Abstract
One obstacle in de novo protein design is the vast sequence space that needs to be searched through to obtain functional proteins. We developed a new method using structural profiles created from evolutionarily related proteins to constrain the simulation search process, with functions specified by atomic-level ligand- protein binding interactions. The approach was applied to redesigning the BIR3 domain of the X-linked inhibitor of apoptosis protein (XIAP), whose primary function is to suppress the cell death by inhibiting caspase-9 activity; however, the function of the wild-type XIAP can be eliminated by the binding of Smac peptides. Isothermal calorimetry and luminescence assay reveal that the designed XIAP domains can bind strongly with the Smac peptides but do not significantly inhibit the caspase-9 proteolytic activity in vitro compared with the wild-type XIAP protein. Detailed mutation assay experiments suggest that the binding specificity in the designs is essentially determined by the interplay of structural profile and physical interactions, which demonstrates the potential to modify apoptosis pathways through computational design.
Keywords: protein design, evolutionary profile, apoptosis pathway, XIAP, isothermal calorimetry
Introduction
The computational design of functional macromolecules useful for disease model systems, diagnostics, therapeutics, and industrial applications is becoming a viable protein engineering method, but success has been hindered by the complex atomic interaction graph that yields such diverse functionality and specificity [1–5]. Here we report a hybrid computational protein–peptide design method using structure-based evolutionary profiles to reduce the inherent complexity of the design simulation search through the identification of protein evolutionary fingerprints, with the biological ligand-binding interaction specified by the physics-based force field. The method is applied to create protein domains to modulate programmed cell death, or apoptosis.
Regulating apoptosis is a powerful medicinal approach, as it can be used to either protect cells from death or cull them. In cancer, for example, promoting apoptosis in oncogenic cells is beneficial to remove them from the body; but blocking apoptosis in cardiovascular disease, such as following reduced blood flow to the heart, or ischemia, may be cardioprotective, and thus delay or reduce myocardial infarction [6]. The BIR3 domain of X-linked inhibitor of apoptosis (XIAP) is an attractive protein design target because it is an inhibitor of caspase-9-dependent cellular apoptosis and has a compact, well-characterized fold that is the subject of active drug discovery [7–9]. The XIAP BIR3 domain inhibits caspase-9 activity through the formation of a stable heterodimeric complex, which blocks caspase-9 from forming a homodimeric proteolytically active state. Caspase-9 is an initiator of the caspase proteolytic cascade involving caspase-3, caspase-6, and caspase- 7 that directly cause cell death and thus completes apoptosis [10]. Interestingly, the protein Smac also binds the XIAP BIR3 domain and subsequently blocks the XIAP-caspase-9 interaction, thus, freeing caspase-9 to homodimerize and initiate apoptosis [7,9,11]. Smac and caspase-9 bind to the same surface region on XIAP and compete for an N-terminal tetrapeptide binding pocket that ultimately governs whether XIAP associates with Smac or caspase-9. The primary goal of this study was to design de novo XIAP “like” protein sequences that were capable of binding the N-terminal Smac tetrapeptide with equal or better affinity than WT-XIAP. These designed proteins were intended to function as “Smac sinks” to remove free cytosolic Smac from the cell, or Smac-like therapeutic compounds, and thus be anti-apoptotic in nature and useful as a reagent in an apoptotic disease model system.
One challenge of computational protein design can be attributed to the fact that the sequence search space is vast compared to the available computational power (20101 permutations for the 101 residue XIAP BIR3 domain) [5]. The problem is exacerbated by imperfect force fields, which cannot accurately describe atomic interactions, or correctly recognize protein folds of given sequences. Borrowing the critical lessons from protein structure prediction experiments, where evolutionary references and fingerprints derived from the ensemble of known protein structures have been the major driving force for the success of high-resolution structure modeling [12,13], we propose to exploit the evolutionary sequence profiles from multiple homologous structures in the PDB to improve the energy landscape of physics-base force fields and the sequence space search of protein design.
In fact, the idea of using evolutionary information to specify the fold of the target protein is not new in computational protein design. For instance, Socolich and coworkers [14] successfully designed a stable fold of WW proteins using constraints from multiple sequence alignments collected by PSI-BLAST search. In recent studies, we proposed a method called EvoDesign [15,16], which utilized the sequence profiles collected from structural alignments to redesign 330 protein domains (87 from the PDB and 243 from the Mycobacterium tuberculosis genome), where 3 out of 5 tested proteins have well-ordered tertiary structure [15]. In particular, the crystal structure of the designed Phox homology domain from the cytokine-independent survival kinases was found to be very close (with 1.32 Å) to the target model of the designed sequence predicted by the I-TASSER-based protein structure prediction [17–19].
Despite the power of the evolutionary design in specifying protein folds, most of the designed proteins are assumed to be non-functional since no biological information (e.g., binding, catalysis, etc.) was incorporated. Here we examine the possibility of introducing function into the evolution-based design simulations by coupling the evolutionary profiles with specific ligand-binding interactions. The binding potentials can be either physical [20] or evolutionary based [21,22]. When applying the method for XIAP, we focused on the use of an atomic ligand–protein interaction potential extended from FoldX [20] to enhance the binding specificity of the XIAP–peptide interactions, which contains van der Waals, solvation, hydrogen-bonding, atomic clash, and entropic interaction terms. In addition, an empirical equation designed for enhancing the association rate of complex formation [23] was introduced to count for the electrostatic contribution between atoms of the interacting molecules (see Fig. 1 for the hybrid pipeline extended from EvoDesign).
Fig. 1.
Flowchart of the extended EvoDesign for hybrid evolution-based protein design. The protocol consists of three modules: (1) structure profile construction by threading the scaffold structure through the PDB using TM-align [24], (2) Monte Carlo sequence search guided by evolutionary structural profiles combined with physics-based binding potentials, (3) sequence selection by clustering with distance matrix defined from BLOSUM62 substitution scores.
A variety of biophysical experiments are designed to examine the folding and ligand-binding affinity of the designed XIAP BIR3 domains. Of particular interest is the novel use of high-resolution hydrogen–deuterium exchange (HDX) mass spectroscopy (MS) in conjunction with a new HDX prediction algorithm to examine the tertiary fold by the comparison of HDX data with I-TASSER-based protein structure prediction [18,19]. Furthermore, the binding specificity of XIAP with two cognate N-terminal Smac motif peptides and the inhibition of caspase-9 function are quantitatively characterized through isothermal calorimetry (ITC) and in vitro luminescence inhibition assay, respectively. The data should provide useful insight into whether and how the physics-based binding potentials can be used to introduce biological activity and specificity into evolutionary protein designs.
Results
Two sequences, Dynamic-Interface XIAP (DI-XIAP) and Fixed-Interface XIAP (FI-XIAP), were designed by the extended EvoDesign pipeline (Fig. 1), using the X-ray structure of the Human XIAP BIR3 domain co-crystallized with a high affinity N-terminus Smac tetrapeptide “AVPF” (PDBID: 2OPZ) [25] as the scaffold. Ten non-homologous proteins with a TM-score >0.5 and the sequence identity <80% to the scaffold were identified by TM-align [24], which have the pairwise protein sequence and structure alignments and the similarity scores listed in Table S1 in Supporting Information (SI, see also Text S1). The pairwise structure alignments were used to construct a profile (Fig. S1A) to guide the sequence design simulations, where the physics-based ligand–protein binding potential from FoldX was extended to constrain the Smac-XIAP interactions (see Materials and Methods). Here, the profile is specified by the substitution scoring matrix derived from the multiple sequence alignments of the templates that are collected based on structural alignments (see (Eq. 1) in Materials and Methods), which is termed “structural profile” afterward. In DI-XIAP, multiple low-energy sequences were generated by the extensive replica-exchange Monte Carlo (REMC) simulation, with the sequence of the global minimum free energy selected by sequence clustering. In FI-XIAP, similar REMC search was implemented but the interface residues in contact with the peptide were taken from the wild-type sequence and kept frozen during the simulation; this choice of two designs is made for examining the impact of extensive versus constrained interface search on the final designs.
The design simulation and selection procedures were fully automated. Only one sequence was selected for each protein from the center of the largest sequence cluster and no experimental optimization was conducted. The DNA and protein sequences designed are listed in Table S2 (see also Text S2). Fig. 2a shows the sequence alignment of the three proteins (WT-XIAP, FI-XIAP, and DI-XIAP) with the functional sites bound with the N-terminal Smac peptide motif or the full-length caspase-9 labeled below the sequences. The overall sequence identity of FI- and DI-XIAP is 52% and 47% to the wild-type XIAP protein, which is higher than all the templates that were used to construct the structure profile (except for 3T6P that has a sequence identity 48.5% by the sequence-based NW-align but with a sequence identity 41.6% by TM-align; see Table S1). Among the 15 (or 30) residues bound with Smac (or caspase-9), 7 (or 14) in DI-XIAP differ from that in WT-XIAP, showing that nearly half of the interface residues were redesigned, with a mutation rate similar to the global sequences. A parameter summary of the FI-, DI-, and WI-XIAP sequences is listed in Table 1.
Fig. 2.
Sequence and predicted structure of designed XIAP proteins. (a) Sequence alignments of WT-, FI- and DI-XIAPs. Secondary structural elements from WT-XIAP X-ray structure (2OPZ) are displayed above the alignments. Interfacial residues between WT-XIAP and the “AVPF” tetrapeptide or caspase-9 are shown below the alignments and colored in black spheres. Red blocked residues indicate differences in the peptide-binding site between WT- and DI-XIAPs. Orange blocked residues are the mutations outside the N-terminal binding pocket known to result in loss of caspase-9 inhibition [8]. (b) Superposition of I-TASSER models of DI- and FI-XIAP on the 17 PDB structures all solved for the same wild-type XIAP sequence (PDB IDs: 1F9XA, 1G3FA, 1NW9A, 1TFQA, 1TFTA, 2JK7A, 2OPZA, 2VSLA, 3CLXA, 3EYLA, 4EC4A, 4HY0A, 1G73D, 2OPYA, 3CM7A, 3G76A, 3CM2A). The wild-type PDB structures are in thin lines with the DI- and FI-XIAP models shown in cartoons (blue to red running from N- to C-terminals). Two arrows mark the borders of the disordered tails. Mutated interface residues are highlighted by red and blue sticks for WT-XIAP and DI-XIAP, respectively. Yellow spheres indicate the “AVPF” tetrapeptide from 2OPZ. (c) Complex structure of designed XIAPs with Caspase-9 crystal structure generated by superposing of the designed XIAP models on the WT-XIAP BIR3 domain of the complex X-ray structure. Insets show the XIAP/Caspase-9 interface, where mutations (G326Q/N, H343Q/K and L344G/A) known to abolish caspase-9 inhibitions are highlighted [8]. Blue and red are side-chain conformations from designed and WT-XIAP proteins for the three mutations.
Table 1.
Parameter summary of the designed proteins with standard deviations
| Parameters | FI-XIAP | DI-XIAP | WT-XIAP |
|---|---|---|---|
| Sequence identity to wild-typea | 52% (100%) | 47% (53%) | |
| RMSD of I-TASSER model to WT (Å)b | 1.12 | 1.01 | 1.28 |
| HDX correlationc | 0.55 | 0.74 | |
| Kds with “AVPF” (nM)d | 352 ± 79 | 167 ± 61 | 80 ± 25 |
| Kds with “AVPIAQKSEKY” (nM)d | 971 ± 191 | 554 ± 93 | 428 ± 72 |
Sequence identity between the designed and wild-type sequences. Values in parenthesis are that in the binding pocket.
Average RMSD of the I-TASSER models in the core region [Y265–Q336] for the designed and wild-type sequences to the 17 PDB structures solved for the same wild-type sequence; while the average RMSD between the 17 PDB structure is 1.29 Å in the core region.
PCCs between the observed and predicted HDX rates on the designed sequences.
Average dissociation constants (Kds) from five repeated ITC experiments. Errors are the average of the standard error from each of the repeated ITC experiments.
The sequence identity between DI- and FI-XIAP is 51%, which seems indicating that the freezing of a few interface residues in FI-XIAP could result in a dramatic change on the global sequence design since nearly half of the sequences are different. In fact, the change is largely due to the sequence selection process, since a number of DI- and FIXIAP sequence pairs in the designed sequence trajectories have a high sequence identity (>80%) but SPICKER clustering does not select them since they were not located at the center of the largest cluster. Meanwhile, the majority of the sequence variations are located in the tail regions, suggesting that many of the difference in the final DI- and FIXIAP selections are not essential to their functions, except for the residues at the core regions.
I-TASSER structure predictions match with HDX data
Prior to gene synthesis, we examined the fold-ability of the designed XIAPs using I-TASSER structure folding simulations [18,19]. In a large-scale experiment to examine the folding of designed sequences [15], it was shown that there is a strong correlation between the confidence score (C-score) of I-TASSER simulations and the folding rate of design proteins, and 80% (or 100%) of designed sequences are foldable for the sequence with an I-TASSER C-score >0 (or >0.8). Such correlation was also confirmed in another design study for the PX domain from cytokine-independent survival kinase, in which the I-TASSER model of the designed sequence with a C-score = 1.31 has a TM-score = 0.91 (or RMSD 1.31 Å) to the finally solved X-ray structure [17]. Here, although all homologous templates with a sequence identity >30% to the target or detected by PSI-BLAST with E-value <0.5 were excluded, the trajectories of the I-TASSER simulations on DI-XIAP (or FI-XIAP) are highly converged with 86% (or 82%) of conformations accumulated in the first SPICKER cluster [26] at an RMSD cutoff of 3.5 Å; this results in a high C-score of folding 0.82 and 0.8 for the DI- and FI-XIAPs, respectively, both being above the threshold of confident folding based on previous benchmark data [15].
In Fig. 2b, we show the first I-TASSER models of DI- and FI-XIAP, superimposed on the wild-type XIAP structures that were solved in 17 PDB entries all for the same sequence. Although nearly half of the designed sequences differ from WT-XIAP, the I-TASSER models are close to the wild-type XIAPs, where the average RMSDs of DI- and FI-XIAPs to the 17 PDB structures are 1.01 Å and 1.12 Å, respectively, in the core region (E16–Q87 on DI-XIAP or Y265–Q336 on WT-XIAP) after removing the tails that are disordered. These distances are even closer than the average distance among the 17 PDB structures (RMSD = 1.29 Å), although none of the PDB structures have been used as template in the I-TASSER simulations. This result is understandable because the DI- and FI-XIAP sequences have been designed with constraints from structural profiles and therefore have the structural features and folding pattern close to the consensus of the XIAP family. When we applied I-TASSER on the WT-XIAP sequence, the average RMSD of the I-TASSER model was 1.28 Å to the 17 PDB structures, which is slightly higher than that of the designed XIAP sequences but lower than the average RMSD between the PDB structures of WT-XIAP (Table 1). For further confirmation, we also submitted the designed sequences to four other state-of-the-art structure prediction programs, including QUARK [27], Rosetta [28], RaptorX [29], and Phyre2 [30], which are built on ab initio and template-based modeling, respectively. As shown in Tables S3 and S4, the models predicted by the different methods are highly consistent with the I-TASSER models, which are all close to the wild-type structure with a TM-score above 0.8 and RMSD below 3.85 Å. These initial computational folding tests gave us confidence on the foldability of the designed sequences; that is, they should probably adopt a similar fold to the wild type despite the low-sequence identity.
To further examine the 3D fold of the designed sequences, we subjected the designed sequences to the HDX experiments [31]. The proteins, purified from bacteria, were incubated briefly in deuterium oxide and the level of backbone amide deuterium incorporation was determined through electron capture dissociation (ECD) MS. The HDX experiments were repeated three times for each design. In Fig. 3, we present the average HDX rate data for both DI-and FI-XIAP proteins. Because the N- and C-tails of the BIR3 domains are disordered as observed in the PDB structures (Fig. 2b), only the HDX levels in the core region (E16–Q87) are presented. From the HDX profile, the loop regions generally have a higher deuterium exchange rate (values approaching 1), indicating that these residues are largely exposed to bulk solvent. In contrast, strand and helix regions have lower scores indicative of being more buried (values approaching 0). However, there are also several loop residues (e.g., 25–30, 50–55, etc.) having low deuterium exchange rates and other regular secondary structure regions (e.g., 30–33) with high deuterium exchange rates, which are not consistent with the simple secondary structure assignments.
Fig. 3.
HDX data of the designed XIAP proteins in the core region (E16–Q87 or Y265–Q336 on WT-XIAP). Up- and down-triangles indicate observed data from c and z* fragment ions, respectively, while open circles are from I-TASSER structure-based HDX predictions. The dashed and solid lines connect the data points to guide the eye. The cartoon above the figure denotes secondary structure assignments based on DI-XIAP model by DSSP. (a) DI-XIAP. (b) FI-XIAP.
The open circles in Fig. 3 show the estimated HDX rates for each residue based on the I-TASSER model. The predicted HDX score is made using an empirical model calculation based on the solvent accessibility of the backbone amide group (see Eq. (8) in Materials and Methods). Despite the simplicity of the model, the estimation is largely consistent with the HDX data, partly confirming the I-TASSER models. The Pearson correlation coefficients (PCCs) between the observed and predicted HDX rates are 0.74 and 0.55, respectively, for DI- and FI-XIAP proteins (Table 1). These correlations approach the limit of the systematical errors of the experimental data. In fact, we compared two sets of HDX profiles on the same ubiquitin protein, one from top-down mass spectrometry [32] and another from NMR spectroscopy [33], and obtained a PCC of 0.72 which is only slightly higher than the I-TASSER-based match for FI-XIAP, but lower than DI-XIAP. The leave-one-out cross-validation on the 394 training data points showed an average PCC of 0.75 that is also consistent with observation on the designed XIAPs.
The same type of top-down ECD experiments was also tried on WT-XIAP. However, the poor ECD fragmentation prevented us from obtaining enough fragments to make figures as for the designed sequences. It is known that the ECD cleavage is highly dependent on the sequence of specific proteins. Although we did not have the HDX data for WT-XIAP, the comparison of structures determined by the top-down HDX-ECD to that determined by NMR has been made on many proteins in our previous experiments in which excellent agreement was achieved [31,34], demonstrating the reliability of the methods. Here, we have used the same conditions as previously used, including sample infusion setup, mass spectrometer, and instrumental settings, for the DI- and FI-XIAPs to ensure that there was no hydrogen/deuterium scrambling during the measurements.
Binding affinity of XIAP with the Smac peptides detected by 2D NMR and ITC assays
To examine the folding and binding ability of the designed proteins with the target peptides, we conducted protein–peptide 2D NMR chemical shift perturbation experiments on FI- and DI-XIAP using two different native Smac peptides (i.e., FI-XIAP with “AVPIAQKSEKY” and DI-XIAP with “AVPF”), which cover both the “best” and “worse” binding affinity with the designs in the ITC experiments (see below). The ratios of peptide to protein were varied from 0:1, 0.5:1, 4:1, and 5:1 to ensure that the proteins were saturated with the peptides. As shown in Fig. 4, the 15N–1H HSQC spectra of the designed proteins present two sets of resonances at sub-stoichiometric ratios of peptide to protein (0.5:1), and saturation was clearly reached by a 4:1 ratio of peptide to protein. There were more than 10 peaks associated with strong chemical shift differences for each of the experiments between the unsaturated and saturated samples (see, e.g., the inset of Fig. 4b). These data are consistent with slow-exchange kinetics of binding and high-affinity interactions. Meanwhile, the significant peak dispersion of the spectra also confirms the well-folded characteristics of the designed sequences.
Fig. 4.
NMR chemical shift perturbation assays on designed XIAP and Smac complexes with different stoichiometric ratios. (a) DI-XIAP with peptide “AVPF.” (b) FI-XIAP with the peptide “AVPIAQKSEKY.” Inset in panel b highlights spectra changes with the three small polygons labeling distinct chemical shift perturbations witnessed upon the addition of the peptides.
To further quantify the binding affinity, ITC experiments were performed on the XIAP proteins with both peptides of “AVPF” and “AVPIAQKSEKY” [25]. The experiment was repeated five times for each sample and all proteins were shown to have a ~1:1 stoichiometry with the peptides. Fig. 5 shows a typical example of the ITC results obtained from the exothermic DI-XIAP/“AVPF”–peptide interaction, with the dissociation constant (Kd) = 105 nM, peptide to protein stoichiometry (n) = 0.87, enthalpy change (ΔH) = −3.2 kcal/mol, and entropy change (ΔS) = 21.3 cal/mol. A summary of all the ITC experiments repeated for the WT-, DI-, and FI-XIAPs with the peptide “AVPF” is listed in Fig. S2, where the average Kds were found to be 80 ± 25 nM for WT-XIAP, 167 ± 61 nM for DI-XIAP, and 352 ± 79 nM for FI-XIAP.
Fig. 5.
ITC binding assay on DI-XIAP and “AVPF” peptide complex. The top panel is the corrected heat rate per injection, and bottom is the heat per mole of injection. Protein concentrations ranged from 60 to 90 μM and peptide from 0.7 to 1.1 mM. Peptide injection volumes were 2 μL, and conditions were 30 mM NaPO4 (pH 7.5) and 150 mM NaCl at 298 K.
For the peptide of “AVPIAQKSEKY,” the binding affinity is general lower, with the average Kds being 428 ± 72 nM for WT-XIAP, 971 ± 191 nM for FI-XIAP, and 554 ± 93 nM for DI-XIAP (Table 1). The lower Kds of the designed proteins with “AVPIAQKSEKY” are probably due to the fact that the designs were optimized for binding with “AVPF” because the co-crystallized XIAP/“AVPF” complex structure has been used as the design scaffold. However, the Kd value of the wild-type XIAP with “AVPIAQKSEKY” is also nearly 5-fold lower than that with “AVPF”; these data are consistent with the results obtained by other experiments on the WT-XIAP with the same peptides [35,36], which suggests that the peptide “AVPIAQKSEKY” is probably more difficult to be associated with the XIAP proteins.
Overall, the magnitudes of the binding affinities are roughly similar for the three proteins, with WT-XIAP having a slightly greater affinity than DI-XIAP, and DI-XIAP with a stronger affinity than FI-XIAP. The binding affinity on “AVPF” is generally stronger than that on “AVPIAQKSEKY” but the relative ordering of affinities is retained. The difference between DI-XIAP and FI-XIAP binding affinity is interesting but understandable, because the sequence space search in the design simulations, as guided by the atomic binding interactions, is more extensive in DI-XIAP (with all residues dynamically changed); therefore, the DI-XIAP design could identify the states of a lower binding free-energy basin compared to the FI-XIAP in which part of the residues in the interface is frozen and the match of the interface design to the global structural profile is probably suboptimal.
Interplay of evolutionary profile and physical potential drives the interface design
The interface design of DI-XIAP is mainly driven by the profile conservation score and the FoldX binding force field. To examine the specific roles of these driving forces, we list in Table 2 the conservation scores of all Smac binding-site residues (a complete list of the conservation scores for all residues is given in Table S5). Here, a conservation score was calculated as the average of the BLOSUM62 substitution scales between the wild-type residue and the residues of all homologous templates at each position of the multiple structure alignment (MSA) built by TM-align as shown in Fig. S1A, where a higher mutation score indicates a higher degree of conservation in evolution at the position. As highlighted in bold font in Table 2, all the binding residues that were mutated in DI-XIAP have a relatively low conservation score (≤2.1), where most of the un-mutated residues have a high conservation score, suggesting that EvoDesign tends to select the evolutionally variable sites to mutate due to the constraints from the evolutionary structural profile. However there are a few exceptions, where two residues (K297 and G306) have a conservation score below 2.1 but were kept un-mutated in DI-XIAP.
Table 2.
Conservation scores (CS) and frequencies for the interface residues in the MSA used to guide EvoDesign
| Residue position | Amino acid in WT (DI) XIAP | CS of WT (DI) amino acid | Frequency of WT (DI) amino acid | Highest frequency (amino acid) in MSA | Highest CS (amino acid) |
|---|---|---|---|---|---|
| 292 | L (T) | −1.0 (3.5) | 0.0 (0.7) | 0.7 (T) | 3.5 (T) |
| 297* | K (K) | 1.5 (1.5) | 0.3 (0.3) | 0.3 (K) | 1.5 (K) |
| 298 | V (V) | 3.6 (3.6) | 0.9 (0.9) | 0.9 (V) | 3.6 (V) |
| 299 | K (K) | 2.9 (2.9) | 0.4 (0.4) | 0.4 (K) | 2.9 (K) |
| 306* | G (G) | 1.2 (1.2) | 0.4 (0.4) | 0.4 (G) | 1.2 (G) |
| 307 | L (L) | 3.5 (3.5) | 0.8 (0.8) | 0.8 (L) | 3.5 (L) |
| 308 | T (A) | −1.2 (−1.0) | 0.0 (0.0) | 0.2 (D/G/R) | 0.7 (Q) |
| 309 | D(S) | 0.0 (0.9) | 0.1 (0.2) | 0.3 (N) | 1.5 (N) |
| 310 | W (W) | 11.0 (11.0) | 1.0 (1.0) | 1.0 (W) | 11.0 (W) |
| 311 | K (E) | 1.7 (3.3) | 0.2 (0.6) | 0.6 (E) | 3.3 (E) |
| 314 | E (D) | 2.0 (6.0) | 0.0 (1.0) | 1.0 (D) | 6.0 (D) |
| 319 | Q (E) | 2.1 (3.8) | 0.1 (0.7) | 0.7 (E) | 3.8 (E) |
| 322 | K (K) | 3.7 (3.7) | 0.7 (0.7) | 0.7 (K) | 3.7 (K) |
| 323 | W (W) | 5.7 (5.7) | 0.6 (0.6) | 0.6 (W) | 5.7 (W) |
| 324 | Y (F) | 1.5 (2.5) | 0.1 (0.5) | 0.5 (F) | 2.5 (F) |
The bold font indicates the locations that were mutated in DI-XIAP (DI), which all have a conservation score ≤2.1.
labels the positions that have a conservation score below 2.1 but were kept un-mutated in DI-XIAP.
To experimentally examine the relevance of these positions to the binding affinity, we made a mutation at G306D that has the lowest conservation score among all the un-mutated binding residues. Here, we chose the aspartate partly because of the fact that the aspartate has a medium size but with a negative charge, which may result in an energetic change that is balanced between the steric and Coulomb interactions compared to glycine that has no side-chain and neutral in charge, while a mutation to a large-sized residue could make the steric violation dominate the energetic changes. Also, G306 is close to a lysine K299 where a salt bridge might form when mutated to aspartic acid, which may potentially enhance the binding between XIAP and the peptide. However, Fig. 6a shows that this mutation drastically reduced the binding affinity by 36 folds with the same Smac peptide of “AVPF.” In Fig. 6b, we present the 3D structure model of the DI-XIAP and Smac complex built from I-TASSER, where the mutated aspartate is sterically overlapped with the Smac peptide atoms, despite the medium size, which probably explains the reduction of the binding affinity. In addition, we also made a single-point saturation mutagenesis analysis of G306 using FoldX to check the binding affinity of all the mutations. The binding affinity changes for mutations G306A, G306C, G306D, G306E, G306F, G306H, G306I, G306K, G306L, G306M, G306N, G306P, G306Q, G306R, G306S, G307T, G306V, G306W, and G306Y are 3.5, 3.8, 5.0, 4.5, 3.6, 3.1, 4.1, 2.0, 4.1, 1.5, 3.5, 4.2, 2.3, 2.3, 4.0, 4.1, 3.8, 5.7, and 3.1 kcal/mol, respectively, compared to G306, which indicates that none of the mutations is favorable to binding in FoldX. Thus, considering that G306 is the most common amino acid at the position in MSA (despite of the low conservation score), these data suggest that the driving force of the DI-XIAP interface design should be attributed to the interplay of both evolutionary profile and the physics-based protein/peptide interactions.
Fig. 6.
Impact of interface mutation on the binding affinity of DI-XIAP. (a) ITC binding assay on mutated DI-XIAP and “AVPF” peptide complex. The top panel is the corrected heat rate per injection, and the bottom is the heat per mole of injection. (b) Complex structure DI-XIAP and “AVPF” peptide created by I-TASSER, where G306D mutation results in a steric overlap with the peptide molecule.
The important impact of the evolutionary profile on the interface design can also be seen by the observation that five (L292 T, K311E, E314D, Q319E, and Y324F) out of the seven mutated interface residues in DI-XIAP have the highest MSA frequency for the mutant residue among all the amino acid types (Table 2). In other two interface mutants (T308A and D309S), however, the residues were not mutated to the amino acids that have the highest MSA frequency, that is, T308 mutated into alanine, which does not appear in the MSA at all and D309 into serine that has a lower frequency (0.2) than asparagine with the highest frequency of 0.3; these data are again consistent with the fact that the interface design of DI-XIAP is governed by both the profile conservation score and the FoldX binding force field.
Since the mutations in the designed sequences were made mainly on the evolutionarily variable residues in the structural profile, one relevant question is if the mutations selected by EvoDesign in the interface involve any critical residues in the binding pocket. Figure S3 presents the 3D structure model of the DI-XIAP/Smac–peptide complex with the mutated interface residues highlighted in red. Compared to the un-mutated interface residues that are shown in blue, there is no obvious tendency where the mutations are positioned. In fact, except for K311E and D309S that are obviously at the border of the binding pocket, most of the mutations in DI-XIAP have the side-chain interacting directly with and/or oriented toward the ligand.
To have a more quantitative estimation of the locations of the mutations related to the binding pocket, we calculated and compared in Table S6 the relative accessible surface area (rASA) of the interface residues in the monomer (rASAm) and complex (rASAc) structures. Based on the classification of Levy [37], three out of the seven mutated residues in DI-XIAP (292L, 309D, 311K) have rASAm >25% and are categorized as “rim,” two (314E, 319Q) have rASAc <25% and are categorized as “support,” and two (308T and 324Y) have rASAm N25% and rASAc b25% and belong to “core” interface residues that are usually more essential to the binding interactions due to the higher portion of the area involved in the interactions. The numbers of “rim,” “support” and “core” residues in the eight residues that were not mutated are four, three, and one, respectively. These numbers further confirm the fact that there are no specific locations on the interface that EvoDesign tends to mutate.
Thus, although we could not conclude that the design in DI-XIAP has changed the binding mode, it is clear that several “core” residues, whose side-chains are in close contact with the peptide, have been changed. This is technically understandable because the homologous proteins for the profile construction have been collected by fold similarity rather than functional similarity. Most of the homologous proteins do not have the same binding pocket/mode as WT-XIAP. Therefore, the conservation score from the resultant structural profile does not necessarily correlate with importance of the binding residues with Smac. Consequently, the mutations in DI-XIAP, which are mainly selected by the conservation score, can be located at both critical and less-critical binding sites.
Inhibition of caspase-9 enzymatic activity relies on the specificity of interface design
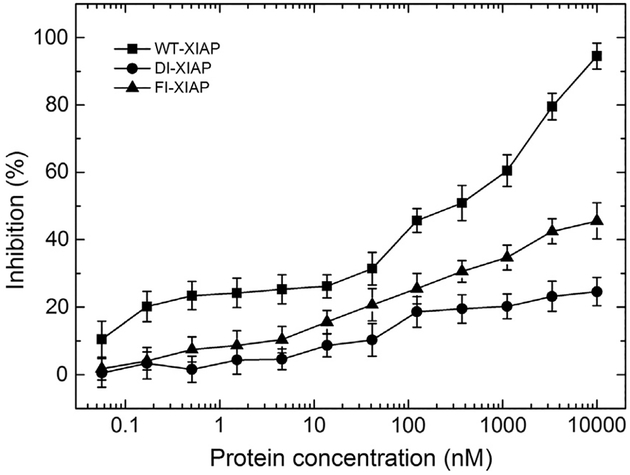
Since the XIAP sequences were designed using a cognate Smac peptide as the binding partner, it is of interest to examine if the designed XIAP proteins would bind caspase-9 and inhibit its function since the latter was not involved in the design simulations. The inhibition of caspase-9 enzymatic activity of FI- and DI-XIAP was tested and compared with WT-XIAP using a commercially available in vitro luminescence XIAP/caspase-9 inhibition assay (Caspase-Glo® 9 Assay). Catalysis of the commercial luminogenic substrate by an active caspase-9 enzyme releases a substrate for luciferase (aminoluciferin), resulting in the luciferase reaction and a detectable luminescence emission in vitro; the luminescence signal generated is proportional to the amount of caspase activity present, and thus, luminescence can be used as a marker for caspase-9 activity. To confirm the data, we repeated the caspase-9 inhibition experiments independently three times. In Fig. 7, we present the average percentage of inhibition of caspase-9 activity, converted from the relative light units (RLU) by Inhibition% = 100 * [1 − (RLU − RLUp)/(RLUn − RLUp)], where RLUn is the luminescence of negative control (no inhibition) and RLUp is the luminescence of positive control (100% inhibition by caspase-9 inhibitor Ac-LEHD-CHO) of that specific experiment. The data show that WT-XIAP strongly inhibits caspase-9 activity, as demonstrated by the increased inhibition% with increasing XIAP concentration. In contrast, inhibition of the designed XIAP domains on the function of the caspase-9 enzyme is significantly reduced, where nearly 60% and 80% of the caspase-9 protease activity remained even when the FI-XIAP and DI-XIAP concentration increases up to 10 k nM, but the caspase-9 activity reduces below 5% at the same concentration of WT-XIAP (Fig. 7).
Fig. 7.
Inhibition assay of wild-type and designed XIAPs on caspase-9 proteolytic activity by the Caspase-Glo® 9 Assay kit from Promega. The percent inhibitions are converted from the relative light units at different concentration of XIAP proteins. Lines connect data points to guide the eye.
The significantly reduced suppression of caspase-9 activity by the new designs is expected, as several key residues involved in the WT-XIAP/caspase-9 interaction were not constrained in the binding interactions in the design process (outside of the Smac/caspase-9 N-termini peptide-binding site, Fig. 2a). As shown in Fig. 2c, three residues, which are known to be critical to the WT-XIAP/caspase-9 binding interactions [8], have been mutated, including G326(Q/N), H343(K/Q), and L344(A/G), where (X/X) represents the (FI-XIAP/DI-XIAP) mutations at those positions. These mutations introduce large polar residues into a non-polar interaction surface area, which disrupt/clash the normal packing (G326Q/N and H343Q/K) or remove the interface contact surface (L344G/A). There are also other mutations in these regions, as highlighted by black dots on caspase-9 interface row in Fig. 2a, which may disrupt the interaction further. These results illustrate that a physiological function not restrained will likely be attenuated or lost during the design process.
It should be mentioned that several studies have used the point mutation technique to identify the single mutants that may interfere the binding inter action of XIAP with caspase-9 [8,38,39]. Depending on the locations, some mutations, for example, E314S, were found to impair binding affinity of XIAP to both caspase-9 and Smac [38]. This residue was also mutated in DI-XIAP but with a different amino acid, that is, E314D. Due to the restraint of the binding force field, this mutation does not impair the interaction with Smac in our case, which partly highlights the specificity of the EvoDesign. Although there are other point mutations that may impair caspase-9 but not Smac, most of which are outside the Smac binding groove [38], we want to emphasize that the principle of the EvoDesign process is fundamentally different from that of the single-point mutation studies. While the point mutation is designed to manually select one or a few residues to change, the de novo design algorithm allows for a complete redesign of the sequences based on automated and comprehensive search simulations guided with specific profiling and binding force field, which has resulted in more than half of the residue changed in the case of DI-XIAP. Among the 30 amino acids interacting with caspase-9, 16 of them do not interact with the Smac peptide, where half of them (G326, E337, I339, N340, N341, H343, L344, T345) were mutated in DI-XIAP. Again, all of the eight mutated residues have a relatively low conservation score ≤2.1, where the majority of the un-mutated residues have a conservation score >2.1 (see Table S7).
Discussion
We have extended the evolution-based method, EvoDesign, for functional protein design, where evolutionary profiles constructed from analogous structures in the PDB have been used as a folding fingerprint to constrain the sequence search simulations, with the physical potentials extended from FoldX for describing the ligand-binding interactions. Compared to the existing evolution-based designs [16] that focus mainly on specifying stable protein folds [14,15], the major technical extension of this work is the incorporation of physics-based ligand- protein binding interactions from FoldX [20], allowing for the introduction of biological functions into designed macromolecular “chassis.”
When applied to the X-linked inhibitor of apoptosis protein (XIAP), two sequences were created by the new binding-specific EvoDesign pipeline, one with all residues dynamically changed (DI-XIAP) and another with binding interface residue frozen (FI-XIAP). To assess the tertiary structure fold, we used five state-of-the-art methods to fold the designed sequences, which generated models all with a close similarity to the consensus of multiple solved structures for the wild-type XIAP sequence in the PDB. An empirical formula estimating solvent accessibility of the backbone amide showed that the computationally predicted models from I-TASSER are in close agreement with the HDX data from the high-resolution ECD MS experiments for both DI-and FI-XIAP proteins.
To examine the function of the design proteins, we incubated DI- and FI-XIAP with two cognate Smac peptides of “AVPF” and “AVPIAQKSEKY,” respectively. At different concentration of the peptides, the 15N–1H NMR chemical shift perturbation assays showed a clear shift of resonance peaks between the unsaturated and saturated samples, illustrating the binding interaction between the peptides and the design proteins. Meanwhile, the excellent peak dispersion in the NMR spectra indicates well-folded feature of the designed proteins. Furthermore, ITC assays showed that the binding affinity of DI-XIAP is stronger than the FI-XIAP to the peptides, both being slightly lower than the wild-type XIAP, but all in a similar mid/high nanomolar magnitude range. The data partly demonstrated the advantage of dynamic interface design procedure in generating tighter ligand-binding interactions. Physically, this is probably due to the fact that the interface residues are liberated in DI-XIAP during the sequence search simulations, which allows the design simulations to identify optimized sequence conformations with a lower folding and binding free energy, compared to FI-XIAP in which the constraint from the frozen interface can limit the optimal match between the interface and the global structure profile.
The binding interactions of the designed XIAPs with caspase-9 were examined by the in vitro luminescence inhibition assays, where dramatically reduced inhibition of the caspase-9 activity was observed in comparison to the wild-type XIAP protein. The data are expected because the caspase-9 binding interaction was not considered in the design simulations and several key residues that are outside the Smac binding pocket but are involved in WT-XIAP/caspase-9 interactions have been mutated. These mutations introduce non-physical polar and steric overlaps, which block the binding interactions with caspase-9 proteins. Overall, the results showed the possibility to introduce biological function into well-designed stable folds by incorporating physics-based ligand-binding interactions into the evolutionary-based design procedure. Apparently a higher-resolution binding-interaction potential with improved accuracy [21,22] will be essential to further enhance the specificity of the functional design. There is clearly room to evolve.
Materials and Methods
Pipeline of evolution-based protein design
The computational design of XIAP BIR3 domain is performed by an extended version of EvoDesign [15,16], which consists of three modules: structural profile construction, Monte Carlo sequence search, and sequence selection. A flowchart of the procedure is depicted in Fig. 1.
Structural profile construction
The recently solved XIAP structure (PDBID: 2OPZ) is a structure of XIAP bound to Smac peptide, which was used as the scaffold to model both bound and apo structures. Ten non-redundant proteins, including 1C9Q, 1E31, 1JD5, 1OXQ, 1QBH, 1SE0, 2QRA, 2VM5, 3M0A, and 3T6P, which have the same fold to the scaffold with a TM-score >0.5 and a sequence identity <80% to the target, were identified from the PDB using the structure alignment program, TM-align [24]. A MSA matrix is then constructed based on the pairwise TM-align alignments, where the designed DI-/FI-XIAP sequences were added to the bottom of two MSAs for reference comparison (see Fig. S1). Here, the bound zinc in 2OPZ has been removed, but it does not affect much of the subsequent design simulations as the cysteine/histidine package is well conserved in the MSA. Next, an L × 20 profile matrix, M(p,a), was calculated from the MSA, which denotes the mutation probability of the amino acid a at the pth position along the sequence, where L is length of the scaffold. The element of the profile matrix is given by
| (1) |
where B(a,x) is the BLOSUM62 substitution matrix and . Here is the frequency of the amino acid x appearing at the pth position of the MSA and H(k) is the Henikoff weight [40] of the kth template sequence in the MSA. The target scaffold is represented by the structural profile in the follow-up design simulations.
Monte Carlo sequence search
Starting from a random sequence, REMC simulations are performed to create a trajectory of artificial sequences (called sequence decoys), where random mutations are made on a set of randomly selected residues at each step of the movements. The energy function of the MC simulation consists of three parts. The first part contains knowledge-based evolutionary terms, which match the ith residue of the decoy sequence with the jth position of the structural profile of the target by a score of
| (2) |
where Ai, ssi, sai, ϕi, and ψi are, respectively, the amino acid, secondary structure (SS), solvent accessibility (SA), and torsion angles (Φ/ψ) of the ith residue of the decoy sequence, and sj, saj, ϕj, and ψj are those at the jth position of the scaffold structure. The SS, SA, and Φ/ψ features of the target are preassigned by DSSP [41] based on the scaffold structure. However, predictions on SS, SA, and Φ/ψ are needed for the sequence decoy at each step of the movements since the sequence and therefore the corresponding secondary features change after each mutation. A quick single-sequence based neural-network predictor was developed, which is much faster (takes <<1 s) than the traditionally used PSI-BLAST based predictors but with a comparable prediction accuracy (72.6% for SS, 70.5% for SA, and 28°/46° for Φ/ψ).
Based on S(i,j), an optimal alignment path between the design and target sequences is obtained by the Needleman–Wunsch dynamic programming [42] with the maximum score assigned as Eevolution, that is,
| (3) |
where k denotes the residue index along the optimal path of dynamic programming alignments. Independent from the sequence alignment, side-chain rotamers of all the residues for each decoy sequence are reconstructed by SCWRL [43] based on the backbone of the scaffold structure and therefore the design does not incorporate indels with respect to the structural template (see Fig. S4 for illustration). We note that the reconstruction of side-chain conformations is performed at each Monte Carlo step when the sequence decoy is updated. The side-chain repacking is implemented on both chains of the bound complex structure, during which the backbone structure is kept frozen. No further relaxation/refinement is performed after SCWRL modeling.
The second energy function, Efoldx(XIAPapo), counts for the physics-based atomic interactions in the apo-form XIAP monomer structure. Efoldx(XIAPapo) contains nine empirical terms accounting for van der Waals interaction, solvation energy, water bridge hydrogen bonding, intra-molecule hydrogen bonding, Coulomb interaction, entropy costs for fixing main-chain and side-chain atoms, and the penalty from atomic steric overlaps [20].
The third energy term counts for the ligand–protein interactions, converted from FoldX:
| (4) |
where Efoldx(complex) counts for the XIAP-Smac complex energy by FoldX. Efoldx(XIAPapo) and Efoldx(Smacapo) are the apo-form monomer energies for XIAP and Smac conformations, respectively. In FoldX, ligand–protein interactions include the inherent contributions of complex structures from van der Waals, solvation, hydrogen-bonding, atomic clash, and entropic interactions, which are similar to the apo-form monomer, but calculated for atom pairs across inter-chains. In addition, an empirical equation that was designed to enhance the association rate of complex formation [23] was introduced to count for the additional electrostatic contribution between atoms of opposite chains, that is,
| (5) |
where the electrostatic energy is calculated through the Debye–Huckel equation of
| (6) |
Here, qi and qj are the charge of the ith and jth charged atoms and rij is the distance; ϵ0 is the permittivity of vacuum. Following Selzer et al. [23], α is set to 6 Å, κ = 0.488, and ϵ= 80.
To balance the energy terms from different resources, Monte Carlo simulations were guided by the sum of the Z-score of three parts of energies, that is,
| (7) |
where 〈E〉 and δE are the average and standard deviations of the energy scores calculated from 1000 random sequences. It is noted that the standard deviations are not a constant and recalculated in each protein design simulation. Because FoldX contains tolerance to large steric clashes, the adoption of the random sequences does not dramatically affect the stability of the standard deviation calculations. As shown in Fig. S5, the standard deviations of different energy terms can quickly converge with the increase of the number of random decoys.
Because the average values do not affect ΔEMC = EMC,new − EMC,old between two MC simulation steps, the actual energy weights for the three terms are w4/δEevolution, w5/δE(XIAPapo), and w6/δE(interface), respectively. The optimized parameters in Eqs. (2) and (7) are as follows: w1 = 1.58, w2 = 2.45, w3 = 1, w4 = −0.5, w5 = 1.22, and w6 = 1.22, which were decided on 625 non-redundant training proteins, that is, w1, w2, and w3 were proportional to the relative accuracy of the SS, SA and Φ/ψ feature predictors; w4 and w5 were adjusted so that the average contribution from evolutional terms and physical terms are comparable based on the design simulations on the training proteins; and w6 is set to be equal to w5 since the terms have the same origin from FoldX. Similarly, the weight parameter for the cross-chain electrostatic contribution (Eq. (5)) is set to be same to that of the Coulomb interaction in FoldX. The target protein XIAP was not included in the training protein set.
Compared with the previous EvoDesign protocol [15], the major difference in scoring function design for the Smac/XIAP complex design is the explicit calculations of the binding interaction with the Efoldx(interface) term in Eq. (7). As shown in Table S8, the average value of δEfoldx(interface) is much smaller than that of δEfoldx(XIAPapo), which can result in neglecting of the binding term in the previous protocol due to the dominant variation of the monomer energy term. On the other hand, the new protocol allows for the appropriate renormalization of different energy terms according to their own deviations and therefore increases the relative weight of physics-based binding interactions. Our simulations show that the change can slightly increase the mutation rate of the interface residues (Fig. S6). It should be also noted that there is a slight inconsistency between the force field of side-chain packing from SCWRL and the physical component of design score from FoldX. However, this inconsistency is largely relieved by the involvement of the evolutionary profiles in EvoDesign. Meanwhile, the extensive REMC simulations cover huge sequence space, which helps to identify the optimal design match even if the force fields are from different origins, given that both tools are well benchmarked and represent reasonable approaches to protein design applications.
Sequence selection
The sequence decoys generated by the REMC simulations are clustered using SPICKER [26] with the distance scale defined by the sum of BLOSUM62 substitution scores overall all the residue pairs that are aligned between the two sequences. All sequences with a distance below a threshold are counted into the same cluster. The choice of distance scale by mutation score instead of sequence identity can help group more homology-related sequences. The threshold parameter is initially set to zero and gradually increased until 40% of the sequences are included in the primary cluster [44]. The sequence with the most neighbors in the primary cluster is chosen as the final design sequence, which represents the lowest free energy state in the MC simulation [26].
Computational time
We use SCWRL for side-chain repacking and FoldX for design scoring, both of which are not very fast. In XIAP/“AVPF” PPI design, it takes about 48 h for a typical 300,000-step REMC simulation run on 20 2.5-Hz Intel (R) Xeon (R) CPU cores in the XSEDE comet server [45].
Biophysical characterizations
Peptides
Two Smac peptide variants were used in the binding assays. The first peptide was the N-terminal tetrapeptide “AVPF” from the 2OPZ crystal structure, and the second consisted of a longer version “AVPIAQKSEKY” (the last two residues are artifacts) from the NMR and crystal structures [7,46].
Expression constructs
DNA sequences of designed FI- and DI-XIAP were optimized based on E. coli K-12 frequent codon usage. The genes were synthesized by Integrated DNA technologies and cloned into an MCSG-7 over-expression vector containing an N-terminal His tag and rTEV protease site via ligation-independent cloning. The following N-terminal artificial cloning residues, “SNA,” remain after rTEV protease cleavage during purification extending the length of the purified proteins from 101 to 104 amino acids. The control WT-XIAP protein, consisting of 116 residues (241–356), was previously cloned into a pet28B (N-terminal 6 × HIS Tag) expression vector [35]. The WT-XIAP expression construct is 139 residues long, which has a C-terminal Cys residue that forms intermolecular dimers in vitro via a disulfide bridge. However, the presence of the disulfide bridge does not affect Smac-XIAP interactions [35]. In our design, a 6-residue segment in the C-terminal containing the cysteine was truncated to create a monomeric protein that simplifies the biophysical characterization of the domains. In Fig. S7, we present the gel filtration results of original WT-XIAP, truncated WT-XIAP, and DI-XIAP, showing that the truncation indeed converts the dimer (original WT-XIAP) into monomer domains (DI-XIAP and truncated WT-XIAP).
Hydrogen/deuterium exchange
Pulsed HDX was conducted using a three-syringe, two-stage continuous-flow setup as described previously [31]. Syringe 1 contains 50 μM XIAP in 10 mM ammonium acetate at pH 7.0. Syringe 2 contains 10 mM ammonium acetate in D2O. The flow rates of the two syringes were 2 and 8 μL/min. After a labeling time of 10 s, the solution was quenched by mixing with the outflow (20 μL/min) from syringe 3, which contained 80% D2O with 0.4% formic acid. The final solution, after the second mixing tee, flows directly into the mass spectrometer. The residence time of the labeled protein under quench conditions was 1.4 s. This short quenching time results in an amide back-exchange level of less of than 1%.
Mass spectrometry
All MS data were acquired on a Bruker 12 T Apex-Qe hybrid FT-ICR mass spectrometer (Bruker Daltonics, Billerica, MA, USA). The parameters for the ion sampling interface and the ion transfer were kept the same as described previously [31] to ensure that no collisional activation-induced H/D scrambling occurs. An ion accumulation time of 0.2 s in the collision cell was used for the acquisition of survey-scan mass spectra, while 0.3 s was used for obtaining ECD data. The ECD experiments were performed on the entire ion population of XIAP within the ICR cell without precursor selection. Top-down ECD experiments on unlabeled XIAP were done by infusing the protein (2.5 μM) in an aqueous solution containing 0.1% formic acid. The ECD parameters are as follows: electron pulse length, 11 ms; electron beam bias, 1.4 V; grid potential, 12 V; and cathode filament heater current, 1.2 A. Up to 600 scans were accumulated for each ECD spectrum over the m/z range 250–2600; this corresponds to an accumulation time of 10 min. Mass calibration was performed using the ECD fragments of ubiquitin.
NMR spectroscopy
A Bruker 600 MHZ spectrometer with cryoprobe was used for NMR experiments at 20 mM NaPO4 (pH 7.5), 150 mM NaCl, and 298 K with protein concentrations ranging from 70 to 150 μM. For 2D NMR chemical shift perturbation assays, Smac peptide was added to 15Nisotopically labeled XIAP-designed proteins in 0.5:1, 4:1, and 5:1 peptide to protein ratios. Saturation was achieved by a 4:1 ratio of peptide to protein. HSQC experiments were performed with 32 scans, 80 increments in the indirect dimension, and 15 N spectral width of 1400 Hz, with offset =118 ppm.
Isothermal calorimetry
ITC assays were conducted in 30 mM NaPO4 (pH 7.5) and 150 mM NaCl at 298 K using TA systems and MicroCal calorimeters using degassed samples. Experiments were conducted in triplicate and the results averaged. Protein concentrations ranged from 60 to 90 μM, peptide concentration ranged from 0.7 to 1.1 mM, and peptide injection volumes were 2 μL.
Cell-free caspase-9 functional assay
The enzymatic activity of active recombinant caspase-9 (Enzo Life Sciences) was evaluated by the Caspase-Glo® 9 Assay kit from Promega, in which catalysis of a substrate by caspase-9 releases a substrate for luciferase (aminoluciferin), resulting in the luciferase reaction and a detectable luminescence emission in vitro. Ten microliters of serial dilutions of designed protein in caspase assay buffer [50 mM of Hepes, 100 mM of NaCl, 1 mM of EDTA, 1 mM DTT with 0.1% of CHAPS and 10% of glycerol (pH 7.4)] were mixed with 2.5 μL of active caspase-9 solution in caspase assay buffer. This mixture was incubated at room temperature for 15 min. Luminogenic Z-LEHD substrate was added with 1:1 ratio to give final caspase-9 concentration of 2.5 unit/reaction (according to the manufacturer’s instructions). This mixture was incubated at 295 K for 1 h without light, and luminescence from substrate cleavage was then determined by a Tecan Infinite M-1000 multimode plate reader.
Structure-based estimation of HDX rate
From a given structure model, the HDX rate of ith residue is estimated by Di = 1 − Si, where Si is calculated by a linear combination of six scoring terms counting for the solvent buried status of hydrogen bonds associated with the amide groups, that is,
| (8) |
Here
| (9) |
with being the fraction of solvent accessibility of ith residue assigned by DSSP [41]; counts for the solvent accessibility of the amide groups, where and are the numbers of water molecules accessible to the amide group with a distance cutoff 3.5 and 4.7 Å, respectively, and and are the maximum number of and , respectively; is the number of hydrogen-bonds associated with the amide group as assigned by HBplus [47] and counts for the feature of amide hydrogen-bonding vector, where θi is angle between the amide proton vector (N → H) and the vector pointing from the amide to the protein center of mass; is the number of residues that have a distance below 3.7 Å to the ith residue divided by the maximum of contacts for a given residue (=14). The weights in Eq. (8) are selected to be c1 = 20, c2 = 15, c3 = 20, c4 = 5, c5 = 5, and c6 = 30, which were decided on a training set of 394 NMR-based HDX data points from alpha lactalbumin (PDB ID: 1HML), kinase interacting forkhead-associated domain (KI-FHA) (PDB ID: 1MZK), ubiquitin (PDB ID: 1UBQ), CopK (PDB ID: 2K0Q), dihydrofolate reductase (PDB ID: 2L28), small archaeal modifier protein 1 (Samp1) (PDB ID: 2L52), and staphylococcal nuclease (PDB ID: 2KQ3), by maximizing the correlation between the predicted and experimental HDX values. None of the training proteins are homologous to the XIAP protein that was tested in this study. As part of the method verification, we made a leave-one-out cross-validation test on the 394 HDX data points, where an average PCC of 0.75 was achieved between the predicted and observed HDX rates. This value was slightly higher than but consistent with the application on the XIAP proteins, suggesting that the weighting parameters selected are reasonably robust for different protein sets.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jeffrey R. Brender and Dr. Yang Cao for insightful comments and discussions. The study is supported in part by the National Institute of General Medical Sciences (GM083107 and GM084222) and National Science Foundation (1564756). The protein design process was performed on the Extreme Science and Engineering Discovery Environment (XSEDE) clusters [45].
Abbreviations used:
- XIAP
X-linked inhibitor of apoptosis protein
- HDX
hydrogen–deuterium exchange
- MS
mass spectroscopy
- DI-XIAP
Dynamic-Interface XIAP
- FI-XIAP
Fixed-Interface XIAP
- REMC
replica-exchange Monte Carlo
- ECD
electron capture dissociation
- PCCs
Pearson correlation coefficients
- ITC
isothermal calorimetry
- MSA
multiple structure alignment
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2018.12.016.
References
- [1].Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D, Principles for designing ideal protein structures, Nature 491 (2012) 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bolon DN, Mayo SL, Enzyme-like proteins by computational design, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 14274–14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yu F, Cangelosi VM, Zastrow ML, Tegoni M, Plegaria JS, Tebo AG, Mocny CS, Ruckthong L, Qayyum H, Pecoraro VL, Protein design: toward functional metalloenzymes, Chem. Rev 114 (2014) 3495–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mandell DJ, Kortemme T, Computer-aided design of functional protein interactions, Nat. Chem. Biol 5 (2009) 797–807. [DOI] [PubMed] [Google Scholar]
- [5].Samish I, MacDermaid CM, Perez-Aguilar JM, Saven JG, Theoretical and computational protein design, Annu. Rev. Phys. Chem 62 (2011) 129–149. [DOI] [PubMed] [Google Scholar]
- [6].Oerlemans MI, Koudstaal S, Chamuleau SA, de Kleijn DP, Doevendans PA, Sluijter JP, Targeting cell death in the reperfused heart: pharmacological approaches for cardioprotection, Int. J. Cardiol 165 (2013) 410–422. [DOI] [PubMed] [Google Scholar]
- [7].Wu G, Chai J, Suber TL, Wu J-W, Du C, Wang X, Shi Y, Structural basis of IAP recognition by Smac/DIABLO, Nature 408 (2000) 1008–1012. [DOI] [PubMed] [Google Scholar]
- [8].Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y, Mechanism of XIAP-mediated inhibition of caspase-9, Mol. Cell 11 (2003) 519–527. [DOI] [PubMed] [Google Scholar]
- [9].Wang S, Design of small-molecule Smac mimetics as IAP antagonists, Curr. Top. Microbiol. Immunol (2010) 89–113. [DOI] [PubMed] [Google Scholar]
- [10].Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC, IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases, EMBO J. 17 (1998) 2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Du C, Fang M, Li Y, Li L, Wang X, Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition, Cell 102 (2000) 33–42. [DOI] [PubMed] [Google Scholar]
- [12].Baker D, Sali A, Protein structure prediction and structural genomics, Science 294 (2001) 93–96. [DOI] [PubMed] [Google Scholar]
- [13].Zhang Y, Progress and challenges in protein structure prediction, Curr. Opin. Struct. Biol 18 (2008) 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R, Evolutionary information for specifying a protein fold, Nature 437 (2005) 512–518. [DOI] [PubMed] [Google Scholar]
- [15].Mitra P, Shultis D, Brender JR, Czajka J, Marsh D, Gray F, Cierpicki T, Zhang Y, An evolution-based approach to de novo protein design and case study on Mycobacterium tuberculosis, PLoS Comput. Biol 9 (2013), e1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mitra P, Shultis D, Zhang Y, EvoDesign: de novo protein design based on structural and evolutionary profiles, Nucleic Acids Res. 41 (2013) W273–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shultis D, Dodge G, Zhang Y, Crystal structure of designed PX domain from cytokine-independent survival kinase and implications on evolution-based protein engineering, J. Struct. Biol 191 (2015) 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y, The I-TASSER suite: protein structure and function prediction, Nat. Methods 12 (2015) 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Roy A, Kucukural A, Zhang Y, I-TASSER: a unified platform for automated protein structure and function prediction, Nat. Protoc 5 (2010) 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L, The FoldX web server: an online force field, Nucleic Acids Res. 33 (2005) W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brender JR, Zhang Y, Predicting the effect of mutations on protein–protein binding interactions through structure-based interface profiles, PLoS Comput. Biol 11 (2015), e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xiong P, Zhang C, Zheng W, Zhang Y, BindProfX: assessing mutation-induced binding affinity change by protein interface profiles with pseudo-counts, J. Mol. Biol 429 (2017) 426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Selzer T, Albeck S, Schreiber G, Rational design of faster associating and tighter binding protein complexes, Nat. Struct. Mol. Biol 7 (2000) 537–541. [DOI] [PubMed] [Google Scholar]
- [24].Zhang Y, Skolnick J, TM-align: a protein structure alignment algorithm based on the TM-score, Nucleic Acids Res. 33 (2005) 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wist AD, Gu L, Riedl SJ, Shi Y, McLendon GL, Structure–activity based study of the Smac-binding pocket within the BIR3 domain of XIAP, Bioorg. Med. Chem 15 (2007) 2935–2943. [DOI] [PubMed] [Google Scholar]
- [26].Zhang Y, Skolnick J, SPICKER: a clustering approach to identify near-native protein folds, J. Comput. Chem 25 (2004) 865–871. [DOI] [PubMed] [Google Scholar]
- [27].Xu D, Zhang Y, Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field, Proteins 80 (2012) 1715–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kim DE, Chivian D, Baker D, Protein structure prediction and analysis using the Robetta server, Nucleic Acids Res. 32 (2004) W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J, Template-based protein structure modeling using the RaptorX web server, Nat. Protoc 7 (2012) 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis, Nat. Protoc 10 (2015) 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pan J, Han J, Borchers CH, Konermann L, Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein, J. Am. Chem. Soc 131 (2009) 12801–12808. [DOI] [PubMed] [Google Scholar]
- [32].Wang G, Abzalimov RR, Bobst CE, Kaltashov IA, Conformer-specific characterization of nonnative protein states using hydrogen exchange and top-down mass spectrometry, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 20087–20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bougault C, Feng L, Glushka J, Kupče E, Prestegard J, Quantitation of rapid proton-deuteron amide exchange using hadamard spectroscopy, J. Biomol. NMR 28 (2004) 385–390. [DOI] [PubMed] [Google Scholar]
- [34].Pan J, Zhang S, Parker CE, Borchers CH, Subzero temperature chromatography and top-down mass spectrometry for protein higher-order structure characterization: method validation and application to therapeutic antibodies, J. Am. Chem. Soc 136 (2014) 13065–13071. [DOI] [PubMed] [Google Scholar]
- [35].Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization, Anal. Biochem 332 (2004) 261–273. [DOI] [PubMed] [Google Scholar]
- [36].Kipp RA, Case MA, Wist AD, Cresson CM, Carrell M, Griner E, Wiita A, Albiniak PA, Chai J, Shi Y, Molecular targeting of inhibitor of apoptosis proteins based on small molecule mimics of natural binding partners, Biochemistry 41 (2002) 7344–7349. [DOI] [PubMed] [Google Scholar]
- [37].Levy ED, A simple definition of structural regions in proteins and its use in analyzing interface evolution, J. Mol. Biol 403 (2010) 660–670. [DOI] [PubMed] [Google Scholar]
- [38].Silke J, Hawkins CJ, Ekert PG, Chew J, Day CL, Pakusch M, Verhagen AM, Vaux DL, The anti-apoptotic activity of XIAP is retained upon mutation of both the caspase 3- and caspase 9-interacting sites, J. Cell Biol 157 (2002) 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun C, Cai M, Meadows RP, Xu N, Gunasekera AH, Herrmann J, Wu JC, Fesik SW, NMR structure and mutagenesis of the third Bir domain of the inhibitor of apoptosis protein XIAP, J. Biol. Chem 275 (2000) 33777–33781. [DOI] [PubMed] [Google Scholar]
- [40].Henikoff S, Henikoff JG, Position-based sequence weights, J. Mol. Biol 243 (1994) 574–578. [DOI] [PubMed] [Google Scholar]
- [41].Kabsch W, Sander C, Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features, Biopolymers 22 (1983) 2577–2637. [DOI] [PubMed] [Google Scholar]
- [42].Needleman SB, Wunsch CD, A general method applicable to the search for similarities in the amino acid sequence of two proteins, J. Mol. Biol 48 (1970) 443–453. [DOI] [PubMed] [Google Scholar]
- [43].Krivov GG, Shapovalov MV, Dunbrack RL Jr., Improved prediction of protein side-chain conformations with SCWRL4, Proteins 77 (2009) 778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bazzoli A, Tettamanzi AG, Zhang Y, Computational protein design and large-scale assessment by I-TASSER structure assembly simulations, J. Mol. Biol 407 (2011) 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Towns J, Cockerill T, Dahan M, Foster I, Gaither K, Grimshaw A, Hazlewood V, Lathrop S, Lifka D, Peterson GD, XSEDE: accelerating scientific discovery, Comput. Sci. Eng 16 (2014) 62–74. [Google Scholar]
- [46].Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW, Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain, Nature 408 (2000) 1004–1008. [DOI] [PubMed] [Google Scholar]
- [47].McDonald IK, Thornton JM, Satisfying hydrogen bonding potential in proteins, J. Mol. Biol 238 (1994) 777–793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.