Abstract
The transcription factor B cell CLL/lymphoma 11B (BCL11B) is indispensable for T lineage development of lymphoid progenitors. Here, we show that chimeric antigen receptor (CAR) expression during early phases of ex vivo generation of lymphoid progenitors suppressed BCL11B, leading to suppression of T cell–associated gene expression and acquisition of NK cell–like properties. Upon adoptive transfer into hematopoietic stem cell transplant recipients, CAR-expressing lymphoid progenitors differentiated into CAR-induced killer (CARiK) cells that mediated potent antigen-directed antileukemic activity even across MHC barriers. CD28 and active immunoreceptor tyrosine–based activation motifs were critical for a functional CARiK phenotype. These results give important insights into differentiation of murine and human lymphoid progenitors driven by synthetic CAR transgene expression and encourage further evaluation of ex vivo–generated CARiK cells for targeted immunotherapy.
Keywords: Immunology, Transplantation
Keywords: Immunotherapy, Leukemias, T cell development
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) currently represents the best curative treatment option for very high–risk leukemias. However, relapse after HSCT remains the major cause for treatment failure. Redirecting T cell responses more specifically to tumor-associated antigens by T cell receptor (TCR) (1–3) or, more recently, chimeric antigen receptor (CAR) engineering has produced impressive clinical results (4–7). Nevertheless, important problems remain or have been underestimated using engineered T cells. This includes (a) that the majority of successful clinical trials using genetic receptor transfer technologies have been conducted with autologous T cells, which requires the collection of a T cell product of sufficient quantity and quality from heavily pretreated patients, (b) the need to manufacture the product on a highly individualized basis, resulting in treatment delays due to the scarcity of production slots, and (c) on-target/off-tumor effects confining receptor engineering to a very narrow choice of suitable target antigens. Thus, a prefabricated T cell product allowing for a wider choice of effectively targetable antigens, being applicable to a wider range of patients, and minimizing the risk of long-term sequela from on-target/off-tumor effects would be highly desirable. In vitro predifferentiated lymphoid progenitors from hematopoietic stem and progenitor cells (HSPCs) that undergo final maturation upon adoptive transfer (AT) in the recipient can be transplanted across major MHC barriers without triggering graft-versus-host disease (GVHD). They give rise to a functional T cell population being both tolerant and MHC restricted to the host even in MHC-mismatched recipients (8–10). TCR-engineered lymphoid precursors with a host-restricted TCR lead to rapid thymic repopulation, and their progeny can mediate potent and long-lasting antileukemia effects (11).
Here, in order to circumvent the limitation of TCR restriction, we used CAR-engineered hematopoietic progenitor cells to generate lymphoid precursors in vitro using the Notch-based OP9-DL1 culture system. Elegant proof-of-principle studies suggested the relevance of this concept for humans; however, exploration in vivo remains complex (12–14). Using a murine and human CD19 CAR (15), we targeted a clinically relevant antigen and were able to assess its immunological impact on lymphoid progenitor development and demonstrate strong evidence for its translational relevance for humans.
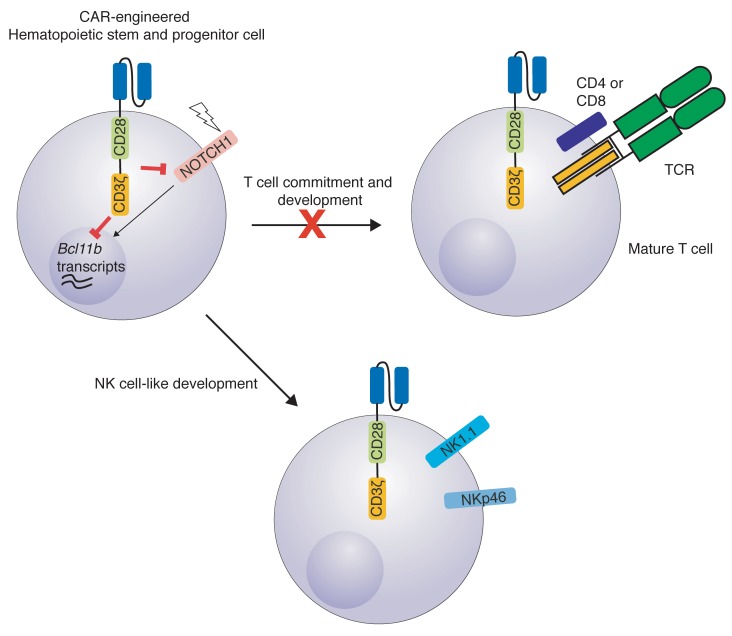
We show that CAR expression during early lymphoid differentiation can affect lymphoid progenitor fate profoundly via suppression of B cell CLL/lymphoma 11B (BCL11B). Bcl11b and Notch1 transcripts are both indispensable for T cell development, both in mice and humans. As a result, T cell development was blocked in favor of a cell population acquiring NK cell–like properties. We termed this cell type CAR-induced killer (CARiK) cells. CARiK cells mediated strong antileukemic effects even across MHC barriers without evoking GVHD. We further demonstrate that this differentiation shift depends on the costimulatory domain and the activity of immune receptor–based activation motifs (ITAMs) used within the CAR construct. Using CAR-engineered hematopoietic stem cells that had been isolated from human umbilical cord blood (UCB), we further show CAR-induced suppression of T cell differentiation in favor of CARiK cell development. These findings encourage efforts to further address the potential of CARiK cells as a cellular product of broader applicability for anticancer immunotherapy.
Results
im1928z1-CAR expression in HSPCs prevents T cell but favors NK-like cell development of lymphoid progenitors in vitro and in vivo.
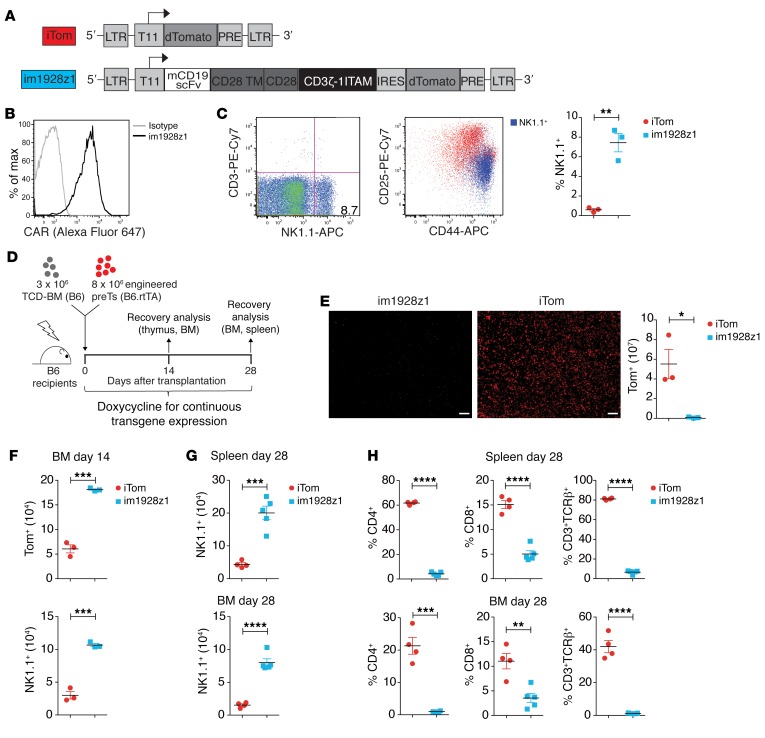
HSPCs transduced with a host HLA-restricted TCR and differentiated into lymphoid progenitors of the T cell lineage have been shown to mediate potent antileukemic activity upon cotransplantation with T cell–depleted BM (TCD-BM) (11). To evaluate the biological consequences of CAR expression in differentiating lymphoid progenitors both in vitro and in vivo, we cloned a previously published murine second-generation CAR directed against mouse CD19 containing a CD28 costimulatory domain and 1 functional ITAM within the CD3ζ signaling domain, termed im1928z1 (Figure 1A, ref. 15, and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI126350DS1). CAR expression was set under the control of a tetracycline-inducible (Tet-On) T11 promoter to enable studying of the impact of time-dependent CAR expression (11, 16). For inducible transgene expression, murine BM-derived Lineage–Sca-1+c-Kit+ (LSK) cells with an rtTA-M2 transactivator knockin were used. The Tet-On system was induced continuously for transgene expression during in vitro and in vivo experiments from the very early beginning unless noted otherwise. Lymphoid progenitors were generated from transduced LSKs using the OP9-DL1 coculture system (Supplemental Figure 1B and ref. 17). In contrast to previously published TCR-engineered lymphoid progenitors, the im1928z1 CAR was highly expressed on generated lymphoid progenitors in vitro (Figure 1B). Cells for AT studies were at least 90% transgene positive, and 50%–60% were at the double-negative (DN) 2 stage (CD25+CD44+/CD4–CD8–) (Figure 1C and Supplemental Figure 1C). Although the OP9-DL1 coculture system is known to allow for limited NK cell development (17), we identified increased frequencies of NK1.1+ cells (mean = 7.4%) with a CD25midCD44+ phenotype within the im1928z1 group. This compared with around 0.6% NK1.1+ cells for controls (Figure 1C).
Figure 1. im1928z1-CAR expression in HSPCs cells prevents T cell, but favors NK-like cell development of lymphoid progenitors in vitro and in vivo.
(A) The lentiviral control and the murine CD19 CAR construct: iTom (inducible dTomato reporter gene only) and im1928z1 (inducible murine CD19 CAR, CD28 costimulation, 1 functional ITAM containing CD3ζ domain) linked to an IRES dTomato cassette. LTR, long terminal repeats; T11, Dox-inducible promotor; scFv, single chain variable fragment; TM, transmembrane domain; IRES, internal ribosome entry site; PRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) Representative data showing im1928z1 expression on in vitro–generated lymphoid progenitors. (C) Representative FACS plots of NK1.1 and CD3 expression on in vitro–generated im1928z1-engineered lymphoid progenitors (left), NK1.1+ population within CD25+CD44+ lymphoid progenitors (middle), and NK1.1+ expression on iTom and im1928z1-transduced lymphoid progenitors before cotransplantation (right) (n = 3 independent cultures were pooled). (D) Irradiated B6 recipients were reconstituted with 3 × 106 B6 TCD-BM and cotransplanted with either 8 × 106 im1928z1-engineered lymphoid progenitors or iTom‑engineered lymphoid progenitors. (E) Thymic sections were imaged for Tom+ cells. Scale bars: 50 μm; Original magnification, × 20. Single cells from harvested thymi were analyzed by FACS for Tom+ progeny of cotransplanted lymphoid progenitors (n = 3 mice, respectively). (F) Lymphoid progenitor–derived progeny in the BM on day 14 (top). Numbers of NK1.1+ cells within the Tom+ population are depicted (bottom) (n = 3 mice per group). (G) Numbers of NK1.1+ and (H) frequencies of CD4+, CD8+, and CD3+TCRβ+ progeny within the Tom+ gate in BM and spleens on day 28 (im1928z1, n = 5; iTom, n = 4). Results from 1 of 2 independent experiments are shown. Statistics was performed using Student’s t test (2 tailed). Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To track the development of CAR-expressing lymphoid progenitors in vivo, irradiated syngeneic C57BL/6 (B6) recipients were transplanted with 3 × 106 TCD-BM cells and adoptively transferred with 8 × 106 im1928z1-engineered lymphoid progenitors (Figure 1D and Supplemental Figure 1D). Cotransplanted lymphoid progenitors have been shown to foster early repopulation of the thymus (8, 11). The im1928z1-generated lymphoid progenitors did, however, completely fail to repopulate the thymus (Figure 1E). Importantly, AT of im1928z1-expressing lymphoid progenitors resulted later in greater numbers of NK1.1+ progeny in BM and spleen, suggesting accentuated NK cell–like development (Figure 1, F and G). In contrast to the iTom controls, im1928z1-generated lymphoid progenitors sparsely matured into CD4+ or CD8+ single-positive (SP) T cells or expressed the CD3/TCR complex (Figure 1H). Therefore, im1928z1 expression suggests T lineage development from early hematopoietic precursors in favor of a population with an NK cell–resembling phenotype.
CARiK cells derived from im1928z1-engineered lymphoid progenitors demonstrate potent antileukemic activity across MHC barriers in vivo.
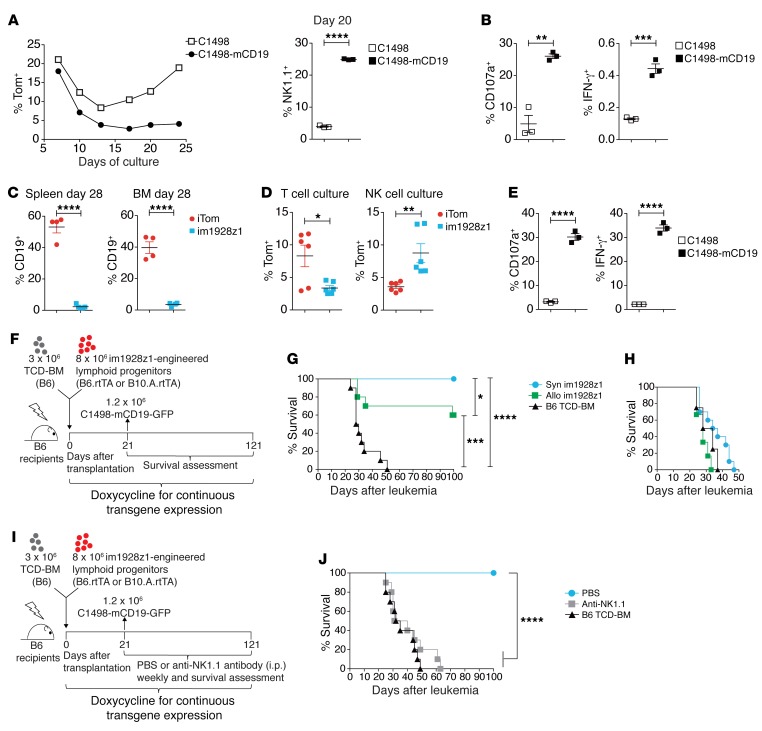
In contrast to TCR expression initiated later during physiologic thymic T cell development, CARs started and continued to be expressed immediately after LSK transduction (14). Therefore, it was intriguing to assess the impact of CAR triggering during the generation process of im1928z1-generated lymphoid progenitors. Early antigen exposure of CAR-transduced LSKs during in vitro differentiation slowed cell expansion and resulted in an increased NK1.1+CD25midCD44+ population (Figure 2A and Figure 1C). In vitro stimulation of im1928z1-generated lymphoid progenitors with CD19-expressing target cells caused prompt degranulation, as seen by expression of CD107a and an IFN-γ response (Figure 2B) suggesting antigen specificity. Next, we assessed recovery of CD19+ B cells after HSCT in cotransplanted recipients to evaluate the activity of matured im1928z1-generated lymphoid progenitors in vivo. We could document induction of profound B cell aplasia following cotransplantation (Figure 2C). CAR-expressing progeny could be recovered from the mouse and further maintained in ex vivo cultures which required NK cell–like high IL-2 doses (1000 U/mL) in contrast to the standard T cell culture conditions (20 U/mL IL-2) (Figure 2D). Upon stimulation with CD19, CD107a expression and copious IFN-γ secretion again demonstrated specific reactivity of this im1928z1-expressing progeny (Figure 2E).
Figure 2. CARiK cells derived from im1928z1-engineered lymphoid progenitors demonstrate potent antileukemic activity across MHC barriers in vivo.
(A) Generation of either stimulated or nonstimulated im1928z1‑generated lymphoid progenitors. Frequencies of Tom+ progenitors (left) and NK1.1+ im1928z1-CARiK cells on day 20 of culture (right). (B) Responses of im1928z1-generated lymphoid progenitors upon stimulation were quantified via CD107a degranulation (left) or IFN-γ production (right). Data from 1 of 2 experiments are shown. (C) CD19+ B cell recovery of irradiated B6 recipients of B6 TCD-BM and either im1928z1-engineered progenitors or iTom controls (n = 4 mice, respectively). (D) Splenocytes were harvested on day 28 and recultured ex vivo under T cell or NK cell culture conditions (n = 6; left). (E) CD107a+ degranulation (middle) and IFN-γ (right) responses to antigen were assessed (n = 3). (A–E) Student’s t test was used for analysis. Data represent mean ± SEM. (F and G) B6 recipients of 3 × 106 B6 TCD-BM (n = 10/group) with or without 8 × 106 syngeneic (syn) or MHC class I and II mismatched (allo) im1928z1-expressing progenitors received 1.2 × 106 C1498-mCD19 cells on day 21 after transplantation and were monitored for survival. Results from 1 of 2 independent experiments are graphed. (H) Survivors were rechallenged with 1.2 × 106 C1498-mCD19 cells on day 100 and reassessed for survival. TCD-BM–only recipients (n = 4) were added for control. (I and J) B6 recipients of 3 × 106 B6 TCD-BM with or without 8 × 106 syngeneic im1928z1-engineered lymphoid progenitors were treated with weekly i.p. injections of an anti-NK1.1 antibody (clone: PK136; 200 μg/dose). PBS was used for control (n = 10 per group). All mice were challenged with 1.2 × 106 C1498-mCD19 cells on day 21 after transplantation (J). Survival curves were compared using Mantel-Cox (log-rank) test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We next studied the antileukemia potential of “off-the-shelf” CAR lymphoid progenitors in a mouse model of post-HSCT relapse (Figure 2F). Recipients of syngeneic B6 TCD-BM, cotransplanted with either syngeneic (B6) or completely MHC mismatched (B10.A-mimicking off-the-shelf) im1928z1-generated lymphoid progenitors, were challenged with a lethal dose of mCD19+ leukemic cells (C1498-mCD19-GFP). Both groups showed a significant survival advantage over controls (Figure 2G). Consistent with prior reports, GVHD was never observed after cotransplantation with MHC-mismatched lymphoid progenitors (data not shown) (9). A second leukemia challenge of 100-day survivors resulted in 100% lethality rate of the recipients, suggesting the absence of long-term activity or functionally relevant memory cell formation (Figure 2H). Depletion of NK1.1+ cells in transplant recipients using a NK1.1-depleting antibody led to a complete loss of antileukemia effects (Figure 2, I and J), demonstrating that the CAR-induced NK1.1+ cells mediate the observed potent antileukemic effects. Due to NK cell–resembling phenotype and functionality of im1928z1-induced NK1.1+ cells, we will further refer to this cell population as CARiK cells.
im1928z1 Expression on HSPCs leads to BCL11B suppression, allowing for CARiK cell development, and concomitantly decreases T cell–associated gene expression.
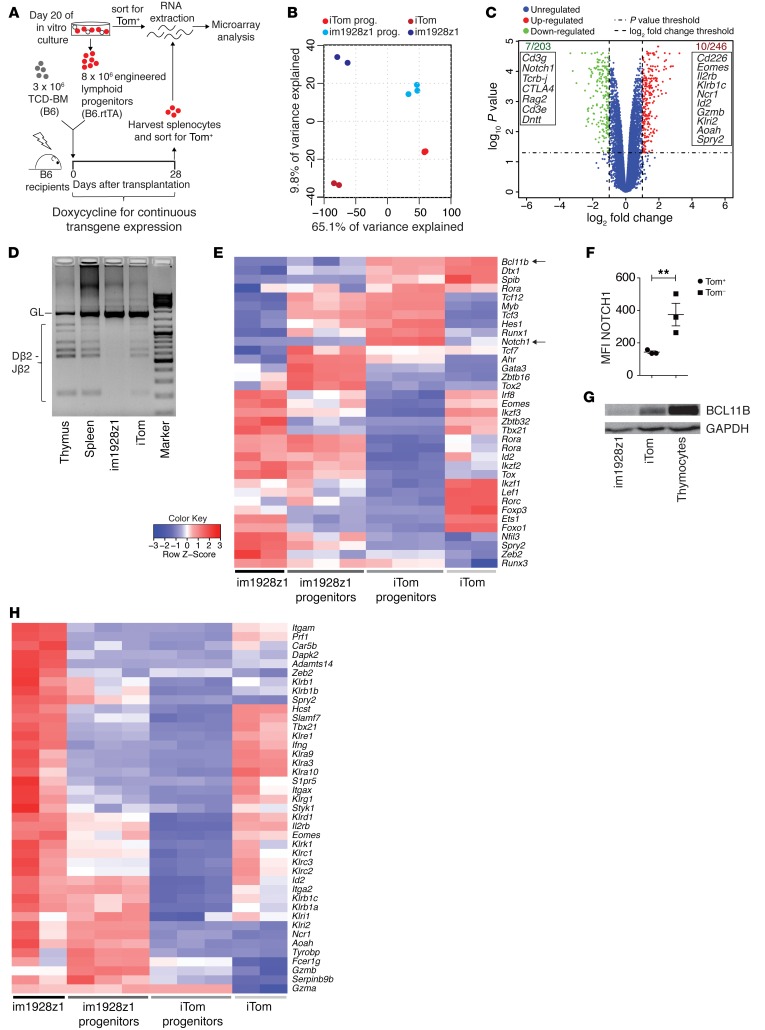
To further substantiate CAR-induced differentiation of early lymphoid progenitors, we asked whether the observed phenotypic and functional data would find an equivalent on the transcriptional level (18, 19). Therefore, we analyzed the gene expression profiles of engineered lymphoid progenitors at the end of in vitro culture immediately prior to cotransplantation and of their progeny after in vivo differentiation 28 days after cotransplantation. (Figure 3A). Principal component analysis (PCA) showed a distinct transcriptional profile of im1928z1-generated lymphoid progenitors and CARiK cells when compared with respective controls (Figure 3B). These differences became more prominent during further in vivo development. Altogether, 449 differently expressed genes were identified in im1928z1-generated lymphoid progenitors. Genes related to the TCR/CD3 complex and enzymes involved in TCR rearrangement (Rag2, Dntt) did show decreased transcriptional activity in im1928z1-generated lymphoid progenitors (Figure 3C). This was accompanied by a complete lack of detectable diversity-joining (D-J) recombination segments within the Tcrb locus (Figure 3D), suggesting the absence of TCR rearrangement in im1928z1-engineered CARiK cells. Complementing the observed phenotype transcripts known to be essential for T cell identity (e.g., Cd8a, Cd8b1, and Cd4) were markedly decreased in lymphoid progenitor–derived im1928z1-CARiK cells (Supplemental Figure 2).
Figure 3. im1928z1 expression on HSPCs leads to BCL11B suppression, allowing for CARiK cell development, and concomitantly decreases T cell–associated gene expression.
(A) For microarray data analysis, RNA from Tom+-sorted im1928z1-generated lymphoid progenitors (n = 3) or iTom‑engineered lymphoid progenitors (n = 3) immediately previous to cotransplantation or from spleen-derived progeny (n = 2, respectively) were isolated on day 28 after transplantation. (B) PCA of total transcriptome profiles from either engineered lymphoid progenitors or their respective progeny is graphed. (C) Volcano plot for comparison of differently regulated transcripts in im1928z1-generated lymphoid progenitors and iTom controls. Gene symbols in the boxes indicate selected transcripts found to be downregulated (green) or upregulated (red) at least 2-fold (P < 0.05) in im1928z1-generated lymphoid progenitors as compared with controls. (D) Recombination of D and J regions of the TCRβ locus in engineered lymphoid progenitors. Genomic DNA of engineered progenitors was isolated on day 20 of culture, and rearrangements were detected by PCR. Splenocytes and thymocytes from WT B6 mice were used as controls. Results from 1 of 2 independent experiments are shown. GL, germ line band. (E) Heatmap showing the relative expression of transcripts for selected TFs. Data are normalized according to expression in each row. (F) NOTCH1 expression on transgene-positive (Tom+) or transgene-negative (Tom–) lymphoid progenitors engineered with im1928z1. Student’s t test was used. Data represent mean ± SEM. **P < 0.01. (G) Western blot analysis for BCL11B in lysates from iTom lymphoid progenitors, im1928z1-generated lymphoid progenitors, or B6 WT thymocytes. Representative data from 1 of 2 independent experiments are shown. (H) Relative expression of selected transcripts for NK cell receptors, integrins, adaptors, effector molecules, and TFs in engineered lymphoid progenitors and their progeny. Data are normalized according to expression in each row.
We assessed the biological consequences of CAR expression in lymphoid progenitor cells and studied transcription factors (TFs), which are known to be involved in T and NK cell development. Our whole-transcriptomic data showed that transcripts of Notch1 and Bcl11b, which are indispensable for T cell development and commitment (20–23), are decreased in im1928z1-engineered cells (Figure 3, C and E). This was reflected in flow cytometric analysis, which showed that the surface NOTCH1 expression was decreased on im1928z1-engineered lymphoid progenitors when compared with that in transgene-negative controls (Figure 3F). Concomitantly, BCL11B protein expression was substantially suppressed when compared with lysates obtained from iTom controls (Figure 3G), indicating that CAR expression on early lymphoid progenitors fundamentally intervenes at the transcription checkpoint of NK cell versus T cell development.
Concurrently, transcripts associated with NK cell–like development and functionality could be documented for im1928z1-expressing cells both in vitro and in vivo (Figure 3H and Supplemental Figure 2). This included a pronounced expression of NK cell–related developmental factors (e.g., Nfil3 and Id2) (24, 25) and the identification of transcripts, such as Car5b, Dapk2, Adamts14, Spry2, Klri2, Ncr1, Aoah, Serpinb9b, and Gzma, that have been reported to be related to NK cells (19). Our results indicate that CAR expression in differentiating lymphoid progenitors enforces expression of NK cell–like transcripts generating CARiK cells, which is paralleled by decreased activity of relevant genes and TFs being closely associated with T cell development.
Transcriptional profile analysis locates CARiK cells at the interface of T lymphocytes and NK cells.
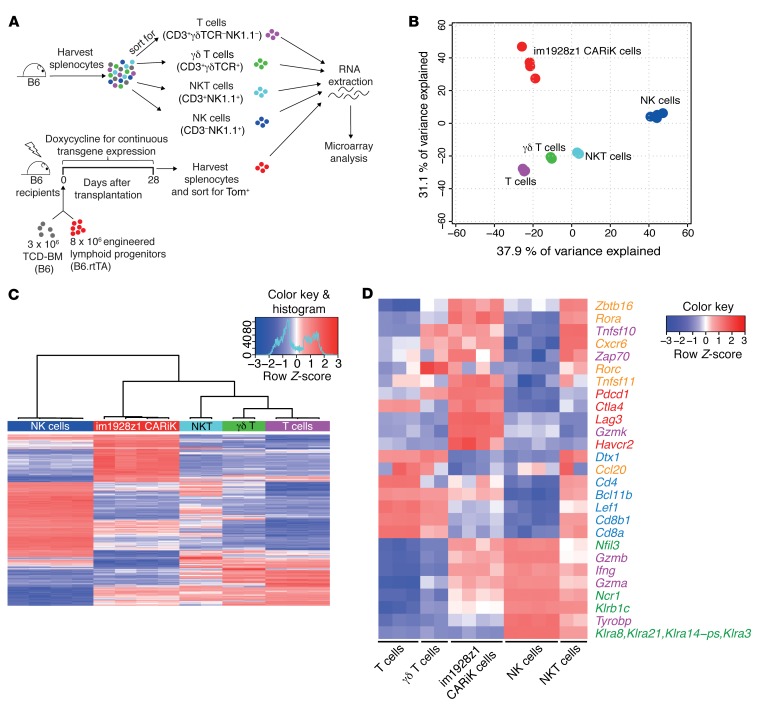
To more distinctively describe the developmental relation of CARiK cells to T and NK cells, we compared transcriptional profiles of CARiK cells that were isolated 28 days after cotransplantation to those of sorted T cells, NKT cells, γδT cells, and NK cells (Figure 4A). PCA analysis revealed a distinct localization from all other lymphoid cell populations (Figure 4B). Hierarchical clustering of the 500 genes with the most variable mRNA expression (adjusted P < 0.05) confirmed that CARiK cells were distinct from both NK cells and the other T lymphoid subsets (Figure 4C).
Figure 4. Transcriptional profile analysis locates CARiK cells at the interface of T lymphocytes and NK cells.
(A) Schematic representation of the experimental setup for transcriptional comparison of CARiK cells and different lymphoid cell populations. Splenocytes of 12-week-old WT B6 mice were harvested and sorted for T cells (CD3+γδTCR–NK1.1–; n = 3), NKT cells (CD3+NK1.1+; n = 2), γδ T cells (CD3+γδTCR+; n = 2), and NK cells (CD3–NK1.1+; n = 4). Tom+ CARiK cells (n = 4) were harvested from recipients on day 28 and consecutively sorted. Extracted RNA samples from all lymphoid subsets were compared by microarray analysis. Experiment was performed once. (B) PCA analysis of transcriptional profiles derived from the sorted lymphoid cell populations. (C) Hierarchical clustering of the 500 most differentially expressed (adjusted P < 0.05) transcripts across CARiK cells and respective lymphoid lineages. (D) Selected transcripts expressed by lymphoid subsets were color coded according to function or lymphoid cell type. Orange: γδ T cells, NKT cells, and innate lymphocytes; purple: cytotoxicity mediators; red: inhibitory receptors; blue: T lymphocytes; green: NK cells.
To further elucidate the lineage of CARiK cells, we grouped key transcripts according to function and association with distinct lymphoid cell types (ref. 26 and Figure 4D). CARiK cells demonstrated decreased expression of transcripts associated with T cell identity, such as for Cd4 and Cd8. Of note, mRNA transcript expression for Bcl11b was further decreased in CARiK cells when compared with other T lymphoid subsets, and NK cell–associated transcripts for Ncr1 and Nfil3 were substantially overexpressed. Nevertheless, CARiK cells expressed transcripts such as Zbtb16, Rorc, and Cxcr6 that are known to be associated with γδT cells, NKT cells, or innate lymphocytes. Expression strength of transcripts important for cytotoxicity mediators such as Ifng, Gzmb, and Gzma was comparable between CARiK cells, NK cells, or NKT cells. High transcriptional activity for inhibitory receptors (Pdcd1, Ctla4, Lag3, Havcr2) suggests a high activation status. Together, these data suggest a CARiK identity at the interface between NK and T cell development.
CAR expression early during lymphoid progenitor cell differentiation is required for CARiK cell generation at the expense of T cell development.
Under standard experimental conditions, CAR expression was induced early on following transduction and maintained thereafter. To assess the role of antigen exposure for further NK cell differentiation in vivo, engineered im1928z1-CARiK cells were cotransplanted into Cd19-KO mice. For these experiments, BM of Cd19-KO donors was used and cotransplanted with im1928z1-generated lymphoid progenitors into Cd19-KO mice to create a complete CD19 antigen-free environment in vivo (Supplemental Figure 3A and ref. 27). Compared with Cd19 WT recipients, similar numbers of NK1.1-expressing progeny arose in BM and spleens, suggesting that decisive signals for killer cell differentiation had been initiated by CAR expression at an early time point (Supplemental Figure 3B).
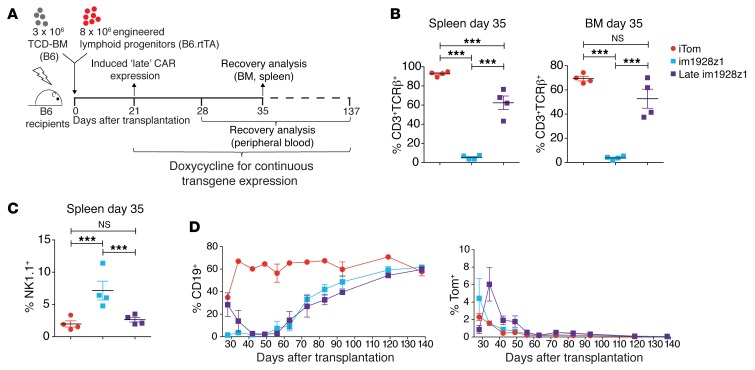
In order to evaluate the timing aspect of CAR expression in lymphoid progenitors, we compared early (day 0, standard experimental setup in Figure 1D) versus delayed (day 21) im1928z1 expression in vivo (Figure 5A and Supplemental Figure 4). In contrast to early im1928z1 expression, delayed im1928z1 induction allowed for limited T cell development, as indicated by CD3 and TCR-β expression of the respective progeny (Figure 5B). This was paralleled by reduced frequencies of NK1.1+ CARiK cells (Figure 5C). B cell suppression occurred to a similar extent in both groups, however, with delayed onset when the CAR was “switched on” late (Figure 5D). In vivo persistence was akin to that of early im1928z1-CARiK cells versus late induced CAR T cells, and reconstitution of the B cell compartment was paralleled by both progressive extinction of the lymphoid progenitor-derived CAR T cells and CARiK cells (Figure 5D). Collectively, the developmental shift of CAR-expressing lymphoid progenitors occurred early during development, leading to functional CARiK cells, whereas delayed CAR expression allowed for the generation of functional CAR T cells.
Figure 5. CAR expression early during lymphoid progenitor cell differentiation is required for CARiK cell generation at the expense of T cell development.
(A–D) Irradiated B6 recipients received 3 × 106 B6 TCD-BM and either 8 × 106 im1928z1-generated lymphoid progenitors or iTom-generated lymphoid progenitors. CAR expression was either induced early (day 0) or late (day 21) after HSCT. Indicated time points refer to the day after transplantation. (B) Frequencies of CD3+TCRβ+ cells were analyzed within the transgene-positive gate on day 35 in both the BM and spleens. (C) Comparative analysis of NK1.1+ cells in spleens of early versus late im1928z1-generated lymphoid progenitor recipients on day 35 after AT. (B and C) Each analysis was done with n = 4 mice. Gating was done on the Tom+ population. Statistics were performed using 1-way ANOVA with Tukey’s post test. Data represent mean ± SEM. ***P < 0.001. (D) CD19+ B cell recovery (left) and frequencies of Tom+ cells (right) in the PB of transplant recipients after early or late im1928z1 induction (n = 3–4 mice per group and time point). Results from 1 of 2 independent experiments are shown.
CARs containing a CD28 costimulatory domain induce killer cells with superior functionality.
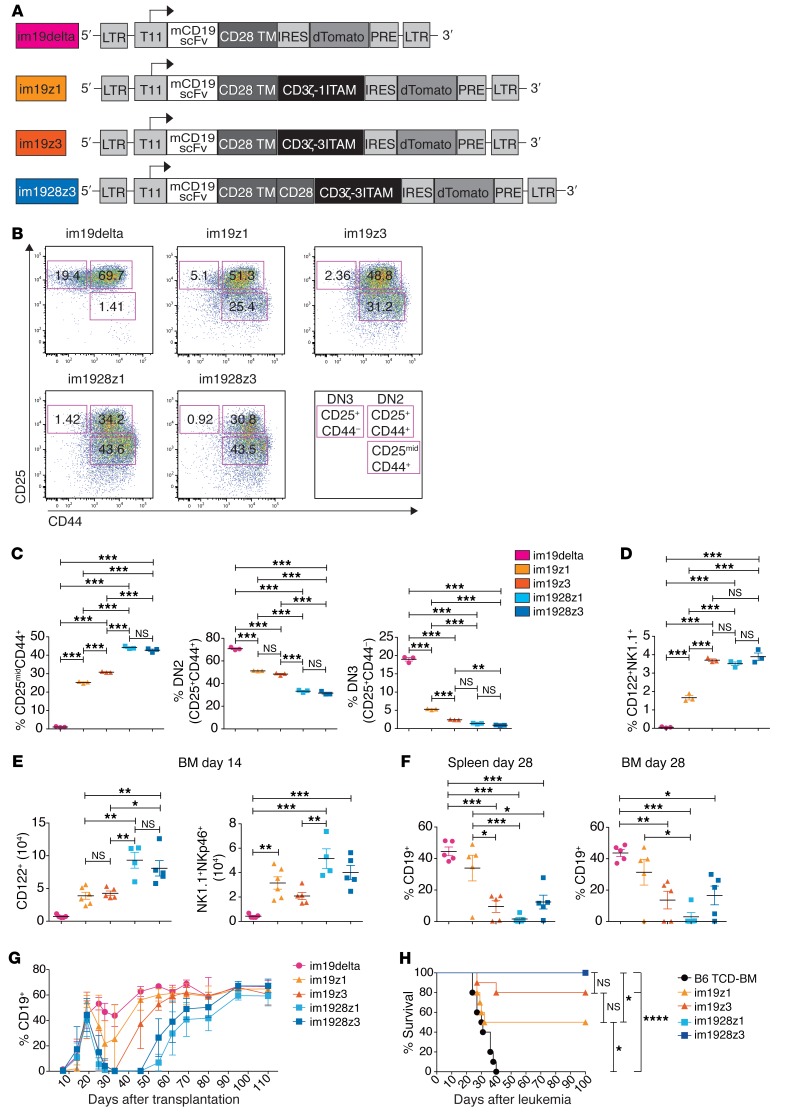
To study the impact of CAR design on lymphoid progenitor development, we generated and compared a more diverse panel of lentiviral CAR vector constructs: im19delta without an ITAM; im19z1 and im19z3 with 1 or 3 ITAMs active, both without the costimulatory domain; and im1928z1 and im1928z3 both containing a CD28 moiety and 1 or 3 active ITAMs within the CD3ζ chain (Figure 6A and Supplemental Figure 5A). CAR constructs were comparatively expressed on resulting lymphoid progenitors (Supplemental Figure 5B). Both im19delta- and iTom-transduced LSKs generated very few NK1.1+ lymphoid progenitors (Supplemental Figure 5C). The number of ITAMs and the presence of the costimulatory moiety CD28 correlated with the occurrence of a CD25midCD44+ population in vitro containing the NK1.1+ CARiK cell subset (Figure 6, B and C, and Figure 1C). The size of this population differed between CAR constructs, was more accentuated when the CAR contained the CD28 costimulatory domain, and seemed independent of 1 versus 3 ITAMs being active. This was paralleled by a significant decrease of the DN2 population and a more pronounced reduction of DN3 cells demonstrating a block of T cell development at this stage (Figure 6, B and C). In contrast, a comparable increase of the CD122+NK1.1+ fraction was observed, with the exception of the 19z1 construct, which produced a less pronounced CARiK shift (Figure 6D). While all signaling-competent CAR variants promoted the generation of CD122+ lymphoid precursors and finally a NK1.1+NKp46+ CARiK cell population in the BM after transplantation, higher numbers were derived from CARs with CD28 costimulation (Figure 6E). In addition, the use of CARiK cells expressing a CAR with CD28 costimulatory domain resulted in prolonged B cell suppression (Figure 6, F and G). This was closely mirrored by strong antileukemic activity upon cotransplantation (Figure 6H), suggesting that a CD28 costimulatory domain allows for enhanced functionality of lymphoid progenitor–derived CARiK cells.
Figure 6. CARs containing CD28 costimulatory domain induce killer cells with superior functionality.
(A) Design of the im19delta, im19z1, im19z3, and im1928z3 constructs. All CAR constructs were linked to an IRES dTomato cassette. (B and C) Representative FACS plots (B) and respective CD25midCD44+, DN2 (CD25+CD44+), and DN3 (CD25+CD44–) populations (C) of lymphoid progenitors engineered with the indicated CAR construct (color coded as indicated) on day 20 of in vitro culture. (D) Frequencies of CD122+NK1.1+ CARiK cells on day 20 of in vitro culture. Tom+ cells were analyzed. (B–D) Data from 1 of 2 independent experiments measured in triplicates are shown. (E–G) Irradiated B6 recipients were reconstituted with 3 × 106 B6 TCD-BM and cotransplanted with 8 × 106 lymphoid progenitors that had been engineered with the indicated CAR constructs. (E) BM cells were analyzed for numbers of CD122+ (left) and NK1.1+NKp46+ cells (right) within on day 14. im19delta, im19z3, and im1928z3 (n = 5 mice); im19z1 (n = 6); im1928z1 (n = 4). (F) CD19+ B cells were quantified in BM (left) and spleens (right) on day 28. n = 5 mice for each group. (G) CD19+ B cells in the peripheral blood were determined in 7- to 14-day intervals. Analysis at each time point was done on n = 4–5 mice per group. (C–F) Analysis was done using 1-way ANOVA with Tukey’s post test. Data represent mean ± SEM. (H) Irradiated B6 recipients were transplanted with 3 × 106 B6 TCD-BM only (n = 10) or additionally with 8 × 106 CAR-expressing lymphoid progenitors (n = 10). Mice were challenged with 1.2 × 106 C1498-mCD19 cells on day 21 after transplantation and monitored for survival. Survival curves were compared using Mantel-Cox (log-rank) test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
CAR-induced developmental shifting from T cell to NK cell–like differentiation translates to humans.
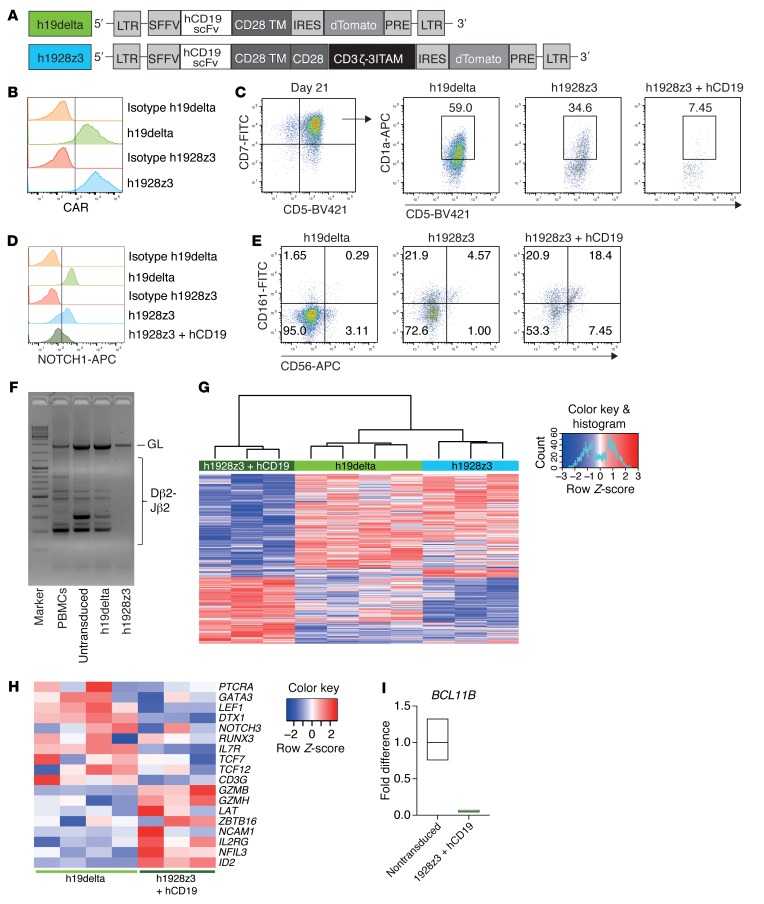
We next sought to evaluate whether the impact of CAR expression on developing human lymphoid progenitors was comparable to that seen in mice by studying human UCB-derived CD34+ HSPCs. We cloned a previously published and clinically tested second generation human CD19 CAR containing the CD28 costimulatory domain (h1928z3) (28) into a lentiviral backbone. The respective human signaling-incompetent h19delta CAR served as control (Figure 7A). CAR-HSPCs were cocultured onto OP9-DL1 monolayers supplemented with human IL-7 (hIL-7), hSCF, hTPO, and hFLT3L to perform differentiation experiments (14). Equivalent to the murine system, CAR expression could already be detected on early hematopoietic progenitor cells (Figure 7B). In human T cell development, the expression of CD1a during early lymphoid development marks an important step for lineage choice (29, 30). We show that CAR expression on human lymphoid progenitors resulted in decreased frequencies of CD1a+ cells in the CD7+CD5+ subpopulation (Figure 7C). This was associated with decreased NOTCH1 cell-surface expression (Figure 7D) and enhanced frequencies of CD56+CD161+ cells (Figure 7E). This phenotype was strongly enhanced by stimulation with the hCD19 antigen during the differentiation process (Figure 7, C–E), demonstrating the importance of signaling strength for the developmental shift. Importantly, rearrangement at the TCRB locus was blocked in h1928z3-expressing lymphoid progenitors (Figure 7F).
Figure 7. CAR-induced developmental shifting from T cell to NK cell–like differentiation translates to humans.
(A) Representation of the lentiviral human CD19 CAR constructs with either CD28 costimulatory and CD3ζ signaling domain (h1928z3) or without signaling domains (h19delta). An IRES dTomato reporter cassette was used. (B–F) Human CD34+ CB-derived HSPCs were engineered with respective CAR constructs and consecutively differentiated on OP9-DL1 stromal cells. FACS analyses were performed within the Tom+ gate on day 21 of coculture. For stimulation, h1928z3 lymphoid progenitors were cocultured with irradiated hCD19+ Daudi cells at a 1:10 ratio from day 4 onwards. Results from 1 of 2 experiments are shown. (B) Expression of the CAR constructs on differentiating human HSPCs analyzed by protein L staining. (C) CD7+CD5+ engineered human lymphoid progenitor cells were evaluated for CD5 and CD1a expression. Numbers represent percentages in the respective gates. (D) Histograms represent NOTCH1 expression on engineered early hematopoietic human progenitors. (E) CAR-modified HSPCs were analyzed for CD161 and CD56 expression. (F) Human CAR-engineered lymphoid progenitors were evaluated for TCRB rearrangement by PCR analysis of genomic DNA on day 18 of culture. Human PBMCs and nontransduced and h19delta-modified progenitors were used as controls. (G) Hierarchical clustering of the 500 most differentially expressed (P < 0.05) transcripts across lymphoid progenitors expressing the h1928z3 CAR that had been either stimulated with hCD19 or not; h19delta CAR served as signaling-deficient control. (H) Heatmap showing the relative expression of exemplary transcripts that related to either T cell or NK cell development. Data are normalized according to expression in each row. (G and H) Experiments were performed once. h1928z3, h1928z3 + hCD19 (n = 3); h19delta (n = 4). (I) qPCR analysis of BCL11B expression in nontransduced or h1928z3-expressing progenitors stimulated with hCD19. Data show mean of triplicates and upper and lower limit from 1 experiment performed.
To further substantiate these findings, we performed microarray analysis experiments of the engineered human lymphoid progenitors. Unlike that seen in mouse, h1928z3 expression on lymphoid progenitors resulted in a transcriptional profile that was comparable to the h19delta control (Figure 7G). However, stimulation of the h1928z3 CAR led to decreased transcript expression of the regulators of T cell development such as PTCRA, GATA3, NOTCH3, and, most pronounced, IL7R (Figure 7H). In contrast, NK cell–related genes, including ID2 and transcripts for functional molecules (GZMB, GZMH), were overexpressed. Similarly to the data obtained from mice, BCL11B expression was significantly decreased in the human CAR-stimulated lymphoid progenitors, as shown by quantitative reverse-transcription PCR (Figure 7I), supporting our findings that signaling strength mediated by the CAR profoundly affects lymphoid differentiation from early hematopoietic progenitors.
Discussion
In general, low-cost production methods of approved agents have to compensate for investments during early phases of drug development. This paradigm has been profoundly challenged by the appearance of clinically highly effective cellular biologicals, such as CAR-engineered T cells for the treatment of B cell malignancies. Since the drug needs to be produced on demand on an individualized basis, production remains expensive, resulting in very high costs for a single treatment. Thus, there is a burning economical and ethical need to develop cell products that can be used for a broader range of patients, independently of their HLA type. Using a syngeneic and an MHC class I/II mismatched murine hematopoietic progenitor cell transplantation model for CD19 expressing leukemia, we intended to study the impact of a TCD-BM graft that had been enriched with prefabricated CAR-expressing lymphoid progenitor cells.
For proof of principle, we initiated the studies using a murine CAR against the clinically relevant antigen CD19 to transduce murine hematopoietic stem cells that we had originally planned to further differentiate into precursor T cells in vitro. The expression of the CAR-encoding gene was additionally set under the control of an inducible promoter system (16) in order to evaluate its impact on T cell fate in a time-dependent manner.
We found that forced CAR expression early in hematopoietic progenitors can profoundly interfere with T cell development in favor of a population with NK cell–like properties. This developmental shift is initiated early during in vitro lymphoid differentiation, which then persists as what we termed — closely reflecting a nomenclature introduced earlier by others (23) — CARiK cells upon cotransfer in vivo. These CARiK cells mediate strong anti-CD19–directed activity, as shown by profound B cell aplasia and leukemia eradication even across a complete MHC class I/ II mismatch. Both the degree of this developmental shift as well as the in vivo persistence of CARiK cells depend heavily on the presence of a CD28 costimulatory moiety in the CAR construct and seem to be influenced to a lesser extent by the number of active ITAMs within its ζ -chain.
We explored the mechanism of these findings by conducting microarray technology–supported whole transcription analysis on both in vitro–generated CAR-engineered lymphoid precursors previous to cotransplantation and their respective progeny after in vivo maturation. We show that CAR expression resulted in decreased transcriptional activity for Bcl11b and Notch1, TFs that have shown to exert gatekeeper function for T cell development. Deletion of Bcl11b has been shown to result in reprogramming of T cells to NK-like cells and was associated with a reduction of Notch transcripts (22, 23, 31). This was paralleled by a distinct NK cell–associated transcriptional profile that became further pronounced after in vivo maturation (18, 19, 24, 32). In order to substantialize these data, we investigated whether these findings would translate to the protein level. Indeed, Western blotting experiments showed a marked reduction of BCL11B in CAR-engineered lymphoid precursors at the end of the in vitro differentiation process already. In addition, NOTCH1 expression on the cell surface was also reduced in concordance with the transcriptional data.
The distinct transcriptional profile on Microarray analysis distinguished CARiK cells from other lymphoid subsets and located their identity at the interface of T lymphocytes and NK cells. Similarly to T progenitor cells that had been generated from induced pluripotent stem (iPS) cells, CARiK cells had overlapping expression profiles with γδT cells, NKT cells, and NK cells (26).
A growing body of evidence suggests that the introduction of either a TCR or a CAR into human hematopoietic progenitor cells can prevent endogenous rearrangement of the TCR-α and TCR-β locus, thereby giving rise to monospecific T cells that express the transgenic antigen receptor only (12, 33, 34). We demonstrated that, as in the murine model, early signals mediated by a CAR that had been transduced into CD34+ hematopoietic human stem cells result in decreased expression of NOTCH1, which leads to a suppression or regular progenitor T cell development in favor of NK-like cell differentiation. As in murine lymphoid progenitor cell development, this was associated with a profound block of TCR gene rearrangement. Although downregulation of BCL11B did not reach significance in the performed microarrays, quantitate reverse-transcription PCR did reveal remarkable BCL11B suppression. Further assessment of its impact in vivo has been frequently hampered by limitations of currently available humanized mouse models. Using in vitro–generated murine precursor T cells that had been engineered with a CAR against human CD19, Zakrzewski and colleagues did find CAR-expressing CD3+CD4+ and CD3+CD8+ peripheral T cells in murine recipients, hinting at normal positive and negative selection processes of transduced progenitor T cells (9). However, the use of a first-generation human CAR in an otherwise murine environment might not have fully met the signaling strength required for a shift in differentiation. We observed that the murine CAR (im1928z1) used in our studies exhibited relevant tonic signaling activity, as assessed in a nuclear factor of activated T cells (NFAT) reporter cell line (data not shown), inducing a decisive NK cell–like differentiation shift in vitro already. Although Notch culture systems might allow for some B cell differentiation, the early developmental shift in the absence of measurable antigen during in vitro culture is further indicative of functional relevant tonic signaling already known for CARs (35–37). In contrast, the human CAR that was used required additional stimulation with CD19 to fully reproduce the differentiation characteristics seen in the murine system.
The observation that in the murine system, CAR-engineered lymphoid progenitor cells continued further differentiation into CARiK cells in both a CD19-competent (WT) and a CD19-deficient host environment suggests that the differentiation program was already initiated early in the in vitro culture. This is supported by the fact that Bcl11b levels were reduced at this point. Since control experiments with a signaling-deficient CAR variant, both in the murine and human system, did again allow for T cell development to the level of CAR-negative controls, the observed shift in differentiation is suggestive of being dependent on CAR-mediated signaling. In contrast to effects caused by early CAR expression, delayed CAR expression largely fails to induce the CARiK cell phenotype. Delayed induction of CAR expression, however, permits normal T cell development to some extent, as indicated by the occurrence of CAR transgene–positive mature CD3+TCRβ+ T cells. This was paralleled by decreased numbers of CARiK cells.
We further show that the CAR-expressing NK cell–like progeny of cotransplanted lymphoid progenitors mediated strong antileukemia activity after HSCT. Early in vivo depletion after HSCT using an NK-depleting antibody led to a complete abrogation of antileukemia effects. In contrast to lymphoid progenitors that had been engineered to express an antileukemic TCR matching the recipient’s MHC type, CAR-induced NK1.1+ progeny did not build a cellular long-term memory (11). In vivo persistence and, as a consequence, CARiK cell–mediated effects were limited, and in its extent, associated with the use of a CD28 costimulatory domain. The anti-CD19–directed activity lasted for about 60 days. This has important implications from a safety perspective, since we found a single injection of the stem cell–derived product to produce durable activity. After that, transgene-positive cells became undetectable in vivo. Leukemia survivors that were rechallenged with a lethal dose of CD19-expressing leukemia cells succumbed to the disease, supporting the notion that no memory cell population had been developed. Although the cell product did not persist, the ready availability of a prefabricated and aliquotted product would permit repeated infusions as needed.
UCB-derived lymphoid progenitor cells (termed proT2) being CD34+CD7+CD5+ have been identified as a potential equivalent of the in vitro–generated murine DN2 precursor T cell population (13, 38). We and others have shown that UCB-derived lymphoid progenitors can be genetically engineered (12, 14). Our data on TCR rearrangement and gene expression in both murine and human systems support earlier studies showing that human and murine lymphoid development are much more similar than originally reported (30) and encourage its further exploitation for clinical use.
The use of genetically engineered HPSCs for AT leads to concerns of safety when it persists in the body for an unduly long duration. Although long-term persistence has been demonstrated to be important in targeting B cell malignancies (5, 7, 39, 40), it has, remained unclear why complete eradication of a target cell population would make further persistence of engineered T cells necessary. Of interest, recently, cures were achieved using third-party gene-edited T cells with limited persistence (41). Targeting antigens beyond CD19/20 might obviate the need for long-term persistence of CAR-engineered immune cells. Our generated CARiK cells with limited in vivo persistence might be of interest when antigen structures are considered for which off tumor/on target effects are of concern. In this scenario, tumor cell eradication might be followed by the recovery of a physiologic cell population sharing the targeted antigen, such as normal myelopoiesis after the treatment of a myeloid malignancy. Several other approaches have been taken to develop CAR cell products for a more universal use. The emergence of potent genome-editing technologies has shown to transform a mature postthymic T cell into a universally applicable cellular carrier of CARs by disruption of both the endogenous TCR and HLA class I expression (41–43). As a promising alternative, iPS cell–derived lymphoid cells (26) offer an attractive platform for immune engineering providing both the generation of T and NK cells (10, 44). Despite impressive progress being achieved, major challenges remain. This includes the necessity to control T and NK lineage specifications, identification of the optimal maturational stage, and preservation of an optimal functional and proliferative potential. Our data predict that the choice of the antigen receptor, including its structural composition, as well as the progenitor cell population going to be engineered can fundamentally direct final cell differentiation, thereby determining biologic properties in both mice and humans.
Methods
Mice.
B6 (H2b) mice were purchased from Charles River Laboratories. B10.A (H2a) mice were purchased from Taconic Laboratories. R26-M2rtTA knockin mice (B6.rtTA, H2b) express a reverse tetracycline-controlled transactivator for doxycycline-inducible (Dox-inducible) transgene expression and were used for transgene expression studies. To create B10.A-R26-M2rtTA (B10.A.rtTA) mice, B6.rtTA mice were backcrossed onto B10.A. Cd19 KO mice on a B6 background (B6 CD19 KO) were a gift from K.H. Lee (Hannover Medical School).
Primary human UCB samples and generation of human-engineered lymphoid progenitors.
UCB samples were processed as described earlier (14). Briefly, purified CD34+ HSPCs were transduced with lentiviral supernatant and transferred onto OP9-DL1 stromal cells in the presence of hIL-7, hSCF, hTPO, and hFLT3L. Human-engineered lymphoid progenitors were harvested every 3–4 days and put onto new OP9-DL1 monolayers supplemented with the respective cytokines. The h1928z3-generated lymphoid progenitors were stimulated by adding irradiated Daudi cells from day 4 of culture.
Cell lines.
The C1498 myeloid leukemia cell line (H2b, B6 origin) was transduced with retroviral supernatant of murine CD19 alone in the pAlpha.SIN.MPSV.wPRE vector (45) or linked with an IRES GFP cassette to generate the murine CD19+ cell lines C1498-mCD19 or C1498-mCD19-GFP. For isolation of stable mCD19 expression, cells were sorted twice for CD19 or CD19 and GFP (FACSAria Ilu, BD Biosciences). C1498, C1498-mCD19, C1498-mCD19-GFP, Daudi, and 58α–β– cells were cultured in RPMI 1640, and 293T cells were cultured in DMEM (Capricorn). Both media were supplemented with 10% heat-inactivated FCS (Capricorn), l-glutamine, HEPES buffer, and penicillin/streptomycin (all Gibco, Thermo Fisher Scientific). OP9-DL1 cells were cultured in complete αMEM (Life Technologies) supplemented with 20% FCS, l-glutamine, HEPES buffer, and penicillin/streptomycin. The hybridoma cell line HB-191 (ATCC) expressing an anti-NK1.1 antibody (clone PK136) was cultured in HybridomMed Dif 1000 (Biochrom). All cell lines were tested for mycoplasm negativity by PCR.
BM and lymphoid progenitor cotransplantation, leukemia challenge, and NK cell in vivo depletion.
Total body irradiation of 8-week-old B6 recipients was performed with 10.5 Gy from a linear accelerator. After 24 hours, mice were cotransplanted with 3 × 106 TCD-BM and 8 × 106 lymphoid progenitors as previously described.(11) All AT studies were performed under permanent administration of Dox-containing water or food for transgene expression unless otherwise noted. For leukemia studies, 1.2 × 106 C1498-mCD19-GFP leukemia cells were injected via the lateral tail vein on day 20 after transplantation. NK1.1+ cell depletion in vivo was performed in CAR lymphoid progenitor–transplanted mice challenged with leukemia cells by i.p. injection of 200 μg anti-NK1.1 antibody weekly and PBS as control. All mice were randomly assigned to experimental groups, and no blinding of investigators was performed.
Lentiviral CAR constructs and production of supernatant.
The murine CD19 CAR construct (im1928z1) and sequence were described previously (15). im1928z1 contains an anti-CD19 scFv, the transmembrane domain and costimulatory domain from mouse CD28, and the CD3ζ signaling domain with 1 functional ITAM. The im19BBz1 construct has the same ITAM configuration, but a 4-1BB costimulatory domain instead of CD28. im1928z3 has the same configuration as im1928z1, but 3 functional ITAMs. Both im19z1 and im19z3 were designed without CD28 costimulatory domain. The im19delta construct contains only the scFv and the CD28 transmembrane domain and lacks the costimulatory and signaling. All CAR sequences were cloned under the control of the Tet-On T11 promotor in combination with an IRES dTomato (Tom) cassette in a shortened version of an all-in-one lentiviral backbone (11, 16). A dTomato-only construct was used as control vector (iTom). A functional human CD19 CAR (h1928z3) (28) and the signaling-deficient h19delta construct for control were cloned in a lentiviral backbone under a SFFV promotor that was linked to a dTomato reporter cassette. Lentiviral supernatants were produced via transient transfection of 293T cells with the viral plasmids pMD2.G, pRSV.Rev, pcDNA.GP.4×CTE (plasmids produced by PlasmidFactory), and the respective CAR plasmid using the calcium phosphate transfection method. The RD114/TR envelope was used for transduction of CD34+ HSPCs. Harvested supernatant was filtered and concentrated via ultracentrifugation. The 58α–β– hybridoma cell line transduced with the M2 transactivator was used for viral titer determination of murine constructs.
Generation of engineered murine lymphoid progenitors.
Murine HSPCs were isolated from BM with antibodies against lineage markers and sorted for c-kit+ and Sca-1+ (LSK) cells. LSKs were then transduced with lentiviral supernatant and cultured as published (11). Briefly, transduced LSK cells were transferred to OP9-DL1 monolayer cells in complete αMEM medium (Life Technologies) supplemented with 10% heat-inactivated FCS, FLT3-L (5 ng/mL), IL-7 (5 ng/mL) (Peprotech), and Dox (2 mg/mL) (MilliporeSigma). Lymphoid progenitors were transferred to new OP9-DL1 monolayers every 3 to 4 days. Transduced lymphoid progenitors were sorted on day 10 of OP9-DL1 coculture for dTomato expression (Tom+) and frozen on days 13 to 17. For AT studies, engineered lymphoid progenitors were thawed and cultured for a further 7 days on OP9-DL1 with cytokines and Dox. Engineered lymphoid progenitors between days 20 and 24 of coculture were used for cotransplantation. Cultures were supplemented with Dox for permanent transgene expression unless otherwise noted. For in vitro antigen stimulation of im1928z1-generated lymphoid progenitors, irradiated C1498-mCD19 or C1498 cells were added in a 1:10 ratio to lymphoid progenitor culture.
Flow cytometry.
Single-cell suspensions of murine origin were stained with the following fluorochrome-conjugated antibodies: CD3ε (Brilliant Violet [BV] 421/PE-Cy7; catalog 100341/100320; clone 145-2c11), CD4 (PE-Cy7/BV 421; catalog 116016/100437; clone RM4-4/GK1.5), CD8α (FITC/APC; catalog 100706/100712; clone 53-6.7), TCR-β (PE-Cy7; catalog 109222; clone H57-597), CD25 (PE-Cy7; catalog 102016; clone PC61), CD44 (APC/FITC; catalog 103012/103006; clone IM7), CD122 (FITC/PE-Cy7; catalog 123215/123207; clone TM-β1), CD19 (FITC/BV 421; catalog 115506/115549; clone 6D5), NK1.1 (APC/FITC; catalog 108710/108706; clone PK136), NKp46 (BV 421; catalog 137612; clone 29A1.4), IFN-γ (APC/PE-Cy7; catalog 505809/505825; clone XMG1.2), CD107a (APC/PE-Cy7; catalog 121613/121619; clone 1D4B), NOTCH1 (APC; catalog 130613; clone HMN-1-12), anti-rat IgG2a isotype (APC; catalog 400119; clone MOPC-21) (all from BioLegend), anti-rat IgG2a isotype (PE-Cy7; catalog 25-4321-81; clone eBR2a) (eBioscience), TCR-β (APC; catalog 553174; clone H57-597), and CD19 (APC; catalog 550992; clone 1D3) (both from BD Biosciences). For lineage–c-kit+Sca-1+ sort, antibodies against lineage markers CD3, CD4, CD8, CD19, NK1.1, Gr-1 (catalog 108406; clone RB6-8C5), and CD11b (catalog 101206; clone M1/70) in FITC and Sca-1 (PE; catalog 108108; clone D7) (all from BioLegend) and c-kit (APC; catalog 17-1172-83; clone ACK2) (eBioscience) were used. For CAR expression detection, cells were stained with goat-anti-rat Fab fragment conjugated with Alexa Fluor 647 (Jackson ImmunoResearch; catalog 112-606-003). Human cells were stained with the following antibodies after blocking with Human TruStain FcX (catalog 422302) purchased from BioLegend: CD34 (PE-Cy7; catalog 343516; clone 581), CD7 (FITC; catalog 343103; clone CD7-6B7), CD5 (BV421; catalog 300626; clone UCHT2), CD1a (APC; catalog 300110; clone HI49), CD56 (APC; catalog 318309; clone HCD56), NOTCH1 (APC; catalog 352107; clone MHN1-519), and CD161 (FITC; catalog 339905; clone HP-3G10). Flow cytometry was performed using a FACSCanto or LSR-II (BD Biosciences), and the data were analyzed with FlowJo software (TreeStar). FACS analysis was based on fluorescence minus one (FMO) controls. Relative numbers from Thymi, BM, and splenocytes were calculated from Tom+ gate.
Western blotting.
Cell lysates of transgene-positive sorted lymphoid progenitors or WT B6 thymocytes were prepared in RIPA buffer as described (46). Equal masses of protein lysates were separated by SDS-PAGE and transferred onto PVDF membranes (Amersham). Membranes were blocked with 5% skim milk, and BCL11B was stained with rat anti-mouse primary antibody (clone 25B6; BioLegend) and detected with HRP-coupled goat anti-rat secondary antibody (Poly4054; BioLegend).
Ex vivo short-term culture.
Splenocytes from cotransplanted mice were harvested 28 days after transplantation and brought into short-term culture under T cell conditions with ConA (5 mg/mL) (MilliporeSigma), IL-7 (5 ng/mL), and IL-2 (20 U/ml) (Peprotech) or cultured with high IL-2 concentrations (1000 U/ml). Cells were split every 2 days and used in functionality assays after 4–6 days of culture.
Intracellular cytokine staining and degranulation assay.
Ex vivo–cultured splenocytes were coincubated with C1498-mCD19 cells and intracellular IFN-γ staining performed as previously described.(47) For detection of degranulation, ex vivo–cultured splenocytes were coincubated on 96-well plates with C1498-mCD19 cells at a 10:1 ratio in the presence of an anti-CD107a antibody. After 1 hour, GolgiStop (BD Bioscience) was added. Cells were harvested after 4 hours and treated with the CytoFix/CytoPerm Kit (BD Bioscience) for FACS analysis.
PCR for rearrangement on the TCR-β locus.
Murine and human engineered lymphoid progenitors were harvested from in vitro culture and sorted for Tom+ cells. Genomic DNA was isolated (QIAGEN) and D-J rearrangement at the Tcrb locus assessed via PCR using TCRB_Jβ2, reverse, TGAGAGCTGTCTCCTACTATCGATT and TCRB_Dβ2, forward, GTAGGCACCTGTGGGGAAGAAACT as primers (5′-3′) as described (23) for mouse TCR rearrangement. Human TCRB rearrangement was assessed using primers as described (48).
Microarray and data analysis.
Murine Tom+ cells were sorted from iTom or im1928z1-generated lymphoid progenitors’ in vitro culture on day 20 or from harvested splenocytes for their progeny on day 28 after transplantation. For the comparison of different lymphocyte subsets and CARiK cells, T cells (CD3+γδTCR–NK1.1–), NKT cells (CD3+NK1.1+), γδ T cells (CD3+γδTCR+), and NK cells (CD3–NK1.1+) were sorted from WT B6-derived splenocytes. Tom+ CARiK cells were sorted on day 28 after cotransplantation. RNA was extracted using QIAGEN RNAeasy (QIAGEN) according to the manufacturer’s manual. For human samples, RNA of Tom+ sorted cells was extracted with the Single Cell Lysis Kit (Invitrogen). Microarray analysis was performed with Affymetrix Clariom S (400 Format) Pico chips. Raw data were analyzed by R/Bioconductor packages oligo and Biobase. Summarized probeset data was log2 transformed, followed by RMA normalization procedure. Normalized data sets were filtered for informative genes (showing at least expression values > log2[10] in more than 2 samples). Data sets were tested across all groups (ANOVA) or pairwise using linear models to assess differential expression in context of the multifactorial designed experiment. For statistical analysis and assessing differential expression, the R/BioConductor package limma was used; this utilizes an empirical Bayes method to moderate the standard errors of the estimated log-fold changes (49). Functional analysis was performed by R package clusterProfiler (50).
Microscopy.
Thymi of mice cotransplanted with engineered lymphoid progenitors were harvested on day 14 after transplantation. Sections from Tissue-Tec OCT (Sakura) embedded thymi were analyzed with a Zeiss Axio Imager 2 microscope (×20 magnification) and acquired with Zen pro software (Zeiss). Images were equally processed with ZEN lite software.
RNA extraction and quantitative reverse-transcription PCR.
RNA from sorted samples on day 18 of OP9-DL1 coculture was extracted using the RNAeasy Mini Kit (QIAGEN) and converted into cDNA using the QuantiTect Reverse Transcription Kit (QIAGEN). Real-time PCR reactions were performed using the QuantiTec SYBR Green PCR kit (QIAGEN) on an Applied Biosystems 7300 Real-Time PCR System (Thermo Fisher Scientific). The QuantiTec Primer assays for BCL11B (QT00080983) and ACTB (QT00095431) were purchased from QIAGEN. Relative expression of BCL11B was normalized to ACTB. Nontransduced lymphoid progenitors were used for controls.
Statistics.
Statistics were performed and graphed with GraphPad Prism 5 software for Mac (GraphPad Software). Survival curves were compared using Mantel-Cox (log-rank) test. Student’s t test (2-tailed) was applied for 2-group comparisons and one-way ANOVA with Tukey’s post test for comparing more than 2 groups. Data were represented as mean ± SEM. P values of less than 0.05 were considered to be significant.
Data availability.
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE104512 and GSE135015).
Study approval.
All animal experiments were approved by the State Government of Lower Saxony, Germany (approval code 33.14-42502-04-15/1781) and performed in accordance with institutional animal care and use guidelines. Human UCB-derived samples were collected after written, informed consent by the child’s parent or guardian. Procedures for the use of UCB for this study were reviewed and approved by the medical ethics committee of Hannover Medical School and handled in accordance with the Declaration of Helsinki.
Author contributions
MM designed research, performed experiments, analyzed and interpreted data, and drafted and edited the manuscript. J Herbst and VS performed experiments. RG, JM, HM, and BEV contributed vital new technology and reagents. AG, MRMVDB, J Huehn, RB, AK, and AS analyzed and interpreted data and edited the manuscript. MGS designed the general concept and research, analyzed and interpreted data, and drafted and edited the manuscript.
Supplementary Material
Acknowledgments
We thank the staff of the Cell Sorting Core Facility and the Department of Transfusion Medicine (both Hannover Medical School) as well as the members of the Genome Analytics Facility of the Helmholtz Centre for Infection Research (Braunschweig) for their assistance. This work was supported by Deutsche Forschungsgemeinschaft (SFB738-A3 to MGS, SFB738-A7 and SFB902-B15 to AK, SFB738-C9 to AS, Cluster of Excellence REBIRTH/EXC62/1), the Integriertes Forschungs- und Behandlungszentrum Transplantation (IFB-Tx) (CBT_6 to MGS and AS), and the PhD program in Molecular Medicine of the Hannover Medical School (HBRS). AG is supported by the SITC-Merck Immunotherapy in Lung Cancer Clinical Fellowship. This research was also supported by NIH R01-CA228358-01 (MRMVDB) and R01-CA228308-01 (MRMVDB), Project 4 of P01-CA023766-38 (MRMVDB), the Lymphoma Foundation, the Susan and Peter Solomon Divisional Genomics Program, and a P30 CA008748 MSK Cancer Center Support Grant/Core Grant. This research was also supported by the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center.
Version 1. 09/03/2019
In-Press Preview
Version 2. 10/22/2019
Electronic publication
Version 3. 12/02/2019
Print issue publication
Funding Statement
To A.K.
To A.S.
To A.S. and M.G.S.
To M.v.d.B.
To M.v.d.B.
To M.G.S.
Footnotes
Conflict of interest: MRMVDB has intellectual property licensing with Seres Therapeutics and Juno Therapeutics. MRMVDB has also received honorariums from Flagship Ventures, Novartis, Evelo, Seres Therapeutics, Jazz Pharmaceuticals, Therakos, Amgen, Merck, the Acute Leukemia Forum, and DKMS (board member) and research support from Seres Therapeutics, from which he has stock options. AG has received research support from Aprea Therapeutics and Infinity Pharmaceuticals.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(12):5108–5122.https://doi.org/10.1172/JCI126350.
See the related Commentary at Adding chimeric antigen receptor–induced killer cells to the medical oncology shelf.
Contributor Information
Marcel Maluski, Email: maluski.marcel@mh-hannover.de.
Arnab Ghosh, Email: ghosha@mskcc.org.
Jessica Herbst, Email: herbst.jessica@mh-hannover.de.
Vanessa Scholl, Email: vanessascholl@gmx.de.
Rolf Baumann, Email: r.baumann@strahlentherapie.de.
Jochen Huehn, Email: jochen.huehn@helmholtz-hzi.de.
Robert Geffers, Email: robert.geffers@helmholtz-hzi.de.
Johann Meyer, Email: meyer.johann@mh-hannover.de.
Holger Maul, Email: h.maul@asklepios.com.
Britta Eiz-Vesper, Email: eiz-vesper.britta@mh-hannover.de.
Andreas Krueger, Email: Andreas.Krueger@kgu.de.
Axel Schambach, Email: Schambach.Axel@mh-hannover.de.
Martin G. Sauer, Email: sauer.martin@mh-hannover.de.
References
- 1.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robbins PF, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21(5):1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapoport AP, et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med. 2015;21(8):914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude SL, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grupp SA, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DW, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385(9967):517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brentjens RJ, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zakrzewski JL, et al. Adoptive transfer of T-cell precursors enhances T-cell reconstitution after allogeneic hematopoietic stem cell transplantation. Nat Med. 2006;12(9):1039–1047. doi: 10.1038/nm1463. [DOI] [PubMed] [Google Scholar]
- 9.Zakrzewski JL, et al. Tumor immunotherapy across MHC barriers using allogeneic T-cell precursors. Nat Biotechnol. 2008;26(4):453–461. doi: 10.1038/nbt1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Themeli M, Rivière I, Sadelain M. New cell sources for T cell engineering and adoptive immunotherapy. Cell Stem Cell. 2015;16(4):357–366. doi: 10.1016/j.stem.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoseini SS, et al. Inducible T-cell receptor expression in precursor T cells for leukemia control. Leukemia. 2015;29(7):1530–1542. doi: 10.1038/leu.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Caeneghem Y, et al. Antigen receptor-redirected T cells derived from hematopoietic precursor cells lack expression of the endogenous TCR/CD3 receptor and exhibit specific antitumor capacities. Oncoimmunology. 2017;6(3):e1283460. doi: 10.1080/2162402X.2017.1283460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awong G, Herer E, Surh CD, Dick JE, La Motte-Mohs RN, Zúñiga-Pflücker JC. Characterization in vitro and engraftment potential in vivo of human progenitor T cells generated from hematopoietic stem cells. Blood. 2009;114(5):972–982. doi: 10.1182/blood-2008-10-187013. [DOI] [PubMed] [Google Scholar]
- 14.Hübner J, et al. Generation of genetically engineered precursor T-cells from human umbilical cord blood using an optimized alpharetroviral vector platform. Mol Ther. 2016;24(7):1216–1226. doi: 10.1038/mt.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kochenderfer JN, Yu Z, Frasheri D, Restifo NP, Rosenberg SA. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood. 2010;116(19):3875–3886. doi: 10.1182/blood-2010-01-265041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinz N, et al. Retroviral and transposon-based tet-regulated all-in-one vectors with reduced background expression and improved dynamic range. Hum Gene Ther. 2011;22(2):166–176. doi: 10.1089/hum.2010.099. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17(6):749–756. doi: 10.1016/S1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 18.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16(3):306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bezman NA, et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13(10):1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med. 2001;194(7):1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14(8):529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329(5987):89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329(5987):85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Male V, et al. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J Exp Med. 2014;211(4):635–642. doi: 10.1084/jem.20132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10(10):1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 26.Themeli M, et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat Biotechnol. 2013;31(10):928–933. doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kochenderfer JN, et al. Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother. 2009;32(7):689–702. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spits H. Development of alphabeta T cells in the human thymus. Nat Rev Immunol. 2002;2(10):760–772. doi: 10.1038/nri913. [DOI] [PubMed] [Google Scholar]
- 30.Dik WA, et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med. 2005;201(11):1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 2005;106(10):3498–3506. doi: 10.1182/blood-2005-02-0496. [DOI] [PubMed] [Google Scholar]
- 32.Leong JW, Wagner JA, Ireland AR, Fehniger TA. Transcriptional and post-transcriptional regulation of NK cell development and function. Clin Immunol. 2017;177:60–69. doi: 10.1016/j.clim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kisielow P, Teh HS, Blüthmann H, von Boehmer H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature. 1988;335(6192):730–733. doi: 10.1038/335730a0. [DOI] [PubMed] [Google Scholar]
- 34.Snauwaert S, et al. In vitro generation of mature, naive antigen-specific CD8(+) T cells with a single T-cell receptor by agonist selection. Leukemia. 2014;28(4):830–841. doi: 10.1038/leu.2013.285. [DOI] [PubMed] [Google Scholar]
- 35.Frigault MJ, et al. Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer Immunol Res. 2015;3(4):356–367. doi: 10.1158/2326-6066.CIR-14-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long AH, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015;21(6):581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomes-Silva D, et al. Tonic 4-1BB costimulation in chimeric antigen receptors impedes T cell survival and is vector-dependent. Cell Rep. 2017;21(1):17–26. doi: 10.1016/j.celrep.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Awong G, et al. Human proT-cells generated in vitro facilitate hematopoietic stem cell-derived T-lymphopoiesis in vivo and restore thymic architecture. Blood. 2013;122(26):4210–4219. doi: 10.1182/blood-2012-12-472803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maude SL, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neelapu SS, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med. 2017;377(26):2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qasim W, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9(374):eaaj2013. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 42.Osborn MJ, et al. Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol Ther. 2016;24(3):570–581. doi: 10.1038/mt.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgiadis C, et al. Long terminal repeat CRISPR-CAR-coupled “universal” T cells mediate potent anti-leukemic effects. Mol Ther. 2018;26(5):1215–1227. doi: 10.1016/j.ymthe.2018.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng J, Tang SY, Toh LL, Wang S. Generation of “off-the-shelf” natural killer cells from peripheral blood cell-derived induced pluripotent stem cells. Stem Cell Reports. 2017;9(6):1796–1812. doi: 10.1016/j.stemcr.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suerth JD, Maetzig T, Galla M, Baum C, Schambach A. Self-inactivating alpharetroviral vectors with a split-packaging design. J Virol. 2010;84(13):6626–6635. doi: 10.1128/JVI.00182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauer MG, Herbst J, Diekmann U, Rudd CE, Kardinal C. SHP-1 acts as a key regulator of alloresponses by modulating LFA-1-mediated adhesion in primary murine T cells. Mol Cell Biol. 2016;36(24):3113–3127. doi: 10.1128/MCB.00294-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koestner W, et al. PD-L1 blockade effectively restores strong graft-versus-leukemia effects without graft-versus-host disease after delayed adoptive transfer of T-cell receptor gene-engineered allogeneic CD8+ T cells. Blood. 2011;117(3):1030–1041. doi: 10.1182/blood-2010-04-283119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramiro AR, Trigueros C, Márquez C, San Millán JL, Toribio ML. Regulation of pre-T cell receptor (pT alpha-TCR beta) gene expression during human thymic development. J Exp Med. 1996;184(2):519–530. doi: 10.1084/jem.184.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 50.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE104512 and GSE135015).