Abstract
Gout is caused by deposition of monosodium urate crystals in joints when plasma uric acid levels are chronically elevated beyond the saturation threshold, mostly due to renal underexcretion of uric acid. Although molecular pathways of this underexcretion have been elucidated, its etiology remains mostly unknown. We demonstrate that gout can be caused by a mutation in LDHD within the putative catalytic site of the encoded d-lactate dehydrogenase, resulting in augmented blood levels of d-lactate, a stereoisomer of l-lactate, which is normally present in human blood in miniscule amounts. Consequent excessive renal secretion of d-lactate in exchange for uric acid reabsorption culminated in hyperuricemia and gout. We showed that LDHD expression is enriched in tissues with a high metabolic rate and abundant mitochondria and that d-lactate dehydrogenase resides in the mitochondria of cells overexpressing the human LDHD gene. Notably, the p.R370W mutation had no effect on protein localization. In line with the human phenotype, injection of d-lactate into naive mice resulted in hyperuricemia. Thus, hyperuricemia and gout can result from the accumulation of metabolites whose renal excretion is coupled to uric acid reabsorption.
Keywords: Genetics, Metabolism
Keywords: Arthritis, Diabetes, Genetic diseases
Introduction
Gout is a common rheumatic disease that affects approximately 4% of adults in the United States (1). This inflammatory arthritis most notably affects joints, but also cartilage, bone, and tendons (2, 3). Gout initially presents as inflammatory episodes with increasing frequency, eventually leading to chronic arthropathy (4), and is often associated with renal and cardiovascular comorbidities (5). Gout occurs as a result of deposition of monosodium urate crystals in joints, formed when plasma uric acid levels are chronically elevated (hyperuricemia) beyond the saturation threshold (6). The majority of hyperuricemia cases result from renal underexcretion of uric acid due to various underlying causes, including decreased renal function, side effects of diuretics or other drugs, and impaired function of one of several renal uric acid transporters facilitating its excretion (7–9). Uric acid is the end-product of purine nucleotide metabolism in humans. When it reaches the bloodstream it is normally excreted from the body primarily via the kidneys, where it undergoes bidirectional transport, ultimately resulting in reabsorption of approximately 90% of the filtered load (10). Although the etiology of underexcretion of uric acid in many cases of hyperuricemia remains unclear, mutations in several genes encoding renal transporters, pivotal in renal uric acid handling, were previously described as a prominent underlying cause (11–14). We now demonstrate that autosomal recessive gout with hyperuricemia and underexcretion of uric acid can be caused by a mutation in lactate dehydrogenase D (LDHD), resulting in excess blood d-lactate that is excreted in exchange for reabsorbed uric acid.
Results and Discussion
Clinical characterization.
Consanguineous Bedouin-Israeli kindred presented with autosomal recessive hyperuricemia (Figure 1A). All affected adults were clinically diagnosed with gout arthropathy and described classic symptoms of the disease, including both upper- and lower-limb joint pain, particularly in small joints of the palms and toes, with acute gout flares every 3 to 6 months (more often in some) that were aggravated by meat consumption. All patients were treated with colchicine and allopurinol, had normal blood creatinine levels with no other evidence of chronic kidney disease or cardiovascular sequelae, and were otherwise healthy.
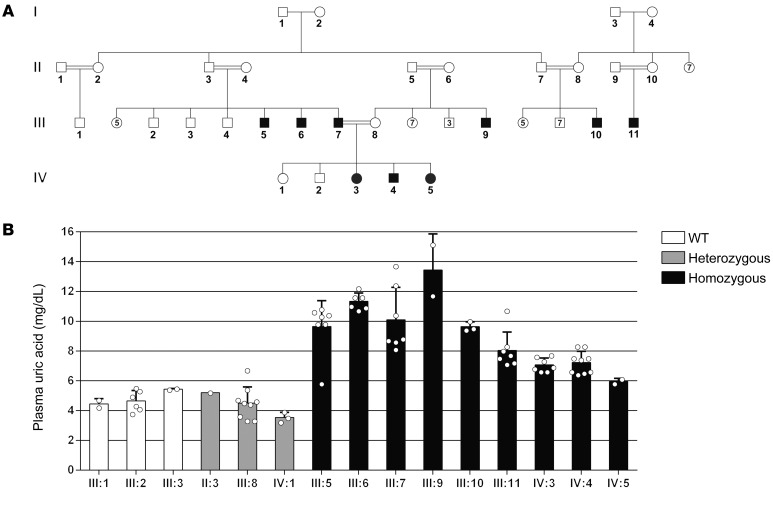
Figure 1. Pedigree and plasma uric acid measurements of a kindred affected with gout.
(A) Diagram of the studied kindred. Black squares and circles denote affected kindred, white squares and circles denote unaffected kindred, and double black lines indicate consanguinity. (B) Average plasma uric acid concentrations in individuals of the studied kindred who were WT, heterozygous, or homozygous for the LDHD mutation. Error bars indicate the SD. P < 0.05 versus homozygous, by Kruskal-Wallis with Dunn’s post hoc test.
Elevated uric acid levels were evident in both adults (III:5, III:6, III:7, III:9, III:10, III:11) and children (IV:3, IV:4, IV:5). Average plasma uric acid levels were 10.34 ± 1.84 mg/dL and 6.75 ± 0.7 mg/dL in the affected adults and children (Figure 1B), whereas the corresponding normal levels are 3.5–7.2 and 2–6.2 mg/dL, respectively. To assess renal handling of uric acid, we performed urine analyses on several of the affected individuals. The average urinary uric acid levels measured in 4 affected individuals (III:5, III:7, IV:3, IV:5) were 24.32 ± 15.3 mg/dL (normal levels, 37–92 mg/dL). The average uric acid levels in a 24-hour urine collection in 2 affected individuals (III:5, III:7) were 125 and 188 ± 68 mg/dL, respectively (normal range, 250–750 mg/dL), whereas creatinine excretion was normal, revealing a substantial reduction in renal clearance of uric acid. In addition, the fractional excretion of uric acid (calculated as the ratio of uric acid clearance to creatinine clearance) in all 5 patients whose samples were assayed (III:5, III:7, III:9, IV:3, IV:5) was 1.4% ± 0.9% (normal value, 7.5%) (8), strongly suggesting that underexcretion of uric acid is the underlying cause of hyperuricemia in this family.
Molecular genetic studies.
We performed linkage analysis and homozygosity mapping, testing all 9 affected family members and 6 unaffected controls (II:3, II:4, III:3, III:8, IV:1, IV:2). We identified a single homozygous locus of approximately 12 Mbp on chromosome 16 (between rs150635 and rs2550904) that was shared between all affected family members and none of the healthy ones (maximal logarithm of the odds [LOD] score of 4.8 at rs7193833). We performed whole-exome sequencing on 2 affected individuals who presented with the most severe hyperuricemia (III:7 and III:9, Figure 1A). Following the variant filtration cascade (see Methods), only a single homozygous variant within the genomic locus identified through linkage analysis was found to be shared by both individuals: c.1108C>T in LDHD, encoding d-lactate dehydrogenase. We performed whole-genome sequencing on one of the affected individuals (III:7) to exclude any insertions or deletions in noncoding regions of the homozygous locus identified in the linkage analysis.
Sanger sequencing validated the segregation of the mutation in the family, as expected for autosomal recessive heredity (Figure 2A). The mutation has not been identified in any of the approximately 240,000 genomes in the Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org), or the approximately 117,000 exomes in the Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org), or our in-house exome database of 180 ethnically matched controls. Mutation screening using restriction analysis, which we performed on 92 unaffected members of the particularly highly inbred tribe of the studied kindred, revealed 7 heterozygous carriers of the mutation and no homozygotes (not shown).
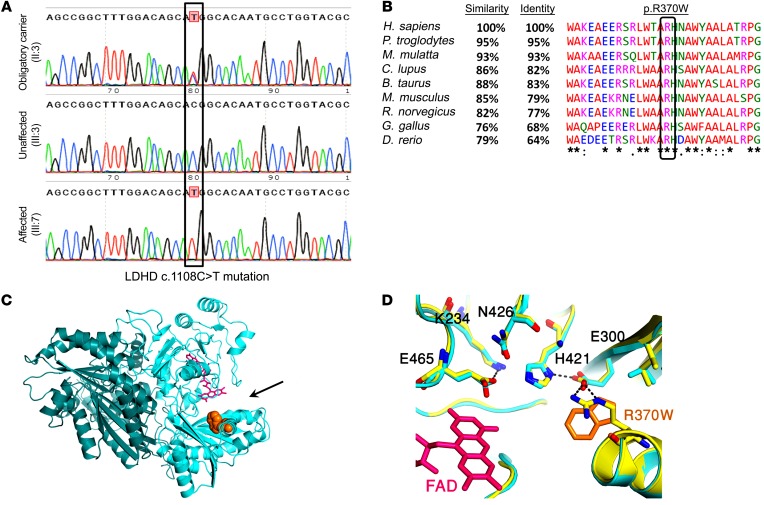
Figure 2. LDHD mutation verification, conservation, and structural prediction.
(A) Sanger sequencing of a heterozygous obligatory carrier (II:3), a healthy family member (III:3), and an affected individual homozygous for the mutation (III:7). The c.1108C>T mutation is outlined by a black rectangle. (B) Alignment of LDHD ortholog sequences using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Identity and similarity were derived from the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The black box marks the highly conserved arginine residue altered by the c.1108C>T mutation. The symbols show conservation: asterisk indicates identical; colon indicates strongly similar; period indicates weakly similar; blank indicates no similarity. (C) Predicted structure of the human d-lactate dehydrogenase dimer, based on its closest structural homolog (putative dehydrogenase from Rhodopseudomonas palustris, Protein Data Bank [PDB] 3PM9 [https://www.rcsb.org/structure/3PM9], 30% protein sequence identity). The 2 monomers of d-lactate dehydrogenase are shown in light blue and dark green; FAD is represented as sticks in pink; the site of the p.R370W substitution is shown in orange; and the black arrow indicates the enzyme’s active site. (D) Zoom-in on the residues comprising the catalytic pocket of 3PM9 (yellow) and an overlay of the model for d-lactate dehydrogenase with its homologous residues (light blue). FAD is presented as sticks in pink; the site of the R370W substitution is shown in orange; the dashed lines indicate hydrogen bonds; nitrogen is labeled in blue; and oxygen is labeled in red.
Lactic acid exists as 2 enantiomers/stereoisomers — d- and l-lactate — based on the orientation around its asymmetric second carbon atom (15). Normally, lactic acid is found in humans entirely as l-lactate, as mammalian cells almost exclusively produce this form (16). However, d-lactate can be generated in miniscule concentrations via the glyoxalase pathway, which facilitates degradation of methylglyoxal, a byproduct of several fundamental metabolic processes including glycolysis, amino acid degradation, and ketone body catabolism. In this pathway, methylglyoxal is converted to d-lactate following a series of enzymatic reactions. The final step of the pathway is the d-lactate-dehydrogenase–dependent conversion of d-lactate to pyruvate (17), which may then serve as a substrate in a variety of downstream metabolic pathways (18, 19).
The LDHD p.R370W missense mutation putatively alters a highly conserved arginine residue to tryptophan (Figure 2B). Prediction of the d-lactate dehydrogenase protein structure, based on its closest homolog (PDB 3PM9), suggested that the altered arginine residue at position 370 resides within the enzyme’s catalytic pocket, approximately 7 Å from its known cofactor, FAD (Figure 2C and ref. 20). As the residues composing the catalytic pocket in the LDHD model are identical to those of the 3PM9 protein, the p.R370W substitution is expected to interfere with the hydrogen bonds in the catalytic pocket and the overall charge interaction networks between the active site’s residues, likely altering the structure of the pocket (Figure 2D), which is essential for its catalytic function. This hypothesis is strongly supported by a recent parallel independent study that proved that 2 separate loss-of-function mutations in the human LDHD gene, p.W374C and p.T463M, caused elevated urinary excretion and plasma concentrations of d-lactate (21). Both mutations alter amino acid residues located in proximity to the residues comprising the putative catalytic pocket of the protein and to the arginine residue altered by the novel p.R370W mutation.
Protein localization analyses.
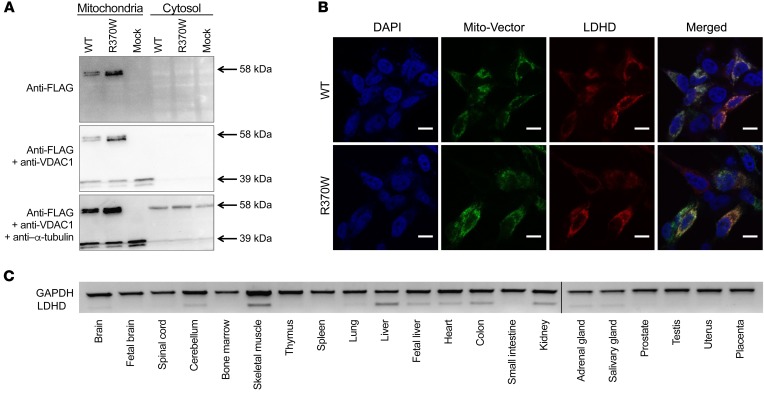
It was previously shown that d-lactate dehydrogenase localizes to mitochondria in mammalian cells (18). To verify that in humans the protein is indeed mitochondrial and to determine whether the mutation affected the subcellular localization of the protein, we transfected HEK293 cells with WT and p.R370W-mutant LDHD constructs containing a C-terminal FLAG epitope. We then isolated the mitochondria and cytosolic fractions of the transfected cells and subsequently performed Western blot analysis. Both the WT and p.R370W-mutant proteins containing the FLAG epitope were detected in the same fraction as VDAC1, which was used as a specific mitochondrial marker, and were not colocalized with the cytosolic marker α-tubulin (Figure 3A). We then confirmed these results using separate constructs in which WT and p.R370W-mutant LDHD were conjugated to red fluorescent protein (tdTomato) and cotransfected with a mitochondrial GFP (Mito-Vector) into SH-SY5Y cells. Confocal microscopic images of the transfected cells showed colocalization of both the WT and p.R370W-mutant constructs with the Mito-Vector protein (Figure 3B). Furthermore, reverse transcription PCR (RT-PCR) demonstrated that, in line with GTEx expression data (https://www.gtexportal.org/home), human LDHD transcripts are abundant in tissues with a high metabolic rate including skeletal muscle, liver, kidney, heart, and colon (Figure 3C).
Figure 3. d-lactate dehydrogenase is localized in mitochondria and expressed in tissues with high metabolic activity.
(A) Western blot analysis of the cytosolic fraction and isolated mitochondria from HEK293T cells transfected with WT or R370W-mutant LDHD constructs containing a FLAG epitope. VDAC1 and α-tubulin were used as mitochondrial and cytosolic markers, respectively. (B) Overexpression of WT or R370W-mutant LDHD conjugated to tdTomato (red), with a mitochondrial GFP (Mito-Vector, green), showing colocalization in SH-SY5Y cells. DNA is stained with DAPI (blue). Scale bars: 10 μm. (C) RT-PCR analysis of 21 healthy human tissues demonstrating the expression pattern of LDHD as compared with the GAPDH control. Lanes were run on the same gel but were noncontiguous (black line).
Mass spectrometry.
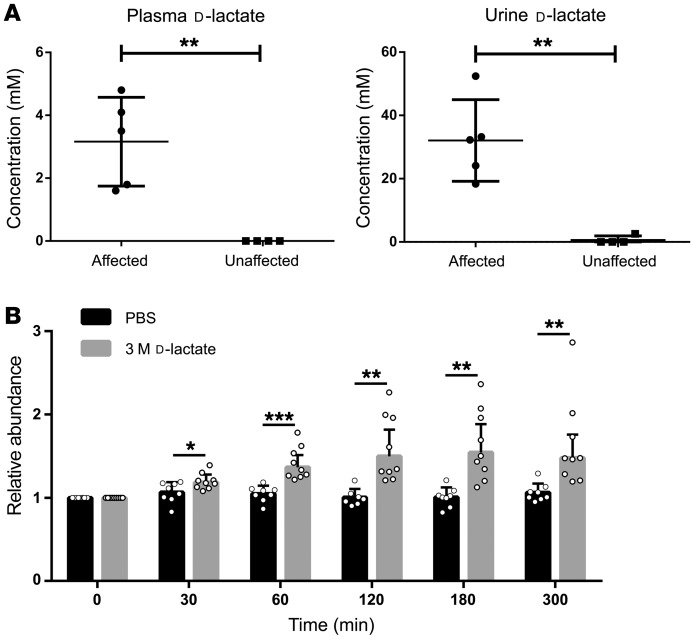
d-lactate dehydrogenase is highly expressed in tissues with increased metabolic activity, and methylglyoxal, which is processed and converted to d-lactate, is a ubiquitous substance that is also abundant in these high-metabolic-rate tissues; thus, d-lactate is constantly produced in humans (22). To our knowledge, there are currently no data describing a particular function of d-lactate in humans. The only known fate of this compound is conversion to pyruvate by d-lactate dehydrogenase or excretion (23, 24). To determine whether this metabolic activity is impaired by the p.R370W mutation as predicted by our structural model, we measured lactate concentrations in plasma and urine samples obtained from 5 affected (III:5, III:7, III:9, IV:3, IV:5) and 4 healthy (II:4, III:1, III:3, III:8) family members. We used a chiral column to differentiate between d-lactate and l-lactate, and a calibration curve was created to determine the absolute concentration of plasma and urine d-lactate and l-lactate, with a minimal detectable concentration of approximately 5 μM. The average levels of l-lactate in plasma were 1.33 ± 0.12 mM in affected individuals and 1.43 ± 0.64 mM in healthy controls (corresponding normal levels are 0.5–2.2 mM). In urine samples, l-lactate was undetectable in both groups. We found that d-lactate levels in all affected individuals were markedly elevated in both plasma and urine. The average concentration of plasma d-lactate in affected individuals was 3.16 ± 0.63 mM and was undetectable in the healthy controls, whereas the corresponding normal levels are less than 0.043 mM (Figure 4A). In the urine samples, the average levels of d-lactate were 32.08 ± 5.77 mM in the affected individuals versus 0.65 ± 0.65 mM in the healthy controls, whereas the corresponding normal levels are 0.0–0.25 mM (Figure 4A). Thus, although we observed no evident differences in l-lactate levels between affected and healthy individuals, we detected markedly elevated levels of both plasma and urine levels of d-lactate in all affected individuals.
Figure 4. d-lactate measurements and in-vivo experiments.
(A) Plasma and urine d-lactate concentrations in affected versus healthy family members. Mass spectrometric data show the average d-lactate concentrations in plasma (left) and urine (right) samples of affected (n = 5) and unaffected (n = 4) individuals. Error bars indicate the SD. **P < 0.01, by Student’s t test. (B) Injection of d-lactate caused an increase in plasma uric acid levels in mice. Relative plasma uric acid levels measured in naive mice before and at several time points after i.p. injection of 200 μL 3 M d-lactate (n = 8) or PBS (n = 8). For each mouse, uric acid levels at each time point were normalized to the level measured before injection (t = 0). Error bars indicate the SD. *P < 0.05, **P < 0.005, and ***P < 0.0005, by Student’s t test.
Excess d-lactate causes hyperuricemia in mice.
To test whether hyperuricemia may be caused by elevated plasma d-lactate levels, as seen in our patients, we measured plasma uric acid levels in mice following injections of d-lactate. Naive C57BL/6 mice were injected with 200 μL PBS or 3 M d-lactate, and blood samples were taken before and at several time points after injection to monitor plasma uric acid levels. In mice injected with PBS, we observed only slight fluctuations in plasma uric acid levels over time; however, we detected a steady increase in plasma uric acid levels in mice injected with d-lactate, peaking approximately 180 minutes after injection, with an average increase in plasma uric acid levels of 54% ± 0.34% (Figure 4B).
Although urate handling in mice in particular and in mammals in general is different from that in humans, these results suggest that elevation in plasma d-lactate can cause elevations in plasma uric acid. A plausible underlying mechanism for this phenomenon could be alterations in renal handling of uric acid as a result of excess in d-lactate. This is in line with lactate being a known anti-uricosuric agent that decreases urinary clearance of uric acid by serving as a substrate for one of several renal organic anion transporters (OATs), including OAT10 and URAT1. These transporters are expressed at the apical membrane of proximal tubule cells or cortical collecting ducts, and their primary function is the inward transport of urate in exchange for the outward shift of monovalent organic anions such as lactate (25, 26); thus, excessive renal secretion of d-lactate in exchange for uric acid reabsorption could culminate in hyperuricemia. Furthermore, 2 additional renal urate transporters, OAT1 and OAT3, were recently shown to affect lactate concentrations in vitro and may therefore play a role in hyperuricemia in the presence of elevated lactate (27).
d-lactate is produced by intestinal microbes, as evidenced by d-lactic acidosis caused in the context of short bowel syndrome (28). Our data suggest the possibility that elevated levels of d-lactate could play a role in mediating the effects of nutrition and the microbiome in the pathogenesis of common forms of hyperuricemia and gout. A clear association between gout and diabetes as well as the comorbidity of these diseases have been reported in various cohorts (29, 30). The pathophysiology of this association is unknown. Although it is possible that hyperuricemia might be causative of diabetes rather than the other way around (31), the causality remains uncertain, because confounders, reverse causality, or common etiological factors might explain the epidemiological results (29). In fact, recent human and mouse data imply that diabetes increases the risk of hyperuricemia (29, 32). Notably, significantly (on average >10-fold) elevated levels of d-lactate have been detected in individuals with diabetes (33). Thus, our data speculatively suggest that the association between diabetes and gout might be mediated at least in part by d-lactate. As lactate is only one substrate of the many renal urate transporters (34–36), it is possible that hyperuricemia may result from the accumulation of additional metabolites whose renal clearance is coupled to uric acid reabsorption. This suggests that metabolome analyses in gout patients with unexplained hyperuricemia may reveal additional hyperuricemia-promoting metabolites, providing a basis for novel, metabolite-targeted, personalized treatment strategies based on the culprit metabolites in patients with gout.
Methods
Complete details on the experimental materials and methods are provided in the supplemental materials; supplemental material available online with this article; https://doi.org/10.1172/JCI129057DS1
Study approval.
This study was approved by the IRB of the Soroka Medical Center (approval no. 5071G) and the Israel Ministry of Health National Helsinki Committee (approval no. 920100319). DNA was extracted from peripheral blood or saliva following written informed consent by all individuals studied or their legal guardians. Animal experiments were approved by the Ben-Gurion University (BGU) Committee for the Ethical Care and Use of Laboratory Animals. The experiments, performed at the BGU rodent facility, were conducted according to the Israel Animal Welfare Law of 1994 and the National Research Council’s 2011 Guidelines for the Care and Use of Laboratory Animals. The animal care and use program at BGU was approved by the Association for the Assessment and Accreditation of Laboratory Animal Care International.
Statistics.
Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software). An unpaired, 2-tailed Student’s t test was used for comparisons between 2 groups, and a Kruskal-Wallis with Dunn’s post hoc test was used for multiple-group comparisons. P values of less than 0.05 were considered statistically significant. All data are presented as the mean ± SD.
Author contributions
MD and OSB initiated the study, contributed to its conception and design, and drafted the manuscript. MD, YY, R. Zarivach, R. Zalk, DH, OW, RK, YP, and OSB acquired and analyzed data. DL analyzed data. LZ, EG, and OSB provided the clinical data.
Supplementary Material
Acknowledgments
The studies were funded by the Morris Kahn Foundation and supported through the National Knowledge Center for Rare/Orphan Diseases sponsored by the Israeli Ministry of Science, Technology and Space. We thank Ifat Abramovich (Eyal Gottlieb Laboratory, Faculty of Medicine, Technion Institute of Technology, Haifa, Israel) for assistance with the mass spectrometry studies.
Version 1. 10/22/2019
Electronic publication
Version 2. 12/02/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(12):5163–5168.https://doi.org/10.1172/JCI129057.
Contributor Information
Max Drabkin, Email: maxdr@post.bgu.ac.il.
Yuval Yogev, Email: yogevyu@post.bgu.ac.il.
Lior Zeller, Email: zellerl@bgu.ac.il.
Raz Zarivach, Email: zarivach@bgu.ac.il.
Ran Zalk, Email: ranzalk@gmail.com.
Daniel Halperin, Email: halperda@post.bgu.ac.il.
Ohad Wormser, Email: wormser@post.bgu.ac.il.
Evgenia Gurevich, Email: Jenygu@clalit.org.il.
Daniel Landau, Email: danny_L@clalit.org.il.
Rotem Kadir, Email: rotemkadir222@gmail.com.
Yonatan Perez, Email: perezyon@post.bgu.ac.il.
References
- 1.Zhu Y, Pandya BJ, Choi HK. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007-2008. Arthritis Rheum. 2011;63(10):3136–3141. doi: 10.1002/art.30520. [DOI] [PubMed] [Google Scholar]
- 2.Dalbeth N, Kalluru R, Aati O, Horne A, Doyle AJ, McQueen FM. Tendon involvement in the feet of patients with gout: a dual-energy CT study. Ann Rheum Dis. 2013;72(9):1545–1548. doi: 10.1136/annrheumdis-2012-202786. [DOI] [PubMed] [Google Scholar]
- 3.Naredo E, et al. Ultrasound-detected musculoskeletal urate crystal deposition: which joints and what findings should be assessed for diagnosing gout? Ann Rheum Dis. 2014;73(8):1522–1528. doi: 10.1136/annrheumdis-2013-203487. [DOI] [PubMed] [Google Scholar]
- 4.Bardin T, Richette P. Definition of hyperuricemia and gouty conditions. Curr Opin Rheumatol. 2014;26(2):186–191. doi: 10.1097/BOR.0000000000000028. [DOI] [PubMed] [Google Scholar]
- 5.Borghi C, et al. Serum uric acid and the risk of cardiovascular and renal disease. J Hypertens. 2015;33(9):1729–1741; discussion 1741. doi: 10.1097/HJH.0000000000000701. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Ruiz F, Dalbeth N, Bardin T. A review of uric acid, crystal deposition disease, and gout. Adv Ther. 2015;32(1):31–41. doi: 10.1007/s12325-014-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzai N, Enomoto A, Endou H. Renal urate handling: clinical relevance of recent advances. Curr Rheumatol Rep. 2005;7(3):227–234. doi: 10.1007/s11926-996-0044-0. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Ruiz F, Calabozo M, Erauskin GG, Ruibal A, Herrero-Beites AM. Renal underexcretion of uric acid is present in patients with apparent high urinary uric acid output. Arthritis Rheum. 2002;47(6):610–613. doi: 10.1002/art.10792. [DOI] [PubMed] [Google Scholar]
- 9. Becker MA. Clinical gout and the pathogenesis of hyperuricemia. In: Koopman WJ, Moreland LW, eds. Arthritis and Allied Conditions: A Textbook of Rheumatology. 15th ed. Philadelphia, Pennsylvania USA: Lippincott Williams & Wilkins; 2005:2303–2339. [Google Scholar]
- 10.Mandal AK, Mount DB. The molecular physiology of uric acid homeostasis. Annu Rev Physiol. 2015;77:323–345. doi: 10.1146/annurev-physiol-021113-170343. [DOI] [PubMed] [Google Scholar]
- 11.Woodward OM, Köttgen A, Coresh J, Boerwinkle E, Guggino WB, Köttgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci USA. 2009;106(25):10338–10342. doi: 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitart V, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40(4):437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 13.Graessler J, et al. Association of the human urate transporter 1 with reduced renal uric acid excretion and hyperuricemia in a German Caucasian population. Arthritis Rheum. 2006;54(1):292–300. doi: 10.1002/art.21499. [DOI] [PubMed] [Google Scholar]
- 14.Jutabha P, et al. Human sodium phosphate transporter 4 (hNPT4/SLC17A3) as a common renal secretory pathway for drugs and urate. J Biol Chem. 2010;285(45):35123–35132. doi: 10.1074/jbc.M110.121301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewaschuk JB, Naylor JM, Zello GA. D-lactate in human and ruminant metabolism. J Nutr. 2005;135(7):1619–1625. doi: 10.1093/jn/135.7.1619. [DOI] [PubMed] [Google Scholar]
- 16.Adeva-Andany M, et al. Comprehensive review on lactate metabolism in human health. Mitochondrion. 2014;17:76–100. doi: 10.1016/j.mito.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Byers FM. Automated enzymic determination of L(+)- and D(−)-lactic acid. Methods Enzymol. 1982;89(C):29–34. [Google Scholar]
- 18.Flick MJ, Konieczny SF. Identification of putative mammalian D-lactate dehydrogenase enzymes. Biochem Biophys Res Commun. 2002;295(4):910–916. doi: 10.1016/S0006-291X(02)00768-4. [DOI] [PubMed] [Google Scholar]
- 19.Thornalley PJ. The glyoxalase system in health and disease. Mol Aspects Med. 1993;14(4):287–371. doi: 10.1016/0098-2997(93)90002-U. [DOI] [PubMed] [Google Scholar]
- 20.Dym O, Pratt EA, Ho C, Eisenberg D. The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme. Proc Natl Acad Sci USA. 2000;97(17):9413–9418. doi: 10.1073/pnas.97.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monroe GR, et al. Identification of human D lactate dehydrogenase deficiency. Nat Commun. 2019;10(1):1477. doi: 10.1038/s41467-019-09458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornalley PJ. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269(1):1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh MS, Uribarri J, Alveranga D, Lazar I, Bazilinski N, Carroll HJ. Metabolic utilization and renal handling of D-lactate in men. Metab Clin Exp. 1985;34(7):621–625. doi: 10.1016/0026-0495(85)90088-5. [DOI] [PubMed] [Google Scholar]
- 24.Connor H, Woods HF, Ledingham JG. Comparison of the kinetics and utilisation of D(-)-and L(+)-sodium lactate in normal man. Ann Nutr Metab. 1983;27(6):481–487. doi: 10.1159/000176723. [DOI] [PubMed] [Google Scholar]
- 25.Bahn A, et al. Identification of a new urate and high affinity nicotinate transporter, hOAT10 (SLC22A13) J Biol Chem. 2008;283(24):16332–16341. doi: 10.1074/jbc.M800737200. [DOI] [PubMed] [Google Scholar]
- 26.Enomoto A, et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 2002;417(6887):447–452. doi: 10.1038/nature742. [DOI] [PubMed] [Google Scholar]
- 27.Vriend J, et al. Organic anion transporters 1 and 3 influence cellular energy metabolism in renal proximal tubule cells. Biol Chem. 2019;400(10):1347–1358. doi: 10.1515/hsz-2018-0446. [DOI] [PubMed] [Google Scholar]
- 28.Oh MS, Phelps KR, Traube M, Barbosa-Saldivar JL, Boxhill C, Carroll HJ. D-lactic acidosis in a man with the short-bowel syndrome. N Engl J Med. 1979;301(5):249–252. doi: 10.1056/NEJM197908023010505. [DOI] [PubMed] [Google Scholar]
- 29.Bardin T, Richette P. Impact of comorbidities on gout and hyperuricaemia: an update on prevalence and treatment options. BMC Med. 2017;15(1):123. doi: 10.1186/s12916-017-0890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh JA, et al. Is gout a risk equivalent to diabetes for stroke and myocardial infarction? A retrospective claims database study. Arthritis Res Ther. 2017;19(1):228. doi: 10.1186/s13075-017-1427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortada I. Hyperuricemia, type 2 diabetes mellitus, and hypertension: an emerging association. Curr Hypertens Rep. 2017;19(9):69. doi: 10.1007/s11906-017-0770-x. [DOI] [PubMed] [Google Scholar]
- 32.Adachi SI, Yoshizawa F, Yagasaki K. Hyperuricemia in type 2 diabetic model KK-Ay/Ta mice: a potent animal model with positive correlation between insulin resistance and plasma high uric acid levels. BMC Res Notes. 2017;10(1):577. doi: 10.1186/s13104-017-2897-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou CK, et al. Elevated urinary D-lactate levels in patients with diabetes and microalbuminuria. J Pharm Biomed Anal. 2015;116:65–70. doi: 10.1016/j.jpba.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 34.Enomoto A, Endou H. Roles of organic anion transporters (OATs) and a urate transporter (URAT1) in the pathophysiology of human disease. Clin Exp Nephrol. 2005;9(3):195–205. doi: 10.1007/s10157-005-0368-5. [DOI] [PubMed] [Google Scholar]
- 35.Eraly SA, et al. Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol Genomics. 2008;33(2):180–192. doi: 10.1152/physiolgenomics.00207.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Shi Y, Zhuang S, Liu N. Recent advances on uric acid transporters. Oncotarget. 2017;8(59):100852–100862. doi: 10.18632/oncotarget.20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.