Abstract
Entomopathogenic fungi routinely kill their hosts before releasing infectious spores, but a few species keep insects alive while sporulating, which enhances dispersal. Transcriptomics- and metabolomics-based studies of entomopathogens with post-mortem dissemination from their parasitized hosts have unraveled infection processes and host responses. However, the mechanisms underlying active spore transmission by Entomophthoralean fungi in living insects remain elusive. Here we report the discovery, through metabolomics, of the plant-associated amphetamine, cathinone, in four Massospora cicadina-infected periodical cicada populations, and the mushroom-associated tryptamine, psilocybin, in annual cicadas infected with Massospora platypediae or Massospora levispora, which likely represent a single fungal species. The absence of some fungal enzymes necessary for cathinone and psilocybin biosynthesis along with the inability to detect intermediate metabolites or gene orthologs are consistent with possibly novel biosynthesis pathways in Massospora. The neurogenic activities of these compounds suggest the extended phenotype of Massospora that modifies cicada behavior to maximize dissemination is chemically-induced.
Keywords: Massospora, Entomophthorales, Zoopagomycota, Entomopathogen, Amphetamine, Cathinone, Tryptamine, Psilocybin, Psilocin, Invertebrate pathology, Magicicada, Okanagana, Platypedia
1. Introduction
Many animal parasites including viruses, horsehair worms, protists, and fungi modulate the behavior of their hosts and thereby increase disease transmission (Berdoy et al., 2000; Thomas et al., 2002; Hoover et al., 2011; Gryganskyi et al., 2017). Each parasite possesses adaptive traits that maximize dispersal, and often the modification of host behavior is interpreted as a dispersal-enhancing “extended phenotype” of the parasite (Dawkins, 1982, Hughes, 2013). “Summit disease” (SD) behavior is an extended phenotype of multiple species of insect-infecting (entomopathogenic) fungi where parasitized insects ascend and affix to elevated substrates prior to death, which facilitates post-mortem dissemination of spores later emitted from their mummified carcasses (Roy et al. 2006; Hughes et al. 2016; Gryganskyi et al., 2017; Elya et al., 2018). A more rarely seen extended phenotype among entomopathogenic fungi is “active host transmission” (AHT) behavior where the fungus maintains or accelerates “normal” host activity during sporulation, enabling rapid and widespread dispersal prior to host death (Roy et al. 2006; Hughes et al. 2016).
The Entomophthorales (Zoopagomycota) are among the most important insect- and non-insect arthropod-destroying fungi and include all known species with AHT behavior (Roy et al., 2006; Spatafora et al. 2016). Massospora, along with the closely related genus Strongwellsea, are the only known genera where AHT is the sole reported form of behavior modification across all known species (Roy et al., 2006). Massospora contains more than a dozen obligate, sexually transmissible pathogenic species that infect at least 21 species of cicadas (Hemiptera) worldwide (Kobayashi, 1951; Ciferri et al., 1957; Soper 1963, 1974, 1981; Ohbayashi et al. 1999). Infectious asexual spores (conidia) are actively disseminated from the posterior end of the infected cicada during mating attempts and flights, which are not diminished despite these conspicuous infections (Soper 1963, 1974; White et al., 1983; Cooley et al. 2018) (Fig. 1). Instead, hypersexual behaviors are observed in Massospora-infected cicadas, where male cicadas, in addition to typical mating behavior, also attract copulation attempts from conspecific males (Murphy and Redden, 2003; Cooley et al., 2018). Massospora effectively hijacks cicadas, turning them into efficient vectors for conidial transmission (Cooley et al. 2018).
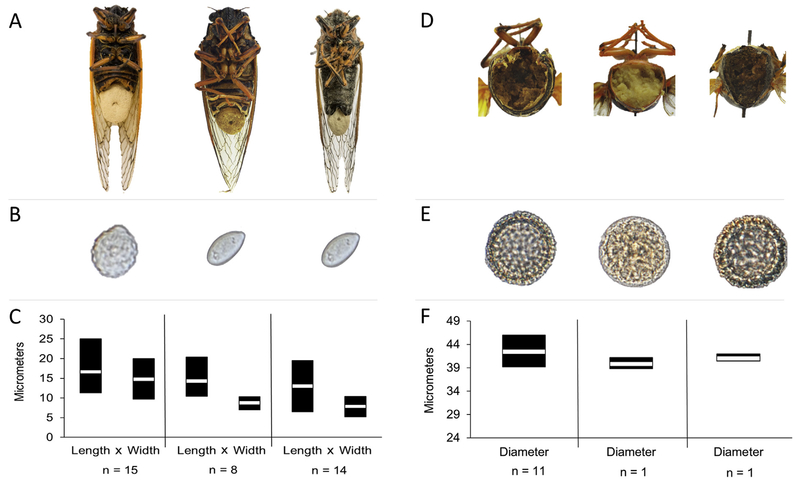
Fig. 1.
Massospora-infected cicadas with associated spore morphology. (A) From left to right: Mas. cicadina-infected periodical cicada (Magicicada septendecim), Mas. levispora-infected Say’s cicada (Okanagana rimosa), and Mas. platypediae infected wing-banger cicada (Platypedia putnami) with a conspicuous conidial “plugs” emerging from the posterior end of the cicada; (B) close-up of conidia for each of three Massospora spp.; (D) posterior cross-section showing internal resting spore infection; and (E) close-up of resting spores for each of three Massospora spp. Specimens in B-F appear in same order as A. Mean (C) conidia and (F) resting spore dimensions for three Massospora species sampled from infected cicadas. Twenty-five conidia or resting spores were measured for each specimen except for Mas. levispora (MI) and Mas. aff. levispora (NM) resting spores, in which 50 spores were measured.
Because of their ephemeral nature, obligate lifestyle, and large genome size, Entomophthorales with both SD and AHT behaviors have been grossly underrepresented in vitro lab investigations as well as in phylogenetic and phylogenomic studies (Spatafora et al. 2016; Gryganskyi et al. 2017). However, recent genomic, transcriptomic, and metabolomic approaches have further helped elucidate important aspects of SD-inducing entomopathogens (Grell et al. 2011; Małagocka et al., 2015, De Fine Licht et al., 2017; Arnesen et al., 2018; Elya et al. 2018; Wrońska et al., 2018). Likewise, -omics based investigations of SD-inducing Hypocrealean entomopathogens have helped unravel infection processes and host responses, especially for the “zombie ant fungus” Ophiocordyceps unilateralis (De Bekker et al. 2014, 2017; Fredericksen et al. 2017). This pathogen is thought to modify host behavior via fungus-derived neurogenic metabolites and enterotoxins, thus providing a chemical basis for this behavioral phenotype (De Bekker et al. 2014, 2017). The mechanisms underlying AHT are completely unexplored, largely due to the inability to axenically culture most Entomophthorales and rear colonies of their insect hosts (such as cicadas) under laboratory conditions. Here, we applied global and targeted liquid chromatography–mass spectrometry (LC-MS)-based metabolomics to freshly collected and archived parasitized cicadas in order to identify candidate small molecules potentially contributing to a hypersexual behavioral phenotype of three Massospora species (Cooley et al. 2018) or more broadly to AHT (Hutchison, 1962; Batko and Weiser, 1965; Roy et al., 2006; Hughes et al. 2016).
2. Methods
2.1. Sampling of healthy and fungus-infected cicudus
A summary of Massospora-infected and healthy cicadas used, including year collected and processed, storage conditions under which cicadas were kept, and specific assays and analyses used are detailed in Table 1. Among periodical cicadas, there are 15 extant “Broods” containing different combinations of four (13-y) and three (17-y) distinct Magicicada spp., respectively (Sota et al., 2013). This study included representatives from six of seven Magicicada spp. spanning 12 (3 13-y and 9 17-y) broods. The term “Brood” refers to populations of periodical cicadas that emerge in the same year and that tend to be geographically contiguous.
Table 1:
Sampling and storage of Massospora-infected cicadas and their asymptomatic cicada counterparts for morphological, molecular, metabolomics, and genomic studies. Sample origin refers to the authors/lab who collected specimens. Numbers before the slash denote conidial isolates whereas those after denote resting spore isolates. Bracketed numbers denote isolates for which sequences were deposited in NCBI Genbank.
| Cicada host/population | Massospora sp. | Year collected/processed | Sample origin | Sample storage | Tissues sampled | Morphology | Phylogenetics | Global Metabolomics | Targeted Metabolomics | Genomics |
|---|---|---|---|---|---|---|---|---|---|---|
| Magicicada septendecula Brood I (WV, USA) | Mas.cicadina | 1978/2017 | JRC/CS | In 95.0% ethanol @4 °C | Fungal spores (FS) | 1/0 | 1/0 | – | – | – |
| Magicicada spp. Brood II (NY & CT, USA) | Mas.cicadina | 1979/2017 | JRC/CS, RAH | Dried and pinned, RT | FS | 3/0 | 1/0 | – | – | – |
| Magicicada septendecula Brood IV or XIX (MO, USA) | Mas.cicadina | 1998/2017 | JRC/CS | Dried and pinned, RT | FS | 1/0 | – | – | – | – |
| Magicicada spp. Brood V (OH & WV, USA) | Mas.cicadina | 2016/2017–2018 | MTK, NA, WJD | Freshly collected to −20 °C | FS | 5/0 | 3/0 [1] | 5/0 | 20/0 | 1/0 |
| Magicicada septendecim Brood VI (NC, USA) | Mas.cicadina | 2017/2017–2018 | TG | Freshly collected to −20 °C | FS | 0/8 | 0/3 | 0/4 | 0/4 | – |
| Magicicada septendecim Brood VIII (PA, USA) | Mas.cicadina | 2002/2017–2018 | JRC/CS | In 95.0% ethanol @4 °C | FS, concentrated ethanol(CE) | 1/0 | – | – | 0/5 | – |
| Magicicada cassini Brood IX (NC, USA) | Mas.cicadina | 2003/2018 | JRC/CS | In 95.0% ethanol @4 °C | CE | – | – | – | 0/1 | – |
| Magicicada cassini Brood X (OH, USA) | Mas.cicadina | 2004/2017 | GK | Dried and pinned, RT | FS | 1/0 | 1/0 | – | – | – |
| Magicicada septendecula Brood XIV (OH, USA) | Mas.cicadina | 2008/2017 | GK | Dried and pinned, RT | FS | 1/0 | 1/0 | – | – | – |
| Magicicada septendecula Brood XIX (IL, USA) | Mas.cicadina | 1998/2017 | JRC/CS | Dried and pinned, RT | FS | 0/3 | 1/0 | – | – | – |
| Magicicada tredecim Brood XXII (MS, USA) | Mas.cicadina | 2001/2018 | JRC/CS | In 95.0% ethanol @4 °C | CE | – | – | – | 3/0 | – |
| Magicicada spp. Brood XXIII (IL & IN, USA) | Mas.cicadina | 1989, 1998, 2002, 2015/2017–2018 | JRC/CS, GK | In 95.0% ethanol @4 °C | FS, CE | 2/0 | 1/0 [1] | – | 0/4 | – |
| Okanagana rimosa (MI) | Mas. levispora | 1998/2017 | JRC/CS | In 95.0% ethanol @4 °C | CE | 8/1 | 3/0 [1] | – | 7/1 | – |
| Platypedia putnami (NM) | Mas. platypediae | 2017/2017–2018 | MTK | Freshly collected to −20 °C | FS | 14/1 | 3/0 [1] | 5/0 | 11/0 | 1/0 |
| Magicicada spp. Brood V | None/healthy | 2016/2017–2018 | MTK | Freshly collected to −20 °C | Healthy abdomen (HA) | – | – | 1 | 1 | – |
| Okanagana rimosa (MI) | None/healthy | 1998/2017–2018 | JRC/CS | In 95.0% ethanol @4 °C | HA | – | – | – | 1 | – |
| Platypedia putnami (NM) | None/healthy | 2017/2017–2018 | MTK | Freshly collected to −20 °C | HA | – | – | 1 | 1 | – |
| Magicicada sp. Brood VII | None/healthy | 2018/2018 | MTK | Freshly collected to −80 °C | HA | – | – | – | 10 | – |
| Platypedia putnami (CA) | None/healthy | 2018/2018 | MTK | Freshly collected to −80 °C | HA | – | – | – | 10 | – |
| Total | 50 | 18 | 16 | 79 | 2 |
Sampling of freshly infected and asymptomatic wing-banger cicadas (Platypedia putnami) and Brood VII periodical cicadas was carried out on private lands in CA (2018), NM (2017), and NY (2018) with permission from individual landowners. Infected Say’s cicada (Okanagana rimosa) and Brood V periodical cicadas (including Magicicada cassini, M. septendeculum, and M. septendecim) were collected from university-owned properties in MI (1998) and WV (2016), respectively. Collection of infected and healthy cicadas from all locations did not require permits. In CA, cicadas were collected under Exemption 5 of California Code of Regulations Section 650, Title 14 (https://nrm.dfg.ca.gov/FileHandler.ashx?DocumentID=157389&inline).
From May 28 to June 6, 2016, newly emerged Brood V 17-y periodical cicadas were visually inspected for Massospora fungal infections on the campus of West Virginia University in Morgantown, West Virginia, USA. Individuals with visible fungal plugs were transferred individually to sterile 20 mL vials, transported to the laboratory, and stored in a −20 C freezer until processing (Table 1). Samples were obtained from 95 infected cicadas, of which 47 were from Mag. cassini, 41 from Mag. septendecim, and seven from Mag. septendecula. A similar number of males and females were sampled across the three Brood V species, with 53 from females and 50 from males.
On June 1, 2016, additional geographically separated Brood V cicada populations were visually inspected for fungal infections. Seven infected specimens of Mug. septendecim and one of Mug. cussini were recovered from Hocking Co., OH. Concurrently, a coauthor provided an additional 34 infected Brood V specimens of Mug. septendecim collected from across five OH counties including Ashland, Cuyahoga, Hocking, Licking, and Lorain counties. Eight female, Massospora-infected, Brood VI specimens of Magicicada septendecim were collected in June 2017 from North Carolina. All samples were collected and stored as previously described (Table 1).
Twenty-three Mas. cicadina-infected specimens of Magicicada collected from 1978 to 2002 were provided from the University of Connecticut, where they had been stored at room temperature, dried and pinned, since their collection. A second, similarly stored, dried and pinned collection from Mount St. Joseph University provided fungal plugs from seven Mas. cicadina-infected specimens of Magicicada collected from 1989 to 2008, three of which were 13-y cicadas and four were 17-y cicadas (Table 1).
In addition to periodic cicadas, two species of annual cicadas also were sampled and included in this study. On June 4, 2017, newly emerged banger-wing cicadas were inspected for Massospora infections 12 km northeast of Gallina, NM along the Continental Divide Trail. Sixteen infected individuals and one outwardly asymptomatic individual (to serve as a negative control) were identified and sampled as described above.
Nine Mas. levispora-infected specimens of Okanagana rimosa were collected by John Cooley from the University of Michigan Biological Station, Cheboygan and Emmet Counties, MI on June 20, 1998 (Cooley, 1999; Cooley et al., 2018). Infected individuals as well as outwardly asymptomatic individuals were placed immediately in individual 20-ml screw-cap vials containing 95% ethanol. Specimens were completely submerged and stored at 4 C until October 2017, when specimens were sent to WVU for processing (Table 1).
Two independent populations of healthy wing-banger cicadas from Trinity County, CA, USA and Brood VII periodical cicadas (Mag. septendecim) from Onondaga Hill, NY were sampled in early June 2018 to serve as negative controls for targeted metabolomics. Five males and five female cicadas were collected for each cicada species, transferred individually to sterile 25–50 mL vials, and stored in a −80 C freezer until processing (Table 1).
Cicadas were processed aseptically, with half of the visible fungal plug removed with a sterile scalpel and transferred to a 1.5 mL microcentrifuge tube for DNA extraction. Controls were processed similarly, where half of the most posterior segments, as well as underlying host tissues, were sampled. A majority of the remaining plug from infected individuals were used for global metabolomics analyses. Dry plugs were transferred aseptically to sterile 1.5 mL microcentrifuge tubes and stored at −80 C. A small sample of spores was retained for morphological studies, although not all specimens were examined.
2.2. Morphological studies of Massospora spp.
Given the scarcity of Massospora sequences from NCBI GenBank, morphological comparisons were necessary to determine if data on putatively assigned Massospora species agreed with that of previously reported measurements for conidial and resting spore stages (Soper 1963, 1974). Table 1 and Supplemental Table 1 provide a comprehensive overview of which infected annual and periodical cicada collections/specimens were used for morphological studies. Specimens chosen for morphological studies included a subset of those populations also used for phylogenetic and metabolomics but also dried and pinned specimens that represented a diversity of Broods and cicada species but were too degraded for other phylogenetic and omics-based approaches.
A portion of select fungal spore masses was harvested with a sterile scalpel and mounted in lactophenol for examination with light field microscopy. Cover slips were fastened with nail polish to allow slides to be archived and re-examined when necessary. Mean conidial/resting spore length x width and cicada host provided the main criteria to differentiate Massospora species. Slides were examined and photographed using a Nikon Eclipse E600 compound microscope (Nikon Instruments, Melville, New York) equipped with a Nikon Digital Sight DS-Ri1 high-resolution microscope camera. A sample of 25 spores from each slide mount were measured using Nikon NIS-Elements BR3.2 imaging software. A total of 51 Massospora-infected cicadas were examined including 26 Mas. cicadina (15 conidial and 11 resting spore isolates), nine Mas. levispora (eight conidial and one resting spore isolates), and 15 Mas. platypediae (14 conidial isolates and one resting spore isolate).
2.3. DNA extraction, amplification, sequencing, and assembly
Fungal genomic DNA was extracted from harvested fungal plugs using a modified Wizard kit (Promega, Madison, WI, USA) as previously described (Short et al. 2015). DNA was suspended in 75 μL Tris-EDTA (TE) buffer and stored at −20 C. Portions of the nuclear 18S (SSU) and 28S (LSU) ribosomal DNA regions were amplified with the primer combinations NS24/NSSU1088R (Hodge et al., 2017)and LR0R/LR5 (Vilgalys and Hester, 1990) using BioLine PCR kits (Bioline USA Inc., Taunton, MA, USA). EFL, an EF-like gene found in some groups of early diverging fungi including the Entomophthorales, was amplified using primers EF1-983F and EF1aZ-1R (James et al., 2006). Eight degenerate primers pairs (Supplemental Table 2), three of which were validated against a positive control, were designed to target three psilocybin biosynthesis enzymes, PsiD (l-tryptophan decarboxylase), PsiK (4-hydroxytryptamine kinase), and PsiM (norbaeocystin methyltransferase), described from hallucinogenic mushrooms, Psilocybe cubensis and Psilocybe cyanescens (Fricke et al., 2017; Reynolds et al. 2018), were used to attempt to amplify Psilocybe psilocybin biosynthesis genes in Mas. platypediae.
PCR conditions were as follows. LSU: initial denaturation at 94 °C for 2 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 51.1 °C for 45s, and elongation at 72 °C for 90s, and final elongation at 72 °C for 5 min. SSU: initial denaturation at 94 °C for 5 min, 36 cycles of denaturation at 94 °C for 30 s, annealing at 49 °C for 30 s, and elongation at 72 °C for 60 s, and final elongation at 72 °C for 7 min. EFL: initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 95 °C for 60 s, annealing at 54 °C for 60 s, and elongation at 72°C for 90s, and final elongation at 72 °C for 7 min. PsiD: initial denaturation at 95 °C for 4 min, 30 cycles of denaturation at 95 °C for 45 s, annealing at 61 °C for 45 s, and elongation at 72 °C for 45 s, and final elongation at 72 °C for 7 min. PsiK: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 45 s, annealing at 52.5 °C for 50 s, and elongation at 72 °C for 50 s, and final elongation at 72 °C for 5 min. PsiM: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 45 s, annealing at 54 °C for 50 s, and elongation at 72 °C for 40 s, and final elongation at 72 °C for 5 min.
Amplified products were mixed with SYBR gold (Invitrogen, Grand Island, NY, USA) and then were loaded onto a 1.0–1.5%, wt/vol agarose gel made with 0.5–1% Tris-borate-EDTA buffer (Amresco, Solon, OH, USA). Psilocybin biosynthesis (Psi) products were cleaned with Zymo Research (Irvine, CA, USA) DNA Clean and Concentrator-5 kit. All products were purified with ExoSAP-IT (Affymetrix, Inc., Santa Clara, CA, USA) and submitted to Eurofins MWG Operon (Huntsville, AL, USA) for Sanger sequencing. Chromatograms were assembled and inspected using Codon Code aligner and phylogenetic analyses conducted using MEGA 7 (Kumar et al. 2016).
The Mas. platypediae (syn. Mas. aff. levispora (NM)) metagenome assembly was generated from 390 Million 2 × 150 bp raw reads. Reads were quality trimmed with sickle using default parameters (Joshi and Fass, 2011). Metagenome assembly was performed with megahit with “–presets meta-sensitive” option and utilizing GPU cores for accelerated matching speed to produce a genome assembly of ~3 Gb (2.1 million contigs; N50 = 2784 bp). The Mas. cicadina assembly was generated with SPAdes using default parameters. Both assemblies were further cleaned for vector and primer contamination with NCBI vecscreen protocol (https://www.ncbi.nlm.nih.gov/tools/vecscreen/) and implemented in a package (https://github.com/hyphaltip/autovectorscreen). Several rounds of BLASTN based screening were applied to remove contaminated contigs or regions. Further screening of contigs was performed during genome submission process to NCBI to remove highly similar matches to plant or insect sequences. These Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accessions QKRY00000000 (Mas. platypediae) and QMCF00000000 (Mas. cicadina). The versions described in this paper are versions QKRY01000000, QMCF01000000.
2.4. Phylogenetic analysis of Massospora spp.
Phylogenetic trees were constructed using LSU, SSU, and EFL sequences generated from 22, 26, and 20 Massospora sequences, respectively, representing three Massospora species, Mas. cicadina, Mas. levispora and Mas. platypediae. Table 1 provides a comprehensive overview of which infected annual and periodical cicada populations/collections were used for phylogenetic studies. Specimens chosen for phylogenetic studies included a subset of those populations also used for morphological studies and metabolomics as well as dried and pinned specimens that represented a diversity of Broods and cicada species but were too degraded for other omics-based approaches. The concatenated set (LSU + SSU) consisted of four sequences, two from Mas. cicadina and one each from Mas. levispora and Mas. platypediae. Nine additional published sequences spanning the known diversity of the Entomophthorales were included to help resolve relationships among Massospora and its close allies (Humber, 2012). One isolate of Conidiobolus pumilus (strain ARSEF 453) served as an outgroup. Sequences were aligned using CLUSTAL-W (Larkin et al. 2007). Alignments were visually inspected, after which an optimized nucleotide substitution model (Tamura 3-parameter + G) was selected using ModelTest (Posada and Crandall, 1998). Partial deletion with site coverage cutoff of 95% was used. Maximum likelihood (ML) trees were estimated using MEGA 7. Phylogenetic support was assessed by bootstrap analysis with 1000 replicates using MEGA 7.
2.5. Phylogenomic analyses of Entomophthoralean fungi
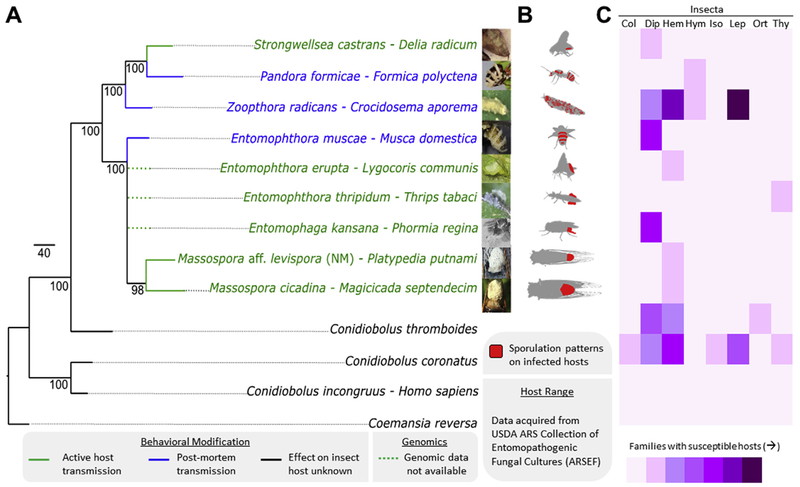
In order to model evolution of behavior-modifying traits across the Entomophthorales and establish whether AHT behavior seen in Massospora and other close relatives, including the only other known AHT-exclusive genus Strongwellsea (Roy et al., 2006, Batko and Weiser, 1965), reflects a single origin or multiple independent origin events, we conducted a phylogenetic analysis of a genome-scale data set of >200 conserved orthologous proteins deduced from the Mas. cicadina and Mas. aff. levispora (NM) metagenomes along with seven other representative taxa (Supplemental Table 3). These taxa span six of 20 described Entomophthorales genera and three of four genera with known active host transmission (Roy et al., 2006; Hodge et al., 2017; Humber, 2012).
Multi-gene phylogenetic analysis of protein coding genes was also performed to assess confidence in the phylogenetic relationships. Since the metagenome assemblies were highly fragmented due to high repetitive sequence content, a complete genome annotation was not undertaken. Instead a targeted similarity search identified contigs encoding homologs of a collection of generally single copy protein coding genes developed for the 1000 Fungal genomes project. These 434 “JGI_1086” markers are available as Hidden Markov Models (https://github.com/1KFG/Phylogenomics_HMMs). Consensus sequences generated from the JGI_1086 markers using hmmemit (HMMER v3) (Eddy, 1998) were used as queries in a translated search of the Massospora metagenome assemblies with TFASTY (Pearson et al. 1997) using a permissive expectation cutoff value of ‘0.001’. These candidate contigs were then annotated with funannotate pipeline (https://github.com/nextgenusfs/funannotate) with informant protein sequences of a collection Zoopagomycota fungi, de novo gene prediction tools Augustus (Stanke and Waack, 2003) and GeneMark ES + ET (Lomsadze et al. 2005).
The predicted proteins for the targeted Massospora contigs were analyzed in the PHYling pipeline with proteins from the annotated genomes of the Kickxellomycotina fungus Coemansia reversa NRRL 1564, the Entomophthoromycotina fungi, Conidiobolus coronatus NRRL 28638, Conidiobolus incongruus B7586, Conidiobolus thromboides FSU 785) and the predicted ORFs from assembled transcriptomes of Entomophthoromycotina Entomophthora muscae, Pandora formicae, Strongwellsea castrans, Zoophthora radicans ATCC 208865. Briefly PHYling (https://github.com/stajichlab/PHYling_unified; https://doi.org/10.5281/zenodo.1257002) uses a preselected marker set of HMMs (https://github.com/1KFG/_Phylogenomics_HMMs; https://doi.org/10.5281/zenodo.1251477) to identify best homologous sequences, aligns, trims, and builds a concatenated phylogenetic tree or individual gene trees. Searches of the annotated genomes of most species identified between 397 (C. coronatus) and 427 (S. castrans) of the 434 marker genes in each species. Searches of the Mas. cicadina and Mas. platypediae targeted annotation set identified 202 and 237 of the 434 marker sets respectively. This lower number indicates a less complete and highly fragmented metagenome assembly but these ~200 protein coding genes provide sufficient phylogenetic information to reconstruct species relationships. The 10-taxon tree was constructed with IQ-TREE (v1.6.3; Lomsadze et al. 2005; Hoang et al., 2017) using the concatenated alignment and 433 partitions and 1000 ultrafast bootstrap support computation (-bb 1000 -st AA -t BIONJ -bspec GENESITE). All marker gene sequences, alignment and partition files are available at https://doi.org/10.5281/zenodo.1306939.
2.6. Fungal metabolite extraction from fungi and insects
Fungal plugs or healthy cicada abdomens were weighed and transferred into sterile 1.5 mL microcentifuge tubes. Metabolite extracts from each fungal sample were normalized to a final concentration of 10 mg of fungal tissue to 1 mL and polar metabolites were extracted in a solution consisting of 80% HPLC grade methanol and 20% water. Samples then were sonicated for 10 min. The sonicated samples were centrifuged at 14,000×g for 15 min at 4 °C to pellet proteins and cellular debris. The supernatant was removed and dried down using lyophilization prior to analysis. Samples were reconstituted in 100 μL of 50% methanol and 50% water for mass spectrometry analysis.
Mycelia from cultures of Conidiobolus coronatus were harvested by scraping 7-d old cultures grown on ½ strength Sabouraud dextrose agar amended yeast extract. Whole Con. coronatus-infected greater wax moth larvae (Galleria mellonella) were harvested at 48 h post inoculation. Both tissues were macerated in an 80% methanol as previously described.
2.7. Analytical procedure and LC-MS conditions for global metabolomics
The Agilent LC-MS system (Agilent Technologies, Santa Clara, CA, USA) included an Agilent 1290 Infinity quaternary ultra-high pressure liquid chromatography (UHPLC) pump. This LC system was coupled to an Agilent 6530 quadrupole time of flight (Q-ToF) mass spectrometer with electrospray ionization. The chromatographic separations were performed using a Merck polymeric bead based ZIC-pHILIC column (100 mm × 2.1 mm, 5 μm).
Mobile phase A consisted of 10 mM ammonium acetate and mobile phase B consisted of 100% acetonitrile. Metabolites were separated on a gradient that went from 95% mobile phase B to 5% mobile phase B over 20-min with 7-min re-equilibration at the end of the chromatographic run.
The mass spectrometer was operated alternately in positive and negative ion mode for the analyses. Samples for global metabolomics consisted of fungal plug extracts (=exposed mass of conidia and host tissue) excised from 5 Mas. cicadina-infected Brood V periodical cicadas (Magicicada spp.; Fig. 1A, Table 1), 5 Mas. platypediae-infected wing-banger cicadas from NM (Platypedia putnami; Fig. 1A, Table 1), and 4 technical reps of excised posterior segments from two healthy brood V periodical cicadas or a healthy P. putnami from NM.
Table 1 provides a comprehensive overview of which healthy and infected annual and periodical cicada populations/collections were used for these studies. Specimens chosen for metabolomics consisted of either freshly collected or well-preserved specimens that had been subjected to cold storage conditions prior to sampling. Data dependent MS/MS was performed on triplicate pooled samples for fragmentation and library searching.
2.8. Data processing, metabolite identification, and statistical data analysis for global metabolomics
The LC-MS/MS data acquired using Agilent Mass Hunter Workstation were processed in Agilent Profinder software version 2.3.1 for batch analysis. The datasets were subjected to spectral peak extraction with a minimum peak height of 600 counts and the charge state for each metabolite was restricted to two. Further, retention time and mass alignment corrections were performed using Agilent Profinder software version 2.3.1 on the runs to remove non-reproducible signals. The resulting features then were exported to Mass Profiler Professional (MPP) software version 2.4.3 (Agilent Technologies, Santa Clara, CA, USA) for statistical analysis. The extracted features were imported into MPP software for statistical analysis. Principle Component Analysis (PCA) was performed to check the quality of the samples after which the data containing filtered features were processed by one-way ANOVA to ascertain differences between control and fungal groups. Only the analytes with p values < 0.05 and fold change (FC) > 5 were treated as statistically significant. Additionally, multiple test corrections using Bonferroni-adjusted p values were applied to reduce false positives and false negatives in the data.
2.9. Targeted metabolomics
Tables 1 and 2 provide a comprehensive overview of which healthy and infected annual and periodical cicada populations/collections were used for targeted metabolomics studies. A total of 79 specimens were used including 59 Massospora-infected individuals and 20 healthy cicadas. In general, only freshly collected or samples subjected to optimal storage from time of collection were utilized for metabolomics studies. For archived samples that had been stored in vials of ethanol, ethanol was removed from the vial, dried down, and reconstituted in HPLC-grade methanol.
Table 2:
Quantification of psilocyibn, psilocin, and cathinone from Massospora fungal plugs against commercially available, DEA-exempt analytical standards using targeted LC-MS. Samples that fell outside the limit of quantification were scored as either present or absent only.
| Sample type | Strain ID | Brood (Loc.a/Year) | Spore type | Psilocybin [m/z 285] |
Psilocin [m/z 205] |
Cathinone [m/z 150] |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Present | Conc. [ng/mL] | ng/plug | Present | Conc. [μg/g] | ng/plug | Present | Conc. [ng/mL] | ng/plug | ||||
| M. cicadina | MC139c | VI (NC; 2017) | Resting spore | ND | – | – | ND | – | – | Yes | BLQ | BLQ |
| M. cicadina | MC140c | VI (NC; 2017) | Resting spore | ND | – | – | ND | – | – | Yes | BLQ | BLQ |
| M. cicadina | MC141c | VI (NC; 2017) | Resting spore | ND | – | – | ND | – | – | Yes | BLQ | BLQ |
| M. cicadina | MC142c | VI (NC; 2017) | Resting spore | ND | – | – | ND | – | – | Yes | BLQ | BLQ |
| M. cicadina | MC10 | V (WV; 2016) | Conidia | – | – | – | – | – | – | Yes | BLQ | BLQ |
| M. cicadina | MC19 | V (WV; 2016) | Conidia | – | – | – | – | – | – | Yes | BLQ | BLQ |
| M. cicadina | MC23c | V (WV; 2016) | Conidia | ND | – | – | ND | – | – | Yes | 24.3/11.6b | 246.2/116.0b |
| M. cicadina | MC33c | V (WV; 2016) | Conidia | ND | – | – | ND | – | – | Yes | 29.9 | 303.0 |
| M. cicadina | MC99 | V (WV; 2016) | Conidia | – | – | – | – | – | – | Yes | 22.2 | 134.7 |
| M. cicadina | MC40 | V (WV; 2016) | Conidia | – | – | – | – | – | – | Yes | 7.3 | 44.3 |
| M. cicadina | G | VIII (PA; 2002) | Resting spore | – | – | – | – | – | – | Yes | 7.9 | 80.1 |
| M. cicadina | M | XXIII (IN; 2002) | Resting spore | – | – | – | – | – | – | Yes | 6.8 | 68.9 |
| M. levispora (MI) | ML03 | (MI; 1998) | Resting spore | ND | – | – | ND | – | – | ND | – | – |
| M. levispora (MI) | ML01 | (MI; 1998) | Conidia | Yes | 1.2 | 7.3 | ND | – | – | ND | – | – |
| M. levispora (MI) | ML02 | (MI; 1998) | Conidia | Yes | 2.3 | 13.9 | ND | – | – | ND | – | – |
| M. levispora (MI) | ML04 | (MI; 1998) | Conidia | Yes | 1.8 | 10.9 | ND | – | – | ND | – | – |
| M. levispora (MI) | ML05 | (MI; 1998) | Conidia | Yes | 2.2 | 13.3 | ND | – | – | ND | – | – |
| M. levispora (MI) | ML06 | (MI; 1998) | Conidia | Yes | BLQ | – | ND | – | – | ND | – | – |
| M. levispora (MI) | ML07 | (MI; 1998) | Conidia | Yes | 3.2 | 19.4 | ND | – | – | ND | – | – |
| M. levispora (MI) | ML08 | (MI; 1998) | Conidia | Yes | 2.8 | 17 | ND | – | – | ND | – | – |
| M. aff. levispora (NM) | NM01 | (NM; 2017) | Conidia | Yes | – | – | Yes | 2.6 | 7.0 | ND | – | – |
| M. aff. levispora (NM) | NM02c,d | (NM; 2017) | Conidia | Yes | – | – | Yes | 1.2 | 3.1 | ND | – | – |
| M. aff. levispora (NM) | NM03c | (NM; 2017) | Conidia | Yes | – | – | Yes | 0.2 | 0.5 | ND | – | – |
| M. aff. levispora (NM) | NM04c | (NM; 2017) | Conidia | Yes | – | – | ND | – | – | ND | – | – |
| M. aff. levispora (NM) | NM05c | (NM; 2017) | Conidia | Yes | – | – | ND | – | – | ND | – | – |
| M. aff. levispora (NM) | NM06c | (NM; 2017) | Conidia | Yes | – | – | Yes | 0.02 | 0.1 | ND | – | – |
| M. aff. levispora (NM) | NM07 | (NM; 2017) | Conidia | Yes | – | – | Yes | 0.3 | 0.8 | ND | – | – |
| M. aff. levispora (NM) | NM08 | (NM; 2017) | Conidia | Yes | – | – | Yes | 4.7 | 12.6 | ND | – | – |
| M. aff. levispora (NM) | NM09 | (NM; 2017) | Conidia | Yes | – | – | Yes | 1.4 | 3.7 | ND | – | – |
| M. aff. levispora (NM) | NM10 | (NM; 2017) | Conidia | Yes | – | – | Yes | 0.7 | 1.8 | ND | – | – |
| M. aff. levispora (NM) | NM14 | (NM; 2017) | Conidia | Yes | – | – | Yes | 0.5 | 1.3 | ND | – | – |
| Healthy cicada | MCNC1 | V (WV; 2016) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy cicada | MCNC2 | V (WV; 2016) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy cicada | MLNC1 | (MI; 1998) | O. rimosa | ND | – | – | ND | – | – | ND | – | – |
| Healthy cicada | MPNC01 | (NM; 2017) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | MM1 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | MM2 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | MM3 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | MM4 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | MM5 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | MF1 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | MF2 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | MF3 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | MF4 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | MF5 | VII (NY; 2018) | M. spetendecim | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | PM1 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | PM2 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | PM3 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | PM4 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy male | PM5 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | PF1 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | PF2 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | PF3 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | PF4 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
| Healthy female | PF5 | (CA; 2018) | P. putnami | ND | – | – | ND | – | – | ND | – | – |
ND, not detected.
BLQ, below limit of quantification.
U.S. state abbreviation.
Indicates samples were included in two independent runs.
Isolates used in global metabolomics studies.
Isolate from which metagenome was sequenced.
Cathinone, psilocybin, and psilocin were monitored individually using QQQ-MS (Table 2). We performed targeted LC-MS/MS absolute quantification of cathinone, psilocybin, and psilocybin’s biologically active bioconversion product psilocin, on individual conidial and resting spore plug extracts from all three cicada-fungal pairs (Table 1) including Mas. levispora (MI)-infected Say’s cicadas (Okanagana rimosa), which were excluded from the global metabolomics on account of their previous long-term storage in ethanol. Given its taxonomic overlap with Mas. aff. levispora (NM), we hypothesized that it might also contain psilocybin. The posterior abdomen of cicada controls used for global metabolomics as well as independently collected healthy Magicicada (n = 10) and Platypedia (n = 10) cicadas were assessed for all three compounds. All samples were quantified against standard curves generated from DEA-exempt analytical standards. Cathinone was quantified in three collections of Mas. cicadina: one from Brood V (2016) in WV, one from brood VI in NC, USA in 2017, and a third from archived collection spanning four broods (VIII, IX, XXII, and XXIII) collected from 2001 to 2003 (Cooley et al. 2018).
The absolute quantification of cathinone, psilocin and psilocybin was performed using a 5500 QTRAP mass spectrometer (SCIEX Inc., Concord, Ontario, Canada). Chromatographic separations were performed using an HPLC system consisting of two Shimadzu LC 20 AD pumps that included a degasser and a Shimadzu SIL 20 AC auto sampler (Shimadzu Corp. Kyoto, Kyoto Prefecture, Japan). Analytes were separated on a 3.0 × 150 mm Imtakt Scherzo SW-C18 column (Imtakt USA, Portland, Oregon). The HPLC column oven was set to 40 °C for the analysis, with an 8 μL injection volume for each sample. Mobile phase A consisted of HPLC grade water with 0.1% formic acid, while mobile B consisted of acetonitrile with 0.1% formic acid. The LC gradient started 5% B for 0.5 min and then increased to 95% B over 5 min followed by a re-equilibration to starting conditions for a period of 2.1 min. The flow rate was set to 0.3 mL/min.
Targeted metabolites were quantified using multiple reaction monitoring. The transition that was monitored for cathinone was m/z 150.1 → 106.3. The transitions that were monitored for psilocybin were m/z 284.5 → 205.2 and 240.1. Psilocin was monitored at m/z 205.2 → 116.3. Mass spectrometer source conditions were set to the following: curtain gas = 15, ion spray voltage = 4800 V, temperature = 600 °C, gas 1 = 45, gas 2 = 50. Calibration samples were prepared in 50% methanol/H2O. Calibration concentrations ranged from 0.05 to 10ng/mL of both psilocin and psilocybin and 1.25–25 ng/mL for cathinone. In order to assess variation in the assay duplicate standard curves were analyzed at the beginning and at the end of the analytical run. Absolute quantification data was processed using Analyst software ver. 1.6.3 (SCIEX Inc., Concord, Ontario, Canada).
2.10. Targeted quantification of cathinone CoA dependent pathway intermediates
Targeted detection of cathinone CoA dependent pathway intermediates including trans-cinnamoyl- CoA, 3-hydroxy-3-phenylpropionyl-CoA, 3-oxy-3-phenylpropionyl-CoA, and benzoyl-CoA was performed using a 5500 QTRAP mass spectrometer (SCIEX Inc., Concord, Ontario, Canada). Chromatographic separations were performed using an HPLC system consisting of two Shimadzu LC 20 AD pumps that included a degasser and a Shimadzu SIL 20 AC auto sampler (Shimadzu Corp. Kyoto, Kyoto Prefecture, Japan). Analytes were separated on a 3.0 × 150 mm Imtakt Imtrada column (Imtakt USA, Portland, Oregon). Mobile phase A consisted of 10 mM ammonium acetate at pH 9.0 and mobile phase B consisted of 100% acetonitrile. CoA intermediates were separated on a gradient that went from 5% mobile phase B to 95% mobile phase B over 15-min with 3-min re-equilibration at the end of the chromatographic run. Absolute quantification data were processed using Analyst software ver. 1.6.3 (SCIEX Inc., Concord, Ontario, Canada).
2.11. Targeted screening of psilocybin and psilocin from Conidiobolus coronatus and 2018-collected healthy Platypedia putnami (CA) and cathinone from Magicicada septendecim (Brood VII)
To test for psilocybin biosynthesis that could result from this single locus uncovered in Conidiobolus coronatus (Entomophthorales), eight representative C. coronatus isolates from a diverse set of hosts (Supplemental Table 4) were acquired from the Agricultural Research Service Collection of Entomopathogenic Fungal Cultures (ARSEF). The eight isolates were used in both live-plating (direct exposure to spores) and injection entomopathogenicity assays on Galleria mellonella, a model insect for entomopathogenicity studies (Panaccione and Arnold, 2017).
To further confirm the fungal origin of monoamine alkaloids from Massospora-infected cicadas, we screened independent healthy populations of the two cicada hosts included in the global metabolomics study, Platypedia putnami from CA and Brood VII Magicicada septendecim from NY, for the presence/absence of cathinone, psilocybin and psilocin. These samples were screened using a high-resolution, accurate mass Orbitrap LC-MS, which allowed for targeted fragmentation of individual alkaloids. Sampling included five outwardly asymptomatic males and females for each cicada species.
Targeted cathinone, psilocybin and psilocin experiments were performed using an Accela 1250 UHPLC coupled to a Q Exactive Orbitrap Mass Spectrometer (Thermo Scientific, San Jose, CA) operating in positive ion mode. An Agilent Zorbax C18 column (2.1 mm diameter by 10 cm length) was loaded with 2 μL of extracts. Separations were performed using a linear gradient ramping from 95% solvent A (water, 0.1% formic acid) to 90% solvent B (acetonitrile, 0.1% formic acid) over 6 min, flowing at 300 μL/min.
The mass spectrometer was operated in targeted-SIM/data-dependent MS2 acquisition mode. Precursor scans were acquired at 70,000 resolution with a 4 m/z window centered on 285.1005 m/z, 205.1341 m/z, and 150.0913 m/z (5e5 AGC target, 100 ms maximum injection time). Charge states 2 and higher were excluded for fragmentation and dynamic exclusion was set to 5.0 s.
2.12. Search for candidate psilocybin and cathinone biosynthetic genes
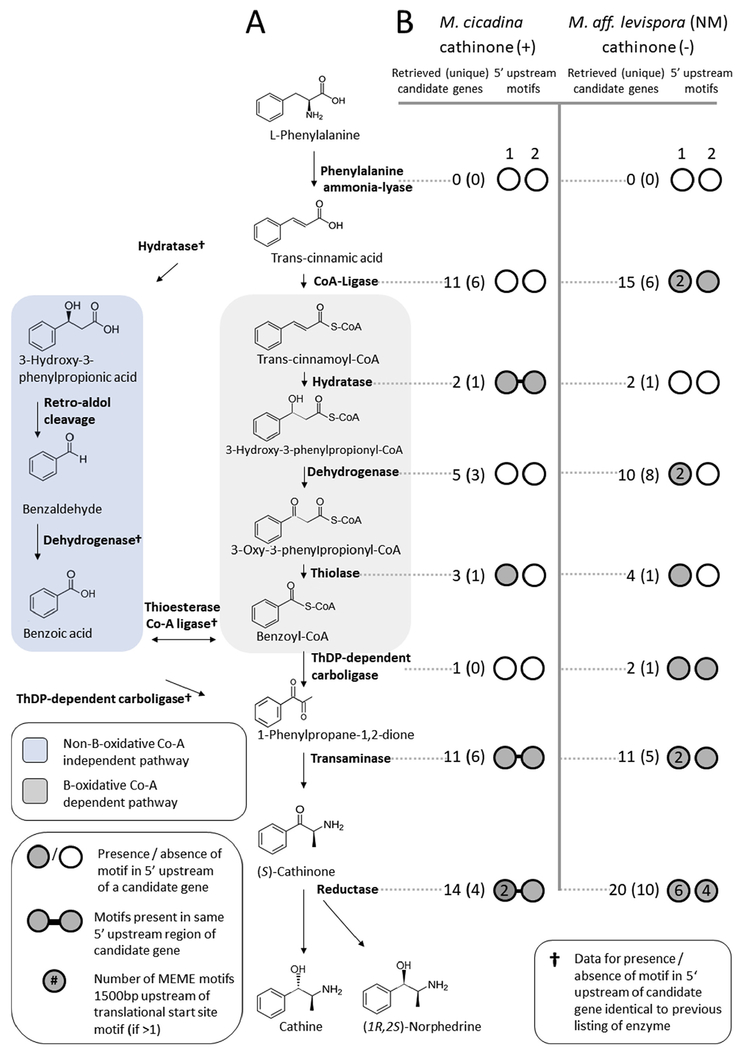
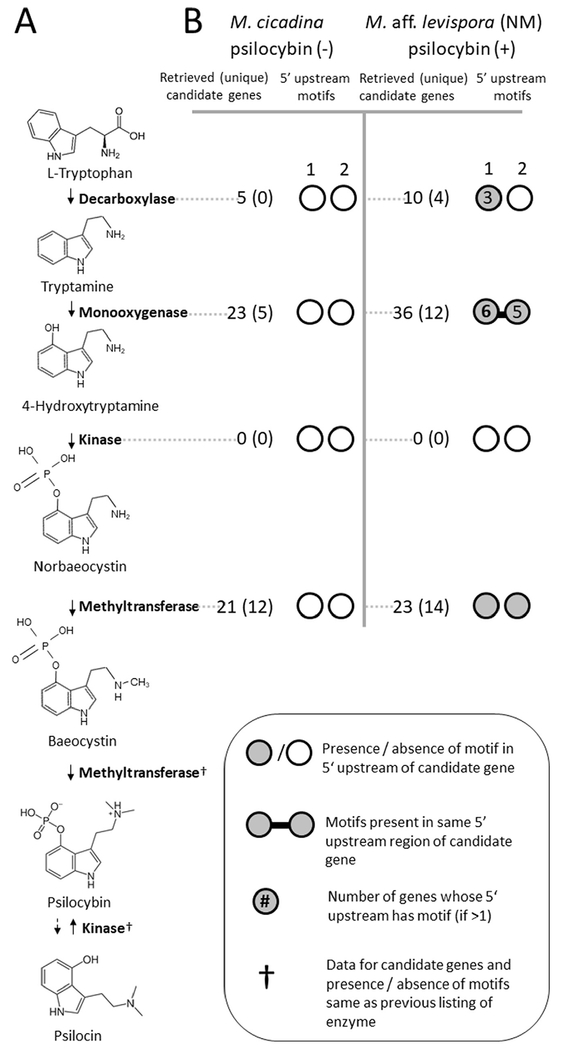
To support our metabolomics findings, we next searched for candidate genes underlying cathinone and psilocybin biosynthesis in assembled Mas. cicadina and Mas. aff. levispora (NM) metagenomes. We used multiple approaches to identify candidate psilocybin and cathinone biosynthetic genes (Supplemental Figure 1). We first performed tBLASTn searches of Massospora metagenome assemblies using four characterized psilocybin biosynthesis proteins from Psilocybe cubensis (Fricke et al., 2017) (norbaeocystin methyltransferase (accession P0DPA9.1); L-tryptophan decarboxylase (accession P0DPA6.1); 4-hydroxytryptamine kinase (accession P0DPA8.1); cytochrome P450 monooxygenase (accession P0DPA7.1)) and all 42 candidate cathinone biosynthesis proteins identified in a transcriptomic study of Catha edulis (retrieved from S1 dataset and Table 2 of (Groves et al. 2015). After failing to retrieve Massospora sequences orthologous to genes from the characterized Psilocybe psilocybin or predicted Catha cathinone pathways in either of our two Massospora metagenomes, we developed eight primer sets with various levels of degeneracy in an attempt to amplify Psilocybe and Psilocybe-like psilocybin biosynthesis genes from multiple fungal plugs of Mas. aff. levispora (NM) in addition to NM-02 from which the metagenomic data was generated (Supplemental Table 2). Fungal plugs from Mas. levispora (MI) were also screened for psilocybin biosynthesis genes using the same primer sets. No DNA sequence data were available in GenBank for cathinone biosynthesis (only RNA sequences were deposited in Genbank Short-Read Archive) so primers were not developed. All primer sets failed to amplify Psi-specific PCR targets in any of the assayed Massospora DNA templates.
We next subtracted aligned sequences between the two assemblies using NUCmer and PROmer from MUMMER v.4.0.0beta2 in order to identify sequence unique to each assembly (Marçais et al. 2018). Finally, we used profile HMM of the protein family (Pfam) domains found in characterized psilocybin and cathinone biosynthetic genes to search the 6-frame amino acid translations of the Massospora metagenomic assemblies with HMMER3 v.3.1b2 ((Eddy, 2011); Supplemental Table 5). We searched with all SAM and SAM-like methyltransferase domains because many are similar to the S-adenosyl-l-methionine (SAM)-dependent methyltransferase domain in the psilocybin pathway’s norbaeocystin methyltransferase. Unless stated otherwise, all BLAST searches were conducted using the BLAST suite v.2.2.25 + with a max. e value threshold = 1e-4 (Altschul et al. 1990) against a local database containing 375 publically available fungal genomes (Supplemental Table 6).
All subsequent steps were performed on the set of sequences retrieved by profile HMM search. We filtered out bacterial, protist and insect sequences by aligning hits to NCBI’s non-redundant protein database (last accessed March 14th, 2018) using BLASTp. We annotated contigs containing filtered hits using WebAUGUSTUS with C. coronatus species parameters using the option “predict any number of (possibly partial) genes” and reporting genes on both strands (Hoff and Stanke, 2013). We manually verified the coordinates of CDS regions by using the hits as queries in a BLASTx search (min similarity = 45%) to the local proteome database. The predicted amino acid sequences of all candidate genes, as well as the nucleotide sequences and coordinate files of annotated contigs are available at https://doi.org/10.5281/zenodo.1306939.
To narrow down our list of candidate genes that may underlie the biosynthesis of these natural products, we employed a two-pronged approach to gather additional lines of evidence that would suggest participation in a common secondary metabolite pathway (Wolf et al., 2015; Slot, 2017). First, we looked for evidence of coordinated regulation from shared nucleotide sequence motifs in the upstream non-coding region of each candidate sequence (Supplemental Figure 1).
To investigate putative ungapped regulatory motifs associated with candidate gene sequences, we used a custom Perl script to extract up to 1500 bp upstream of each gene’s start codon and submitted each set of extracted sequences to command line MEME v.4.12.0 (max evalue = 5e-2; min width = 25; max width = 100; max motifs = 10) (Bailey and Elkan, 1994). Motifs were manually inspected and those consisting solely of homopolymer repeats were discarded.
We also searched for evidence of gene clustering among homologs of the candidate sequences across a phylogenetically diverse database of 375 fungal genomes (Supplemental Figure 1; Supplemental Table 7). Because the Massospora assemblies were themselves too fragmented to assess if candidate genes were found in clusters, we searched for evidence of clusters containing candidate gene homologs retrieved from the local fungal genome database using BLASTp and a custom Perl script (available at https://github.com/egluckthaler/cluster_retrieve). Homologs were considered clustered if separated by 6 or fewer intervening genes. Only clusters with at least 3 genes were retained. Genes at clustered loci of interest were annotated with eggNOG-mapper v.0.99.1 (Huerta-Cepas et al., 2017) based on orthology data from the fungal-specific fuNOG database (Huerta-Cepas et al., 2016). The two candidate regulatory motifs recovered upstream of candidate cathinone biosynthesis genes in the Mas. cicadina assembly (cathinone +) were found upstream of the same predicted hydratase, transaminase and reductase genes (Supplemental Figure 2, Supplemental Table 8).
Since genes encoding the synthesis of specialized secondary metabolites are occasionally inherited through non-vertical means, we assessed the potential for horizontal gene transfer of cathinone and psilocybin biosynthesis genes by constructing maximum likelihood phylogenetic trees for 22 high quality candidate pathway genes from Mas. cicadina and Mas. aff. levispora (NM) (Supplemental Figure 3 & 4; Supplemental Tables 3 and 9). High quality candidate genes were defined as sharing putative regulatory motifs in their 5’ upstream non-coding regions with either a transaminase or methyltransferase gene (which encode enzymes that produce cathinone or psilocybin, respectively), or homologous to genes in the cluster of interest in C. coronatus.
To investigate the evolution of the 22 high quality candidate gene sequences (Supplemental Table 9), we first divided sequences into groups based on the profile HMM that initially retrieved them. Sequences from each group were used as queries to retrieve hits from the local fungal genome database (Supplemental Table 7) which was modified at this point to also include all candidate genes identified from both Massospora assemblies, as well as the meta-transcriptomes of Conidiobolus incongruus B7586, Entomophthora muscae (ARSEF 13376), Zoophthora radicans ATCC 208865, Strongwellsea sp. (HHDFL040913–1), and Pandora formicae (Małagocka et al. 2015) using BLASTp.
The Strongwellsea sp. transcriptome was based on total RNA extracted from a single infected Delia radicum caught in an organically grown cabbage field in Denmark: Sørisgaard (55.823706, 12.171149). The fly had the characteristic Strongwellsea-caused open hole on the side of the abdomen and was snap-frozen in liquid nitrogen as quickly as possible. Illumina sequencing yielded ca. 20 million 150 bp paired-end reads that were assembled using Trinity 2.2.0 (Grabherr et al., 2011). Transcripts were filtered so that only the highest expressed isoform for each transcript and transcripts assigned to kingdom Fungi using MEGAN4 (Huson et al., 2011) were retained.
Each set of queries and associated hits (with two exceptions, see below) were then aligned with MAFFT v.7.149b using the -auto strategy (Katoh and Standley, 2013), trimmed with TrimAL v.1.4 using the –automated1 strategy (Capella-Gutiérrez et al., 2009), and had sequences composed of >70% gaps removed. Sets of sequences retrieved using candidates with the ‘p450’ or ‘aldo_ke-t_red’ Pfam HMMs were too large to construct initial alignments. Instead, all retrieved hits and queries were first sorted in homolog groups by submitting the results of an all vs. all BLASTp to OrthoMCL v.2.0 (inflation = 1.5) (Li et al. 2003). All sequences in the homolog groups containing the query candidate genes were then aligned and trimmed as above. Each trimmed alignment was then used to construct a phylogenetic tree with FASTTREE v.2.1.10 using default settings that was then midpoint rooted (Price et al., 2010). For each high quality query sequence present in the tree, a strongly supported clade containing the query sequence and closely related Entomophthoralean sequences was extracted. Sequences in the extracted clade were re-aligned and trimmed (as above) and used to build final maximum likelihood trees using RAxML v8.2.11 with 100 rapid bootstraps (Stamatakis, 2014); Supplemental Figure 3 & 4). Final RAxML trees were rooted based on the outgroup in the clade extracted from the midpoint rooted FASTTREE (Supplemental Table 10).
3. Results
3.1. Morphology of Massospora specimens
Morphological studies were conducted to permit species-level identification of collected specimens based on previously published studies (Soper 1963, 1974). Conidia (n = 37), and resting spore (n = 13) measurements were acquired from freshly collected Mas. cicadina-infected periodical cicadas (Magicicada spp.) and Mas. platypediae-infected wing-banger annual cicadas (Platypedia putnami), as well as from archived Mas. levispora-infected Okanagana rimosa and Mas. cicadina-infected periodical cicadas (Fig. 1, Table 1). Morphology of conidia were studied in 15, 14, and 8 specimens of Mas. cicadina (Mc), Mas. platypediae (Mp), and Mas. levispora (Ml), respectively, while morphology of resting spores was examined in 11, 1 and 1 specimens (Fig. 1, Supplemental Table 1). The conidial specimens of Mas. cicadina-infected periodical cicadas spanned eight Broods and six of the seven Magicicada species collected over 38 y and distributed across the eastern U.S., while Mas. platypediae- and Mas. levispora-infected cicadas were each from single annual cicada populations collected in New Mexico (2017) and Michigan (1998), USA, respectively (Supplemental Table 1).
Mean conidia (Mc = 15.4 μm × 16.0 μm and Ml = 8.8 μm × 14.3 μm) and resting spore (Mc = 42.4 μm and Ml = 39.8 μm) dimensions for Mas. cicadina and Mas. levispora overlapped with previously reported measurements (Supplemental Table 1). In contrast, spore dimensions (8 × 13.2 μm) in the majority of Mas. platypediae specimens fell completely outside the reported range for this species (Supplemental Table 1). Interestingly, all the Mas. platypediae measurements fell within the reported range for Mas. levispora, including a single resting spore specimen, which had not been previously observed for Mas. platypediae (Supplemental Table 1).
3.2. Phylogenetic relationships among Massospora morphotypes
To infer evolutionary relationships among collected specimens, phylogenetic analyses were performed. Prior to this study, Massospora was represented in NCBI Genbank by a total of four DNA sequences from a single isolate (ARSEF 374) of Mas. cicadina (NCBI Genbank accessions EF392377, EF392548, EF392492, and DQ177436). A total of 18 isolates representing Mas. cicadina (n = 12), Mas. levispora (n = 3), and Mas. platypediae (n = 3) were used for single gene phylogenetic studies, four of which were included as representative isolates in the combined phylogeny (Table 1). Reference LSU, SSU, and EFL DNA sequences for Mas. cicadina, Mas. platypediae and Mas. levispora (MI) were deposited in NCBI Genbank (MH483009 - MH483011, MH483015-MH483023 and MH547099).
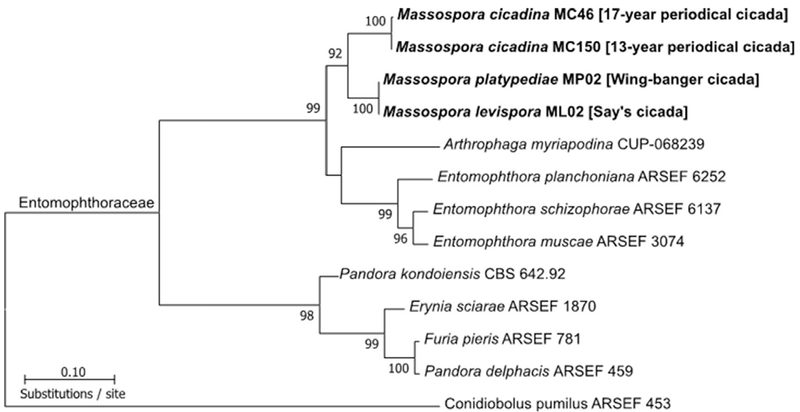
Phylogenetic analysis of the partitioned individual alignments (data not shown) and combined dataset (LSU + SSU) using maximum likelihood (ML) resolved all sampled Massospora in a strongly supported monophyletic group sister to a clade containing Entomophthora and Arthrophaga (Fig. 2). EFL was excluded from the concatenated dataset on account of its heterogeneous distribution (some taxa have EF1 and others have EFL; see James et al. 2006) across the Entomophthorales taxa included in the phylogenetic analysis. However, representative sequences were deposited into Genbank for all three Massospora species (see above). Massospora cicadina isolates formed a single lineage that encompassed all sampled cicada broods and Magicicada species (data not shown). The previously deposited Mas. cicadina ARSEF 374 was one exception as it aligned with Entomophthora, indicating a wrongly assigned species to previously deposited DNA sequences in Genbank. To confirm this finding, we successfully resampled and amplified the SSU region (deposited as Genbank accession MH483023) from the physical specimen associated with ARSEF 374, which was 100% identical to the other Mas. cicadina sequences from this study. Mas. levispora and Mas. platypediae did not form distinct clades; instead, they appeared as a single lineage sister to Mas. cicadina (Fig. 2). Because more populations of Mas. platypediae and Mas. levispora are needed to confirm these phylogenetic findings, subsequent mentions of Mas. platypediae from cicada host Platypedia putnami from New Mexico (NM), USA will be referred to hereafter as Mas. aff. levispora (NM), while Mas. levispora sensu stricto from its cicada host Okanagana rimosa in Michigan (MI), USA will retain the designation of Mas. levispora (MI).
Fig. 2.
Concatenated LSU + SSU maximum likelihood (ML) tree consisting of Massospora species and related species in the Entomophthorales. Given that Mas. levispora and Mas. platypediae were not genealogically exclusive, Mas. platypediae will be hereafter referred to as Mas. aff. levispora (NM). Bootstrap support is indicated near each node and only values greater than 70% are shown.
3.3. Discovery of monoamine alkaloids using global metabolomics
Each of the three sample types (Mas. cicadina and Mas. aff. levispora (NM) fungal plugs and healthy cicada abdomens) produced unique metabolome profiles, while those of conidial plugs from the two Massospora species were more similar compared to the cicada control. In total, 1176 small molecule spectra (free amino acids, phospholipids, ceramides, alkaloids, etc.) were differentially detected between the two Massospora spp. including two notable behavior-modifying monoamine alkaloids. Small molecule spectra included metabolites identifiable only based on m/z and retention time (e.g. 198.0605@2.268) or chemical formula (e.g. C22 H17 N18 O5). All detected metabolites are listed in Supplemental Table 11. Alkaloids were the initial target of this inquiry given the extensive behavior-modifying properties of these compounds (Brookhart et al. 1987; Frischknecht and Waser, 1978; Witt, 1971). In addition, fungal-specific metabolites, previously described from the literature and the metabolomics databases, were investigated.
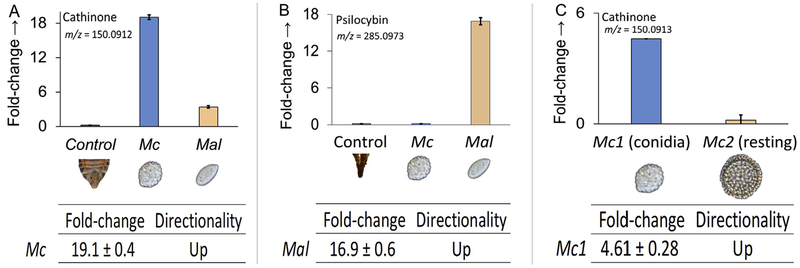
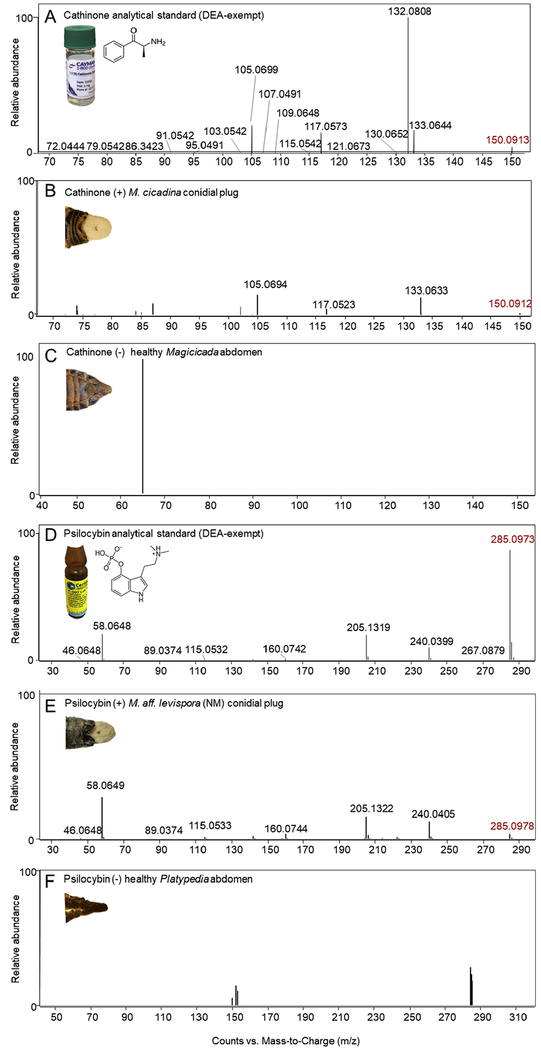
The amphetamine cathinone (m/z 150.0912), previously reported from the plant Catha edulis (Groves et al., 2015) was detected only in the Mas. cicadina samples and not in the Mas. aff. levispora (NM) conidial plugs nor the healthy cicada controls (Fig. 3). Cathinone was also detected in resting spore specimens in a second global metabolomics study between Mas. cicadina resting spores, an independently collected population sampled from North Carolina (NC), USA in 2017, and the initial conidial plugs from West Virginia (WV), USA in 2016. However, the conidial plugs contained cathinone levels 4.61 ± 0.28 higher compared to resting spores (Fig. 3). Fragmentation patterns for cathinone in Mas. cicadina conidial and resting spore plugs matched the experimentally observed fragmentation patterns of a commercially available DEA-exempt analytical standard used in this study; three fragments with m/z less than 10 ppm mass error of those obtained from the analytical standard were recovered (Fig. 4A–C). Several cathinone pathway intermediates, including 1-phenylpropane-1,2-dione, the metabolite immediately preceding cathinone, were putatively identified from the global metabolomics output using MS/MS screening but fragmentation patterns did not fall within the accepted mass error limits when compared to experimentally observed fragmentation patterns of a commercially available analytical standard. Other pathway intermediates, initially identified from the Mas. cicadina resting spore global metabolomics output, including benzaldehyde, were not explored further due to their involvement in several divergent biosynthetic pathways (Lomascolo et al. 1999).
Fig. 3.
Global metabolomics fold-change comparisons of (A) cathinone and (B) psilocybin among healthy cicada controls (posterior sections), Mas. cicadina (Mc), and Mas. aff. levispora (NM) (Mal) conidial plugs, and (C) cathinone between Mas. cicadina conidial plugs (Mc1) and resting spore plugs (Mc2). Value is average of five biological replicates. Fold change ± standard error based on comparisons with control.
Fig. 4.
Representative LC-MS/MS spectra for (A) cathinone DEA-exempt analytical standard; (B) cathinone-positive Mas. cicadina conidial plugs from infected Magicicada spp.; (C) cathinone-negative Magicicada septendecim abdominal sample; (D) psilocybin DEA-exempt analytical standard; (E) psilocybin-positive Mas. aff. levispora (NM) conidial plugs from infected Platypedia putnami; (F) psilocybin-negative Platypedia putnami abdominal sample. M/z in red denote precursor fragmentwhile m/z in black denote observed fragments. All labeled m/z values from Massospora plugs (B and E) are within 10 ppm mass error compared to analytical standard.
Over 10 CoA species were monitored including all five hypothesized cathinone CoA-dependent pathway intermediates, trans-cinnamoyl-CoA, 3-hydroxy-3-phenylpropionyl-CoA, 3-oxy-3-phenylpropionyl-CoA, benzoyl-CoA (Groves et al., 2015). The most abundant CoAs detected were acetyl-CoA and malonyl-CoA, yet none of the cathinone CoA-dependent pathway intermediates monitored were detected (data not shown).
Psilocybin (m/z 285.0973), a psychoactive tryptamine produced by about 200 Agaricalean mushroom species (Kosentka et al., 2013), was among the most abundant alkaloids detected in Mas. aff. levispora (NM) conidial plugs but was not detected in Mas. cicadina or the cicada controls (Fig. 3). Fragmentation patterns for psilocybin in Mas. aff. levispora (NM) plugs matched the experimentally observed fragmentation patterns of a commercially available DEA-exempt analytical standard used in this study; eight fragments with m/z less than 10 ppm mass error of those obtained from the analytical standard were recovered (Fig. 4D–F). One intermediate in the psilocybin biosynthetic pathway in Psilocybe cubensis mushrooms (Fricke et al., 2017), 4-hydroxytryptamine, was identified using MS/MS screening for both Mas. cicadina and Mas. aff. levispora (NM) but fragmentation patterns could not be compared to an analytical standard since 4-HT is not commercially available for purchase. Analytical standards for other pathway intermediates either do not exist or lack DEA-exempt formulations.
Several other classes of alkaloids, including bisbenzylisoquinoline, monoterpene, strychnos, and Veratrum alkaloids, were observed in both/either Mas. cicadina or Mas. aff. levispora (NM) at higher fold-change compared to the cicada controls (see underlined compounds, Supplemental Table 11). In addition to alkaloids, a dozen or so basidiomycete- and ascomycete-specific metabolites were uncovered including trichothecenes and triterpene glycosides (see bolded compounds, Supplemental Table 11). However, unlike cathinone and psilocybin, none of these other alkaloids could be confirmed via fragmentation, indicating that many of these IDs are likely isobaric compounds (having the same precursor m/z as a previously characterized compound).
3.4. Quantification of cathinone, psilocybin, and psilocin using targeted metabolomics
Cathinone was present in 4 of 4 Brood VI Mas. cicadina resting spore plugs and 6 of 24 Brood V Mas. cicadina conidial plugs and was quantifiable in concentrations from 44.3-303.0 ng/infected cicada in four of six Brood V fungal plugs (Table 2). From the archived collection, cathinone was present and quantifiable in concentrations of 68.9 and 80.1 ng/infected cicada from 2 of 13 resting spore plugs spanning two separate broods (Table 2). A Brood V Mas. cicadina conidial plug extraction that had 4-months earlier yielded a total of 246 ng of cathinone and served as a positive control in the absolute quantification of archived samples was found to contain 116 ng, a notable reduction from the previous time point (Table 2). This reduction in quantity is not unexpected given the labile nature of amphetamines (Kalix, 1988).
Psilocybin was detected in 11 of 11 assayed Mas. aff. levispora (NM) plugs and 7 of 8 assayed Mas. levispora (MI) plugs while psilocin was detected in 9 of 11 plugs and 0 of 8 plugs, respectively (Table 2). Of these, psilocybin was quantifiable only in Mas. levispora (MI), with concentrations from 7.3 to 19.4 ng/infected cicada across 6 fungal plugs (Table 2). Psilocin was detected only in Mas. aff. levispora (NM) in concentrations from 0.1 to 12.6 μg/infected cicada across 9 fungal plugs (Table 2).
Similar to data from cicada controls in the global metabolomics study, results of screening of independent healthy populations of Platypedia putnami from CA and Brood VII Magicicada septendecim from NY failed to detect any of the three monitored compounds from macerated posterior abdominal sections presumably including the portion of the abdomen that contains the cicada bacteriome (see discussion).
3.5. Searches for genetic mechanisms of psilocybin and cathinone biosynthesis in Massospora
In each metagenome, we identified multiple genes representative of all classes of enzymes in each alkaloid pathway (psilocybin and cathinone) with two exceptions (Fig. 5; Fig. 6; Supplemental Figure 1; Supplemental Tables 12 and 13). Although at least one candidate homolog could be identified for 7 of the 8 genes predicted to play a role in cathinone biosynthesis in both the Mas. cicadina (cathinone +) and Mas. aff. levispora (NM) (cathinone −) metagenomes, we failed to detect genes encoding phenylalanine ammonia-lyase (PAL) in either metagenome (Supplemental Table 14). Similarly, in both the Mas. aff. levispora (NM) (psilocybin +) and Mas. cicadina (psilocybin −) metagenomes, we identified at least one candidate homolog for 3 of the 4 genes participating in psilocybin biosynthesis, the exception being 4-hydroxytryptamine kinase (PsiK) in either metagenome (Supplemental Table 14).
Fig. 5.
(A) Predicted cathinone biosynthesis pathways from Catha edulis (modified from Groves et al. 2015); (B) Putative regulatory motifs found upstream of candidate cathinone biosynthesis genes from Mas. cicadina and Mas. aff. levispora (NM) assemblies. Sequences were retrieved using profile HMMs of enzymes predicted for each pathway step. Candidate gene co-regulation was predicted by shared nucleotide motifs within 1500 bp upstream of predicted translational start sites. Only motifs associated with transaminase genes (predicted to catalyze the production of cathinone) are shown.
Fig. 6.
(A) Psilocybin biosynthesis pathway from basidiomycete fungi; (B) Putative regulatory motifs found upstream of candidate psilocybin biosynthesis genes from both Mas. cicadina and Mas. aff. levispora (NM) assemblies. Sequences were retrieved using profile HMMs of enzymes predicted for each pathway step. Candidate gene co-regulation was predicted by shared nucleotide motifs within 1500 bp upstream of predicted translational start sites. Only motifs associated with methyltransferase genes (which catalyzes the formation of psilocybin) are shown.
Only two of the seven candidate cathinone regulatory motifs in the Mas. aff. levispora (NM) assembly (cathinone −) were found upstream of transaminase genes, while the rest were not (Supplemental Figure 2, Supplemental Table 8). Two of five motifs found upstream of candidate psilocybin biosynthesis genes in the Mas. aff. levispora (NM) assembly (psilocybin +) were present upstream of methyltransferase genes, while the single motif found in the Mas. cicadina assembly (psilocybin −) was not (Supplemental Figure 2, Supplemental Table 8).
Searches of a local database of 375 publicly available fungal genomes identified 2396 gene clusters containing homologs of at least three candidate cathinone genes, but none contained a transaminase, which is predicted to be required for cathinone synthesis (Supplemental Table 14). We also found 309 clusters containing homologs of at least three candidate psilocybin genes, 175 of which contain a methyltransferase, which catalyzes the formation of psilocybin (Supplemental Tables 6 and 14). One notable locus in Conidiobolus coronatus (Entomophthorales) contained four aromatic-L-amino-acid decarboxylases and one methyltransferase (both functions are required for psilocybin biosynthesis), in addition to one helix-loop-helix transcription factor, one inositol phosphatase, and various amino acid, sugar and inorganic ion transporters (Supplemental Table 15).
Based on the above findings, eight representative C. coronatus isolates from a diverse set of hosts were used in both live-plating (direct exposure to spores) and injection entomopathogenicity assays on Galleria mellonella (Supplemental Table 4), a model insect for entomopathogenicity studies (Panaccione and Arnold, 2017). By 24-h post-inoculation, all fungus-inoculated individuals were dead and by 48-h all live plated individuals were symptomatic and many had died. Symptomatic individuals were collected at 48-h post-inoculation and immediately placed at −80 C prior to maceration to permit screening of psilocybin and psilocin from both infected larvae and pure cultures of each isolate using a high resolution, accurate mass Orbitrap LC-MS. Neither compound was detected in macerated infected larvae or week-old fungal cultures of these same isolates on Sabouraud Dextrose Yeast Agar. However, C. coronatus isolate ARSEF 8715 (NRRL 28638), from which the genome and candidate locus was obtained, was not available and therefore was not included in the study.
All gene trees of these candidates had topologies consistent with vertical inheritance among the Entomophthorales with highly similar homologs present in other Entomophthoralean fungi. The phylogenies of candidate psilocybin pathway sequences gleaned from the preliminary metagenomic data are consistent with an origin of psilocybin biosynthesis independent from mushroom-forming basidiomycetes, but other evolutionary origins cannot be ruled out until the exact genes underlying this pathway are confirmed.
3.6. Phylogenomic resolution of Entomophthorales fungi and their behavior-modifying lifestyles
Mapping of behavioral traits and external sporulation patterns onto the inferred phylogeny from the concatenated genome-wide alignment revealed three possible parsimonious reconstructions of AHT in Entomophthorales fungi (Fig. 7). These topologies include either two independent gains of AHT, one of which is followed by a single loss, or a single gain in the most recent shared ancestor of Massospora, Entomophthora, and Strongwellsea followed by three losses, the latter of which suggests this behavior modification may have been the key innovation that led to the diversification of the Entomophthorales. At the same time, AHT entomopathogens exhibit a high degree of host specificity compared to their summit disease pathogens (Supplemental Table 16).
Fig. 7.
Phylogenomic relationships among behavior-modifying Entomophthoralean fungi, the behavioral and morphological host modifications they impose, and their host specificity. (A) RAxML phylogenetic tree ofEntomopthoralean fungi (Zoopagomycota) based on the concatenated alignment of >200 conserved orthologous protein sequences with overlaid behavioral modification in infected hosts. Actual number of protein sequences per taxon denoted in gray. (B) External sporulation patterns, (C) Heat map showing both the host range within and across select insect orders: Col: Coleoptera, Dip: Diptera, Hem: Hemiptera, Hym: Hymenoptera, Iso: Isoptera, Lep: Lepidoptera, Ort: Orthoptera, and Thy: Thysanoptera. The species Z. radicans and C. thromboides may represent broader species complexes, of which each phylogenetic species may have a narrower host spectrum. Photos used with permission from co-authors as well as Florian Freimoser, Joanna Maiagocka, Judith Pell, and Ruth Ahlburg
4. Discussion
One of the least explored frontiers in host-parasite interactions is the molecular basis for behavioral modification of hosts by fungal pathogens that promotes disease transmission. Recent studies of entomopathogenic Ophiocordyceps (Ascomycota) have identified neurogenic metabolites and enterotoxins that are possibly contributing to host manipulation (Hughes, 2013; De Bekker et al., 2017). Here, we report the surprising discovery that two species of Massospora, a genus of obligate entomopathogenic fungi infecting cicadas, produce psychoactive compounds during infection. These compounds are cathinone, an amphetamine only previously known from plants, and psilocybin, a tryptamine only previously known from basidiomycete mushrooms. Prior work has demonstrated that Massospora can modify host behavior, and we provide evidence for a chemical basis for this manipulation. This is the first evidence of amphetamine production in fungi and of psilocybin production outside the Basidiomycota.
The discovery of psilocybin in both Mas. levispora and Mas. platypediae, coupled with the morphological and phylogenetic data, indicate two possible scenarios: (1) Mas. levispora and Mas. platypediae comprise a single annual-cicada infecting Massospora species, which occupies a broader geographic and host range than previously reported, or (2) Mas. levispora and Mas. platypediae are two related species capable of infecting wing-banger cicadas. Interestingly, all known Massospora spp. are reported from a single genus of cicadas except for M. cicadina which, in addition to Magicicada spp. in the U.S., has been reported from the cicada Platypleura kaempferi in Japan (Kobayashi, 1951) and Diceroprocta biconica in Cuba (Weiser and Stary, 1967). These observations were based solely on cicadas infected with reticulated resting spores (a feature shared across all Massospora spp.) and, as such, their identification as M. cicadina is questionable (Soper, 1974). Nevertheless, modern collections of these Massospora spp. are needed to resolve these relationships.
Although some individuals from every freshly-collected infected cicada population yielded alkaloids, these compounds were not detected from every individual. Failure to detect cathinone, psilocybin, or psilocin from all Massospora-infected specimens is not unexpected given the natural variation in alkaloid production across a given population as has been shown with ergot alkaloids in individual morning glory seeds (Nowak et al., 2016). Indeed, these findings also may indicate specimen-age related degradation or other aspects of disease progression not captured during this study. For example, some specimens had only one or two of their abdominal sclerites sloughed off while others had lost 5–6 sclerites, possibly indicating variation in duration of conidial infections. Another consideration is the fact that a majority of samples were subjected to targeted metabolomics quantification and concentrations of individual compounds, although present, could have been below the limits of detection.
Without exposure studies involving healthy cicadas (nymphs and adults), the mechanism(s) by which monoamine alkaloids modify cicada behaviors cannot be fully resolved. However, previous arthropod exposure studies have provided extensive insight into the effects of amphetamine and psilocybin on various behaviors (Brookhart et al., 1987; Frischknecht and Waser, 1978; Witt, 1971). For example, low doses (1–12 μg) of amphetamine similar to the cathinone levels detected in Mas. cicadina increased intraspecific aggression in ants (Frischknecht and Waser, 1978) and significantly depleted biogenic amines, altering feeding behavior in adult blow flies (Phormia regina) by decreasing responsiveness to weak gustatory stimuli (Brookhart et al. 1987). Psilocybin ingestion had less observable impact on behavior compared to amphetamine for the limited number of arthropods observed (Witt, 1971), but still psilocybin may antagonize certain mycophagous insects that feed broadly on dung-loving basidiomycetes, thus conferring an evolutionary advantage to psilocybin-producing mushrooms (Reynolds et al., 2018). Psilocybin may also confer protection against predation, competition, and/or parasitism for a select few insects that exhibit indifference to psilocybin. For example, the dark-winged fungus gnat (Sciaridae), can successfully complete its lifecycle in fruit bodies of psilocybin-containing Psilocybe cyanescens (Awan et al. 2018). Likewise, leafcutter ants (Acromyrmex lobicornis) have been observed actively foraging on Psilocybe coprophila fruit bodies in Argentina, transporting basidiocarps back into the nest, possibly for defense purposes (Masiulionis et al., 2013). Although the effect of psilocybin on Mas. levispora (MI) differs somewhat in that the production of these compounds seem to benefit the parasite more than the host, the psilocybin may offer the insect some protection against attacks by the parasitoid fly Emblemasoma auditrix (Diptera: Sarcophagidae) (Soper et al., 1976).
While these secondary metabolites from Massospora, especially cathinone, may help improve endurance required to engage with other conspecifics given their debilitating infections, activity-level effects of amphetamine and psilocybin cannot easily explain the display of female-typical courtship behaviors by male cicadas (Cooley et al., 2018), except possibly in P. putnami (Murphy and Redden, 2003). Infected P. putnami males were found to produce significantly fewer crepitations (wing tapping behaviors required for mating) per observation period (Murphy and Redden, 2003). Fewer crepitations are associated with female-typical behavior, and the manifestation of these behaviors in males may indicate impaired temporal processing, a known effect of psilocybin in vertebrates (Wittmann et al., 2007). However, the atypical mating behaviors of other cicada species may be better explained by Massospora-mediated hormonal changes incidental to amphetamine and psilocybin production. For example, 2-hydroxyestradiol, a catechol estrogen and metabolite of estradiol and 3-O-acetylecdysone 2-phosphate, a metabolite of ecdysone, a steroidal prohormone of the major insect molting hormone 20-hydroxyecdysone, were both detected based on m/z and found to be upregulated in male Platypedia putnami with Mas. aff. levispora (NM)-infections compared to a mixed sex group Magicicada septendecim with Mas. cicadina-infections and healthy controls using global metabolomics (data not shown). These hormones may accumulate as a result of disruption of typical pathways (i.e. castration of testis) or they may be fungus-derived. Comparisons of Mas. aff. levispora (NM) fragmentation patterns with experimentally observed analytical standards are needed to validate these preliminary findings. Nevertheless, the differentially expressed “hypersexualization” between conidial- and resting spore-infected cicadas seems to support the latter hypothesis since changes in male behaviors cannot be incidental effects of castration or infection, or the observed behaviors would be seen in hosts accumulating either spore type. The basis of hypersexualization remains unknown; the compounds we identified maybe involved wholly or partly in this phenomenon, and/or may serve other roles outside this observed phenotype.
Culturable fungi have yielded many antimicrobial drugs, medications, and industrial enzymes over the last century. The discovery of amphetamine production in fungi as well psilocybin production in the Zoopagomycota is consistent with the breadth of diverse secondary metabolite synthesis pathways that have evolved in fungi. However, the discovery of these alkaloids with limited genetic evidence of a common biosynthetic pathway raises questions about their proposed enzymatic route. The lack of evidence of a fungal kinase, an enzyme class which catalyzes the phosphotransfer step in psilocybin biosynthesis and of phenylalanine ammonia lyase (PAL), an enzyme class that catalyzes a reaction converting L-phenylalanine to ammonia and trans-cinnamic acid in the early steps of cathinone biosynthesis, coupled with the lack of many of these same pathway metabolites seems to support novel or altered biosynthetic pathways for both compounds. One possibility is that the cicadas themselves might be producing these compounds upon infection with Massospora since these compounds are not produced in non-infected individuals. Another possibility is that Massospora may be interacting with resident microbes already present in/associated with healthy cicadas, such as secondary-metabolite-rich Hypocreales that have recently been described from the bacteriomes of certain cicadas (Matsuura et al., 2018). Further investigations with higher quality genome assemblies of fungi and hosts are needed to further test these hypotheses.
Convergent pathways for de novo metabolite synthesis are known from fungi, such as pyridoxine biosynthesis in the phytopathogenic fungus Cercospora nicotianae (Ehrenshaft et al., 1999). Likewise, psychoactive cannabinoids, well known from Cannabis sativa, have been recently confirmed from liverwort, providing a marked example of convergent evolution of bioactive plant secondary metabolites (Chicca et al. 2018). Similarly, β-carboline alkaloids, are found in both plants and fungi, including in the Entomophthoralean fungus, Conidiobolus coronatus, where it is thought to interfere with insect serotonin levels and larval development (Wrońska et al., 2018).
The discovery of cathinone in Massospora-infected cicadas also raises interesting questions about the biological origin of amphetamine production in Catha plants and the possibility of cryptic fungal partners involved in its biosynthesis. Over the last few decades, fungal endophytes have been identified as the source of ergot alkaloid production in fescue (Festuca spp.) and morning glories (Ipomoea spp.) (Lyons et al., 1986; Kucht et al. 2004). Culture-based studies of Catha edulis (Alhubaishi and Abdel-Kader 1991; Mahmoud, 2000) and ephedrine-producing Ephedra nebrodensis (Peláez et al., 1998), have uncovered a diverse assortment of fungi, none of which have been screened for cathinone production. Interestingly, at least two cicada genera have been reported feeding on Ephedra plants (Sanborn and Phillips, 2011; Wang et al., 2017), which may indicate a tolerance to these compounds that are known to be toxic to other insects including bees (Detzel and Wink, 1993).
The results of this study provide a strong impetus for enhanced screening of Ephedraceous and Celastraceous plants for alkaloid-producing endophytes that may also have entomopathogenic capabilities. Interestingly, several recent studies have elucidated such tripartite relationships between insects, entomopathogenic fungi with known endophytic capability, and plant hosts whereby plants supply fungi with photosynthate in exchange for insect-derived nitrogen (Behie et al. 2012, 2017; Barelli et al., 2016).
Similarly, despite the very recent discovery of Hypocrealean fungal symbionts from the fat bodies surrounding the bacteriomes and/or the well-developed surface sheath cells of more than a dozen cicada species native to Japan (Matsuura et al., 2018), our results do not support the presence of cryptic domesticated monoamine alkaloid-producing fungal symbionts in the posterior region of healthy or outwardly asymptomatic wing-banger and periodical cicadas. Further studies should focus on resolving whether such domesticated symbionts are present in any North American cicada and how these fungi, if present, may interact with obligate pathogens such as Massospora.
The obligate lifestyle and ephemeral nature of many Entomophthoralean fungi have long constrained our understanding of these early diverging entomopathogens despite up to a century of observational studies into their behavior-modifying capabilities. We anticipate these discoveries will foster a renewed interest in early diverging fungi and their pharmacologically important secondary metabolites, which may serve as the next frontier for novel drug discovery and pave the way for a systems biology approach for obligate fungal pathogens spanning the known diversity of Kingdom Fungi.
Supplementary Material
Acknowledgments
Special thanks to the Drug Enforcement Administration (DEA) for guidance on discovery and handling of Schedule 1 controlled substances. Genomes and transcriptome sequencing of C. thromboides, C. coronatus, and Z. radicans were supported by the Department of Energy’s Joint Genomes Institute under the JGI Community sequencing program project 1978 ‘Genomics of the early diverging lineages of fungi and their transition to terrestrial, plant-based ecologies’ supported by the Office of Science off the US Department of Energy under contract DE-AC02-05CH11231.
Funding
G.R.B. was supported by Protea Biosciences and NSF grant DBI-1349308, J.C.S. by NSF DEB 1638999, J.E.S. by NSF DEB 1441715, W.J.D. and T.Y.J. by NSF DEB 1441677, D.G.P by NIH 2R15GM114774-2, H.H.D.F.L. by the Villum Foundation (grant number 10122), T.G. by NSF grant CHE-1608149, J.R.C. and C.S. by NSF DEB 1655891, and M.T.K. by funds from West Virginia Agricultural and Forestry Experiment Station and NSF DBI-1349308.
Footnotes
Scientific article No. 3361 of the West Virginia Agricultural and Forestry Experiment Station, Morgantown, West Virginia, USA, 26506.
Conflicts of interest
Authors declare no competing interests.
Data and materials availability
Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accessions QKRY00000000 (Mas. platypediae or Mas. aff. levispora (NM)) and QMCF00000000 (Mas. cicadina). The versions described in this paper are QKRY01000000 and QMCF01000000. All phylogenomic marker and candidate cathinone and psilocybin gene sequences are available at https://doi.org/10.5281/zenodo.1306939.
Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.funeco.2019.06.002.
References
- Alhubaishi AAA, Abdel-Kader MIA, 1991. Phyllosphere and phylloplane fungi of qat in Sana’a, Yemen Arab Republic. J. Basic Microbiol 31 (2), 83–89. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J. Mol. Biol 215 (3), 403–410. [DOI] [PubMed] [Google Scholar]
- Arnesen JA, Małagocka J, Gryganskyi A, Grigoriev IV, Voigt K, Stajich JE, De Fine Licht HH, 2018. Early diverging insect pathogenic fungi of the order Entomophthorales possess diverse and unique subtilisin-like serine proteases. G3:Genes|Genomics|Genetics 8, 3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan AR, Winter JM, Turner D, Shaw WM, Suz LM, Bradshaw AJ, Ellis T, Dentinger B, 2018. Convergent Evolution of Psilocybin Biosynthesis by Psychedelic Mushrooms. bioRxiv, p. 374199. [Google Scholar]
- Bailey TL, Elkan C, 1994. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Bipolymers, pp. 28–36. [PubMed] [Google Scholar]
- Batko A, Weiser J, 1965. On the taxonomic position of the fungus discovered by Strong, Wells, and Apple: Strongwellsea castrans gen. et sp. nov. (Phycomycetes; Entomophthoraceae). J. Invertebr. Pathol 7 (4), 455–463. [Google Scholar]
- Barelli L, Moonjely S, Behie SW, Bidochka MJ, 2016. Fungi with multifunctional lifestyles: endophytic insect pathogenic fungi. Plant Mol. Biol 90 (6), 657–664. [DOI] [PubMed] [Google Scholar]
- Behie SW, Zelisko PM, Bidochka MJ, 2012. Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science 336 (6088), 1576–1577. [DOI] [PubMed] [Google Scholar]
- Behie SW, Moreira CC, Sementchoukova I, Barelli L, Zelisko PM, Bidochka MJ, 2017. Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat. Commun 8, 14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdoy M, Webster JP, Macdonald DW, 2000. Fatal attraction in rats infected with Toxoplasma gondii. Proc. R. Soc. Lond. B Biol. Sci 267 (1452), 1591–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookhart GL, Edgecomb RS, Murdock LL, 1987. Amphetamine and reserpine deplete brain biogenic amines and alter blow fly feeding behavior. J. Neurochem 48 (4), 1307–1315. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T, 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15), 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicca A, Schafroth MA, Reynoso-Moreno I, Erni R, Petrucci V, Carreira EM, Gertsch J, 2018. Uncovering the psychoactivity of a cannabinoid from liverworts associated with a legal high. Sci. Adv 4 (10), eaat2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri R, Machado AA, Vital AF, 1957. A new species of the genus Massospora with an Allomyces species. Atti dell’Istituto Botanico della Universitá e Laboratorio Crittogamico di Pavia, Series 5 (14), 15–22. [Google Scholar]
- Cooley JR, 1999. Sexual Behavior in North American Cicadas of the Genera Magicicada and Okanagana. Doctoral dissertation, University of Michigan. [Google Scholar]
- Cooley JR, Marshall DC, Hill KB, 2018. A specialized fungal parasite (Massospora cicadina) hijacks the sexual signals of periodical cicadas (Hemiptera: Cicadidae: Magicicada). Sci. Rep 8 (1), 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, 1982. The Extended Phenotype-The Gene as the Unit of Selection.
- De Bekker C, Quevillon LE, Smith PB, Fleming KR, Ghosh D, Patterson AD, Hughes DP, 2014. Species-specific ant brain manipulation by a specialized fungal parasite. BMC Evol. Biol 14 (1), 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bekker C, Ohm RA, Evans HC, Brachmann A, Hughes DP, 2017. Ant-infecting Ophiocordyceps genomes reveal a high diversity of potential behavioral manipulation genes and a possible major role for enterotoxins. Sci. Rep 7 (1), 12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fine Licht HH, Jensen AB, Eilenberg J, 2017. Comparative transcriptomics reveal host-specific nucleotide variation in entomophthoralean fungi. Mol. Ecol 26 (7), 2092–2110. [DOI] [PubMed] [Google Scholar]
- Detzel A, Wink M, 1993. Attraction, deterrence or intoxication of bees (Apis mellifera) by plant allelochemicals. Chemoecology 4 (1), 8–18. [Google Scholar]
- Eddy SR, 1998. Profile hidden Markov models. Bioinformatics 14 (9), 755–763. [DOI] [PubMed] [Google Scholar]
- Eddy SR, 2011. Accelerated profile HMM searches. PLoS Comput. Biol 7 (10), e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenshaft M, Bilski P, Li MY, Chignell CF, Daub ME, 1999. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc. Natl. Acad. Sci. Unit. States Am 96 (16), 9374–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elya C, Lok TC, Spencer QE, McCausland H, Martinez CC, Eisen MB, 2018. Robust manipulation of the behavior of Drosophila melanogaster by a fungal pathogen in the laboratory. eLife 7, e34414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen MA, Zhang Y, Hazen ML, Loreto RG, Mangold CA, Chen DZ, Hughes DP, 2017. Three-dimensional visualization and a deep-learning model reveal complex fungal parasite networks in behaviorally manipulated ants. Proc. Natl. Acad. Sci. Unit. States Am 201711673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke J, Blei F, Hoffmeister D, 2017. Enzymatic synthesis of psilocybin. Angew. Chem. Int. Ed 56 (40), 12352–12355. [DOI] [PubMed] [Google Scholar]
- Frischknecht HR, Waser PG, 1978. Actions of hallucinogens on ants (Formica pratensis)–II. Effects of amphetamine, LSD and delta-9-tetrahydrocannabinol. Gen. Pharmacol. Vasc. Syst 9 (5), 375–380. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol 29 (7), 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell MN, Jensen AB, Olsen PB, Eilenberg J, Lange L, 2011. Secretome of fungus-infected aphids documents high pathogen activity and weak host response. Fungal Genet. Biol 48 (4), 343–352. [DOI] [PubMed] [Google Scholar]
- Groves RA, Hagel JM, Zhang Y, Kilpatrick K, Levy A, Marsolais F, Lewinsohn E, Sensen CW, Facchini PJ, 2015. Transcriptome profiling of khat (Catha edulis) and Ephedra sinica reveals gene candidates potentially involved in amphetamine-type alkaloid biosynthesis. PLoS One 10 (3), e0119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryganskyi AP, Mullens BA, Gajdeczka MT, Rehner SA, Vilgalys R, Hajek AE, 2017. Hijacked: Co-option of host behavior by entomophthoralean fungi. PLoS Pathog. 13 (5), e1006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS, 2017. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol 35 (2), 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge KT, Hajek AE, Gryganskyi A, 2017. The first entomophthoralean killing millipedes, Arthrophaga myriapodina n. gen. n. sp., causes climbing before host death. J. Invertebr. Pathol 149, 135–140. [DOI] [PubMed] [Google Scholar]
- Hoff KJ, Stanke M, 2013. WebAUGUSTUS–a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res 41 (W1), W123–W128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover K, Grove M, Gardner M, Hughes DP, McNeil J, Slavicek J, 2011. A gene for an extended phenotype. Science 333 (6048), 1401–1401. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P, 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol 34 (8), 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P, 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol 33 (6), 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, 2013. Pathways to understanding the extended phenotype of parasites in their hosts. J. Exp. Biol 216 (1), 142–147. [DOI] [PubMed] [Google Scholar]
- Hughes DP, Arafljo JPM, Loreto RG, Quevillon L, De Bekker C, Evans HC, 2016. From so simple a beginning: the evolution ofbehavioral manipulation by fungi In: Advances in Genetics, vol. 94 Academic Press, pp. 437–469. [DOI] [PubMed] [Google Scholar]
- Humber RA, 2012. Entomophthoromycota: a new phylum and reclassification for entomophthoroid fungi. Mycotaxon 120 (1), 477–492. [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC, 2011. Integrative Analysis of Environmental Sequences Using MEGAN4. Genome Research gr-120618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison JA, 1962. Studies on a new Entomophthora attacking calyptrate flies. Mycologia 54 (3), 258–271. [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, 2006. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443 (7113), 818. [DOI] [PubMed] [Google Scholar]
- Joshi NA, Fass JN, 2011. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33)[Software]. [Google Scholar]
- Kalix P, 1988. Khat: a plant with amphetamine effects. J. Subst. Abus. Treat 5 (3), 163–169. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30 (4), 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, 1951. Notes on fungi.(1) on the newly found genus Massospora from Japan. J. Jpn. Bot 26, 117–119. [Google Scholar]
- Kosentka P, Sprague SL, Ryberg M, Gartz J, May AL, Campagna SR, Matheny PB, 2013. Evolution of the toxins muscarine and psilocybin in a family of mushroom-forming fungi. PLoS One 8 (5), e64646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucht S, Groß J, Hussein Y, Grothe T, Keller U, Basar S, Konig WA, Steiner U, Leistner E, 2004. Elimination of ergoline alkaloids following treatment of Ipomoea asarifolia (Convolvulaceae) with fungicides. Planta 219 (4), 619–625. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol 33 (7), 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, 2007. Clustal W and clustal X version 2.0. Bioinformatics 23 (21), 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS, 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13 (9), 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomascolo A, Stentelaire C, Asther M, Lesage-Meessen L, 1999. Basidiomycetes as new biotechnological tools to generate natural aromatic flavours for the food industry. Trends Biotechnol 17 (7), 282–289. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M, 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33 (20), 6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons PC, Plattner RD, Bacon CW, 1986. Occurrence of peptide and clavine ergot alkaloids in tall fescue grass. Science 232 (4749), 487–489. [DOI] [PubMed] [Google Scholar]
- Mahmoud AL, 2000. Mycotoxin-producing potential of fungi associated with qat (Catha edulis) leaves in Yemen. Folia Microbiol 45 (5), 452–456. [DOI] [PubMed] [Google Scholar]
- Małagocka J, Grell MN, Lange L, Eilenberg J, Jensen AB, 2015. Transcriptome of an entomophthoralean fungus (Pandora formicae) shows molecular machinery adjusted for successful host exploitation and transmission. J. Invertebr. Pathol 128, 47–56. [DOI] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A, 2018. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol 14 (1), e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulionis VE, Weber RW, Pagnocca FC, 2013. Foraging of Psilocybe basidiocarps by the leaf-cutting ant Acromyrmex lobicornis in Santa Fé, Argentina: SpringerPlus; 2 (1), 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y, Moriyama M, Lukasik P, Vanderpool D, Tanahashi M, Meng XY, McCutcheon JP, Fukatsu T, 2018. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl. Acad. Sci. Unit. States Am 201803245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Redden GA, 2003. Fungal Infection and Gender Confusion in the Wing-Banger Cicada Platypedia Putnami, pp. 65–68. Norse Scientist (NKU). [Google Scholar]
- Nowak J, Woźniakiewicz M, Klepacki P, Sowa A, Kościelniak P, 2016. Identification and determination of ergot alkaloids in Morning Glory cultivars. Anal. Bioanal. Chem 408 (12), 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi T, Sato H, Igawa S, 1999. An epizootic on Meimuna boninensis (distant)(Homoptera: Cicadidae), caused by Massospora sp.(Zygomycetes: Entomophthorales) and Nomuraea cylindrospora (Deuteromycotina: Hyphomycetes in the Ogasawara (Bonin) Islands, Japan. Appl. Entomol. Zool 34 (3), 339–343. [Google Scholar]
- Panaccione DG, Arnold SL, 2017. Ergot alkaloids contribute to virulence in an insect model of invasive aspergillosis. Sci. Rep 7 (1), 8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Wood T, Zhang Z, Miller W, 1997. Comparison of DNA sequences with protein sequences. Genomics 46 (1), 24–36. [DOI] [PubMed] [Google Scholar]
- Peláez F, Collado J, Arenal F, Basilio A, Cabello A, Matas MD, Garcia JB, Del Val AG, Gonzalez V, Gorrochategui J, Hernández P, 1998. Endophytic fungi from plants living on gypsum soils as a source of secondary metabolites with antimicrobial activity. Mycol. Res 102 (6), 755–761. [Google Scholar]
- Posada D, Crandall KA, 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14 (9), 817–818. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP, 2010. FastTree 2–approximately maximum likelihood trees for large alignments. PLoS One 5 (3), e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds HT, Vijayakumar V, Gluck-Thaler E, Korotkin HB, Matheny PB, Slot JC, 2018. Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evol. Lett 2 (2), 88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy HE, Steinkraus DC, Eilenberg J, Hajek AE, Pell JK, 2006. Bizarre interactions and endgames: entomopathogenic fungi and their arthropod hosts. Annu. Rev. Entomol 51, 331–357. [DOI] [PubMed] [Google Scholar]
- Sanborn AF, Phillips PK, 2011. Elevation of a subspecies of Tibicen (Hemiptera: Cicadoidea: Cicadidae) to a full species. SW. Nat 56 (3), 363–368. [Google Scholar]
- Short DP, Double M, Nuss DL, Stauder CM, MacDonald W, Kasson MT, 2015. Multilocus PCR Assays Elucidate Vegetative Incompatibility Gene Profiles of Cryphonectria Parasitica in the United States. Applied and Environmental Microbiology. AEM-00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot JC, 2017. Fungal gene cluster diversity and evolution In: Advances in Genetics, vol. 100 Academic Press, pp. 141–178. [DOI] [PubMed] [Google Scholar]
- Soper RS, 1963. Massospora levispora, a new species of fungus pathogenic to the cicada, Okanagana rimosa. Can. J. Bot 41 (6), 875–878. [Google Scholar]
- Soper SR, 1974. The genus Massospora, entomopathogenic for cicadas. Part I. Taxonomy of the genus. Mycotaxon 1, 13–40. [Google Scholar]
- Soper SR, 1981. New cicada pathogens: Massospora cicadettae from Australia and Massospora pahariae from Afghanistan. Mycotaxon 13, 50–58. [Google Scholar]
- Soper RS, Shewell GE, Tyrrell D, 1976. Colcondamyia auditrix nov. sp. (Diptera: Sarcophagidae), a parasite which is attracted by the mating song of its host, Okanagana rimosa (Homoptera: Cicadidae). Can. Entomol 108 (1), 61–68. [Google Scholar]
- Sota T, Yamamoto S, Cooley JR, Hill KB, Simon C, Yoshimura J, 2013. Independent divergence of 13-and 17-y life cycles among three periodical cicada lineages. Proc. Natl. Acad. Sci. Unit. States Am 110 (17), 6919–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108 (5), 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9), 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Waack S, 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Thomas F, Schmidt-Rhaesa A, Martin G, Manu C, Durand P, Renaud F, 2002. Do hairworms (Nematomorpha) manipulate the water seeking behaviour of their terrestrial hosts? J. Evol. Biol 15 (3), 356–361. [Google Scholar]
- Vilgalys R, Hester M, 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol 172 (8), 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He Z, Wei C, 2017. A new cicada species of Psalmocharias Kirkaldy feeding on an Ephedra plant from China (Hemiptera: Cicadidae). Zootaxa 4290 (2), 367–372. [Google Scholar]
- Weiser J, Stary P, 1967. La chicharra Diceroprocta biconica Walker, neuvo hu sped en Cuba del Hongo Massospora cicadina Peck 1870. Poeyana (Havana) 35, 1–6. [Google Scholar]
- White JA, Ganter P, McFarland R, Stanton N, Lloyd M, 1983. Spontaneous, field tested and tethered flight in healthy and infected Magicicada septendecim L. Oecologia 57 (3), 281–286. [DOI] [PubMed] [Google Scholar]
- Witt PN, 1971. Drugs alterweb-building of spiders: a review and evaluation. Behav. Sci 16 (1), 98–113. [DOI] [PubMed] [Google Scholar]
- Wittmann M, Carter O, Hasler F, Cahn BR, Grimberg U, Spring P, Hell D, Flohr H, Vollenweider FX, 2007. Effects of psilocybin on time perception and temporal control of behaviour in humans. J. Psychopharmacol 21 (1), 50–64. [DOI] [PubMed] [Google Scholar]
- Wolf T, Shelest V, Nath N, Shelest E, 2015. CASSIS and SMIPS: promoter-based prediction of secondary metabolite gene clusters in eukaryotic genomes. Bioinformatics 32 (8), 1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrońska AK, Bogus MI, Kaczmarek A, Kazek M, 2018. Harman and norharman, metabolites of entomopathogenic fungus Conidiobolus coronatus (Entomopthorales), disorganize development of Galleria mellonella (Lepidoptera) and affect serotonin-regulating enzymes. PLoS One 13 (10), e0204828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.