Abstract
Innate-like tissue-resident γδ T cell compartments capable of protecting against carcinogenesis are well-established in mice. Conversely, the degree to which they exist in humans, their potential properties, and their contributions to host benefit are mostly unresolved. Here we demonstrate that healthy human breast harbours a distinct γδ T cell compartment, primarily expressing T cell receptor (TCR) Vδ1 chains, by comparison to Vδ2 chains that predominate in peripheral blood. Breast-resident Vδ1+ cells were functionally skewed toward cytolysis and IFN-γ production, but not IL-17 which has been linked with inflammatory pathologies. Strikingly, breast-resident Vδ1+ cells could be activated innately via the NKG2D receptor, whereas neighbouring CD8+ αβ T cells required TCR signalling. A comparable population of Vδ1+ cells was found in human breast tumors, and when paired tumor and non-malignant samples from eleven triple negative breast cancer patients were analyzed, progression-free and overall survival correlated significantly with Vδ1+ cell representation, but not with either total γδ T cells or Vδ2+ T cells. As expected, progression-free survival also correlated with αβ TCRs. However, whereas TCRαβ repertoires in most cases focussed, typical of antigen-specific responses, this was not so for Vδ1+ cells, consistent with their innate-like responsiveness. Thus, maximal patient benefit may accrue from the collaboration of innate-like responses mounted by tissue-resident Vδ1+ compartments and adaptive responses mounted by αβ T cells.
Introduction
γδ T cells comprise a highly conserved third lineage of lymphocytes that uses somatic gene rearrangement to encode the defining antigen receptor (1, 2). Although this is a hallmark of adaptive immunity, subsets of murine γδ T cells also display innate-like activity, manifest in rapid responses to self-encoded “stress antigens” such as ligands for the NKG2D receptor (3–6). This is known as lymphoid stress-surveillance (7).
Given that NKG2D ligands are upregulated by over-activity of epidermal growth factor receptor (EGFR) signalling and DNA damage (8, 9), it is natural that lymphoid stress-surveillance might contribute to cancer immunosurveillance (10). Indeed, γδ T cell-deficient mice show greatly increased susceptibility to cancer in several systems (4, 11–13), and many attempts are ongoing to exploit their activities clinically (14). Such approaches may enhance the efficacy of current immunotherapies such as checkpoint blockade and in particular, chimeric antigen receptor (CAR) T cells which have shown limited success in treating solid tumors. Moreover, the capacity of some γδ T cell subsets to secrete chemokines and cytokines and/or to present antigen argues strongly for their potential to promote the therapeutic potentials of other cell types (12, 15–17).
In mice, signature γδ T cell compartments are associated with discrete tissues such as epidermis, dermis, lung, uterus, and intestinal epithelium (18–25), seemingly offering optimal capacity to detect and respond to malignant transformation of neighbouring cells. Accordingly, γδ T celldeficient mice have increased susceptibility to skin carcinogens owing to the lack of dendritic epidermal γδ T cells (5). Whether local γδ T cell compartments populate all tissues is unresolved.
Nonetheless, the prospect of a mouse breast-associated compartment was supported by the fact that the representation, albeit variable, of γδ T cells in lactating mammary glands was at least fourfold higher than in draining lymph nodes (LNs). Moreover, those cells employed a variety of γδTCRs, distinguishing them from skin and gut-resident γδ T cell compartments (26).
There has been long-standing interest in the degree to which tissue-associated γδ T cell compartments might be conserved in humans, and whether or not they contribute to cancer immunosurveillance. On the one hand, humans harbour no obvious counterparts to dendritic epidermal γδ T cells; on the other hand, jawless vertebrates possess skin-resident and gutintraepithelial cells with many parallels to γδ T cells, suggesting that such compartments have been conserved for over half a billion years (27). We therefore hypothesized that sub-optimal methods for the detection and/or extraction of T cells from human tissues might have confounded attempts to identify and characterize conserved extra-lymphoid γδ T cell compartments. This hypothesis is consistent with inefficiencies and biases reported both for extracting TCRαβ+ tissue-resident memory T (TRM) cells (28) and for visualizing tumor infiltrating lymphocytes (TILs) in situ (29), and derives support from our recent characterization of a large intraepithelial γδ T cell compartment in the human gut (30).
In this regard, the care of women in a large breast cancer risk surveillance and treatment practice offered a rare opportunity to analyze the status of γδ T cells in healthy tissue obtained from reduction mammoplasty or risk-reducing mastectomy; from malignant tissue from wide local resection; and from paired malignant and non-malignant tissues from therapeutic mastectomies.
Furthermore, the importance of investigating the possible existence of local γδ T cells was underlined by evidence that TIL densities were positive prognostic indicators in some types of breast cancer (31, 32). Despite this, the role of immunotherapy in breast cancer remains unclear with disappointing response rates to current immunotherapies, such as checkpoint blockade (33, 34).
Results
Vδ1 T cells compose a major human breast-resident γδ subset and are Tc1-skewed
Our initial goal was to assess the status of T cells resident within healthy human breast. However, the obtainment and characterization of lymphocytes from healthy human tissues has commonly been confounded by poor and irreproducible yields and low cell viability. To redress this problem, Clark and colleagues developed a “grid” explant culture system that permitted the recovery and characterization of large numbers of healthy skin-resident and lung-resident T cells without substantively changing their phenotype (35–38).
We therefore applied grids to disaggregated breast tissue from 29 healthy subjects undergoing reduction mammoplasties or risk-reducing mastectomies. Compared to the limited and variable recovery and poor viability of lymphocytes examined directly ex vivo, grids facilitated in every case the recovery and maintenance of CD3- NK cells / Innate Lymphoid cells (ILC), γδ T cells, and CD4+ and CD8+ αβ T cells, albeit there was some enrichment of NK/ILC and γδ T cells (table S1; Fig. 1A).
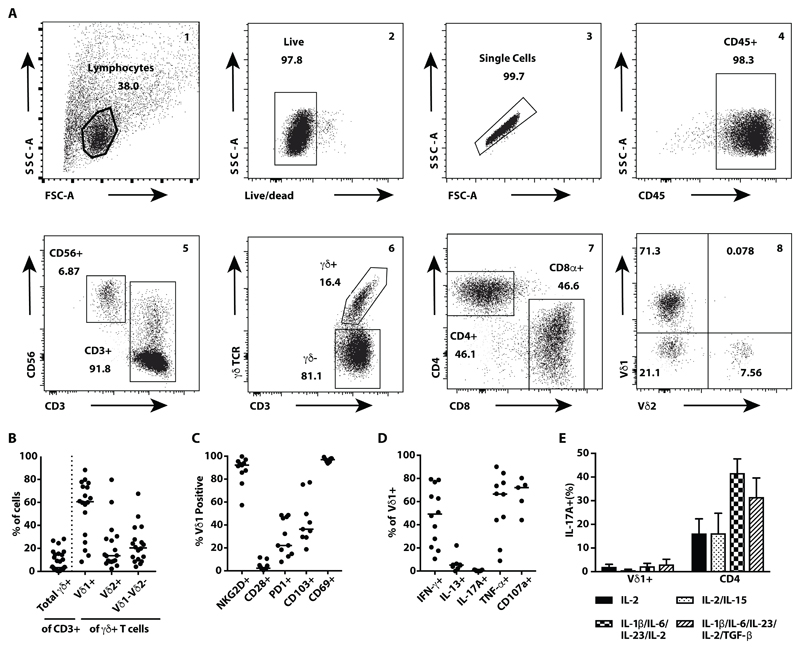
Fig. 1. Healthy human breast tissue harbours tissue-resident Vδ1+, Tc1-skewed γδ T cells.
(A) Representative flow cytometry plots showing the gating strategy to identify lymphocytes including γδ T cell subsets isolated from human breast tissue and following grid culture. Lymphocytes were gated on size and scatter (1), followed by live-dead exclusion (2), a singlet gate (3) and CD45 (4) prior to sub-setting (5–8). (B) Summary dot plots showing TCRγδ+ cells isolated from healthy human breast tissue, expressed as a percentage of recovered CD3+ cells (n=29); median indicated. In a subset of these, Vδ chain usage was quantified and expressed as percentage of pan TCRγδ+ (n=18); medians indicated. (C) Expression of cell surface markers NKG2D, CD28, PD-1, CD103, and CD69 on Vδ1+ T cells (n=9-11); medians indicated. (D) Functional phenotype of tissue-resident Vδ1+ T cells. Dot plots showing intracellular cytokine staining for IFN-γ (n=12), IL-13 (n=8), IL-17A (n=9) and TNF (n=10), and cell surface CD107a (n=5) expression, following in vitro stimulation of bulk CD3+ cultures with PMA and ionomycin [4h]; medians indicated. (E) Summary data showing the percentages of breast-resident Vδ1+ or CD4 αβ T cells stained intracellularly for IL-17A. Cells were isolated by explant culture and then grown in two forms of IL-17-skewing media, followed by in vitro activation with PMA and ionomycin [4h] (n=3, except for Vδ1+ cells grown in TGF-β containing medium where n=2); mean with SEM indicated.
As is common for PB γδ T cells (39), γδ T cell representation in breast showed considerable inter-individual variation (Fig. 1B). However, breast-extractable cells were clearly distinct from PB γδ T cells by TCRVδ chain usage. Whereas most PB cells express Vδ2 paired to Vγ9, most donors’ breast γδ T cells were predominantly Vδ1+ (median=60.5% of γδ T cells), as are most human skin-resident and gut-resident γδ T cells (40) (Fig. 1B). In all cases, there was some representation of Vδ2+ T cells (median=13.6% of γδ T cells), and of cells expressing neither Vδ1 nor Vδ2 (median=20.45% of γδ T cells), which in some cases were almost exclusively Vδ3+ (fig. S1A).
The phenotypic consistency of γδ T cells in grid-cultures and counterpart cells examined directly ex vivo was apparent from the expression patterns of several biologically important surface markers, albeit that CD69 was expressed by more cells in grid culture (fig. S1B; fig. S1C, top two rows). By multiparameter analysis of a subset of donors, we could deduce a consensus Vδ1+ T cell phenotype that resembled that of extralymphoid γδ T cell subsets in other tissues of mice and humans (4, 41–44): namely, uniform positivity for the activating NK cell receptor NKG2D, and for CD69, and largely lacking the lymphoid T cell co-stimulator, CD28 (Fig. 1C; fig. S1C, bottom row). On average, ~20% of breast Vδ1+ T cells expressed PD-1, whereas slightly more expressed the epithelial interaction integrin, CD103 (α Eβ 7), albeit with high inter-individual variation (Fig. 1C; fig. S1C, bottom row).
To assess the cells’ functional potential, they were incubated with phorbol myristate acetate (PMA) and ionomycin which jointly mimic TCR signalling, and analyzed for intracellular cytokine production, and for surface expression of CD107a, a marker of degranulation and exocytosis of cytolytic mediators such as granzymes and perforin. Breast-associated Vδ1+ T cells combined CD107a expression with Tumor Necrosis Factor (TNF) and IFN-δ production (Fig. 1D), a Tc1 phenotype that among CD8+ TCRαβ+ TILs is considered highly patient-beneficial (45–48). Cells from some donors expressed IL-13 (median=5.2% of γδ T cells), which was recently linked to T cell tumor surveillance (49), but there was no production of IL-17A, an effector cytokine commonly produced by mouse γδ T cells, in which species it has been associated with tumor promotion (50–52).
Because IL-17 production by human γδ T cells is reportedly difficult to observe (53), we tested whether breast-resident γδ T cells would respond in culture to IL-17-skewing conditions; namely, IL-1β, IL-6, IL-23, IL-2 +/- TGFβ (54). However, whereas breast-resident CD4+ αβ T cells extracted and maintained together with Vδ1+ T cells in the identical breast explant cultures (table S1) produced IL-17A [median=16.5% (using IL-2) and 12.6% (using IL-2+IL-15)] of CD4+ T cells] and markedly increased production in IL-17-skewing conditions (medians=41.5% and 35.5%, respectively) (Fig. 1E), breast Vδ1+ T cells produced negligible IL-17A under all circumstances (Fig. 1E).
To place the phenotypes of breast Vδ1+ T cells into context, similar analyses were performed on co-extracted Vδ2+ and Vδ1-Vδ2-γδ T cells, on CD4+ and CD8+ αβ T cells, and on CD3- lymphocytes that will include NK and ILC. The greatest similarities to Vδ1+ T cells were shown by Vδ2+ and Vδ1-Vδ2- γδ cells and by CD8+ αβ T cells, although some such cells expressed CD28, and CD8+ αβ T cells were more uniformly CD103+ (fig. S1D). NK/ILC were also similar to Vδ1+ T cells except that they uniformly lacked PD-1. Finally, and as anticipated, CD4+ αβ T cells lacked NKG2D expression, and were mostly CD28+ and CD103- (fig. S1D), consistent with a recently-described tissue resident CD4+ phenotype (55).
Upon activation, breast-associated CD8+ αβ T cells were functionally most similar to Vδ1+ T cells in their Tc1 phenotype, showing an even greater frequency of IFN-γ-producing cells (fig. S1E). Likewise, breast-explanted CD4+ αβ T cells included more IL-13 producers than did Vδ1+ T cells. In sum, the healthy breast harboured a complex lymphoid ecosystem of multiple cell-types with related but distinct phenotypes.
Human breast Vδ1+ T cells are innate-like
Murine skin-resident γδ T cells can respond in vivo to NKG2D ligand upregulation without overt TCR stimulation (5). In relation to cancer, this is potentially important because NKG2D ligands are upregulated by DNA damage (8) and EGFR over-activity (9). We therefore investigated whether breast-explanted Vδ1+ T cells would respond to plate-bound recombinant MICA protein, a NKG2D ligand commonly expressed by tumors. MICA clearly provoked a subset of Vδ1+ T cells to produce TNF, IFN-γ, and to upregulate CD107a in a response inhibited by anti-NKG2D (Fig. 2A). Conversely, CD8+ αβ T cells within the identical grid cultures did not make significant responses to MICA relative to controls (Fig. 2A; fig. S2A), whereas both cell types showed increased responses when MICA was provided as a co-stimulator to anti-CD3 (Fig. 2B) (56, 57). The innate-like responsiveness of other breast γδ T cells was challenging to examine because even in cases where they composed a greater proportion of tissue γδ T cells, non-Vδ1+ cells were usually too few to assay reliably (fig. S2B). Despite this, we observed some NKG2D-dependent, innate-like responses for some Vδ1- Vδ2- T cells, but not for breast Vδ2+ T cells (fig. S2B).
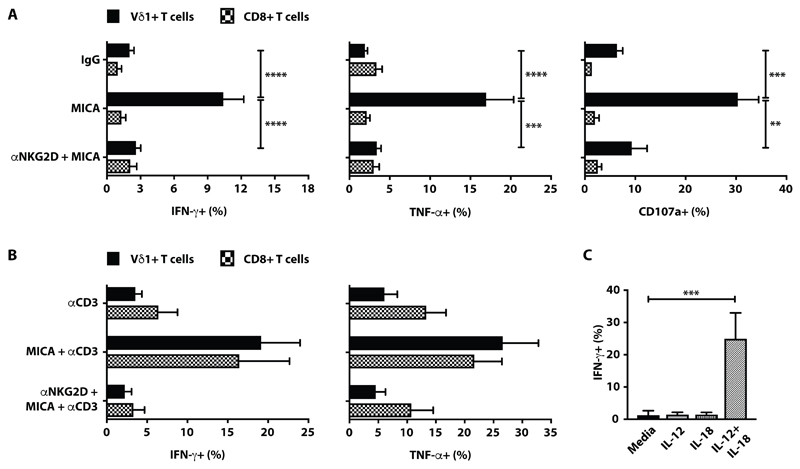
Fig. 2. Breast tissue-resident Vδ1+ T cells are innate-like.
(A) Summary data showing intracellular staining for IFN-γ (n=19 for Vδ1+ and n=15 for CD8+), TNF (n=17 for Vδ1+ and n=13 for CD8+) and cell-surface CD107a (n=10 for Vδ1+ and n=5 for CD8+) expression, following in vitro activation of breast-resident Vδ1+ cells or of Vδ1+ and CD8+ αβ T cells from within the same cultures, exposed to plate-bound recombinant MICA (10μg/ml) in the presence of brefeldin A (BFA). Plotted as percentage of parent Vδ1+ or CD8+ gate. (B) Summary data showing intracellular staining for IFN-γ (n=6 for Vδ1+ and n=6 for CD8+) and TNF (n=5 for Vδ1+ and n=5 for CD8+) following in vitro activation of breast tissue-resident Vδ1+ and CD8+ αβ T cells with low-dose plate-bound anti-CD3 antibody (50ng/ml) with or without plate-bound recombinant MICA (10μg/ml) in the presence of BFA. Where indicated, MICA stimulated cells were pre-treated with anti-human NKG2D antibody. Plotted as percentage of parent Vδ1+ or CD8+ gate. (C) Summary data for breastresident Vδ1+ T cells, showing intracellular IFN-γ production following in vitro activation with IL- 12 (n=3) or IL-18 (n=3) or IL-12 + IL-18 (n=9) and with medium alone (n=9). For all panels, mean with SEM indicated. *p<0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, Kruskal-Wallis with post-hoc Dunn’s test corrected for multiple testing.
It was also reported that innate-like γδ T cells make strong, TCR-independent responses to combinations of a STAT-signalling cytokine and an IL-1 family member (58). Indeed, breast-explanted Vδ1+ T cells produced IFN-γ in response to IL-12 + IL-18 but not to either alone, whereas CD4+ and CD8+ αβ T cells responded significantly less well to IL-12 + IL-18 (fig. S2C). In sum, the healthy breast harboured a mixture of innate-like Vδ1+ T cells and primarily adaptive αβ T cells
Innate-like γδ T cells in human breast cancers
The identification of innate-like γδ T cells in healthy human breast formed a backdrop to examining the tissue-associated lymphoid compartment in breast cancer subjects. Although breast cancers vary in lymphoid infiltrates (32, 59), γδ T cells were invariably recovered and were largely comparable to those from healthy tissue in terms of TCR usage: Vδ1 was predominant, although the tumor samples included some examples where either Vδ2+ cells or Vδ1-Vδ2- cells predominated (fig. S3A). As with γδ T cells from healthy breast, cells isolated from tumors using grids were phenotypically comparable with those examined directly ex vivo, albeit that grid cells again showed higher expression of CD69, and to some extent, NKG2D (fig. S3B). The similarities of TCR usage and surface marker expression for γδ T cells from tumor and non-malignant tissue were particularly apparent in paired samples from 26 subjects (Fig. 3A; fig. S3C). Moreover, this comparability extended to other lymphocyte subsets simultaneously harvested from the paired samples (fig. S3D).
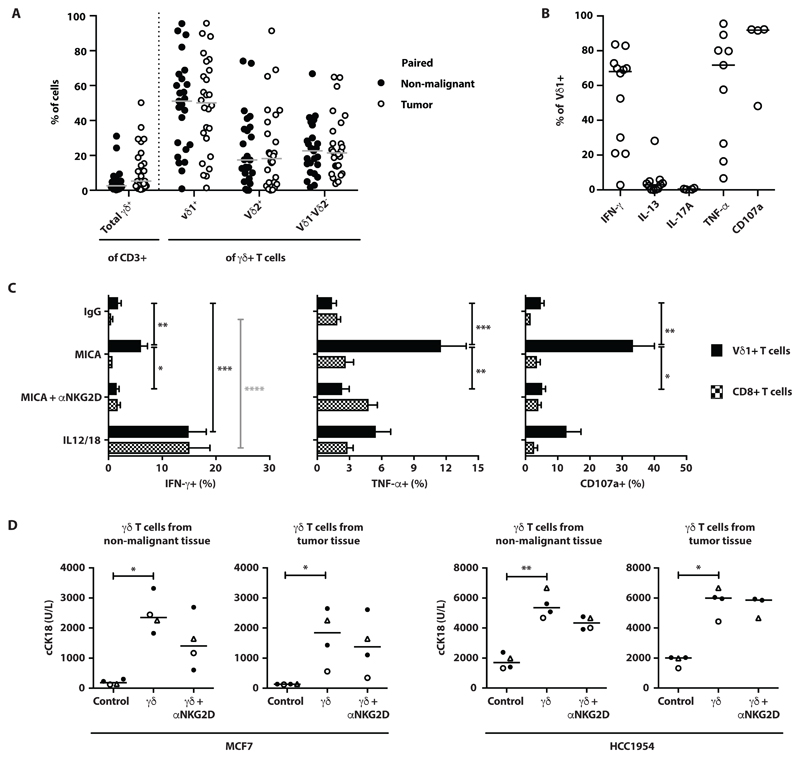
Fig. 3. Innate-like Vδ1+ Tc1 cells are a major subset of breast tumor infiltrating lymphocytes.
(A) γδ T cells were consistently isolated from human breast tumor samples obtained from mastectomies following grid culture. Dot plots showing numbers of TCRγδ+ cells isolated from paired non-malignant breast tissue (•) or tumor (o) expressed as a percentage of CD3+ T cells. γδ T cells were further phenotyped for Vδ1+, Vδ2+ and Vδ1-Vδ2- TCR usage expressed as a percentage of TCRγδ+ T cells (n=25); medians indicated. (B) Breast tumor infiltrating Vδ1+ T cells are functionally skewed. Dot plots showing intracellular cytokine staining for IFN-γ (n=12), IL-13 (n=11), IL-17A (n=4) TNF (n=9), and cell surface CD107a expression (n=4), following in vitro activation with PMA (10ng/ml) and ionomycin in the presence of Brefeldin A (BFA); medians indicated. (C). Summary data showing intracellular staining for IFN-γ (n=9-12 depending on activating condition), TNF (n=7), and cell-surface CD107a (n=4-7, depending on activating condition) expression following in vitro activation of Vδ1+ and CD8+ αβ T cells with plate-bound recombinant MICA (10μg/ml) in the presence of BFA, or in vitro activation with IL-12 (100ng/ml) and IL-18 (100ng/ml). Where indicated, MICA stimulated cells were pre-treated with blocking anti-human NKG2D antibody; mean with SEM indicated. (D) Tumor cell lines, MCF7 and HCC1954, were incubated with negatively sorted γδ T cells (γδ) derived from non-malignant breast tissue or breast tumor, at E:T of 5:1, in the presence (γδ + αNKG2D) or absence (γδ) of a blocking anti- NKG2D antibody for 48 hours. Cell lines without effector γδ T cells were used as negative controls (Control). Dot plots show concentrations of caspase-cleaved cytokeratin 18 (cCK18). Each data point represents the mean of two technical replicates; the median values for those data-points are indicated by a horizontal line (note that there were only 3 donors for the killing assay of HCC1954 cells by tumor-derived γδ T cells in the presence of anti-NKG2D). Δ and ○ are 2 donors for which there were paired non-malignant breast tissue and breast tumor. *p<0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, Kruskal-Wallis with post-hoc Dunn’s test corrected for multiple testing.
Likewise, the functional potential of Vδ1+ TILs was comparable to those of non-malignant breast Vδ1+ T cells, in being Tc1-skewed and IL-17-deficient (Fig. 3B). Vδ1+ TILs were again responsive to NKG2D ligands and IL-12+IL-18 in the absence of overt γδ TCR signalling, whereas co-isolated CD8+ αβ TILs did not show significant responses to MICA relative to controls, although they did respond better to IL-12+IL-18 than did counterparts from healthy breast (Fig. 3C).
Given their strong cytolytic responsiveness, we tested the capacity of breast-derived γδ T cells to kill two breast tumor cell lines, MCF7 and HCC1954, for which tumor cell lysis was distinguished from lymphocyte death by quantitating cytokeratin 18 release (60). Note that because γδ T cells are not MHC-restricted, it was possible to assess their functional responses to non-autologous tumor lines. γδ T cells from healthy breast donors (n=4) reproducibly killed MCF7 cells at an effector:target (E:T) ratio of 5:1 (fig. S3E), and using this ratio we found that cells from healthy breast and from tumor samples (n=4) showed comparable capacity to kill MCF7 and HCC1954 (Fig. 3D). However, whereas NKG2D receptor blockade reduced killing by γδ T cells from healthy breast, killing by TCRγδ+ TILs was less affected (Fig. 3D). We also observed primary tumor cell killing by autologous TCRγδ+ TILs for the one patient from whom primary tumor cells could be grown and stably maintained (fig. S3F). In sum, primary γδ T cells obtained from breast cancers were functionally competent; could respond innately via NKG2D engagement; and could lyse breast tumor cells, albeit that this was not overtly NKG2D-dependent.
Vδ1+ TILs and durable remission
Given the functional Tc1 skew of γδ TILs, we wished to examine their status in relation to clinical outcome in an aggressive subset of breast cancer where time-to-events is relatively short. To this end, we sought patients with Triple Negative Breast Cancer (TNBC) from whom we could access sufficient paired tumor and non-malignant tissues and for whom accurate clinical follow-up data were available. Those criteria were met by 11 patients treated at Guy’s and St Thomas’ Hospitals, London (BTBC Study REC 13/LO/1248), for whom there were bio-banked, formalin-fixed paraffin-embedded (FFPE) samples. Patients were otherwise unselected. All patients had localized or loco-regional TNBC (AJCC Stage I-III) and had surgery with curative intent. Five of eleven remained in complete remission at last follow up (range 48-63 months; table S2; demarcated in blue in the figures that follow), whilst six had relapsed with distant metastatic disease within 18 months (range 6-18 month; table S2; demarcated in red in the figures that follow).
FFPE blocks were needle-dissected to delineate tumor and non-malignant tissue for genomic DNA extraction. Given the difficulty of immunohistochemical approaches in detecting γδ T cells in FFPE (61), we employed quantitative genomic DNA sequencing of rearranged TCRα and TCRγ chain genes to infer absolute counts of αβ and γδ T cells. Indeed, this approach has been shown to be more sensitive than immunohistochemistry for detecting TILs (29), and is used clinically, for example to assess minimal residual disease.
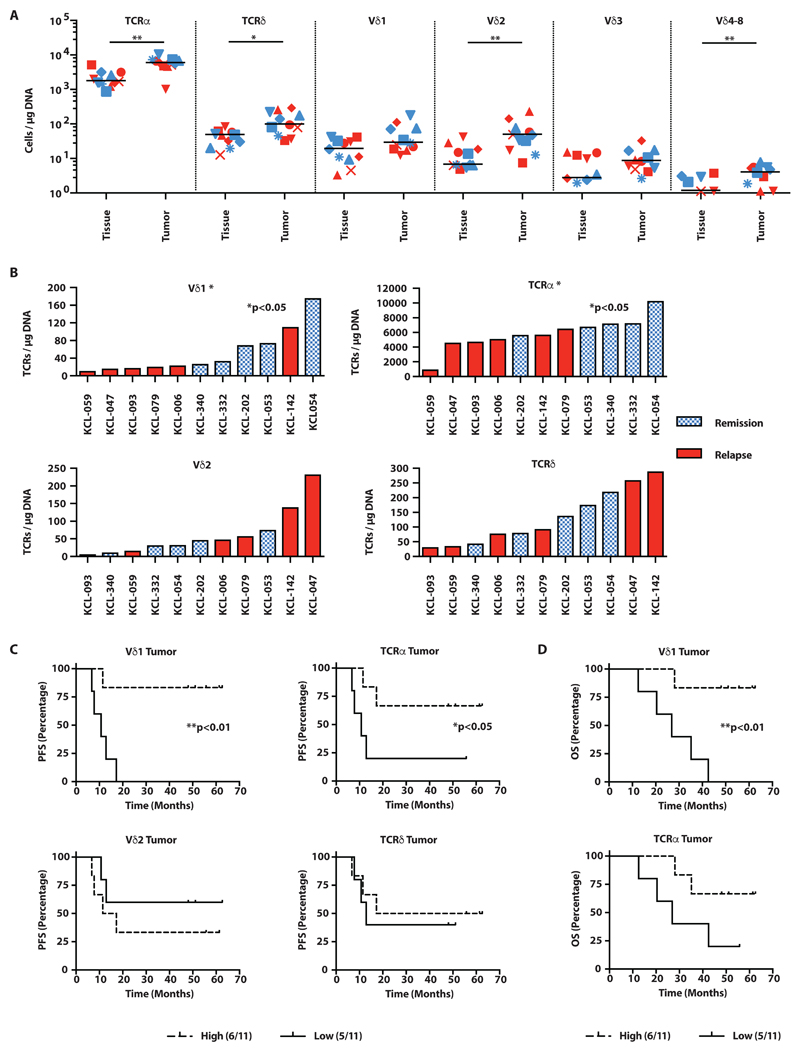
We found that both αβ and γδ T cells were significantly more abundant per microgram of DNA extracted from tumor tissue versus paired non-malignant tissue (Fig. 4A, fig. S4A). However, it was striking that in cases of remission, the numbers of TCRα, TCRγδ, TCRVδ1, and TCRVδ2 DNAs were invariably enriched in paired malignant versus non-malignant tissue, whereas the pattern in relapsed cases was essentially random (fig. S4A). Moreover, in addition to enrichment relative to healthy tissue, there were conspicuously more TCRα+ and Vδ1+ TCRs per microgram of total tumor DNA in remission cases versus relapse (Fig. 4B). Thus, in indicating positive clinical outcome, the dynamics of small numbers of Vδ1+ T cells were as potent as the much larger-scale dynamics of αβ T cells, defined subsets of which have been shown to predict survival in TNBC (62). Conversely, this was not so for either total TCRγδ or Vδ2 TCRs (Fig. 4B), the latter possibly reflective of cells infiltrating from the PB.
Fig. 4. Vδ1+ T cells in TNBC are predictive of disease-free and overall survival.
(A) Overall landscape of T cell subsets in non-malignant breast tissue (“Tissue”) and matched tumor tissue (“Tumor”), determined by quantitative sequencing of rearranged TCR genes from patients undergoing mastectomy for TNBC. Absolute TCR copies (a surrogate for T cell numbers) are plotted per microgram of input DNA. Individual patients plotted: red=patients with relapsed disease, blue=patients in remission; median bar plotted, *p<0.05, **p≤0.01, Wilcoxon matched pairs signed rank test. (B) Intratumoral presence of αβ T cells and γδ T cells in patients with subsequent relapsed disease versus those who remained in remission. *p<0.05 K-S test. (C) PFS (months from surgery) split on median T cell subsets found in 11 TNBC tumors. *p<0.05, **p≤0.01, Gehan-Breslow-Wilcoxon test. (D) OS (months from surgery) split on median T cell subsets found in 11 TNBC tumors. *p<0.05, **p≤0.01, Gehan-Breslow-Wilcoxon test.
Striking manifestations of the correlations were evident from Kaplan-Meier plots of progression-free survival (PFS), where limited representation (less than median values) of intra-tumoral TCRα and Vδ1 TCRs were predictive of poor PFS, whereas neither total TCRγδ nor Vδ2+ TCRs predicted outcome (Fig. 4C). Furthermore, there was a positive and significant correlation of intra-tumor TCRα with Vδ1 TCRs (Spearman r=0.75, p≤0.01) (fig. S4B), whereas neither total TCRγδ nor Vδ2 TCRs correlated with TCRα: moreover, no TCRα-TCRδ correlation existed in non-malignant tissue (fig. S4B). Additionally, Vδ1 TCRs predicted overall survival (OS), although TCRα did not (Fig. 4D).
In situ evidence of innate-like Vδ1+ TILs
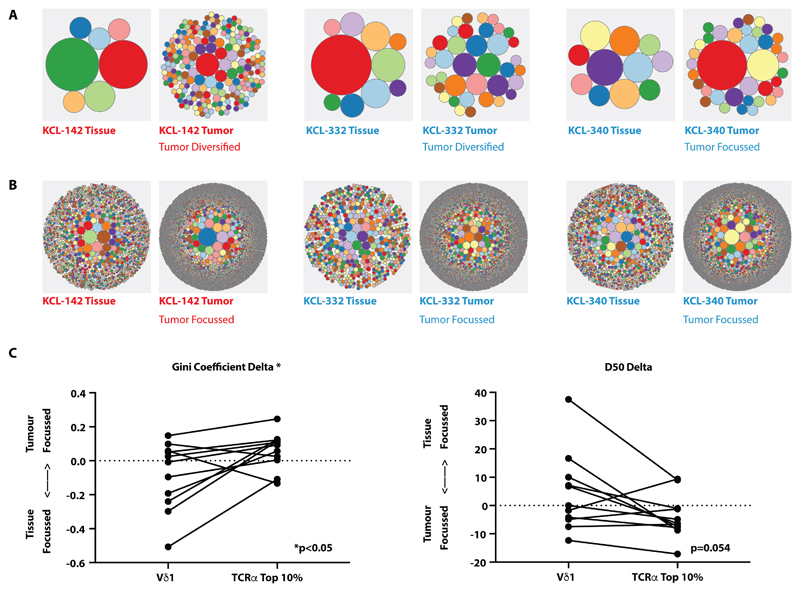
Having established significant positive correlates of TCRαβ+ T cells with PFS and of Vδ1+ T cells with PFS and OS, we assessed their TCR repertoires. When represented as circular tree-plots, the Vδ1+ repertoires in tumors compared to paired non-malignant tissue showed no clear overall focussing (examples shown in Fig. 5A), as quantitated by D50 (the smallest number of clones accounting for 50% of the total number of sequences observed) from paired non-malignant and tumor tissues (table S3). Focussing would have suggested an adaptive TCR-driven response, as we observed for the TCRα repertoires of most tumors versus paired normal tissue (examples shown in Fig. 5B), consistent with previous reports (62, 63).
Fig. 5. Lack of tumor Vδ1+ TCR focussing vis-à-vis focussing of TCRα.
(A) Examples of circular tree plots of the Vδ1+ repertoire in paired non-malignant tissue (“Tissue”) and tumor tissue (“Tumor”), where each circle represents a unique TCR clonotype and the size of the circle is proportional to the representation of the specified clone. Plots were generated from total Vδ1+ TCRs. (B) Examples of circular tree plots of the TCRα repertoire in paired non-malignant tissue (“Tissue”) and tumor tissue (“Tumor”). Plots were generated from total TCRα+ TCRs. (C) Vδ1+ and TCRα+ sequences in non-malignant and tumor tissue were down-sampled (within each patient) to equivalent numbers to calculate diversity metrics. The degree of repertoire focussing was assessed by the delta of the Gini coefficient and the delta D50 of sequences of Vδ1 chains and the top 10% in abundance of TCRα sequences in tumor versus paired non-malignant repertoires. To test if the degree of repertoire focusing was different between Vδ1+ and TCRα+ compartments in individual patients, the Wilcoxon matched pairs signed rank test was used to compare ΔGini and ΔD50. All sequences were analyzed based on amino acid sequence. n=11.
To further analyze the data, we also calculated the Gini coefficient (a statistical measure of distribution where 0=fully polyclonal and 1=monoclonal) for Vδ1 TCRs from paired non-malignant and tumor tissues (table S3). Note that the tumor TCRs were down-sampled so that equivalent numbers of TCRs were compared within each patient. The same treatment was then applied to TCRα, although we considered only the most abundant 10% of TCRα TCRs, given recent evidence that the most relevant antigen-reactive αβ T cells commonly sit within this fraction (64). For each patient, we then calculated the delta of the Gini coefficient of paired tumor tissues and nonmalignant tissues for both Vδ1 and for TCRα: note that tumor focussing would be reflected by positive ΔGini coefficient (Fig. 5C). We then likewise calculated the delta of the D50 values for paired tumor and non-malignant tissues for both Vδ1 and for TCRαβ: note that tumor focussing would be reflected by negative ΔD50 (Fig. 5C). These analyses confirmed quantitatively that TCRα repertoire focussing occurred in all but two tumor samples (KCL-059 and KCL-202), whereas Vδ1 repertoires showed no bias either toward focussing or toward diversification (Fig. 5C).
As a complementary approach, we also applied repertoire metrics to non-down-sampled (raw) TCR reads, using normalized measures of clonality (normalized Shannon entropy, Gini coefficient, D50), as previously employed by others (29, 63). These methods too suggested tumor repertoire focussing for TCRα (significance was reached for D50), whereas there was no such finding for Vδ1 (fig. S5, table S4). Collectively these data strongly suggest that the Vδ1+ cell responses were not driven by clonotypic antigens.
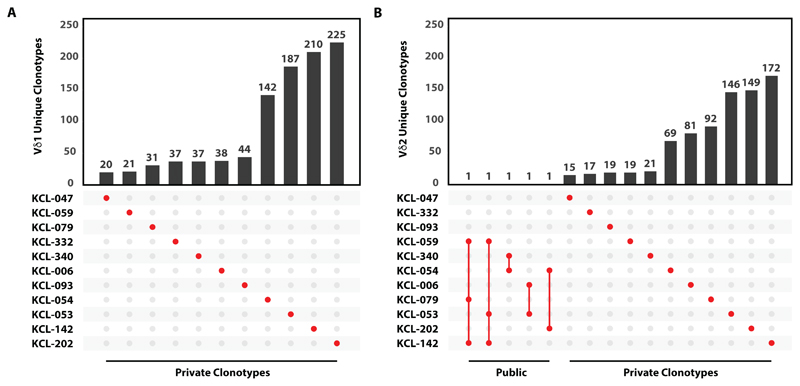
Given that among TCR gene rearrangements, TCRδ harbours the highest potential for junctional diversity (65), it was not surprising that no public Vδ1 sequences were observed across different donors’ tumors (Fig. 6A). However, there was some Vδ1 sequence overlap between tumors and tissue from the same donor (fig. S6). Although the sample size was small, the lack of public sequences would also be consistent with TCR-agnostic, innate-like regulation of tissue resident Vδ1+ T cells. By contrast, assessment of a comparably-sized sample showed that some Vδ2 TCRs were shared across multiple donors (Fig. 6B). Interestingly, most shared sequences reflected TCRs reactive to phospho-antigens that can be upregulated in tumors (66), with a conserved hydrophobic residue in position 97 (table S5) specifically associated with phospho-antigen-mediated selection of the Vδ2+ repertoire (67, 68).
Fig. 6. No detectable public intratumoral Vδ1+ clonotypes.
(A) Intersections of Vδ1+ clonotypes between 11 patient tumor samples. Vertical bars represent the number of unique TCRs whilst the dot matrix represents sharing of TCRs across patients. A shared or public clonotype would be represented by at least two red dots (sharing between two patients) joined by a vertical red line. Private sequences are presented by an unconnected single red dot. (B) Intersections of Vδ2+ clonotypes between 11 patient tumor samples. All sequences were analyzed based on amino acid sequence.
Discussion
The past decade has witnessed a sea-change in cancer immunology, with the realization that tumors are often antigenic, and that tumor-reactive T cells can provide patient-beneficial responses, particularly if de-repressed by checkpoint blockade (69, 70). Hence, there is considerable interest in the immune-ecology of tumors. Over the same period, it became clear that several extra-lymphoid tissues in which tumors commonly form ordinarily harbor large myeloid and lymphocyte compartments, including γδ T cells that become tissue-resident during the cells’ development, and systemic αβ T cells that become TRM cells following priming in secondary lymphoid organs (71, 72).
Although positive clinical outcomes in TNBC have been associated with CD8+ TCRαβ+ TRM cells (62), there has been little investigation of whether a human breast-resident γδ T cell compartment exists that might influence breast cancer outcomes (73). This study addresses this point by first establishing a tissue-resident γδ T cell compartment in healthy human mammary tissue. This may be evolutionarily conserved, since TCRγδ+ lymphocytes were reported in alveolar mammary epithelia of lactating cows (74), and were associated with lactating mammary glands in mice (26). To characterise human breast-resident γδ T cells in sufficient numbers, we employed grid cultures previously used to elucidate key features of human skin and lung T cells (35–38). While this is a limitation, there was strong phenotypic consistency with breast γδ T cells examined directly ex vivo. This permitted our description of the compartment as mostly γδ1+, NKG2D+, CD69+, partly CD103+, and with a Tc1 phenotype lacking IL-17 production. This is very distinct from peripheral blood γδ T cells but shares features with human intestinal epithelial γδ T cells (40).
Additionally, human breast γδ T cells were innately responsive to NKG2D activators, whereas co-located αβ T cells required coincident TCR stimulation. Thus, healthy human breast Vδ1+ cells have an inherent potential to detect and respond rapidly to local cells en route to malignancy. Some of these signature properties may be shared with local Vδ1-δ2- T cells, that are often Vδ3+, although these cells were most often present in very small numbers.
Within breast tumors, Vδ1+ T cells and αβ T cells were frequently more abundant than in paired healthy tissue, particularly in patients in remission. This likely reflects an inflexion point at which an activating immune response to the tumors occurred. Furthermore, when extracted from breast tumors, the comparatively expanded Vδ1+ and αβ T cell populations were functionally competent, retaining the innate-responsiveness and Tc1 potential of cells from healthy breast. These observations evoked evidence that PD-1+ TILs responded functionally to re-stimulation (75), and that their presence could be associated with favourable outcome (62). Indeed, although only few patients were available for in-depth analysis in this study, they were sufficient to show significant positive correlations of tumor-derived Vδ1+ and αβ T cells with clinical outcome, with Vδ1 TCRs correlating with both PFS and OS. It is therefore attractive to hypothesize that maximum patient benefit accrues from a collaboration of the innate responsiveness of local Vδ1+ cells with the antigen-specific modus operandi of αβ T cells, particularly CD8+ TCRαβ+ TRM cells (62).
At least two patient-beneficial facets of collaboration between Vδ1+ and TCRαβ+ T cells may be envisioned. First, by recognizing tumors via innate stimuli, Vδ1+ T cell responses may not be limited either by the number of neo-antigen-generating somatic mutations, or by immune-evasive suppression of peptide antigen presentation (47, 76–79). The innate stimuli may include ligands for several NK receptors (43), including but not limited to NKG2D.
Second, the cytolytic, Tc1 phenotype of Vδ1+ cells may be augmented by the cells’ capacity to promote tissue immunogenicity via chemokine secretion and possibly via direct antigen presentation (17, 80, 81). Indeed, a critical role of tissue-resident Vδ1+ cells may be to orchestrate multicomponent local immune responses to defined challenges, while remaining tolerant to others. In this context, tissue/tumor immunogenicity might be effectively enhanced in the clinic by agents promoting the activities of local Vδ1+ T cells, or by the cells’ adoptive transfer, in concert with the activation/de-repression of adaptive T cells. Because γδ T cells are not MHC-restricted, they might be adoptively transferred from heterologous donors, and because they are naturally tissue-resident, local Vδ1+ cells may cope with hypoxic environments that prove hostile to systemic lymphocytes (82, 83).
Such considerations may pertain to other human tissues harbouring local γδ T cell compartments, such as the gut, and may underpin reportedly strong correlations of γδ T cells with favourable clinical outcomes across a broad spectrum of human tumors (84). Nonetheless, it is important to note that in this study neither total γδ TCRs nor Vδ2+ TCRs correlated with clinical outcome. This emphasizes the fact that γδ T cells comprise biologically distinct subsets, as is the case for αβ T cells or ILCs. Indeed, even amongst intratumoral CD8+ T cells, which are traditionally associated with patient benefit, most benefit in TNBC was attributable to a discrete subset of local CD8+ TRM cells (62). In mice, functionally different γδ T cell subsets have been reported to either mediate or repress tumor immunosurveillance (85, 86). Most often, IL-17 has been implicated in tumor promotion by γδ T cells (50–52), whereas IL-17 producing cells are seemingly rare in humans, wherein the predominant phenotype is cytolytic, TNF/IFN-δ-producing (87) as described here.
In mice, the innate responsiveness of γδ T cells and their suppression of IL-17 production were induced developmentally by subset-specific, tissue-specific selecting elements of the butyrophilin-like (Btnl/BTNL) family, members of which can also regulate human γδ T cells (30, 40, 58, 88). It is therefore possible that such elements act locally to select and regulate human breast-resident Vδ1+ T cells, in which regard the mammary gland is one of reportedly few tissues expressing BTNL9 (89).
Our study did not focus on interactions of breast Vδ1+ T cells with other breast-resident immune cells including B cells (32). Likewise, spatial relationships between breast γδ T cells and tumor-associated tertiary lymphoid structures were not determined (31, 32). In practical terms, clinical studies have suggested that human breast cancer, including TNBC, can be vulnerable to immune attack (33, 34), and yet the efficacy of immunotherapies in this indication has been relatively poor. We strongly believe that this may be redressed by shifting therapies away from their unique focus on conventional, adaptive T cell responses, and by learning from the natural ecology of the local breast T cell compartment. In particular, we believe this may promote the immunogenicity of tumor tissues that drives and sustains adaptive responses.
Materials and Methods
Study design
The aim of this study was to ascertain whether the human breast might contain a tissue-resident γδ T cell compartment and to determine if this might be protective in breast cancers. This study was undertaken first by demonstrating the reproducible presence of γδ T cells in healthy breast tissue with a particular focus on Vδ chain usage. Then we determined their functional potential and whether they might be consistent with protective tumor immunosurveillance. These approaches were subsequently applied to γδ T cells in breast tumors. Co-located αβ T cells isolated using the same protocol and maintained in the same culture conditions were used as controls. Having established the presence of γδ T cells in human breast tissue and tumors and their innate-like immunosurveillance capacity in vitro, we examined the presence of these cells in situ (via their DNA rearrangements), and correlated this to prognosis in clinical samples. We also sought in situ evidence, particularly TCR repertoire clonality, for cells functioning in an innate-like immunosurveillance capacity, as was established ex vivo. Detailed study design, sample sizes, replicates, and inclusion/exclusion criteria are provided in the figure legends or in Materials and Methods. The sample sizes and experimental repetitions were sufficient to permit rigorous statistical analysis as described in the Figure legends and Materials and Methods. All antibodies and key reagents are listed in table S6. Primary data are reported in data file S1.
Clinical material
Human breast samples were obtained from adult female patients undergoing breast reduction or risk-reducing mastectomy (29 patients) or breast tumor resection (90 patients) after informed consent as part of a non-interventional clinical trial (BTBC study: REC No: 13/LO/1248, IRAS ID 131133; Principal Investigator: Professor Andrew Tutt. Study title: Analysis of functional immune cell stroma and malignant cell interactions in breast cancer in order to discover and develop diagnostics and therapies in breast cancer sub-types). This study had local research ethics committee approval and was conducted adhering to the principles of the Declaration of Helsinki. Specimens were collected from surgery into sterile saline and transported immediately to cut-up. A histopathologist or pathology-trained technician identified and collected tumor material and ipsilateral non-adjacent normal breast tissue from surgical specimens from which lymphocytes were subsequently isolated as described below. Demographics of the patients are detailed in table S2. In addition to the patients above, for TCR sequencing experiments, tumor and paired non-malignant tissue DNA was extracted from bio-banked FFPE blocks from eleven patients with AJCC Stage I-III TNBC who had mastectomies for which we could access sufficient material and accurate clinical follow-up data. No other criteria were applied. The eleven cases were also part of the BTBC clinical trial described above.
Primary lymphocyte extraction and culture
For direct ex vivo isolation, fresh breast tumor or tissue was coarsely minced with scalpels and then dissociated using the MACS human tumor dissociation kit on a gentleMACS Dissociator as per manufacturer’s instructions (Miltenyi Biotec). Samples were washed twice with sterile RPMI-1640 and used immediately for downstream assays. Lymphocytes were also harvested using a “grid” explant system adapted from a protocol first described by Clark and colleagues (35). Briefly, fresh breast tumor or tissue was minced using scalpels and placed onto rat tail collagen (100μg/ml, BD Biosciences)-coated Cellfoam grids (Cytomatrix Pty Ltd). Each grid was placed into a separate well of a 24-well tissue culture plate and cultured in complete medium [Iscove’s Modified Dulbecco’s Medium (IMDM, Life Technologies), 10% heat-inactivated foetal bovine serum (FBS, Life Technologies), L-glutamine (292μg/ml, Life Technologies), penicillin (100units/ml, Life Technologies), streptomycin (100μg/ml, Life Technologies) and 2-mercaptoethanol (3.5μl/L, Life Technologies)] supplemented with recombinant human (rh) IL-2 (100 IU/ml, Proleukin, Novartis Pharmaceuticals UK Ltd), and rhIL-15 (10 ng/ml, BioLegend). The grids were maintained for 3 weeks in culture at 37°C/5% CO2, and the lymphocytes harvested by washing the wells/grids with 0.01 mM HEPES/HBSS.
Flow cytometry and fluorescence-activated cell sorting
Cells were washed in sterile PBS to remove traces of serum and stained for 20 minutes at room temperature with LIVE/DEAD Fixable Aqua (Thermo Fisher Scientific) in PBS. Subsequent surface staining was carried out in FACS buffer (PBS, 2% FCS, 1mM EDTA) for 20 minutes at 4°C (see table S6) before washing twice with FACS buffer and fixing with CellFIX (BD) for 10 minutes at room temperature. For intracellular cytokine staining (ICS), fixed cells were washed twice with Intracellular Staining Permeabilization Wash Buffer (BioLegend) and stained for 20 minutes at room temperature before two further washes with Intracellular Staining Permeabilization Wash Buffer. Samples were acquired on a BD FACSCanto II or BD LSRFortessa and were analyzed using FlowJo software (Flowjo LLC). For FACS, cells were not fixed and sorted on a BD FACSAriaII as detailed below. Antibodies are listed in table S6 and were used at 1:50 dilution unless otherwise specified.
In vitro lymphocyte activation assays
Directly isolated and grid explant-isolated lymphocytes were stimulated with PMA (10ng/ml, Sigma) and ionomycin (1μg/ml, Sigma) in the presence of brefeldin A (BFA, 20μg/ml, Sigma) for 4 hours at 37°C/5% CO2 before surface marker and intracellular cytokine staining and acquisition on a BD FACS Canto or Fortessa. For plate-bound NKG2D ligand assays, lymphocytes were harvested from explant cultures 24 hours prior to activation and rested in complete media without cytokine supplementation. After resting, lymphocytes were transferred to 96-well flat bottom cell culture plates (Corning) coated with rhMICA (10μg/ml, R&D), anti-CD3 (50ng/ml, Biolegend), anti-CD3 (50ng/ml, Biolegend) and rhMICA (10μg/ml, R&D), or mouse IgG2a (50ng/ml, Biolegend) at 100,000 cells per well in 100μl of complete medium. Plates were incubated for 6 hours at 37°C/5% CO2 in the presence of IL-15 (10ng/ml, BioLegend) and brefeldin A (20μg/ml, Sigma). Where CD107a was used as a functional read-out, anti-human CD107a antibody (1:400 final concentration, BioLegend) was also added at the start of the assay along with monensin at 1x (BioLegend). For NKG2D-blocking conditions, anti-NKG2D antibody (10μg/ml, clone 1D11, BioLegend) was added to lymphocytes just prior to plating.
For cytokine activation assays, lymphocytes were incubated with IL-12 (100ng/ml, PeproTech) and/or IL-18 (100ng/ml, Medical and Biological Laboratories) for a total of 24 hours at 37°C/5% CO2 with BFA (20μg/ml, Sigma) added for the last 4 hours before surface and intracellular cytokine staining for flow cytometry. For IL-17-skewing assays, breast tissue explants were cultured in complete medium and in conditions as described above with the addition of rhIL-2 +/- rhIL-15, rhIL-1b, rhIL-6, rhIL-23 and rhTGFβ for 3 days. These cells were then activated with PMA and ionomycin, in the presence of BFA (20μg/ml) for 4 hours at 37°C/5% CO2 before surface and intracellular cytokine staining for flow cytometry.
Cytotoxicity assays
Grid explant-derived γδ T cells from breast tissues and tumors were isolated by fluorescence-activated cell sorting via depletion of TCRαβ+ and NKp46+ cells. Target cells, MCF7 and HCC1954 (CRUK/Francis Crick Institute Cell Service), were seeded at 10,000 cells/well in 96-well flat bottom plates (Corning) 24 hours prior. 50,000 negatively sorted γδ T cells were added to target cells in the presence or absence of blocking NKG2D antibody (10μg/ml, BioLegend). Cells were incubated at 37°C for 48 hours, after which supernatants were collected and stored at -20°C until further analysis. Target cell apoptosis was measured using ELISA for the epithelial cell-specific caspase-cleaved cytokeratin 18 (Diapharma), according to manufacturer’s instructions.
Cell lines and culture conditions
Target MCF7 and HCC1954 cell lines were sourced from Cancer Research UK Cell Services (Clare Hall, London) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technologies), supplemented with 10% heat-inactivated foetal bovine serum (FBS, Life Technologies), penicillin (100units/ml, Life Technologies), streptomycin (100μg/ml, Life Technologies) at 37°C/5% CO2. HCC1954 were maintained in RPMI-1640 (Gibco) supplemented with 10% heat-inactivated (FBS, Life Technologies), penicillin (100units/ml, Life Technologies), streptomycin (100μg/ml, Life Technologies) at 37°C/5% CO2.
DNA extraction
DNA was extracted from FFPE paired tumor tissue and normal tissue blocks of 11 patients with TNBC treated with mastectomy as part of the BTBC study. Tumor tissue was needle micro-dissected after sectioning. The QIAmp DNA FFPE Tissue kit (Qiagen) was used per manufacturer’s instructions to extract DNA. DNA was quantified using a Qubit fluorometer and material from patients with >1μg of DNA from both tumor and normal tissue were sent for quantitative TCRα/δ locus sequencing by Adaptive Biotechnologies.
TCR Sequencing
TCR sequencing was performed by Adaptive Biotechnologies. The Adaptive Biotechnologies platform uses genomic DNA and can quantitate T cell numbers. Reads were aligned and annotated by Adaptive Biotechnologies and data was output as .csv files (available from https://osf.io/d4eu6/) for downstream analysis from the immunoSEQ Analyzer (https://clients.adaptivebiotech.com/login). Output was filtered on in-frame CDR3s as well as TCRA to TCRA V-J family joins for TCRαβ and TCRD to TCRD V-J family joins for TCRγδ T cells. We normalized the absolute counts of TCRs to 1μg of input DNA for each sample to enable normalized comparison of infiltrating T cell numbers across all samples. All analyses were carried out using CDR3 amino acid sequences as opposed to nucleotide sequences.
Clonal repertoire analysis
To compare clonality metrics within each patient between paired tumor and non-malignant tissue we down sampled TCRs from each pair of samples. For Vδ1, TCRs from tumor and non-malignant tissue were down sampled to the number of clones in the smaller sample with probability of drawing a clonotype equal to its frequency in the full sample. Down sampling with replacement was performed 200 times. For TCRαβ, clonotypes were ordered in decreasing frequency and the top 10% of total TCRs in each pair of tumor and non-malignant tissue were used for down sampling as described above. Clonality metrics were then applied to the down sampled data and the median values plotted. As an alternative we also applied normalized measures of clonality (normalized Shannon entropy, Gini coefficient, D50) to the raw data. Treemaps were generated using the MacroFocus Treemap programme (https://www.treemap.com). We visualized shared clonotypes patients using the UpSet R package (https://ieeexplore.ieee.org/abstract/document/6876017 and https://vcg.github.io/upset).
Statistical Analysis
Statistical significance was determined by Kolmogorov-Smirnov test, log rank test, Wilcoxon matched pairs signed rank test, or Mann Whitney test, as indicated in figure legends using Prism 7 software (GraphPad). All findings were considered significant at a P value threshold of <0.05. Where results of statistical test are shown; *p<0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001 unless otherwise indicated.
Supplementary Material
One sentence summary.
An innate-like γδ T cell compartment in healthy human breast is described, and shown to correlate with remission in Triple Negative Breast Cancer
Acknowledgements
We thank Angela Clifford, Sheeba Irshad and Thanussuyah Alaguthurai for consenting and recruiting patients and current and former colleagues for helpful advice and discussions. We thank the flow cytometry service units of the Peter Gorer Department of Immunobiology and the Guy’s and St Thomas’ Hospital Trust Biomedical Research Centre (BRC) for outstanding support.
Funding: The work was supported by a Wellcome Trust [WT] Investigator Award [106292/Z/14/Z] to AH, and by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CRUK) (FC001093), the UK Medical Research Council (FC001093), and the WT (FC001093). FK is funded by a BRC fellowship; Y.W. by a National Institute for Health Research fellowship; AG and AT by Breast Cancer Now funding at King’s College London. The work was conducted as part of the CRUK Cancer Immune Therapy Accelerator (CITA) and the CRUK City of London Major Centre.
Footnotes
Author contributions: F.K.C., Y.W., R.T.W and A.H. conceived the study and designed experiments. F.K.C. and Y.W. performed and analyzed experiments. C.N-L., J.O. and P.G. processed patient samples and provided technical assistance. A.G., A.L., D.B. and P.V. assisted with data analysis. A.T. recruited patients. F.K.C., Y.W. and A.H. wrote the manuscript. A.T. and A.H. supervised the study.
Competing interests: A.H. is a co-founder and equity holder in Gamma Delta Therapeutics, plc.
Data and materials availability: All data associated with this study are present in the paper or Supplementary Materials.
References
- 1.Hayday AC, Saito H, Gillies SD, Kranz DM, Tanigawa G, Eisen HN, Tonegawa S. Structure, organization, and somatic rearrangement of T cell gamma genes. Cell. 1985;40:259–269. doi: 10.1016/0092-8674(85)90140-0. [DOI] [PubMed] [Google Scholar]
- 2.Brenner MB, McLean J, Dialynas DP, Strominger JL, Smith JA, Owen FL, Seidman JG, Ip S, Rosen F, Krangel MS. Identification of a putative second T-cell receptor. Nature. 1986;322:145–149. [PubMed] [Google Scholar]
- 3.Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 4.Girardi M, Oppenheim DE, Steele CR, Lewis JM, Glusac E, Filler R, Hobby P, Sutton B, Tigelaar RE, Hayday AC. Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294:605–609. doi: 10.1126/science.1063916. [DOI] [PubMed] [Google Scholar]
- 5.Strid J, Roberts SJ, Filler RB, Lewis JM, Kwong BY, Schpero W, Kaplan DH, Hayday AC, Girardi M. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nature Immunology. 2008;9:146–154. doi: 10.1038/ni1556. [DOI] [PubMed] [Google Scholar]
- 6.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of Ligands for the NKG2D Activating Receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayday AC. γδ T Cells and the Lymphoid Stress-Surveillance Response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vantourout P, Willcox C, Turner A, Swanson CM, Haque Y, Sobolev O, Grigoriadis A, Tutt A, Hayday A. Immunological Visibility: Posttranscriptional Regulation of Human NKG2D Ligands by the EGF Receptor Pathway. Science Translational Medicine. 2014;6:231ra49–231ra49. doi: 10.1126/scitranslmed.3007579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci USA. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girardi M, Glusac E, Filler RB, Roberts SJ, Propperova I, Lewis J, Tigelaar RE, Hayday AC. The Distinct Contributions of Murine T Cell Receptor (TCR)γδ +and TCRαβ +T Cells to Different Stages of Chemically Induced Skin Cancer. Journal of Experimental Medicine. 2003;198:747–755. doi: 10.1084/jem.20021282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Yang W, Pan M, Scully E, Girardi M, Augenlicht LH, Craft J, Yin Z. Gamma delta T cells provide an early source of interferon gamma in tumor immunity. J Exp Med. 2003;198:433–442. doi: 10.1084/jem.20030584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Eltoum I-EA, Guo B, Beck BH, Cloud GA, Lopez RD. Protective Immunosurveillance and Therapeutic Antitumor Activity of γδ T Cells Demonstrated in a Mouse Model of Prostate Cancer. J Immunol. 2008;180:6044–6053. doi: 10.4049/jimmunol.180.9.6044. [DOI] [PubMed] [Google Scholar]
- 14.Fisher JP, Heuijerjans J, Yan M, Gustafsson K, Anderson J. γδ T cells for cancer immunotherapy: A systematic review of clinical trials. oncoimmunology. 2014;3:e27572. doi: 10.4161/onci.27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girardi M, Lewis J, Glusac E, Filler RB, Geng L, Hayday AC, Tigelaar RE. Resident skin-specific gammadelta T cells provide local, nonredundant regulation of cutaneous inflammation. J Exp Med. 2002;195:855–867. doi: 10.1084/jem.20012000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandes M, Willimann K, Moser B. Professional antigen-presentation function by human gammadelta T Cells. Science. 2005;309:264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Wu W, Wong WM, Ward E, Thrasher AJ, Goldblatt D, Osman M, Digard P, Canaday DH, Gustafsson K. Human gamma delta T cells: a lymphoid lineage cell capable of professional phagocytosis. The Journal of Immunology. 2009;183:5622–5629. doi: 10.4049/jimmunol.0901772. [DOI] [PubMed] [Google Scholar]
- 18.Stingl G, Gunter KC, Tschachler E, Yamada H, Lechler RI, Yokoyama WM, Steiner G, Germain RN, Shevach EM. Thy-1+ dendritic epidermal cells belong to the T-cell lineage. Proc Natl Acad Sci USA. 1987;84:2430–2434. doi: 10.1073/pnas.84.8.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asarnow DM, Kuziel WA, Bonyhadi M, Tigelaar RE, Tucker PW, Allison JP. Limited diversity of gamma delta antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell. 1988;55:837–847. doi: 10.1016/0092-8674(88)90139-0. [DOI] [PubMed] [Google Scholar]
- 20.Goodman T, Lefrancois L. Expression of the gamma-delta T-cell receptor on intestinal CD8+ intraepithelial lymphocytes. Nature. 1988;333:855–858. doi: 10.1038/333855a0. [DOI] [PubMed] [Google Scholar]
- 21.Kyes S, Carew E, Carding SR, Janeway CA, Hayday A. Diversity in T-cell receptor gamma gene usage in intestinal epithelium. Proc Natl Acad Sci USA. 1989;86:5527–5531. doi: 10.1073/pnas.86.14.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itohara S, Farr AG, Lafaille JJ, Bonneville M, Takagaki Y, Haas W, Tonegawa S. Homing of a gamma delta thymocyte subset with homogeneous T-cell receptors to mucosal epithelia. Nature. 1990;343:754–757. doi: 10.1038/343754a0. [DOI] [PubMed] [Google Scholar]
- 23.Sumaria N, Roediger B, Ng LG, Qin J, Pinto R, Cavanagh LL, Shklovskaya E, Fazekas de St Groth B, Triccas JA, Weninger W. Cutaneous immunosurveillance by self-renewing dermal γδ T cells. J Exp Med. 2011;208:505–518. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy-Grand D, Vassalli P, Eberl G, Pereira P, Burlen-Defranoux O, Lemaître F, Di Santo JP, Freitas AA, Cumano A, Bandeira A. Origin, trafficking, and intraepithelial fate of gut-tropic T cells. J Exp Med. 2013;210:1839–1854. doi: 10.1084/jem.20122588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vantourout P, Hayday A. Six-of-the-best: unique contributions of. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reardon C, Lefrancois L, Farr A, Kubo R, O'Brien R, Born W. Expression of gamma/delta T cell receptors on lymphocytes from the lactating mammary gland. J Exp Med. 1990;172:1263–1266. doi: 10.1084/jem.172.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, Cooper MD. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501:435–438. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, Southern PJ, Masopust D. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 2015;161:737–749. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page DB, Yuan J, Redmond D, Wen YH, Durack JC, Emerson R, Solomon S, Dong Z, Wong P, Comstock C, Diab A, et al. Deep Sequencing of T-cell Receptor DNA as a Biomarker of Clonally Expanded TILs in Breast Cancer after Immunotherapy. Cancer Immunology Research. 2016;4:835–844. doi: 10.1158/2326-6066.CIR-16-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melandri D, Zlatareva I, Chaleil RAG, Dart RJ, Chancellor A, Nussbaumer O, Polyakova O, Roberts NA, Wesch D, Kabelitz D, Irving PM, et al. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nature Publishing Group. 2018;19:1352–1365. doi: 10.1038/s41590-018-0253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, Perez EA, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Annals of Oncology. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buisseret L, Garaud S, de Wind A, Van den Eynden G, Boisson A, Solinas C, Gu-Trantien C, Naveaux C, Lodewyckx J-N, Duvillier H, Craciun L, et al. Tumor-infiltrating lymphocyte composition, organization and PD-1/PD-L1 expression are linked in breast cancer. oncoimmunology. 2017;6:e1257452. doi: 10.1080/2162402X.2016.1257452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nanda R, Chow LQM, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, Karantza V, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. Journal of Clinical Oncology. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im S-A, Shaw Wright G, Henschel V, et al. IMpassion130 Trial Investigators, Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 35.Clark RA, Chong BF, Mirchandani N, Yamanaka K-I, Murphy GF, Dowgiert RK, Kupper TS. A Novel Method for the Isolation of Skin Resident T Cells from Normal and Diseased Human Skin. J Investig Dermatol. 2006;126:1059–1070. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 36.Clark RA, Chong B, Mirchandani N, Brinster NK, Yamanaka K-I, Dowgiert RK, Kupper TS. The Vast Majority of CLA +T Cells Are Resident in Normal Skin. J Immunol. 2006;176:4431–4439. doi: 10.4049/jimmunol.176.7.4431. [DOI] [PubMed] [Google Scholar]
- 37.Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS ONE. 2011;6:e16245. doi: 10.1371/journal.pone.0016245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark RA, Watanabe R, Teague JE, Schlapbach C, Tawa MC, Adams N, Dorosario AA, Chaney KS, Cutler CS, LeBoeuf NR, Carter JB, et al. Skin effector memory T cells do not recirculate and provide immune protection in alemtuzumab-treated CTCL patients. Science Translational Medicine. 2012;4:117ra7. doi: 10.1126/scitranslmed.3003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esin S, Shigematsu M, Nagai S, Eklund A, Wigzell H, Grunewald J. Different percentages of peripheral blood gamma delta + T cells in healthy individuals from different areas of the world. Scandinavian Journal of Immunology. 1996;43:593–596. doi: 10.1046/j.1365-3083.1996.d01-79.x. [DOI] [PubMed] [Google Scholar]
- 40.Di Marco Barros R, Roberts NA, Dart RJ, Vantourout P, Jandke A, Nussbaumer O, Deban L, Cipolat S, Hart R, Iannitto ML, Laing A, et al. Epithelia Use Butyrophilin-like Molecules to Shape Organ-Specific γδ T Cell Compartments. Cell. 2016;167:203–218. doi: 10.1016/j.cell.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribot JC, deBarros A, Silva-Santos B. Searching for “signal 2”: costimulation requirements of γδ T cells. Cell Mol Life Sci. 2011;68:2345–2355. doi: 10.1007/s00018-011-0698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chodaczek G, Papanna V, Zal MA, Zal T. Body-barrier surveillance by epidermal γδ TCRs. Nature Immunology. 2012;13:272–282. doi: 10.1038/ni.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almeida AR, Correia DV, Fernandes-Platzgummer A, da Silva CL, da Silva MG, Anjos DR, Silva-Santos B. Delta One T Cells for Immunotherapy of Chronic Lymphocytic Leukemia: Clinical-Grade Expansion/Differentiation and Preclinical Proof of Concept. Clin Cancer Res. 2016;22:5795–5804. doi: 10.1158/1078-0432.CCR-16-0597. [DOI] [PubMed] [Google Scholar]
- 44.Hunter S, Willcox CR, Davey MS, Kasatskaya SA, Jeffery HC, Chudakov DM, Oo YH, Willcox BE. Human liver infiltrating γδ T cells are composed of clonally expanded circulating and tissue-resident populations. J Hepatol. 2018;69:654–665. doi: 10.1016/j.jhep.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 46.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, Chen P-L, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–404. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crawford G, Hayes MD, Seoane RC, Ward S, Dalessandri T, Lai C, Healy E, Kipling D, Proby C, Moyes C, Green K, et al. Epithelial damage and tissue γδ T cells promote a unique tumor-protective IgE response. Nature Immunology. 2018;19:859–870. doi: 10.1038/s41590-018-0161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, Wang Z, Wang C, Zhang Z, Xia W, Chen Z, et al. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40:785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coffelt SB, Kersten K, Doornebal CW, Weiden J, Vrijland K, Hau C-S, Verstegen NJM, Ciampricotti M, Hawinkels LJAC, Jonkers J, de Visser KE. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, Ameh S, Sandel D, Liang XS, Mazzilli S, Whary MT, et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell. 2019;176:998–1013. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caccamo N, La Mendola C, Orlando V, Meraviglia S, Todaro M, Stassi G, Sireci G, Fournié JJ, Dieli F. Differentiation, phenotype, and function of interleukin-17-producing human V 9V 2 T cells. Blood. 2011;118:129–138. doi: 10.1182/blood-2011-01-331298. [DOI] [PubMed] [Google Scholar]
- 54.Muranski P, Restifo NP. Essentials of Th17 cell commitment and plasticity. Blood. 2013;121:2402–2414. doi: 10.1182/blood-2012-09-378653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beura LK, Fares-Frederickson NJ, Steinert EM, Scott MC, Thompson EA, Fraser KA, Schenkel JM, Vezys V, Masopust D. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. Journal of Experimental Medicine. 2019;216:1214–1229. doi: 10.1084/jem.20181365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nature Immunology. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 57.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 58.Wencker M, Turchinovich G, Di Marco Barros R, Deban L, Jandke A, Cope A, Hayday AC. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nature Immunology. 2013;15:80–87. doi: 10.1038/ni.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solinas C, Marcoux D, Garaud S, Vitória JR, Van den Eynden G, de Wind A, De Silva P, Boisson A, Craciun L, Larsimont D, Piccart-Gebhart M, et al. BRCA gene mutations do not shape the extent and organization of tumor infiltrating lymphocytes in triple negative breast cancer. Cancer Letters. 2019;450:88–97. doi: 10.1016/j.canlet.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 60.Goliwas KF, Richter JR, Pruitt HC, Araysi LM, Anderson NR, Samant RS, Lobo-Ruppert SM, Berry JL, Frost AR. Methods to Evaluate Cell Growth, Viability, and Response to Treatment in a Tissue Engineered Breast Cancer Model. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-14326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jungbluth AA, Frosina D, Fayad M, Pulitzer MP, Dogan A, Busam KJ, Imai N, Gnjatic S. Immunohistochemical Detection of γ/δ T Lymphocytes in Formalin-fixed Paraffin-embedded Tissues. Appl Immunohistochem Mol Morphol. 2018;00:1–3. doi: 10.1097/PAI.0000000000000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, Salgado R, Byrne DJ, Teo ZL, Dushyanthen S, Byrne A, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 63.Sherwood AM, Emerson RO, Scherer D, Habermann N, Buck K, Staffa J, Desmarais C, Halama N, Jaeger D, Schirmacher P, Herpel E, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62:1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheper W, Kelderman S, Fanchi LF, Linnemann C, Bendle G, de Rooij MAJ, Hirt C, Mezzadra R, Slagter M, Dijkstra K, Kluin RJC, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 2019;25:89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 65.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179:323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gober H-J, Kistowska M, Angman L, Jenö P, Mori L, De Libero G. Human T Cell Receptor γδ Cells Recognize Endogenous Mevalonate Metabolites in Tumor Cells. J Exp Med. 2003;197:163–168. doi: 10.1084/jem.20021500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davodeau F, Peyrat MA, Hallet MM, Houde I, Vié H, Bonneville M. Peripheral selection of antigen receptor junctional features in a major human gamma delta subset. Eur J Immunol. 1993;23:804–808. doi: 10.1002/eji.1830230405. [DOI] [PubMed] [Google Scholar]
- 68.Yamashita S. Recognition mechanism of non-peptide antigens by human T cells. International Immunology. 2003;15:1301–1307. doi: 10.1093/intimm/dxg129. [DOI] [PubMed] [Google Scholar]
- 69.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perdiguero EG, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald H-R. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Masopust D, Soerens AG. Tissue-Resident T Cells and Other Resident Leukocytes. Annu Rev Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zumwalde NA, Haag JD, Sharma D, Mirrielees JA, Wilke LG, Gould MN, Gumperz JE. Analysis of Immune Cells from Human Mammary Ductal Epithelial Organoids Reveals Vδ2+ T Cells That Efficiently Target Breast Carcinoma Cells in the Presence of Bisphosphonate. Cancer Prev Res (Phila) 2016;9:305–316. doi: 10.1158/1940-6207.CAPR-15-0370-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamaguchi T, Hiratsuka M, Asai K, Kai K, Kumagai K. Differential Distribution of T Lymphocyte Subpopulations in the Bovine Mammary Gland During Lactation. Journal of Dairy Science. 1999;82:1459–1464. doi: 10.3168/jds.S0022-0302(99)75373-7. [DOI] [PubMed] [Google Scholar]
- 75.Egelston CA, Avalos C, Tu TY, Simons DL, Jimenez G, Jung JY, Melstrom L, Margolin K, Yim JH, Kruper L, Mortimer J, et al. Human breast tumor-infiltrating CD8+ T cells retain polyfunctionality despite PD-1 expression. Nature Communications. 2018;9:4297. doi: 10.1038/s41467-018-06653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGranahan N, Furness AJS, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TBK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, Birkbak NJ, Veeriah S, Van Loo P, Herrero J, Swanton C. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171:1259–1271. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, Lund T, Tanić M, Reading JL, Joshi K, Henry JY, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brandes M, Willimann K, Moser B. Professional antigen-presentation function by human gammadelta T Cells. Science. 2005;309:264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- 81.Brandes M, Willimann K, Bioley G, Lévy N, Eberl M, Luo M, Tampé R, Lévy F, Romero P, Moser B. Cross-presenting human gammadelta T cells induce robust CD8+ alphabeta T cell responses. Proc Natl Acad Sci USA. 2009;106:2307–2312. doi: 10.1073/pnas.0810059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Facciabene A, Peng X, Hagemann IS, Balint K, Barchetti A, Wang L-P, Gimotty PA, Gilks CB, Lal P, Zhang L, Coukos G. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and Treg cells. Nature. 2011;475:226–230. doi: 10.1038/nature10169. [DOI] [PubMed] [Google Scholar]
- 83.Cox TR, Rumney RMH, Schoof EM, Perryman L, Høye AM, Agrawal A, Bird D, Latif NA, Forrest H, Evans HR, Huggins ID, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He W, Hao J, Dong S, Gao Y, Tao J, Chi H, Flavell R, O'Brien RL, Born WK, Craft J, Han J, et al. Naturally activated V gamma 4 gamma delta T cells play a protective role in tumor immunity through expression of eomesodermin. The Journal of Immunology. 2010;185:126–133. doi: 10.4049/jimmunol.0903767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hao J, Dong S, Xia S, He W, Jia H, Zhang S, Wei J, O'Brien RL, Born WK, Wu Z, Wang P, et al. Regulatory Role of Vγ1 γδ T Cells in Tumor Immunity through IL-4 Production. J Immunol. 2011;187:4979–4986. doi: 10.4049/jimmunol.1101389. [DOI] [PubMed] [Google Scholar]
- 87.Hayday AC. γδ T Cell Update: Adaptate Orchestrators of Immune Surveillance. The Journal of Immunology. 2019;203:311–320. doi: 10.4049/jimmunol.1800934. [DOI] [PubMed] [Google Scholar]
- 88.Harly C, Guillaume Y, Nedellec S, Peigné C-M, Mönkkönen H, Mönkkönen J, Li J, Kuball J, Adams EJ, Netzer S, Déchanet-Merville J, et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood. 2012;120:2269–2279. doi: 10.1182/blood-2012-05-430470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oncomine. https://www.oncomine.org/2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.