonghua Zhong Liu Za Zhi. 2007;29:444–448. (In Chinese) [PubMed] [Google Scholar]
- 33.Luo CH, Liu QQ, Zhang PF, Li MY, Chen ZC, Liu YF. Prognostic significance of annexin II expression in non-small cell lung cancer. Clin Transl Oncol. 2013;15:938–946. doi: 10.1007/s12094-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, Yuan J, Zhang J, Tian R, Ji W, Zhou Y, Yang Y, Song W, Zhang F, Niu R. Anxa2 binds to STAT3 and promotes epithelial to mesenchymal transition in breast cancer cells. Oncotarget. 2015;6:30975–30992. doi: 10.18632/oncotarget.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han Y, Ye J, Dong Y, Xu Z, Du Q. Expression and significance of annexin A2 in patients with gastric adenocarcinoma and the association with E-cadherin. Exp Ther Med. 2015;10:549–554. doi: 10.3892/etm.2015.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang YK, Liu H, Wang XZ, Zhu S. Annexin A2 and CD105 expression in pancreatic ductal adenocarcinoma is associated with tumor recurrence and prognosis. Asian Pac J Cancer Prev. 2014;15:9921–9926. doi: 10.7314/APJCP.2014.15.22.9921. [DOI] [PubMed] [Google Scholar]
- 37.Xiu D, Liu L, Qiao F, Yang H, Cui L, Liu G. Annexin A2 coordinates STAT3 to regulate the invasion and migration of colorectal cancer cells in vitro. Gastroenterol Res Pract. 2016;2016:3521453. doi: 10.1155/2016/3521453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Yao M, Wu W, Qiu L, Sai W, Yang J, Zheng W, Huang J, Yao D. Up-regulation of annexin A2 expression predicates advanced clinicopathological features and poor prognosis in hepatocellular carcinoma. Tumour Biol. 2015;36:9373–9383. doi: 10.1007/s13277-015-3678-6. [DOI] [PubMed] [Google Scholar]
- 39.Kling T, Ferrarese R, Ó hAilín D, Johansson P, Heiland DH, Dai F, Vasilikos I, Weyerbrock A, Jörnsten R, Carro MS, Nelander S. Integrative modeling reveals annexin A2-mediated epigenetic control of mesenchymal glioblastoma. EBioMedicine. 2016;12:72–85. doi: 10.1016/j.ebiom.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo S, Xie C, Wu P, He J, Tang Y, Xu J, Zhao S. Annexin A2 is an independent prognostic biomarker for evaluating the malignant progression of laryngeal cancer. Exp Ther Med. 2017;14:6113–6118. doi: 10.3892/etm.2017.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Myrvang HK, Dekker LV. Annexin A2 complexes with S100 proteins: Structure, function and pharmacological manipulation. Br J Pharmacol. 2015;172:1664–1676. doi: 10.1111/bph.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang CY, Chen CL, Tseng YL, Fang YT, Lin YS, Su WC, Chen CC, Chang KC, Wang YC, Lin CF. Annexin A2 silencing induces G2 arrest of non-small cell lung cancer cells through p53-dependent and -independent mechanisms. J Biol Chem. 2012;287:32512–32524. doi: 10.1074/jbc.M112.351957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung H, Kim JS, Kim WK, Oh KJ, Kim JM, Lee HJ, Han BS, Kim DS, Seo YS, Lee SC, et al. Intracellular annexin A2 regulates NF-κB signaling by binding to the p50 subunit: Implications for gemcitabine resistance in pancreatic cancer. Cell Death Dis. 2015;6:e1606. doi: 10.1038/cddis.2014.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang HJ, Yao DF, Yao M, Huang H, Wu W, Yan MJ, Yan XD, Chen J. Expression characteristics and diagnostic value of annexin A2 in hepatocellular carcinoma. World J Gastroenterol. 2012;18:5897–5904. doi: 10.3748/wjg.v18.i41.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Zhao P, Xu XL, Cai L, Song ZS, Cao DY, Tao KS, Zhou WP, Chen ZN, Dou KF. Annexin A2 promotes the migration and invasion of human hepatocellular carcinoma cells in vitro by regulating the shedding of CD147-harboring microvesicles from tumor cells. PLoS One. 2013;8:e67268. doi: 10.1371/journal.pone.0067268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Abd N, Fawzy A, Elbaz T, Hamdy S. Evaluation of annexin A2 and as potential biomarkers for hepatocellular carcinoma. Tumour Biol. 2016;37:211–216. doi: 10.1007/s13277-015-3524-x. [DOI] [PubMed] [Google Scholar]
- 47.Du R, Liu B, Zhou L, Wang D, He X, Xu X, Zhang L, Niu C, Liu S. Downregulation of annexin A3 inhibits tumor metastasis and decreases drug resistance in breast cancer. Cell Death Dis. 2018;9:126. doi: 10.1038/s41419-017-0143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang K, Li J. Overexpression of ANXA3 is an independent prognostic indicator in gastric cancer and its depletion suppresses cell proliferation and tumor growth. Oncotarget. 2016;7:86972–86984. doi: 10.18632/oncotarget.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Köllermann J, Schlomm T, Bang H, Schwall GP, von Eichel-Streiber C, Simon R, Schostak M, Huland H, Berg W, Sauter G, et al. Expression and prognostic relevance of annexin A3 in prostate cancer. Eur Urol. 2008;54:1314–1323. doi: 10.1016/j.eururo.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Pan QZ, Pan K, Wang QJ, Weng DS, Zhao JJ, Zheng HX, Zhang XF, Jiang SS, Lv L, Tang Y, et al. Annexin A3 as a potential target for immunotherapy of liver cancer stem-like cells. Stem Cells. 2015;33:354–366. doi: 10.1002/stem.1850. [DOI] [PubMed] [Google Scholar]
- 51.Tong M, Fung TM, Luk ST, Ng KY, Lee TK, Lin CH, Yam JW, Chan KW, Ng F, Zheng BJ, et al. ANXA3/JNK signaling promotes self-renewal and tumor growth, and its blockade provides a therapeutic target for hepatocellular carcinoma. Stem Cell Reports. 2015;5:45–59. doi: 10.1016/j.stemcr.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma XL, Jiang M, Zhao Y, Wang BL, Shen MN, Zhou Y, Zhang CY, Sun YF, Chen JW, Hu B, et al. Application of serum annexin A3 in diagnosis, outcome prediction and therapeutic response evaluation for patients with hepatocellular carcinoma. Ann Surg Oncol. 2018;25:1686–1694. doi: 10.1245/s10434-018-6402-0. [DOI] [PubMed] [Google Scholar]
- 53.Dreier R, Schmid KW, Gerke V, Riehemann K. Differential expression of annexins I, II and IV in human tissues: An immunohistochemical study. Histochem Cell Biol. 1998;110:137–148. doi: 10.1007/s004180050275. [DOI] [PubMed] [Google Scholar]
- 54.Gaudio E, Paduano F, Ngankeu A, Ortuso F, Lovat F, Pinton S, D'Agostino S, Zanesi N, Aqeilan RI, Campiglia P, et al. A Fhit-mimetic peptide suppresses annexin A4-mediated chemoresistance to paclitaxel in lung cancer cells. Oncotarget. 2016;7:29927–29936. doi: 10.18632/oncotarget.9179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer. 2008;98:426–433. doi: 10.1038/sj.bjc.6604128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi CH, Chung JY, Chung EJ, Sears JD, Lee JW, Bae DS, Hewitt SM. Prognostic significance of annexin A2 and annexin A4 expression in patients with cervical cancer. BMC Cancer. 2016;16:448. doi: 10.1186/s12885-016-2459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao HS, Sun C, Li XX, Wang Y, Jin KZ, Zhang XP, Hu ZQ. Annexin A4-nuclear factor-kappaB feedback circuit regulates cell malignant behavior and tumor growth in gallbladder cancer. Sci Rep. 2016;6:31056. doi: 10.1038/srep31056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen W, Chen L, Cai Z, Liang D, Zhao B, Zeng Y, Liu X, Liu J. Overexpression of annexin A4 indicates poor prognosis and promotes tumor metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9343–9355. doi: 10.1007/s13277-016-4823-6. [DOI] [PubMed] [Google Scholar]
- 59.Peng B, Guo C, Guan H, Liu S, Sun MZ. Annexin A5 as a potential marker in tumors. Clin Chim Acta. 2014;427:42–48. doi: 10.1016/j.cca.2013.09.048. [DOI] [PubMed] [Google Scholar]
- 60.Guo WX, Man XB, Yuan HX, Shi J, Xue J, Wu MC, Cheng SQ. Proteomic analysis on portal vein tumor thrombus-associated proteins for hepatocellular carcinoma. Zhonghua Yi Xue Za Zhi. 2007;87:2094–2097. (In Chinese) [PubMed] [Google Scholar]
- 61.Sun X, Liu S, Wang J, Wei B, Guo C, Chen C, Sun MZ. Annexin A5 regulates hepatocarcinoma malignancy via CRKI/II-DOCK180-RAC1 integrin and MEK-ERK pathways. Cell Death Dis. 2018;9:637. doi: 10.1038/s41419-018-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava M, Torosyan Y, Raffeld M, Eidelman O, Pollard HB, Bubendorf L. ANXA7 expression represents hormone-relevant tumor suppression in different cancers. Int J Cancer. 2007;121:2628–2636. doi: 10.1002/ijc.23008. [DOI] [PubMed] [Google Scholar]
- 63.Du Y, Meng J, Huang Y, Wu J, Wang B, Ibrahim MM, Tang J. Guanine nucleotide-binding protein subunit beta-2-like 1, a new Annexin A7 interacting protein. Biochem Biophys Res Commun. 2014;445:58–63. doi: 10.1016/j.bbrc.2014.01.119. [DOI] [PubMed] [Google Scholar]
- 64.Song L, Mao J, Zhang J, Ibrahim MM, Li LH, Tang JW. Annexin A7 and its binding protein galectin-3 influence mouse hepatocellular carcinoma cell line in vitro. Biomed Pharmacother. 2014;68:377–384. doi: 10.1016/j.biopha.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 65.Huang Y, Wang Q, Du Y, Bai L, Jin F, Zhang J, Fan S, Wang H, Song L, Gao Y, et al. Inhibition of annexin A7 gene and protein induces the apotosis and decreases the invasion, migration of the hepatocarcinoma cell line. Biomed Pharmacother. 2014;68:819–824. doi: 10.1016/j.biopha.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 66.Wang XY, Gao F, Sun YR, Bai LL, Ibrahim MM, Wang B, Tang JW. In vivo and in vitro effect of hepatocarcinoma lymph node metastasis by upregulation of Annexin A7 and relevant mechanisms. Tumour Biol. 2016;37:911–924. doi: 10.1007/s13277-015-3691-9. [DOI] [PubMed] [Google Scholar]
- 67.Morgan RO, Jenkins NA, Gilbert DJ, Copeland NG, Balsara BR, Testa JR, Fernandez MP. Novel human and mouse annexin A10 are linked to the genome duplications during early chordate evolution. Genomics. 1999;60:40–49. doi: 10.1006/geno.1999.5895. [DOI] [PubMed] [Google Scholar]
- 68.Kim J, Kim MA, Jee CD, Jung EJ, Kim WH. Reduced expression and homozygous deletion of annexin A10 in gastric carcinoma. Int J Cancer. 2009;125:1842–1850. doi: 10.1002/ijc.24541. [DOI] [PubMed] [Google Scholar]
- 69.van der Heijden AG, Mengual L, Lozano JJ, Ingelmo-Torres M, Ribal MJ, Fernández PL, Oosterwijk E, Schalken JA, Alcaraz A, Witjes JA. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur J Cancer. 2016;64:127–136. doi: 10.1016/j.ejca.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Zhu J, Wu J, Pei X, Tan Z, Shi J, Lubman DM. Annexin A10 is a candidate marker associated with the progression of pancreatic precursor lesions to adenocarcinoma. PLoS One. 2017;12:e0175039. doi: 10.1371/journal.pone.0175039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Peng X, Hu Z, Zhao Q, He J, Li J, Zhong X. Effects of over-expression of ANXA10 gene on proliferation and apoptosis of hepatocellular carcinoma cell line HepG2. J Huazhong Univ Sci Technolog Med Sci. 2012;32:669–674. doi: 10.1007/s11596-012-1015-5. [DOI] [PubMed] [Google Scholar]
- 72.Li Y, Xu A, Jia S, Huang J. Recent advances in the molecular mechanism of sex disparity in hepatocellular carcinoma. Oncol Lett. 2019;17:4222–4228. doi: 10.3892/ol.2019.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu C, Ren YF, Dong J, Ke MY, Ma F, Monga SPS, Wu R, Lv Y, Zhang XF. Activation of SRY accounts for male-specific hepatocarcinogenesis: Implication in gender disparity of hepatocellular carcinoma. Cancer Lett. 2017;410:20–31. doi: 10.1016/j.canlet.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 74.Zheng B, Zhu YJ, Wang HY, Chen L. Gender disparity in hepatocellular carcinoma (HCC): Multiple underlying mechanisms. Sci China Life Sci. 2017;60:575–584. doi: 10.1007/s11427-016-9043-9. [DOI] [PubMed] [Google Scholar]
- 75.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 76.Kanda T, Takahashi K, Nakamura M, Nakamoto S, Wu S, Haga Y, Sasaki R, Jiang X, Yokosuka O. Androgen receptor could be a potential therapeutic target in patients with advanced hepatocellular carcinoma. Cancers (Basel) 2017;9:pii43. doi: 10.3390/cancers9050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh SH, Chen PJ. Gender disparity of hepatocellular carcinoma: The roles of sex hormones. Oncology. 2010;78(Suppl 1):S172–S179. doi: 10.1159/000315247. [DOI] [PubMed] [Google Scholar]
- 78.Torosyan Y, Simakova O, Naga S, Mezhevaya K, Leighton X, Diaz J, Huang W, Pollard H, Srivastava M. Annexin-A7 protects normal prostate cells and induces distinct patterns of RB-associated cytotoxicity in androgen-sensitive and -resistant prostate cancer cells. Int J Cancer. 2009;125:2528–2539. doi: 10.1002/ijc.24592. [DOI] [PubMed] [Google Scholar]
- 79.Grewal T, Wason SJ, Enrich C, Rentero C. Annexins-insights from knockout mice. Biol Chem. 2016;397:1031–1053. doi: 10.1515/hsz-2016-0168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in the (TCGA) and (Oncomine) repositories, (https://portal.gdc.cancer.gov/, http://www.oncomine.org/).