Abstract
Currently there are no effective antifibrotic therapies for liver cirrhosis, a major killer worldwide. To obtain a cellular resolution of directly-relevant pathogenesis and to inform therapeutic design, we profile the transcriptomes of over 100,000 human single cells, yielding molecular definitions for non-parenchymal cell types present in healthy and cirrhotic human liver. We uncover a novel scar-associated TREM2+CD9+ macrophage subpopulation, which expands in liver fibrosis, differentiates from circulating monocytes and is pro-fibrogenic. We also define novel ACKR1+ and PLVAP+ endothelial cells which expand in cirrhosis, are topographically scar-restricted and enhance leucocyte transmigration. Multi-lineage ligand-receptor modelling of interactions between the novel scar-associated macrophages, endothelial cells and PDGFRα+ collagen-producing mesenchymal cells reveals intra-scar activity of several pro-fibrogenic pathways including TNFRSF12A, PDGFR and NOTCH signalling. Our work dissects unanticipated aspects of the cellular and molecular basis of human organ fibrosis at a single-cell level, and provides the conceptual framework required to discover rational therapeutic targets in liver cirrhosis.
Recent estimates suggest that 844 million people worldwide have chronic liver disease, with two million deaths per year and a rising incidence1. Iterative liver injury secondary to any cause leads to progressive fibrosis ultimately resulting in liver cirrhosis. Importantly, the degree of liver fibrosis predicts adverse patient outcomes2. Hence, effective antifibrotic therapies for patients with chronic liver disease are urgently required3,4.
Liver fibrosis involves a complex interplay between multiple non-parenchymal cell (NPC) lineages including immune, endothelial and mesenchymal cells spatially located within areas of scarring, termed the fibrotic niche. Despite progress in our understanding of liver fibrogenesis accrued using rodent models, there remains a significant 'translational gap' between putative targets and effective patient therapies3,4. This is in part due to limited definition of the functional heterogeneity and interactome of cell lineages that contribute to the fibrotic niche of human liver cirrhosis, which is imperfectly recapitulated by rodent models3.
Single-cell RNA sequencing (scRNA-seq) is delivering a step change in our understanding of disease pathogenesis, allowing the interrogation of individual cell populations at unprecedented resolution5. Here, we studied the mechanisms regulating human liver fibrosis using scRNA-seq.
Results
Single-cell atlas of human liver NPC
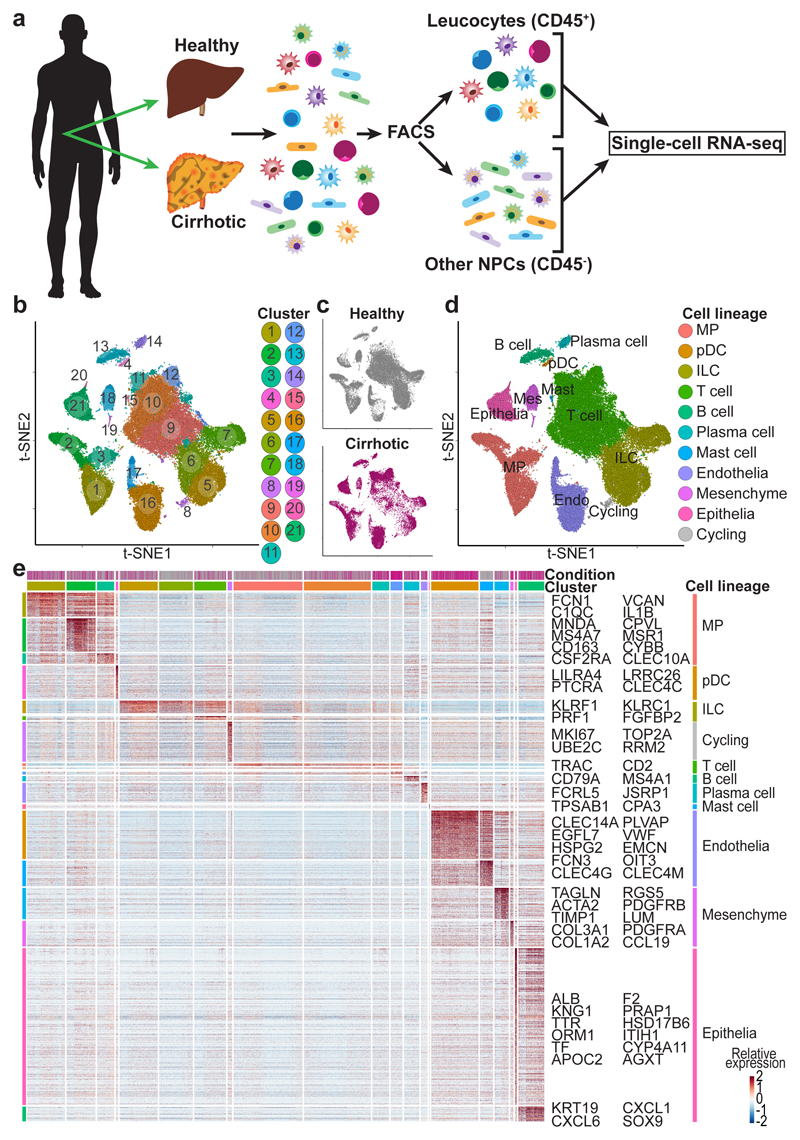
Hepatic NPC were isolated from healthy and cirrhotic human livers spanning a range of aetiologies of cirrhosis (Fig. 1a, Extended Data Fig. 1a). Leucocytes (CD45+) or other NPC (CD45-) fractions (Extended Data Fig. 1b) were FACS-sorted prior to scRNA-seq. To discriminate between liver-resident and circulating leucocytes, we also performed scRNA-seq on CD45+CD66b- peripheral blood mononuclear cells (PBMC) (Extended Data Fig. 1c, g-i). The combined tissue and PBMC dataset was partitioned into clusters (Extended Data Fig. 1d) and annotated using signatures of known lineage markers (Extended Data Fig. 1d-e; Supplementary Table 2). To generate an atlas of liver-resident cells, contaminating circulating cells were removed from the liver tissue datasets, by excluding cells from the tissue samples which mapped transcriptionally to blood-derived clusters 1 and 13 (Extended Data Fig. 1d). Liver-resident cells expressed higher levels of tissue-residency markers such as CXCR4 compared to PBMC (Extended Data Fig. 1f).
Figure 1. Single cell atlas of human liver NPC.
a, Overview: isolation, FACS-sorting and sc-RNASeq of leucocytes (CD45+) and other NPC fractions (CD45-). b, Clustering 66,135 cells from 5 healthy and 5 cirrhotic human livers. c, Annotation by injury condition. d, Cell lineage inferred from expression of marker gene signatures. Endo, endothelial cell; ILC, innate lymphoid cell; Mast, mast cell; Mes, mesenchymal cell; MP, mononuclear phagocyte; pDC, plasmacytoid dendritic cell. e, Heatmap: cluster marker genes (top, colour coded by cluster and colour coded by condition) and exemplar genes and lineage annotation labelled (right). Cells columns, genes rows.
Re-clustering the 66,135 liver-resident cells from 10 livers (n=5 healthy and n=5 cirrhotic) revealed 21 populations (Fig. 1b), each containing cells from both healthy and cirrhotic livers (Fig. 1c; Extended Data Fig. 2), across 10 cell lineages (Fig. 1d, Extended Data Fig. 2a, b). Subpopulation markers were identified across all clusters and lineages (Fig. 1e; Supplementary Tables 3, 4). QC metrics were highly reproducible between individual samples and condition (Extended Data Fig. 2c-f, Supplementary Table 1). Expression of collagens type I and III, the main fibrillar collagens within the fibrotic niche, was restricted to cells of the mesenchymal lineage (Fig. 1e).
We proceeded to annotate all human liver NPC lineages (below, Supplementary Notes 1-3, Extended Data Fig. 3), and provide an open-access gene browser (http://www.livercellatlas.mvm.ed.ac.uk) allowing assessment of NPC gene expression between healthy and cirrhotic livers.
Distinct macrophage subpopulations inhabit the fibrotic niche
Previous studies in rodents have highlighted macrophage subpopulations orchestrating both liver fibrosis progression and regression6–8. Clustering of mononuclear phagocytes (MP) identified ten clusters; annotated as scar-associated macrophages (SAMΦ), Kupffer cells (KC), tissue monocytes (TMo), conventional dendritic cells (cDC) and cycling (proliferating) cells (Fig. 2a, Extended Data Fig. 4a, Supplementary Note 2). Clusters MP(4) and MP(5), named SAMΦ(1) and SAMΦ(2) respectively, were expanded in cirrhotic livers (Fig. 2b), as confirmed by quantification of the MP cell composition of each liver individually (Fig. 2c).
Figure 2. Identifying scar-associated macrophage subpopulations.
a, Clustering 10,737 mononuclear phagocytes (MP) from 5 healthy and 5 cirrhotic human livers. TMo, tissue monocyte; SAMΦ, scar-associated macrophage; KC, Kupffer cell; cDC, conventional dendritic cell. b, Annotation by injury condition. c, Fractions of MP subpopulations in 5 healthy versus 5 cirrhotic livers, Mean±SEM, Wald test. d, Heatmap: MP cluster marker genes (top, colour coded by cluster and condition), exemplar genes labelled (right). Cells columns, genes rows. e, Scaled gene expression of SAMΦ and TMo cluster markers across MP cells from healthy (n=5) and cirrhotic (n=5) livers. f, Flow cytometry: TREM2+CD9+ MP fraction in healthy (n=2) versus cirrhotic (n=3) liver, Mean±SEM. g, Primary human hepatic stellate cells (HSC) treated with conditioned media from SAMΦ (n=3) or TMo (n=3); qPCR of stated genes, expression relative to mean expression of control HSC (n=6), Mean±SEM, Kruskal-Wallis and Dunn. h, Representative immunofluorescence image, cirrhotic liver: TREM2 (red), CD9 (white), collagen 1 (green), DAPI (blue), scale bar 50μm.
Clusters MP(6) and MP(7) were enriched in CD163, MARCO and TIMD4 (Extended Data Fig. 4b); tissue staining confirmed these as Kupffer cells (KC; resident liver macrophages), facilitating annotation of these clusters as KC(1) and KC(2) respectively (Extended Data Fig. 4c). A lack of TIMD4 expression distinguished KC(2) from KC(1) (Extended Data Fig. 4b); cell counting demonstrated TIMD4+ cell numbers to be equivalent between healthy and cirrhotic livers, but showed a loss of MARCO+ cells, consistent with selective reduction in MARCO+TIMD4- KC(2) in liver fibrosis (Fig. 2c, Extended Data Fig. 4d, e).
Scar-associated clusters SAMΦ(1) and SAMΦ(2) expressed the unique markers TREM2 and CD9 (Fig. 2d, e). These macrophages displayed a hybrid phenotype, with features of both tissue monocytes and KC (Fig. 2d, e), analogous to monocyte-derived macrophages in murine liver injury models7. Flow cytometry confirmed expansion of TREM2+CD9+ macrophages in human fibrotic livers (Fig. 2f, Extended Data Fig. 4f). Conditioned media from FACS-sorted SAMΦ promoted fibrillar collagen expression by primary human hepatic stellate cells (HSC) (Fig. 2g), indicating that SAMΦ have a pro-fibrogenic phenotype. Tissue staining demonstrated the presence of TREM2+CD9+MNDA+ SAMΦ topographically localised in collagen-positive scar regions (Fig. 2h, Extended Data Fig. 4g-i), and significantly expanded in cirrhotic livers (Extended Data Fig. 4j, k). Cell counting of stained cirrhotic livers morphologically segmented into regions of fibrotic septae and parenchymal nodules, confirmed SAMΦ accumulation within the fibrotic niche (Extended Data Fig. 4l).
Local proliferation plays a significant role in macrophage expansion at sites of fibrosis in rodent models7,9. Cycling MP cells (Fig. 2a) subclustered into cDC1, cDC2, KC and SAMΦ subpopulations (Extended Data Fig. 4m, Supplementary Table 8). Cycling SAMΦ expanded in cirrhosis (Extended Data Fig. 4m), highlighting the potential role of macrophage proliferation in promoting SAMΦ accumulation in the fibrotic niche.
Pro-fibrogenic phenotype of scar-associated macrophages
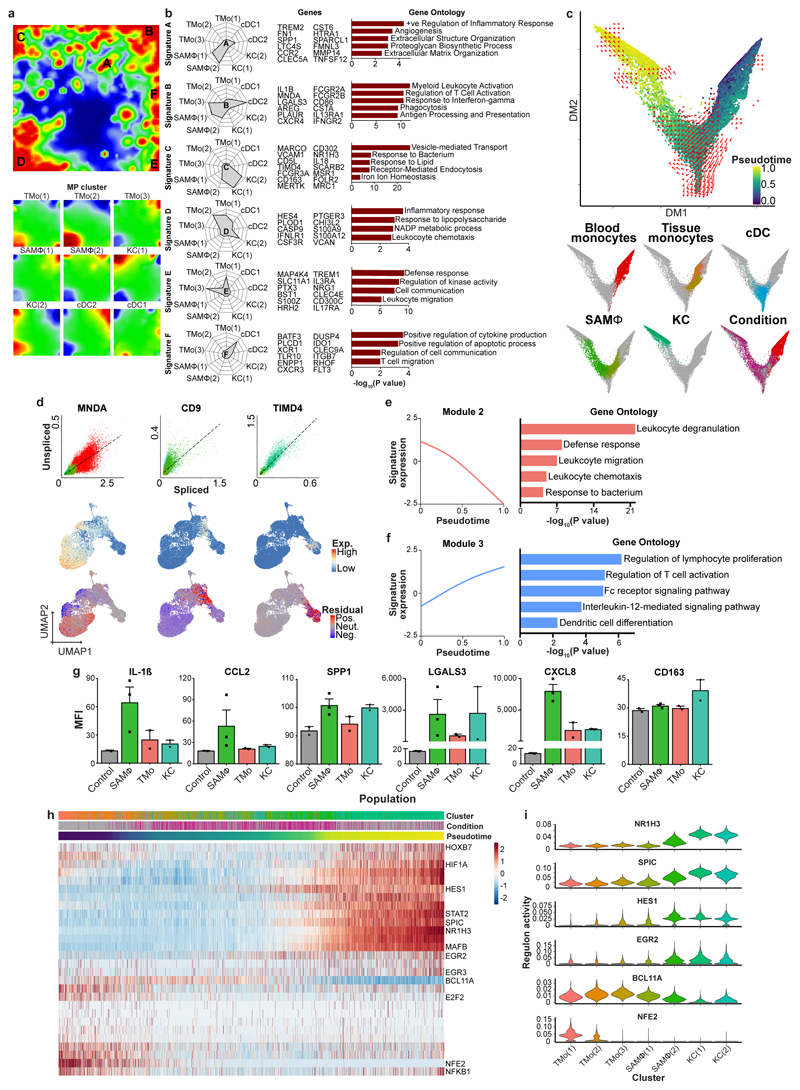
To delineate the functional profile of SAMΦ we visualised co-ordinately expressed gene groups across the MP subpopulations using self-organising maps (Extended Data Fig. 5a). We identified six optimally-differentiating metagene signatures, denoted as A-F (Extended Data Fig. 5a, Supplementary Table 9). Signatures A and B defined SAMΦ and were enriched for ontology terms relevant to tissue fibrosis (Extended Data Fig. 5b). These SAMΦ-defining signatures included genes such as TREM2, IL1B, SPP1, LGALS3, CCR2, and TNFSF12, some of which are known to regulate the function of scar-producing myofibroblasts in fibrotic liver diseases10–13. The remaining MP subpopulations were defined by signature C (KC), signatures D, E (TMo) and signature F (cDC1); ontology terms matched known functions for the associated cell type (Extended Data Fig. 5b, Supplementary Table 9).
In mice, under homeostatic conditions, embryologically-derived self-renewing tissue-resident KC predominate14–16. However, following injury, macrophages derived from circulating monocytes accumulate in the liver and regulate fibrosis7,8. The ontogeny of human hepatic macrophage subpopulations is unknown. TREM2+CD9+ SAMΦ demonstrated a monocyte-like morphology (Fig. 2h, Extended Data Fig. 4g-i) and a distinct topographical distribution from KC (Extended Data Fig. 4l). To assess SAMΦ origin, we performed in silico trajectory analysis on a combined dataset of peripheral blood monocytes and liver-resident MPs. We visualised the transcriptional profile of these cells (Fig. 3a, Extended Data Fig. 5c), mapped them along a pseudotemporal trajectory and interrogated their directionality via spliced and unspliced mRNA ratios (RNA velocity17). These analyses suggested a differentiation trajectory from peripheral blood monocytes into either SAMΦ or cDC, with no differentiation from KC to SAMΦ, and no progression from SAMΦ to KC (Fig. 3a; Extended Data Fig. 5c). Additional RNA velocity analyses17 showed downregulation (negative velocity) of the monocyte gene MNDA in SAMΦ, upregulation (positive velocity) of the SAMΦ marker gene CD9 in tissue monocytes, and a lack of KC gene TIMD4 velocity in SAMΦ (Extended Data Fig. 5d). Furthermore, assessment of the probabilities of cells in this dataset transitioning into SAMΦ, indicated a higher likelihood of tissue monocytes than KC differentiating into SAMΦ (Fig. 3b). Overall, these data suggest that SAMΦ are monocyte-derived, and represent a terminally-differentiated cell state within the fibrotic niche.
Figure 3. Fibrogenic phenotype of scar-associated macrophages.
a, UMAP visualisation, 23,075 cells from liver-resident MP (5 healthy and 5 cirrhotic) and blood monocytes (5 PBMC), annotating monocle pseudotemporal dynamics (purple to yellow). RNA velocity field (red arrows) visualised using Gaussian smoothing on regular grid. Right: Annotation of MP subpopulation, injury condition. b, Transition probabilities per SAMΦ subpopulation, indicating for each cell the likelihood of transition into either SAMΦ(1) or SAMΦ(2), calculated using RNA velocity (yellow high; purple low; grey below threshold of 2x10-4). c, Heatmap: spline curves fitted to genes differentially expressed across blood monocyte-to-SAMΦ (right arrow) and blood monocyte-to-cDC (left arrow) pseudotemporal trajectories, grouped by hierarchical clustering (k=3). Gene co-expression modules (colour) labelled right with exemplar genes from each module. d, Spline curve fitted to averaged expression of all genes in module 1, along monocyte-to-SAMΦ pseudotemporal trajectory, selected GO enrichment (right), Fisher’s exact test.
To further characterise SAMΦ phenotype, we identified differentially expressed genes along monocyte differentiation trajectories. We defined three gene co-expression modules, with module 1 representing upregulated genes during blood monocyte-to-SAMΦ differentiation (Fig. 3c). Module 1 contained multiple pro-fibrogenic genes including SPP1, LGALS3, CCL2, CXCL8, PDGFB and VEGFA10–13, and displayed ontology terms consistent with promoting tissue fibrosis and angiogenesis (Fig. 3c, d; Supplementary Table 10). Module 2 contained genes that were downregulated during monocyte-to-SAMΦ differentiation (Fig. 3c; Extended Data Fig. 5e), whilst Module 3 encompassed a group of upregulated genes during monocyte-to-cDC differentiation (Fig. 3c; Extended Data Fig. 5f; Supplementary Table 10). SAMΦ isolated from cirrhotic human livers (Fig. 2f, Extended Data Fig. 4f) demonstrated enhanced protein secretion of several of the mediators identified by transcriptional analysis (Extended Data Fig. 5g) and promoted fibrillar collagen expression by primary human HSC (Fig. 2g), confirming that SAMΦ have a pro-fibrogenic phenotype.
To enable cross-species comparison, we performed scRNA-seq on liver MP cells isolated from control mice or mice treated with chronic carbon tetrachloride (CCl4), a mouse model of liver fibrosis7. MP cells from fibrotic livers were isolated 24 hours after the final CCl4 injection, a time of active fibrogenesis7. Five MP cell clusters were defined (Extended Data Fig. 6a-d, Supplementary Table 11), and injury-specific cluster mMP(2) was differentiated by high expression of Cd9, Trem2, Spp1 and Lgals3 (Extended Data Fig. 6a-d). We confirmed expansion of this CD9+ SAMΦ (mSAMΦ) population in liver fibrosis (Extended Data Fig. 6e, f) and co-culture of mSAMΦ with quiescent primary murine HSC promoted fibrillar collagen expression in HSC (Extended Data Fig. 6g). Canonical correlation analysis between human and mouse MP datasets18 demonstrated that human SAMΦ (hSAMΦ) and mSAMΦ clustered together (h&mMP(2); Extended Data Fig. 6h, i) and that this cluster was enriched for SAMΦ markers CD9, TREM2 and SPP1 (Extended Data Fig. 6j), confirming that mSAMΦ represent a corollary population to human SAMΦ (hSAMΦ).
To identify potential transcriptional regulators of human SAMΦ we defined sets of genes co-expressed with known transcription factors (regulons) along the tissue monocyte-macrophage pseudotemporal trajectory and in KC (Extended Data Fig. 5g, h, Supplementary Table 12). This identified regulons and corresponding transcription factors associated with distinct macrophage phenotypes, highlighting HES1 and EGR2 activity in SAMΦ.
To determine whether SAMΦ expand in earlier stage human liver disease, we analysed cohorts of patients with non-alcoholic fatty liver disease (NAFLD). Application of differential gene expression signatures of human SAMΦ, KC and TMo to a deconvolution algorithm19 enabled assessment of hepatic monocyte-macrophage composition in whole liver microarray data across the spectrum of early-stage NAFLD20 (Extended Data Fig. 7a). This demonstrated SAMΦ expansion in patients with non-alcoholic steatohepatitis (NASH) (Extended Data Fig. 7a, b), an increased frequency of SAMΦ with worsening histological NASH activity (NAS) and fibrosis scores (Extended Data Fig. 7c), but no association with other patient demographics (Extended Data Fig. 7d). In a separate NASH biopsy cohort, SAMΦ expansion increased with NASH activity (Extended Data Fig. 7e) and positively correlated with the degree of fibrosis across the full severity spectrum of NAFLD-induced liver fibrosis (Extended Data Fig. 7f).
In summary, these data demonstrate that TREM2+CD9+ SAMΦ derive from the recruitment and differentiation of circulating monocytes, are conserved across species, display a pro-fibrogenic phenotype and expand early in the course of liver disease progression.
Distinct endothelial subpopulations inhabit the fibrotic niche
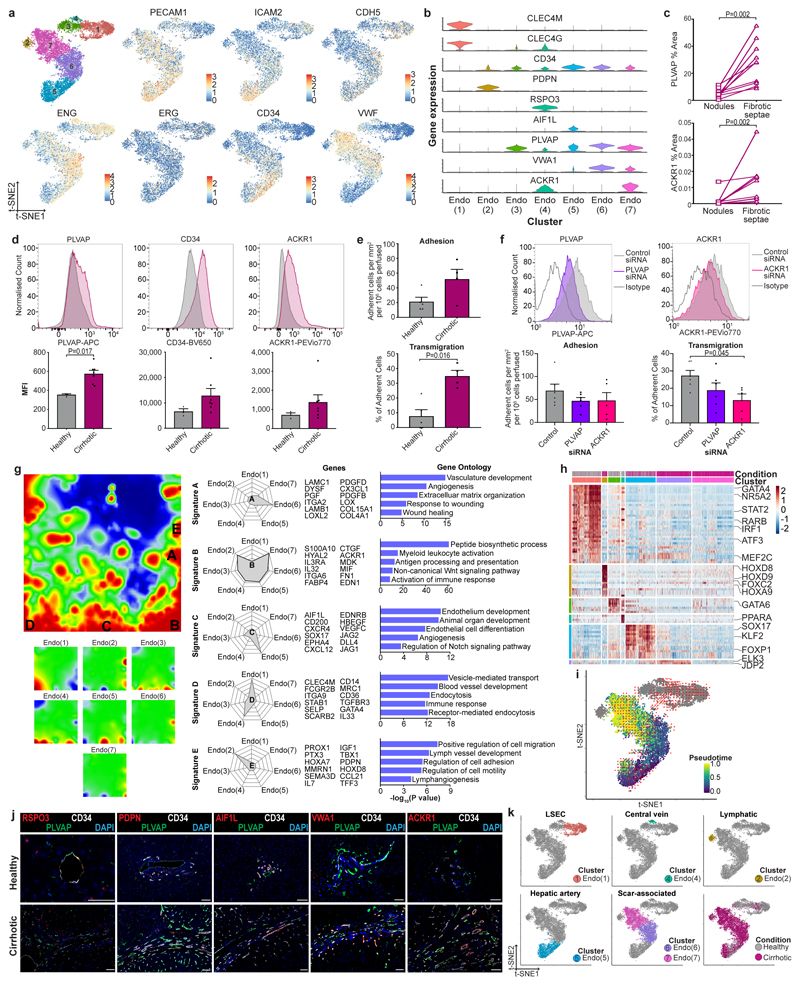
In rodent models, hepatic endothelial cells are known to regulate fibrogenesis. Clustering of human liver endothelial cells identified seven subpopulations (Fig. 4a). Classical endothelial cell markers did not discriminate between the seven clusters, although Endo(1) was distinct in lacking CD34 expression (Extended Data Fig. 8a). To fully annotate endothelial subpopulations (Supplementary Note 3, Extended Data Fig. 8k), we identified differentially expressed markers (Fig. 4c, Supplementary Table 13), determined functional expression profiles (Extended Data Fig. 8g, Supplementary Table 14), performed transcription factor regulon analysis (Extended Data Fig. 8h, Supplementary Table 15) and assessed spatial distribution via tissue staining (Fig. 4d, Extended Data Fig. 8j).
Figure 4. Identifying scar-associated endothelial subpopulations.
a, Clustering 8,020 endothelial cells from 4 healthy and 3 cirrhotic human livers, annotating injury condition (right). b, Fractions of endothelial subpopulations in healthy (n=4) versus cirrhotic (n=3) livers, Mean±SEM, Wald. c, Heatmap: endothelial cluster marker genes (colour coded top by cluster and condition), exemplar genes labelled right. Cells columns, genes rows. d, Representative immunofluorescence images: CD34 (red), CLEC4M (white), PLVAP (green), DAPI (blue), scale bar 50μm. e, Digital pixel quantification: CLEC4M staining healthy (n=5) versus cirrhotic liver (n=8), PLVAP staining healthy (n=11) versus cirrhotic liver (n=11), ACKR1 staining healthy (n=10) versus cirrhotic liver (n=10), scale bars 50μm, Mean±SEM, Mann-Whitney two-tailed.
Disease-specific endothelial cells Endo(6) and Endo(7), CD34+PLVAP+VWA1+ and CD34+PLVAP+ACKR1+ respectively (Fig. 4a-c, Extended Data Fig. 8b), expanded in cirrhotic liver tissue (Fig. 4e) and were restricted to the fibrotic niche (Fig. 4d, e, Extended Data Fig. 8c), allowing annotation as scar-associated endothelia SAEndo(1) and SAEndo(2) respectively. In contrast, CD34-CLEC4M+ Endo(1) (annotated as liver sinusoidal endothelial cells), were reduced in fibrotic livers (Fig. 4b, e). Metagene signature analysis demonstrated that Endo(6) (SAEndo(1)) cells expressed pro-fibrogenic genes including PDGFD, PDGFB, LOX, LOXL2; associated ontology terms included extracellular matrix organization (signature A; Extended Data Fig. 8g). Endo(7) (SAEndo(2)) cells displayed an immunomodulatory phenotype (signature B; Extended Data Fig. 8g). The most discriminatory marker for this cluster, ACKR1, has a role in regulating leucocyte recruitment21. We confirmed increased expression of PLVAP, CD34 and ACKR1 on endothelial cells isolated from cirrhotic livers (Extended Data Fig. 8d). Flow-based adhesion assays22 demonstrated that cirrhotic endothelial cells display enhanced leucocyte transmigration (Extended Data Fig. 8e), which was attenuated by ACKR1 knockdown (Extended Data Fig. 8f).
PDGFRA expression defines scar-associated mesenchymal cells
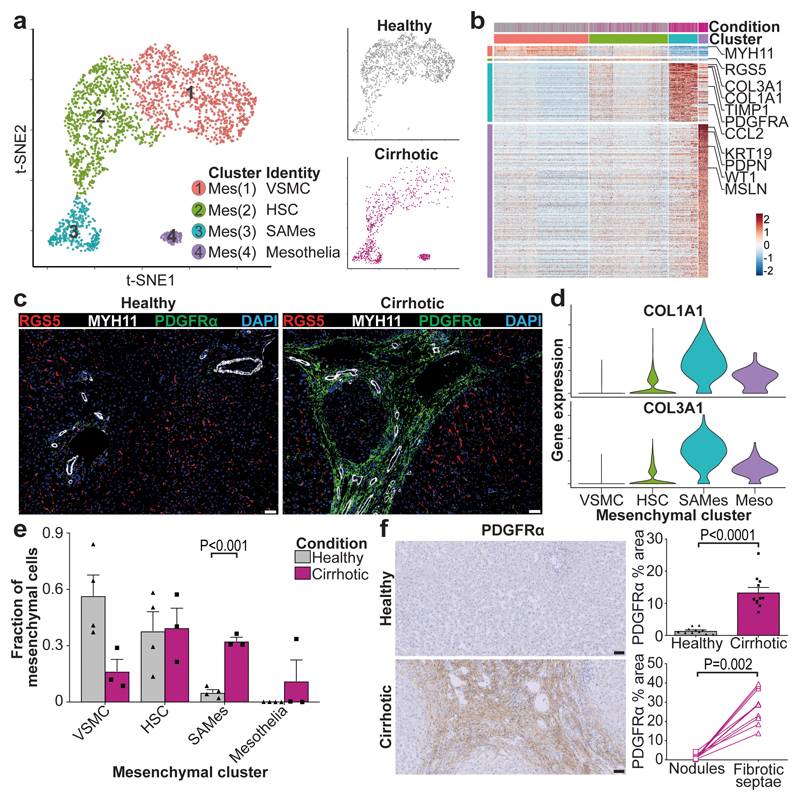
Clustering of human liver mesenchymal cells identified four populations (Fig. 5a, b, Extended Data Fig. 9a, Supplementary Table 16). Cluster Mes(1), distinguished by MYH11 expression (Fig. 5b, Extended Data Fig. 9a), was identified as vascular smooth muscle cells (VSMC) (Fig. 5c). Mes(4) demonstrated expression of mesothelial markers (Fig. 5b, Extended Data Fig. 9a). Cluster Mes(2) expressed high levels of RGS5 (Fig. 5b, Extended Data Fig. 9a), and RGS5 staining identified this population as hepatic stellate cells (HSC) (Fig. 5c). RGS5+ cells were absent from the fibrotic niche (Fig. 5c). Cluster Mes(3) (distinguished by PDGFRA expression) expressed high levels of fibrillar collagens and pro-fibrogenic genes (Fig. 5b, d, Extended Data Fig. 9a). PDGFRα+ cells expanded in cirrhotic livers (Fig. 5a, e, f) and were mapped to the fibrotic niche (Fig. 5f), enabling annotation as scar-associated mesenchymal cells (SAMes).
Figure 5. Identifying scar-associated mesenchymal cell populations.
a, Clustering 2,318 mesenchymal cells (Mes) from 4 healthy and 3 cirrhotic human livers, annotating injury condition (right). VSMC, vascular smooth muscle cell; HSC, hepatic stellate cell; SAMes, scar-associated mesenchymal cell. b, Heatmap: Mesenchymal cluster marker genes (top, colour coded by cluster and condition), exemplar genes labelled (right). Cells columns, genes rows. c, Representative immunofluorescence images: RGS5 (red), MYH11 (white), PDGFRα (green), DAPI (blue), scale bars 50μm d, Scaled gene expression of fibrillar collagens across mesenchymal cells from healthy (n=4) and cirrhotic (n=3) livers. Meso, mesothelial cell. e, Fraction of mesenchymal subpopulations in healthy (n=4) versus cirrhotic (n=3) livers, Mean±SEM, Wald test. f, PDGFRα immunohistochemistry, digital pixel quantification of healthy (n=11) versus cirrhotic (n=11) liver (top right), Mean±SEM, Mann-Whitney two-tailed. PDGFRα pixel quantification in fibrotic septae and parenchymal nodules in 10 cirrhotic livers (bottom right), Wilcoxon two-tailed, scale bars 50μm.
To study SAMes heterogeneity, further clustering (Extended Data Fig. 9b) identified two populations of SAMes (Extended Data Fig. 9c, d, Supplementary Table 17). OSR1 expression distinguished cluster SAMesB (Extended Data Fig. 9c), and labelled a subpopulation of periportal cells in healthy liver and scar-associated cells in the fibrotic niche (Extended Data Fig. 9e, f). Cluster SAMesB also expressed other known portal fibroblast markers23 (Extended Data Fig. 9g).
In rodent liver fibrosis models, HSC differentiate into scar-producing myofibroblasts 24–26. Pseudotemporal ordering and RNA velocity analyses demonstrated a trajectory from human HSC to SAMes (Extended Data Fig. 9h). Assessment of gene co-expression modules along the HSC-to-SAMes differentiation continuum indicated upregulation of fibrogenic genes including COL1A1, COL1A2, COL3A1, TIMP1 and downregulation of genes including RGS5, IGFBP5, ADAMTS1 and GEM, which are known to be downregulated in murine HSC in response to liver injury25 (Extended Data Fig. 9i).
Resolving the multi-lineage interactome in the fibrotic niche
Having defined the scar-associated macrophage, endothelial and mesenchymal populations, we confirmed the close topographical association of these cells within the fibrotic niche (Extended Data Fig. 10a, b), and used CellPhoneDB27 to perform an unbiased ligand-receptor interaction analysis between these populations.
Numerous statistically significant paracrine and autocrine interactions were detected between ligands and cognate receptors expressed by SAMΦ, SAEndo and SAMes (Supplementary Table 18, Extended Data Fig. 10f-m). To interrogate how scar-associated NPC regulate fibrosis and to identify tractable therapeutic targets, we focused functional analyses on interactions with SAMes (Fig. 6a, e, Extended Data Fig. 10d). In keeping with our data demonstrating that SAMΦ promote fibrillar collagen expression in HSC (Fig. 2g), SAMΦ expressed epidermal growth factor receptor (EGFR) ligands which are known to regulate mesenchymal cell activation28 (Fig. 6a). Additionally, SAMΦ expressed mesenchymal cell mitogens TNFSF12 and PDGFB, signaling to cognate receptors TNFRSF12A and PDGFRA on SAMes (Fig. 6a). We confirmed localization of these ligand-receptor pairs within the fibrotic niche (Fig. 6b). Both TNFSF12 and PDGF-BB induced primary human HSC proliferation, which was inhibited by blockade of TNFSF12A and PDGFRA respectively (Fig. 6c, d). Conditioned media from primary human SAMΦ promoted primary human HSC proliferation ex vivo (Extended Data Fig. 10c), demonstrating a functional role for SAMΦ in regulating SAMes expansion.
Figure 6. Multi-lineage interactions in the fibrotic niche.
a, Dotplot: ligand-receptor interactions between SAMΦ (n=10 human livers) and SAMes (n=7 human livers). X-axis, ligand (red) and cognate receptor (blue); y-axis, cell populations expressing ligand (red) and receptor (blue); circle size, P value (permutation test); colour (red, high; yellow, low), means of average ligand and receptor expression levels in interacting subpopulations. b, Representative immunofluorescence images, fibrotic niche. Left, TREM2 (red), PDGFB (white), PDGFRα (green), DAPI (blue), arrows TREM2+PDGFB+ cells. Right, TNFRSF12A (red), TNFSF12 (white), PDGFRα (green), DAPI (blue), arrows TNFRSF12A+PDGFRα+ cells, scale bars 50μm. c to d, HSC proliferation assay: y-axis, area under curve (AUC) of % change in HSC number over time (hours), Mean±SEM, one-way ANOVA and Tukey. c, Control, TNFSF12, anti-TNFRSF12A, isotype control, all n=3. d, Vehicle, PDGF-BB, Crenolanib, all n=3. e, Dotplot: ligand-receptor interactions between SAEndo (n=7 human livers) and SAMes (n=7 human livers). X-axis, ligand (red) and cognate receptor (blue); y-axis, cell populations expressing ligand (red) and receptor (blue); circle size, P value (permutation test); colour (red, high; yellow, low), means of average ligand and receptor expression levels in interacting subpopulations. f, Representative immunofluorescence image, fibrotic niche. NOTCH3 (red), DLL4 (white), PDGFRα (green), DAPI (blue), arrows NOTCH3+PDGFRα+ cells, scale bar 50μm. g, Endothelial cell JAG1 flow cytometry: healthy (n=3) or cirrhotic (n=9) liver, representative histogram (left), mean fluorescence intensity (MFI, right), Mean±SEM, Mann-Whitney two-tailed. h, Cirrhotic endothelial cell and HSC co-culture, Notch inhibitor Dibenzazepine (DBZ). Representative immunofluorescence images (left), Collagen 1 (magenta), PECAM1 (green), DAPI (blue). Digital pixel analysis (right); collagen 1 area, n=3, Mean±SEM, RM one-way ANOVA and Tukey. n, HSC gene knockdown: control (n=7) or NOTCH3 (n=7) siRNA, qPCR of stated gene, expression relative to mean expression of control siRNA, Mean±SEM, Mann-Whitney two-tailed.
SAEndo expressed high levels of non-canonical Notch ligands JAG1, JAG2 and DLL4 interacting with Notch receptor NOTCH3 on SAMes (Fig. 6e). NOTCH3 was identified on PDGFRα+ SAMes within the fibrotic niche (Fig. 6f), whilst primary endothelial cells from cirrhotic human liver demonstrated increased expression of JAG1 (Fig. 6g). Co-culture of primary human HSC and endothelial cells from cirrhotic livers promoted fibrillar collagen production by HSC, which was inhibited by addition of the Notch-signalling inhibitor Dibenzazepine (DBZ) (Fig. 6h). Furthermore, knockdown of NOTCH3 expression in primary human HSC resulted in reduced fibrillar collagen expression (Fig. 6i).
In summary, our unbiased dissection of the key ligand-receptor interactions between scar-associated NPC highlights TNFRSF12A, PDGFRA and Notch signaling as important regulators of mesenchymal cell function within the human liver fibrotic niche.
Discussion
Here, using scRNA-seq and spatial mapping, we resolve the fibrotic niche of human liver cirrhosis, identifying pathogenic subpopulations of TREM2+CD9+ macrophages, ACKR1+ and PLVAP+ endothelial cells and PDGFRα+ collagen-producing myofibroblasts. We dissect a complex, pro-fibrotic interactome between multiple scar-associated cell lineages and identify highly relevant intra-scar pathways that are potentially druggable. In this era of precision medicine, this unbiased multi-lineage approach should inform the design of highly-targeted combination therapies which will very likely be necessary to achieve effective antifibrotic potency3,4.
Application of our novel scar-associated cell markers could also potentially inform molecular pathology-based patient stratification, which is fundamental to the prosecution of successful antifibrotic clinical trials. Our work illustrates the power of single-cell transcriptomics to decode the cellular and molecular basis of human organ fibrosis, providing a conceptual framework for the discovery of relevant therapeutic targets to treat patients with a broad range of fibrotic diseases.
Methods
Study subjects
Local approval for procuring human liver tissue and blood samples for scRNA-seq, flow cytometry and histological analysis was obtained from the NRS BioResource and Tissue Governance Unit (Study Number SR574), following review at the East of Scotland Research Ethics Service (Reference 15/ES/0094). All subjects provided written informed consent. Healthy background non-lesional liver tissue was obtained intraoperatively from patients undergoing surgical liver resection for solitary colorectal metastasis at the Hepatobiliary and Pancreatic Unit, Department of Clinical Surgery, Royal Infirmary of Edinburgh. Patients with a known history of chronic liver disease, abnormal liver function tests or those who had received systemic chemotherapy within the last four months were excluded from this cohort. Cirrhotic liver tissue was obtained intraoperatively from patients undergoing orthotopic liver transplantation at the Scottish Liver Transplant Unit, Royal Infirmary of Edinburgh. Blood from patients with a confirmed diagnosis of liver cirrhosis were obtained from patients attending the Scottish Liver Transplant Unit, Royal Infirmary of Edinburgh. Patients with liver cirrhosis due to viral hepatitis were excluded from the study. Patient demographics are summarised in Extended Data Fig. 1a. Isolation of primary hepatic macrophage subpopulations and endothelial cells from healthy and cirrhotic livers for cell culture and analysis of secreted mediators was performed at the University of Birmingham, UK. Local ethical approval was obtained (Reference 06/Q2708/11, 06/Q2702/61) and all patients provided written, informed consent. Liver tissue was acquired from explanted diseased livers from patients undergoing orthotopic liver transplantation, resected liver specimens or donor livers rejected for transplant at the Queen Elizabeth Hospital, Birmingham. For histological assessment of NAFLD biopsies, anonymised unstained formalin-fixed paraffin-embedded liver biopsy sections encompassing the complete NAFLD spectrum were provided by the Lothian NRS Human Annotated Bioresource under authority from the East of Scotland Research Ethics Service REC 1, reference 15/ES/0094.
Human tissue processing
Importantly, to minimise artefacts29, we developed a rapid tissue processing pipeline, obtaining fresh non-ischaemic liver tissue taken by wedge biopsy prior to the interruption of the hepatic vascular inflow during liver surgery or transplantation, and immediately processing this for FACS. This enabled a workflow time of under three hours from patient to single-cell droplet encapsulation.
For human liver scRNA-seq and flow cytometry analysis, a wedge biopsy of non-ischaemic fresh liver tissue (2-3 grams) was obtained by the operating surgeon, prior to interruption of the hepatic vascular inflow. This was immediately placed in HBSS (Gibco) on ice. The tissue was then transported directly to the laboratory and dissociation routinely commenced within 20 minutes of the liver biopsy. To enable paired histological assessment, a segment of each liver specimen was also fixed in 4% neutral-buffered formalin for 24 hours followed by paraffin-embedding. Additional liver samples, obtained via the same method, were fixed in an identical manner and used for further histological analysis. For human macrophage cell sorting and endothelial cell isolation, liver tissue (40 grams) was used from cirrhotic patients undergoing orthotopic liver transplantation or control samples from donor liver or liver resection specimens.
Animals
Adult male C57BL/6JCrl mice aged 8-10 weeks were purchased from Charles River. Mice were housed under specific pathogen-free conditions at the University of Edinburgh. All experimental protocols were approved by the University of Edinburgh Animal Welfare and Ethics Board in accordance with UK Home Office regulations. Liver fibrosis was induced with 4 weeks (9 injections) of twice-weekly intraperitoneal carbon tetrachloride (CCl4) at a dose of 0.4 μl/g body weight, diluted 1:3 in olive oil as previously described7. Mice were randomly assigned to receive CCl4 or to serve as healthy controls. No sample size calculation or blinding was performed. Liver tissue was harvested 24 hours following the final CCl4 injection, a time of active fibrogenesis7. Comparison was made to age-matched uninjured mice.
Preparation of single-cell suspensions
For human liver scRNA-seq, liver tissue was minced with scissors and digested in 5mg/ml pronase (Sigma-Aldrich, P5147-5G), 2.93mg/ml collagenase B (Roche, 11088815001) and 1.9mg/ml DNase (Roche, 10104159001) at 37°C for 30 minutes with agitation (200–250 r.p.m.), then strained through a 120μm nybolt mesh along with PEB buffer (PBS, 0.1% BSA, and 2mM EDTA) including DNase (0.02mg/ml). Thereafter all processing was done at 4°C. The cell suspension was centrifuged at 400g for 7 minutes, supernatant removed, cell pellet resuspended in PEB buffer and DNase added (0.02mg/ml), followed by additional centrifugation (400g, 7 minutes). Red blood cell lysis was performed (BioLegend, 420301), followed by centrifugation (400g, 7 minutes), resuspension in PEB buffer and straining through a 35μm filter. Following another centrifugation at 400g for 7 minutes, cells were blocked in 10% human serum (Sigma-Aldrich, H4522) for 10 minutes at 4°C prior to antibody staining.
For both human liver macrophage flow cytometry analysis and cell sorting and mouse liver macrophage flow cytometry, cell sorting and scRNA-seq, single-cell suspensions were prepared as previously described, with minor modifications30. In brief, liver tissue was minced and digested in an enzyme cocktail 0.625 mg/ml collagenase D (Roche, 11088882001), 0.85 mg/ml collagenase V (Sigma-Aldrich, C9263-1G), 1 mg/ml dispase (Gibco, Invitrogen, 17105-041), and 30 U/ml DNase (Roche, 10104159001) in RPMI-1640 at 37°C for 20 minutes (mouse) or 45 minutes (human) with agitation (200–250 r.p.m.), before being passed through a 100μm filter. Following red blood cell lysis (BioLegend, 420301), cells were washed in PEB buffer and passed through a 35μm filter. Before the addition of antibodies, cells from human samples were blocked in 10% human serum (Sigma-Aldrich, H4522) and mouse samples were blocked in anti-mouse CD16/32 antibody (1:100; Biolegend, 101302) and 10% normal mouse serum (Sigma, M5905) for 10 minutes at 4 °C.
For human PBMC scRNA-seq, 4.9ml peripheral venous blood samples were collected in EDTA-coated tubes (Sarstedt, S-Monovette® 4.9ml K3E) and placed on ice. Blood samples were transferred into a 50ml Falcon tube. Following red cell lysis (Biolegend, 420301), blood samples were then centrifuged at 500g for 5 minutes and supernatant was removed. Pelleted samples were then resuspended in staining buffer (PBS plus 2% BSA; Sigma-Aldrich) and centrifugation was repeated. Samples were then blocked in 10% human serum (Sigma-Aldrich, H4522) in staining buffer on ice for 30 minutes. Cells were then resuspended in staining buffer and passed through a 35μm filter prior to antibody staining.
Flow cytometry and cell sorting
Incubation with primary antibodies was performed for 20 minutes at 4°C. All antibodies, conjugates, lot numbers and dilutions used in this study are presented in Supplementary Table 19. Following antibody staining, cells were washed with PEB buffer. For both human macrophage flow cytometry analysis and cell sorting, cells were then incubated with streptavidin-BV711 for 20 minutes at 4°C (Biolegend 405241; Dilution 1:200). For human and mouse cell sorting (FACS) and mouse flow cytometry analysis, cell viability staining (DAPI; 1:1000 dilution) was then performed, immediately prior to acquiring the samples.
Human cell sorting for scRNA-seq was performed on a BD Influx (Becton Dickinson, Basel, Switzerland). Viable single CD45+ (leucocytes) or CD45- (other non-parenchymal cells) cells were sorted from human liver tissue (Extended Data Fig. 1b) and viable CD45+ CD66b- (PBMC) cells were sorted from peripheral blood (Extended Data Fig. 1c) and processed for droplet-based scRNA-seq.
To generate conditioned media from cirrhotic liver macrophage subpopulations, cells were sorted on a BD FACSAria™ Fusion (Becton Dickinson, Basel, Switzerland). Sorted SAMΦ (viable CD45+Lin-HLA-DR+CD14+CD16+CD163-TREM2+CD9+), TMo (viable CD45+Lin- HLA-DR+CD14+CD16+CD163-TREM2-CD9-) and KC (viable CD45+Lin-HLA-DR+CD14+CD16+CD163+CD9-) were plated in 12-well plates (Corning, 3513) in DMEM (Gibco, 41965039) containing 2% FBS (Gibco, 10500056) at 1x106 cells/ml for 24 hours at 37°C 5%CO2. Control wells contained media alone. Conditioned media was collected, centrifuged at 400g for 10 minutes and supernatant stored at -80°C.
For human macrophage flow cytometry analysis, following surface antibody staining, cells were stained with Zombie NIR fixable viability dye (Biolegend, 423105) according to manufacturers’ instructions. Cells were washed in PEB then fixed in IC fixation buffer (Thermo-Fisher, 00-8222-49) for 20 minutes at 4°C. Fixed samples were stored in PEB at 4°C until acquisition. Flow cytometry acquisition was performed on 6-laser Fortessa flow cytometer (Becton Dickinson, Basel, Switzerland). The gating strategy is shown (Extended Data Fig. 4f, Fig. 2f).
Mouse macrophage cell sorting for scRNA-seq and co-culture experiments was performed on a BD FACSAriaII (Becton Dickinson, Basel, Switzerland). For scRNA-seq, viable CD45+ Lin(CD3, NK1.1, Ly6G, CD19)- cells were sorted from healthy (n=3) and CCl4-treated (n=3) mice and processed for droplet-based scRNA-seq. For transwell co-culture, viable CD45+ Lin- CD11b+ F4/80+ TIMD4- CD9+ (mSAMΦ) or CD9- (mTMo) cells were sorted from CCl4-treated mice (Extended Data Fig. 6e). Flow cytometry analysis on macrophages from healthy and CCl4-treated mice was also performed on a BD FACSAriaII (Becton Dickinson, Basel, Switzerland), using the same gating strategy (Extended Data Fig. 6e). All flow cytometry data was analysed using Flowjo software (Treestar, Ashland, TN).
Luminex Assay
Detection of CCL2, Galectin-3, IL-1β, CXCL8 and Osteopontin (SPP1) and CD163 proteins in conditioned media from human liver macrophage subpopulations was performed using a custom human luminex assay (R&D systems), according to the manufacturers protocol. Data was acquired using a Bio-Plex® 200 (Bio-Rad, UK) and is presented a median fluorescence intensity (MFI) for each analyte.
Cell Culture
Human hepatic stellate cell activation
Primary human hepatic stellate cells (HSC) were purchased (ScienCell, 5300) and cultured in stellate cell medium (SteCM, ScienCell, 5301) on Poly-L-Lysine (Sigma, P4832) coated T75 tissue culture flasks, according to the suppliers protocol. All experiments were performed using cells between passage 3 and 5. For assessment of fibrillar collagen gene expression, HSC were plated at 75,000 cells per well in 24 well-plates (Costar, 3524) in HSC media consisting of DMEM (Gibco, 21969-035) supplemented with 20 µM HEPES (Sigma, H3375,), 2 mM L-Glutamine (Gibco, 25030-024), 1% Penicillin Streptomycin (Gibco, 15140-122, Gibco) and 2% Foetal Bovine Serum (FBS, Gibco, 10270). HSC were serum starved overnight (in HSC media without FBS), washed with PBS, then 250μl of conditioned media from primary human macrophage subpopulations added for 24 hours. HSC were harvested for RNA.
Human hepatic stellate cell proliferation
For proliferation assays, following serum starvation HSC were harvested using TrypLE Express (Gibco, Cat. no. 12604013), re-suspended in HSC media at 2.5x104/ml with Incucyte NucLight Rapid Red (Essen Biosciences, 4717) at a dilution of 1 in 500 and seeded into 384-well plates (Greiner Bio-One, 781090) at 25μl per well. HSC were then treated with control media or (i) PDGF-BB (10ng/ml; Peprotech, 100-14B) or TNFSF12 (100ng/ml; Peprotech, 310-06-5) with or without the PDGFRα inhibitor Crenolanib31 (1μM; Cayman chemicals, CAY1873), anti-TNRSF12A (2μg/ml; Life Technologies, 16-9018-82, clone ITEM-4), mouse IgG2b kappa isotype control antibody (2μg/ml; Life Technologies, 16-4732-82, clone eBMG2b) or vehicle control as indicated or (ii) conditioned media from human hepatic macrophage subpopulations as indicated. The final volume was 50μl for all conditions. Cells were then incubated in an Incucyte ZOOM live cell analysis system (Essen Biosciences) humidified at 37°C with 5% CO2 with imaging every 3 hours using the 10x optic for either 87 hours (recombinant cytokines/inhibitors) or 39 hours (macrophage conditioned media). Analysis was performed with the Incucyte proprietary analysis software (version 2018A) by using machine learning to distinguish the individual nuclei (stained red by the NucLight Rapid Red dye) and perform nuclear counts of the images at each 3 hour time point over the period of culture. Data are expressed as area under curve (AUC) for % change in nuclear number from baseline versus time (hours), calculated in GraphPad Prism (GraphPad Software, USA).
Gene knockdown in human hepatic stellate cells
Knockdown of NOTCH3 in human HSC was performed using siRNA. HSC were plated at 75,000 cells per well in a 12 well plate (Costar, 3513) followed by serum starvation overnight (in HSC medium without FBS). siRNA duplexes with Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher, 13778075) were prepared in OptiMEM (ThermoFisher, 31985070) according to the manufacturer’s recommendations, and used at a concentration of 50nM. Cells were exposed to the duplex for 48 hours, in HSC media containing 2% FBS. Cells were harvested for RNA and RT-qPCR. Knockdown efficiency was assessed by NOTCH3 RT-qPCR. The best siRNA for knockdown was determined empirically using the FlexiTube GeneSolution kit (Qiagen, GS4854). HSC treated with control siRNA (Qiagen, 1027280) and siRNA for NOTCH3 (Qiagen, Hs_NOTCH3_3, SI00009513; knockdown 83%) were then assessed for fibrillar collagen gene expression.
Mouse hepatic stellate cell activation
Primary murine HSC culture were isolated from healthy mice as described26. Briefly, after cannulation of the inferior vena cava, the portal vein was cut to allow retrograde step-wise perfusion with pronase (Sigma, P5147) and collagenase D (Roche, 11088882001) containing solutions, before ex vivo digestion in a solution containing pronase, collagenase D and Dnase1 (Roche, 10104159001). HSC were isolated from the digest solution by Histodenz (Sigma, D2158-100G) gradient centrifugation. HSC were plated at a density of 400,000 cells per well in a 24 well plate (Costar, 3524) in HSC media containing 10% FBS. Following overnight culture, cells were washed with PBS and cultured in HSC media containing 2% FBS. For macrophage co-culture, transwell inserts (0.4μm polyester membrane; Costar, 3470) were then placed above adherent HSC. FACS-sorted CD9+ mSAMΦ or CD9- mTMo from CCl4-treated mice were resuspended in HSC media containing 2% FBS at 400,000 cells/ml and 200,000 cells added to the top of the transwell insert. Co-culture proceeded for 48 hours and HSC were harvested for RNA. Quiescent HSC (harvested at start of co-culture) were used as a control population.
Human liver endothelial cell isolation
Human liver endothelial cells (LEC) were isolated from cirrhotic explant livers and non-fibrotic control donor liver as previously described32. Endothelial cells were cultured on plasticware coated with rat-tail collagen (Sigma, C3867) in complete LEC medium consisting of endothelial basal media (ThermoFisher, 11111044) containing 10% heat inactivated human serum (tcsBiosciences, CS100-500), 100U penicillin, 100 µg/mL streptomycin, 2mM glutamine (Sigma, G6784), VEGF (10 ng/mL; Peprotech, 100-20) and 10 ng/mL HGF (10 ng/mL; Peprotech, 100-39). LEC expression of PLVAP, CD34, ACKR1 and JAG1 was assessed using flow cytometry.
Flow-based adhesion assays
Flow-based adhesion assays were performed as described22,32. Briefly, LEC from healthy and cirrhotic liver were seeded into a rat-tail collagen coated Ibidi slide VI0.4 (Ibidi, 80606) at a density to give a monolayer and incubated overnight. Peripheral blood was collected from healthy donors in EDTA-coated tubes. Peripheral blood monocuclear cells (PBMC) were isolated using a lympholyte density gradient (Cedarlane laboratories) then washed in PBS containing 1mM Ca2+, 0.5 mM Mg2+ and 0.15% bovine serum albumin (PBS/BSA). Monocytes were enriched from PBMC using pan-monocyte isolation kit (Miltenyi biotech, 130-096-537) according to the manufacturer’s protocol. For flow-based adhesion assay, cells were resuspended at 106 cells per millilitre in endothelial basal media (ThermoFisher, 11111044) containing 0.15% BSA, then perfused over the LEC monolayer for 5min at 0.28ml/min. Non-adherent cells were washed off during 5min perfusion of 0.15% BSA human basal endothelial medium and 10 random non-overlapping images were randomly recorded from each channel. Total adherent (bright-phase; expressed as cell number/mm2/1 million cells perfused) and transmigrating cells (dark-phase; expressed as percentage total adherent cells) on an LEC monolayer from each patient were counted and quantified as previously described22.
Gene knockdown in endothelial cells
Knockdown of ACKR1 and PLVAP gene expression in human cirrhotic LEC was performed using siRNA as previously described32. In brief, siRNA duplexes for PLVAP, ACKR1, or negative control (Qiagen, 1027280) with Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher, 13778075) were prepared in OptiMEM (ThermoFisher, 31985070) according to the manufacturer’s recommendations, and used at a concentration of 25nM. Cells were exposed to the duplex for 4 hours at 37°C after which time the media was replaced with endothelial basal media containing 10% heat-inactivated human serum for 24 hours. The media was then replaced with complete LEC media and incubated at 37°C with 5% CO2 for a further 24 hours. Knockdown efficacy was assessed by flow cytometry and mean fluorescence intensity (Extended Data Fig. 8f). The best siRNA for knockdown was determined empirically using the FlexiTube GeneSolution kit (Qiagen, GS83483 (PLVAP) and GS2532 (ACKR1)). For flow-based adhesion assays, siRNAs for PLVAP (Qiagen, Hs_PLVAP_1, SI00687547; knockdown 50.6%), ACKR1 (Qiagen, Hs_Fy_5, SI02627667; knockdown 37.7%) or control siRNA were selected. 90,000 LEC from cirrhotic patients (n=6) were seeded into channels of a rat-tail collagen coated Ibidi slide VI0.4, gene knockdown performed, followed by flow-based adhesion assay as described above.
Endothelial and hepatic stellate cell co-culture
HSC (15,000 cells) were seeded into an Ibidi slide VI0.4 with and without primary human LEC (15,000 cells) from individual cirrhotic patients (n=3) in LEC complete medium. After 2h, all growth factor supplements were removed and cells were cultured for a further 72 hours in endothelial basal media containing 10% heat-inactivated human serum ± Notch-signalling inhibitor Dibenzazepine (10μM, Bio-Techne, 4489/10) or vehicle (DMSO) control. Cells were fixed in 4% PFA for 30 minutes, permeabilised with 0.3% Triton PBS for 5 minutes, blocked with 10% goat serum in PBS for 30 minutes followed by primary antibody incubation (mouse anti-PECAM1 and rabbit anti-collagen 1; see Supplementary Table 19) for 1 hour. Cells were washed in 0.1% Triton PBS followed by addition of fluorescently-conjugated secondary antibodies (1:500 dilution) for 1 hour. Cells were mounted with Pro-long Gold anti-fade DAPI, images were taken on the Confocal Microscope Zeiss LSM780, and collagen 1 area staining quantified using IMARIS.
RNA extraction and RT-qPCR
RNA was isolated from HSC using the RNeasy Plus Micro Kit (Qiagen, 74034) and cDNA synthesis performed using QuantiTect Reverse Transcription Kit (Qiagen, 205313) according to the manufacturers’ protocol. Reactions were performed in triplicate in 384-well plate format and were assembled using the QIAgility automated pipetting system (Qiagen). RT-qPCR for human HSC was performed using PowerUp SYBR Green Master Mix (ThermoFisher, A25777) with the following primers (all Qiagen): GAPDH (QT00079247), COL1A1 (QT00037793), COL3A1 (QT00058233), NOTCH3 (QT00003374). RT-qPCR for mouse HSC was performed using TaqMan Fast Advanced Master Mix (ThermoFisher, 4444557) with the following primers: Gapdh (ThermoFisher, Mm99999915_g1) and Col3a1 (ThermoFisher, Mm00802300_m1). Samples were amplified on an ABI 7900HT FAST PCR system (Applied Biosystems, ThermoFisher Scientific). Data was analysed using ThermoFisher Connect cloud qPCR analysis software (ThermoFisher Scientific). The 2−ΔΔCt quantification method, using GAPDH for normalization, was used to estimate the amount of target mRNA in samples, and expression calculated relative to average mRNA expression levels from control samples.
Immunohistochemistry, immunofluorescence, smFISH
Formalin-fixed paraffin-embedded human liver tissue was cut into 4 μm sections, dewaxed, rehydrated, then incubated in 4% neutral-buffered formalin for 20 minutes. Following heat-mediated antigen retrieval in pH6 sodium citrate (microwave; 15 minutes), slides were washed in PBS and incubated in 4% hydrogen peroxide for 10 minutes. Slides were then washed in PBS, blocked using protein block (GeneTex, GTX30963) for 1 hour at room temperature before incubation with primary antibodies for 1 hour at room temperature. A full list of primary antibodies and conditions are shown in Supplementary Table 19. Slides were washed in PBST (PBS plus 0.1% Tween20; Sigma-Aldrich, P1379) then incubated with ImmPress HRP Polymer Detection Reagents (depending on species of primary; rabbit, MP-7401; mouse, MP-6402-15; goat, MP-7405; all Vector Laboratories) for 30 minutes at room temperature. Slides were washed in PBS followed by detection. For DAB staining, sections were incubated with DAB (DAKO, K3468) for 5 minutes and washed in PBS before a haematoxylin (Vector Laboratories, H3404) counterstain. For multiplex immunofluorescence staining, following the incubation with ImmPress and PBS wash, initial staining was detected using either Cy3, Cy5, or Fluorescein tyramide (Perkin-Elmer, NEL741B001KT) at a 1:1000 dilution. Slides were then washed in PBST followed by further heat treatment with pH6 sodium citrate (15 minutes), washes in PBS, protein block, incubation with the second primary antibody (incubated overnight at 4°C), ImmPress Polymer and tyramide as before. This sequence was repeated for the third primary antibody (incubated at room temperature for 1 hour) and a DAPI-containing mountant was then applied (ThermoFisher Scientific, P36931).
For AMEC Staining (only CLEC4M immunohistochemistry), all washes were carried out with TBST (dH2O, 2oomM Tris, 1.5M NaCl, 1% Tween20 (all Sigma-Aldrich) pH7.5) and peroxidase blocking was carried out for 30mins in 0.6% hydrogen peroxide in Methanol. Sections were incubated with AMEC (Vector Laboratories, SK-4285) for 20 minutes and washed in TBST (dH2O, 200mM Tris, 1.5M NaCl, 1% Tween20 (all Sigma-Aldrich)) before a haematoxylin (Vector Laboratories, SK-4285) counterstain.
For combined single-molecule fluorescent in situ hybridization (smFISH) and immunofluorescence, detection of TREM2 was performed using the RNAscope® 2.5 LS Reagent Kit - BrownAssay (Advanced Cell Diagnostics (ACD)) in accordance with the manufacturer’s instructions. Briefly, 5 μm tissue sections were dewaxed, incubated with endogenous enzyme block, boiled in pretreatment buffer and treated with protease, followed by target probe hybridization using the RNAscope® LS 2.5 Hs-TREM2 (420498, ACD) probe. Target RNA was then detected with Cy3 tyramide (Perkin-Elmer, NEL744B001KT) at 1:1000 dilution. The sections were then processed through a pH6 sodium citrate heat-mediated antigen retrieval, hydrogen peroxidase treatment and protein block (all as for multiplex immunofluorescence staining as above). MNDA antibody was applied overnight at 4°C, completed using a secondary ImmPress HRP Anti-Rabbit Peroxidase IgG (Vector Laboratories, MP7401), visualised using a Flourescein tyramide (Perkin-Elmer, NEL741B001KT) at a 1:1000 dilution and stained with DAPI.
Brightfield and fluorescently-stained sections were imaged using the slide scanner AxioScan.Z1 (Zeiss) at 20X magnification (40X magnification for smFISH). Images were processed and scale bars added using Zen Blue (Zeiss) and Fiji software33.
Cell counting and image analysis
Automated cell counting was performed using QuPath software34. Briefly, DAB-stained whole tissue section slide-scanned images (CZI files) were imported into QuPath. Cell counts were carried out using the positive cell detection tool, detecting haematoxylin-stained nuclei and then thresholding for positively-stained DAB cells, generating DAB-positive cell counts/mm2 tissue. Identical settings and thresholds were applied to all slides for a given stain and experiment. For cell counts of fibrotic septae vs parenchymal nodules, the QuPath segmentation tool was used to segment the DAB-stained whole tissue section into fibrotic septae or non-fibrotic parenchymal nodule regions using tissue morphological characteristics (Fig. 2j). Positive cell detection was then applied to the fibrotic and non-fibrotic regions in turn, providing cell DAB-positive cell counts/mm2 in fibrotic septae and non-fibrotic parenchymal nodules for each tissue section.
Digital morphometric pixel analysis was performed using the Trainable Weka Segmentation (TWS) plugin35 in Fiji software33. Briefly, each stained whole tissue section slide-scanned image was converted into multiple TIFF files in Zen Blue software (Zeiss). TIFF files were imported into Fiji and TWS plugin trained to produce a classifier which segments images into areas of positive staining, tissue background and white space35. The same trained classifier was then applied to all TIFF images from every tissue section for a particular stain, providing a percentage area of positive staining for each tissue section. For digital morphometric quantification of positive staining of fibrotic septae versus parenchymal nodules, TIFF images were segmented into fibrotic septae or non-fibrotic parenchymal nodule regions using tissue morphological characteristics, followed by analysis using the TWS plugin in Fiji software.
Histological assessment of NASH sections
Haematoxylin and eosin and picro-sirius red stained sections from each case were whole-slide imaged using a NanoZoomer imager (Hamamatsu Photonics, Japan). Images of stained sections were independently scored by a consultant liver transplant histopathologist (T.J.K.) at the national liver transplant centre with experience in trial scoring by applying the ordinal NAFLD activity score36. For observer-independent quantification of picro-sirius red positive staining, images were split using ndpisplit37 into tiles of x5 magnification before the application of a classifier that had been trained by the liver histopathologist using the machine learning WEKA plugin in FIJI33,35, as previously described38. All analysis was undertaken blind to all other data.
Droplet-based scRNA-seq
Single cells were processed through the Chromium™ Single Cell Platform using the Chromium™ Single Cell 3’ Library and Gel Bead Kit v2 (10X Genomics, PN-120237) and the Chromium™ Single Cell A Chip Kit (10X Genomics, PN-120236) as per the manufacturer’s protocol. In brief, single cells were sorted into PBS + 0.1% BSA, washed twice and counted using a Bio-Rad TC20. 10,769 cells were added to each lane of the 10X chip. The cells were then partitioned into Gel Beads in Emulsion in the Chromium™ instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, fragmentation and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina HiSeq 4000.
Computational Analysis
In total, we analysed 67,494 human cells from healthy (n=5) and cirrhotic (n=5) livers, 30,741 PBMCs from cirrhotic patients (n=4) and compared our data with a publicly-available reference dataset of 8,381 PBMCs from a healthy donor.
Pre-processing scRNA-seq data
We aligned to the GRCh38 and mm10 (Ensembl 84) reference genomes as appropriate for the input dataset, and estimated cell-containing partitions and associated UMIs, using the Cell Ranger v2.1.0 Single-Cell Software Suite from 10X Genomics. Genes expressed in fewer than three cells in a sample were excluded, as were cells that expressed fewer than 300 genes or mitochondrial gene content >30% of the total UMI count. We normalised by dividing the UMI count per gene by the total UMI count in the corresponding cell and log-transforming. Variation in UMI counts between cells was regressed according to a negative binomial model, prior to scaling and centering the resulting value by subtracting the mean expression of each gene and dividing by its standard deviation (En), then calculating ln(104*En+1).
Dimensionality reduction, clustering, and DE analysis
We performed unsupervised clustering and differential gene expression analyses in the Seurat R package v2.3.039. In particular we used SNN graph-based clustering, where the SNN graph was constructed using from 2 to 11 principal components as determined by dataset variability shown in principal components analysis (PCA); the resolution parameter to determine the resulting number of clusters was also tuned accordingly. To assess cluster similarity we used the BuildClusterTree function from Seurat.
In total, we present scRNA-seq data from ten human liver samples (named Healthy 1-5 and Cirrhotic 1-5), five human blood samples (n=4 cirrhotic named Blood 1-4 and n=1 healthy named PBMC8K; pbmc8k dataset sourced from single-cell gene expression datasets hosted by 10X Genomics), and two mouse liver samples (n=3 uninjured and n=3 fibrotic). For seven human liver samples (Healthy 1-4 and Cirrhotic 1-3) we performed scRNA-seq on both leucocytes (CD45+) and other non-parenchymal cells (CD45-); for the remaining three human livers (Healthy 5, Cirrhotic 4-5) we performed scRNA-seq on leucocytes only (Extended Data Fig. 2e, f).
Initially, we combined all human scRNA-seq datasets (liver and blood) and performed clustering analysis with the aim of isolating a population of liver-resident cells, by identifying contaminating circulatory cells within datasets generated from liver digests and removing them from downstream analysis. Specifically, we removed from our liver datasets cells that fell into clusters 1 and 13 of the initial dataset in Extended Data Fig. 1d.
Using further clustering followed by signature analysis, we interrogated this post-processed liver-resident cell dataset for robust cell lineages. These lineages were isolated into individual datasets, and the process was iterated to identify robust lineage subpopulations. At each stage of this process we removed clusters expressing more than one unique lineage signature in more than 25% of their cells from the dataset as probable doublets. This resulted in removal of 1,351 cells. Where the cell proliferation signature identified distinct cycling subpopulations, we re-clustered these again to ascertain the identity of their constituent cells.
The murine scRNA-seq datasets were combined, clustered, and interrogated for cell lineages in a similar manner as their human counterparts.
All heatmaps, t-SNE and UMAP visualisations, violin plots, and dot plots were produced using Seurat functions in conjunction with the ggplot2, pheatmap, and grid R packages. t-SNE and UMAP visualisations were constructed using the same number of principal components as the associated clustering, with perplexity ranging from 30 to 300 according to the number of cells in the dataset or lineage. We conducted differential gene expression analysis in Seurat using the standard AUC classifier to assess significance. We retained only those genes with a log-fold change of at least 0.25 and expression in at least 25% of cells in the cluster under comparison.
Defining cell lineage signatures
For each cell we obtained a signature score across a curated list of known marker genes per cell lineage in the liver (Supplementary Table 2). This signature score was defined as the geometric mean of the expression of the associated signature genes in that cell. Lineage signature scores were scaled from 0 to 1 across the dataset, and the score for each cell with signature less than a given threshold (the mean of said signature score across the entire dataset) was set as 0.
Batch effect and quality control
To investigate agreement between samples we extracted the average expression profile for a given cell lineage in each sample, and calculated the Pearson correlation coefficients between all possible pairwise comparisons of samples per lineage40.
Imputing dropout in T cell and ILC clusters
To impute dropout of low-abundance transcripts in our T cell and ILC clusters so that we might associate them with known subpopulations, we down-sampled to 7,380 cells from 36,900 and applied the scImpute R package v0.0.841, using as input both our previous annotation labels and k-means spectral clustering (k=5), but otherwise default parameters.
Analysing functional phenotypes of scar-associated cells
For further analysis of function we adopted the self-organising maps (SOM) approach as implemented in the SCRAT R package v1.0.042. For each lineage of interest we constructed a SOM in SCRAT using default input parameters and according to its clusters. We defined the signatures expressed in a cell by applying a threshold criterion (ethresh = 0.95 × emax) selecting the highest-expressed metagenes in each cell, and identified for further analysis those metagene signatures defining at least 30% of cells in at least one cluster within the lineage. We smoothed these SOMs using the disaggregate function from the raster R package for visualisation purposes, and scaled radar plots to maximum proportional expression of the signature. Gene ontology enrichment analysis on the genes in these spots was performed using PANTHER 13.1 (pantherdb.org).
Inferring injury dynamics and transcriptional regulation
To generate cellular trajectories (pseudotemporal dynamics) we used the monocle R package v2.6.143. We ordered cells in a semi-supervised manner based on their Seurat clustering, scaled the resulting pseudotime values from 0 to 1, and mapped them onto either the t-SNE or UMAP visualisations generated by Seurat or diffusion maps as implemented in the scater R package v1.4.044 using the top 500 variable genes as input. We removed mitochondrial and ribosomal genes from the geneset for the purposes of trajectory analysis. Differentially-expressed genes along this trajectory were identified using generalised linear models via the differentialGeneTest function in monocle.
When determining significance for differential gene expression along the trajectory, we set a q-value threshold of 1e-20. We clustered these genes using hierarchical clustering in pheatmap, cutting the tree at k=3 to obtain gene modules with correlated gene expression across pseudotime. Cubic smoothing spline curves were fitted to scaled gene expression along this trajectory using the smooth.spline command from the stats R package, and gene ontology enrichment analysis again performed using PANTHER 13.1.
We verified the trajectory and its directionality using the velocyto R package v0.6.017, estimating cell velocities from their spliced and unspliced mRNA content. We generated annotated spliced and unspliced reads from the 10X BAM files via the dropEst pipeline, before calculating gene-relative velocity using kNN pooling with k=25, determining slope gamma with the entire range of cellular expression, and fitting gene offsets using spanning reads. Aggregate velocity fields (using Gaussian smoothing on a regular grid) and transition probabilities per lineage subpopulations were visualised on t-SNE, UMAP, or diffusion map visualisations as generated previously. Gene-specific phase portraits were plotted by calculating spliced and unspliced mRNA levels against steady-state inferred by a linear model; levels of unspliced mRNA above and below this steady-state indicate increasing and decreasing expression of said gene, respectively. Similarly we plotted unspliced count signal residual per gene, based on the estimated gamma fit, with positive and negative residuals indicating expected upregulation and downregulation respectively.
For transcription factor analysis, we obtained a list of all genes identified as acting as transcription factors in humans from AnimalTFDB45. To further analyse transcription factor regulons, we adopted the SCENIC v0.1.7 workflow in R46, using default parameters and the normalised data matrices from Seurat as input. For visualisation, we mapped the regulon activity (AUC) scores thus generated to the pseudotemporal trajectories from monocle and the clustering subpopulations from Seurat.
Analysing inter-lineage interactions within the fibrotic niche
For comprehensive systematic analysis of inter-lineage interactions within the fibrotic niche we used CellPhoneDB27. CellPhoneDB is a manually curated repository of ligands, receptors, and their interactions, integrated with a statistical framework for inferring cell-cell communication networks from single-cell transcriptomic data. In brief, we derived potential ligand-receptor interactions based on expression of a receptor by one lineage subpopulation and a ligand by another; as input to this algorithm we used cells from the fibrotic niche as well as liver sinusoidal endothelial cells and Kupffer cells as control, and we considered only ligands and receptors expressed in greater than 5% of the cells in any given subpopulation. Subpopulation-specific interactions were identified as follows: 1) randomly permuting the cluster labels of all cells 1000 times and determining the mean of the average receptor expression of a subpopulation and the average ligand expression of the interacting subpopulation, thus generating a null distribution for each ligand-receptor pair in each pairwise comparison between subpopulations, 2) calculating the proportion of these means that were "as or more extreme" than the actual mean, thus obtaining a p-value for the likelihood of subpopulation specificity for a given ligand-receptor pair, 3) prioritising interactions that displayed specificity to subpopulations interacting within the fibrotic niche.
Canonical correlation analysis
To compare human and murine populations of monocytic phagocytes, we used canonical correlation analysis as implemented in Seurat18. We map the genes in the human dataset to their murine orthologues using biomaRt, discarding any genes for which no orthologues can be found. We then calculate the shared low-dimensional subspace on the union of genes that are variably expressed in both datasets (n=159), and align using six canonical components as determined by evaluating the biweight midcorrelation. Results are visualised on t-SNEs as previously described.
Deconvolution of whole liver microarray data
To assess macrophage composition early-stage NAFLD, we performed deconvolution analysis on publicly available microarray data from annotated liver biopsy specimens taken across the NAFLD disease spectrum (GEO accession GSE48452)20. Tissue MP cells from our human scRNA-seq data were manually clustered into the main annotated MP populations. Signature gene expression profiles of SAMΦ, TMo, KC were used to deconvolve the monocyte-macrophage composition of liver biopsy samples from GSE48452 using Cibersort47, as previously described19. The monocyte-macrophage composition of each biopsy sample was then compared to the associated histological and demographic features, available from the GEO database.
Statistics and Reproducibility
To assess whether our identified subpopulations were significantly overexpressed in injury, we posited the proportion of injured cells in each cluster as a random count variable using a Poisson process, as previously described40. We modelled the rate of detection using the total number of cells in the lineage profiled in a given sample as an offset, with the condition of each sample (healthy vs cirrhotic) provided as a covariate factor. The model was fitted using the R command glm from the stats package. The P value for the significance of the proportion of injured cells was assessed using a Wald test on the regression coefficient.
Remaining statistical analyses were performed using GraphPad Prism (GraphPad Software, USA). Comparison of changes between two groups was performed using a Mann-Whitney test (unpaired; two-tailed) or using a Wilcoxon matched-pairs signed rank test (paired; two-tailed). Comparison of changes between multiple groups was performed using a Kruskal-Wallis and Dunn, one-way ANOVA and Tukey or repeated measures one-way ANOVA and Tukey tests. Correlations were preformed using Pearson correlation and best fit line plotted using linear regression. P values<0.05 were considered statistically significant. All immunofluorescence stains were repeated in a minimum of 3 patients and representative images are displayed.
Extended Data
Extended Data Figure 1. Strategy for isolation of human liver non-parenchymal cells.
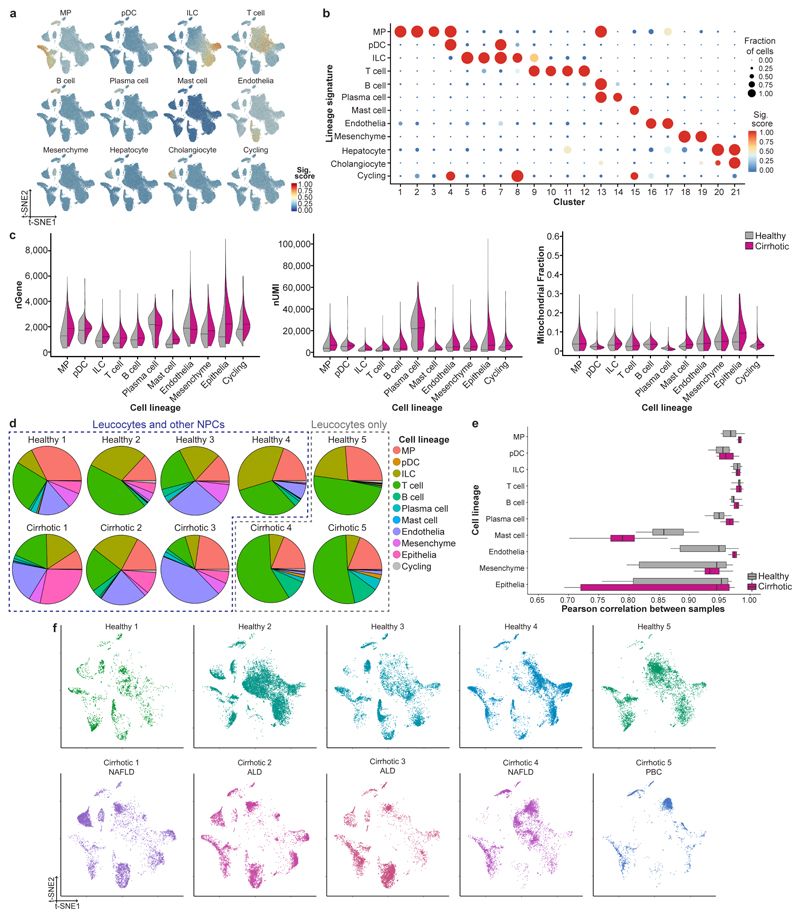
a, Patient demographics and clinical information, Mean±SEM. b, Flow cytometry gating strategy for isolating leucocytes (CD45+) and other non-parenchymal cells (CD45-) from human liver, representative plots from 10 livers. c, Flow cytometry plots: gating strategy for isolating peripheral blood mononuclear cells (PBMC), representative plots from 4 patients. d, Clustering 103,568 cells from 5 healthy human livers, 5 cirrhotic human livers, 1 healthy PBMC and 4 cirrhotic PBMC (left), annotating source (PBMC versus liver; middle) and cell lineage inferred from known marker gene signatures (right). Endo, endothelial cell; ILC, innate lymphoid cell; Mast, mast cell; Mes, mesenchymal cell; MP, mononuclear phagocyte; pDC, plasmacytoid dendritic cell. e, Dotplot: annotating PBMC and liver clusters by lineage signatures. Circle size indicates cell fraction expressing signature greater than mean; colour indicates mean signature expression (red, high; blue, low). f, CXCR4 gene expression in single cells derived from blood or liver tissue, divided by cell lineage. Representative immunofluorescence micrograph, human liver: CXCR4 (green), DAPI (blue), arrows CXCR4- cells in the lumen of a blood vessel. Scale bar 50μm. g, Violin plots: number of unique genes (nGene), number of total Unique Molecular Identifiers (nUMI) and mitochondrial gene fraction expressed in 5 PBMC samples; Black line, median. g, Pie charts: proportion of cell lineage per PBMC sample. h, Box and whisker plot: agreement in expression profiles across 5 PBMC samples. Pearson correlation coefficients between average expression profiles for cell in each lineage, across all pairs of samples. Black bar, median value; box edges, 25th and 75th percentiles; whiskers, full range.
Extended Data Figure 2. Quality control and annotation of human liver-resident cells.
a, Lineage signature expression across 66,135 liver-resident cells from 5 healthy and 5 cirrhotic human livers (red, high; blue, low). b, Dotplot: annotating liver-resident cell clusters by lineage signature. Circle size indicates cell fraction expressing signature greater than mean; colour indicates mean signature expression (red, high; blue, low). c, Violin plot: number of unique genes (nGene; left), number of total Unique Molecular Identifiers (nUMI; middle) and mitochondrial gene fraction (right) across 66,135 liver-resident cells from 5 healthy versus 5 cirrhotic livers; Black line, median. d, Pie charts: proportion of cell lineage per liver sample. e, Box and whisker plot: agreement in expression profiles across 5 healthy and 5 cirrhotic liver samples. Pearson correlation coefficients between average expression profiles for cell in each lineage, across all pairs of samples. Black bar, median value; box edges, 25th and 75th percentiles; whiskers, range. f, t-SNE visualisation: liver-resident cells per liver sample; Cirrhotic samples annotated by aetiology of underlying liver disease; NAFLD, Non-alcoholic fatty liver disease; ALD, Alcohol-related liver disease; PBC, Primary biliary cholangitis.
Extended Data Figure 3. Annotating human liver lymphoid cells.
a, Clustering and annotating 36,900 T cells and innate lymphoid cells (ILC) (left) from 5 healthy and 5 cirrhotic human livers, annotating injury condition (right). cNK, cytotoxic NK cell. b, Fractions of T cell and ILC subpopulations in healthy (n=5) versus cirrhotic (n=5) livers, Mean±SEM, Wald. c, Selected gene expression in 36,900 T cells and ILC. d, Heatmap: T cell and ILC cluster marker genes (colour coded top by cluster and condition), exemplar genes labelled right. Cells columns, genes rows. e, t-SNE visualisations: downsampled T cell and ILC dataset (7,380 cells from 5 healthy and 5 cirrhotic human livers) pre- and post-imputation; annotating data used for visualisation and clustering, inferred lineage, injury condition. No additional heterogeneity was observed following imputation. f, Clustering 2,746 B cells and plasma cells (left) from 5 healthy and 5 cirrhotic human livers, annotating injury condition (right). g, Heatmap: B cell and plasma cell cluster marker genes (colour coded top by cluster and condition), exemplar genes labelled right. Cells columns, genes rows. h, Fractions of B cell and plasma cell subpopulations in healthy (n=5) versus cirrhotic (n=5) livers, Mean±SEM.
Extended Data Figure 4. Annotating human liver mononuclear phagocytes.
a, Clustering and selected genes expressed in 10,737 mononuclear phagocytes (MP) from 5 healthy and 5 cirrhotic human livers. b, Scaled gene expression of Kupffer cell (KC) cluster markers across MP cells from healthy (n=5) and cirrhotic (n=5) livers. c, Representative immunofluorescence images, healthy versus cirrhotic liver: TIMD4 (red), CD163 (white), MARCO (green), DAPI (blue), arrows CD163+MARCO+TIMD4- cells, scale bars 50μm. d, TIMD4 immunohistochemistry, cell counts healthy (n=12) versus cirrhotic (n=9) liver, Mean±SEM, scale bars 50μm. e, MARCO immunohistochemistry, cell counts healthy (n=8) versus cirrhotic (n=8) liver, Mean±SEM, Mann-Whitney two-tailed, scale bars 50μm. f, Flow cytometry: gating strategy for identifying KC, TMo and SAMΦ in healthy (n=2) and cirrhotic (n=3) liver. SAMΦ are detected as TREM2+CD9+ cells within the TMo and SAMΦ gate (see Fig. 2f). g, Representative immunofluorescence image, cirrhotic liver: TREM2 (red), MNDA (white), collagen 1 (green), DAPI (blue), scale bars 50μm. h, Representative image, cirrhotic liver: TREM2 (smFISH; red), MNDA (immunofluorescence; green), DAPI (blue), scale bars 50μm. i, Representative immunofluorescence image, cirrhotic liver: CD9 (red), MNDA (white), collagen 1 (green), DAPI (blue), scale bars 50μm. j,TREM2 immunohistochemistry, cell counts healthy (n=10) versus cirrhotic (n=9) liver, Mann-Whitney two-tailed, scale bars 50μm. k,CD9 immunohistochemistry, cell counts healthy (n=12) versus cirrhotic (n=10) liver, Mann-Whitney two-tailed, scale bars 50μm. l, Exemplar tissue segmentation of cirrhotic liver section into fibrotic septae (orange) and parenchymal nodules (purple) (top); TREM2 (n=9), CD9(n=11), TIMD4 (n=9) and MARCO (n=7) immunohistochemistry and cell counts in parenchymal nodules versus fibrotic septae, Wilcoxon two-tailed. m, Clustering and annotation of 208 cycling MP cells from healthy (n=5) and cirrhotic (n=5) livers, scaled gene expression of MP subpopulation markers across 4 clusters of cycling MP cells (top), fractions of cycling MP subpopulations in 5 healthy versus 5 cirrhotic livers (bottom), Mean±SEM, Wald.
Extended Data Figure 5. Phenotypic characterisation of mononuclear phagocytes in healthy and cirrhotic human livers.
a, Self-Organising Map (SOM; 60x60 grid): smoothed scaled metagene expression of 10,737 mononuclear phagocyte (MP) cells from healthy (n=5) and cirrhotic (n=5) livers. 20,952 genes, 3,600 metagenes, 44 signatures. A-F label metagene signatures overexpressed in one or more MP subpopulations (top). Smoothed mean metagene expression profile for each MP subpopulation (bottom). b, Radar plots (left): metagene signatures A-F showing distribution of signature expression across MP subpopulations from 10,737 MP cells, exemplar genes (middle) and selected Gene Ontology (GO) enrichment (right), Fisher’s exact test. c, Diffusion map visualisation, blood monocytes and liver-resident MP lineages (23,075 cells from healthy (n=5) and cirrhotic (n=5) liver samples and PBMCs (n=4), annotating monocle pseudotemporal dynamics (purple to yellow). RNA velocity field (red arrows) visualised using Gaussian smoothing on regular grid (top). Annotation of MP subpopulation, injury condition (bottom). d, Unspliced-spliced phase portraits (top row), 23,075 cells coloured and visualised as in Fig 3a, monocyte (MNDA), SAMΦ (CD9) and KC marker genes (TIMD4). Cells plotted above or below the steady-state (black dashed line) indicate increasing or decreasing expression of gene, respectively. Spliced expression profile for stated genes (middle row; red high, blue low). Unspliced residuals for stated genes (bottom row), positive (red) indicating expected upregulation, negative (blue) indicating expected downregulation. MNDA displays negative velocity in SAMΦ, CD9 displays positive velocity in monocytes and SAMΦ, TIMD4 velocity is restricted to KC. e, Cubic smoothing spline curve fitted to averaged expression of all genes in module 2 from blood monocyte-SAMΦ pseudotemporal trajectory (see Fig. 3c), selected GO enrichment (right), Fisher’s exact test. f, Cubic smoothing spline curve fitted to averaged expression of all genes in module 3 from blood monocyte-cDC pseudotemporal trajectory (see Fig. 3c), selected GO enrichment (right), Fisher’s exact test. g, Luminex assay: quantification of levels of stated proteins in culture medium from FACS-isolated scar-associated macrophages (SAMΦ, n=3), tissue monocytes (TMo, n=2), Kupffer cells (KC, n=2), and control (media alone, n=2), Mean±SEM, MFI median fluorescence intensity. h, Heatmap: transcription factor regulons across MP pseudotemporal trajectory and in KC (colour coded top by MP cluster, condition and pseudotime), selected regulons labelled right. Cells columns, regulons rows. i, Scaled regulon expression of selected regulons across MP clusters from healthy (n=5) and cirrhotic (n=5) livers.
Extended Data Figure 6. Characterisation of macrophages in mouse liver fibrosis.
a, Clustering and annotating 3,250 mouse mononuclear phagocytes (mMP) from healthy (n=3) and fibrotic (4 weeks carbon tetrachloride treatment, n=3) livers. mTMo, tissue monocyte; mSAMΦ, scar-associated macrophage; mKC, Kupffer cell; mcDC, conventional dendritic cell. b, Annotating mMP cells by injury condition. c, Heatmap: mMP cluster marker genes (top, colour coded by cluster and condition), exemplar genes labelled (right). Cells columns, genes rows. d, Selected genes expressed in 3,250 mMP e, Flow cytometry: gating strategy for identifying KC, TMo and SAMΦ in healthy (n=2) and cirrhotic (n=3) liver. Flow cytometry plots: gating strategy for identifying mKC, CD9- mTMo and CD9+ mSAMΦ in fibrotic mice (n=8 from 2 independent experiments). f, Quantifying mouse macrophage subpopulations by flow cytometry: healthy (n=6) and fibrotic (n=8) mouse livers from 2 independent experiments, macrophage subpopulation (x-axis) as a percentage of total viable CD45+ cells (y-axis), Mean±SEM, Mann-Whitney two-tailed. g, Hepatic stellate cell activation assay: co-culture of hepatic stellate cells (HSC) from uninjured mouse liver and FACS-isolated macrophage subpopulations (MΦ) from fibrotic mouse liver (left). Co-culture with CD9- mTMo or CD9+ mSAMΦ isolated from 8 fibrotic mice (2 independent experiments), qPCR of Col3a1 in HSC, expression relative to mean expression of quiescent HSC (right), Wilcoxon two-tailed. h, Clustering 3,250 mouse mononuclear phagocytes (mMP) and 10,737 human mononuclear phagocytes (hMP) into 5 clusters using canonical correlation analysis (CCA). Annotation of cross-species clusters (identity). i, Annotation of human and mouse macrophage subpopulations from 3,250 mMP and 10,737 hMP. j, Selected genes expressed in 3,250 mMP and 10,737 hMP.
Extended Data Figure 7. Scar-associated macrophage expansion in human NASH.
a to d,Deconvolution: publicly available whole liver microarray data (n=73) assessed for frequency of scar-associated macrophages (SAMΦ), Kupffer cells (KC) and tissue monocytes (TMo) using Cibersort algorithm. a, Macrophage composition: x-axis, GEO accession number; y-axis, fraction of monocyte-macrophages; Top, annotated by liver phenotype; NASH, Non-alcoholic steatohepatitis. b, Frequency of SAMΦ in control (n=14), heathy obese (n=27), steatosis (n=14) and NASH (n=18) livers, Mean±SEM, Kruskal-Wallis and Dunn. c, Frequency of SAMΦ in patients with histological NAFLD activity score (NAS) of 0 (n=37), 1-3 (n=19) and 4-7 (n=17) (left). Frequency of SAMΦ in patients with histological fibrosis score of 0 (n=46), 1 (n=20) and 2-4 (n=5) (right), Mean±SEM, Kruskal-Wallis and Dunn. d, Frequency of SAMΦ in female (n=58) and male (n=15) patients (left). Frequency of SAMΦ in patients aged 23-39 (n=22), 40-49 (n=29) and 50-80 (n=22) (middle). Frequency of SAMΦ in patients with a body mass index (BMI) of 17-30 (n=18), 31-45 (n=28) and 46-70 (n=27) (right). e, CD9 and TREM2 staining in NASH liver biopsy sections (left), Scale bars, 50μm. Cell counting (right): CD9 staining, NAS 1-3 (n=13) versus NAS 4-8 (n=21), Mean±SEM, Mann-Whitney two-tailed. TREM2 staining, NAS 1-3 (n=12) versus NAS 4-8 (n=16), Mean±SEM. f, Correlation of cell counts with picrosirius red (PSR) digital morphometric pixel quantification in NAFLD liver biopsy tissue; CD9 staining (top; n=39); TREM2 staining (bottom; n=32); Pearson correlation and linear regression.
Extended Data Figure 8. Phenotypic characterisation of endothelial cells in healthy and cirrhotic human livers.
a, Clustering and selected genes expressed in 8,020 endothelial cells from 4 healthy and 3 cirrhotic human livers. b, Scaled gene expression of endothelial cluster markers across endothelial cells from healthy (n=4) and cirrhotic (n=3) livers. c, Digital pixel quantification: PLVAP immunostaining of cirrhotic liver sections (n=10) in parenchymal nodules versus fibrotic septae (top), Wilcoxon two-tailed. ACKR1 immunostaining of cirrhotic liver sections (n=10) in parenchymal nodules versus fibrotic septae (bottom), Wilcoxon two-tailed. d, Flow cytometry: endothelial cells from healthy (n=3, grey) or cirrhotic (n=7, red) livers, representative histogram for stated marker (top), mean fluorescence intensity (MFI, bottom), Mean±SEM, Mann-Whitney two-tailed. e, Flow-based adhesion assay: peripheral blood monocytes assessed for adhesion (top) and % of adherent cells which transmigrate (bottom); endothelial cells from healthy (n=5) or cirrhotic (n=4) livers, Mean±SEM, Mann-Whitney two-tailed. f, Endothelial cell gene knockdown: cirrhotic endothelial cells treated with siRNA to PLVAP (n=6), ACKR1 (n=5) or control siRNA (n=6). Representative flow cytometry histograms for stated marker (top); comparison to isotype control antibody. Flow-based adhesion assay (bottom), peripheral blood mononuclear cells assessed for adhesion (bottom left) and % of adherent cells which transmigrate (bottom right) following siRNA treatment of endothelial cells, Mean±SEM, Kruskal-Wallis and Dunn. g, Self-Organising Map (SOM; 60 x 60 grid; top left): smoothed scaled metagene expression of endothelia lineage. 21,237 genes, 3,600 metagenes, 45 signatures. A-E label metagene signatures overexpressed in one or more endothelial subpopulations. SOM: smoothed mean metagene expression profile for each endothelial subpopulation (bottom left). Radar plots (middle): metagene signatures A-E showing distribution of signature expression across endothelial subpopulations, exemplar genes (middle) and Gene Ontology (GO) enrichment (right), Fisher’s exact test. h, Heatmap: endothelial subpopulation transcription factor regulon expression (colour coded top by cluster and condition) across 8,020 endothelial cells from 4 healthy and 3 cirrhotic human livers. Exemplar regulons labelled right. Cells in columns, regulons in rows. i, t-SNE visualisation, endothelial lineage (8,020 cells from healthy (n=4) and cirrhotic (n=3)), annotating monocle pseudotemporal dynamics (purple to yellow; grey indicates lack of inferred trajectory). RNA velocities (red arrows) visualised using Gaussian smoothing on regular grid. (purple to yellow). j, Representative immunofluorescence images healthy versus cirrhotic liver: RSPO3, PDPN, AIF1L, VWA1 or ACKR1 (red), CD34 (white), PLVAP (green), DAPI (blue), scale bars, 50μm. k, Annotation of 8,020 endothelial cells by subpopulation and injury condition. LSEC, Liver sinusoidal endothelial cells.
Extended Data Figure 9. Characterisation of mesenchymal cells in healthy and cirrhotic human livers.
a, Selected genes expressed in 2,318 mesenchymal cells from 4 healthy and 3 cirrhotic human livers. b, Clustering 319 scar-associated mesenchymal cells (SAMes) into 2 further subclusters. c, Heatmap: SAMes subcluster marker genes (top, colour coded by cluster and condition), exemplar genes labelled (right). Cells columns, genes rows. d, Fractions of SAMes subpopulations in healthy (n=4) versus cirrhotic (n=3) livers, Mean±SEM, Wald. e, Representative immunofluorescence image, portal region of healthy liver: OSR1 (red), Collagen 1 (green), DAPI (blue), Scale bar 50μm. f, Representative immunofluorescence image, fibrotic niche of cirrhotic liver: OSR1 (red), Collagen 1 (green), DAPI (blue), Scale bar 50μm. g, Scaled gene expression of selected genes across 2,318 mesenchymal cells from healthy (n=4) and cirrhotic (n=3) livers. h, t-SNE visualisation, 1,178 Hepatic Stellate Cells (HSC) and SAMes from healthy (n=4) and cirrhotic (n=3) livers annotated by monocle pseudotemporal dynamics (purple to yellow). RNA velocity field (red arrows) visualised using Gaussian smoothing on regular grid. i, Heatmap: cubic smoothing spline curves fitted to genes differentially expressed across HSC-to-SAMes pseudotemporal trajectories, grouped by hierarchical clustering (k=2). Colour coded by pseudotime and condition (top). Gene co-expression modules (colour) and exemplar genes labelled right.
Extended Data Figure 10. Characterisation of the cellular interactome in the fibrotic niche.
a to b, Representative immunofluorescence images, fibrotic niche cirrhotic liver. a, TREM2 (red), PLVAP (white), PDGFRα (green), DAPI (blue), Scale bars 50μm. b, TREM2 (red), ACKR1 (white), PDGFRα (green), DAPI (blue), Scale bars 50μm. c, Proliferation assay: Human hepatic stellate cells (HSC) treated with conditioned media from primary hepatic macrophage subpopulations SAMΦ (n=2), tissue monocytes (TMo, n=2), Kupffer cells (KC, n=2) or control media (n=2). y-axis, area under curve (AUC) of % change in HSC number over time (hours), Mean±SEM. d, Circle plot: potential interaction magnitude from ligands expressed by scar-associated macrophages (SAMΦ) and endothelial cells (SAEndo) to receptors expressed on scar-associated mesenchyme (SAMes). e, Circle plot: potential interaction magnitude from ligands expressed by SAMes to receptors expressed on SAMΦ and SAEndo. f, Dotplot: ligand-receptor interactions between SAMes (n=7 human livers), SAMΦ (n=10 human livers) and SAEndo (n=7 human livers). X-axis, ligand (red) and cognate receptor (blue); y-axis, ligand (red) and receptor (blue) expressing cell populations; circle size, P value (permutation test); colour (red, high; yellow, low), means of average ligand and receptor expression levels in interacting subpopulations. g, Representative immunofluorescence images, fibrotic niche in cirrhotic liver. Top; CCL2 (red), CCR2 (white), PDGFRα (green), DAPI (blue), arrows CCL2+PDGFRα+ cells. Bottom; ANGPT1 (red), TEK(white), PDGFRα (green), DAPI (blue), arrows ANGPT1+PDGFRα+ cells. Scale bars 50μm. h, Circle plot: potential interaction magnitude from ligands expressed by to receptors expressed on SAEndo. i, Dotplot: ligand-receptor interactions between SAMΦ (n=10 human livers) and SAEndo (n=7 human livers). X-axis, ligand (red) and cognate receptor (blue); y-axis, ligand (red) and receptor (blue) expressing cell populations; circle size, P value (permutation test); colour (red, high; yellow, low), means of average ligand and receptor expression levels in interacting subpopulations. j, Representative immunofluorescence image, fibrotic niche in cirrhotic liver. TREM2 (red), FLT1 (white), VEGFA (green), DAPI (blue), arrows TREM2+VEGFA+ cells, Scale bar 50μm. k, Circle plot: potential interaction magnitude from ligands expressed by SAEndo to receptors expressed on SAMΦ. l, Dotplot: ligand-receptor interactions between SAEndo (n=7 human livers) and SAMΦ (n=10 human livers). X-axis, ligand (red) and cognate receptor (blue); y-axis, ligand (red) and receptor (blue) expressing cell populations; circle size, P value (permutation test); colour (red, high; yellow, low), means of average ligand and receptor expression levels in interacting subpopulations. m, Representative immunofluorescence images, fibrotic niche in cirrhotic liver. Top; TREM2 (red), CD200 (white), CD200R (green), DAPI (blue), arrows TREM2+CD200R+ cells. Bottom; TREM2 (red), DLL4 (white), NOTCH2 (green), DAPI (blue), arrows TREM2+NOTCH2+ cells. Scale bars, 50μm.
Supplementary Material
Acknowledgements
This work was supported by an MRC Clinician Scientist Fellowship (MR/N008340/1) to P.R., a Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 103749) to N.C.H., an AbbVie Future Therapeutics and Technologies Division grant to N.C.H., a Guts UK– Children's Liver Disease Foundation grant (ref. R43927) to N.C.H., a Tenovus Scotland grant (ref. E18/05) to R.D. and N.C.H., and British Heart Foundation grants (RM/17/3/33381; RE/18/5/34216) to N.C.H. R.V-T. was funded by EMBO and Human Frontiers long-term fellowships. C.J.W. was funded by a BBSRC New Investigator Award (BB/N018869/1). P.N.N., C.J.W. and N.T.L. are funded by the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. This paper presents independent research supported by the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. J.P.I. is funded by the NIHR Bristol Biomedical Research Centre, University Hospitals Bristol Foundation Trust and the University of Bristol. This work was also supported by Wellcome Sanger core funding (WT206194). We thank the patients who donated liver tissue and blood for this study. We thank J. Davidson, C. Ibbotson, J. Black and A. Baird of the Scottish Liver Transplant Unit and the research nurses of the Wellcome Trust Clinical Research Facility for assistance with consenting patients for this study. We thank the liver transplant coordinators and surgeons of the Scottish Liver Transplant Unit and the surgeons and staff of the Hepatobiliary Surgical Unit, Royal Infirmary of Edinburgh for assistance in procuring human liver samples. We thank S. Johnston, W. Ramsay and M. Pattison (QMRI Flow Cytometry and Cell Sorting Facility, University of Edinburgh) for technical assistance with fluorescence activated cell sorting (FACS) and flow cytometry. We thank Jon Henderson for technical support and Gillian Muirhead for assistance with liver endothelial cell isolation. This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications).
Footnotes
Data and materials availability
Our expression data will be freely available for user-friendly interactive browsing online at http://www.livercellatlas.mvm.ed.ac.uk. CellPhoneDB is available at www.CellPhoneDB.org. All raw sequencing data will be deposited in the Gene Expression Omnibus (GEO accession GSE136103).
Code availability
R scripts enabling the main steps of the analysis are available from the corresponding authors on reasonable request.
Author Contributions
P.R. performed experimental design, tissue procurement, data generation, data analysis and interpretation, and manuscript preparation; R.D. performed experimental design, data generation and data analysis; E.D., K.P.M., B.E.P.H., M.B., J.A.M. and N.T.L. performed data generation and analysis; J.R.P. generated the interactive online browser; M.E. and R.V-T. assisted with CellPhoneDB analyses and critically appraised the manuscript; T.J.K. performed pathological assessments and provided intellectual contribution; N.O.C., J.A.F. and P.N.N. provided intellectual contribution; C.J.W. performed tissue procurement, data generation, interpretation and intellectual contribution; J.R.W-K. performed computational analysis with assistance from J.R.P and R.S.T. and advice from C.P.P., J.M. and S.A.T.; J.R.W-K. also helped with manuscript preparation and C.P.P., J.M. and S.A.T. critically appraised the manuscript; E.M.H., D.J.M. and S.J.W. procured human liver tissue and critically appraised the manuscript. J.P.I., F.T. and J.W.P. provided intellectual contribution and critically appraised the manuscript; N.C.H. conceived the study, designed experiments, interpreted data and prepared the manuscript.
Author Information
The authors declare no competing financial interests.
References
- 1.Marcellin P, Kutala BK. Liver diseases: A major, neglected global public health problem requiring urgent actions and large-scale screening. Liver Int. 2018 doi: 10.1111/liv.13682. [DOI] [PubMed] [Google Scholar]
- 2.Angulo P, et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramachandran P, Henderson NC. Antifibrotics in chronic liver disease: tractable targets and translational challenges. Lancet Gastroenterol Hepatol. 2016;1:328–340. doi: 10.1016/S2468-1253(16)30110-8. [DOI] [PubMed] [Google Scholar]
- 4.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nature Medicine. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358:58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffield JS, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramachandran P, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci. 2012;109:E3186–E3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlmark KR, et al. Hepatic recruitment of the inflammatory Gr1 + monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology. 2009;50:261–274. doi: 10.1002/hep.22950. [DOI] [PubMed] [Google Scholar]
- 9.Minutti CM, et al. Local amplifiers of IL-4Rα–mediated macrophage activation promote repair in lung and liver. Science (80-.) 2017;356:1076–1080. doi: 10.1126/science.aaj2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pradere J-P, et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology. 2013;58:1461–1473. doi: 10.1002/hep.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson NC, et al. Galectin-3 regulates myofibroblast activation and hepatic fibrosis. Proc Natl Acad Sci. 2006;103:5060–5065. doi: 10.1073/pnas.0511167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seki E, et al. CCR2 promotes hepatic fibrosis in mice. Hepatology. 2009;50:185–197. doi: 10.1002/hep.22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syn WK, et al. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology. 2011;53:106–115. doi: 10.1002/hep.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott CL, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7 doi: 10.1038/ncomms10321. 10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez Perdiguero E, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science (80-.) 2016;353 doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Manno G, et al. RNA velocity of single cells. Nature. 2018;1 doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schelker M, et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02289-3. 2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahrens M, et al. DNA Methylation Analysis in Nonalcoholic Fatty Liver Disease Suggests Distinct Disease-Specific and Remodeling Signatures after Bariatric Surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Pruenster M, et al. The Duffy antigen receptor for chemokines transports chemokines and supports their promigratory activity. Nat Immunol. 2009;10:101–8. doi: 10.1038/ni.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shetty S, Weston CJ, Adams DH, Lalor PF. A Flow Adhesion Assay to Study Leucocyte Recruitment to Human Hepatic Sinusoidal Endothelium Under Conditions of Shear Stress. J Vis Exp. 2014 doi: 10.3791/51330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwaisako K, et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci. 2014;111:E3297–E3305. doi: 10.1073/pnas.1400062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson NC, et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Minicis S, et al. Gene Expression Profiles During Hepatic Stellate Cell Activation in Culture and In Vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 26.Mederacke I, Dapito DH, Affò S, Uchinami H, Schwabe RF. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc. 2015;10:305–315. doi: 10.1038/nprot.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minutti CM, et al. A Macrophage-Pericyte Axis Directs Tissue Restoration via Amphiregulin-Induced Transforming Growth Factor Beta Activation. Immunity. 2019;50:645–654.e6. doi: 10.1016/j.immuni.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Searle BC, Gittelman RM, Manor O, Akey JM. Detecting sources of transcriptional heterogeneity in large-scale RNA-seq data sets. Genetics. 2016;204:1391–1396. doi: 10.1534/genetics.116.193714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bain CC, et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat Commun. 2016;7 doi: 10.1038/ncomms11852. ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinrich MC, et al. Crenolanib Inhibits the Drug-Resistant PDGFRA D842V Mutation Associated with Imatinib-Resistant Gastrointestinal Stromal Tumors. Clin Cancer Res. 2012;18:4375–4384. doi: 10.1158/1078-0432.CCR-12-0625. [DOI] [PubMed] [Google Scholar]
- 32.Patten DA, et al. SCARF-1 promotes adhesion of CD4+ T cells to human hepatic sinusoidal endothelium under conditions of shear stress. Sci Rep. 2017;7 doi: 10.1038/s41598-017-17928-4. 17600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-17204-5. 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arganda-Carreras I, et al. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 36.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 37.Deroulers C, et al. Analyzing huge pathology images with open source software. Diagn Pathol. 2013;8:92. doi: 10.1186/1746-1596-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kendall TJ, et al. Hepatic elastin content is predictive of adverse outcome in advanced fibrotic liver disease. Histopathology. 2018;73:90–100. doi: 10.1111/his.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017 doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li WV, Li JJ. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03405-7. 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camp JG, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang HM, et al. AnimalTFDB 2.0: A resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 2015;43:D76–D81. doi: 10.1093/nar/gku887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aibar S, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015 doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.