Abstract
Cystic fibrosis (CF) is caused by mutations in the CF transmembrane conductance regulator (CFTR), with approximately 90% of patients harboring at least one copy of the disease-associated variant F508del. We utilized a yeast phenomic system to identify genetic modifiers of F508del-CFTR biogenesis, from which ribosomal protein L12 (RPL12/uL11) emerged as a molecular target. In the present study, we investigated mechanism(s) by which suppression of RPL12 rescues F508del protein synthesis and activity. Using ribosome profiling, we found that rates of translation initiation and elongation were markedly slowed by RPL12 silencing. However, proteolytic stability and patch-clamp assays revealed RPL12 depletion significantly increased F508del-CFTR steady-state expression, interdomain assembly, and baseline open-channel probability. We next evaluated whether Rpl12-corrected F508del-CFTR could be further enhanced with concomitant pharmacologic repair (e.g., using clinically approved modulators lumacaftor and tezacaftor) and demonstrated additivity of these treatments. Rpl12 knockdown also partially restored maturation of specific CFTR variants in addition to F508del, and WT Cftr biogenesis was enhanced in the pancreas, colon, and ileum of Rpl12 haplosufficient mice. Modulation of ribosome velocity therefore represents a robust method for understanding both CF pathogenesis and therapeutic response.
Keywords: Cell Biology, Genetics
Keywords: Genetic diseases, Protein misfolding, Translation
Introduction
Cystic fibrosis (CF) is caused by abnormalities of the CF transmembrane conductance regulator (CFTR), with the most common variant being F508del (deletion of phenylalanine at position 508). The F508del defect alters protein biogenesis with subsequent ER retention and proteasomal degradation, leading to profound multiorgan disease (1, 2). Understanding is incomplete with regard to mechanism or mechanisms that underlie abnormal CFTR processing, including factors responsible for variation in overall disease severity and response to repair by small molecules.
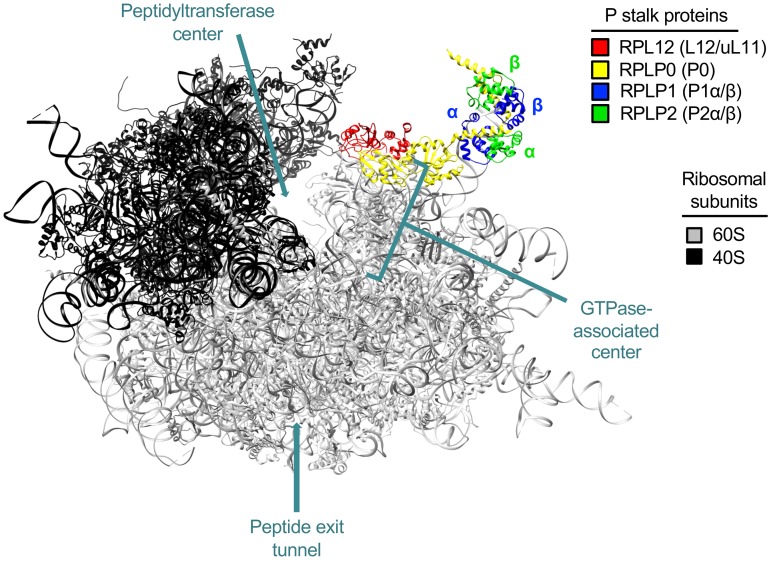
Our previous phenomic screen using the Saccharomyces cerevisiae deletion strain library identified a number of unexpected genes that directly influence F508del-CFTR maturation (3). These include human ribosomal proteins such as ribosomal protein L12 (RPL12/uL11) and RPLP2 (P2), located in the 60S subunit P stalk (Figure 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI124282DS1) as well as several proteins surrounding the GTPase-associated center (GAC) and the peptide exit tunnel. The P stalk interacts with eukaryotic elongation factors (eEFs) during translation and, in particular, RPL12 with eEF2 (4–6). An important unresolved question relevant to F508del and other class II (i.e., misprocessing) CFTR variants is whether kinetics of translation — specifically elongation — directly mediate biogenesis or molecular phenotype. CFTR folding occurs cotranslationally beginning with nucleotide-binding domain 1 (NBD1) (7), and overall assembly is precisely coordinated during synthesis (8). Interestingly, synonymous SNPs (sSNPs) have been shown to alter the speed of translation, induce changes in CFTR mRNA secondary structure, and confer functionally significant effects on CFTR folding, protein yield, and activity (9). Moreover, sSNPs that revert translational velocity by altering transfer ribonucleic acid (tRNA) availability or affect mRNA folding at the mutated codon regulate CFTR stability and channel function (9, 10).
Figure 1. RPL12 (uL11) is an integral component of the 60S subunit P stalk.
RPL12 (red) resides in the GAC of the human 80S ribosome (PDB file 4V6X) (4) and, together with RPLP0 (yellow), forms the base of the P stalk to serve as a platform for further assembly by α and β subunits of RPLP1 (blue) and RPLP2 (green), respectively.
The critical relationship between polypeptide folding and translation rate is an emerging and topical area with ramifications affecting not only CF disease mechanism, but also clinical phenotype and intervention strategy. The objective of the present study, therefore, was to examine the underlying mechanism or mechanisms of RPL12-dependent rescue of the F508del-CFTR biogenesis defect. RPL12 was selected among several ribosomal proteins for this analysis because it was previously identified as exerting the most robust impact on F508del processing (3). Ribosome profiling, together with RNA-Seq and tRNA microarrays, were adopted for direct measurement of CFTR translational dynamics and demonstrated marked slowing of initiation and elongation when RPL12 was repressed by siRNA. Short-circuit current (ISC) and single-channel patch-clamp established that RPL12 depletion strongly rescues F508del gating activity — a marker of enhanced protein folding — which was further verified by limited proteolysis showing stabilization of transmembrane domain-2 (TMD2) in both WT- and F508del-CFTR.
The RPL12 mechanism appears distinct from CFTR corrector molecules such as lumacaftor (VX-809) or tezacaftor (VX-661) as well as intragenic F508del suppressors. Most importantly, gastrointestinal and other tissues collected from Rpl12+/– mice exhibited enhanced processing of WT-Cftr. Taken together, our results point to translation rate as a crucial modulator of CFTR biogenesis and trafficking. For CFTR processing variants (i.e., class II), including those unresponsive to approved modulators, RPL12 may also represent a useful therapeutic target for pharmacologic correction. To our knowledge, no other human disease mechanism has been previously shown to be alleviated by slowing ribosome velocity.
Results
Unanticipated participants identified during CFTR biogenesis.
The yeast oligomycin efflux pump Yor1 is a homologue of CFTR with a conserved phenylalanine (F670) corresponding to F508 in CFTR (Supplemental Figure 1A). Deletion of F670 in Yor1 leads to protein misfolding, defective maturation, premature degradation, and other features similar to those found in F508del-CFTR (11). Evaluating the Yor1-F670del phenotype on a background of the entire yeast deletion/knockout strain library using oligomycin-sensitive growth suppression confirmed a number of cellular proteins whose human homologues participate during F508del-CFTR biogenesis (Supplemental Figure 1, B and C) together with several contributors we believe to be novel. The present study focuses on mechanisms of aberrant protein rescue by suppression of RPL12 (uL11), a component of the 60S subunit P stalk (Figure 1), which exhibits unexpected and pronounced beneficial effects on F508del maturation (i.e., at levels comparable to those of the best available small molecule correctors).
RPL12 knockdown slows translation elongation in a context-dependent manner.
Previous studies have shown that RPL12 depletion slows translation in yeast (12) and decreases total protein synthesis in HeLa cells, as monitored by 35S-methionine labeling (3). We therefore tested to determine whether suppression of RPL12 would influence translation kinetics in human CF bronchial epithelia (CFBE41o–) stably transduced to express WT- or F508del-CFTR. We found that RPL12 silencing decreased total 35S-methionine and 35S-cysteine incorporation in CFBE cells (Supplemental Figure 2, A and B), but enhanced CFTR expression by approximately 3-fold (Supplemental Figure 2C). Translation dynamics were then formally evaluated by ribosome profiling, complemented by RNA-Seq and tRNA microarrays.
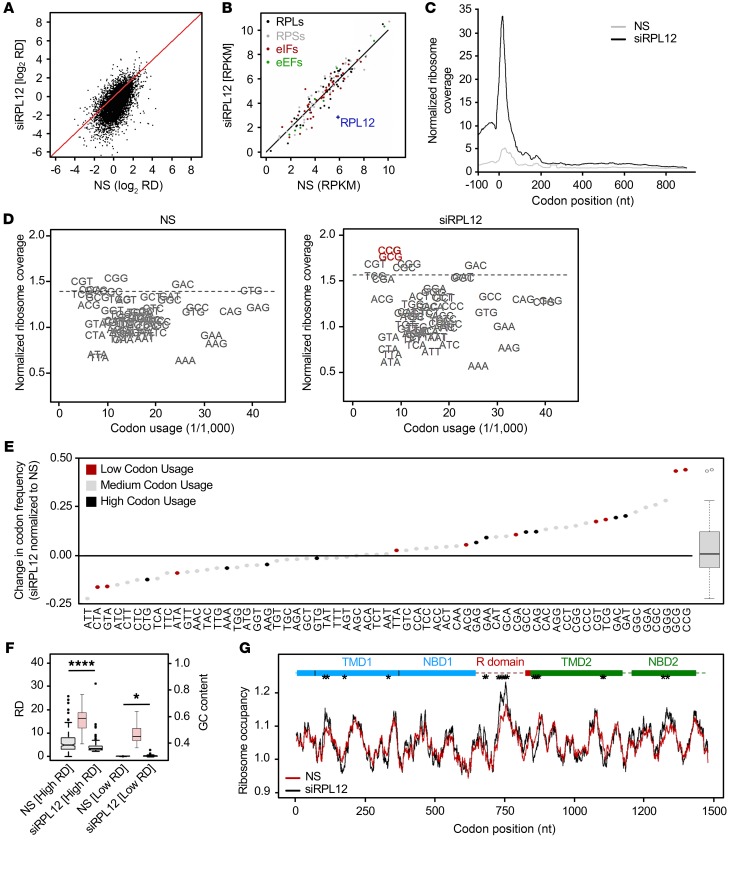
To characterize the effect of RPL12 repression on protein synthesis, we computed ribosome density (RD, known also as TE value; ref. 13) per transcript under RPL12-depleted conditions and compared those values to the RD observed in control cells — i.e., nontreated (NT) or treated with nonspecific (NS) siRNA. Assuming that each ribosome produces 1 nascent chain, the RD value is informative with regard to both protein production and translation efficiency. In samples treated with RPL12-directed siRNA (siRPL12), RPL12 mRNA levels were reduced by approximately 80% (Supplemental Figure 2D), and total cellular protein production was decreased by approximately 20%, as judged by 35S labeling (Supplemental Figure 2, A and B) and RD across the entire transcriptome (Figure 2A). Of note, as revealed by RNA-Seq, we did not observe gross alterations in cellular transcript abundance within NS siRNA–treated cells (Supplemental Figure 2E), indicating that NS siRNA did not lead to changes in gene expression on a transcriptome-wide basis. Strikingly, while translation efficiency (RD) was globally reduced following RPL12 depletion (Figure 2A and Supplemental Figure 2, A and B), mRNA expression levels of translation apparatus components (e.g., ribosomal proteins and initiation and elongation factors) were insensitive to RPL12 knockdown (Figure 2B). This result suggested that RPL12 suppression did not influence ribosome biogenesis or stoichiometry of ribosomal proteins.
Figure 2. RPL12 suppression slows translation rate in CFBE.
(A) Comparison of RD values for CFBE F508del-CFTR cells treated with RPL12-directed siRNA (siRPL12) or NS siRNA from ribosome-profiling data. Diagonal denotes the position of equivalent RD values. (B) Expression of specific translational machinery proteins assessed by ribosome profiling. RPLs, ribosomal proteins (large subunit); RPSs, ribosomal proteins (small subunit); eIFs, eukaryotic initiation factors. (C) Cumulative metagene profiles of RPFs as a function of position for all protein-coding genes detected over the threshold from ribosome profiling. Each transcript was weighted equally. Zero designates the first nucleotide of the start codon. (D) Ribosome-dwelling occupancy for cells treated with siRPL12 or NS siRNA as determined from ribosome profiling and compared with genome codon usage. Dashed line denotes the upper boundary (90% confidence interval). (E) Difference in ribosome occupancy (y axis) between NS and siRPL12-treated cells quantified from D for single codons (represented as dots) and for all codons (box plot on right). Top 10 most (black) or least (red) utilized codons are annotated. (F) Box plot of 200 most (high RD; P = 3.0 × 10–7) and least (low RD; P = 0.045) expressed genes in NS and siRPL12-treated cells. GC content of high- and low-RD groups is represented in red (right y axis). *P < 0.05; ****P < 0.0001, Wilcoxon’s rank sum test. (G) Profiles of ribosome-dwelling occupancy computed from D for CFTR mRNA in NS and siRPL12-treated cells. Single codons exhibiting an occupancy variance greater than the 95th percentile (*) are considered statistically significant.
Because RPL12 silencing decreased RD (Figure 2A) without inducing significant transcriptome-wide changes in mRNA levels (Supplemental Figure 2F), we conclude that the RPL12 depletion effect is primarily exerted on translation. Thus, we compared the distribution of ribosome-protected fragments (RPFs) along the nucleotide sequence of transcripts in order to detect specific differences between NS and RPL12 siRNA treatments. Both initiation and elongation were reduced in response to RPL12 suppression, with initiation being more influenced, as indicated by the larger peak at the start codon (Figure 2C).
We next asked whether RPL12 suppression exhibits a uniform effect on all codons. From ribosome-profiling data, we calculated residence frequency of each codon in the ribosomal A-site as previously described (14). The likelihood that a particular codon occupies the A-site is inversely proportional to the translational velocity of that codon, i.e., higher ribosome dwell time, or ribosome occupancy, at a codon indicates slower speed of translation. We calibrated RPFs on the ribosomal A-site as described previously (9, 14) and found ribosome residence at specific codons differed by a factor of 2. On average, the lowest decoding rate was observed for guanine-cytosine–rich (GC-rich) codons (Figure 2D), which correlates with similar studies conducted in yeast (15). Following RPL12 knockdown, subsets of codons exhibited increases or decreases in ribosome occupancy compared with control cells treated with NS siRNA (Figure 2, D and E), suggesting reduced or accelerated elongation rates at these codons, respectively. In particular, RPL12 repression significantly diminished the rate of elongation for GC-rich codons (P = 7.64 × 10–5; Wilcoxon’s rank sum test). For approximately one-third of all codons, the ribosome dwell time did not change significantly (Figure 2, D and E). When comparing low- versus high-abundance codons, we did not observe a preferential decrease in elongation speed, meaning enhanced ribosome occupancy was equally distributed among both rarely and frequently utilized codons (P = 0.907 or P = 0.999, respectively, Kolmogorov-Smirnov test; Figure 2E). Off-target effects of NS siRNA also were not observed, as translational velocity for each codon was indistinguishable from the values obtained in NT cells (Supplemental Figure 2G).
Since RPL12 depletion influences translation in a codon-specific manner, we analyzed whether high- versus low-RD genes were altered by RPL12 silencing. In control samples, we extracted the first 200 genes with highest or lowest RD values and compared them to RD values in samples treated with RPL12 siRNA. Both transcript groups exhibiting elevated and low-level RD values were influenced by RPL12 repression, although the effect was larger for the former (i.e., those with high RD values; Figure 2F). Notably, the GC content of the transcripts with high RDs was significantly greater (Figure 2F), which mirrored the preferential slowing of elongation at GC-rich codons (Figure 2E). We also compared GC content of genes most or least affected by RPL12 silencing, i.e., displaying significant RD depletion or no change in RD, respectively (Supplemental Figure 2, H and I). Following RPL12 knockdown, transcripts with the greatest decrease in RD values exhibited higher GC content than those unaltered by RPL12 siRNA (P = 0.008, Kolmogorov-Smirnov test; Supplemental Figure 2I). Genes enriched in the intermediate group play roles in rRNA processing, mRNA stability, ubiquitin ligase activity, and many other critical pathways, indicating that essential cellular functions remain intact in response to RPL12 silencing. In addition, none of the genes identified within the high RD group represent components of the integrated stress-response pathway, arguing against cellular toxicity attributable to RPL12 repression.
Findings with regard to GC content and RD may be explained in part by codon:anticodon pairing. Enhancement of codon:anticodon interactions in the A-site result in decelerated translocation of peptidyl-tRNA to the P-site, which is facilitated by GTP hydrolysis (16). Since RPL12 and other P stalk proteins interact with GTP-bound translation factors, we therefore postulate that the slowing of GC-rich codons in response to RPL12 suppression could be due to a reduced rate of translocation. Interestingly, inhibition of RPL12 had the largest effect on 2 specific codons: CCG (encoding proline) and GCG (encoding alanine) (outliers in the box plot; Figure 2, D and E). Among all 20 amino acids, proline exhibits the slowest incorporation rate, with peptidyl transfer being the rate-limiting step (17) despite its cognate tRNAs being comparatively well expressed (Supplemental Figure 3, A and B). Proline is encoded by 3 other codons (CCU, CCC, and CCA), which also demonstrated higher ribosomal occupancy in the RPL12-depleted background, albeit to a lesser extent than CCG (Figure 2, D and E). For alanine (GCG codon), the effects observed might be similarly due to slower translocation and/or peptide bond formation (18), although stronger inhibition of tRNA release from the E-site may also be a contributing factor.
A major determinant of ribosomal speed for any particular codon is the concentration of the cognate tRNA. In mammalian systems, tRNA abundance varies between proliferating and differentiating cells (19) as well as among different organs (20) and subregions of tissues (21) despite similar codon usage. Consequently, codon usage by itself does not provide a reliable estimate of translation rate (9). Hence, we analyzed whether RPL12 suppression preferentially reduced the speed of codons read by low- or high-abundance tRNAs using tRNA-tailored comparative microarrays (9). Plotting tRNA concentration against genomic codon usage showed poor correlation (R2 = 0.11; Supplemental Figure 3C), indicating the complex relationship among codon usage, tRNA abundance, and translational speed. Consequently, we divided tRNAs into 3 equal groups based on their abundance (i.e., low, medium, and high). Following RPL12 knockdown, we observed a clear trend toward preferential slowing of codons pairing to higher-abundance tRNAs (P = 0.0227; Supplemental Figure 3, A–C). To address the effect of RPL12 knockdown on the translational profile of F508del, we calculated ribosome occupancy for each codon in CFTR. We did not observe a global reduction in translational speed across the entire CFTR transcript, but rather in specific regions, including TMD1, the R domain, and NBD2 (Figure 2G). Taken together, these findings reveal RPL12 suppression results in reduced ribosome velocity through a mechanism dependent on GC content of codons pairing to high-abundance tRNAs, rather than simply through interactions attributable to codon usage bias (i.e., differential occurrence of codons encoding the same amino acid).
Silencing RPL12 enhances F508del-CFTR plasma membrane stability and gating.
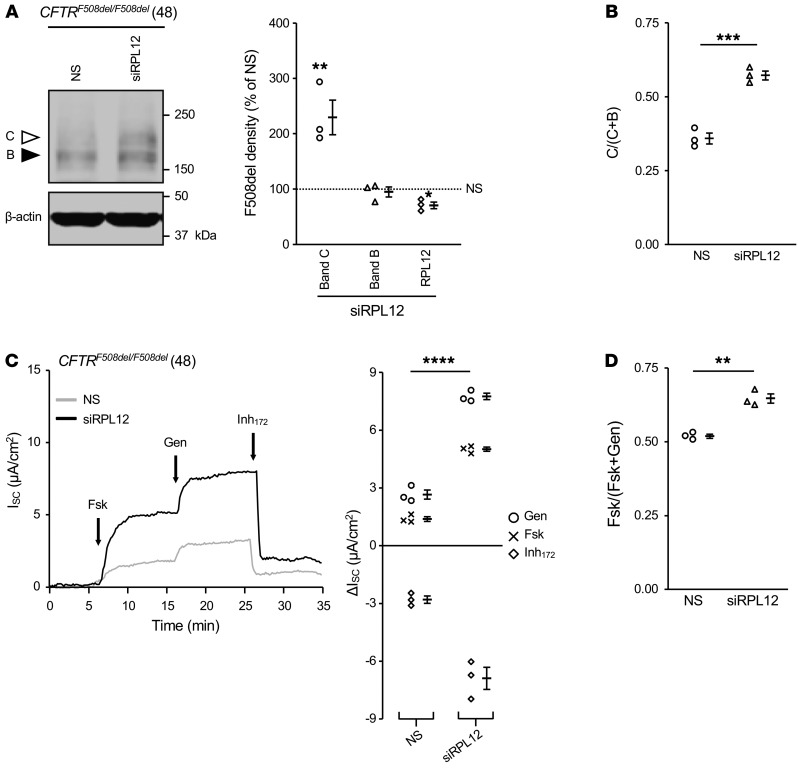
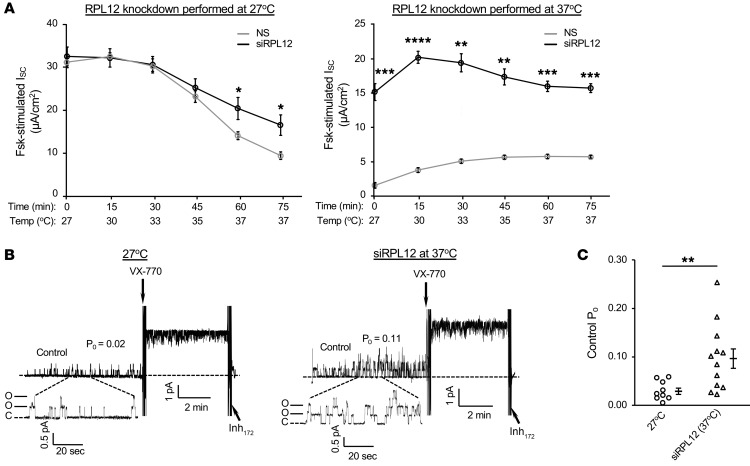
Following treatment with RPL12 siRNA, strong enhancement of both the mature CFTR glycoform (band C) and ISC were observed in primary human bronchial epithelia (HBE) monolayers collected from an F508del homozygous individual (Figure 3). To further address the impact of RPL12 depletion on CFTR channel activity and stability, we monitored functional integrity of F508del-CFTR using the following 2 parameters: (a) ion transport stability in the plasma membrane (PM) at 37°C and (b) single-channel gating characteristics. In order to evaluate whether surface stability of F508del-CFTR was affected by RPL12 knockdown, polarizing monolayers of CFBE cells stably expressing F508del were cultured at 27°C with or without RPL12 siRNA, mounted in Ussing chambers, and tested for transepithelial ion transport following a 37°C temperature shift. Chronic incubation at low temperature (27°C) was utilized to partially rescue F508del to the cell surface (22) as a means to study PM stability in the setting of RPL12 repression. Notably, F508del-CFTR function was partially maintained following RPL12 suppression conducted at 27°C (~56% functional after a 75-minute temperature ramp) in comparison with that in cells treated with NS siRNA (~33% active; Figure 4A). In addition, when RPL12 silencing was performed at 37°C, F508del-dependent ion transport exhibited approximately 7-fold higher levels than the NS siRNA control and remained nearly stable for the entire thermal ramp protocol (Figure 4A), establishing that RPL12-dependent modulation of translational velocity enhances F508del-CFTR activity at the cell surface.
Figure 3. Silencing RPL12 enhances F508del-CFTR functional expression in primary HBE.
(A) Following RPL12 knockdown, endogenous expression of mature, fully glycosylated F508del protein (band C, white arrowhead) is compared with levels of immature glycoform (band B, black arrowhead). Amounts of band B, band C, and RPL12 are quantified on right. Data are represented as mean ± SEM obtained from siRPL12-treated cells normalized to NS siRNA controls (dotted line set to 100%) (n = 3). Asterisks represent statistical comparison between siRPL12 and NS siRNA. *P < 0.0167; **P < 0.01 (unequal variance t test on log2-transformed data with post hoc Bonferroni’s correction; α = 0.0167). (B) Conversion ratio of band C to bands B and C (i.e., test of maturation efficiency) from A. Data include mean ± SEM (n = 3). ***P < 0.001 (2-sample t test). (C) F508del-mediated transepithelial ion transport (ISC) is augmented by RPL12 depletion (n = 3). Asterisks represent statistical comparison of forskolin+genistein stimulation (i.e., total constitutive plus acutely potentiated CFTR function). ****P < 0.0001, 2-sample t test. Fsk, forskolin; Gen, genistein. 48, deidentified patient code. (D) siRPL12 application enhances levels of cAMP-dependent CFTR ion transport (i.e., ratio of forskolin- to forskolin+genistein). Data are represented as mean ± SEM (n = 3). **P < 0.01, 2-sample t test. NS, NS siRNA; siRPL12, RPL12-targeting siRNA; Forskolin (5 μM), activator of PKA; genistein (50 μM), stimulator of CFTR-gating activity (i.e., potentiator); Inh172, 10 μM, inhibitor of CFTR. HBE were isolated from a CF individual with CFTRF508del/F508del genotype, cultured at air-liquid interface, and transfected twice per week with siRPL12 or NS siRNA (100 nM) for 3 weeks.
Figure 4. Surface function and baseline PO of F508del-CFTR are enhanced following repression of RPL12.
(A, left panel) Ussing chamber analysis in CFBE shows RPL12 knockdown at low temperature (27°C). Following a temperature ramp to approximately 35–37°C, F508del-CFTR channels retained significantly more residual function with RPL12 repressed compared with that in NS control (n = 3–4). Asterisks represent statistical comparison between siRPL12 and NS siRNA. *P < 0.025, unequal variance t test with post-hoc Bonferroni’s correction; α = 0.025. (A, right panel) Silencing RPL12 at 37°C led to an approximately 7-fold higher F508del-CFTR ion transport activity than the NS control and appeared thermally stable for at least 15 minutes at 37°C (n = 3–4). Asterisks represent statistical comparison between siRPL12 and NS siRNA. **P < 0.01; ***P < 0.001; ****P < 0.0001, unequal variance t test with post-hoc Bonferroni correction; α = 0.025. (B, left panel) HEK-293T cells, amenable to membrane-patch analysis, were transfected with F508del-CFTR cDNA (without RPL12 knockdown) and cultured at 27°C for 24 hours to promote maturation (n = 9). In the setting of 27°C rescue, F508del-CFTR PO is significantly blunted compared with WT, yet can be stimulated by VX-770 (49, 58). (B, right panel) HEK-293T cells transfected with F508del-CFTR and siRPL12 (at 37°C) were analyzed by excised inside-out patch (n = 12). (C) Mean PO comparison of 27°C-corrected and siRPL12-rescued (at 37°C) F508del-CFTR prior to VX-770 addition. All channels were activated with PKA catalytic subunit (110 U/mL) and MgATP (1.5 mM). VX-770 (200 nM) was added with holding potential of 60 mV. Data are shown as mean ± SEM. **P < 0.01, 2-sample t test. siRPL12, RPL12-targeting siRNA;forskolin, 5 μM; Inh172, 10 μM.
We next determined whether RPL12 suppression-induced changes in translation rate would lead to differences in folding and/or membrane stability indirectly detectable by the inside-out patch-clamp technique. An established assay was applied for monitoring CFTR single channels using human embryonic kidney (HEK) cells transiently expressing F508del-CFTR cDNA (23, 24). Low temperature–corrected F508del-CFTR exhibited a mean open probability (PO) of 0.02, whereas F508del-CFTR rescued by RPL12 knockdown at 37°C displayed a significantly higher PO of 0.11 (Figure 4, B and C), consistent with Ussing chamber analyses (Figure 4A). Correction by either low temperature or RPL12 depletion resulted in F508del-CFTR channels that were strongly stimulated at 37°C by the CFTR potentiator VX-770. The findings therefore demonstrate RPL12 siRNA-rescued F508del-CFTR exhibits improvement in PO (in support of enhanced CFTR folding) compared with 27°C correction.
CFTR folding is improved by RPL12 depletion.
RPL12 depletion has been shown to augment WT- and F508del-CFTR functional expression at both the ER and cell surface (by cell-based ELISA) as well as enhance the half-life and maturation efficiency of F508del protein (by cycloheximide- and 35S pulse-chase, respectively) (3). We therefore hypothesized that slowing translation by inhibition of RPL12 improves CFTR folding, which consequently increases CFTR expression and function. To quantitatively probe CFTR folding, we performed limited proteolysis on newly synthesized WT- or F508del-CFTR stably expressed in CFBE cells. Both constructs were radioactively pulse labeled, followed by proteinase K digestion of cell lysates generated immediately after the pulse or chase. Protease-resistant fragments of TMD1, TMD2, NBD1, and NBD2 (Figure 5A) were immunoprecipitated with domain-specific antibodies to probe for conformational differences.
Figure 5. RPL12 depletion stabilizes CFTR interdomain assembly.
(A) WT-CFTR with domains annotated: TMD1 (dotted); NBD1 (black); regulatory domain (R) (white); TMD2 (light gray); NBD2 (dark gray). N, amino; C, carboxy. (B) CFBE expressing WT- or F508del-CFTR and treated with siRPL12 or NS siRNA were pulse labeled for 15 minutes and chased for 0–2 hours. RPL12 silencing was confirmed by immunoblotting (with tubulin-loading control). Lysates were digested with proteinase K and protease-resistant CFTR domain fragments immunoprecipitated/resolved by SDS-PAGE. Immature, ER-resident CFTR (band B, black arrowhead), fully glycosylated, mature CFTR (band C, white arrowhead), early domain fragments (T1a-c, N1a, T2a-b, N2a), and late domain fragments (T1d-f, T2b-c) are designated (see Methods). V, empty vector; asterisk indicates background. Brief chase applied here shows F508del-CFTR exits the ER slowly (i.e., band B at 2 hours remains significantly elevated following siRPL12 treatment), yet is stabilized in post-ER compartments, as evidenced by moderate band C accumulation (see also ref. 59). Increased proteolytic fragments predominantly reflect the ER-localized glycoform. (C) Quantification of RPL12 knockdown (n = 3–4). (D) Comparison of total radiolabeled CFTR (bands B and C) following RPL12 repression (n = 4–5). (E and F) Fold increase of domain-specific fragment intensities for WT- (E) or F508del-CFTR (F). Bands are corrected for total radiolabeling (n = 4–5). Note NS-treated F508del signal in the 2-hour chase is less than 2-fold over background in D and F. (C–F) Data are shown as mean ± SEM from siRPL12-treated cells normalized to NS siRNA (dotted line set to 100% or 1 as shown). Asterisks represent statistical comparison between siRPL12 and NS. Unequal variance t test on log2-transformed data with post-hoc Bonferroni’s correction. α = 0.025 (C and D); α = 0.0125 (E and F). **P < 0.01; ***P < 0.001 (C and D). *P < 0.0125; **P < 0.001; ***P < 0.0001 (E and F).
As introduced above, our results establish that depletion of RPL12 reduced total protein synthesis, yet simultaneously augmented CFTR expression approximately 3-fold (Figure 5, B–D, and Supplemental Figure 2, A, B and C). More specifically, both WT- and F508del-CFTR band B (immature, ER-retained protein) and band C (mature, Golgi-modified glycoform) appeared proportionally augmented by RPL12 depletion (Figure 5B and Supplemental Figure 3D). The amount of F508del-CFTR band C increased substantially from a nondetectable threshold to an approximately 3-fold higher level (Figure 5, B and D).
To determine the basis for enhanced CFTR protein production, we analyzed folding of CFTR structural domains through protease resistance at different time points (7, 25): (a) immediately after synthesis following the radiolabel pulse (early fragments: T1a-c, N1a, T2a-b, and N2a; Figure 5B) and (b) after a 2-hour chase (late fragments: T1d-f, T2b-c; Figure 5B). During the process of folding, CFTR becomes more compact and therefore more protease resistant, leading to larger proteolytic fragments at later time points. NBD1 is an exception, because it acquires full protease resistance early and does not change during the chase period (see ref. 7 and Figure 5B). Following the initial synthesis of both WT- and F508del-CFTR, RPL12 knockdown robustly stabilized TMD2 compared with other domains (Figure 5, B, E, and F). This suggests that TMD2 improvement drives the increase in total CFTR and that this particular domain is the primary target of RPL12 silencing–induced rescue of F508del.
Deletion of F508 in CFTR confers misfolding of NBD1 and abrogates subsequent interdomain assembly (26). We can therefore detect NBD1 misfolding as absence of the typical protease-resistant N1a fragments (Figure 5B and ref. 25). At the 2-hour chase time point, our data reveal that RPL12 inhibition modestly enhanced NBD1-N1a expression, but significantly increased intensities of all late fragments within TMD1, TMD2, and NBD2 (Figure 5, B, E, and F, and Supplemental Figure 3, E and F), which may account for increased overall stability of the WT and F508del channels. When analyzing against the total amount of radiolabeled protein, all 4 domains of WT- and F508del-CFTR appeared stabilized (Figure 5, E and F). However, the data support TMD2 as a driver for CFTR stabilization (Figure 5F). Because TMD2 stability is augmented, stabilization of TMD1 and NBD2, but not NBD1, are likely improved through interdomain assembly.
Rpl12 knockdown is additive with second-site suppressors that promote F508del biogenesis.
To further investigate mechanisms by which Rpl12 inhibition augments F508del-CFTR stability and function, we assessed interactions between Rpl12 knockdown and intragenic suppressors of F508del. Fischer rat thyroid (FRT) cells, which utilize Flp-In technology to control for site of genomic integration and maintain constant levels of mRNA independently of the particular variant being tested, allow stringent comparisons of this type (27, 28). Comparative tRNA-based microarray analysis demonstrated that the tRNA levels were often similar in CFBE and FRT cells (Supplemental Figure 3, A and B), and hence, we expect codon-specific effects of Rpl12 suppression similar to those observed in CFBE cells (Figure 2, D and E).
The R555K mutation has been shown previously to partially overcome instability of F508del-CFTR NBD1 (29). Our present results demonstrate R555K augments yield of the F508del mature glycoform, cell-surface expression, and channel activity (Figure 6, A–C; compare with F508del-CFTR baseline in Supplemental Figure 4, A–D). Following knockdown of Rpl12 using 4 distinct siRNAs, levels of F508del-R555K-CFTR bands B and C were further increased (~200%–300% increase; Figure 6A). We also observed enhancement of F508del-R555K PM density and ISC in response to Rpl12 repression, as compared with the levels achieved by R555K alone on the F508del background (Figure 6, B and C). Importantly, these results are not specific to the R555K suppressor. We detected very similar effects with the R1070W mutation (Figure 6, D–F), which rescues the F508del-CFTR biogenesis defect by stabilizing an interface between NBD1 and ICL4 (29). These findings further establish a stabilizing effect of Rpl12 depletion on F508del-CFTR that appears additive with R555K or R1070W and suggests a mechanism distinct from either second-site suppressor.
Figure 6. In FRT epithelia, Rpl12 inhibition is additive with concomitant F508del-CFTR second-site suppression by R555K or R1070W mutations.
(A) F508del-R555K immature (band B, black arrowhead) and mature (band C, white arrowhead) protein levels are markedly enhanced following approximately 50%–70% knockdown of Rpl12. Representative immunoblot (left panel) is quantified on right (n = 3). (B) Rpl12 siRNAs significantly increase F508del-R555K-HRP trafficking to the PM (n = 9). (C) ISC measurements using F508del-R555K demonstrate an increase following Rpl12 inhibition (n = 4). (D) Augmentation of F508del-R1070W-CFTR band B and band C expression following an approximately 50%–60% reduction in Rpl12 protein levels. Representative immunoblot (left panel) is quantified on right (n = 3). (E) Rpl12 knockdown significantly increases F508del-R1070W-HRP at the cell surface (n = 12). (F) ISC measurements using F508del-R1070W demonstrate an increase in CFTR-dependent ion transport following Rpl12 suppression (n = 4). Data in A and D are represented as mean ± SEM obtained from siRPL12-treated cells normalized to NS siRNA (dotted line set to 100%). Asterisks represent statistical comparison between siRPL12 and NS control. *P < 0.0167; **P < 0.01, unequal variance t test on log2-transformed data with post-hoc Bonferroni’s correction; α = 0.0167. Data in B and E data are represented as mean ± SEM. *P < 0.0083; **P < 0.001; ****P < 0.00001, unequal variance t test with post-hoc Bonferroni’s correction; α = 0.00833. Data in C and F data are represented as mean ± SEM. Asterisks represent statistical comparison of forskolin+VX-770 stimulation (i.e., total constitutive plus activated CFTR function). *P < 0.0125; **P < 0.001; ***P < 0.0001; ****P < 0.00001, unequal variance t test with post-hoc Bonferroni’s correction; α = 0.0125. RPL12_6, _7, _9, and _11 denote RPL12 siRNAs with crossreactivity against rat mRNA; forskolin, 5 μM; VX-770, 5 μM; Inh172, 10 μM.
CFTR processing mutants are differentially corrected by Rpl12 suppression.
In primary airway cells, the ratio of F508del band C to total CFTR was increased by RPL12 silencing (Figure 3A), a finding consistent with enhanced maturational efficiency. In the FRT model, F508del overexpression with siRPL12 treatment resulted in strong proportional increases in both band B and band C (Supplemental Figure 4A). This may reflect inability of cellular quality control to efficiently process high levels of immature CFTR in recombinant cells. In either case, suppression of the ribosomal protein markedly restored F508del-CFTR function in both cell models. Such findings represent significant CFTR “amplifier” type effects, i.e., when ER-localized CFTR is increased, it is suitable for further rescue by pharmacologic agents or other interventions (30, 31).
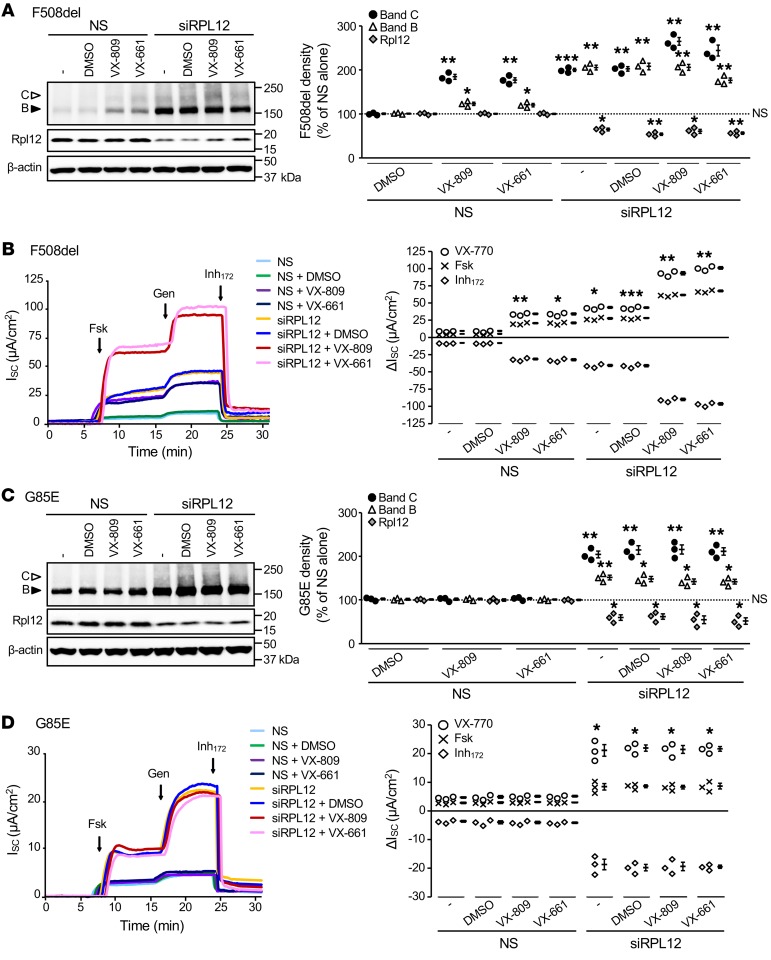
From both a mechanistic and therapeutic perspective, we next evaluated whether Rpl12-dependent rescue could be augmented by simultaneous treatment with FDA-approved CFTR correctors, lumacaftor (VX-809), or tezacaftor (VX-661). In FRT epithelia, silencing of Rpl12 enhanced F508del-CFTR band C expression to the same extent as VX-809 or VX-661, with concurrent treatments resulting in additional increase in protein biogenesis (Figure 7A). Moreover, combination of either small molecule and Rpl12 knockdown increased F508del-mediated ion transport by approximately 10-fold above baseline (Figure 7B). To determine whether the Rpl12 effect was specific for only F508del, we tested a small cohort of other CFTR “processing” variants for responsiveness to Rpl12 depletion alone or with concomitant pharmacocorrection (Figures 7 and 8). For G85E-CFTR, a variant highly resistant to pharmacologic rescue, modest enhancement of band B and C expression was observed, whereas no effect was noted due to VX-809 or VX-661 alone or in combination with Rpl12 siRNA (Figure 7C). Furthermore, with Rpl12 suppressed, G85E ion transport was increased by approximately 4-fold, while addition of VX-809 or VX-661 resulted in no activation (Figure 7D). For both F508del- and G85E-CFTR, strong potentiator-stimulated responses were observed, demonstrating that Rpl12-corrected channels that reach the cell surface likely still harbor gating abnormalities (Figure 7, B and D).
Figure 7. Rpl12 knockdown and VX-809 differentially rescue trafficking of F508del- and G85E-CFTR stably expressed in FRT cells.
(A) Expression of immature (band B, black arrowhead) and mature (band C, white arrowhead) F508del-CFTR protein are augmented following an approximately 40% reduction in Rpl12 expression (n = 3). (B) F508del-CFTR ion transport activity is increased by Rpl12 knockdown (100 nM siRNA, 4 days) or addition of a corrector molecule (VX-809 or VX-661, 3 μM, 48 hours), with additivity observed following combined application (n = 3). (C) Augmentation of G85E-CFTR band B and band C expression was observed following an approximately 30% reduction of Rpl12 protein (n = 3). (D) ISC measurement reveals an approximately 4-fold increase in channel function with Rpl12 suppressed and no effect following application of either VX-809 or VX-661 (3 μM, 48 hours) alone or in combination with siRPL12 (100 nM, 4 days) (n = 3). Representative findings in left panels are quantified on right. Data shown in A and C represent mean ± SEM normalized to NS siRNA without drug treatment (dotted line set to 100%). Asterisks represent statistical comparison versus NS siRNA without pharmacocorrection. *P < 0.0167; **P < 0.01; ***P < 0.001 (unequal variance t test on log2-transformed data with post hoc Bonferroni’s correction; α = 0.0167). Data shown in B and D represent mean ± SEM. Asterisks represent forskolin+VX-770 stimulation (i.e., total constitutive plus activated CFTR function) versus NS siRNA without drug treatment. *P < 0.00625; **P < 0.001; ***P < 0.0001 (unequal variance t test with post hoc Bonferroni’s correction; α = 0.00625). VX-809 or VX-661 augmented F508del-CFTR band C (P < 0.0167) and activity (P < 0.001) above siRPL12 alone. siRPL12, RPL12 siRNA with crossreactivity against rat mRNA; forskolin, 5 μM; genistein, 50 μM; Inh172, 10 μM.
Figure 8. In FRT cells, A455E-CFTR functional expression is partially rescued by siRPL12, whereas P67L-CFTR is unaffected.
(A) Following an approximately 30% knockdown of Rpl12, A455E-CFTR band B (black arrowhead) and band C (open arrowhead) are significantly increased (n = 3). (B) A455E-mediated transepithelial ion transport is augmented by Rpl12 depletion (100 nM siRNA, 4 days) and additive to treatment with small molecule correctors (VX-809 or VX-661, 3 μM, 48 hours) (n = 3). (C) P67L-CFTR band C is significantly enhanced as is band B (to a lesser extent) following addition of VX-809 or VX-661 (n = 3). (D) P67L-CFTR ion transport is markedly increased by either CFTR corrector (3 μM, 48 hours), yet unaltered by siRPL12 (100 nM, 4 days) (n = 3). Representative measurements in left panels are quantified on right. Data shown in A and C are represented as mean ± SEM normalized to NS siRNA without drug treatment (dotted line set to 100%). Asterisks represent statistical comparison versus NS siRNA without pharmacocorrection. *P < 0.0167; **P < 0.01; ***P < 0.001; ****P < 0.0001, unequal variance t test on log2-transformed data with post-hoc Bonferroni’s correction; α = 0.0167. Data shown in B and D are represented as mean ± SEM. Asterisks represent forskolin+VX-770 stimulation (i.e., total constitutive plus activated CFTR function) versus NS siRNA without drug treatment. *P < 0.00625; **P < 0.001; ***P < 0.0001, unequal variance t test with post-hoc Bonferroni’s correction; α = 0.00625. VX-809 or VX-661 augmented A455E-CFTR band C (P < 0.01) and activity (P < 0.001) above siRPL12 alone. Forskolin, 5 μM; genistein, 50 μM; Inh172, 10 μM.
We tested 2 additional and well-described CFTR maturational processing mutations, P67L and A455E. The profile of Rpl12-dependent rescue for A455E-CFTR resembled that of F508del-CFTR, as assessed by correction of band C and transepithelial ion transport (Figure 8, A and B). On the other hand, P67L-CFTR, which responds robustly to small molecule correction, showed no functional improvement following Rpl12 knockdown (Figure 8, C and D).
Activity of the A455E- and G85E-CFTR variants was augmented by Rpl12 depletion, and in both cases, the amino acid exchange led to 2 alterations: (a) addition of a large, charged side chain, which likely hampered folding, and (b) an inversion of the rate of translation at each particular codon — i.e., codons pairing to low-abundance tRNAs are exchanged and read by high-abundance tRNAs and are therefore likely to be translated at greater velocity in the mutant proteins (Supplemental Figure 3B). In contrast, the P67L mutation (CCT to CTT) possesses a “kink-inducing” proline rather than a hydrophobic leucine and changes the velocity in the opposite direction — i.e., the concentration of tRNA pairing to the mutated codon is approximately 10-fold lower (Supplemental Figure 3B) and is therefore likely to be translated more slowly than the original codon. Together, our results suggest Rpl12 suppression reduces translation rate, which may affect certain CFTR-processing mutations and also elicit site-specific and/or codon-specific contributions to the overall folding pathway.
Rpl12 mediates CFTR folding in vivo.
Knockdown of Rpl12 by as little as approximately 30% partially corrects F508del-CFTR biogenesis in murine (Figure 7) or human (Figure 3) cell-culture models, but it has remained unclear whether Rpl12 depletion could be tolerated in an intact animal. Consequently, we constructed an Rpl12+/– (heterozygous) mouse model on a pure C57BL/6 background (Supplemental Figure 5), which demonstrated normal growth and reproductive phenotype for at least 6 months postnatally. From these mice, 4 tissue types (lung, ileum, colon, and pancreas) were evaluated and found to exhibit Rpl12 protein levels that were approximately 70% of those of Rpl12+/+ littermates (Figure 9). Strikingly, WT-Cftr band C was significantly increased in 3 of the Rpl12+/– tissues examined, with the pancreas being most prominent (Figure 9). These findings indicate that in mammalian tissues, Rpl12 suppression leads to a significant (P < 0.0167) enhancement of Cftr biogenesis and band C formation in vivo.
Figure 9. Rpl12 protein levels are reduced and WT-Cftr maturation enhanced in Rpl12+/– mice.
Data for (A) pancreas, (B) colon, (C) ileum, and (D) lung. Expression of WT-Cftr band B, band C, and Rpl12 from Rpl12+/+ mice are set at 100% (dotted line) and compared with those of Rpl12+/– littermates. Data are represented as mean ± SEM (n = 3 independent animals and tissue samples per condition in each panel). *P < 0.0167; **P < 0.01, unequal variance t test on log2-transformed data with post-hoc Bonferroni’s correction; α = 0.0167. Low baseline levels of band C in the lungs are consistent with absent respiratory phenotype in mice lacking Cftr (reviewed in refs. 60, 61). Regulatory steps responsible for Cftr quality control, protein folding, transcriptional regulation, and posttranslational modification in mammalian tissues are not sufficiently understood to account for levels of band B noted here, although earlier studies conducted on WT C57BL/6 mice support these findings (62, 63).
Discussion
Genome-wide screening in yeast (Supplemental Figure 1) provides an evolutionarily conserved means to discover unexpected contributors to CFTR maturation and strongly implicates translational velocity in this process. The ribosome controls kinetics of elongation (8, 32) and serves as a hub for vital auxiliary folding and related functions that significantly influence native protein structure (33, 34). Building on previous studies that have established that (a) CFTR domains and subdomains within NBDs fold and assemble in an organized, cotranslational manner (7, 29, 35) and (b) translational speed and mRNA codon composition impact CFTR biosynthesis (3, 8–10), the present findings provide direct evidence that translation rate acts as a substantial contributor to CFTR functional expression. Our results point to a perspective with relevance to the CF disease mechanism. We establish that inhibiting the ribosomal stalk protein, RPL12 (uL11), slows translation in a codon-specific manner and influences CFTR folding through a pathway distinct from — and additive to — existing forms of pharmacologic correction (Supplemental Figure 6).
Application of ribosome profiling, high-throughput RNA-Seq, and tRNA microarray technologies (13, 36, 37) enabled us to assemble a comprehensive and positional map of translating ribosomes with single codon resolution as well as extract specific features of translational kinetics. We show that suppression of RPL12 alters the elongation rate of a subset of codons with GC-rich signature that also pair to high-abundance tRNAs. These changes in elongation velocity augment F508del-CFTR expression, steady-state protein levels, and channel function. Highly affected regions (i.e., codons slowed by RPL12 knockdown) were present within TMD1, the R domain, and NBD2 (Figure 2G). The subtle decrease in translation rate within these regions most likely altered the folding trajectory and stabilized subdomain topologies, as evidenced by the robust enhancement of TMD2 protease resistance (Figure 5, B and F). In the same manner, TMD1 and NBD2 became more protease resistant in contrast to NBD1, the stability of which was much less influenced by RPL12 depletion. As a cumulative result, the amount of newly synthesized F508del-CFTR increased and remained stable due to improved folding. The enhanced quantity and quality of several other mutant forms of the protein (as well as WT-CFTR) led to cell-surface rescue and improved function of these CFTR variants. Our findings also corroborate observations in yeast that RPL12 deletion slows translation (11, 12) and complement these data with the discovery that the overall effect is global, albeit dependent on the specific protein defect and/or codon context. For what we believe is the first time, we used this approach to establish impact of disease-relevant mutations on CFTR biogenesis from a mechanistic perspective, including effects on elongation.
Slowing translational velocity has been suggested previously to enhance F508del-CFTR processing (38), although, as shown here, the effect of RPL12 depletion appears distinct. F508del correction facilitated by RPL12 repression does not restore NBD1 folding, but bypasses this defect in part through stabilization of TMD2. Improvement in TMD2 associated with RPL12 suppression is additive, with TMD1 rescue achieved by VX-809 (Figure 7, A and B) as well as with NBD1 stabilization mediated by R555K (Figure 6, A–C). The R1070W mutation appears less additive with Rpl12 knockdown because of the overlapping target domain (i.e., TMD2) (Figure 6, D–F).
In a similar fashion, we tested 3 rare CFTR variants (G85E, P67L, and A455E) with mutations that localize to different structural domains in CFTR (Supplemental Figure 7). Each of these variants affects CFTR folding directly due to a point mutation in the primary amino acid structure, but each variant is also predicted to confer altered rates of translation based on respective codon substitutions (Supplemental Figure 3B). Rescue of G85E-CFTR is particularly exciting, as this mutant is impervious to all available corrector drugs, which is likely due to defective membrane insertion of the TMD1 TM helices (39). Whereas Rpl12 depletion enhanced both folding and function of G85E-CFTR, it did not render this variant sensitive to correction by VX-809 (Figure 7, C and D). Our findings therefore suggest that TMD1 is not restored to a conformation susceptible to pharmacocorrection. We show that Rpl12 suppression also rescues A455E-CFTR, but not the P67L mutant, which indicates that TMD2 stabilization alone may not be sufficient to overcome the P67L defect (Figure 8).
An alternative viewpoint is offered by the missense-codon–induced change in ribosome velocity. CFTR amino acid exchanges that accelerate elongation at an involved codon (e.g., G85E, A455E), but not a mutation that slows translation (e.g., P67L), were rescued by Rpl12 repression. These data therefore suggest the possibility that, in addition to the effect of the amino acid exchange on CFTR function, mutations that invert the codon translation rate may also affect protein biogenesis through an effect or effects that alter translational kinetics of nascent CFTR synthesis. Recent findings, for example, suggest that sSNPs altering translation speed or CFTR mRNA structure can have significant effects on CFTR conformation, function, and F508del-CFTR pharmacocorrection (9). Thus, mutations that cause regional changes in translational velocity within key elements of the CFTR polypeptide may have direct effects on the final folded state.
The mechanism by which RPL12 suppression improves mutant CFTR folding/trafficking appears evolutionarily conserved in yeast, murine, and human cells. Components of the P stalk and GAC interface with initiation, elongation, and termination factors, but it is eEF2 in particular that has an established interaction with RPL12 in multiple eukaryotic models (Supplemental Figure 6). In yeast, depletion of other ribosomal proteins in close physical proximity to Rpl12 (e.g., P2, uL6, uL13, eL40) strongly overcome the F670del defect in Yor1 (Figure 1 and Supplemental Figure 1). In the context of F508del-CFTR, silencing similar domains (e.g., RPL12, P2) and eEF2 correct CFTR functional expression (3), implicating a prominent role for the P stalk and kinetics of elongation during CFTR rescue. RPL12 siRNA–induced slowing of overall CFTR translation (Figure 2, C and G), which occurs most significantly through GC-rich codons pairing to high-abundance tRNAs (Figure 2, D–F), further points to the importance of translational velocity during both CFTR domain folding and interdomain assembly (Figure 5) (7). Since saturating concentrations of correctors (VX-809, VX-661) and second-site suppression (R1070W) are believed to act by stabilizing the NBD/TMD interface (29, 40) — and because these measures are additive with Rpl12 silencing — the mechanism of rescue mediated by Rpl12 depletion appears distinct (Figures 6 and 7). Enhancement of bands B and C for certain CFTR variants (A455E; Figure 8, A and B), but not others (P67L; Figure 8, C and D), is of considerable scientific and therapeutic interest. Increased levels of band B generated in this fashion provide an excellent substrate for lumacaftor, tezacaftor, or other small molecule correctors (i.e., the so-called CFTR “amplifier” effect noted above).
Our murine studies demonstrate that reduced Rpl12 levels confer increased Cftr band C in epithelial tissues (Figure 9), but we have not yet shown enhanced Cftr ion transport in this setting. In our experience, small changes in murine intestinal ISC can be variable, even when bowel samples (ileum, colon) are taken from a single animal. In addition, Cftr+/+ and Cftr+/– mice can exhibit comparable intestinal ISC (in semblance to sweat chloride, nasal potential difference, or rectal biopsy measurements from CFTR+/+ versus CFTR+/– human subjects) (41, 42), further indicating that increases of endogenous WT-CFTR protein may elicit only minimal ISC activation in full-thickness intestinal tissues. Moreover, as in WT HBE monolayers in vitro, a modest increase in endogenous CFTR protein is often difficult to detect by ISC (in contrast to appearance of CFTR in an otherwise CFTR-null cell model). Based on earlier work and considerations such as these, we note the bioelectric driving force (established by basolateral ion movement) or other factors may therefore be rate limiting with regard to WT murine intestinal ISC. Studies to crossbreed Rpl12-deficient animals with humanized F508del CF mice — where apical CFTR is rate limiting for anion transport — are in progress and will provide a future test of CFTR activation conferred by Rpl12 suppression in vivo.
In FRT, CFBE, or primary airway epithelia, no evidence of cytotoxicity was observed following RPL12 knockdown as judged by alamarBlue assay, nuclear/cytoplasmic ratio, or transepithelial resistance of monolayers (3), as well as negligible transcriptome-wide effects and maintenance of key cellular pathways (Figure 2B and Supplemental Figure 2, F, H, and I). Moreover, our present ribosome profiling studies indicate absence of an unfolded protein response (UPR) — i.e., gene targets downstream of XBP1, ATF6, and ATH4 signaling cascades are not increased by RPL12 inhibition. Like other transgenic murine models deficient in specific ribosomal proteins (e.g., Rpl29, ref. 43; Rpl38, ref. 44; or Rpl24, refs. 44, 45), mice tolerate 1 functional copy of Rpl12 in terms of weight gain, reproduction, longevity, and activity level. In addition, gross anatomic defects were not observed during dissections for experiments such as those shown in Figure 9. Ultimately, therapeutic usefulness of the approach will depend on whether animals encoding CFTR variants demonstrate increased survival and/or other benefit following Rpl12 suppression in vivo.
In summary, our results emphasize the power of high-throughput yeast phenomics to discover mechanisms influencing a genetic disease such as CF. The present work provides insight regarding ribosomal components that could serve as molecular targets to restore F508del or other CFTR variants and function apart from previously established interventions. Furthermore, our findings have implications with regard to variation of clinical CF disease phenotype and response to corrector compounds, i.e., “precision” intervention (2, 46, 47). For example, the relation of CFTR variants to translation velocity might be considered more prominently when assessing response to pharmacologic rescue. This is extendable to numerous human congenital diseases that are poorly understood in terms of molecular pathogenesis.
To our knowledge, no human disease has previously been suggested to be directly influenced by translational rate. Hence, our study provides a basis from which to develop disease-ameliorating strategies that modulate ribosome velocity. Moreover, the pronounced heterogeneity of disease progression among individuals with CF that harbor the same genotype, including their varied responsiveness to modulator treatment, is not well understood. In vitro and in vivo findings presented here provide strong rationale for future studies to examine the impact of translational velocity as a contributor to pathogenesis and the clinical course of CF.
Methods
Reagents and antibodies.
Lumacaftor (VX-809), ivacaftor (VX-770), and tezacaftor (VX-661) were obtained from SelleckChem. siRNAs (QIAGEN) for targeted sequences are listed in Supplemental Table 1. Anti-CFTR mouse monoclonal antibodies — purchased through the CFTR Antibody Distribution Program (University of North Carolina, Chapel Hill, North Carolina, USA) — were directed toward the R domain (UNC570) or NBD2 (UNC596). Anti-Cftr rabbit polyclonal antibody (catalog ab59394, Abcam) was used to detect mouse Cftr. Endogenous human RPL12 and murine Rpl12 were probed using a crossreactive rabbit polyclonal antibody (catalog ab157130, Abcam). Additional antibodies included anti–β-actin mouse monoclonal (catalog AC-15, Sigma), anti–α-tubulin mouse monoclonal (catalog 302211, Synaptic Systems), goat anti–mouse-HRP (catalog P0447, Agilent), goat anti–mouse-HRP (catalog 1706516, Bio-Rad), goat anti–rabbit-HRP (catalog 31460, Thermo Fisher), and goat anti–rabbit-HRP (catalog 1706515, Bio-Rad). Polyclonal rabbit antisera TMD1-C, TMD2-C, and Mr. Pink were raised against CFTR residues 362-381, 1172-1186, and purified human NBD1, respectively (25). Mr. Pink was shown to recognize various epitopes on NBD1, including all forms of the domain (i.e., folded, misfolded, or denatured).
Cell culture and media.
Parental CFBE41o– cells (i.e., no CFTR transgene overexpression) were a gift of Dieter Gruenert (UCSF, San Francisco, California, USA) and Karl Kunzelmann (University of Regensburg, Regensburg, Germany). Generation and maintenance of CFBE cell lines constitutively overexpressing WT- or F508del-CFTR have been described previously and were cultured in 1× minimal essential medium (Gibco, Thermo Fisher Scientific) supplemented with Earle’s salts, l-glutamine, and 10% FBS (48). CFBE-expressing F508del-CFTR were used to provide a human respiratory epithelial background for experiments investigating aspects of F508del correction, cell-surface stability, ribosome profiling, and tRNA abundance. Parental FRT cells were a gift of Michael Welsh (University of Iowa, Iowa City, Iowa, USA). FRT lines were stably transduced with WT-, F508del-, G85E-, A455E-, or P67L-CFTR cDNA using the Flp-In system (Invitrogen) and cultured in F12 Ham Coon’s modified nutrient mixture F-12 Ham (F6636, MilliporeSigma) pH 7.3 supplemented with 5% FBS (27, 28). FRTs were selected clonally and shown to utilize a single transgene insertion by Southern blot and express similar levels of mRNA (by quantitative reverse-transcriptase PCR [RT-qPCR]), allowing valid mechanistic comparisons between different CFTR variants. For CFTR single-channel analysis, HEK-293T cells (ATCC) maintained in 1× DMEM (Gibco, Thermo Fisher Scientific) supplemented with 4.5 g/L d-glucose, l-glutamine, 110 mg/L sodium pyruvate, and 5% FBS were transfected with F508del-CFTR cDNA using Lipofectamine 2000 (Invitrogen). Primary HBE were harvested from a CF patient homozygous for F508del-CFTR and grown as described (49).
siRNA transfection.
Knockdown of RPL12 (uL11) was achieved based on a published protocol (3). Briefly, the target gene was silenced in nonpolarized CFBE, FRT, or HEK cells by forward transfection using 100 nM siRNA crossreactive with human and rat RPL12 (QIAGEN) and Lipofectamine RNAiMAX (Invitrogen). In polarized CFBE or FRT, 100 nM siRNA duplexes were reverse transfected as described (50), followed by 4-day growth to allow monolayer establishment and polarity. Primary HBE were forward transfected with 100 nM siRNA for 6 to 8 hours on day 1 after seeding, with transfections repeated twice per week. Polarized monolayers were cultured at air-liquid interface for a total of 3 weeks.
Ribosome profiling, RNA-Seq, and tRNA microarrays.
CFBE cells stably expressing F508del-CFTR that were NT or transfected with siRPL12 or NS siRNA were flash-frozen and processed for ribosome profiling, RNA-Seq, and tRNA microarrays as described (9). Sequenced reads were trimmed using fastx-toolkit (0.0.13.2; quality threshold: 20), adapters were cut using cutadapt (1.2.1; minimal overlap: 1 nt), and processed reads were uniquely mapped to the human genome (GRCh37) using Bowtie (0.12.9), allowing a maximum of 2 mismatches (parameter settings: -l 16 -n 1 -e 50 -m 1 --strata -- best y). Uniquely mapped reads from ribosome profiling or RNA-Seq were normalized to reads per million mapped reads (RPM) or reads per kilobase per million mapped reads (RPKM). Each RNA-Seq data set was treated with RNA Spike-In Mix (Thermo Fisher) to determine detection threshold. RD for each gene was computed as follows: RD = RPF/mRNA, where RPF and mRNA are expressed in RPM.
To exclude influence of initiation, we omitted the first 70 codons (Figure 2C) from analysis. To calibrate reads, RPFs were binned into groups of equal read length and each group aligned via P-site positioning over the start codon (14, 51). The majority of sequence reads were 27–29 nt as expected.
Ribosome dwelling time, or ribosome occupancy, per codon was calculated as described (9, 14). Reads over each position in a gene (i) were normalized to the average number of footprints. Ratios were then averaged across all genes to determine ribosome-dwelling occupancy of a given codon in the transcriptome. The first 51 nt were excluded from calculations to avoid ribosome depletion due to elongation run-off near transcript start sites during cell harvesting. CFTR mRNA ribosome occupancy profiles were calculated from ribosome-dwelling values at each codon following treatment with NS or RPL12 siRNA. Codons were binned in groups of 5 based on ribosome occupancy value. Variance between 2 profiles was determined by computing mean coefficient of variance across each bin. Values outside the 95th percentile were considered significantly different.
tRNA microarrays were performed with probes covering the full-length sequence of 41 cytoplasmic tRNA species complementary to 49 nuclear encoding tRNA families with sequences previously described (9). Total RNA was extracted, propagated as above (TriReagent), and deacetylated (45 minutes, 37°C, 100 mM Tris-HCl, pH 9.0). For comparative arrays, tRNAs were labeled with fluorescent stem-loop RNA/DNA hairpin oligonucleotides bearing a Cy3 fluorescent dye (Microsynth) and hybridized on the same array with HeLa tRNAs labeled with an Atto-653 fluorescent RNA/DNA hairpin. Absolute tRNA levels in HeLa were previously determined (9). To assess biological functions affected by RPL12 knockdown, we identified GO terms with the DAVID gene functional tool. P values were adjusted for FDR as reported (52).
Patch-clamp analysis.
HEK-293T cells were transfected with siRPL12 (day 1 after seeding), followed by F508del-CFTR cDNA (day 2). Inside-out patch-clamp studies were performed (day 3) using excised membranes (23, 24). Cells transfected with F508del-CFTR, but without siRPL12, were cultured at 27°C for 24 hours to allow comparison with an established method of F508del rescue. CFTR channels were activated with protein kinase A (PKA) catalytic subunit (110 U/1 mL) and Mg-ATP (1.5 mM), then stimulated further by VX-770 (200 nM). Patches were held at 60 mV for unitary current recordings with analog filtering (110 Hz), then digitally filtered (10 Hz) with Clampfit 9.2 (Axon Instruments). Mean PO was calculated by Clampfit when the number of channels was ≤ 8. An estimate of n was derived using Itotal = n × PO × i when the number of channels was greater than 8. In this setting, PO for F508del-CFTR can be best estimated by maximal activation of the channel using VX-770 (as reported in ref. 53).
Pulse-chase analysis.
NS or siRPL12-transfected CFBE cells stably expressing WT- or F508del-CFTR at approximately 80% confluence were analyzed as described previously (25). Cells were washed with HBSS (Thermo Fisher) and depleted of cysteine and methionine for 15 minutes in MEM (MP Biomedicals) with 10 mM HEPES (pH 7.4). Cells were pulse-labeled (15 minutes, 37°C) with 143 μCi/dish EasyTag and 35S-Express Protein Labeling Mix (PerkinElmer), then chased in MEM with 10 mM HEPES (pH 7.4), 5% FBS, and 5 mM methionine and cysteine. Afterward, cells were transferred onto ice, washed twice with ice-cold HBSS, and lysed with ice-cold nondenaturing lysis buffer (150 mM NaCl, 1% nonylphenyl polyethylene glycol [NP-40; Fluka Biochemika], 50 mM Tris-HCl, pH 8.0). Lysates were centrifuged (10 minutes, 15,000 g, 4°C) and supernatant divided into aliquots for limited proteolysis, direct IP of CFTR, and immunoblotting. PMSF (1 mM) and protease inhibitors (chymostatin, leupeptin, antipain, pepstatin A [MilliporeSigma; 0.01 mg/mL each]) were added to the latter 2 aliquots.
Limited proteolysis.
Lysates were subjected to limited proteolysis with Proteinase K (25 μg/mL; MilliporeSigma), as previously described (7, 25). Following incubation (15 minutes, 4°C), proteolysis was arrested by PMSF (2.5 mM).
IP and SDS-PAGE.
CFTR was immunoprecipitated from cell lysates (150 mM NaCl, 1% NP-40, 50 mM Tris-HCl pH 8.0) using polyclonal Mr. Pink, whereas protease-resistant fragments were immunoprecipitated from proteolyzed lysates using TMD1-C (for TMD1 fragments), Mr. Pink (for NBD1 fragments), TMD2-C (for TMD2 fragments), and UNC596 (for NBD2 fragments). Lysates were transferred to protein A Sepharose beads (GE Healthcare) preincubated with antisera (15 minutes, 4°C). After 3-hour IP, complexes were washed twice (20 minutes, room temperature [RT]) in the following buffers: TMD1-C in 10 mM Tris-HCl (pH 8.6), 300 mM NaCl, 0.05% SDS, and 0.05% Tx100; Mr. Pink in 10 mM Tris-HCl (pH 8.6), 300 mM NaCl, 0.1% SDS, and 0.05% Tx100; TMD2-C in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM EDTA; and UNC596 in 30 mM Tris-HCl (pH 7.5), 20 mM MES, 100 mM NaCl, and 0.5% Tx100. Washed pellets were resuspended in 10 mM Tris-HCl (pH 6.8) with 1 mM EDTA.
Sample loading buffer was added to a final concentration of 200 mM Tris-HCl (pH 6.8), 3% SDS, 10% glycerol, 0.004% bromophenol blue, 1 mM EDTA, and 25 mM DTT. Equal amounts of protein from each sample — determined by BCA assay (Thermo Fisher) — were heated (5 minutes, 55°C) and run through SDS-PAGE (e.g., 7.5% for full-length CFTR, 12% for CFTR fragments, 15% for RPL12). Gels were dried and exposed to film (Carestream Biomax MR), Phosphorimager (GE Healthcare), or transferred onto PVDF membranes (GE Healthcare) for immunoblotting (see below). Radioactive band intensities were quantified using Phosphorimager with ImageQuant software (Molecular Dynamics).
Immunoblotting.
FRT and HBE cells were lysed with native lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris-HCl, pH 8.0) combined with 1% protease inhibitor (Halt; Thermo Fisher). Mouse tissues were homogenized in 1× PBS on ice followed by lysis in RIPA (Thermo Fisher) with 1% protease inhibitor. Total protein was quantified using BCA. Equal amounts of protein (cells, 30 μg; tissues, 60 μg) were mixed with 4× loading buffer, heated (10 minutes, 37°C), resolved by SDS-PAGE, and transferred onto PVDF. Membranes were blocked (5% milk, 30 minutes, RT), incubated with primary antibody (2 hours, RT), washed 3 times (1× PBS-T, 10 minutes each, RT), blocked with goat serum (1:1,000 in 5% milk, 30 minutes, RT), bound by secondary antibody (1 hour, RT), and followed by 3 washes as above.
For FRT, HBE, and mouse tissues, SuperSignal West Femto or West Pico (Thermo Fisher) substrates were employed. In CFBE samples prepared by IP, signals were detected using Clarity Western ECL substrate (Bio-Rad). All samples were quantified on a ChemiDocXRS (Bio-Rad) (3). Antibodies used to visualize CFTR in CFBE were UNC570 (1:2,500; primary) and goat anti-mouse HRP (catalog 1706516, Bio-Rad) (1:5,000; secondary), whereas UNC570 and UNC596 (1:5,000 each; primary) with goat anti-mouse HRP (catalog P0447, Agilent) (1:10,000; secondary) were used to detect CFTR in FRT and HBE cells. In mouse tissues, Cftr was probed with rabbit anti-CFTR antibody (catalog ab59394, Abcam) (1:1,000; primary) and goat anti-rabbit HRP (catalog 31460, Thermo Fisher) (1:5,000; secondary). Human and murine RPL12 were detected using rabbit anti-RPL12 antibody (catalog ab157130, Abcam) (1:5,000; primary) and either 31460 (1:10,000; secondary) or goat anti-rabbit HRP (catalog 1706515, Bio-Rad) (1:5,000; secondary). Mouse anti-actin (catalog AC-15, MilliporeSigma) (1:5,000; primary) and P0447 (1:10,000; secondary) were employed for Western blotting of β-actin, and α-tubulin was detected using α-tubulin mouse monoclonal (catalog 302211, Synaptic Systems) (1:10,000; primary) and goat anti-rabbit HRP (catalog 1706515, Bio-Rad) (1:5,000; secondary). Note that terms such as “biogenesis” or “band C production” are applied to designate augmented levels of mature CFTR. “Maturational efficiency” denotes specific enhancement of band B to band C conversion, as indicated by pulse-chase (or in some cases, an increase in the ratio of band C to bands C+B).
HRP cell-surface assay.
FRT cells expressing F508del-, F508del-R555K-, or F508del-R1070W-CFTR cDNA with HRP conjugated to extracellular loop 4 (ECL4) of the CFTR polypeptide (generated as above) were used to test CFTR PM density (27). HRP-based fluorescence was normalized to protein content (BCA assay).
ISC measurement.
CFBE, FRT, or HBE cells were cultured on permeable filters at air-liquid interface and mounted in P2300 Ussing chambers (MC8 apparatus, Physiologic Instruments), as previously described (3, 48). Briefly, cells were equilibrated (5 minutes) in regular Ringer’s solution, followed by exchange to low-chloride Ringer’s solution on the apical surface. After addition of amiloride (100 μM; sodium channel inhibitor), CFTR agonists were sequentially added: forskolin (5 μM; activator of PKA) and VX-770 (5 μM; potentiator of CFTR gating) or genistein (50 μM; potentiator). Each experiment concluded with addition of the CFTR-specific Inhibitor172 (Inh172) (MilliporeSigma) (10 μM). Changes in CFTR-dependent transepithelial ion transport were calculated using the highest value of a stable current plateau achieved for several minutes.
For studies of CFTR surface localization at 37°C, a similar protocol was employed with the following modifications. Each experiment was initiated at 27°C, and CFTR-dependent ion transport was stimulated with forskolin. Following ISC stabilization (5 minutes), bath temperature was increased step-wise to 37°C. Once the inner chamber temperature reached 37°C, CFTR activity was measured for 15 minutes or until the current reached less than half the original baseline value.
Generation of Rpl12+/– mouse and breeding.
Pretargeted C57BL/6 embryonic stem cells carrying a loxP-flanked Rpl12 “knockout-first” allele with lacZ-tagged insertion and neo-driven selection cassette (Rpl12tm1a(EUCOMM)Hmgu) were purchased from the European Conditional Mouse Mutagenesis (EUCOMM) program (Helmholtz Zentrum München, Neuherberg, Germany) (Supplemental Figure 5). This construct was injected into blastocysts to generate chimeric founders, which were then crossed to WT C57BL/6 mice (catalog 000664, Jackson Laboratory). In subsequent offspring, germline transmission of the modified Rpl12 allele was validated by PCR performed on tail biopsies as previously described (54).
All animals were congenic C57BL/6 and maintained in temperature- and humidity-controlled vivaria (12-hour light/ 12-hour dark cycle; water and food ad libitum). Routine health monitoring indicated no evidence of pathogenic infection. All heterozygous Rpl12 mice were viable, fertile, exhibited normal growth characteristics, and did not display abnormal embryonic or postnatal phenotypes. Equal numbers of female and male animals (4 months old) were analyzed in this study. Rpl12+/+ littermates were utilized as controls.
Molecular graphics and data deposition.
Molecular graphics for Figure 1 and Supplemental Figures 1, 6, and 7 were performed with the UCSF Chimera package (55), the GNU Image Manipulation Program (56), and freely accessible vectors from the Library of Science and Medical Illustrations (57). Sequencing data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE74365 and GSE104329).
Statistics.
Unless stated otherwise, statistical analysis was performed by 2-tailed Student’s t test with a mean of at least 3 independent experiments. A 95% confidence interval (P ≤ 0.05) was considered significant. When comparing more than 2 groups within a given time point, we applied post hoc Bonferroni’s correction, with significance level adjusted to α = 0.05/number of pair-wise comparisons.
Study approval.
Primary HBE from a CF patient with CFTRF508del/F508del genotype were obtained under a protocol and consent form approved by the Institutional Review Board at UAB (X080625002). Mouse experiments were conducted in the Transgenic and Genetically Engineered Model Systems Core at UAB under a protocol approved by the Institutional Animal Use and Care Committee (140409618).
Author contributions
KEO, RR, MM, RAK, KLK, AR, IB, JSH, JLH, ZI, and EJS designed studies. KEO, RR, MM, WW, MJW, and JM conducted experiments. KEO, RR, MM, WW, JM, CMS, RAK, IB, JLH, ZI, and EJS acquired and analyzed data. KEO, RR, MM, IB, JLH, ZI, and EJS wrote the manuscript.
Supplementary Material
Acknowledgments
We thank David Bedwell (UAB) and David Schneider (UAB) for helpful discussions and technical guidance. Funding was provided by the NIH (F31HL131231, R01HL136414, P30DK072482), the Cystic Fibrosis Foundation (OLIVER17F0, BRAAKM14XXO, HARTMA15G0, IGNATO17XXO, SORSCH13XXO, SORSCH14XXO), the Burroughs Wellcome Fund (Collaborative Research Travel Grant, 1018774), Deutsche Forschungsgemeinschaft FOR1805 (IG74/14-1, IG74/14-2), the German Cystic Fibrosis Foundation (muko e.V. 1603), and the Netherlands Science Organization (graduate school program grant).
Version 1. 10/28/2019
Electronic publication
Version 2. 12/02/2019
Print issue publication
Footnotes
Conflict of interest: JLH is founder of and holds equity in Spectrum PhenomX LLC, which aims to commercialize quantitative high-throughput cell array phenotyping (Q-HTCP) technology.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(12):5236–5253.https://doi.org/10.1172/JCI124282.
Contributor Information
Kathryn E. Oliver, Email: kolive3@emory.edu.
Robert Rauscher, Email: robert.rauscher.bio@gmail.com.
Marjolein Mijnders, Email: marjolein.mijnders@gmail.com.
Wei Wang, Email: weiwang@uab.edu.
Jessica Maya, Email: jmaya2895@gmail.com.
Kevin L. Kirk, Email: 24kirkhawk@gmail.com.
Andras Rab, Email: andras.rab@emory.edu.
Ineke Braakman, Email: i.braakman@uu.nl.
Jeong S. Hong, Email: jeong.s.hong@emory.edu.
John L. Hartman, IV, Email: jhartman@uab.edu.
Zoya Ignatova, Email: zoya.ignatova@chemie.uni-hamburg.de.
References
- 1.Cheng SH, et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63(4):827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 2.Veit G, et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell. 2016;27(3):424–433. doi: 10.1091/mbc.e14-04-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veit G, et al. Ribosomal stalk protein silencing partially corrects the ΔF508-CFTR functional expression defect. PLoS Biol. 2016;14(5):e1002462. doi: 10.1371/journal.pbio.1002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497(7447):80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 5.Ito K, et al. Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1α. Nucleic Acids Res. 2014;42(22):14042–14052. doi: 10.1093/nar/gku1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23(5):1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20(2):277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Kim SJ, et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348(6233):444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 9.Kirchner S, et al. Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 2017;15(5):e2000779. doi: 10.1371/journal.pbio.2000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartoszewski RA, et al. A synonymous single nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem. 2010;285(37):28741–28748. doi: 10.1074/jbc.M110.154575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louie RJ, et al. A yeast phenomic model for the gene interaction network modulating CFTR-ΔF508 protein biogenesis. Genome Med. 2012;4(12):103. doi: 10.1186/gm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wawiórka L, et al. Functional analysis of the uL11 protein impact on translational machinery. Cell Cycle. 2016;15(8):1060–1072. doi: 10.1080/15384101.2016.1154245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15(3):205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 14.Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. Measurement of average decoding rates of the 61 sense codons in vivo. Elife. 2014;3:e03735. doi: 10.7554/eLife.03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol. 2011;18(11):1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PubMed] [Google Scholar]
- 17.Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106(1):50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson M, et al. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci USA. 2011;108(1):79–84. doi: 10.1073/pnas.1012612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingold H, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2(12):e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimura R, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345(6195):455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denning GM, Anderson MP, Amara JF, Marshall J, Smith AE, Welsh MJ. Processing of mutant cystic fibrosis transmembrane conductance regulator is temperature-sensitive. Nature. 1992;358(6389):761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Okeyo GO, Tao B, Hong JS, Kirk KL. Thermally unstable gating of the most common cystic fibrosis mutant channel (ΔF508): “rescue” by suppressor mutations in nucleotide binding domain 1 and by constitutive mutations in the cytosolic loops. J Biol Chem. 2011;286(49):41937–41948. doi: 10.1074/jbc.M111.296061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Roessler BC, Kirk KL. An electrostatic interaction at the tetrahelix bundle promotes phosphorylation-dependent cystic fibrosis transmembrane conductance regulator (CFTR) channel opening. J Biol Chem. 2014;289(44):30364–30378. doi: 10.1074/jbc.M114.595710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoelen H, et al. The primary folding defect and rescue of ΔF508 CFTR emerge during translation of the mutant domain. PLoS ONE. 2010;5(11):e15458. doi: 10.1371/journal.pone.0015458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mijnders M, Kleizen B, Braakman I. Correcting CFTR folding defects by small-molecule correctors to cure cystic fibrosis. Curr Opin Pharmacol. 2017;34:83–90. doi: 10.1016/j.coph.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Mutyam V, et al. Discovery of clinically approved agents that promote suppression of cystic fibrosis transmembrane conductance regulator nonsense mutations. Am J Respir Crit Care Med. 2016;194(9):1092–1103. doi: 10.1164/rccm.201601-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabusap CM, et al. Analysis of cystic fibrosis-associated P67L CFTR illustrates barriers to personalized therapeutics for orphan diseases. JCI Insight. 2016;1(14):e86581. doi: 10.1172/jci.insight.86581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendoza JL, et al. Requirements for efficient correction of ΔF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148(1-2):164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung WJ, et al. Increasing the endoplasmic reticulum pool of the F508del allele of the cystic fibrosis transmembrane conductance regulator leads to greater folding correction by small molecule therapeutics. PLoS ONE. 2016;11(10):e0163615. doi: 10.1371/journal.pone.0163615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molinski SV, et al. Orkambi and amplifier co-therapy improves function from a rare CFTR mutation in gene-edited cells and patient tissue. EMBO Mol Med. 2017;9(9):1224–1243. doi: 10.15252/emmm.201607137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16(3):274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 33.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16(6):589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334(6063):1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okiyoneda T, et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329(5993):805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meriin AB, et al. A novel approach to recovery of function of mutant proteins by slowing down translation. J Biol Chem. 2012;287(41):34264–34272. doi: 10.1074/jbc.M112.397307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong X, Bragin A, Widdicombe JH, Cohn J, Skach WR. Structural cues involved in endoplasmic reticulum degradation of G85E and G91R mutant cystic fibrosis transmembrane conductance regulator. J Clin Invest. 1997;100(5):1079–1088. doi: 10.1172/JCI119618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loo TW, Clarke DM. Corrector VX-809 promotes interactions between cytoplasmic loop one and the first nucleotide-binding domain of CFTR. Biochem Pharmacol. 2017;136:24–31. doi: 10.1016/j.bcp.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 41.Cuthbert AW, Halstead J, Ratcliff R, Colledge WH, Evans MJ. The genetic advantage hypothesis in cystic fibrosis heterozygotes: a murine study. J Physiol (Lond) 1995;482(Pt 2):449–454. doi: 10.1113/jphysiol.1995.sp020531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilschanski M, et al. Mutations in the cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. Am J Respir Crit Care Med. 2006;174(7):787–794. doi: 10.1164/rccm.200509-1377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oristian DS, Sloofman LG, Zhou X, Wang L, Farach-Carson MC, Kirn-Safran CB. Ribosomal protein L29/HIP deficiency delays osteogenesis and increases fragility of adult bone in mice. J Orthop Res. 2009;27(1):28–35. doi: 10.1002/jor.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sosnay PR, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45(10):1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliver KE, Han ST, Sorscher EJ, Cutting GR. Transformative therapies for rare CFTR missense alleles. Curr Opin Pharmacol. 2017;34:76–82. doi: 10.1016/j.coph.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bebok Z, et al. Failure of cAMP agonists to activate rescued deltaF508 CFTR in CFBE41o- airway epithelial monolayers. J Physiol (Lond) 2005;569(Pt 2):601–615. doi: 10.1113/jphysiol.2005.096669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Goor F, et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci USA. 2009;106(44):18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramachandran S, et al. Efficient delivery of RNA interference oligonucleotides to polarized airway epithelia in vitro. Am J Physiol Lung Cell Mol Physiol. 2013;305(1):L23–L32. doi: 10.1152/ajplung.00426.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 2016;14(4):686–694. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 53.Wang W, Hong JS, Rab A, Sorscher EJ, Kirk KL. Robust stimulation of W1282X-CFTR channel activity by a combination of allosteric modulators. PLoS ONE. 2016;11(3):e0152232. doi: 10.1371/journal.pone.0152232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Connor AK, Kesterson RA, Yoder BK. Generating conditional mutants to analyze ciliary functions: the use of Cre-lox technology to disrupt cilia in specific organs. Methods Cell Biol. 2009;93:305–330. doi: 10.1016/S0091-679X(08)93015-6. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56. GNU image manipulation program (GIMP), v. 2.8.22. The GIMP Team Web Site. https://www.gimp.org Updated May 11, 2017. Accessed June 30, 2017.
- 57. Library of Science and Medical Illustrations. Somersault18:24 Web Site. http://www.somersault1824.com/science-illustrations Updated January 20, 2017. Accessed June 30, 2017.
- 58.Kopeikin Z, Yuksek Z, Yang HY, Bompadre SG. Combined effects of VX-770 and VX-809 on several functional abnormalities of F508del-CFTR channels. J Cyst Fibros. 2014;13(5):508–514. doi: 10.1016/j.jcf.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 59.van Willigen M, et al. Folding-function relationship of the most common cystic fibrosis-causing CFTR conductance mutants. Life Sci Alliance. 2019;2(1):e201800172. doi: 10.26508/lsa.201800172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Semaniakou A, Croll RP, Chappe V. Animal models in the pathophysiology of cystic fibrosis. Front Pharmacol. 2018;9:1475. doi: 10.3389/fphar.2018.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilke M, et al. Mouse models of cystic fibrosis: phenotypic analysis and research applications. J Cyst Fibros. 2011;10 Suppl 2:S152–S171. doi: 10.1016/S1569-1993(11)60020-9. [DOI] [PubMed] [Google Scholar]
- 62.Li D, et al. Tanshinone IIA sulfonate protects against cigarette smoke-induced COPD and down-regulation of CFTR in mice. Sci Rep. 2018;8(1):376. doi: 10.1038/s41598-017-18745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noel S, Panin N, Beka M, Dhooghe B, Huaux F, Leal T. Vardenafil reduces macrophage pro-inflammatory overresponses in cystic fibrosis through PDE5- and CFTR-dependent mechanisms. Clin Sci. 2017;131(11):1107–1121. doi: 10.1042/CS20160749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.