Abstract
The proximal tubule has a remarkable capacity for repair after acute injury, but the cellular lineage and molecular mechanisms underlying this repair response are incompletely understood. Here, we developed a Kim1-GFPCreERt2 knockin mouse line (Kim1-GCE) in order to perform genetic lineage tracing of dedifferentiated cells while measuring the cellular transcriptome of proximal tubule during repair. Acutely injured genetically labeled clones coexpressed KIM1, VIMENTIN, SOX9, and KI67, indicating a dedifferentiated and proliferative state. Clonal analysis revealed clonal expansion of Kim1+ cells, indicating that acutely injured, dedifferentiated proximal tubule cells, rather than fixed tubular progenitor cells, account for repair. Translational profiling during injury and repair revealed signatures of both successful and unsuccessful maladaptive repair. The transcription factor Foxm1 was induced early in injury, was required for epithelial proliferation in vitro, and was dependent on epidermal growth factor receptor (EGFR) stimulation. In conclusion, dedifferentiated proximal tubule cells effect proximal tubule repair, and we reveal an EGFR/FOXM1-dependent signaling pathway that drives proliferative repair after injury.
Keywords: Nephrology, Stem cells
Keywords: Adult stem cells
Introduction
Acute kidney injury (AKI) has a wide spectrum of outcomes ranging from full recovery to failed repair and transition to chronic kidney disease. According to a recent report from the CDC examining trends in hospitalizations for AKI in the US from 2000 to 2014, the rate of AKI hospitalizations increased by 230% over this time frame, going from 3.5 to 11.7 per 1000 persons (1). Furthermore, it has been reported that Medicare patients aged 66 years and older who were hospitalized for AKI had a 35% cumulative probability of a recurrent AKI hospitalization within one year and 28% were diagnosed as having chronic kidney disease in the year following an AKI hospitalization (2). These troubling statistics point toward a pressing need to identify therapeutic interventions to prevent and treat AKI.
The proximal tubular epithelium makes up the bulk of the kidney cortex and is responsible for reabsorption of a large portion of the glomerular filtered load in order to maintain solute and volume homeostasis. Due to its high metabolic activity, it is also the renal compartment most vulnerable to injury. It is well known that the tubular epithelium has regenerative potential; however, this repair capacity is not unlimited and may be dependent on the degree of injury (3). Based on studies from our lab and others, acute injury with proximal tubule death is followed by a wave of tubular proliferation, peaking at 48 hours after injury, to restore tubular cell mass. Lineage analysis indicates that the source of the repairing cells derives from within the tubule rather than a circulating or interstitial progenitor (4). Several lines of evidence indicate that surviving epithelia dedifferentiate, and these dedifferentiated epithelia have an equivalent capacity for repair (5–8). In contrast, a separate body of work has suggested a different model: that a fixed population of WNT-responsive and/or PAX2-positive intratubular progenitors selectively proliferate and differentiate into proximal tubule cells (9–11).
The phosphatidylserine receptor KIM1 (gene name Havcr1) is induced in acutely injured proximal tubule and binds to apoptotic cells and fragments to clear the tubular lumen of debris (12). Its expression is undetectable in healthy kidney, and expression falls back to baseline after proximal tubule repair is complete (13). Because KIM1 is not expressed at baseline, but is rapidly induced in all injured cells, we reasoned that it cannot mark a fixed progenitor population and that genetic lineage analysis of injured Kim1+ cells could address the issue of whether injured, dedifferentiated proximal tubule epithelia are responsible for repair versus a fixed intratubular progenitor. Importantly, PAX2+ putative intratubular progenitors do not express KIM1 after injury (10), excluding the possibility that our genetic strategy would label this proposed progenitor population. We created a Kim1-GFPCreERt2 (hereafter referred to as Kim1-GCE) knockin mouse line, and traced the fate of individual clones labeled soon after injury. We also performed ribosomal pull-down RNA-sequencing during injury and repair to define the repair process in molecular terms. We took advantage of this powerful tool to perform transcriptional profiling to identify the transcriptional signature of the injured tubular epithelial cells. We show that injured, dedifferentiated proximal tubule undergoes proliferative expansion after injury, define transcriptional patterns of these cells during repair, and identify an EGFR-FOXM1 signaling pathway that regulates proximal tubule proliferation.
Results
Characterization of mouse model.
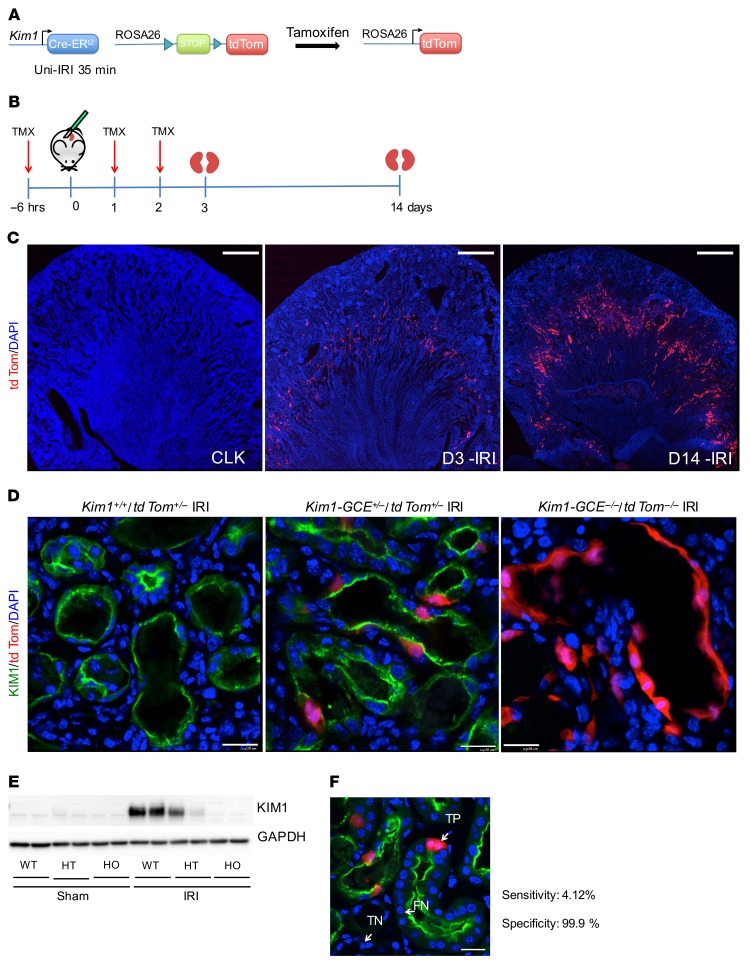
Kim1-GCE mice were generated by gene targeting (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI125519DS1). The resulting line knocks out endogenous Kim1 expression and replaces it with a GFPCreERt2 cassette (Figure 1A). To evaluate recombination specificity, bigenic Kim1-GCE+/–; R26tdTomato+/– mice received tamoxifen 6 hours before surgery and on days 1 and 2 after surgery. After unilateral ischemia/reperfusion injury (Uni-IRI), tdTomato expression was analyzed at days 3 and 14 after surgery (Figure 1B). There was no tdTomato expression at baseline, but in injured kidneys, tdTomato expression was localized to the outer segment of the outer medulla. Recombination efficiency at day 3 was unexpectedly low, but there was significantly increased tdTomato expression at day 14, suggesting expansion of the labeled tubular epithelial cells (Figure 1C).
Figure 1. Kim1-GCE mouse model.
(A) Kim1-GCE was crossed to the Rosa26tdTomato reporter mouse to allow permanent labeling of injured tubular epithelial cells upon tamoxifen-mediated recombination. (B) Uni-IRI was performed to validate the mouse model with kidneys harvested at day 3 and day 14 after injury. (C) Immunofluorescent staining showing endogenous tdTomato expression in the outer segment of the outer medulla at day 3 with increased expression at day 14. There is absence of tdTomato expression in the contralateral kidney after tamoxifen administration indicating no leaky expression. (D) Immunostaining with KIM1 antibody showing coexpression with tdTomato-labeled cells in Kim1-GCE heterozygous mice. There is absence of KIM1 expression in Kim1-GCE homozygous mice, as expected since this a knockin to the ATG site. (E) Western blot for KIM1 showing half the amount of protein expressed in Kim1-GCE heterozygous as compared with WT mice and absence of KIM1 protein in Kim1-GCE homozygous consistent with immunofluorescent staining. (F) Immunostaining showing examples of TP, TN, and FN for determination of sensitivity and specificity for the mouse model. n= 3–4 mice. Scale bars: 500 μM (C); 20 μM (D and F).
To further evaluate recombination specificity, we performed immunofluorescent staining for KIM1. All tdTomato-positive cells also expressed KIM1 at day 3, although only a minority of Kim1-positive cells coexpressed tdTomato (Figure 1D). Mice homozygous for the GFPCreERt2 allele did not express KIM1 protein, as expected (Figure 1, D and E). To provide a quantitative assessment of the specificity and sensitivity of the model, we counted the number of tdTomato-expressing cells that were positive for KIM1 (true positive [TP]) and also determined the number of true negatives (TNs), false positives (FPs), and false negatives (FNs) as described in Methods. We determined that the mouse model is 99.9% specific and 4.12% sensitive (Figure 1F). The mechanism behind this low recombination efficiency remains unexplained; however, the very high specificity indicates that the line faithfully reports KIM1 expression without any leaky expression, simply in a minority of cells, which appears to be stochastic.
Lineage analysis reveals clonal expansion of injured proximal tubule after injury.
Since KIM1 is not expressed in healthy kidney, but is induced in all injured proximal tubule (rather than a subset) very early after injury, it cannot be a marker of a putative fixed intratubular progenitor cell. We therefore used the Kim1-GCE line to determine whether injured and dedifferentiated cells labeled by Kim1-GCE undergo proliferative repair or not. Tamoxifen was administered 12 hours after injury (Bi-IRI) at low dose (1 mg) to generate single-cell clones, reducing the possibility of de novo recombination in adjacent cells due to residual tamoxifen (Figure 2A). Kidneys were collected at day 2 and 14 after injury. Blood urea nitrogen (BUN) was measured on day 2 and rose between 80 and 150 mg/dL indicating successful IRI. For the 14-day clonal analysis group, BUN measurement indicated renal recovery, as reflected by reduced BUN from day 2 to day 14 (Supplemental Figure 1B). Lineage analysis revealed that, at day 2 after injury, clones were predominantly single-cell clones in separate tubules, but by day 14, there were coherent clones of adjacent tdTomato+ cells (Figure 2B). Careful quantitation revealed that at day 2, 87% of the clones were single cell. By day 14, the number of single-cell clones had decreased to 52.2% (P < 0.0005) and the number of multicellular clones (>5 cells) had increased from 0.8 % to 10 % (P < 0.05, Figure 2C). Maximum clone size was 10 cells, similar to a report tracking PAX2-labeled clones (10). These results indicate that differentiated tubular epithelial cells that become injured are capable of proliferative repair, arguing against the existence of a fixed intratubular progenitor population (6).
Figure 2. Lineage tracing of injured tubular epithelial cells.
(A) Kim1-GCE;tdTom mice heterozygous for both alleles were subjected to Bi-IRI or Uni-IRI and low-dose tamoxifen (TMX) (1 mg) administered 12 hours after surgery. (B) Immunostaining showing single tdTom cells labeled at day 2 after injury and clusters of tdTom cells at day 14 in Bi-IRI and Uni-IRI. (C) Quantification of clone size at day 2 and day 14 after injury. (D) Immunostaining for PAX2, VIMENTIN, and KI67 showing coexpression with tdTom cells at day 2. By day 14, there is persistent PAX2 and VIMENTIN expression in tdTom cells. KI67 is absent from tdTom cells at day 14, since the cells have completed repair. Quantification showing percentages of coexpression of the tdTom cells with each of the markers. For A–C, n = 4–6 mice per experiment. For D, n = 3–4 mice. Scale bars: 10 μM. *P < 0.05; ***P < 0.001; ****P < 0.0001, 2-way ANOVA with post hoc Dunnett’s multiple comparisons test (C) and Student’s t test (D).
We also performed lineage analysis after severe injury to evaluate whether the proliferative response would be similar. Given the high mortality with more severe injury in the Bi-IRI model, we performed Uni-IRI with a prolonged ischemia time of 24 minutes to induce severe injury. Quantitation showed that at day 14, the number of single-cell clones was 51%, which was similar to our quantitation for day 14 in the Bi-IRI model (Figure 2C). However, we observed a doubling in the number of clones with more than 5 cells as compared with moderate injury (20% vs. 10%), indicating that dedifferentiated, injured tubular epithelial cells augment their proliferative response in the event of more severe injury. We asked whether dedifferentiation markers SOX9 and VIMENTIN could be detected in collecting duct, but by immunostaining, there was no obvious coexpression (Supplemental Figure 1C). Further evaluation of dedifferentiation in distal segments was beyond the scope of the current study.
Lineage tracing reveals a failed repair population.
We next sought to characterize the repair process in more detail. We confirmed that labeled, injured proximal tubule clones undergo a burst of proliferation based on the finding that nearly 60% of tdTomato+ cells coexpressed KI67 at day 2, but only 5% expressed KI67 at day 14 (Figure 2D). This is in good agreement with reports of bulk tubular proliferation at this time point (4, 14). Pax2 and Vimentin are genes that have also been characterized as markers of dedifferentiated proximal tubule cells (14, 15). Two days after injury, we could detect expression of PAX2 and VIMENTIN in about 40% and 20% of tdTomato-labeled cells, respectively. Unlike KI67, this fraction continued to express these markers at day 14, suggesting some degree of incomplete repair in those populations (Figure 2D).
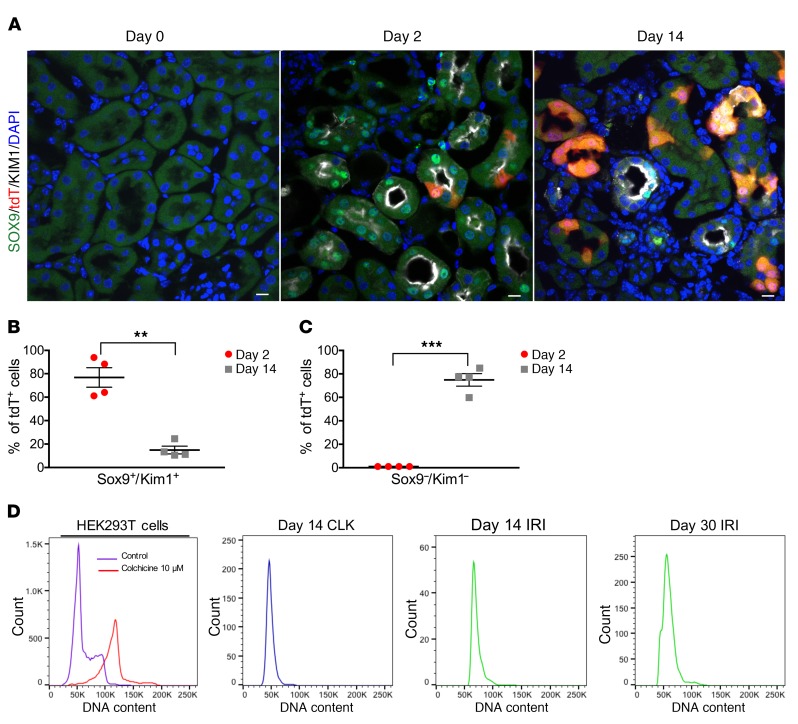
To further investigate the question of whether tdTomato-labeled proximal tubule cells underwent complete repair, or not, we next examined temporal expression of KIM1 protein and SOX9, which has recently been identified as both a marker of proximal tubule injury and a transcriptional regulator of repair (11, 16). At day 2, about 80% of tdTomato+ cells coexpressed both SOX9 and KIM1, indicating that these cells are injured and dedifferentiated (Figure 3, A and B). This population fell to about 15% by day 14, indicating that while the majority of tdTomato-labeled cells had successfully repaired, as reflected by their downregulation of KIM1 and SOX9, about 15% had persistent injury and thus failed to repair by day 14. To approach the question from the opposite perspective, we also quantified the number of tdTomato cells that expressed neither KIM1 nor SOX9: the population of cells to undergo successful repair. At day 2, these cells were nearly undetectable, but by day 14, close to 80% of tdTomato cells were negative for both KIM1 and SOX9 (Figure 3C). These results indicate that injured proximal tubules proliferate after injury and that the majority have largely completed repair by day 14, but that about 15% remain injured and dedifferentiated at this time point, likely reflecting failed repair.
Figure 3. SOX9 immunostaining reveals a population of proximal tubule cells that have failed to repair with no evidence for endocycle.
(A) Immunostaining for SOX9 shows absence at baseline (day 0), but expression in tdTom cells upon injury (day 2). At day 14, there are a few tdTom cells that have persistent SOX9 expression, suggesting that these are cells that have failed repair. Scale bars: 10 μM. (B) Quantification of percentages of tdTom cells that express both SOX9 and KIM1 at day 2 and day 14. (C) Quantification of the percentages of tdTom cells that do not express SOX9 and KIM1 at day 2 and day 14. (D) DNA content analysis. Far left, HEK293T cells treated with colchicine used as a positive control for polyploidy. Sorted tdTomato+ cells from CLK and IRI kidneys at the designated time points show no polyploidy. For A–C, n = 4 mice per time point were used for analysis. For D, representative experiments are shown from n = 4 independent experiments for each time point. **P < 0.01; ***P < 0.001, Student’s t test.
A recent study proposed that injured proximal tubules progress through the cell cycle without dividing, resulting in a single polyploid nucleus in a process termed endoreplication or endocycle (10). Since our clonal analysis indicated cell division in injured proximal tubule and not endocycle (if endocycle were occurring, then all clones would remain single cells), we sought direct evidence for increased DNA content in tdTomato-labeled cells after injury. We performed Uni-IRI and collected the kidneys at day 14 and day 30 after injury. After kidney dissociation, the single cell suspension was fixed and stained with DAPI. Cell-cycle analysis was performed in the gated tdTomato population. As a positive control, we treated HEK293T cells with colchine (see Methods) to induce polyploidy through endocycle (17, 18). We did not observe evidence of polyploidy at either time point (Figure 3D). Therefore, our studies do not support a role for endocycle in proximal tubule repair.
Transcriptional profiling of proximal tubular epithelial cells during injury and repair.
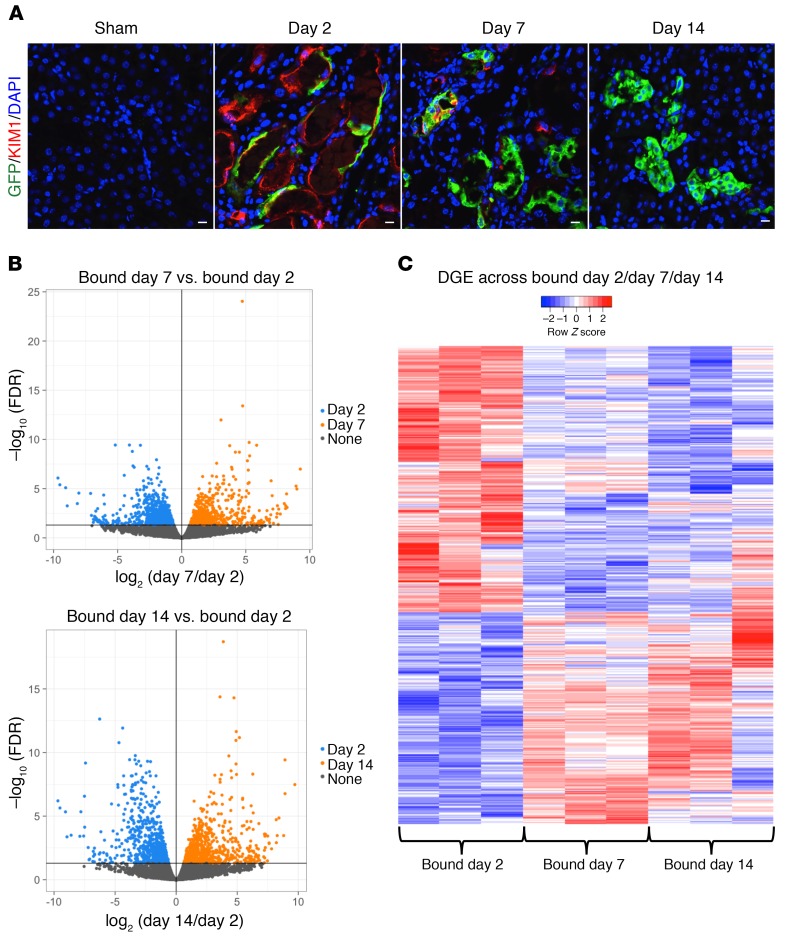
Recent work has carefully measured global kidney transcriptional changes over the full course of murine IRI (19). Although this is a powerful resource, it is limited in that relevant cell-specific gene expression signatures may be lost within the integrated expression profiles of the other cell types in the sample. We therefore sought to generate RNA-Seq profiles of injured proximal tubule cells during the course of injury and repair by ribosomal pull-down. We generated bigenic heterozygous Kim1-GCE; R26-LSL-EGFPL10a mice in order to perform translating ribosome affinity purification (TRAP) (20, 21). We isolated mRNA from injured proximal tubule cells as well as in sham controls at days 2, 7, and 14 after IRI. Tamoxifen was administered via gavage 6 hours before surgery and on day 1 after surgery. The isolated polysomal RNA (bound fraction) of 3 biological replicates for each time point was submitted for next-generation sequencing. The increased TRAP RNA yield across time points (Supplemental Figure 2A) was consistent with proliferative expansion of labeled cells during repair, which was corroborated by immunofluorescent staining for GFP (Figure 4A). We verified the TRAP protocol by determining GFP expression by quantitative PCR (qPCR) in the bound versus unbound fraction, since KIM1 is not expressed in uninjured kidney (i.e., sham) (Supplemental Figure 2B). We observed strong enrichment for GFP in the sham and day 2 IRI-bound fraction and not in the unbound fraction. The detectable GFP expression in the sham-bound fraction was due to leaky EGFPL10a expression in podocytes and the collecting duct, as previously described (22), but this was about 20-fold lower compared with EGFP expression at day 2 after IRI.
Figure 4. Transcriptional profiling of injured tubular epithelial cells.
(A) Immunostaining for GFP in bigenic Kim1-GCE;EGFPL10a kidney sections shows absent GFP expression in sham and coexpression with tdTom cells at day 2 after injury. There are increases in GFP expression at day 7 and day 14, since there is clonal expansion of the surviving tubular epithelial cells. Scale bars: 10 μM. (B) Volcano plots of the DGE list for bound day 7 versus bound day 2 and bound day 14 versus day 2. (C) Heatmap of the DGE list across all 3 time points. n = 3 mice for each time point.
We used the edgeR package to perform the differential expression analysis and filtered out the low expressing genes. Principal component analysis (PCA) showed that the biological replicates clustered together across time points, indicating a high degree of similarity (Supplemental Figure 2C). The biological replicates for day 14 clustered close to the sham group, suggesting that the day 14 group is returning toward the baseline transcriptional state. Since KIM1 is not expressed in sham kidney, we compared day 7 to day 2 and day 14 to day 2 to identify the transcriptional signature during injury and repair. We identified 1457 differentially expressed genes when comparing bound fractions of day 7 to day 2 and 1478 differentially expressed genes when comparing day 14 to day 2 (Figure 4B). Plotting the differential gene expression (DEG) list (Supplemental Table 1) across the 3 time points in a heatmap, we observed that about half of the genes were strongly upregulated at day 2 and that their expression subsequently fell at days 7 and 14. The other half were genes that were strongly downregulated at day 2 and whose expression rose on days 7 and 14 when the cell was returning to homeostasis (Figure 4C).
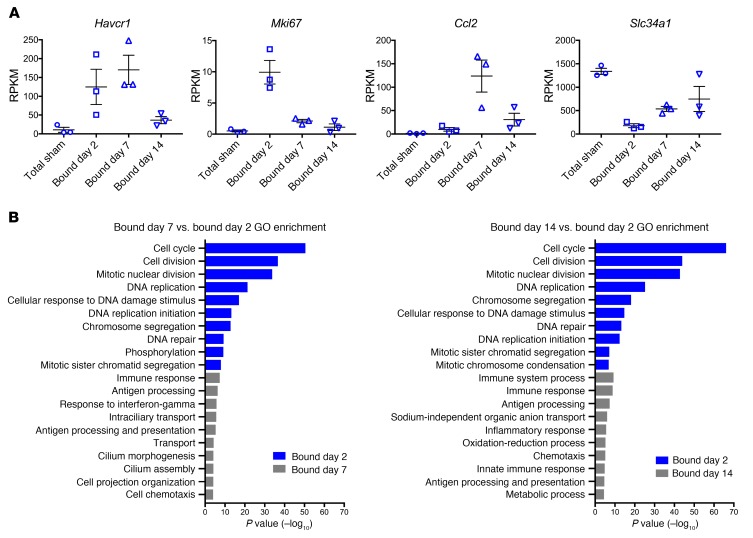
To corroborate these results, we selected known markers such as Havcr1 (which encodes KIM1), Ki67, Ccl2, and Slc34a1 during injury and repair and plotted their expression over time points (Figure 5A). These genes reflected the 4 different patterns we observed. Havcr1 was highly upregulated at days 2 and 7 and decreased by day 14, when the majority of repair was complete, as expected. Ki67 was highly expressed at day 2 after injury, since this is the peak of proliferation, but it was much lower at days 7 and 14. Ccl2, an inflammatory marker, was not upregulated until day 7, but then fell by day 14. Finally, the sodium-phosphate exchanger Slc34a1 was markedly downregulated during dedifferentiation at day 2, but expression recovered over time as the tubular epithelial cell redifferentiated.
Figure 5. Validation and DAVID GO Analysis of differentially expressed genes in injured tubular epithelial cells.
(A) RPKM values across different time points after injury of known upregulated and downregulated genes. (B) GO analysis of the 2 comparisons: bound day 7 versus day 2 and bound day 14 versus day 2. For all experiments, n = 3 replicates.
We also took a bioinformatic approach to evaluating for endocycle, which is viewed as a state in which cells oscillate between G and S phases. We would expect to see an enrichment in genes related to G and S phases at day 14 after injury if endocycle were occurring. Using a previously published marker gene list for the phases of the cell cycle (23), we scored the cell-cycle expression of each of our TRAP samples as described in the Methods section. We did not observe an enrichment of genes related to the G2M and S phases in the day 14 injury samples, and in fact, these samples were similar in cell-cycle expression to the sham samples, indicating that they had returned to baseline (Supplemental Figure 3).
To further dissect the significance of differentially expressed genes during injury and repair, we performed Database for Annotation, Visualization, and Integrated Discovery (DAVID) gene ontology (GO) analysis focusing on biological process. The top 10 terms for each comparison (day 7 vs. day 2 and day 14 vs. 2) are shown in Figure 5B. As expected, the top GO terms were related to cell-cycle and DNA repair, since these are key events in the injury response. GO terms for day 7 reflect an immune response, and interestingly, there were terms related to cilium morphogenesis and cilium assembly, suggesting that cilium may play a role during the repair phase. Day 14 GO terms were related to cell transport and metabolic processes, suggesting that tubular epithelial cells were redifferentiating. We selected some of the most highly expressed genes and performed ISH and qPCR to validate their expression in injured kidney. Candidate genes included the following: Slc22a7, Rrm2, Ctss, and Sprr2f (Figure 6, A and B). Slc22a7 is an organic anion transport with a role in creatinine transport (24, 25). Using ISH, we observed that Slc22a7 was expressed at baseline in the S3 segment of the proximal tubule. Upon injury, Slc22a7 was essentially undetectable. The loss of this marker of differentiation reflected epithelial injury–induced dedifferentiation. At 14 days after injury, there was reexpression of Slc22a7, since the regenerating cells were now returning to a differentiated state. We observed a similar trend of downregulation during injury and reexpression during repair by qPCR.
Figure 6. Validation of candidate genes Slc22a7, Rrm2, Ctss, and Sprr2f.
(A) ISH in kidney sections from adult, male C57BL/6 mouse at 3 different time points after Bi-IRI. Scale bars: 500 μM (upper panels); 50 μM (lower panels). (B) qPCR in whole kidney lysates for the candidate genes. Representative results from n = 3–4 independent samples per time point. *P < 0.05; ***P < 0.001; ****P < 0.0001, 1-way ANOVA with post hoc Dunnett’s multiple comparisons test.
Rrm2 encodes the regulatory subunit of ribonucleotide reductase, which catalyzes the synthesis of deoxyribonucleotides from ribonucleotides (26). Using ISH, we found that Rrm2 had a very low level of expression at baseline and could only be detected in scattered individual tubule cells in the cortex. At day 2 after injury, Rrm2 expression was substantially upregulated, primarily in the outer medulla, which is consistent with the need for DNA synthesis to support cell division in this segment, which is damaged the most after IRI. At day 14 after surgery, Rrm2 expression was decreased and had a pattern similar to that at baseline. Ctss has multiple roles, including extracellular matrix degradation and antigen processing and presentation (27, 28). Ctss was not detected in uninjured kidney, but it was expressed in the outer segment of the outer medulla during injury (day 2), and expression even increased by day 14 day, as shown by ISH and qPCR. Ctss is involved in EGFR degradation (29); therefore, one can hypothesize that persistent upregulation of Ctss in the tubular epithelium may prevent further EGFR activation, which could potentially promote renal fibrosis as previously reported (30, 31). Sprr2f belongs to the Sprr family of proteins, which are expressed at high levels in the epidermis and function to maintain epithelial integrity (32). At baseline, there was complete absence of Sprr2f; however, on day 2 after injury, there was marked expression throughout the cortex, though it was localized to specific tubular segments that were dilated and thus may represent localized tubular damage. Sprr2f has antioxidative functions, so it could mediate reactive oxygen species detoxification after IRI (33, 34). Fourteen days after surgery, Sprr2f expression was decreased, though individual tubule segments still expressed it strongly, potentially reflecting failed repair.
Differential expression of transcription factors and secreted proteins during injury and repair.
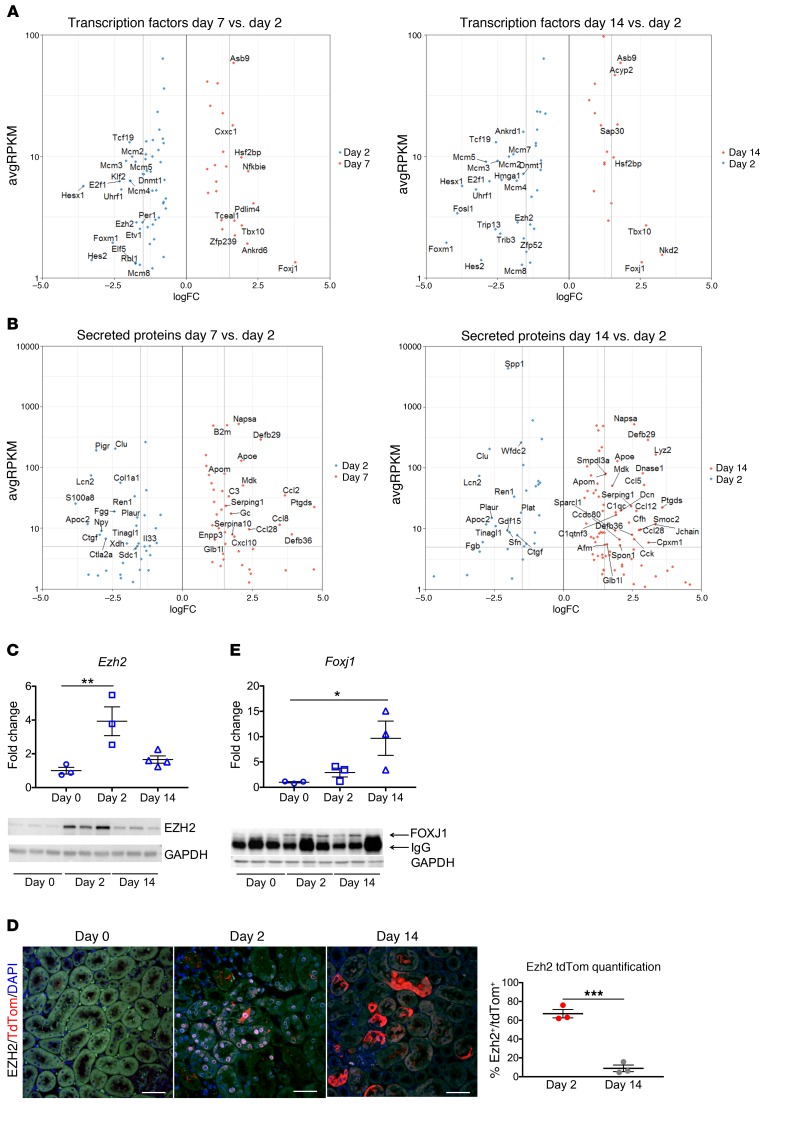
Transcription factors regulate cell state genes, we therefore identified dynamic transcription factor expression during injury and repair. We took our list of DEG and cross-referenced it against the Riken Transcription Factor Database (35). We compared the lists for day 7 versus day 2 and day 14 versus day 2 and identified 87 and 66 transcription factors, respectively (Supplemental Table 1). Figure 7A illustrates a portion of the identified transcription factors. Among the transcription factors, we evaluated Ezh2, Foxm1, and Foxj1 in more detail. We also crossreferenced the DEG list for the 2 comparisons against a database of curated secreted proteins (36), and the results are shown in Figure 7B and Supplemental Table 1.
Figure 7. Transcription factors and secreted proteins identified during translational profiling of injured tubular epithelial cells.
(A and B) Scatter plots showing some of the upregulated and downregulated transcription factors and secreted proteins when comparing bound day 7 versus day 2 and bound day 14 versus day 2. (C) Ezh2 mRNA and protein expressions by qPCR and Western blot, respectively, showing upregulation at day 2 and downregulation by day 14. (D) Immunostaining and quantification for EZH2 shows coexpression in tdTomato-labeled cells at day 2 and almost complete absence by day 14 when repair is complete. (E) Foxj1 mRNA and protein expression by qPCR and Western blot. There is increased Foxj1 mRNA expression at day 2 after injury, with further upregulation by day 14. At the protein level, FOXJ1 expression is increased at day 2 compared with day 0 and continues to be expressed by day 14. Scale bars: 50 μM. n = 3–4 samples per time point. *P < 0.05; **P < 0.01; ***P < 0.001, 1-way ANOVA with post hoc Dunnett’s multiple comparisons test.
Ezh2 belongs to the polycomb repressive complex 2 (PRC2), which participates in methylation of histone 3 (H3K27me), leading to transcriptional repression (37, 38). Ezh2 has been described as having an important role in coordinating cell differentiation in embryonic stem (ES) cells (39, 40), mesenchymal stem cells (41–43), hematopoietic stem cells (44, 45), and also in various types of cancer (46, 47). Inhibition of Ezh2 has been reported to prevent renal fibroblast activation (48), though more recently the same group reported that Ezh2 acts in epithelial cells to promote fibrosis (49). We confirmed by qPCR that Ezh2 was upregulated at day 2 after Bi-IRI as compared with baseline (day 0) and that its expression downtrended by day 14 (Figure 7C). At the protein level, there was very low expression at baseline (Figure 7C), which was not detectable by immunofluorescence (Figure 7D). However, 2 days after Bi-IRI, EZH2 was highly expressed both by Western blot and immunofluorescence staining (Figure 7, C and D). Approximately 65% of tdTomato-positive cells also expressed EZH2. On the other hand, by day 14 after injury, only 10% of tdTomato cells were still expressing EZH2 (Figure 7D). The temporal pattern of EZH2 expression suggests that it may be involved with the transient repression of terminal differentiation genes during injury and repair.
Foxj1 is a transcription factor essential for the assembly of motile cilia (50). Its role in renal injury has been explored in different models in zebra fish, where it was found to be induced upon epithelial injury and required for cilia maintenance (51). We observed Foxj1 upregulation at day 2 after injury, with continued expression at day 14 (Figure 7E). By Western blot, FOXJ1 expression was almost undetectable at day 0, but increased at day 2 and day 14 after Bi-IRI (Figure 7E).
Foxm1 is upregulated during tubular epithelial injury in murine kidney.
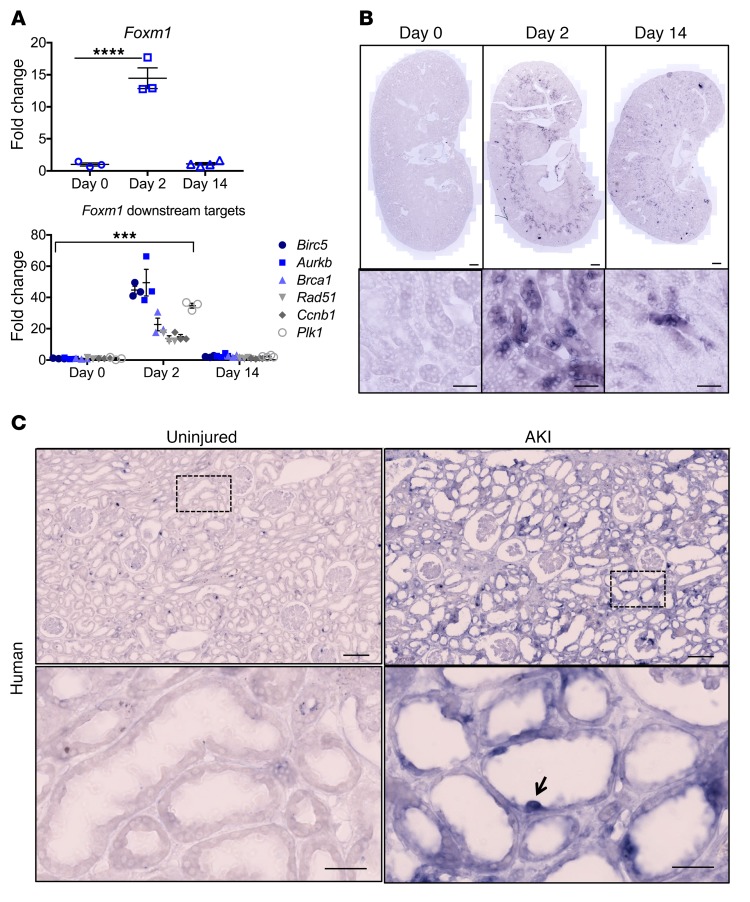
One of the most highly upregulated transcription factors in our data set was Foxm1, which in other cellular contexts drives cell-cycle progression (52, 53). It is expressed mostly in high cycling organs, such as testes and thymus, and is absent in terminally differentiated cells (52). Foxm1 is also expressed during development in several organs, including the kidney, and is upregulated in various types of cancers (52, 54, 55). It plays a key role in the G2/M transition and for chromosome segregation and cytokinesis (56). It also participates in DNA break repair (57). Foxm1 has been found to be reactivated after injury in certain organs, such as lung (58), liver (59, 60), and pancreas (61). We sought to validate Foxm1 expression after injury by qPCR and observed that Foxm1 was upregulated 15-fold in day 2 injured kidney compared with day 0 and that its expression returned almost back to baseline at day 14 when most of the repair had occurred (Figure 8A). We also evaluated by qPCR downstream targets of Foxm1 related to cell-cycle (Ccnb1, Plk1, Aurkb) and DNA repair (Birc5, Brca, Rad51) and found them to be significantly upregulated at day 2 and their expression returning close to baseline at day 14 (Figure 8A). We next designed Foxm1 antisense and sense probes and performed ISH. There was no detectable Foxm1 expression at day 0, but it was clearly expressed in the outer stripe of the outer medulla at day 2, with subsequent downregulation in most (but not all) tubule segments by day 14 (Figure 8B). We next performed ISH on an uninjured and an acutely injured human kidney. FOXM1 was undetectable in healthy kidney, but could be detected in dedifferentiated, flattened epithelia in the AKI kidney (Figure 8C).
Figure 8. Foxm1 is expressed after kidney injury in mouse and human.
(A) mRNA expression of Foxm1 and its downstream targets after injury. (B) ISH in uninjured and injured mouse kidneys sections showing increased expression in the outer segment of the outer medulla at day 2 and significant downregulation at day 14. Scale bars: 500 μM (upper panels); 50 μM (lower panels). (C) ISH in human samples from uninjured and injured kidney showing absent FOXM1 expression in the uninjured kidney and expression in cells from injured tubules. Scale bars: 200 μM (upper panels); 50 μM (lower panels). For A and B, n = 3–4 mice per time point. For C, n = 1 for each condition. ***P < 0.001; ****P < 0.0001, 2-way ANOVA with post hoc Dunnett’s multiple comparisons test.
FOXM1 knockdown in human proximal tubular epithelial cells impairs proliferation.
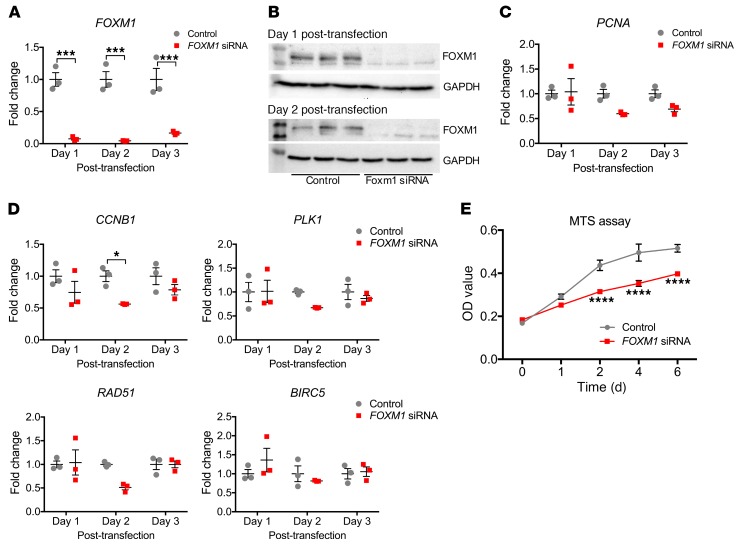
Given the known role of FOXM1 in cellular proliferation, we next asked whether the absence of FOXM1 in primary human proximal tubular epithelial cells (hRPTECs) causes a proliferation defect during cell culture. We transfected early passage hRPTECs with FOXM1 siRNA or negative control. Figure 9A shows that FOXM1 siRNA reduced FOXM1 mRNA expression by close to 95% at day 1 and day 2 after transfection, and this was supported by Western blot showing absence of the FOXM1 protein in the siRNA-treated hRPTECs compared with control (Figure 9B). We evaluated PCNA mRNA expression as a surrogate marker for proliferation and observed that it was downregulated in the FOXM1 siRNA–treated hRPTECs, consistent with a proliferative defect (Figure 9C). We also checked known downstream targets of FOXM1, including cell-cycle regulators CCNB1 and PLK1 and DNA repair genes RAD51 and BIRC5. Expression of 3 out of 4 of these was reduced with FOXM1 knockdown compared with controls 2 days after transfection (Figure 9D). We also measured cell proliferation directly. Consistent with the prior results, FOXM1 knockdown hRPTECS had a lower rate of proliferation than the scrambled siRNA controls (Figure 9E).
Figure 9. Foxm1 drives proximal tubular epithelial proliferation.
(A) qPCR for FOXM1 showing efficient FOXM1 siRNA knockdown in hRPTECs at different time points after transfection. (B) Western blot for FOXM1 in hRPTECs corroborating siRNA knockdown. (C) PCNA mRNA expression in control and FOXM1 siRNA–treated hRPTECs. (D) qPCR for FOXM1 downstream genes in hRPTECs treated with FOXM1 siRNA versus control. (E) MTS assay in hRPTECs shows decrease proliferation in FOXM1 siRNA–treated cells compared with control. n = 3 replicates for each time point, except MTS assay, which was done on n = 6 per each day evaluated. *P < 0.05; ***P < 0.001; ****P < 0.0001, 2-way ANOVA with post hoc Bonferroni’s multiple comparisons test.
FOXM1 is downstream of the EGFR pathway in hRPTECs and after IRI in vivo.
The EGFR pathway is known to play an important role in tubular epithelial proliferation after injury (62, 63); and FOXM1 regulates keratinocyte cell-cycle progression in an EGFR-dependent fashion (64). Therefore, we asked whether FOXM1 expression is regulated by EGFR in kidney. We treated hRPTECs with the EGFR inhibitor erlotinib or vehicle. Both FOXM1 mRNA expression and FOXM1 downstream targets substantially decreased with the use of erlotinib (Figure 10A). This result was confirmed also by Western blot for FOXM1, which showed absence of the FOXM1 protein and absence of the phospho-EGFR protein, confirming that the EGFR inhibition was in fact induced (Figure 10B).
Figure 10. Foxm1 is downstream of the Egfr pathway in tubular epithelial proliferation.
(A) mRNA expression for FOXM1 and several of its downstream targets in hRPTECs after treatment with erlotinib. (B) Western blot in lysates of hRPTECs treated with erlotinib versus vehicle. There is complete absence of FOXM1 protein upon inhibition of EGFR with erlotinib, indicating that FOXM1 is downstream of the EGFR pathway. Lack of phosphor-EGFR expression confirms inhibition of EGFR by erlotinib. (C–E) qPCR for Foxm1, Plk1, and Ki67 2 days after IRI in mice of different strains treated with erlotinib and vehicle. For cell culture experiments, n = 3 replicates per group. For in vivo experiments, n = 3–5 mice per group. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, 2-way ANOVA with post hoc Bonferroni’s multiple comparisons test.
We next investigated whether EGFR regulates Foxm1 expression after injury in vivo. Surprisingly, in C57BL/6 mice, erlotinib (100 mg/kg) had no effect on either Foxm1 expression, its target gene Plk1, or Ki67 (Figure 10C). In contrast, in mixed C57/129 mice (the background of Kim1-GCE), we observed partial inhibition of Foxm1 and Plk1 by erlotinib, but no significant effect on Ki67 (Figure 10D). Since FVB/NJ mice have been reported to be sensitive to Egfr inhibition in a renal fibrosis model (30), we also tested this strain. Using the same dose of erlotinib (80 mg/kg) or vehicle prior to Bi-IRI and on day 1 after injury, we observed a nearly complete inhibition of Foxm1 induction by erlotinib. Both Plk1 and Ki67 had similar (>90%) reduction in gene expression (Figure 10E). Thus the Egfr/Foxm1 signaling pathway exists in vivo, though in a strain-dependent fashion.
Discussion
There are two primary conclusions from the current study. First, lineage analysis of cells that express KIM1 after injury shows a proliferative expansion of these dedifferentiated cells during repair. These results do not support a model whereby a fixed tubular progenitor population exclusively repairs injured tubule because KIM1 is not expressed in uninjured kidney and it is not expressed in putative PAX2+ intratubular progenitors (10). Second, we identify an EGFR/FOXM1 pathway that regulates proximal tubule proliferation. Our comprehensive transcriptional analysis of proximal tubule during injury and repair will serve as a resource for understanding dedifferentiation, redifferentiation, and failed repair at the molecular level.
It has been proposed that after acute injury, a majority of injured epithelial cells upregulate cell-cycle markers, but fail to undergo mitosis — so-called endocycle — and that this explains why past studies relying on cell-cycle markers reached incorrect conclusions (10). Our studies here refute this claim, because our genetic labeling strategy excluded putative PAX2+ progenitors since they are resistant to injury and do not upregulate KIM1 in the first place (10). Rather, we observed clonal expansion of Kim1-labeled injured cells that indeed completed mitosis, as judged by an expansion in the size of coherent clones during repair. Consistent with this conclusion, we could find no evidence for increased DNA content in Kim1-labeled cells, as would be expected had they undergone endocycle. Instead, our results are consistent with the traditional view that injured proximal tubule cells have the capacity to contribute to tubule repair through proliferation (14, 65–67). Our prior epithelial lineage analysis studies after injury support this conclusion as well (4–6)
With respect to our transcriptional analysis of injury and repair, one limitation of our approach is that we do not have a matched baseline proximal tubule–specific transcriptome to compare with, since KIM1 is not expressed in uninjured kidney. As a consequence, we compared acutely injured proximal tubule transcriptomes with those during repair at 7 and 14 days after injury. We recognize that a portion of labeled proximal tubule cells (about 20%) remain injured at day 14. This did not prevent us from defining a large number of transcription factors and secreted proteins with dynamic expression over the injury and repair time course.
During the acute injury phase (day 2), we noted a strong keratinocyte differentiation gene signature. This included the small proline-rich (Sprr) genes, such as Sprr2f. This family of proteins are induced during keratinocyte differentiation and provide structural integrity to the cornified cell envelope of stratified epithelial cells (32). It is unclear what their role might be in tubular repair, since tubular epithelium is a simple epithelium. It is possible that SPRR proteins are being expressed as part of the plasticity of tubular epithelium during injury and confer a transient keratinocyte-like phenotype to provide protection in the face of damage and inflammation. Interestingly, some genes from the keratin family were found to be differentially expressed during injury; these included Krt4, Krt5, Krt18, Krt19, and Krt20. Krt18 and Krt20 have been recently recognized as being upregulated after ischemia/reperfusion, but their role has not yet been defined (19). Keratins also act as scaffolds that endow epithelial cells with the ability to sustain mechanical and nonmechanical stresses (68); their expression during tubular injury may therefore protect injured epithelia from the harsh postischemic tubular environment.
We identified the transcription factor Foxm1 as strongly upregulated in proximal tubule during injury. Foxm1 is a proliferation-specific transcription factor with expression mostly in high cycling organs, such as testes and thymus (52). It is also expressed during development in different organs, including the kidney, and various types of cancers (52, 54, 55). Multiple studies have provided proof of its role in cellular proliferation and have identified downstream targets that are critical for the G2/M transition and for chromosome segregation and cytokinesis (56). Foxm1 is also reactivated after injury in lung, liver, and pancreas. In a butylated hydroxytoluene (BHT) model of lung injury, Foxm1 was found to be expressed in pulmonary epithelial, endothelial, and smooth muscle cells (58). Foxm1 was induced in hepatocytes after liver injury with carbon tetrachloride and partial hepatectomy (59, 60). One study found that absence of Foxm1 led to impaired β cell proliferation after pancreatectomy (61). In kidney, Foxm1 has only been reported in the context of renal cell carcinoma (69). Based on ISH staining, Foxm1 expression localized predominantly in the S3 segment, which is the area more susceptible to injury, reinforcing the notion that proliferation is a central component of the renal repair response. Our results here indicate that Foxm1 may be an important regulator of injury-induced proximal tubule proliferation. Our in vitro studies using hRPTECs confirmed that absence of FOXM1 leads to decreased proliferation, along with downregulation of downstream targets involved in the cell cycle. That EGFR inhibition abolished FOXM1 expression makes sense, since EGF is a potent epithelial mitogen with known importance in regulating the repair response. Furthermore, our in vivo studies confirmed a role for Egfr regulation of Foxm1 after IRI. The precise signaling pathway linking EGFR to FOXM1 will require further study.
In conclusion, surviving, injured epithelial cells are capable of proliferation after injury. Our results do not support the existence of a fixed intratubular progenitor population. We also identify an EGFR/FOXM1 signaling circuit that regulates proximal tubule proliferation after acute injury.
Methods
Creation of GFPCreERt2 knockin to the Havcr1 locus.
A targeting vector was constructed to insert the EGFPCreERT2-SV40pA (GCE) transgene and a FRT-flanked PGKneobpA selection cassette into the 5′ UTR of the Havcr1 gene. The GCE transgene comprises an EGFP and a tamoxifen inducible Cre-recombinase fusion gene (CreERT2). A negative selectable marker thymidine kinase (MC1TK) cassette was also included in the targeting vector to select against nonhomologous recombination. The genomic sequence of the mouse Havcr1 gene and surrounding sequence was downloaded from the University of California, Santa Cruz, Genome Browser (http://genome.ucsc.edu). Repetitive sequences were masked. The lengths of the homology arms were dictated by the repetitive DNA sequence surrounding the target site, resulting in a 1765 bp kb 5′ homology arm and a 2791 bp 3′ homology arm.
A cloning strategy with suitable restriction enzymes and appropriate primers was chosen based on the sequences of the homology arms and all transgene cassettes. Homology arms were amplified from BAC clones containing the Havcr1 locus, RP23-58M12, and RP23-82L5 (BACPAC Resources Center) using Platinum Pfx DNA polymerase (Thermo Fisher Scientific). All DNA oligonucleotides were ordered from Integrated DNA Technologies. Both homology arms and the transgene cassettes were cloned into pBluescript KS(–). Linearized targeting construct was electroporated into v6.5 (C57BL/6 × 129/Sv F1 hybrid) ES cells, and transformants were selected by culture in G418 and ganciclovir. Resistant clones were screened by long-range PCR. Positive clones were expanded and underwent injection into albino B6 (C57BL/6J-Tyrc-2J/J) blastocysts using standard procedures.
Animals.
Kim1-GCE mice were created as described above. Rosa26tdTomato (JAX stock 007909), EGFP-L10a (JAX stock 024750), C57BL/6J (JAX stock 000664), and FVB/NJ (JAX stock 001800) were purchased from Jackson Laboratories.
Surgery.
For bilateral IRI, 8- to 12-week-old male mice were anesthetized with isoflurane and buprenorphine SR was administered for pain control. Body temperature was monitored and maintained at 36.5–37.5°C throughout the procedure. Bilateral flank incisions were made and the kidneys exposed. Ischemia was induced by clamping the renal pedicle with a nontraumatic microaneurysm clamp (Roboz) for 20 minutes. For the FVB/NJ mice, 21 minutes of ischemia were used. The clamps were removed at time completion and kidneys returned to the peritoneal cavity. The peritoneal layer was closed with absorbable suture and the flank incisions closed with wound clips. For unilateral IRI in the lineage-tracing experiment, the left kidney was clamped for 24 minutes.
Human kidney samples.
Kidney parenchyma was obtained from discarded human donor kidney with donor anonymity preserved. The injured kidney came from a 62-year-old man with a serum creatinine of 3.3 mg/dL at time of collection. The healthy kidney was from a 38-year-old woman with a serum creatinine of 0.6 mg/dL.
TRAP.
Kidneys were harvested and TRAP was performed as previously described (21). RNA integrity and quantity were determined using the Agilent RNA PicoChip Kit and the Agilent 2100 Bioanalyzer System (Agilent Technologies). The Clontech SMARTer Universal Low Input RNA Kit (Takara Bio USA) was used for cDNA library preparation. cDNA libraries and next-generation sequencing were performed at the Genome Technology Access Center at Washington University in St. Louis. Twelve samples were sequenced with an Illumina HiSeq3000, obtaining 25–30 million reads per sample.
RNA-Seq data analysis.
DEG analysis was performed using the edgeR package (70) and setting a cutoff CPM of more than 0.4 and an FDR of less than 5%. GO was performed using DAVID (71, 72) and analyzed using the functional annotation tool. Scattered plots for transcription factors and secreted proteins were created using ggplot.
Bioinformatic cell-cycle analysis.
We obtained the marker gene list for G2M and S phase from a previous study (17). PCA was performed based on the expression of the G2M and S phase markers. We then assigned a cell-cycle score (from –1 to 1) on each TRAP sample according to its gene expression of G2/M and S phase markers using the CellCycleScoring function in Seurat R package. We assigned each sample to a cell-cycle phase based on the following criteria: (a) if Sscore (S phase score) > 0 and Sscore > G2Mscore (G2M phase score), S phase; (b) G2Mscore > 0 and G2Mscore > Sscore, G2/M phase; and (c) If Sscore and G2Mscore < 0, G1 phase. The samples were colored by cell-cycle phase or cell-cycle score and visualized in the PCA map.
Kidney tissue dissociation and cell fixation.
Kidneys were harvested at day 14 and day 30 after injury. Single-cell dissociation was performed as previously described (73) with some modifications. Briefly, kidney was minced with a razor blade in a petri dish on ice and divided into 2 Eppendorf tubes containing 1 L of cold protease enzyme solution (0.5 mg/mL Bacilus Licheniformis protease [MilliporeSigma P5380], 5 mM CaCl2 in DPBS]. The sample was incubated at 6°C in a thermoshaker with trituration using a 1 mL pipette (15 seconds every 5 minutes) for 20 minutes. After this time, the cell suspension was put on ice for 5 minutes. The supernatant was carefully removed from both tubes, passed through a 40 μM filter, and rinsed with 3 mL of 10% FBS to stop the reaction. The remaining tissue at the bottom of the tube was resuspended in cold active protease solution containing 10 mg/mL protease and 5 mM CaCl2 in DPBS. The cell suspension was incubated at 6°C for 20 minutes with trituration. After 20 minutes, the cell suspension from both tubes was transferred to a C tube and run in the D-01 program on the gentleMACS Dissociator (Miltenyi Biotec) in a cold room. Single cell dissociation was examined under the microscope. The cell suspension was passed through a 40 μM filter and rinsed with 3 mL of 10% FBS to stop the reaction. The single cell suspensions from both cold protease digestion steps were combined and centrifuged at 1200 g for 5 minutes at 4°C. Supernatant was discarded. The cells were rinsed with 2 mL of DPBS/0.01% BSA and pelleted (1200 g, 5 minutes), and this step was repeated twice more. The cells were then fixed in 1% PFA in PBS for 1 hour on ice. After 1 hour, the cells were centrifuged at 1200 g for 5 minutes and supernatant discarded. Cells were resuspended in 2 mL of DPBS/0.01% BSA and centrifuged at 1200 g for 5 minutes. Supernatant was discarded leaving a small volume (~200 μL) to resuspend the cells. Next, we fixed the cells in 1 mL 80% cold ethanol by adding the ethanol dropwise with gentle vortexing, and cells were incubated overnight at 4°C.
DNA content/cell-cycle analysis.
Cells were fixed as described above. Cells were pelleted at 2000 g for 5 minutes and the supernatant removed, followed by rinsing with DPBS/0.01% BSA and centrifugation at 1200 g for 5 minutes twice. DNA content was determined by staining with a DAPI solution containing 1 μg/mL DAPI in PBS with 0.01% Triton-X 100 for at least 15 minutes just prior to cell-cycle analysis. The contralateral (CLK) kidney without injury was used as a negative control for gating in the tdTomato population, since there were no Kim1+ cells detected in the CLK kidney. HEK293T cells treated and not treated with colchicine as described below were used a positive control for polyploidy. The DNA content analysis was done in a BD LSRFortessa X-20 analyzer using a violet laser (405 nm) to detect DAPI and a yellow/green laser (561 nm) for tdTomato detection. FACSDiva software was used during data acquisition. At least 50,000 events were collected, and further DNA content analysis was performed using FlowJo, version 10.5.3 (TreeStar).
Real-time PCR.
Kidney tissue was snap-frozen in liquid nitrogen at the time of harvesting. RNA was extracted using the Direct-zol MiniPrep Plus Kit (Zymo) following the manufacturer’s instructions. The extracted RNA (600 ng) was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies). Quantitative PCR (RT-PCR) was performed using iTaq Universal SYBR Green Supermix (Bio-Rad). Expression levels were normalized to GAPDH and data analyzed using the 2-ΔΔCt method. Primers used are listed in Supplemental Table 2.
Tissue preparation and histology.
Mice were perfused via the left ventricle with ice-cold PBS. Kidneys were harvested and fixed in 4% paraformaldehyde on ice for 1 hour, then incubated in 30% sucrose at 4°C overnight. The next day, tissues were embedded in OCT medium (Sakura Finetek). Kidney sections were cut at 6 μm and mounted on Superfrost slides. Immunofluorescent staining was performed as follows: kidney sections were washed with 1× PBS for 10 minutes and permeabilized with 0.25% Triton X for 10 minutes. Blocking was done with 5%BSA in PBS for 1 hour. Primary antibodies were incubated for 1 hour at room temperature and sections rinsed with 1× PBS for 5 minutes × 3. Secondary antibodies (1:200) were incubated for 1 hour at room temperature and rinsed with 1× PBS for 5 minutes × 3. DAPI was used for counterstaining. The following antibodies were used: KIM1 (catalog AF1817, R&D Systems), KI67 (catalog 14-5698, eBioscience), EGFP (catalog GFP-1020, Aves Labs), VIMENTIN (catalog Ab92547, Abcam), SOX9 (catalog Ab185230, Abcam), EZH2 (catalog 5246, Cell Signaling), and DBA (catalog FL-1031, Vector Labs).
Sensitivity and specificity quantification.
Kidney sections from Kim1-GCE+/–;tdTom+/– mice were stained with KIM1 antibody (AF1817 R&D Systems), and ×400 images (n = 10) were taken randomly. A TP cell was defined as tdTomato cell that expresses KIM1. A FN cell was a non-tdTomato cell that did not express KIM1. A TN was a non-tdTomato cell that did not express KIM1. A FP was a tdTomato cell that did not express KIM1. Sensitivity was then defined as TP/(TP + FN) and specificity as TN/(FP + TN).
Western blot.
Kidney tissue was snap-frozen in liquid nitrogen upon harvesting. Tissue was homogenized in RIPA lysis buffer containing protease inhibitors (Roche). Protein concentration was measured using the BCA assay (Thermo Fisher). For hRPTECs, cells were washed with 1× PBS and lysates prepared in RIPA buffer with protease inhibition. Using 10% polyacrylamide gel, 10–20 μg of protein was separated by SDS electrophoresis and transferred to an Immobilon PVDF membrane (Millipore). Membrane was blocked with 5% milk in TBST and probed overnight at 4°C with the primary antibody. After washing the membrane with TBST, it was incubated for 1 hour at room temperature with HRP-conjugated secondary antibody (Dako). The membrane was developed using the ECL Detection System (GE Healthcare). Primary antibodies were as follows: KIM1 (catalog AF1817 R&D Systems), EZH2 (catalog 5246, Cell Signaling Technology), FOXM1 (catalog 5436, Cell Signaling Technology), phospho-EGFR (catalog 3777, Cell Signaling Technology), and GAPDH (catalog A300-641A, Bethyl Laboratories).
ISH.
Kidneys were perfused with RNase-free PBS and fixed with 4% PFA for 1 hour at 4°C and then switched to 30% sucrose and kept overnight at 4°C. All solutions were prepared in RNase-free PBS. Tissue was embedded in OCT and sectioned into 15 μM sections. ISH was performed as previously described, with some minor modifications (19). Briefly, sections were incubated with 4% PFA at 4°C overnight. After rinsing, sections were incubated with Proteinase K (10 μg/mL) for 20 minutes, acetylated with 0.375% acetic anhydride, and hybridized with sense or antisense digoxigenin-labeled (DIG-labeled) riboprobe (final 500 ng/mL) overnight at 68°C. The next day, sections were washed with a series of stringency washes at 68°C and blocked with 2% Roche blocking reagent for 1 hour at room temperature. Subsequently, sections were incubated with anti–DIG-AP antibody (11093274910, Roche, 1:4000) at 4°C overnight, followed by development with BM Purple (Roche), which varied from 6 hours to 10 days, depending on staining intensity. Finally, sections were fixed with 4% PFA and mounted with ProLong Gold (Invitrogen). ISH images were generated using a Zeiss Axio Scan Z1 slide scanner. DIG-labeled riboprobes were generated from template PCR, and the primer sequences are listed in Supplemental Table 2.
Cell culture experiments.
Primary human proximal tubular cells were purchased from Lonza (CC-2553) and cultured with Renal Epithelium Cell Growth Medium 2 (PromoCell) supplemented with 10 ng/mL EGF, 5% v/v fetal calf serum, provided with the medium kit. Cells were maintained in a humidified 5% CO2 atmosphere at 37°C. Experiments were carried out on early passage cells.
FOXM1 siRNA transfection. hRPTECs were grown to 50%–60% confluency, at which point they were transfected with 10 nmol/L FOXM1 siRNA (Silencer Select siRNA s5248, Thermo Fisher) or negative control siRNA (Silencer Select siRNA 4390843, Thermo Fisher) using Lipofectamine RNAiMAX (Life Technologies) following the manufacturer’s protocol. Cells were harvested at day 1 and day 2 after transfection for protein and RNA isolation in order to validate knockdown. The siRNA sequences were as follows: sense, GCUCAUACCUGGUACCUAUTT; antisense, AUAGGUACCAGGUAUGAGCTG.
MTS assay.
For the MTS experiments, hRPTECs were transfected with FOXM1 siRNA or negative control as above. One day after transfection, cells were trypsinized (Gibco, Thermo Fisher Scientific) and counted using a hematocytometer. Cells were seeded at a density of 1250 cells per well in a 96-well plate in Renal Epithelium Cell Growth Medium 2 (PromoCell). Six replicates were prepared per group. Proliferation was measured using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) per the manufacturer’s protocol. Optical density readings were taken 2 hours after first seeding for day 0 and subsequently on days 1, 2, 4, and 6.
Erlotinib treatment.
hRPTECs were starved overnight by culturing on Renal Epithelium Cell Growth Medium 2 without any supplements. Following starvation, hRPTECs were switched to full medium and treated with the EGFR inhibitor erlotinib (10483, Cayman Chemicals) at concentrations of 10 μM for 24 hours. Cells treated with DMSO served as controls. hRPTECs were then harvested for downstream analysis.
For the in vivo studies, erlotinib hydrochloride (E-4007, LC Laboratories) was administered by oral gavage per the experimental protocol at 80 mg/kg body weight (BW) or 100 mg/kg BW. Erlotinib was dissolved in 0.5% methylcellulose and 1% Tween-80. Control mice received vehicle.
Colchicine treatment.
HEK293T cells (ATCC CRL-3216) were cultured in DMEM with 10% FBS and 1× penicillin/streptomycin. Cells were treated with 10 μM colchicine dissolved in DMSO for 1 hour. Cells treated with DMSO served as control. After 1 hour, cells were returned to cultured medium and harvested after 48 hours.
Data availability.
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE121191). To increase rigor and reproducibility, the raw sequencing data generated as part of this study were also uploaded into the (Re)Building a Kidney (RBK) consortium database and are fully accessible at https://doi.org/10.25548/16-E1GE
Statistical analysis.
Data are presented as mean ± SEM. Unpaired 2 tailed Student’s t test was used to compare 2 groups, and a P value of less than 0.05 was considered significant. For multiple group comparisons, 1-way or 2-way ANOVA followed by post hoc correction with Dunnett’s test or Bonferroni’s test where appropriate was applied. Statistics were performed using GraphPad Prism 7.0.
Study approval.
All mouse experiments were approved by the Animal Care and Use Committee at Washington University in St. Louis. Human kidney specimen collection and use was approved by the IRB of Washington University in St. Louis. Informed consent was not obtained, as samples were obtained from discarded human donor kidney with donor anonymity preserved.
Author contributions
MCP and FFK designed and carried out experiments, analyzed results and reviewed the manuscript. ML, SI, HW, EK, MMA, AH, and AK carried out some of the experiments, analyzed data, and reviewed the manuscript. BDH conceived of the work, designed experiments, analyzed results, and wrote the manuscript together with MCP.
Supplementary Material
Acknowledgments
Primary support for this work was from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) ReBuilding a Kidney consortium grant DK107374. Additional support was from NIH/NIDDK grants DK103740, an Established Investigator Award of the American Heart Association (all to BDH), and F32 DK103441 (to MCP). We also thank Erica Lantelme at the Washington University Pathology FACS core for her assistance with cell-cycle analysis and The Genome Technology Access Center in the Department of Genetics for assistance with next-generation sequencing.
Version 1. 11/11/2019
Electronic publication
Version 2. 12/02/2019
Print issue publication
Footnotes
AK’s present address is: Department of Kidney Development, Institute of Molecular Embryology and Genetics, Kumamoto University, Kumamoto, Japan.
Conflict of interest: BDH is a scientific cofounder of and holds equity in Chinook Therapeutics, a biotechnology company seeking to develop drugs to treat kidney diseases. His laboratory also receives research support from Chinook Therapeutics and Janssen Research & Development, LLC. He has consulted for Chinook Therapeutics, Janssen Research & Development, LLC, and Indalo Therapeutics.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(12):5501–5517.https://doi.org/10.1172/JCI125519.
Contributor Information
Monica Chang-Panesso, Email: mchang-panesso@wustl.edu.
Haojia Wu, Email: haojiawu@wustl.edu.
Shiyo Ikeda, Email: ikedayoichiro.tky@gmail.com.
Eirini Kefaloyianni, Email: ikefalog@gmail.com.
Mai M. Abdelmageed, Email: abdelmm@wustl.edu.
Andreas Herrlich, Email: aherrlich@wustl.edu.
Akio Kobayashi, Email: a.kobayashi.001@gmail.com.
Benjamin D. Humphreys, Email: bhumphre@dom.wustl.edu.
References
- 1.Pavkov ME, Harding JL, Burrows NR. Trends in hospitalizations for acute kidney injury - United States, 2000-2014. MMWR Morb Mortal Wkly Rep. 2018;67(10):289–293. doi: 10.15585/mmwr.mm6710a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saran R, et al. US Renal Data System 2017 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am J Kidney Dis. 2018;71(3 Suppl 1):A7. doi: 10.1053/j.ajkd.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takaori K, et al. Severity and frequency of proximal tubule injury determines renal prognosis. J Am Soc Nephrol. 2016;27(8):2393–2406. doi: 10.1681/ASN.2015060647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphreys BD, et al. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2(3):284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Humphreys BD, Czerniak S, DiRocco DP, Hasnain W, Cheema R, Bonventre JV. Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci U S A. 2011;108(22):9226–9231. doi: 10.1073/pnas.1100629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD. Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci U S A. 2014;111(4):1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang-Panesso M, Humphreys BD. Cellular plasticity in kidney injury and repair. Nat Rev Nephrol. 2017;13(1):39–46. doi: 10.1038/nrneph.2016.169. [DOI] [PubMed] [Google Scholar]
- 8.Berger K, et al. Origin of regenerating tubular cells after acute kidney injury. Proc Natl Acad Sci U S A. 2014;111(4):1533–1538. doi: 10.1073/pnas.1316177111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinkevich Y, et al. In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell Rep. 2014;7(4):1270–1283. doi: 10.1016/j.celrep.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazzeri E, et al. Endocycle-related tubular cell hypertrophy and progenitor proliferation recover renal function after acute kidney injury. Nat Commun. 2018;9(1):1344. doi: 10.1038/s41467-018-03753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang HM, Huang S, Reidy K, Han SH, Chinga F, Susztak K. Sox9-positive progenitor cells play a key role in renal tubule epithelial regeneration in mice. Cell Rep. 2016;14(4):861–871. doi: 10.1016/j.celrep.2015.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118(5):1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ichimura T, et al. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998;273(7):4135–4142. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 14.Witzgall R, Brown D, Schwarz C, Bonventre JV. Localization of proliferating cell nuclear antigen, vimentin, c-Fos, and clusterin in the postischemic kidney. Evidence for a heterogenous genetic response among nephron segments, and a large pool of mitotically active and dedifferentiated cells. J Clin Invest. 1994;93(5):2175–2188. doi: 10.1172/JCI117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imgrund M, et al. Re-expression of the developmental gene Pax-2 during experimental acute tubular necrosis in mice 1. Kidney Int. 1999;56(4):1423–1431. doi: 10.1046/j.1523-1755.1999.00663.x. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, et al. Sox9 activation highlights a cellular pathway of renal repair in the acutely injured mammalian kidney. Cell Rep. 2015;12(8):1325–1338. doi: 10.1016/j.celrep.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 17.Eigsti OJ. A cytological study of colchicine effects in the induction of polyploidy in plants. Proc Natl Acad Sci U S A. 1938;24(2):56–63. doi: 10.1073/pnas.24.2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizzoni M, Palitti F. Regulatory mechanism of cell division. I. Colchicine-induced endoreduplication. Exp Cell Res. 1973;77(1):450–458. doi: 10.1016/0014-4827(73)90599-5. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight. 2017;2(18):94716. doi: 10.1172/jci.insight.94716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grgic I, et al. Translational profiles of medullary myofibroblasts during kidney fibrosis. J Am Soc Nephrol. 2014;25(9):1979–1990. doi: 10.1681/ASN.2013101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, et al. Cell-specific translational profiling in acute kidney injury. J Clin Invest. 2014;124(3):1242–1254. doi: 10.1172/JCI72126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen H, et al. Characterization of organic anion transporter 2 (SLC22A7): a highly efficient transporter for creatinine and species-dependent renal tubular expression. Drug Metab Dispos. 2015;43(7):984–993. doi: 10.1124/dmd.114.062364. [DOI] [PubMed] [Google Scholar]
- 25.Lepist EI, et al. Contribution of the organic anion transporter OAT2 to the renal active tubular secretion of creatinine and mechanism for serum creatinine elevations caused by cobicistat. Kidney Int. 2014;86(2):350–357. doi: 10.1038/ki.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson S, Martin DW. Ribonucleotide reductase in cultured mouse lymphoma cells. Cell cycle-dependent variation in the activity of subunit protein M2. J Biol Chem. 1981;256(18):9436–9440. [PubMed] [Google Scholar]
- 27.Wilkinson RD, Williams R, Scott CJ, Burden RE. Cathepsin S: therapeutic, diagnostic, and prognostic potential. Biol Chem. 2015;396(8):867–882. doi: 10.1515/hsz-2015-0114. [DOI] [PubMed] [Google Scholar]
- 28.Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, Rudensky AY. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol. 2005;174(3):1205–1212. doi: 10.4049/jimmunol.174.3.1205. [DOI] [PubMed] [Google Scholar]
- 29.Huang CC, Lee CC, Lin HH, Chang JY. Cathepsin S attenuates endosomal EGFR signalling: A mechanical rationale for the combination of cathepsin S and EGFR tyrosine kinase inhibitors. Sci Rep. 2016;6:29256. doi: 10.1038/srep29256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, et al. EGFR signaling promotes TGFβ-dependent renal fibrosis. J Am Soc Nephrol. 2012;23(2):215–224. doi: 10.1681/ASN.2011070645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overstreet JM, et al. Selective activation of epidermal growth factor receptor in renal proximal tubule induces tubulointerstitial fibrosis. FASEB J. 2017;31(10):4407–4421. doi: 10.1096/fj.201601359RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carregaro F, Stefanini AC, Henrique T, Tajara EH. Study of small proline-rich proteins (SPRRs) in health and disease: a review of the literature. Arch Dermatol Res. 2013;305(10):857–866. doi: 10.1007/s00403-013-1415-9. [DOI] [PubMed] [Google Scholar]
- 33.Vermeij WP, Backendorf C. Skin cornification proteins provide global link between ROS detoxification and cell migration during wound healing. PLoS ONE. 2010;5(8):e11957. doi: 10.1371/journal.pone.0011957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vermeij WP, Florea BI, Isenia S, Alia A, Brouwer J, Backendorf C. Proteomic identification of in vivo interactors reveals novel function of skin cornification proteins. J Proteome Res. 2012;11(6):3068–3076. doi: 10.1021/pr300310b. [DOI] [PubMed] [Google Scholar]
- 35.Kanamori M, Konno H, Osato N, Kawai J, Hayashizaki Y, Suzuki H. A genome-wide and nonredundant mouse transcription factor database. Biochem Biophys Res Commun. 2004;322(3):787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- 36.Meinken J, Walker G, Cooper CR, Min XJ. MetazSecKB: the human and animal secretome and subcellular proteome knowledgebase. Database (Oxford) 2015;2015:bav077. doi: 10.1093/database/bav077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 38.Viré E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 39.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 40.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Jin Q, Lee JE, Su IH, Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A. 2010;107(16):7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YH, et al. Myocyte enhancer factor-2 interacting transcriptional repressor (MITR) is a switch that promotes osteogenesis and inhibits adipogenesis of mesenchymal stem cells by inactivating peroxisome proliferator-activated receptor gamma-2. J Biol Chem. 2011;286(12):10671–10680. doi: 10.1074/jbc.M110.199612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu YL, et al. EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1C-dependent calcium signaling. J Biol Chem. 2011;286(11):9657–9667. doi: 10.1074/jbc.M110.185124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su IH, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 45.Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 46.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16(12):803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 47.Wen Y, Cai J, Hou Y, Huang Z, Wang Z. Role of EZH2 in cancer stem cells: from biological insight to a therapeutic target. Oncotarget. 2017;8(23):37974–37990. doi: 10.18632/oncotarget.16467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X, et al. Enhancer of Zeste Homolog 2 inhibition attenuates renal fibrosis by maintaining Smad7 and phosphatase and tensin homolog expression. J Am Soc Nephrol. 2016;27(7):2092–2108. doi: 10.1681/ASN.2015040457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Xiong C, Tolbert E, Zhao TC, Bayliss G, Zhuang S Targeting histone methyltransferase enhancer of zeste homolog-2 inhibits renal epithelial-mesenchymal transition and attenuates renal fibrosis. FASEB J. 2018:fj201800237R. doi: 10.1096/fj.201800237R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet. 2008;40(12):1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- 51.Hellman NE, et al. The zebrafish foxj1a transcription factor regulates cilia function in response to injury and epithelial stretch. Proc Natl Acad Sci U S A. 2010;107(43):18499–18504. doi: 10.1073/pnas.1005998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle. 2011;10(3):396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res. 2013;118:97–398. doi: 10.1016/B978-0-12-407173-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 54.Ye H, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17(3):1626–1641. doi: 10.1128/MCB.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim IM, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 56.Laoukili J, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 57.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta. 2014;1839(11):1316–1322. doi: 10.1016/j.bbagrm.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalinichenko VV, et al. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell types following lung injury. J Biol Chem. 2003;278(39):37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33(6):1404–1414. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99(26):16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ackermann Misfeldt A, Costa RH, Gannon M. Beta-cell proliferation, but not neogenesis, following 60% partial pancreatectomy is impaired in the absence of FoxM1. Diabetes. 2008;57(11):3069–3077. doi: 10.2337/db08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J, Chen JK, Harris RC. Deletion of the epidermal growth factor receptor in renal proximal tubule epithelial cells delays recovery from acute kidney injury. Kidney Int. 2012;82(1):45–52. doi: 10.1038/ki.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, You H, Li Y, Xu Y, He Q, Harris RC. EGF Receptor-Dependent YAP Activation Is Important for Renal Recovery from AKI. J Am Soc Nephrol. 2018;29(9):2372–2385. doi: 10.1681/ASN.2017121272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoll SW, et al. The EGF receptor ligand amphiregulin controls cell division via FoxM1. Oncogene. 2016;35(16):2075–2086. doi: 10.1038/onc.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humes HD, Cieslinski DA, Coimbra TM, Messana JM, Galvao C. Epidermal growth factor enhances renal tubule cell regeneration and repair and accelerates the recovery of renal function in postischemic acute renal failure. J Clin Invest. 1989;84(6):1757–1761. doi: 10.1172/JCI114359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogetseder A, Karadeniz A, Kaissling B, Le Hir M. Tubular cell proliferation in the healthy rat kidney. Histochem Cell Biol. 2005;124(2):97–104. doi: 10.1007/s00418-005-0023-y. [DOI] [PubMed] [Google Scholar]
- 67.Vogetseder A, Picard N, Gaspert A, Walch M, Kaissling B, Le Hir M. Proliferation capacity of the renal proximal tubule involves the bulk of differentiated epithelial cells. Am J Physiol, Cell Physiol. 2008;294(1):C22–C28. doi: 10.1152/ajpcell.00227.2007. [DOI] [PubMed] [Google Scholar]
- 68.Gu LH, Coulombe PA. Keratin function in skin epithelia: a broadening palette with surprising shades. Curr Opin Cell Biol. 2007;19(1):13–23. doi: 10.1016/j.ceb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Z, Zhang G, Kong C. FOXM1 participates in PLK1-regulated cell cycle progression in renal cell cancer cells. Oncol Lett. 2016;11(4):2685–2691. doi: 10.3892/ol.2016.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 72.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adam M, Potter AS, Potter SS. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development. 2017;144(19):3625–3632. doi: 10.1242/dev.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE121191). To increase rigor and reproducibility, the raw sequencing data generated as part of this study were also uploaded into the (Re)Building a Kidney (RBK) consortium database and are fully accessible at https://doi.org/10.25548/16-E1GE