Abstract
The APOBEC3 family of cytosine deaminases are part of the innate immune response to viral infection, but also have the capacity to damage cellular DNA. Detection of mutational signatures consistent with APOBEC3 activity, together with elevated APOBEC3 expression in cancer cells, has raised the possibility that these enzymes contribute to oncogenesis. Genome deamination by APOBEC3 enzymes also elicits DNA damage response signaling and presents therapeutic vulnerabilities for cancer cells. Here, we discuss implications of APOBEC3 activity in cancer and the potential to exploit their mutagenic activity for targeted cancer therapies.
Keywords: APOBEC3, cytosine deaminase, DNA damage response, mutational patterns, synthetic lethality
The benefit of endogenous DNA mutators
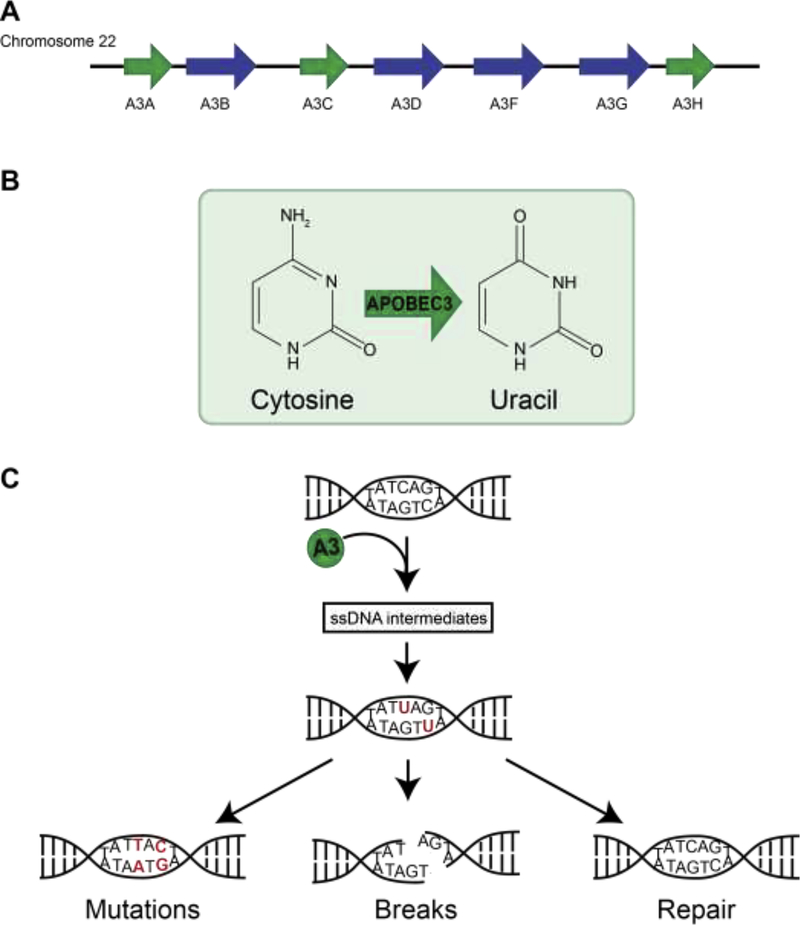
Seven APOBEC3 (apolipoprotein B mRNA editing catalytic polypeptide-like 3) cytosine deaminases (A3A-A3H) are encoded by the human genome as separate but highly homologous genes on chromosome 22 [1] (Fig 1A). APOBEC3 enzymes deaminate cytosine bases in single-stranded DNA (ssDNA), leaving uracil in its place (Fig 1B). Genomic uracil may be excised by uracil-DNA glycosylases (UDG) resulting in an abasic site (AP-site) or may be replicated by polymerases. Repair or replication of uracil-DNA bases results in a variety of molecular outcomes including accurate replacement of cytosine, mutagenic C→T transitions, and DNA breaks (Fig 1C).
Figure 1. APOBEC3 enzymes are single-strand-specific DNA cytosine deaminases.
(A) The APOBEC3 gene cluster is located on chromosome 22. Green arrows depict single deaminase domain family members. Blue arrows depict double deaminase domain family members.
(B) APOBEC3 enzymes deaminate cytosine resulting in a DNA uracil base.
(C) Molecular outcomes of APOBEC3-mediated deamination events include low-fidelity replication of uracil resulting in mutations, processing of uracils resulting in ssDNA nicks or dsDNA breaks, and high-fidelity excision and repair with no resulting DNA lesion.
The APOBEC3 subfamily is part of a larger APOBEC gene family which originated from the founder gene, APOBEC1, and includes activation-induced deaminase (AID), APOBEC2, and APOBEC4 [1]. Despite their mutagenic potential, evidence suggests that the APOBEC3 gene cluster arose from evolutionarily advantageous amplification events. While humans and non-human primates encode seven APOBEC3 genes, fewer APOBEC3 genes are present in lower mammals [1]. Within the human locus, APOBEC3 genes have an increased ratio of amino acid altering mutations compared to silent mutations, indicating that they are under positive selection [1–4]. Since DNA mutators pose a threat to organisms with DNA genomes, evolutionary selection of APOBEC3 enzymes suggests that they confer a survival advantage. AID deaminates genes at the immunoglobulin (Ig) locus to promote class-switch recombination and somatic hypermutation, processes which are essential for development of adaptive immunity [5]. The exact function of each APOBEC3 family member has not been determined, but the benefit of APOBEC3 enzymes appears to be through their role as innate immune effectors.
The most well described endogenous function of APOBEC3 enzymes is as part of the innate immune response to viral infections. The first APOBEC3 enzyme to be identified was A3G [1, 6, 7]. A3G was found to be a DNA deaminase and restriction factor for HIV-1 [6, 8–10]. The mechanism by which A3G limits HIV infection has been elegantly defined. Briefly, A3G is packaged into the HIV virion and, upon infection of a target cell, viral replication produces a cDNA intermediate on which A3G acts [2, 10–12]. Deamination of proviral HIV cDNA by A3G results in widespread mutations, loss of genetic integrity, and decreased infectivity [2, 8–11]. The HIV-1 protein Vif (virus infectivity factor) blocks A3G by directing polyubiquitination and degradation of the enzyme [13]. Investigation of other APOBEC3 family members revealed that A3D, A3F and A3H also have the capacity to restrict HIV infection through a mechanism similar to A3G [14, 15]. Subsequent studies identified APOBEC3 enzymes as restriction factors for additional viruses including hepatitis B virus, human T-cell leukemia virus, parvoviruses, human papillomavirus, and possibly gamma-herpesviruses as well as endogenous retroviruses [2, 16–23]. Although deamination is central to most APOBEC3-mediated virus restriction, the manner in which APOBEC3 enzymes access and act on viral genomes varies depending on the virus and type of genome. For example, a polymorphism resulting in deletion of the entire A3B coding sequence is common in Southeast Asians, but this population does not have an increased rate of HIV-1 infection nor do the characteristics of HIV-1 infections in patients with an A3B deletion differ from those with intact A3B gene [24]. These data combined with ex vivo evaluation indicate that, while A3B may be capable of mutating HIV-1 genomes, it does not serve as an HIV-1 restriction factor in the same manner as A3G. Similarly, the anti-viral function of A3A was first identified by its activity against parvoviruses [16]. A3A but not other APOBEC3 family members inhibited recombinant adeno-associated virus production [16, 25]. While the anti-viral activity of several APOBEC3 enzymes may confer an evolutionary advantage, the lack of anti-viral activity in some family members suggests additional roles for APOBEC3 enzymes that have not yet been defined. The anti-viral activity of APOBEC3 enzymes has been reviewed extensively [2, 11, 13, 26, 27]; here we focus on the implications of APOBEC3 activity on cellular genomes.
Variability among APOBEC enzymes
Despite their genetic homology, several important characteristics distinguish APOBEC3 enzymes from each other. Clues to the functional variability of APOBEC3 family members may be gleaned from their differences in virus restriction capacity, cellular expression, structure, and biochemical activity.
Tissue expression.
A3A and A3G were defined as interferon-stimulated genes (ISGs) in peripheral blood mononuclear cells (PBMC) during studies of HIV-1 restriction [28–30]. More comprehensive evaluation of APOBEC3 expression in human tissues showed that basal mRNA levels of most APOBEC3 genes are elevated in T cells relative to other PBMCs. The expression level of APOBEC3 genes in non-hematopoietic tissues is generally low [31]. Interestingly, A3A is an outlier in that it is most highly expressed in myeloid-lineage cells and is most sensitive to induction by type I interferon [25, 30–32]. High levels of A3A and A3B expression are also found in human papillomavirus (HPV)-associated malignant tissues [33–35]. Interestingly, expression of the HPV oncoproteins E6 and E7 appear to be independently capable of upregulating A3A and A3B expression [34, 35]. Further, E7 was shown to block ubiquitination of A3A, resulting in stabilization of the enzyme [36]. These data suggest pathways of APOBEC3 regulation that may be modified by virus infection. Additional regulators of APOBEC3 expression in healthy and malignant tissue remain to be identified.
Deaminase domains.
The A3A, A3C, and A3H enzymes have only one cytosine deaminase domain in contrast to the other APOBEC3 family members which have two (Fig 1A) [1]. Of the double-domain APOBEC3 enzymes, the C-terminal deaminase domain (CTD) is catalytically active whereas the N-terminal domain (NTD) is not [37]. The structural and functional relevance of the NTD appears to be in regulating catalytic activity through interaction with ssDNA substrates and oligomerization of the enzyme [38, 39]. Enzyme oligomerization occurs during packaging into HIV-1 virions, which may be a method to ensure that APOBEC3 enzymes will be present on viral genomes, and promotes a deaminase-independent method of viral restriction through steric inhibition of viral reverse transcriptase [40–42].
Subcellular localization.
Access to replicating virus and cellular DNA is in part determined by the subcellular localization of APOBEC3 enzymes. The A3B protein is nuclear, A3A and A3C are detected throughout the cell, and the remaining family members localize to the cytoplasm [43, 44]. All APOBEC3 enzymes are small enough to diffuse passively into the nucleus, thus determinants of localization appear to be complex and distinct for each enzyme. A3B has a putative NLS, although mutation of this region does not necessarily alter nuclear localization [45, 46]. Rather, the localization determinant appears to lie within the first 60 amino acids of the protein; when amino acids in the N-terminus of A3G are substituted for corresponding amino acids from A3B, A3G is expressed throughout the cell although this only occurs for the NTD of A3G [45]. When the CTD is added, A3G again localizes to the cytoplasm suggesting a second region contributes to localization [45]. Consistent with this finding, a chimeric construct containing the NTD of A3B joined to A3A exhibited pan-cellular localization [38], which was proposed to be due to loss of a localization signal within the A3B CTD. A recent report identified another region of the A3B NTD which drives nuclear localization [47]. Although this region is not conserved among double-domain APOBEC3 enzymes, it was shown to be sufficient to relocalize cytoplasmic A3G and pan-cellular A3D to the nucleus in chimeric proteins [47]. These and other studies suggest complex intra- and inter-protein interactions of APOBEC3 enzymes are responsible for subcellular localization [43, 48, 49]. The variation in localization among the APOBEC3 family members suggests differences in endogenous function and macromolecular interactions.
Context preference.
The context preference in which cytosine bases are deaminated differ among APOBEC3 enzymes. A3A and A3B act on a cytosine with a thymidine in the −1 position (TC context), whereas A3G acts on cytosines in a CC context [16, 49, 50]. Current data regarding APOBEC3 activity on the cellular genome have centered on A3A and A3B, both of which have a TC context preference in keeping with mutational signatures identified in cancer genomes. The context preference of A3A can be distinguished from that of A3B, as shown in yeast models, by the −2 nucleotide position: A3A preferentially deaminates cytosines following a −2 pyrimidine base, whereas A3B deaminates cytosines preceded by a −2 purine base [51]. Additionally, these family members have at least partial nuclear localization, which would provide access to cellular genomic DNA. In the following sections, we focus on A3A and A3B as agents of genome instability.
Evidence for A3 activity in human cancer
As the anti-viral activity of APOBEC3 deaminases has been examined, the potential for deamination of the cellular genome has become evident. In contrast to APOBEC3 family members, the endogenous activity of AID requires deamination of the cellular genome [5]. AID can be oncogenic; off-target activity is associated with chromosomal translocations, specifically IgH-Myc, that drive B cell lymphomas [52, 53]. Although there is no known function of APOBEC3 enzymes that necessitates targeting of the cellular genome, mutations in human cancer genomes attributable to APOBEC3 activity have been extensively reported. Recent investigations have focused on the mechanism by which APOBEC3 enzymes act on cellular DNA in comparison to their targeting of viral genomes. While current data do not suggest that APOBEC3 enzymes are oncogenes, it has become clear that some members of the APOBEC3 family are active on cancer genomes and may impact tumor development or progression.
APOBEC3 activity elicits cellular DNA damage responses.
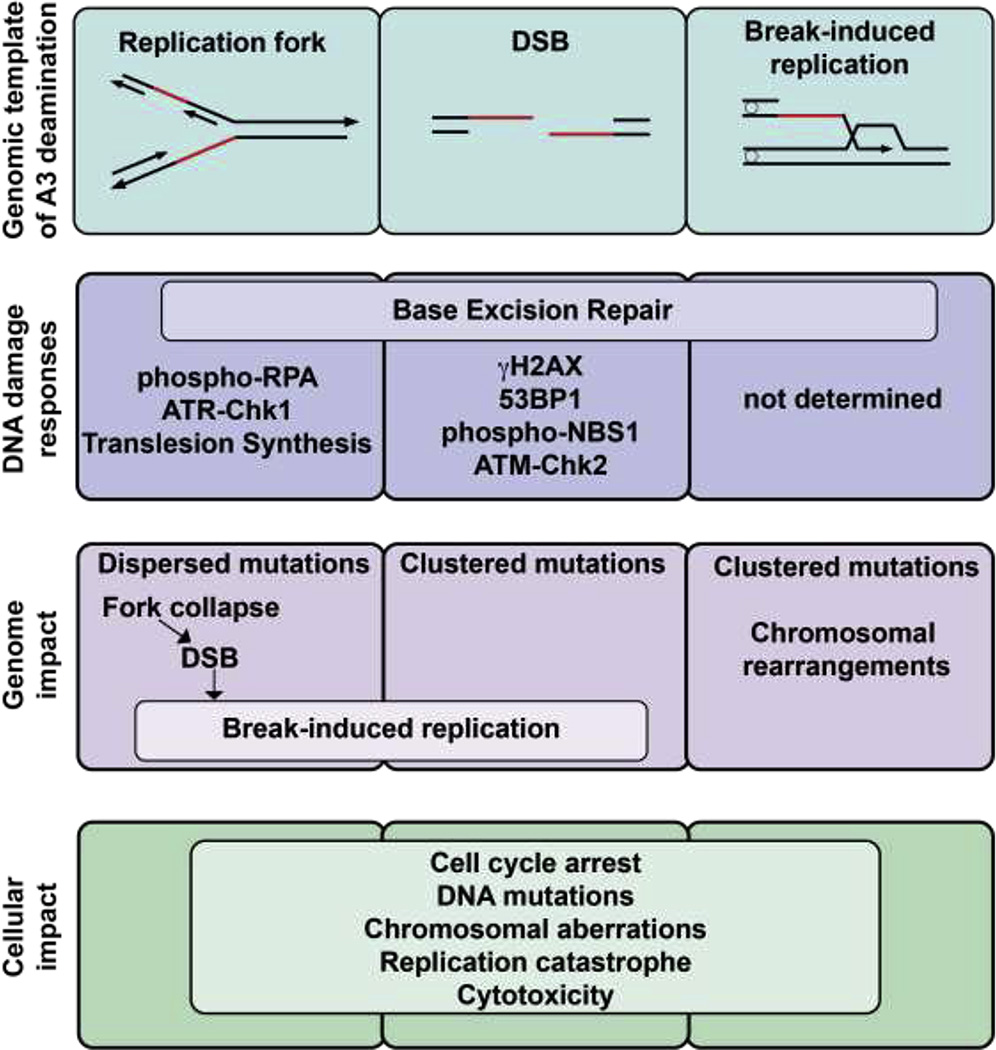
A preliminary indication that APOBEC3 enzymes have the capacity to act on the cellular genome came from the observation of DNA damage signaling in response to ectopic A3A expression in cancer cell lines (Fig 2) [54]. A3A expression resulted in dose-dependent increases in phosphorylated H2AX (γH2AX), a marker of double-stranded DNA breaks (DSB) [54, 55]. Several additional markers of DNA damage response (DDR) signaling are activated upon A3A expression, including responses to ssDNA (phospho-RPA), DSB recognition and repair (53BP1, phospho-NBS1), and cell cycle checkpoints (ATR-Chk1, ATM-Chk2) (Fig 2) [54–57]. Complementary studies in cancer cell lines and primary PBMCs demonstrated editing of foreign and cellular DNA by A3A [58, 59]. Transgene expression and plasmid stability were limited upon expression of A3A [59]. Differential DNA denaturing (3D)-PCR, a technique based on the capacity for DNA with fewer inter-strand hydrogen bonds (e.g. A/T-rich regions) to amplify at lower melting temperatures, demonstrated PCR amplification of reporter genes at lower temperatures when A3A was expressed in cells [59]. Similarly, editing of mitochondrial and nuclear DNA evaluated by 3D-PCR was most robust in cell lines transfected with A3A expression vectors as compared to those transfected with other APOBEC3 enzymes [58]. Notably, genomic mutations induced by A3A were only recovered by 3D-PCR in the presence of a uracil glycosylase inhibitor (UGI), indicating that cells exposed to A3A are dependent on DNA repair processes for genome protection [58, 59]. These early studies suggested that APOBEC3 enzymes, and specifically A3A, are able to access and act on cellular DNA resulting in mutations and cellular DDR activation.
Figure 2. APOBEC3 deamination at cellular ssDNA templates activates DNA damage responses and causes genotoxicity.
Single-stranded substrates of APOBEC3 deamination include single stranded intermediates at replication forks, DSBs, and those generated during break-induced replication. DNA mutations and breaks induced by APOBEC3 enzymes elicit DNA damage responses which may be specific to the template on which APOBEC3 acts. Replication fork collapse leading to DSBs, break-induced replication, and various patterns of mutations result from APOBEC3 activity on cellular DNA. DNA damage responses aid in recognition and repair of APOBEC3-induced lesions leading to cell cycle arrest, although APOBEC3 activity may overwhelm cellular responses leading to widespread mutations, replication fork collapse, DNA breaks, and cell death.
The potential for A3B to incite DDR signaling was evaluated by overexpression of the enzyme in 293T cells [60]. Cells with ectopic A3B expression exhibited increased γH2AX staining and longer tails by COMET assay, a technique used to quantify DNA breaks within a cell, suggesting that A3B also causes DNA breaks [60]. When compared with A3A, DNA damage caused by A3B appeared to occur at a slower rate and be less severe. In conjunction with experimental models of ssDNA mutations in yeast that led to the identification of APOBEC3 mutagenesis in human cancer genomes [61], these findings led to the concerning, although likely, possibility that APOBEC3 enzymes could contribute to genome instability. Notably, many studies in which induction of DNA damage responses by A3A and A3B were observed utilized overexpression models of the enzymes which may produce exaggerated or non-physiologic results. Knock-down of individual endogenous APOBEC3 genes have enabled assessment of the impact of each enzyme on genome instability though further studies of endogenous APOBEC3 activity are warranted.
Mutational signatures are attributable to APOBEC3 activity.
Further evidence for the involvement of APOBEC3 enzymes in human cancers came when mutational signatures were defined in the sequences of 21 breast cancer genomes [62]. The original five signatures were later expanded to several dozen signatures which enabled nuanced identification of responsible mutagens and mutational processes [63, 64]. A mutational pattern attributed to APOBEC3 activity was identified in the original evaluation of mutational signatures and has persisted throughout refinements of signature definitions as the repertoire of signatures has expanded. This mutational pattern not only implicated APOBEC3 enzymes as mutagens acting on cancer genomes, but helped to define the interaction between APOBEC3 enzymes and the cellular genome.
The APOBEC3 mutational signature is defined by predominantly C→T transitions in a TC context. Two APOBEC3 family members, A3A and A3B, that deaminate TC dinucleotides also have at least partial localization to the nucleus and are highly expressed in subsets of human tumors. Thus, A3A and A3B have been investigated as etiologic agents of cancer mutagenesis. APOBEC3 signature mutations largely occurred in close proximity to one another, which was termed “kataegis,” [62] indicating highly processive mutagenic activity on short stretches of the genome. Based on analysis of mutational patterns, the APOBEC3 signature has been observed in approximately one quarter of cancer genomes examined, making it one of the most prevalent signatures in all human cancer genomes [63, 65, 66].
APOBEC3 expression in cancer.
In a landmark study by Burns, et al., A3B transcript levels were analyzed in breast cancer cell lines and primary breast cancers [60]. Breast cancer cell lines with high A3B expression had more genomic uracil and more C→T mutations than paired cell lines in which A3B was knocked down. In aggregate, genomes from primary breast cancers with high endogenous A3B expression had more mutated cytosines than those with low A3B expression [60]. This was the first study to link A3B expression and activity in human cancer cells. Subsequent TCGA-based analyses of APOBEC3 transcript expression revealed a correlation between A3B expression and APOBEC3 mutational hallmarks in a variety of cancers including lung, bladder, cervix, ovary, and head and neck [65, 67, 68]. Later refinement of the APOBEC3 context preference, enabled distinction of A3A from A3B-induced deamination based on the −2 nucleotide surrounding a mutated cytosine [51]. Evaluation of signatures using a more granular definition of A3 enzyme activity indicated that A3A, rather than A3B, is the predominant mutagen in TCGA samples with APOBEC3 mutational signatures [51]. Analysis of >1000 human cell lines also revealed that the most common sequence context in which cytosine deamination occurs is with a pyrimidine in the −2 position, consistent with A3A activity [69]. However, the reported enrichment of A3A mutagenesis is not correlated with A3A mRNA levels in tumors from the TCGA database [51]. The frequency with which APOBEC3 mutational patterns are seen in cancer is striking, but the lack of correlation with APOBEC3 expression raises the possibility that expression levels alone do not determine APOBEC3 activity.
APOBEC3 expression in healthy tissue is generally low, with few notable exceptions. Interestingly, APOBEC3 mutational signatures have been frequently reported in genomes from solid tumors such as breast, bladder, cervical, and ovarian cancer as well as multiple myeloma [60, 61, 63, 65, 67, 68, 70]. These tumors arise in tissues that do not normally have high APOBEC3 expression levels [31] which implies dysregulated APOBEC3 expression. An exception to the typically low expression of APOBEC3 enzymes in healthy tissue is that of A3A expression in myeloid lineage cells [30, 31, 59]. The reason for this high level of A3A expression in myeloid cells is unclear, although it has been proposed that A3A may impede HIV-1 infection of monocytes by viral restriction [29]. Efforts to correlate high A3A expression with APOBEC3 mutational signatures in hematologic malignancies are limited both by the availability of samples in published databases, and the relatively lower mutational burden found in leukemias and lymphomas as compared with solid tumors. Despite these hurdles, evaluation of RNA-sequencing data from primary acute myeloid leukemias (AML) demonstrated a subset of AML that express unusually high levels of A3A [56]. This finding suggests that A3A has the potential to act on AML genomes. Given that A3A is an ISG and expression can be stimulated in lymphocytes [30], A3A may also be highly expressed in malignant lymphocytes and thus responsible for mutational burdens within lymphoblastic leukemias and lymphomas. Further studies are warranted to determine the expression and activity of A3A in hematologic malignancies.
Current data are limited in the evaluation of APOBEC3 protein levels and regulation of APOBEC3 expression. Antibodies generated against specific APOBEC3 proteins are of inconsistent quality and have high potential to cross-react with other family members given their homology. A conventional model of expression correlating to activity has been weakly supported by existing data [60, 67]. However, APOBEC3 enzymes are cytotoxic during prolonged expression in cell culture experiments [60] and thus may also be cytotoxic in human cancers when highly expressed. An alternative model in which APOBEC3 enzymes are transiently expressed and intermittent activity on the genome results in mutational “footprints” has been proposed [71, 72]. Consistent with this hypothesis, a recent analysis of the temporal acquisition of mutational patterns in cancer cell lines indicated intermittent APOBEC3 activity over time which resulted in significant variability in APOBEC3 mutational patterns within subclones from the same parental origin [69]. Thus, expression captured at the time of tumor biopsy may not reflect prior levels of expression and might therefore not be expected to correlate precisely with mutational hallmarks.
APOBEC3 polymorphisms alter cancer risk.
Additional evidence for APOBEC3 association with human cancers was revealed by genome-wide association studies (GWAS) in Chinese women with breast cancer [73]. A deletion in the A3B gene, which results in the loss of the complete coding region of A3B thus tethering the open reading frame of A3A to the 3’ untranslated region of A3B, is prevalent in East Asian populations (>30%). Studies of Japanese and Chinese women demonstrated an increased risk of breast cancer in patients with heterozygous or homozygous A3B gene deletion [73, 74]. These findings were replicated in a study of women with European ancestry, in which the germline A3B deletion was shown to be far less common (~10%) but still correlated with an increased risk of breast cancer [75]. However, a meta-analysis including several ethnic populations found a limited correlation between the A3B deletion allele and breast cancer risk [76]. An evaluation of mutational signatures in breast cancer genomes from patients with the A3B deletion demonstrated an increase in APOBEC3 mutational hallmarks [77], which are consistent with the activity of A3A based on its distinct mutational signature including a pyrimidine in the −2 position preceding a mutated cytosine [51]. A mechanistic assessment of this phenomenon demonstrated that the A3B deletion resulted in increased stability of the A3A transcript, increased A3A protein levels, and increased damage to nuclear DNA [78]. Alternatively, another APOBEC3 with TC dinucleotide preference may be responsible for mutations in genomes with the A3B deletion. Several haplotypes of the A3H gene have been identified, and haplotype I appears to localize to the nucleus [47]. A3H expression also correlates with TC mutations in breast tumors with the A3B deletion allele [79]. While the A3B deletion appears not to limit overall APOBEC3 activity in cancer genomes, the impact of the deletion on cancer risk remains unclear.
An additional polymorphism, rs17000526, located upstream of the APOBEC3 gene cluster has been associated with increased risk of bladder cancer, increased A3B expression, and high mutational burden within tumor genomes [80]. These data implicate APOBEC3 enzymes in oncogenesis, and also suggest that individual APOBEC3 enzymes may be relevant in certain tumors. While increased A3B expression has been reported recurrently in breast cancer, the finding of APOBEC3 mutations in breast cancers with an A3B deletion indicates that there is a limit to inferences that can be drawn from APOBEC3 expression levels alone.
Cellular Targets of A3 Deamination
The repertoire of viruses restricted by the APOBEC3 enzymes reflects their single-strand-specific deaminase activity [12, 16, 27, 81]. APOBEC3 family members have been reported to restrict viruses with ssDNA genomes, retroviruses with cDNA intermediates during replication, and ssDNA generated from retroelements during retrotransposition [8, 10, 12, 16, 82]. The double-stranded human genome contains regions that are found transiently but predictably in a ssDNA state. Exposure of ssDNA occurs at replication forks during DNA replication, at DNA breaks in preparation for recombination, and at the non-transcribed strand during gene transcription. These structures result in varying lengths of ssDNA and durations of exposure. A yeast model aimed at identifying a mechanism to explain mutational patterns revealed clustering of mutations on ssDNA generated during DNA replication and repair [61]. Similar clustering patterns were detected in cancer genome sequences [61] and are analogous to kataegis, a proposed hallmark of the APOBEC3 mutational signature [62]. Investigations of the cellular ssDNA templates on which APOBEC3 enzymes act to generate these clustered mutations have focused on intermediates of DNA replication and repair processes (Fig 2).
Replication structures.
Although only short stretches of ssDNA are exposed for brief periods as Okazaki fragments on the lagging strand during DNA replication, replicating DNA appears to be the major target of APOBEC3 activity within the cellular genome. This conclusion was made by several investigators simultaneously through use of varied model systems and experimental approaches [55, 83–86].
By assessing mutational asymmetries between complementary DNA strands in genome sequences from model organisms, several groups defined the lagging strand of replication forks as the primary ssDNA template on which APOBEC3 enzymes act [83–86]. Yeast genomes exposed to ectopic APOBEC3 expression accumulated strandbiased mutations surrounding replication origins consistent with APOBEC3 deamination of the lagging strand [86]. Similarly, next generation sequencing of E. coli strains identified a bias for cytosine mutations on the lagging strand when A3G was expressed in bacteria [84]. No significant clustering was observed in bacterial genomes, although relatively low numbers of mutations were analyzed in this study [84]. An evaluation of asymmetrically mutated regions within cancer genomes found that the APOBEC3 mutational signature is most prevalent on the lagging-strand template [85]. Interestingly, this analysis also did not identify significant clustering on the lagging strand attributable to APOBEC3 deamination despite high mutational burdens observed in the genomes analyzed.
The lack of clustered APOBEC3 mutations in replication-associated regions is not necessarily mutually exclusive of APOBEC3-induced kataegis; several studies have demonstrated that the majority of APOBEC3 mutations occur dispersed throughout the genome rather than in clusters [61, 62, 65, 83, 87]. Thus, both clustered and nonclustered TC mutations may be caused by APOBEC3 deamination. Clustered mutations in cancer genomes are associated with DNA rearrangement breakpoints, whereas dispersed APOBEC3 mutations mostly occur on the lagging strand during DNA replication (Fig 2) [83, 85]. These findings suggest that the template or genomic context in which APOBEC3 enzymes are acting likely drives the degree to which mutations are clustered.
Concordant with these computational studies, A3A-induced DNA damage and DDR signaling is significantly increased in actively replicating cells or those in S phase as compared to quiescent or G1 phase cells [55]. These findings further strengthen the conclusion that APOBEC3 enzymes act on ssDNA at replication structures.
Several investigations simultaneously determined that replication stress can potentiate A3-induced mutations. Conditional A3A expression in cells treated with lowdose hydroxyurea to stall replication forks demonstrated persistent DNA damage in cells in which A3A expression was induced [55]. Consistent with this finding, an increased mutagenic rate on both leading and lagging strands by A3A and A3B was observed in replication-deficient yeast strains [86]. Additionally, replication stress was reported to correlate with increased A3B expression and activity in breast cancer cell lines [88]. These studies suggest that APOBEC3 enzymes may promote mutagenesis by capitalizing on the replication stress frequently incurred by cancer cells [89].
Double-stranded DNA breaks.
Repair of DSBs occurs through one of many repair pathways. Homologous recombination, the most high-fidelity DSB repair pathway, is initiated by exonuclease-mediated resection leaving exposed ssDNA on either side of the DSB, and these ssDNA templates are used to recombine with a sister chromatid. Several studies have suggested that resected ends of DSBs are substrates for APOBEC3 deamination [61, 62, 87]. The first description of APOBEC3 mutational signatures identified clusters of C→T transitions in close proximity to clusters of G→A mutations, which was termed “alternating processivity” [62]. These patterns are consistent with the concept of strand-coordination, which was defined by similar mutational patterns found at resected ends of DSBs in yeast genomes [61]. Furthermore, these strand-coordinated clusters were additionally identified in human cancer genome sequences and co-localized with DNA rearrangement breakpoints [61, 62]. Prospective evaluations of APOBEC3 activity at DSB resected ends were performed in yeast in which I-SceI induction of a DSB in yeast genomes resulted in increased kataegis-like mutational patterns around the break upon expression of A3G [87]. From these data emerge a model of APOBEC3 enzymes acting processively on ssDNA resected ends at a DSB (Fig 2).
An alternative model was suggested by which DNA break-induced replication (BIR) was observed to provide a template for kataegis [90]. During BIR, only one end of a DSB can be repaired and is done so by a migrating bubble in which replication of the lagging strand is delayed behind the leading strand and ssDNA accumulates, providing a substrate for clustered mutations (Fig 2). Although the contribution of APOBEC3 deamination to this model of kataegis has not yet been demonstrated, one can imagine a cycle of APOBEC3 activity at replication forks leading to collapsed forks, BIR, and further template on which APOBEC3 deamination occurs.
The potential for APOBEC3 enzymes to cause DSBs has been proposed due to increased γH2AX in response to ectopic APOBEC3 expression in cell lines [54, 60]. One example is the finding that replication stress induced by hydroxyurea treatment of cancer cells expressing A3A led to increased γH2AX [55]. An explanation for this observation is that A3A activity at stalled replication forks results in deamination events that cause ssDNA breaks, collapse of forks, and ultimately DSBs (Fig 2). A more granular pathway was proposed to define the mechanism by which A3A activity leads to DSBs, in which A3A deamination causes replicative polymerase stalling, accumulation of ssDNA, and eventual replication catastrophe resulting in DNA breaks [57]. Intriguingly, single-stranded DNA breaks (SSB) induced by CRISPR-Cas9 nickase mutants enabled A3Bmediated indel mutations [91]. Investigation into the mechanism of this finding led to molecular elucidation of APOBEC3-mediated DSBs as an intermediate step in indel formation. Specifically, processing of SSBs results in generation of genomic ssDNA. Cytosine deamination of ssDNA by A3B led to excision of uracil and an AP site. Subsequent breakage of ssDNA at the AP site caused a DSB [91]. This is one possible route by which APOBEC3 deamination results in DSBs, although there are likely other means of APOBEC3-induced DSB formation which depend on the structure of deaminated DNA.
Transcription.
The transcription bubble provides a ssDNA template on the nontranscribed strand, which is the main substrate for the B cell-specific APOBEC family member, AID [92, 93]. In vitro data suggested that A3A was capable of deaminating the non-transcribed strand during active transcription, though less efficiently than AID [94]. However, whole genome assessment of yeast expressing APOBEC3 did not reveal transcriptional strand-biased mutations [86]. Similarly, an analysis of strand-biased mutations in human cancer genomes found few APOBEC3-attributable mutations in regions of transcript-strand asymmetry [85].
A more precise evaluation of APOBEC3 activity at transcribed genes in yeast models demonstrated deaminase-induced mutations enriched at promoters of actively transcribed genes, rather than at transcription bubbles [95]. These regions were upstream of the TSS where assembly of the pre-initiation complex results in DNA melting which produces two ssDNA templates. Cytosine deamination was evident on both templates in yeast genomes evaluated [95]. Additionally, a recent study of deaminase activity in yeast found A3B activity on the non-transcribed strand at tRNA genes, which occurred in a transcription-dependent manner and was enhanced by the presence of R-loops [96]. In this study, A3B activity at tRNA genes was more abundant than at protein-coding genes during transcription [96]. While APOBEC3 enzymes are likely capable of acting on ssDNA formed during transcription, this appears to be less prevalent, or perhaps less genotoxic, than APOBEC3 activity at intermediates of DNA replication and repair. The enzymatic, genomic, and cellular factors that influence APOBEC3 activity on a given DNA substrate are still largely unknown.
Exploiting APOBEC3 Activity for Cancer Therapy
Elucidating the cellular targets of deamination is an essential step towards understanding the role of APOBEC3 mutagenesis in genome instability and cancer, and has also defined specific DDR pathways that are activated in response to APOBEC3 activity. Deficiencies in DDR pathways enable tolerance to genome instability of cancer cells. However, DDR deficiencies also present the potential for therapeutic vulnerabilities due to the lack of redundancy in pathways regulating genome maintenance [97]. The classic example of therapeutic opportunity presented by DDR deficiency is that of poly (ADP-ribose) polymerase-1 (PARP1) inhibition in homologous recombination-deficient tumors. PARP1 is a key enzyme in single-strand break repair, an essential compensatory DNA repair pathway in cells deficient in HR. Thus, inhibition of PARP1 in an HR-deficient context results in genotoxicity and cell death. This concept of cell death due to loss of two genes, neither of which alone would cause cytotoxicity, is referred to as synthetic lethality [98]. DDR components have become enticing targets for synthetic lethality in cancer [99].
Amidst the focus on the oncogenic potential of APOBEC3 enzymes, their capacity for activating DDR pathways has generated interest in possible therapeutic vulnerabilities. Indeed, cancer cells with high levels of APOBEC3 activity are susceptible to inhibition of ATR-Chk1 signaling (Fig 3) [56, 57]. This was demonstrated in several models of APOBEC3 expression in cancer including A3A in AML [56] and osteosarcoma [57], as well as evaluation of A3A and A3B expression in 293T cells [100].
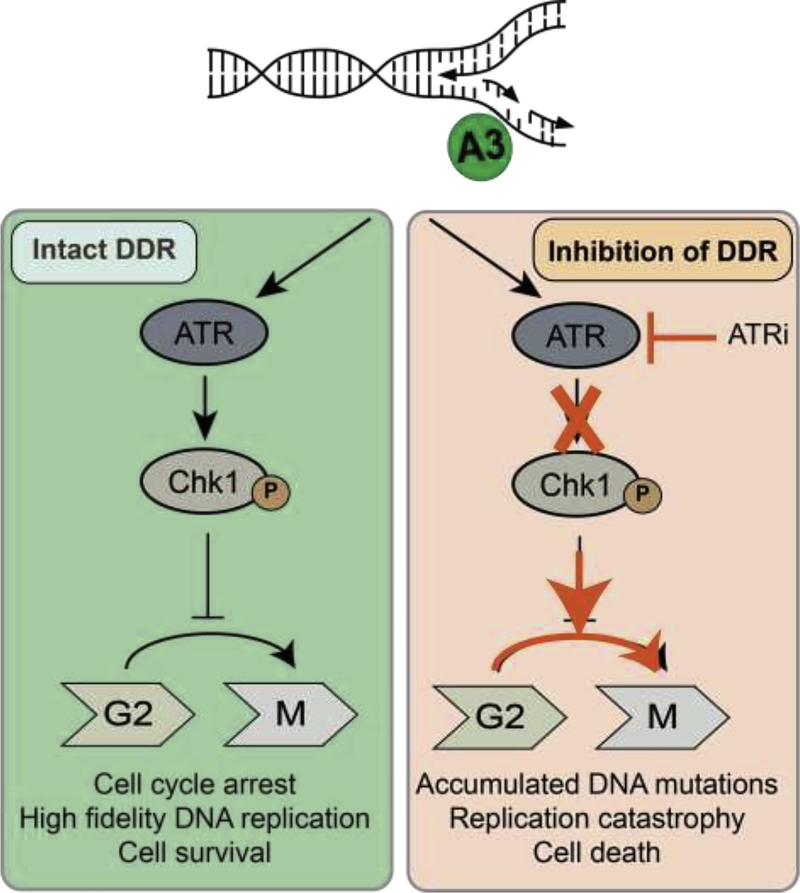
Figure 3. APOBEC3 activity presents a therapeutic vulnerability in cancer cells.
APOBEC3 activity at replication structures recruits ATR and activates downstream signaling via phosphorylation of Chk1. When ATR signaling is intact (left panel), cell cycle arrest prior to mitosis enables repair of replication structures prior to cell division thus ensuring viable genome replication. If ATR signaling is inhibited (right panel), the DNA replication checkpoint is not activated and cells proceed to mitosis while accumulating mutations and breaks at replication forks. ATR inhibition in combination with APOBEC3 activity leads to mutations, collapse of replication forks, DNA breaks, and ultimately genotoxicity. Cellular dependence on ATR signaling when APOBEC3 enzymes are active exemplifies the potential for targeted therapeutics in tumors with high APOBEC3 activity.
A3A activity on replicating genomes causes replication stress and robust activation of the DNA replication checkpoint regulated by ATR kinase signaling via phosphorylation of downstream Chk1 [56, 57]. In leukemia cell lines with conditional A3A expression, A3A triggered ATR-dependent Chk1 phosphorylation and arrest of cells at the G2-M transition [56]. This cell cycle arrest was abrogated by inhibition of either ATR or Chk1 which led to widespread A3A-induced DNA damage and eventual cell death [56]. Buisson, et al. demonstrated similar findings in which A3A induced replication stress by generation of AP sites that elicited ATR activation in order to prevent DSB formation [57]. ATR inhibition in the presence of A3A expression resulted in replication catastrophe, DSBs, and cell death.
Interestingly, although A3A has been shown to activate cellular responses to DSBs via phosphorylation of H2AX, ATM, and Chk2 [54, 56], A3A activity does not sensitize cancer cells to inhibition of ATM-Chk2 signaling. In a leukemia model, induction of A3A caused Chk2 phosphorylation, but neither chemical nor genetic inhibition of ATM resulted in cell death [56]. Consistently, high A3B expression did not correlate with baseline Chk2 phosphorylation in a panel of cancer cell lines, nor did A3B knockdown lead to a decrease in phospho-Chk2 indicating minimal A3B-related ATM-Chk2 activation [57]. The activation of ATM by A3A may be indirect and related to the collapse of replication forks rather than direct induction of DSBs by A3A, thus an intact ATR signaling pathway would circumvent cytotoxicity potentiated by ATM inhibition.
While these models of A3A and A3B expression in cancer cells revealed selective sensitivity to ATR pathway inhibition, an evaluation of A3B expression in p53-deficient 293T cells demonstrated sensitivity to all DDR inhibitors tested, including ATMi [100]. Interestingly, an evaluation in breast cancer cell lines found that ATR-Chk1 activation was associated with increased transcription and activity of endogenous A3B that was abrogated by genetic or chemical inhibition of ATR-Chk1 [88]. These discrepancies may be due to the cellular contexts in which A3A and A3B were assessed, and suggest complex mechanisms by which APOBEC3 enzymes act on the genome which may differ between enzymes within the family. Future studies will benefit from evaluation of endogenous APOBEC3 activity using in vivo cancer models.
DDR activation has been evaluated as a proxy for understanding the mechanisms by which APOBEC3 enzymes cause DNA damage. A3A causes replication stress by cytosine deamination of the lagging strand, leading to AP sites which cause stalled polymerases and unstable replication structures, thus creating more ssDNA which is exposed for longer durations. This ssDNA becomes an additional template on which APOBEC3 enzymes can act, thus exacerbating DNA damage in a “feed-forward loop” [57]. APOBEC3 enzymes restrict viruses by deaminase-dependent and deaminase-independent mechanisms [25, 101]. The latter may be related to DNA binding or steric hindrance of reverse transcription [102]. An alternative or additional mechanism of APOBEC3-induced DNA damage may be one in which APOBEC3 binds replication structures causing steric hindrance of polymerases, thereby promoting stalled replication and ultimately DSBs. One mechanism by which A3A activity on replicating DNA results in increased genomic uracil is deamination [55], however it is possible that APOBEC3 enzymes also cause deaminase-independent replication stress and DNA damage in cancer cells.
Perspectives and Challenges
Although the preponderance of evidence points toward APOBEC3 enzymes as agents of mutagenesis in cancer, the studies to date have largely relied on retrospective, indirect correlations between APOBEC3 mutational signatures and APOBEC3 mRNA expression. Prospective studies are needed to confirm that APOBEC3 enzymes are active on human cancer genomes and are responsible for the proposed mutational hallmarks. Furthermore, the impact of these mutations must be investigated. For example, several studies have suggested that APOBEC3 activity on cancer genomes contributes to oncogene activation [33] and clonal diversity [103–105], although APOBEC3 mutational “hotspots” have been identified most frequently in genes unrelated to cancer [106]. It is additionally possible that mutations caused by APOBEC3 deamination can contribute to the development of chemotherapy resistance [107, 108], analogous to AID-induced mutations in BCR-ABL fusions that result in imatinib-resistant CML [109]. Chemical inhibitors of APOBEC3 enzymes are in development, and are proposed to avert development of clonal heterogeneity and chemoresistance [110]. Another possibility is the generation of neo-epitopes by APOBEC3 mutation of cancer genomes, leading to enhanced immune surveillance of tumors. APOBEC3 signature mutations in bladder cancer are associated with high mutational burdens, high neo-antigen load, and improved survival [80, 111]. While APOBEC3 enzymes are undoubtedly active in human cancers, the effect of their mutagenic activity remains unclear.
A general lack of understanding of the measures that regulate APOBEC3 activity on human genomes raises many questions: what genes or genetic hotspots are susceptible to APOBEC3 mutagenesis? What drives the expression or activity of APOBEC3 enzymes in normal conditions and how is that altered in cancer? How do tumors tolerate these highly active DNA mutators? While some studies have addressed these questions, a complete picture has yet to emerge. In addition, focused investigations of APOBEC3-related therapeutic vulnerabilities in specific tumor contexts are warranted, specifically in DDR-deficient tumors in which APOBEC3 activity alone may result in death of cancer cells. In this context, chemical agonism of APOBEC3 activity may be warranted. A correlation between APOBEC3 expression and programmed cell death protein ligand-1 (PD-L1) expression was observed [112–114], which contributed to the hypothesis that APOBEC3 activity may result in generation of neo-antigens and therefore impact tumor susceptibility to immunotherapeutic agents. The promise of APOBEC3 enzymes as biomarkers for targeted cancer therapy will be more fully realized as our understanding of APOBEC3 expression, regulation, and resulting cellular responses improves.
Acknowledgements
We thank members of the Weitzman Lab for insightful discussions and input. We thank Rahul Kohli, Sebastien Landry, Daphne Avgousti, and Rachel DeWeerd for critical reading of the manuscript. A.M.G. was supported by a Young Investigator Award from the Alex’s Lemonade Stand Foundation, and by the National Institutes of Health (K12 CA076931 and K08 CA212299). Research on APOBEC enzymes in the Weitzman lab was supported by grants to M.D.W. from the National Institutes of Health (CA181259 and CA185799). The authors declare that there are no conflicts of interest.
REFERENCES
- [1].Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N, An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22, Genomics, 79 (2002) 285–296. [DOI] [PubMed] [Google Scholar]
- [2].Harris RS, Dudley JP, APOBECs and virus restriction, Virology, 479–480 (2015) 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McLaughlin RN Jr., Gable JT, Wittkopp CJ, Emerman M, Malik HS, Conservation and Innovation of APOBEC3A Restriction Functions during Primate Evolution, Mol Biol Evol, 33 (2016) 1889–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sawyer SL, Emerman M, Malik HS, Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G, PLoS Biol, 2 (2004) E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T, Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme, Cell, 102 (2000) 553–563. [DOI] [PubMed] [Google Scholar]
- [6].Sheehy AM, Gaddis NC, Choi JD, Malim MH, Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein, Nature, 418 (2002) 646–650. [DOI] [PubMed] [Google Scholar]
- [7].Harris RS, Petersen-Mahrt SK, Neuberger MS, RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators, Mol Cell, 10 (2002) 1247–1253. [DOI] [PubMed] [Google Scholar]
- [8].Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D, Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts, Nature, 424 (2003) 99–103. [DOI] [PubMed] [Google Scholar]
- [9].Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L, The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA, Nature, 424 (2003) 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH, DNA deamination mediates innate immunity to retroviral infection, Cell, 113 (2003) 803–809. [DOI] [PubMed] [Google Scholar]
- [11].Malim MH, Bieniasz PD, HIV Restriction Factors and Mechanisms of Evasion, Cold Spring Harb Perspect Med, 2 (2012) a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR, Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome, Nat Struct Mol Biol, 11 (2004) 435–442. [DOI] [PubMed] [Google Scholar]
- [13].Malim MH, Natural resistance to HIV infection: The Vif-APOBEC interaction, C R Biol, 329 (2006) 871–875. [DOI] [PubMed] [Google Scholar]
- [14].Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS, Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1, J Virol, 85 (2011) 11220–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V, HIV-1 Vif adaptation to human APOBEC3H haplotypes, Cell Host Microbe, 14 (2013) 411–421. [DOI] [PubMed] [Google Scholar]
- [16].Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD, APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons, Curr Biol, 16 (2006) 480–485. [DOI] [PubMed] [Google Scholar]
- [17].Cheng AZ, Yockteng-Melgar J, Jarvis MC, Malik-Soni N, Borozan I, Carpenter MA, McCann JL, Ebrahimi D, Shaban NM, Marcon E, Greenblatt J, Brown WL, Frappier L, Harris RS, Epstein-Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity, Nat Microbiol, 4 (2019) 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ooms M, Krikoni A, Kress AK, Simon V, Munk C, APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1, J Virol, 86 (2012) 6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP, Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo, Proc Natl Acad Sci U S A, 102 (2005) 8321–8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vartanian JP, Guetard D, Henry M, Wain-Hobson S, Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions, Science, 320 (2008) 230–233. [DOI] [PubMed] [Google Scholar]
- [21].Suspene R, Aynaud MM, Koch S, Pasdeloup D, Labetoulle M, Gaertner B, Vartanian JP, Meyerhans A, Wain-Hobson S, Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo, J Virol, 85 (2011) 7594–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C, APOBEC3 proteins inhibit human LINE-1 retrotransposition, J Biol Chem, 281 (2006) 22161–22172. [DOI] [PubMed] [Google Scholar]
- [23].Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR, APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells, Nucleic Acids Res, 34 (2006) 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Imahashi M, Izumi T, Watanabe D, Imamura J, Matsuoka K, Ode H, Masaoka T, Sato K, Kaneko N, Ichikawa S, Koyanagi Y, Takaori-Kondo A, Utsumi M, Yokomaku Y, Shirasaka T, Sugiura W, Iwatani Y, Naoe T, Lack of association between intact/deletion polymorphisms of the APOBEC3B gene and HIV-1 risk, PLoS One, 9 (2014) e92861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD, Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase, PLoS Pathog, 5 (2009) e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Daugherty MD, Malik HS, Rules of engagement: molecular insights from host-virus arms races, Annu Rev Genet, 46 (2012) 677–700. [DOI] [PubMed] [Google Scholar]
- [27].Harris RS, Liddament MT, Retroviral restriction by APOBEC proteins, Nat Rev Immunol, 4 (2004) 868–877. [DOI] [PubMed] [Google Scholar]
- [28].Stopak KS, Chiu YL, Kropp J, Grant RM, Greene WC, Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells, J Biol Chem, 282 (2007) 3539–3546. [DOI] [PubMed] [Google Scholar]
- [29].Peng G, Greenwell-Wild T, Nares S, Jin W, Lei KJ, Rangel ZG, Munson PJ, Wahl SM, Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression, Blood, 110 (2007) 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH, Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets, J Virol, 83 (2009) 9474–9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS, Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction, Nucleic Acids Res, 38 (2010) 4274–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Covino DA, Gauzzi MC, Fantuzzi L, Understanding the regulation of APOBEC3 expression: Current evidence and much to learn, J Leukoc Biol, 103 (2018) 433–444. [DOI] [PubMed] [Google Scholar]
- [33].Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR, APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development, Cell Rep, 7 (2014) 1833–1841. [DOI] [PubMed] [Google Scholar]
- [34].Vieira VC, Leonard B, White EA, Starrett GJ, Temiz NA, Lorenz LD, Lee D, Soares MA, Lambert PF, Howley PM, Harris RS, Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B, MBio, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D, APOBEC3A functions as a restriction factor of human papillomavirus, J Virol, 89 (2015) 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Westrich JA, Warren CJ, Klausner MJ, Guo K, Liu CW, Santiago ML, Pyeon D, Human Papillomavirus 16 E7 Stabilizes APOBEC3A Protein by Inhibiting Cullin 2-Dependent Protein Degradation, J Virol, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bransteitter R, Prochnow C, Chen XS, The current structural and functional understanding of APOBEC deaminases, Cell Mol Life Sci, 66 (2009) 3137–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Caval V, Bouzidi MS, Suspene R, Laude H, Dumargne MC, Bashamboo A, Krey T, Vartanian JP, Wain-Hobson S, Molecular basis of the attenuated phenotype of human APOBEC3B DNA mutator enzyme, Nucleic Acids Res, 43 (2015) 9340–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xiao X, Yang H, Arutiunian V, Fang Y, Besse G, Morimoto C, Zirkle B, Chen XS, Structural determinants of APOBEC3B non-catalytic domain for molecular assembly and catalytic regulation, Nucleic Acids Res, 45 (2017) 7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Adolph MB, Love RP, Chelico L, Biochemical Basis of APOBEC3 Deoxycytidine Deaminase Activity on Diverse DNA Substrates, ACS Infect Dis, 4 (2018) 224–238. [DOI] [PubMed] [Google Scholar]
- [41].Adolph MB, Webb J, Chelico L, Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase, PLoS One, 8 (2013) e64196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG, Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G, Nucleic Acids Res, 35 (2007) 7096–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lackey L, Demorest ZL, Land AM, Hultquist JF, Brown WL, Harris RS, APOBEC3B and AID have similar nuclear import mechanisms, J Mol Biol, 419 (2012) 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lackey L, Law EK, Brown WL, Harris RS, Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination, Cell Cycle, 12 (2013) 762–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stenglein MD, Matsuo H, Harris RS, Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization, J Virol, 82 (2008) 9591–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Pak V, Heidecker G, Pathak VK, Derse D, The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B, J Virol, 85 (2011) 8538–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Salamango DJ, McCann JL, Demir O, Brown WL, Amaro RE, Harris RS, APOBEC3B Nuclear Localization Requires Two Distinct N-Terminal Domain Surfaces, J Mol Biol, 430 (2018) 2695–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA, Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces, Structure, 18 (2010) 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bogerd HP, Wiegand HL, Doehle BP, Cullen BR, The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains, Virology, 364 (2007) 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Langlois MA, Beale RC, Conticello SG, Neuberger MS, Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities, Nucleic Acids Res, 33 (2005) 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, Getz G, Gordenin DA, An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers, Nat Genet, 47 (2015) 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC, Role of genomic instability and p53 in AID-induced c-myc-Igh translocations, Nature, 440 (2006) 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC, AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations, Cell, 135 (2008) 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Landry S, Narvaiza I, Linfesty DC, Weitzman MD, APOBEC3A can activate the DNA damage response and cause cell-cycle arrest, EMBO Rep, 12 (2011) 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Green AM, Landry S, Budagyan K, Avgousti DC, Shalhout S, Bhagwat AS, Weitzman MD, APOBEC3A damages the cellular genome during DNA replication, Cell Cycle, 15 (2016) 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Green AM, Budagyan K, Hayer KE, Reed MA, Savani MR, Wertheim GB, Weitzman MD, Cytosine Deaminase APOBEC3A Sensitizes Leukemia Cells to Inhibition of the DNA Replication Checkpoint, Cancer Res, 77 (2017) 4579–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Buisson R, Lawrence MS, Benes CH, Zou L, APOBEC3A and APOBEC3B Activities Render Cancer Cells Susceptible to ATR Inhibition, Cancer Res, 77 (2017) 4567–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Suspene R, Aynaud MM, Guetard D, Henry M, Eckhoff G, Marchio A, Pineau P, Dejean A, Vartanian JP, Wain-Hobson S, Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism, Proc Natl Acad Sci U S A, 108 (2011) 4858–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS, APOBEC3 proteins mediate the clearance of foreign DNA from human cells, Nat Struct Mol Biol, 17 (2010) 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS, APOBEC3B is an enzymatic source of mutation in breast cancer, Nature, 494 (2013) 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions, Mol Cell, 46 (2012) 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR, Breast C Cancer Working Group of the International Cancer Genome, Mutational processes molding the genomes of 21 breast cancers, Cell, 149 (2012) 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian I. Pancreatic Cancer Genome, Consortium IBC, Consortium IM-S, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR, Signatures of mutational processes in human cancer, Nature, 500 (2013) 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, Campbell PJ, Vineis P, Phillips DH, Stratton MR, Mutational signatures associated with tobacco smoking in human cancer, Science, 354 (2016) 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA, An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers, Nat Genet, 45 (2013) 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Petljak M, Alexandrov LB, Understanding mutagenesis through delineation of mutational signatures in human cancer, Carcinogenesis, 37 (2016) 531–540. [DOI] [PubMed] [Google Scholar]
- [67].Burns MB, Temiz NA, Harris RS, Evidence for APOBEC3B mutagenesis in multiple human cancers, Nat Genet, 45 (2013) 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A, Vogel RI, Nikas JB, Law EK, Brown WL, Li Y, Zhang Y, Maurer MJ, Oberg AL, Cunningham JM, Shridhar V, Bell DA, April C, Bentley D, Bibikova M, Cheetham RK, Fan JB, Grocock R, Humphray S, Kingsbury Z, Peden J, Chien J, Swisher EM, Hartmann LC, Kalli KR, Goode EL, Sicotte H, Kaufmann SH, Harris RS, APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma, Cancer Res, 73 (2013) 7222–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Petljak M, Alexandrov LB, Brammeld JS, Price S, Wedge DC, Grossmann S, Dawson KJ, Ju YS, Iorio F, Tubio JMC, Koh CC, Georgakopoulos-Soares I, Rodriguez-Martin B, Otlu B, O’Meara S, Butler AP, Menzies A, Bhosle SG, Raine K, Jones DR, Teague JW, Beal K, Latimer C, O’Neill L, Zamora J, Anderson E, Patel N, Maddison M, Ng BL, Graham J, Garnett MJ, McDermott U, Nik-Zainal S, Campbell PJ, Stratton MR, Characterizing Mutational Signatures in Human Cancer Cell Lines Reveals Episodic APOBEC Mutagenesis, Cell, 176 (2019) 1282–1294 e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I, Dawson KJ, Iorio F, Nik-Zainal S, Bignell GR, Hinton JW, Li Y, Tubio JM, McLaren S, S OM, Butler AP, Teague JW, Mudie L, Anderson E, Rashid N, Tai YT, Shammas MA, Sperling AS, Fulciniti M, Richardson PG, Parmigiani G, Magrangeas F, Minvielle S, Moreau P, Attal M, Facon T, Futreal PA, Anderson KC, Campbell PJ, Munshi NC, Heterogeneity of genomic evolution and mutational profiles in multiple myeloma, Nat Commun, 5 (2014) 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Roberts SA, Gordenin DA, Hypermutation in human cancer genomes: footprints and mechanisms, Nat Rev Cancer, 14 (2014) 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Roberts SA, Gordenin DA, Clustered and genome-wide transient mutagenesis in human cancers: Hypermutation without permanent mutators or loss of fitness, Bioessays, 36 (2014) 382–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Long J, Delahanty RJ, Li G, Gao YT, Lu W, Cai Q, Xiang YB, Li C, Ji BT, Zheng Y, Ali S, Shu XO, Zheng W, A common deletion in the APOBEC3 genes and breast cancer risk, J Natl Cancer Inst, 105 (2013) 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Komatsu A, Nagasaki K, Fujimori M, Amano J, Miki Y, Identification of novel deletion polymorphisms in breast cancer, Int J Oncol, 33 (2008) 261–270. [PubMed] [Google Scholar]
- [75].Xuan D, Li G, Cai Q, Deming-Halverson S, Shrubsole MJ, Shu XO, Kelley MC, Zheng W, Long J, APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry, Carcinogenesis, 34 (2013) 2240–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Klonowska K, Kluzniak W, Rusak B, Jakubowska A, Ratajska M, Krawczynska N, Vasilevska D, Czubak K, Wojciechowska M, Cybulski C, Lubinski J, Kozlowski P, The 30 kb deletion in the APOBEC3 cluster decreases APOBEC3A and APOBEC3B expression and creates a transcriptionally active hybrid gene but does not associate with breast cancer in the European population, Oncotarget, 8 (2017) 76357–76374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, Ramakrishna M, Shlien A, Simonic I, Xue Y, Tyler-Smith C, Campbell PJ, Stratton MR, Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer, Nat Genet, 46 (2014) 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S, A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage, Nat Commun, 5 (2014) 5129. [DOI] [PubMed] [Google Scholar]
- [79].Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, Feng Y, Adolph MB, Chelico L, Law EK, Carpenter MA, Harris RS, The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis, Nat Commun, 7 (2016) 12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Middlebrooks CD, Banday AR, Matsuda K, Udquim KI, Onabajo OO, Paquin A, Figueroa JD, Zhu B, Koutros S, Kubo M, Shuin T, Freedman ND, Kogevinas M, Malats N, Chanock SJ, Garcia-Closas M, Silverman DT, Rothman N, Prokunina-Olsson L, Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors, Nat Genet, 48 (2016) 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Conticello SG, Creative deaminases, self-inflicted damage, and genome evolution, Ann N Y Acad Sci, 1267 (2012) 79–85. [DOI] [PubMed] [Google Scholar]
- [82].Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV, APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition, Elife, 3 (2014) e02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Seplyarskiy VB, Soldatov RA, Popadin KY, Antonarakis SE, Bazykin GA, Nikolaev SI, APOBEC-induced mutations in human cancers are strongly enriched on the lagging DNA strand during replication, Genome Res, 26 (2016) 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bhagwat AS, Hao W, Townes JP, Lee H, Tang H, Foster PL, Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli, Proc Natl Acad Sci U S A, 113 (2016) 2176–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Haradhvala NJ, Polak P, Stojanov P, Covington KR, Shinbrot E, Hess JM, Rheinbay E, Kim J, Maruvka YE, Braunstein LZ, Kamburov A, Hanawalt PC, Wheeler DA, Koren A, Lawrence MS, Getz G, Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair, Cell, 164 (2016) 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hoopes JI, Cortez LM, Mertz TM, Malc EP, Mieczkowski PA, Roberts SA, APOBEC3A and APOBEC3B Preferentially Deaminate the Lagging Strand Template during DNA Replication, Cell Rep, 14 (2016) 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS, DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis, Elife, 2 (2013) e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, McGranahan N, Rogers R, Law EK, Gromova I, Kschischo M, Walton MI, Rossanese OW, Bartek J, Harris RS, Venkatesan S, Swanton C, DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer, Genome Biol, 17 (2016) 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Halazonetis TD, Gorgoulis VG, Bartek J, An oncogene-induced DNA damage model for cancer development, Science, 319 (2008) 1352–1355. [DOI] [PubMed] [Google Scholar]
- [90].Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A, Break-induced replication is a source of mutation clusters underlying kataegis, Cell Rep, 7 (2014) 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lei L, Chen H, Xue W, Yang B, Hu B, Wei J, Wang L, Cui Y, Li W, Wang J, Yan L, Shang W, Gao J, Sha J, Zhuang M, Huang X, Shen B, Yang L, Chen J, APOBEC3 induces mutations during repair of CRISPR-Cas9-generated DNA breaks, Nat Struct Mol Biol, 25 (2018) 45–52. [DOI] [PubMed] [Google Scholar]
- [92].Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW, Transcription-targeted DNA deamination by the AID antibody diversification enzyme, Nature, 422 (2003) 726–730. [DOI] [PubMed] [Google Scholar]
- [93].Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC, Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand, Nat Immunol, 4 (2003) 452–456. [DOI] [PubMed] [Google Scholar]
- [94].Love RP, Xu H, Chelico L, Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A, J Biol Chem, 287 (2012) 30812–30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Taylor BJ, Wu YL, Rada C, Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes, Elife, 3 (2014) e03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Saini N, Roberts SA, Sterling JF, Malc EP, Mieczkowski PA, Gordenin DA, APOBEC3B cytidine deaminase targets the non-transcribed strand of tRNA genes in yeast, DNA Repair (Amst), 53 (2017) 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Curtin NJ, DNA repair dysregulation from cancer driver to therapeutic target, Nat Rev Cancer, 12 (2012) 801–817. [DOI] [PubMed] [Google Scholar]
- [98].Kaelin WG Jr., The concept of synthetic lethality in the context of anticancer therapy, Nat Rev Cancer, 5 (2005) 689–698. [DOI] [PubMed] [Google Scholar]
- [99].Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FM, Therapeutic opportunities within the DNA damage response, Nat Rev Cancer, 15 (2015) 166–180. [DOI] [PubMed] [Google Scholar]
- [100].Nikkila J, Kumar R, Campbell J, Brandsma I, Pemberton HN, Wallberg F, Nagy K, Scheer I, Vertessy BG, Serebrenik AA, Monni V, Harris RS, Pettitt SJ, Ashworth A, Lord CJ, Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells, Br J Cancer, 117 (2017) 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Morse M, Huo R, Feng Y, Rouzina I, Chelico L, Williams MC, Dimerization regulates both deaminase-dependent and deaminase-independent HIV-1 restriction by APOBEC3G, Nat Commun, 8 (2017) 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gillick K, Pollpeter D, Phalora P, Kim EY, Wolinsky SM, Malim MH, Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination, J Virol, 87 (2013) 1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Burrell RA, McGranahan N, Bartek J, Swanton C, The causes and consequences of genetic heterogeneity in cancer evolution, Nature, 501 (2013) 338–345. [DOI] [PubMed] [Google Scholar]
- [104].McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C, Clonal status of actionable driver events and the timing of mutational processes in cancer evolution, Sci Transl Med, 7 (2015) 283ra254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, Rosenberg J, Mosquera JM, Robinson B, Elemento O, Sboner A, Beltran H, Demichelis F, Rubin MA, Clonal evolution of chemotherapy-resistant urothelial carcinoma, Nat Genet, 48 (2016) 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, Lawrence MS, Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features, Science, 364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Evgin L, Huff AL, Kottke T, Thompson J, Molan AM, Driscoll CB, Schuelke M, Shim KG, Wongthida P, Ilett EJ, Smith KK, Harris RS, Coffey M, Pulido JS, Pandha H, Selby PJ, Harrington KJ, Melcher A, Vile RG, Suboptimal T-cell Therapy Drives a Tumor Cell Mutator Phenotype That Promotes Escape from First-Line Treatment, Cancer Immunol Res, 7 (2019) 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Law EK, Sieuwerts AM, LaPara K, Leonard B, Starrett GJ, Molan AM, Temiz NA, Vogel RI, Meijer-van Gelder ME, Sweep FC, Span PN, Foekens JA, Martens JW, Yee D, Harris RS, The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer, Sci Adv, 2 (2016) e1601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM, Hofmann WK, Jumaa H, Groffen J, Heisterkamp N, Martinelli G, Lieber MR, Casellas R, Muschen M, The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia, Cancer Cell, 16 (2009) 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Olson ME, Harris RS, Harki DA, APOBEC Enzymes as Targets for Virus and Cancer Therapy, Cell Chem Biol, 25 (2018) 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, Castro MAA, Gibb EA, Kanchi RS, Gordenin DA, Shukla SA, Sanchez-Vega F, Hansel DE, Czerniak BA, Reuter VE, Su X, de Sa Carvalho B, Chagas VS, Mungall KL, Sadeghi S, Pedamallu CS, Lu Y, Klimczak LJ, Zhang J, Choo C, Ojesina AI, Bullman S, Leraas KM, Lichtenberg TM, Wu CJ, Schultz N, Getz G, Meyerson M, Mills GB, McConkey DJ, Network TR, Weinstein JN, Kwiatkowski DJ, Lerner SP, Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer, Cell, 171 (2017) 540–556 e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wang S, Jia M, He Z, Liu XS, APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer, Oncogene, 37 (2018) 3924–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Boichard A, Tsigelny IF, Kurzrock R, High expression of PD-1 ligands is associated with kataegis mutational signature and APOBEC3 alterations, Oncoimmunology, 6 (2017) e1284719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Venkatesan S, Rosenthal R, Kanu N, McGranahan N, Bartek J, Quezada SA, Hare J, Harris RS, Swanton C, Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution, Ann Oncol, 29 (2018) 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]