Abstract
Oxaliplatin is the first-line chemotherapy for metastatic colorectal cancer. Unlike other platinum anticancer agents, oxaliplatin does not result in significant renal impairment and ototoxicity. Oxaliplatin, however, has been associated with acute and chronic peripheral neuropathies. Despite the awareness of these side-effects, the underlying mechanisms are yet to be clearly established. Therefore, in this study, we aimed to understand the factors involved in the generation of chronic neuropathy elicited by oxaliplatin treatment. We established a rat model of oxaliplatin-induced neuropathic pain (4 mg kg-1 intraperitoneally). The paw withdrawal thresholds were assessed at different time-points after the treatment, and a significant decrease was observed 3 and 4 weeks after oxaliplatin treatment as compared to the vehicle treatment (4.4 ± 1.0 vs. 16.0 ± 4.1 g; P < 0.05 and 4.4 ± 0.7 vs. 14.8 ± 3.1 g; P < 0.05, respectively). We further evaluated the role of different mitogen-activated protein kinases (MAPKs) pathways in the pathophysiology of neuropathic pain. Although the levels of total extracellular signal-regulated kinase (ERK) 1/2 in the dorsal root ganglia (DRG) were not different between oxaliplatin and vehicle treatment groups, phosphorylated ERK (p-ERK) 1/2 was up-regulated up to 4.5-fold in the oxaliplatin group. Administration of ERK inhibitor PD98059 (6 μg day-1 intrathecally) inhibited oxaliplatin-induced ERK phosphorylation and neuropathic pain. Therefore, upregulation of p-ERK by oxaliplatin in rat DRG and inhibition of mechanical allodynia by an ERK inhibitor in the present study may provide a better understanding of intracellular molecular alterations associated with oxaliplatin-induced neuropathic pain and help in the development of potential therapeutics.
Introduction
Oxaliplatin, a platinum-based drug, is used as the first-line chemotherapy for metastatic colorectal cancer. Unlike other platinum anticancer agents, oxaliplatin does not result in significant renal impairment and ototoxicity. However, oxaliplatin is associated with acute and chronic peripheral neuropathies [1, 2]. Oxaliplatin-induced acute neuropathy is characterized by acral paresthesia that is enhanced by exposure to cold. Furthermore, cumulative oxaliplatin dose can cause chronic neuropathy, which includes pain, paresthesia, hypoesthesia, dysesthesia, and changes in proprioception. Therefore, oxaliplatin-induced neuropathic pain is a major clinical side-effect that can influence the treatment as well as the quality of life.
Pain results from the activation of a subset of sensory neurons termed nociceptors. Under physiological conditions, activation of unmyelinated (C-fiber) and myelinated (Aδ-fiber) nociceptive afferent fibers indicates potential tissue damage, which is reflected in the high thresholds of nociceptors for mechanical, thermal, and chemical stimuli; these neurotransmissions are attributed to ion channels, neurotransmitters, and intracellular signaling [3, 4]. These conditions change dramatically in neuropathic pain states, including chemotherapy-induced peripheral neuropathy (CIPN). Understanding the changes that occur in neuropathic pain is vital to identify new therapeutic targets and develop novel analgesics [4]. Recently, it has been reported that oxaliplatin-induced acute paresthesia is induced by voltage-dependent sodium channel (NaV1.6) dysfunction [5–7] and upregulation of transient receptor potential (TRP) channels, TRPM8 and TRPA1 [8–11], which are temperature-sensitive channels. However, the pathophysiology of oxaliplatin-induced neuropathic pain as a chronic neuropathy has not yet been clearly established.
Mitogen-activated protein kinases (MAPKs) signaling cascade is known to be involved in the regulation of cellular functions such as cell differentiation, proliferation, and apoptosis [12, 13]. MAPKs, such as extracellular signal-regulated kinase (ERK), p38 kinase, and c-jun N-terminal kinase (JNK), have been linked with the development of pain [12, 13]. Furthermore, it has recently been reported that the modulation of MAPKs activation is associated with oxaliplatin-induced apoptosis in cultured dorsal root ganglion (DRG) neurons [14, 15].
Therefore, the aim of the current study was to understand the factors involved in the generation of chronic neuropathy elicited by oxaliplatin treatment. We investigated whether MAPKs were modulated by oxaliplatin in the rat DRG and found that oxaliplatin treatment up-regulates ERK phosphorylation in rat DRG and induced chronic neuropathic pain. We also demonstrated that administration of an ERK inhibitor inhibits oxaliplatin-induced neuropathic pain. Thus, our study suggests a novel mechanism by which oxaliplatin treatment can influence MAPKs signaling and contribute to chronic neuropathy.
Materials and methods
Animals
Six-week-old male Sprague Dawley rats (Kudo, Japan) weighing approximately 200–250 g were used in the study. All rats were individually housed in a temperature- and humidity-controlled environment with a 12-hour light-dark cycle and were permitted free access to food and water. The study was conducted in strict accordance with the guidelines for Proper Conduct of Animal Experiments (Science Council of Japan). The experiments were approved by the Experimental Animal Care and Use Committee of University of Miyazaki (Permit Number: 2015–528). All efforts were made to minimize the number of animals used and their suffering.
Pharmacological treatments
In the first series of experiments, we investigated the intracellular molecular alterations in DRG. Oxaliplatin (4 mg kg-1 of body weight; Sigma-Aldrich, St. Louis, MO, USA) or vehicle (5% glucose) was injected intraperitoneally (i.p.) twice a week for 4 weeks [16]. Oxaliplatin was prepared in 5% glucose to a final concentration of 2 mg ml-1. von Frey test was conducted before and 1 week after each oxaliplatin or vehicle treatment. On day 28, at the end of the last behavioral test, L4-L6 DRGs were dissected from each group and intracellular molecules including ERK were measured using western blot analysis.
In the second series of experiments, we investigated the effects of ERK inhibitor on oxaliplatin-induced neuropathy. Rats were anesthetized with an intraperitoneal injection of a combination anesthetic (0.375 mg kg-1 of medetomidine, 2.0 mg kg-1 of midazolam, and 2.5 mg kg-1 of butorphanol). A PE-10 polyethylene catheter (Becton-Dickinson, Sparks, MD, USA) was inserted into the subarachnoid space through the atlanto-occipital membrane and pushed to the region of lumbar enlargement [17]. An osmotic pressure pump (ALZET model 2004, DURECT, Cupertino, CA, USA; total volume of the pump: 200 μl, drug infusion: 0.25 μl hour-1 for 4 weeks) was connected to the catheter and placed subcutaneously on the back [18]. Immediately after surgery, the operating surgeon regularly observed animals until they were ambulatory. Furthermore, the animals' appearance, movement, and appetite were observed daily for one week after surgery. ERK inhibitor PD98059 (6 μg day-1; Sigma-Aldrich, St. Louis, MO, USA) or vehicle [20% dimethyl sulfoxide (DMSO)] was injected intrathecally (i.t.) for 4 weeks with the osmotic pressure pump. PD98059 was dissolved in 20% DMSO to a final concentration of 1 μg μl-1. One week after the pump placement, oxaliplatin (4 mg kg-1) or vehicle (5% glucose) was injected i.p. twice a week for 3 weeks. The sham-operated (Sham) mice underwent a similar surgical procedure except for pump placement and drug treatments. von Frey test was conducted before pump placement, and before and 1 week after each oxaliplatin or vehicle treatment. On day 28 of pump placement, at the end of the last behavioral test, L4-L6 DRGs were dissected from each group and intracellular levels of various molecules were measured using western blot analysis.
von Frey test
Mechanical sensitivity was examined by testing the paw withdrawal threshold using the von Frey (VF) filaments (Stoelting, Wood Dale, IL, USA). Briefly, each rat was placed in a 20 cm × 20 cm suspended chamber on a metallic mesh floor. After an acclimation period of 30 minutes, a series of calibrated VF filaments were applied perpendicularly to the plantar surface of the right and left hind paws with sufficient force to bend the filament for 5 seconds. Brisk withdrawal or paw flinching was considered as a positive response. In the absence of a response, the filament of next greater force was applied. After a positive response, the filament of next lower force was applied. The tactile stimulus producing a 50% likelihood of withdrawal response was calculated using the up-down method [19]. von Frey test was conducted before an osmotic pressure pump placement, and before and 1 week after each intraperitoneal oxaliplatin or vehicle treatment for 4 weeks.
Western blot analysis
On day 28 of intraperitoneal treatment or pump placement, all rats were euthanized with sevoflurane exposure and the L4-L6 DRGs from each group were quickly dissected for further analysis. Briefly, the collected DRGs were mechanically homogenized in ice-cold lysis buffer composed of 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, and 20 mM Tris-HCl, pH 7.5, with added Protease and Phosphatase Inhibitor Cocktails (Roche Diagnostics, Mannheim, Germany) and centrifuged at 12000 rpm and 4°C for 10 minutes. The supernatant was collected and stored at -80°C until use. The total protein content was determined in each sample using the Bradford method-based protein assay kit, with bovine serum albumin (BSA) as standard (Aproscience, Naruto, Japan). The supernatants were solubilized in 2× SDS electrophoresis sample buffer and heated at 98°C for 5 minutes. Equal amount of proteins (7.0–7.5 μg per lane) were separated by SDS-12% polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Merck Millipore, Burlington, MA, USA). The membrane was then incubated with a blocking solution [5% BSA in Tween-Tris-buffered saline (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween-20)] and further incubated overnight at 4°C in Can Get Signal Solution-1 (TOYOBO, Osaka, Japan) with rabbit anti-ERK polyclonal antibody (1:2000, K-23, Santa Cruz, Dallas, TX, USA), mouse anti-p-ERK monoclonal antibody (1:2000, E-4, Santa Cruz), rabbit anti-p38 monoclonal antibody (1:2000, D13E1, Cell Signaling Technology, MA, USA), rabbit anti-p-p38 monoclonal antibody (Thr180/Tyr182) (1:2000, D3F9, Cell Signaling Technology), rabbit anti-JNK polyclonal antibody (1:2000, D-2, Santa Cruz), mouse anti-p-JNK monoclonal antibody (1:2000, Santa Cruz), rabbit anti-BDNF polyclonal antibody (1:2000, ab226843, Abcam, Cambridge, UK) or mouse anti-β-actin monoclonal antibody (1:2000, A1978, Sigma-Aldrich, St. Louis, MO, USA). After repeated washing, the immunoreactive bands were developed using Can Get Signal Solution-2 with horseradish peroxidase-conjugated anti-rabbit antibody (1:5000, GE Healthcare Japan Corporation, Tokyo, Japan) or anti-mouse antibody (1:5000, Santa Cruz), then visualized using an enhanced chemiluminescence detection system reagent (Amersham ECL-prime, GE Healthcare Japan Corporation), and captured in a LAS-3000 Luminoimage analyzer (Fuji Film, Tokyo, Japan). A commercially available molecular weight marker (Amersham ECL rainbow marker–full range, GE Healthcare Japan Corporation), consisting of proteins of molecular weight 12 to 225 kDa, was used as a reference for each molecular weight. The densities of protein blots were quantified using ImageJ [20] and the protein levels were normalized to β-actin levels.
Statistical analysis
For behavioral experiments, the hindpaw data within each group were analyzed using one-way repeated measures analysis of variance (ANOVA) followed by Bonferroni post hoc analysis. Comparisons between two means of the hindpaw data and western blot data were performed by Welch’s test and Student’s t-test, respectively. The results were presented as mean ± SEM (for von Frey test) or ± SD (for western blot analysis). P < 0.05 was considered as significant. The statistics software used was JMP 11 (SAS Institute, Inc., Cary, NC, USA) for Macintosh.
Results
Mechanical allodynia in a rat model of oxaliplatin-induced neuropathic pain
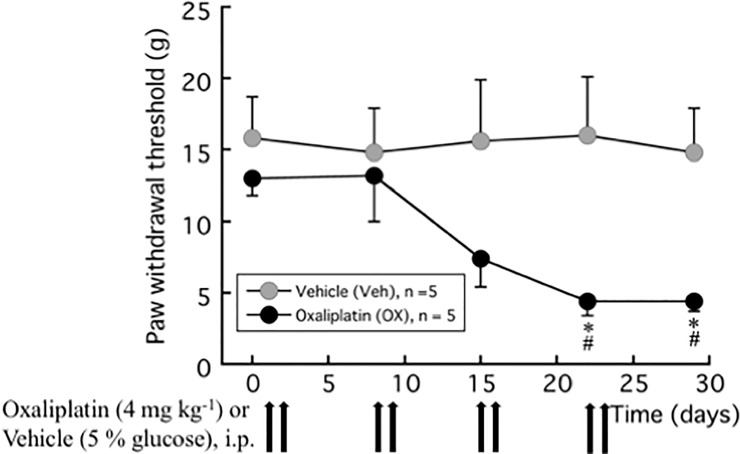
The oxaliplatin treatment (4 mg kg-1, twice a week for 4 weeks) induced increased pain behavior in the rat model [16]. Fig 1 shows that the paw withdrawal thresholds measured with VF filaments to the non-noxious mechanical stimulus at 3 and 4 weeks after oxaliplatin treatment were significantly lower than the vehicle treatment (4.4 ± 1.0 g vs. 16.0 ± 4.1 g; P = 0.046 and 4.4 ± 0.7 g vs. 14.8 ± 3.1 g; P = 0.027, respectively).
Fig 1. Paw withdrawal test (von Frey test) for mechanical allodynia induced by oxaliplatin.
Oxaliplatin (4 mg kg-1) was administered i.p. twice a week for 4 weeks (days 1, 2, 8, 9, 15, 16, 22, and 23). We confirmed the incidence of mechanical allodynia on day 28. The von Frey test was performed before and 1 week after each oxaliplatin or vehicle (5% glucose) treatment. The hindpaw data within each group were analyzed using one-way repeated measures ANOVA followed by Bonferroni post hoc analysis. For comparisons between groups at the same time, Welch’s test was used. All data are calculated as mean ± SEM of 5 animals. * P < 0.05, compared with Time 0 (baseline). # P < 0.05, compared with the vehicle at the same time.
Oxaliplatin treatment up-regulates ERK phosphorylation in rat DRG neurons
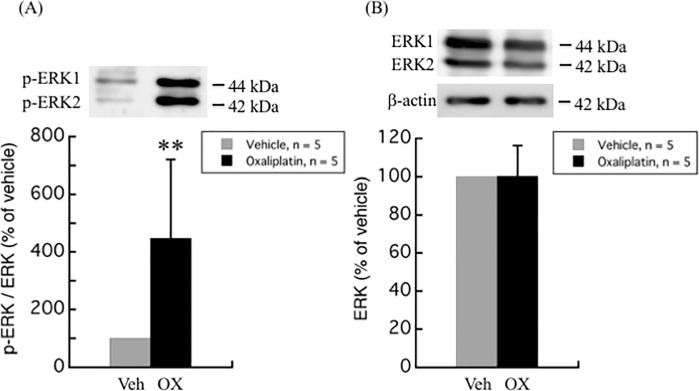
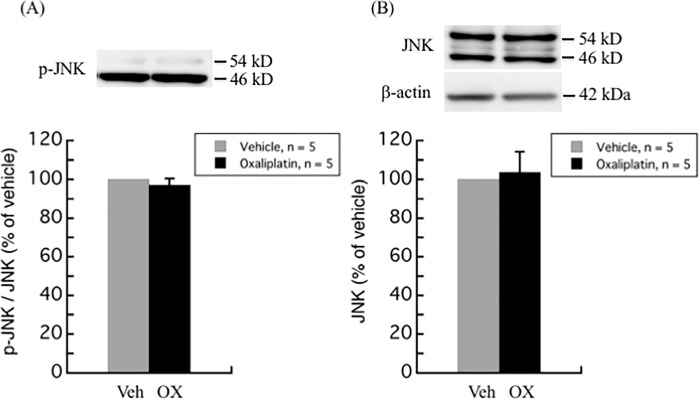
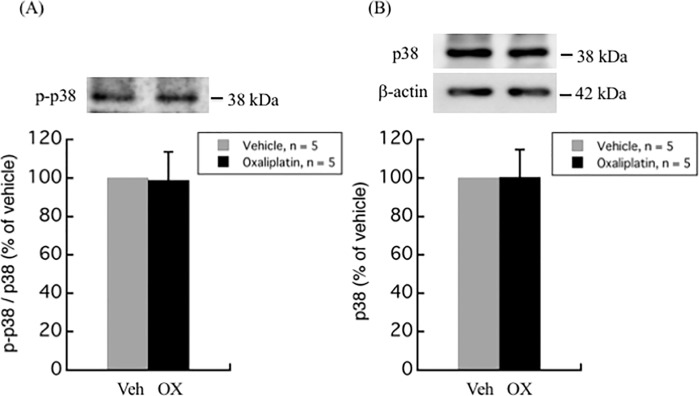
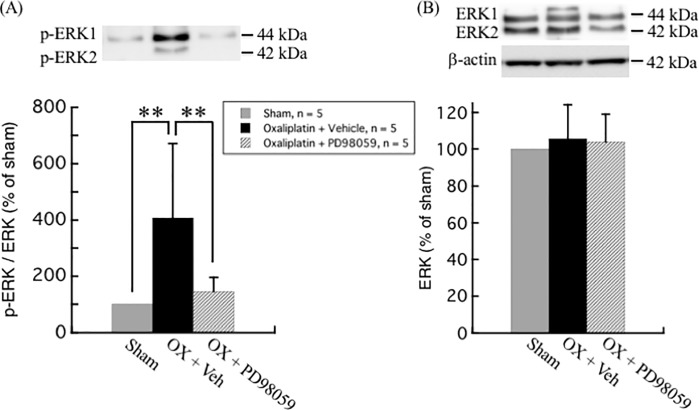
The modulation of MAPKs activation, including ERK, p38 kinase, and JNK pathways, have not only been linked with the development of pain [12, 13], but also with the oxaliplatin-induced apoptosis in DRG [14, 15]. Western blot analyses of different MAPKs in the DRG of oxaliplatin-treated rats as compared to vehicle-treated rats are illustrated in Figs 2–4. Although no difference was observed in the total ERK1/2 levels between oxaliplatin and vehicle treatment groups, p-ERK1/2 was found to be up-regulated up to 4.5-fold (447.6 ± 273.6%, P = 0.0029) in DRG of oxaliplatin-induced neuropathic pain rat model (Fig 2). On the other hand, no change was observed in the phosphorylation and protein levels of p38 and JNK between oxaliplatin and vehicle treatment groups (Figs 3 and 4).
Fig 2. Upregulation of ERK phosphorylation by oxaliplatin in rat DRG.
(A) The ratio of p-ERK to ERK expression was significantly increased in DRG of oxaliplatin treated rats. (B) No difference was observed in the protein levels of ERK between oxaliplatin and vehicle treatment groups. Comparisons between two groups of the blots were performed by Student’s t-test. All data are calculated as mean ± SD of 5 animals. ** P < 0.001, compared to the vehicle.
Fig 4. JNK phosphorylation and protein levels in rat DRG.
There was no difference in JNK phosphorylation and protein levels between oxaliplatin and vehicle treatment groups. Comparisons between two groups of the blots were performed by Student’s t-test. All data are calculated as mean ± SD of 5 animals.
Fig 3. p38 phosphorylation and protein levels in rat DRG.
There was no difference in p38 phosphorylation and protein levels between oxaliplatin and vehicle treatment groups. Comparisons between two groups of the blots were performed by Student’s t-test. All data are calculated as mean ± SD of 5 animals.
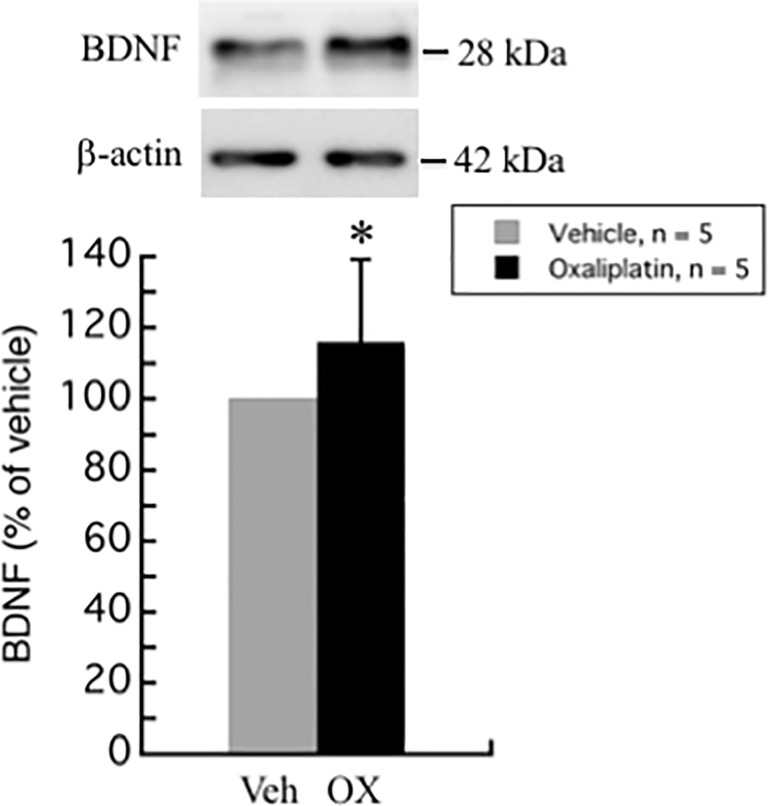
Oxaliplatin treatment increases brain-derived neurotrophic factor (BDNF) levels in rat DRG
BDNF is not only a nerve growth factor, but also a neurotransmitter of nociceptive fibers in the dorsal horn of the spinal cord. In the spinal nerve ligation (SNL) model of neuropathic pain, BDNF expression was found to be up-regulated in the rat spinal dorsal horn [21]. BDNF expression was also up-regulated in DRG in lumbar 5 ventral root transection model of neuropathic pain [21].
In our study, BDNF levels were increased in DRG of oxaliplatin-induced neuropathic pain rat model (115.8 ± 23.4%, P = 0.047) (Fig 5).
Fig 5. Upregulation of BDNF by oxaliplatin in rat DRG.
The expression of BDNF was significantly increased in DRG of oxaliplatin treated rats. Comparisons between two groups of the blots were performed by Student’s t-test. All data are calculated as mean ± SD of 5 animals. * P < 0.05, compared to the vehicle.
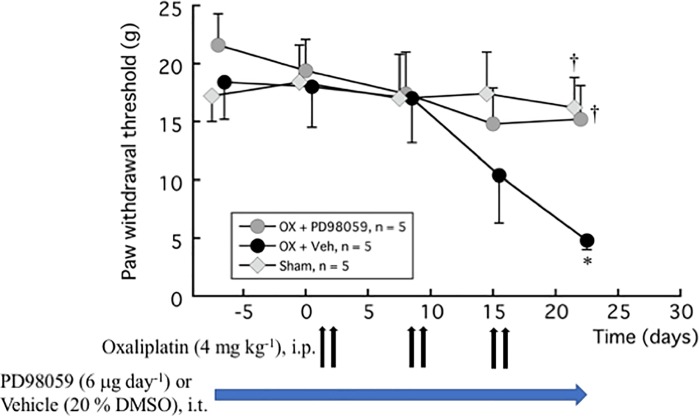
Effects of ERK inhibitor on oxaliplatin-induced neuropathic pain
Fig 6 shows that ERK inhibitor PD98059 (6 μg day-1) injected intrathecally inhibited oxaliplatin-induced mechanical allodynia. The paw withdrawal thresholds in the oxaliplatin and PD98059 treatment group were mostly maintained from baseline and were significantly higher than the oxaliplatin and vehicle treatment group at 3 weeks after oxaliplatin treatment (OX + PD98059: 15.2 ± 2.9 g and Sham: 16.2 ± 2.6 g vs. OX + Veh: 4.8 ± 0.8 g; P = 0.021 and P = 0.01, respectively). Concomitantly, PD98059 also inhibited oxaliplatin-induced upregulation of ERK phosphorylation in DRG (OX + PD98059: 147.6 ± 47.6%,vs. OX + Veh: 402.9 ± 251.2%, P = 0.0094) (Fig 7).
Fig 6. Inhibition of oxaliplatin-induced mechanical allodynia by ERK inhibitor PD98059.
PD98059 (6 μg day-1) or vehicle (20% DMSO) was injected i.t. for 4 weeks using an osmotic pressure pump. One week after the pump placement, oxaliplatin (4 mg kg-1) or vehicle (5% glucose) was injected i.p. twice a week for 3 weeks. von Frey test was done before the pump placement, and before and 1 week after each oxaliplatin or vehicle treatment (days 1, 2, 8, 9, 15, and 16). We confirmed the incidence of mechanical allodynia on day 22 (day 28 from pump placement). The hindpaw data within each group were analyzed using one-way repeated measures ANOVA followed by Bonferroni post hoc analysis. Welch’s test was used to compare between groups. All data are calculated as mean ± SEM of 5 animals. * P < 0.05, compared with Time -7 days (baseline). † P < 0.05, compared with OX + Veh at the same time.
Fig 7. Effect of ERK inhibitor PD98059 on oxaliplatin-induced upregulation of ERK phosphorylation in rat DRG.
(A) The ratio of p-ERK to ERK expression was significantly increased in DRG of oxaliplatin treated rats, which was inhibited by PD98059. (B) No difference was observed in the protein level of ERK among sham, oxaliplatin + vehicle (20% DMSO), and oxaliplatin + PD98059 treatment groups. Comparisons between two groups of the blots were performed by Student’s t-test. All data are calculated as mean ± SD of 5 animals. ** P < 0.01, compared with OX + Veh.
Discussion
Platinum-based drugs are the first-line chemotherapy for different cancers. Platinum derivatives such as oxaliplatin and cisplatin act as cytotoxins on tumor cells by forming platinum-DNA adducts, thus leading the tumor cells to programmed cell death. These platinum derivatives induce Chemotherapy-Induced Peripheral Neuropathy (CIPN) as one of the clinical side-effects [1, 2]. In an in-vitro study, treatment of cultured DRG neurons from E15 rat embryos with toxic doses of oxaliplatin or cisplatin induced a dose-dependent neuronal apoptosis by phosphorylating and inactivating the anti-apoptotic protein Bcl-2 and increasing the levels of the pro-apoptotic protein Bax [14].
Furthermore, studies have shown that these platinum derivatives modulate different MAPKs [13]. MAPKs are vital for intracellular signal transduction and play critical roles in regulation of neural plasticity and inflammatory responses [12, 13]. This family of kinases consists of three key members: ERK, p38, and JNK. Accumulating evidence shows that the activation of MAPKs can induce the synthesis of pronociceptive mediators via distinct molecular and cellular mechanisms, resulting in the enhancement and prolongation of pain [12, 13]. The platinum derivatives phosphorylate and activate p38 while they reduce the levels of active and total JNK. Both oxaliplatin and cisplatin have shown to activate ERKs during early stages (4–8 hours after treatment), although they behave differently at later stages [14]. Moreover, by using specific inhibitors of the different MAPKs, it has been demonstrated that the platinum-induced neuronal apoptosis is mediated by early p38 and ERK1/2 activation [14]. In in-vivo studies, oxaliplatin has shown to increase p38 phosphorylation at 0.5 and 4 hours after the treatment [22], or protein kinase C (PKC) phosphorylation, ERK1/2 phosphorylation, and c-fos expression on day 14 after the treatment [23], in the spinal cord of oxaliplatin-induced neuropathy mouse model. These results and our findings suggest a role for MAPKs including ERK in the generation and development of oxaliplatin-induced peripheral neuropathy.
Electrophysiological studies in patients undergoing oxaliplatin-treatment demonstrated nerve hyperexcitability in both peripheral motor [24] and sensory axons [25]. Furthermore, electrophysiological in-vitro studies in isolated peripheral nerve segments indicated that the hyperexcitability is characterized by an increase in the duration of the compound A-fiber action potential and the emergence of after-activity persisting over several tens of milliseconds [26–29]. A modulating effect on both voltage-dependent sodium channels and delayed rectifier potassium channels has been demonstrated during oxaliplatin administration to the myelinated axons in frog nerves [30] and to the neuronal cells in cell culture [31]. Thus, abnormal Na+ channel and/or K+ channel function has been highlighted as a possible mechanism of oxaliplatin-induced neuropathy. However, focusing on Na+ channel subtypes, NaV1.7, NaV1.8, and NaV1.9, expressed in the DRG, conditional knockout mice established using the NaV1.7Advill line, which eliminates NaV1.7 expression in all the DRG neurons, and the NaV1.7Wnt1 line, which lacks NaV1.7 expression in the DRG and sympathetic ganglion neurons, both NaV1.7Advill and NaV1.7Wnt1 mice developed mechanical and cold allodynia normally following oxaliplatin treatment [32]. In addition, global deletion of NaV1.3, NaV1.8, or NaV1.9 also did not attenuate either mechanical or cold allodynia in oxaliplatin-induced neuropathy. Furthermore, in our study, the expression levels of NaV1.7, NaV1.8, and NaV1.9 were not altered in the DRG of oxaliplatin treated rats compared to non-treated rats (S1 Fig and S2 Fig). These findings suggest that the expression of the voltage-dependent Na+ channel subtypes NaV1.3, NaV1.7, NaV1.8, and NaV1.9 is not required for the development of oxaliplatin-induced neuropathy.
So, how does abnormal Na+ current lead to the development of nerve hyperexcitability? Firstly, Na+ channel subtypes other than NaV1.3, NaV1.7, NaV1.8, and NaV1.9 could contribute to oxaliplatin-induced nerve hyperexcitability. Indeed, a recent study revealed that the expression of NaV1.6 was dramatically increased in the DRG in oxaliplatin-induced CIPN model rats [33]. Furthermore, the agomir of miR-30b, a microRNA implicated in neuropathic pain, cancer, and neurodegenerative diseases, can downregulate NaV1.6 and alleviate oxaliplatin-induced mechanical allodynia and cold hypersensitivity [33]. Secondly, the cytokines and chemokines associated with altering intracellular signaling could modify Na+ current, which is conducive to nerve hyperexcitability. Recently accumulated evidence has shown that oxaliplatin treatment increases pro- and anti-inflammatory cytokines and chemokines [34–40]. Oxaliplatin injection enhanced the mRNA levels of cytokines including tissue necrosis factor-α (TNF-α) and interleukin-1β (IL-1β), and chemokines including monocyte chemoattractant protein-1 (MCP-1, also referred to as C-C chemokine ligand (CCL) 2) and monocyte inflammatory protein-1 (MIP-1α, also referred to as CCL3) in the spinal dorsal horn [34]. Melatonin attenuates pain hypersensitivity by inhibition of TNF-α in oxaliplatin-induced neuropathy [33]. CCL2 and its receptor CCR2 have also been shown to be increased in the DRG after oxaliplatin administration, in parallel with the development of mechanical hypersensitivity [35, 36]. Wang et al. have reported that oxaliplatin treatment up-regulates NF-κB and induces neuronal hyperexcitability in DRG [37]. This neuronal hyperexcitability was inhibited by NF-κB inhibitors. Oxaliplatin-induced pain has also been shown to be accompanied with the upregulation of PI3K-mTOR and mTOR-mediated signals as well as IL-1β, IL-6, and TNF-α in DRG. As PI3K or mTOR signal was inhibited, mechanical and cold hypersensitivity were attenuated in oxaliplatin treated rats, and the levels of proinflammatory cytokines also decreased [38]. The upregulation of pro-inflammatory cytokines and membrane pro-inflammatory cytokine receptors in the midbrain periaqueductal gray, which has an inhibitory or excitatory control on pain transmission via the rostral ventromedial medulla, projecting to the spinal dorsal horn, of oxaliplatin treated rats is likely to impair the descending inhibitory pathways in regulation of pain transmission and thereby, contribute to the development of neuropathic pain after the administration of chemotherapeutic oxaliplatin [39]. These reports suggest that the mechanism of development of oxaliplatin-induced neuropathy resembles inflammatory pain. Furthermore, Huang et al. and Liu et al. have reported an increase in the levels of TNF-α, NF-κB, and phosphorylation of ERK in the spinal cord and DRG of an oxaliplatin-induced peripheral neuropathy rat model and a lumber disk herniation rat model [36, 40]. Previous studies have demonstrated that TNF-α enhances TTX-R Na+ currents via TNFR1 and the p38 pathway in the cultured DRG neurons within 1 minute of the onset of TNF-α application (peak effect within 3–5 minutes) [41], as well as via the p38 pathway in the uninjured DRG neurons after L5-ventral root transection (VRT) in vivo [42]. The phosphorylated ERK1 lowers the activation threshold, making it easier to open NaV1.7 channel in response to weak stimuli [43]. We have also reported that veratridine-induced 22Na+ influx was inhibited by the inhibitors of ERK and p38, indicating that the basal constitutive activities of ERK and p38 may prime NaV1.7 to open [44]. These findings suggest that oxaliplatin-induced neuroinflammation and inflammatory mediators would evolve abnormal Na+ channel currents via MAPK including ERK phosphorylation, and thus, lead to the development of neuropathy.
Conclusions
Oxaliplatin administration induces chronic mechanical allodynia in rats. The phosphorylation of ERK is upregulated in the DRG of oxaliplatin-induced neuropathic pain rat model, whereas other MAPKs, p38, and JNK are not altered. ERK inhibitor impedes mechanical allodynia by inhibiting oxaliplatin-induced upregulation of ERK phosphorylation. Thus, the findings from the present study may provide a better understanding of the intracellular molecular alterations in the development of oxaliplatin-induced neuropathic pain and help in designing effective therapeutics.
Supporting information
Typical western blots of NaV1.7, NaV1.8, and NaV1.9 are shown. These images suggested that there was no difference in NaV1.7, NaV1.8, and NaV1.9 protein levels between oxaliplatin and vehicle treatment groups.
(TIFF)
Typical polymerase chain reaction (PCR) gel images of NaV1.7, NaV1.8, and NaV1.9 are shown. These images suggested that there was no difference in NaV1.7, NaV1.8, and NaV1.9 mRNA expression levels between oxaliplatin and vehicle treatment groups.
(TIFF)
Acknowledgments
This study is attributed to the Department of Anesthesiology, Faculty of Medicine, University of Miyazaki. The authors would like to thank Noriko Hidaka, Mio Kurogi, and Toshiko Watanabe for their technical and secretarial assistance in this study. The authors would like to thank Editage (www.editage.jp) for English language editing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a Grant for Clinical Research from University of Miyazaki Hospital. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Extra JM, Marty M, Brienza S, Misset JL. Pharmacokinetics and safety profile of oxaliplatin. Semin Oncol. 1998;25(2 Suppl 5): 13–22. [PubMed] [Google Scholar]
- 2.Lehky TJ, Leonard GD, Wilson RH, Grem JL, Floeter MK. Oxaliplatin-induced neurotoxicity: acute hyperexcitability and chronic neuropathy. Muscle Nerve. 2004;29: 387–392. 10.1002/mus.10559 [DOI] [PubMed] [Google Scholar]
- 3.Baron R, Binder A, Wasner G. Neuropathic pain: diagnosis, pathophysiological mechanisms, and treatment. Lancet Neurol. 2010;9:807–819. 10.1016/S1474-4422(10)70143-5 [DOI] [PubMed] [Google Scholar]
- 4.St John Smith E. Advances in understanding nociception and neuropathic pain. J Neurol. 2018;265:231–238. 10.1007/s00415-017-8641-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grolleau F, Gamelin L, Boisdron-Celle M, Lapied B, Pelhate M, Gamelin E. A possible explanation for a neurotoxic effect of the anticancer agent oxaliplatin on neuronal voltage-gated sodium channels. J Neurophysiol. 2001;85: 2293–2297. 10.1152/jn.2001.85.5.2293 [DOI] [PubMed] [Google Scholar]
- 6.Sittl R, Lampert A, Huth T, Schuy ET, Link AS, Fleckenstein J, et al. Anticancer drug oxaliplatin induces acute cooling-aggravated neuropathy via sodium channel subtype Na(V)1.6-resurgent and persistent current. Proc Natl Acad Sci U S A. 2012;109: 6704–6709. 10.1073/pnas.1118058109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deuis JR, Zimmermann K, Romanovsky AA, Possani LD, Cabot PJ, Lewis RJ, et al. An animal model of oxaliplatin-induced cold allodynia reveals a crucial role for Nav1.6 in peripheral pain pathways. Pain. 2013;154: 1749–175 10.1016/j.pain.2013.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauchan P, Andoh T, Kato A, Kuraishi Y. Involvement of increased expression of transient receptor potential melastatin 8 in oxaliplatin-induced cold allodynia in mice. Neurosci Lett. 2009;458: 93–95. 10.1016/j.neulet.2009.04.029 [DOI] [PubMed] [Google Scholar]
- 9.Nassini R, Gees M, Harrison S, De Siena G, Materazzi S, Moretto N, et al. Oxaliplatin elicits mechanical and cold allodynia in rodents via TRPA1 receptor stimulation. Pain. 2011;152: 1621–1631. 10.1016/j.pain.2011.02.051 [DOI] [PubMed] [Google Scholar]
- 10.Descoeur J, Pereira V, Pizzoccaro A, Francois A, Ling B, Maffre V, et al. Oxaliplatin-induced cold hypersensitivity is due to remodelling of ion channel expression in nociceptors. EMBO Mol Med. 2011;3:266–278. 10.1002/emmm.201100134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kono T, Satomi M, Suno M, Kimura N, Yamazaki H, Furukawa H, et al. Oxaliplatin-induced neurotoxicity involves TRPM8 in the mechanism of acute hypersensitivity to cold sensation. Brain Behav. 2012;2: 68–73. 10.1002/brb3.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obata K, Noguchi K. MAPK activation in nociceptive neurons and pain hypersensitivity. Life Sci. 2004;74: 2643–2653. 10.1016/j.lfs.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 13.Ji RR, Gereau RW 4th, Malcangio M, Strichartz GR. MAP kinase and pain. Brain Res Rev. 2009;60: 135–148. 10.1016/j.brainresrev.2008.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scuteri A, Galimberti A, Maggioni D, Ravasi M, Pasini S, Nicolini G, et al. Role of MAPKs in platinum-induced neuronal apoptosis. Neurotoxicology. 2009;30: 312–319. 10.1016/j.neuro.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 15.Scuteri A, Galimberti A, Ravasi M, Pasini S, Donzelli E, Cavaletti G, et al. NGF protects dorsal root ganglion neurons from oxaliplatin by modulating JNK/Sapk and ERK1/2. Neurosci Lett. 2010;486: 141–145. 10.1016/j.neulet.2010.09.028 [DOI] [PubMed] [Google Scholar]
- 16.Kawashiri T, Egashira N, Watanabe H, Ikegami Y, Hirakawa S, Mihara Y, et al. Prevention of oxaliplatin-induced mechanical allodynia and neurodegeneration by neurotropin in the rat model. Eur J Pain. 2011;15:344–350. 10.1016/j.ejpain.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Pai M, Brederson JD, Wilcox D, Hsieh G, Jarvis MF, et al. Monosodium iodoacetate-induced joint pain is associated with increased phosphorylation of mitogen activated protein kinases in the rat spinal cord. Mol Pain. 2011;7: 39 10.1186/1744-8069-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi A, Shinoda M, Sessle BJ, Honda K, Imamura Y, Hitomi S, et al. Mechanisms involved in extraterritorial facial pain following cervical spinal nerve injury in rats. Mol Pain. 2011;7: 12 10.1186/1744-8069-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka N, Yamaga M, Tateyama S, Uno T, Tsuneyoshi I, Takasaki M. The effect of pulsed radiofrequency current on mechanical allodynia induced with resiniferatoxin in rats. Anesth Analg. 2010;111: 784–790. 10.1213/ANE.0b013e3181e9f62f [DOI] [PubMed] [Google Scholar]
- 20.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siniscalco D, Giordano C, Rossi F, Maione S, de Novellis V. Role of neurotrophins in neuropathic pain. Curr Neuropharmacol. 2011;9: 523–529. 10.2174/157015911798376208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsubaki M, Takeda T, Tani T, Shimaoka H, Suzuyama N, Sakamoto K, et al. PKC/MEK inhibitors suppress oxaliplatin-induced neuropathy and potentiate the antitumor effects. Int J Cancer. 2015;137: 243–250. 10.1002/ijc.29367 [DOI] [PubMed] [Google Scholar]
- 23.Yeo JH, Yoon SY, Kim SJ, Oh SB, Lee JH, Beitz AJ, et al. Clonidine, an alpha-2 adrenoceptor agonist relieves mechanical allodynia in oxaliplatin-induced neuropathic mice; potentiation by spinal p38 MAPK inhibition without motor dysfunction and hypotension. Int J Cancer. 2016;138: 2466–2476. 10.1002/ijc.29980 [DOI] [PubMed] [Google Scholar]
- 24.Lehky TJ, Leonard GD, Wilson RH, Grem JL, Floeter MK. Oxaliplatin-induced neurotoxicity: acute hyperexcitability and chronic neuropathy. Muscle Nerve. 2004;29: 387–392. 10.1002/mus.10559 [DOI] [PubMed] [Google Scholar]
- 25.Park SB, Goldstein D, Lin CS, Krishnan AV, Friedlander ML, Kiernan MC. Acute abnormalities of sensory nerve function associated with oxaliplatin-induced neurotoxicity. J Clin Oncol. 2009;27: 1243–1239. 10.1200/JCO.2008.19.3425 [DOI] [PubMed] [Google Scholar]
- 26.Adelsberger H, Quasthoff S, Grosskreutz J, Lepier A, Eckel F, Lersch C. The chemotherapeutic oxaliplatin alters voltage-gated Na+ channel kinetics on rat sensory neurons. Eur J Pharmacol. 2000;40: 25–32. [DOI] [PubMed] [Google Scholar]
- 27.Webster RG, Brain KL, Wilson RH, Grem JL, Vincent A. Oxaliplatin induces hyperexcitability at motor and autonomic neuromuscular junctions through effects on voltage-gated sodium channels. Br J Pharmacol. 2005;146: 1027–39. 10.1038/sj.bjp.0706407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagiava A, Tsingotjidou A, Emmanouilides C, Theophilidis G. The effects of oxaliplatin, an anticancer drug, on potassium channels of the peripheral myelinated nerve fibres of the adult rat. Neurotoxicology. 2008;29: 1100–1106. 10.1016/j.neuro.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 29.Sittl R, Carr RW, Fleckenstein J, Grafe P. Enhancement of axonal potassium conductance reduces nerve hyperexcitability in an in vitro model of oxaliplatin-induced acute neuropathy. Neurotoxicology. 2010;31: 694–700. 10.1016/j.neuro.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 30.Benoit E, Brienza S, Dubois JM. Oxaliplatin an anticancer agent that affects both Na+ and K+ channels in frog peripheral myelinated axons. Gen Physiol Biophys. 2006;25: 263–276. [PubMed] [Google Scholar]
- 31.Wu SN, Chen BS, Wu YH, Peng H, Chen LT. The mechanism of the actions of oxaliplatin on ion currents and action potentials in differentiated NG108-15 neuronal cells. Neurotoxicology. 2009;30: 677–685. 10.1016/j.neuro.2009.04.010 [DOI] [PubMed] [Google Scholar]
- 32.Minett MS, Falk S, Santana-Varela S, Bogdanov YD, Nassar MA, Heegaard AM, et al. Pain without nociceptors? Nav1.7-independent pain mechanisms. Cell Rep. 2014;6: 301–312. 10.1016/j.celrep.2013.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Shao J, Wang J, Liu Y, Zhang Y, Zhang M, et al. MiR-30b-5p attenuates oxaliplatin-induced peripheral neuropathic pain through the voltage-gated sodium channel Nav1.6 in rats. Neuropharmacology. 2019;153:111–120. 10.1016/j.neuropharm.2019.04.024 [DOI] [PubMed] [Google Scholar]
- 34.Wang YS, Li YY, Cui W, Li LB, Zhang ZC, Tian BP, et al. Melatonin Attenuates Pain Hypersensitivity and Decreases Astrocyte-Mediated Spinal Neuroinflammation in a Rat Model of Oxaliplatin-Induced Pain. Inflammation. 2017;40: 2052–2061. 10.1007/s10753-017-0645-y [DOI] [PubMed] [Google Scholar]
- 35.Illias AM, Gist AC, Zhang H, Kosturakis AK, Dougherty PM. Chemokine CCL2 and its receptor CCR2 in the dorsal root ganglion contribute to oxaliplatin-induced mechanical hypersensitivity. Pain. 2018;159:1308–1316. 10.1097/j.pain.0000000000001212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Huang J, Jiang Y, Huang X, Xing W, He Y, et al. Oxaliplatin regulates chemotherapy induced peripheral neuropathic pain in the dorsal horn and dorsal root ganglion via the calcineurin/NFAT Pathway. Anticancer Agents Med Chem. 2018;18:1197–1207. 10.2174/1871520618666180525091158 [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Zhang XS, Tao R, Zhang J, Liu L, Jiang YH, et al. Upregulation of CX3CL1 mediated by NF-κB activation in dorsal root ganglion contributes to peripheral sensitization and chronic pain induced by oxaliplatin administration. Mol Pain. 2017;13: 1744806917726256 10.1177/1744806917726256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan Z, Su Z, Wang H, Pang X. Involvement of pro-inflammation signal pathway in inhibitory effects of rapamycin on oxaliplatin-induced neuropathic pain. Mol Pain. 2018;14: 1744806918769426 10.1177/1744806918769426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu D, Zhao H, Gao H, Zhao H, Liu D, Li J. Participation of pro-inflammatory cytokines in neuropathic pain evoked by chemotherapeutic oxaliplatin via central GABAergic pathway. Mol Pain. 2018;14: 1744806918783535 10.1177/1744806918783535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu ZH, Miao GS, Wang JN, Yang CX, Fu ZJ, Sun T. Resolvin D1 Inhibits Mechanical Hypersensitivity in Sciatica by Modulating the Expression of Nuclear Factor-κB, Phospho-extracellular Signal-regulated Kinase, and Pro- and Anti-inflammatory Cytokines in the Spinal Cord and Dorsal Root Ganglion. Anesthesiology. 2016;124: 934–944. 10.1097/ALN.0000000000001010 [DOI] [PubMed] [Google Scholar]
- 41.Jin X, Gereau RW 4th. Acute p38-mediated modulation of tetrodotoxin-resistant sodium channels in mouse sensory neurons by tumor necrosis factor-α. J Neurosci. 2006;26: 246–255. 10.1523/JNEUROSCI.3858-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu JT, Xin WJ, Wei XH, Wu CY, Ge YX, Liu YL, et al. p38 activation in uninjured primary afferent neurons and in spinal microglia contributes to the development of neuropathic pain induced by selective motor fiber injury. Exp Neurol. 2007;204: 355–365. 10.1016/j.expneurol.2006.11.016 [DOI] [PubMed] [Google Scholar]
- 43.Stamboulian S, Choi JS, Ahn HS, Chang YW, Tyrrell L, Black JA, et al. ERK1/2 mitogen-activated protein kinase phosphorylates sodium channel Na(v)1.7 and alters its gating properties. J Neurosci. 2010;30: 1637–1647. 10.1523/JNEUROSCI.4872-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemoto T, Miyazaki S, Kanai T, Maruta T, Satoh S, Yoshikawa N, et al. NaV1.7-Ca2+ influx-induced increased phosphorylations of extracellular signal-regulated kinase (ERK) and p38 attenuate tau phosphorylation via glycogen synthase kinase-3β: priming of NaV1.7 gating by ERK and p38. Eur J Pharmacol. 2010;640: 20–28. 10.1016/j.ejphar.2010.04.048 [DOI] [PubMed] [Google Scholar]