Abstract
5T4 (trophoblast glycoprotein, TPBG) is a transmembrane tumor antigen expressed on more than 90% of primary renal cell carcinomas (RCC) and a wide range of human carcinomas but not on most somatic adult tissues. The favorable expression pattern has encouraged the development and clinical testing of 5T4-targeted antibody and vaccine therapies. 5T4 also represents a compelling and unexplored target for T-cell receptor (TCR)-engineered T-cell therapy. Our group has previously isolated high-avidity CD8+ T-cell clones specific for an HLA-A2-restricted 5T4 epitope (residues 17–25; 5T4p17). In this report, targeted single-cell RNA sequencing was performed on 5T4p17-specific T-cell clones to sequence the highly variable complementarity-determining region 3 (CDR3) of T-cell receptor α chain (TRA) and β chain (TRB) genes. Full-length TRA and TRB sequences were cloned into lentiviral vectors and transduced into CD8+ T-cells from healthy donors. Redirected effector T-cell function against 5T4p17 was measured by cytotoxicity and cytokine release assays. Seven unique TRA-TRB pairs were identified. All seven TCRs exhibited high expression on CD8+ T-cells with transduction efficiencies from 59 to 89%. TCR-transduced CD8+ T-cells demonstrated redirected cytotoxicity and cytokine release in response to 5T4p17 on target-cells and killed 5T4+/HLA-A2+ kidney-, breast-, and colorectal-tumor cell lines as well as primary RCC tumor cells in vitro. TCR-transduced CD8+ T-cells also detected presentation of 5T4p17 in TAP1/2-deficient T2 target-cells. TCR-transduced T-cells redirected to recognize the 5T4p17 epitope from a broadly shared tumor antigen are of interest for future testing as a cellular immunotherapy strategy for HLA-A2+ subjects with 5T4+ tumors.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02419-4) contains supplementary material, which is available to authorized users.
Keywords: 5T4, Trophoblast glycoprotein, Renal cell carcinoma, T-cell receptor, Adoptive cell therapy, Antigen processing
Introduction
Renal cell carcinoma (RCC) accounts for 90% of malignant neoplasms arising in the kidney in adults and is the eighth most common cancer in the United States, with an estimated 73,820 new cases in 2019 [1]. Despite the development of targeted molecular therapies, including tyrosine kinase inhibitors (TKIs) and mammalian target of rapamycin (mTOR) inhibitors, metastatic RCC is generally considered an incurable disease with a 5-year survival rate of only 12% [1]. However, metastatic RCC can be uniquely sensitive to systemic immunotherapy. Both high-dose interleukin-2 (IL-2) [2] and, more recently, combination immune checkpoint blockade with antibodies targeting programmed cell death-protein 1 (PD1) and cytotoxic T-lymphocyte associated protein 4 (CTLA4) [3] have been associated with complete radiographic responses in 5–9% of RCC patients. The anti-tumor effects produced by these agents are thought to be mediated by tumor-reactive T-cell responses.
While the antigens associated with immunotherapy mediated regression of RCC are not well defined, previous studies have identified 5T4 (trophoblast glycoprotein, TPBG) as an RCC-associated antigen of therapeutic interest. 5T4 is highly expressed by placental trophoblasts and a wide range of human carcinomas, including renal, prostate, pancreatic, ovarian, breast, cervical, gastric, and non-small cell lung cancer [4, 5]. Greater than 90% of RCC tumors over-express 5T4 and the expression is maintained on metastatic lesions [6]. 5T4 expression on tumors has been associated with a stem-cell phenotype subpopulation in human non-small cell lung cancers, nasopharyngeal carcinoma [7–9] and with epithelial–mesenchymal transition that may be related to cancer cell motility and metastatic spread [4]. 5T4 protein is undetectable or expressed at a very low level by healthy adult tissues [5, 6, 10].
The favorable expression pattern of 5T4 has encouraged the development and clinical testing of 5T4-targeted antibody–drug conjugates (ADCs) engineered with superantigen [11] or chemotherapy payloads [12] and a recombinant modified vaccinia virus Ankara (MVA) expressing the full-length 5T4 gene (MVA-5T4). MVA-5T4 is the most extensively studied 5T4-target therapy and has been applied to > 580 subjects with colorectal, prostate, and renal cancer [4]. Early phase clinical testing demonstrated MVA-5T4 was able to elicit 5T4-specific serological and T-cell responses in vaccinated cancer subjects [13]. 5T4-targeting by ADC or MVA-5T4 vaccine has not been associated with off-tumor on-target toxicities affecting healthy tissues. However, despite encouraging early phase data, none of these agents have gained regulatory approval as a cancer therapy.
Engineering T-cells to express foreign TCRs or chimeric antigen receptors (CARs) targeting tumor-associated antigens represents a therapy platform with the potential to massively expand tumor-reactive T-cells in cancer subjects. The recent clinical success of engineered T-cells expressing CARs specific for CD19 achieving complete remissions of refractory acute lymphocytic leukemia [14] and non-Hodgkin lymphoma [15] has created intense interest to extend engineered T-cells as a therapeutic modality to solid tumor targets. TCR-engineered T-cell therapy targeting the cancer/testis antigen NY-ESO-1 in melanoma and synovial sarcoma [16, 17], and more recently TCR engineered T-cells targeting human papillomavirus (HPV) antigens E6 or E7 in HPV+ cancers [18, 19] associated with partial tumor responses in some patients establish proof-of-concept for the therapeutic use of TCR engineered T-cells targeting a single tumor antigen to result in significant tumor regression.
5T4 represents a compelling and unexplored target for TCR-engineered T-cell therapy. Our group has previously isolated high-avidity CD8+ T-cell clones from both healthy and kidney cancer donors specific for an HLA-A2-restricted 5T4 epitope (residues 17–25; 5T4p17) [10]. In this study, we sequenced the CDR3s from the TRA and TRB genes isolated from these high-avidity 5T4p17-specific clones to identify unique TCRs recognizing 5T4p17. We have assessed 5T4p17-specific TCR-transduced T-cells from healthy donors for redirected recognition of 5T4p17 on target cells, including HLA-A2+ human tumor-cell lines and short-term in vitro cultures of primary RCC tumors expressing the 5T4 antigen.
Materials and methods
CDR3 domain sequencing for TRA and TRB genes from 5T4p17-specific CD8+ T-cell clones
Genomic DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) from 19 CD8+ T-cell clones specific for 5T4p17 presented by HLA-A2. High throughput-bulk sequencing of the T-cell receptor β chain was performed using the hsTCRB ImmunoSeq kit (Adaptive Biotechnologies, Seattle, WA) at survey level resolution [20] on the Illumina MiSeq platform (v3 150 cycle) in the Genomics Core Facility at the Fred Hutchinson Cancer Research Center. Repertoire analyses were conducted using the LymphoSeq R package (created by D. G. Coffey; http://bioconductor.org/packages/LymphoSeq).
Targeted single-cell TRA and TRB sequencing were conducted according to methods previously reported [21]. For each clone, 8 or 16 single CD8+CD3+DAPI− cells were sorted into a 96-well PCR plate. Targeted-reverse transcription of CDR3-regions was conducted on the mRNA transcripts of TRA and TRB using the One-step RT PCR kit (Qiagen, Hilden, Germany). The cDNA library was PCR-amplified, barcoded [21], pooled and purified with Agencourt AMPure XP beads (Beckman Coulter, Brea, CA). Sequencing was performed for pair-end 250 bp (MiSeq reagent kit v2, 500-cycles, Illumina, San Diego, CA). FASTQ files were de-multiplexed, and CDR3 regions with associated V(D)J region-information were extracted with the MiXCR package [22]. Net charges of CDRregions were computed by the R package “Peptides” [23].
Cloning full-length TRA and TRB sequences
Reference V- and C-gene open-reading-frames of TRA and TRB were obtained from the International Immunogenetics Information System (IMGT) [24, 25]. Codon optimized Vα and Vβ DNA fragments with corresponding CDR3 sequences were then synthesized by the GeneArt Strings DNA Fragments service (Invitrogen, Carlsbad, CA). Each DNA fragment included the following Gibson overhang sequences attached to both ends: Vα 5′: AGGAGACGTGGAAGAAAACCCCGGTCCC; Vα 3′: ACATCCAGAACCCCGACCCTGCAGTGTACCAGCTGCGGGAC; Vβ 5′: TCCCCGAGCTCAATAAAAGAGCCCACAACCCCTCACTCGGCGCGCCGGCCACC; Vβ 3′: GTGTTCCCCCCAGAGGTGGCCGTGTTCGAG. The stop codon of constant region of TCR-β gene (TRBC) was deleted and the self-cleaving porcine teschovirus-1 2A sequence.
(P2A: GGTTCCGGAGCCACGAACTTCTCTCTGTTAAAGCAAGCAGGAGACGTGGAAGAAAACCCCGGTCCC) was incorporated between TRA and TRB, to ensure equal expression of α and β chains. Full-length TCRs were assembled using the Gibson Assembly® Ultra kit (SGI-DNA, La Jolla, CA). TCRs were then cloned into lentiviral vectors pRRLSIN, with the murine stem cell virus (MSCV) promoter to drive expression in human primary T-cells (pRRLSIN, pRSV-REV, pMD2-G, and pMDLg/pRRE were generous gifts of Dr. Philip Greenberg, Seattle, WA) [26]. Constructs were transformed into One Shot® TOP10 Chemically Competent E. coli (Invitrogen). Plasmid DNA was extracted using the Endotoxin-free Mini- and Midi-Prep DNA isolation kits (Qiagen).
Lentiviral packaging and T-cell transduction
Lenti-X 293T virus packaging cells (Clontech Laboratories, Mountain View, CA) were seeded at 60% confluency in RPMI-HEPES supplemented with 10% fetal bovine serum, 2 mmol/L L-glutamine, and 1% penicillin/streptomycin (termed LCL medium). 5T4p17-specific TCR encoding lentivirus vectors were co-transfected with packaging plasmids (pRRSIN-TCR: 1.5 μg, pRSV-REV: 1 μg, pMD2-G: 0.5 μg, and pMDLg/pRRE 1 μg) using the Effectene transfection reagent (Qiagen). Media was changed the next day; from day 2 post-transfection, and lentivirus containing supernatants were harvested each day for 2 days and passed through 0.45 μm filters. Viral supernatants were concentrated using Lenti-X viral concentrator (Clontech Laboratories), aliquoted and applied to T-cells for transduction or stored at − 80 °C.
CD8+ T-cells were isolated from healthy donor PBMCs using a human CD8+ T-cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) and activated for 4 h with CD3/CD28 Dynabeads® human T-cell expander (Thermo Fisher Scientific, Waltham, MA). Viral supernatant was applied with 6 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO) to T-cells and centrifuged at 1455 × g for 90 min at 35 °C. Transduction was repeated the following day. T-cell cultures were maintained in cytotoxic T-lymphocyte culturing media [27] with 50 U/mL recombinant human IL-2 (PeproTech, Rocky Hill, NJ) for 7 days.
Endogenous TRA knockout
Healthy donor CD8+ T-cells were stimulated for 2 days with CD3/CD28 Dynabeads® human T-cell expander (Thermo Fisher Scientific). crRNA–tracrRNA duplex was prepared the morning of electroporation. Constant region of TRA (TRAC) targeting crRNA (Alt-R® CRISPR-Cas9 crRNA: AGAGTCTCTCAGCTGGTACA [28], Integrated DNA technology, Inc., Coralville, IA) and tracrRNA (Alt-R® CRISPR-Cas9 tracrRNA, Integrated DNA technology, Inc.) were reconstituted to 200 µM with Nuclease-Free Duplex Buffer (Integrated DNA technology, Inc.) and mixed at equimolar concentrations. Oligos were then annealed by heating at 95 °C for 5 min in PCR thermocycler and slowly cooled to room temperature. Alternatively, control crRNA (Alt-R® CRISPR-Cas9 Control Kit, human, Integrated DNA technology, Inc.) was mixed with the tracrRNA the same manner. Duplexes and Cas9 Nuclease V3 (Integrated DNA technology, Inc.) were gently mixed and incubated at room temperature for at least 10 min. 1 million T-cells were resuspended in 20 µL primary-cell nucleofection solution (P3 Primary Cell Nucleofector™ Solution, Lonza, Basel, Switzerland). T-cells were added with Alt-R Cas9 Electroporation Enhancer (Integrated DNA technology, Inc.) to 4 μM, and incubated with 5 µL duplex-Cas9 nuclease mix for 2 min. T-cells were electroporated using a 4D nucleofector (Lonza) with EH115 program. After nucleofection, T-cells were cultured at 106 cells per well in 200 µL prewarmed complete T-cell media. 6 h post nucleofection, T-cells were transduced with the corresponding lentivirus as described in the previous section.
Flow cytometry and T-cell expansion
To assess 5T4p17-specific TCR expression, lentiviral-transduced T-cell cultures were stained by DAPI, 1:20 dilution of APC-Cy7-labeled anti-CD3 mAb, (clone SK7; BD Biosciences, San Jose, CA), 1:20 dilution of FITC-labeled anti-CD8 mAb (clone RPA-T8; BD Biosciences) as well as 10 μg/mL of APC-labeled 5T4p17/HLA-A2 tetramer (TET, generated by the Immune Monitoring Core Laboratory at our center) and analyzed by flow cytometry (BD FACSymphony™, BD biosciences, San Jose, CA). Viable TET+CD8+CD3+ T-cells were flow-sorted (BD FACSAria™ II, BD biosciences) and expanded as described [29] in cytotoxic T-lymphocyte culturing media supplemented with 50 U/mL IL-2 (PeproTech) and 5 ng/mL Interleukin-15 (PeproTech).
Cytotoxicity and ELISA assays
For some experiments, T2- and LCL-targets were infected with wild-type MVA or MVA-5T4 (Oxford Biomedica, Oxford, UK) at a 10:1 multiplicity of infection in serum-free RPMI media at 37 °C for 1 h. Media were then adjusted to 10% serum. To confirm 5T4 expression on surface after MVA-5T4 infection, target cells were stained with a 5T4-specific mAb (clone 524744; R&D systems, Minneapolis, MN) at 1 μg/mL followed by a 1:50 dilution of a secondary PE-labeled anti-mouse IgG1 mAb (clone A85-1; BD Biosciences) for analysis by flow cytometry. T-cell and target-cell co-culture supernatants were harvested after 4 h to perform chromium-release assays as previously described [30]. Supernatants after 16 h of T-cell target-cell co-culture were assayed for interferon-γ (IFN-γ) or tumor necrosis factor-α (TNF-α) by enzyme-linked immunosorbent assays (ELISA) according to the manufacturer’s protocol (BosterBio, Pleasanton, CA).
HLA-A2 stabilization assay
Synthetic peptide 5T4p17 RLARLALVL, and alanine-substituted variant sequences ALARLALVL (R1A), RLAALALVL (R4A), RLARAALVL (L5A), RLARLAAVL (L7A), RLARLALAL (V8A) at a purity > 90% (Genscript, Piscataway, NJ) were dissolved in 100% dimethyl sulfoxide (Invitrogen) and stored at 4 °C. Aliquots of 2 × 105 T2 cells were cultured in serum-free RPMI media supplemented with human β2-microglobulin (BosterBio) at 5 μg/mL and the test peptide. After overnight incubation at 37 °C, cells were stained with an APC-labeled HLA-A2-specific mAb (clone BB7.2, BD Biosciences) at dilution of 1:50 with DAPI and analyzed by flow cytometry.
Results
Sequencing TCRs from 5T4p17-specific CD8+ T-cell clones reveals similarity in CDR3 region and V(D)J gene usage
Our group has previously characterized high-avidity 5T4p17-specific CD8+ T-cell clones isolated from seven T-cell lines stimulated in vitro with varying concentrations of 5T4p17. These lines were derived from leukapheresis products of four HLA-A2+ donors (three healthy donors, HD_A, HD_B, and HD_C; and one donor with 5T4+ metastatic kidney cancer, KCD_D, 107 responder T-cells for each condition) [10]. To assess the TCR-β gene usage, we sequenced CDR3 regions from the TCR-β gene (TRB-CDR3) expressed by 19 5T4p17-specific CD8+ T-cell clones. Two clones had two TRB-CDR3 sequences—HD_A, clone #1 and HD_B, clone #20. In each case, one TRB-CDR3 sequence was identical to the other clones isolated from the same T-cell line (Table 1). Our hypothesis to account for two TRB-CDR3 sequences in these two “clones” was flow sorting of doublets representing a 5T4p17-specific CD8+ T-cell that stained tetramer-positive adherent to an unrelated passenger T-cell. All three clones isolated from the HD_C/0.1 μg/mL-stimulated T-cells had a nonproductive TRB rearrangement (“null” allele) and a shared in-frame TRB-CDR3 sequence (CASSYMGPEAFF; Table 1).
Table 1.
High throughput bulk sequencing of the T-cell receptor β chain for 5T4p17-specific CD8+ T-cell clones isolated from healthy or kidney cancer donors
| Donor | [5T4p17] stimulation (µg/mL) | Clone | TRB-CDR3 sequence | Frequency (%) | TRA/B single-cell sequencing |
|---|---|---|---|---|---|
| HD_A | 10 | 1 | CASSELPAGGTNEQFF | 62.70 | − |
| CASSIYGETGTEAFF | 35.20 | ||||
| 2 | CASSELPAGGTNEQFF | 97.94 | + | ||
| 0.1 | 7 | CASSFFSNTGELFF | 99.20 | − | |
| 8 | CASSFFSNTGELFF | 98.17 | − | ||
| 12 | CASSFFSNTGELFF | 98.52 | − | ||
| 15 | CASSFFSNTGELFF | 97.94 | + | ||
| HD_B | 10 | 17 | CASSQVSGYEQYF | 95.48 | − |
| 19 | CASSQVSGYEQYF | 99.54 | + | ||
| 0.1 | 20 | CASSLTSQGFQPQHF | 65.25 | + | |
| CASSPQGDNEQFF | 34.06 | ||||
| 21 | CASSPQGDNEQFF | 99.02 | + | ||
| 24 | CASSPQGDNEQFF | 95.96 | − | ||
| HD_C | 10 | 1 | CASMDLAFKQYF | 97.50 | − |
| 3 | CASMDLAFKQYF | 98.41 | + | ||
| 6 | CASMDLAFKQYF | 98.41 | − | ||
| 11 | CASMDLAFKQYF | 98.74 | − | ||
| 0.1 | 17 | Null | 63.66 | + | |
| CASSYMGPEAFF | 34.79 | ||||
| 18 | Null | 62.21 | − | ||
| CASSYMGPEAFF | 36.81 | ||||
| 20 | Null | 62.80 | − | ||
| CASSYMGPEAFF | 36.48 | ||||
| KCD_D | 10 | 6 | CASSFLTDTQYF | 96.75 | + |
The bolded sequences were shared among clones from the same donor and peptide stimulation concentration
TRB T-cell receptor beta, CDR3 complementarity-determining region 3, TRA T-cell receptor alpha, HD healthy donor, KCD kidney cancer donor
Based on this interpretation, our analysis of 19 5T4p17-specific CD8+ T-cell clones identified seven unique TRB-CDR3 sequences, with only one unique sequence corresponding to each of the seven originating T-cell lines. This result indicates the precursor frequency for each T-cell line with unique TRB-CDR3 could be as low as one cell out of the initial input 107 CD8+ T-cells. To further infer precursor frequency, we sequenced TRB-CDR3 from flow-sorted CD8+ T-cells purified from available donor leukapheresis products (HD_A, HD_B, and KCD_D) as well as two additional PBMC samples and a tumor-infiltrating lymphocyte sample also from donor KCD_D (Supplementary Table 1 and Supplementary Fig. 1). KCD_D was a 60-year-old male subject with metastatic clear-cell RCC, treated with high-dose IL-2 and multiple lines of targeted drug. Our analyses did not detect the corresponding TRB-CDR3 sequences associated with 5T4p17-specificity from any of the samples collected from donors HD_A, HD_B, or KCD_D. These data suggest an upper boundary for precursor frequencies for 5T4p17-specific clones at < 1 in 1–1.5 × 105 CD8+ T-cells.
To identify the TCR-α gene CDR3 region (TRA-CDR3) pairing with each of the unique TRB-CDR3, targeted single-cell RNA-Seq of the CDR3 regions of TRA and TRB genes was then performed. We discovered seven unique single TRA-CDR3 sequences that each paired with one of the seven TRB-CDR3s (Supplementary Table 2). Single-cell RNA-Seq analysis of HD_C, clone #17 confirmed both the nonproductive- and productive-rearrangement of TRB within each cell. Single-cell RNA-Seq analysis of HD_B, clone 20 resolved two separate cell populations, each expressing one of the identified TRB-CDR3 sequences consistent with our hypothesis of a flow-sorted doublet.
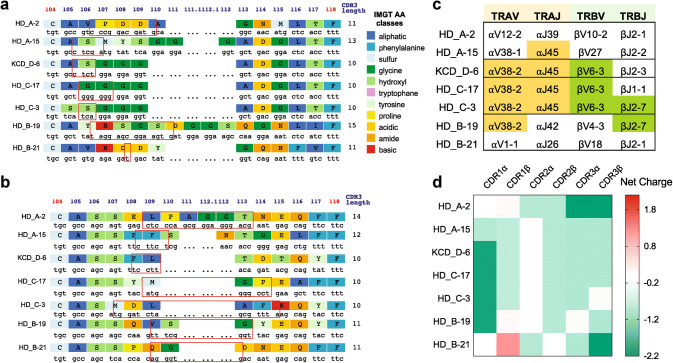
Our analysis did not reveal public TCR sequences shared between donors. However, we did observe CDR3 sequences with a high degree of homology. The TRA-CDR3 region of KCD_D-6 and HD_C-17 differed by only one amino acid at position 106 (serine of KCD_D-6, glycine of HD_C-17), while KCD_D-6 and HD_C-3 differed only at position 105 (alanine of KCD_D-6, serine of HD_C-3, Fig. 1a). V(D)J gene analysis with the IMGT V-quest [31, 32] also revealed common gene-segment usage among subsets of 5T4p17-specific TCRs. TRAV38-2 and TRAJ45 were both shared by 4 of 7 TCRs, and TRBV6-3 and TRBJ2-7 were shared by 3 and 2 of 7 TCRs, respectively (Fig. 1c).
Fig. 1.
Characteristics of 5T4p17 -specific CDR3 regions of TRA and TRB. IMGT junction analysis is shown for aTRA-CDR3 regions and bTRB-CDR3 regions. Red boxes indicate N nucleotides and/or D regions (TRB) between V gene and J gene encoded sequence. c V- and J-gene usage for TRA and TRB are shown. Color shading identifies common gene use between TCRs. d The net charge for CDR1, 2, and 3 amino acid sequences from 5T417–25-specific TCRs are shown
TCRs use a conserved mechanism to bind to HLA-A2 despite lack of consensus motifs in CDR1/CDR2 regions. Two unique positive-charged residues (R65 and K66) on the HLA-A2 α1-helix are crucial elements to interact with the negatively charged residues (Asp and Glu) in CDR1/CDR2 domains of HLA-A2-specific TCRs [33]. Therefore, V(D)J gene-segment usage for HLA-A2 specific tumor antigen recognition is biased [34]. Although the most frequently used V gene-segments in 5T4p17-specific clones (TRAV38 and TRBV6-3) are not among the most common HLA-A2 biased V genes, the CDR1 amino acid sequences of TRAV38 and TRBV6-3 are negatively charged at − 2 and − 0.9, respectively, consistent with preferential binding with HLA-A2 (Supplementary Table 2 and Fig. 1d). The amino acid sequences of 5T4p17-specific CDR3 regions are also negatively charged (Fig. 1d), except for CDR3α on HD_B-19 and CDR3β on HD_C-3, indicating a direct interaction of CDR3s and the positively charged 5T4p17 epitope (RLARLALVL, with two positively charged arginine residues).
5T4p17-specific TCRs exhibit robust and stable expression on CD8+ T-cells from healthy donors
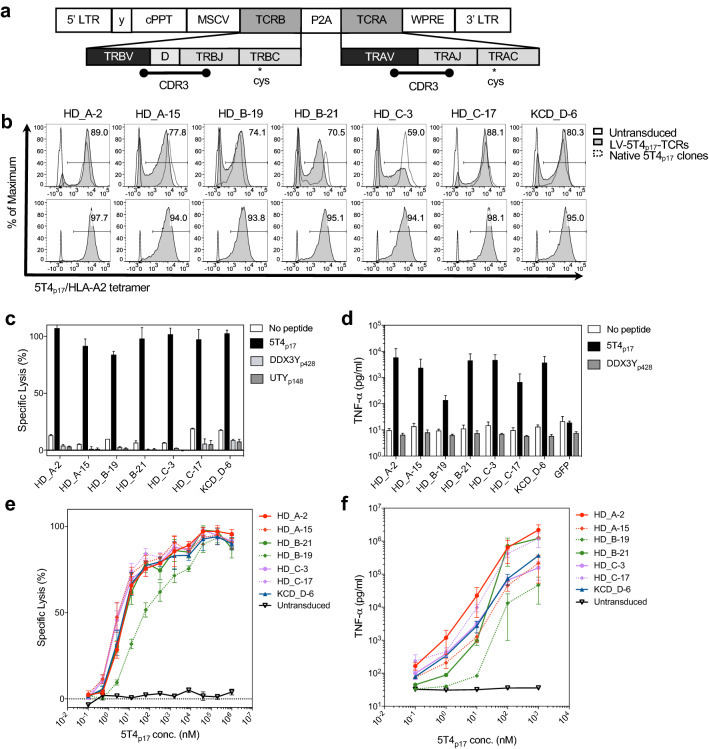
We assembled the full sequences of TRA and TRB genes by adding reference V gene- and C gene-segments to the CDR3s (Supplementary Table 3; Supplementary Table 4). Codon optimized full-length TCRα and β sequences were designed to incorporate cysteine point mutations in TRAC and TRBC, respectively (Supplementary Table 5), to crosslink lentiviral encoded 5T4p17-specific α and β chains and minimize mispairing with endogenous TCR counterparts (Fig. 2a) [35]. A P2A autocleavage site linked the full-length TRA and TRB genes with the orientation of TRB followed by TRA [36] to ensure equimolar production. CD8+ T-cells from peripheral blood of HLA-A2+ healthy donors were isolated and transduced with lentivirus encoding 5T4p17-specific TCRs. At day 7 post-transduction, 5T4p17-specific TCRs were detected by tetramer staining on 59–89% of transduced T-cells (Fig. 2b). No significant differences were observed in the transduction efficiency from three healthy donors (Supplementary Fig. 2). The mean fluorescent intensity of 5T4p17-specific TCR expression on flow-sorted tetramer+ cells following 12 days expansion was maintained at levels similar to the primary transduced cells (Fig. 2b).
Fig. 2.
5T4p17-specific TCRs expression on CD8+ T-cells from healthy donors and redirected effector functions against peptide-pulsed T2 targets. a Vector structure of 5T4p17-specific TCR assembly: TRB and TRA are transcribed under control of the MSCV promotor; P2A cleaves the primary transcripts to equal molar quantity of TRA and TRB in the cytosol. The enlarged graph indicates the gene segments and CDR3s, contributing to TRA and TRB sequence assembly. The relative location for the introduced transmembrane cysteines in TRAC and TRBC are indicated. b Cell surface expression of 5T4p17-specific TCRs stained by 5T4p17/HLA-A2 tetramer at day 7 post-transduction is shown in comparison to tetramer staining of the native T-cell clone expressing the corresponding TCR (upper panels). Cell surface expression of 5T4p17-specific TCRs on healthy donor T-cells is shown after sorting for 5T4p17/HLA-A2 tetramer+ cells and 12 days expansion (lower panels). CD8+ T-cells expressing 5T4p17-specific TCRs were tested for recognition of c T2 cells pulsed with 10 nM of 5T4p17 or control HLA-A2-binding peptides DDX3Y428–436 or UTY148–156 in a 4-h cytotoxicity assay. The effector: target ratio (E:T) was 10:1. d TNF-α release was measured by ELISA in culture supernatants harvested after 18 h co-culture of effector T-cells with T2 cells pulsed with 10 nM of 5T4p17 or the control peptide DDX3Y428–436 at 10:1 E:T. e Effector T-cells were tested for recognition of T2 cells pulsed with 5T4p17 peptide in a 4-h cytotoxicity assay at 10:1 E:T. f TNF-α release was measured by ELISA in culture supernatants harvested after 18 h co-culture of effector T-cells with T2 cells pulsed with 5T4p17 peptide at 10:1 E:T
CD8+ T-cells expressing lentiviral-transduced TCRs exhibit redirected effector functions for 5T4p17/HLA-A2
TCR transduced T-cells exhibited potent cytolytic activity for T2 targets pulsed with 10 nM 5T4p17 peptide without cross-reactivity for control HLA-A2 binding peptides (DDX3Y428–436 FLLDILGAT and UTY148–156 KAFQDVLYV) (Fig. 2c) [37]. When tested for recognition of limiting dilutions of 5T4p17, six of the 7 TCR-transduced T-cell lines exhibited similar half-maximum lysis of 5T4p17 peptide-pulsed T2 targets at concentrations of 1–10 nM peptide (Fig. 2e). After overnight co-culture with T2 target-cells pulsed with 10 nM 5T4p17, TCR transduced CD8+ T-cells released robust amounts of TNF-α (Fig. 2d) again without cross-reactivity to control HLA-A2 binding peptides. The half-maximum release of TNF-α was observed with ~ 10 nM concentration of 5T4p17 peptide for six of the 7 TCRs (Fig. 2f). Five of the seven TCR-transduced T-cell lines produced detectable TNF-α release above background at a 5T4p17 peptide concentration as low as 0.1 nM. Comparing with other TCRs, the HD_B-19 TCR-transduced T-cell lines required approximately tenfold higher peptide concentration for equivalent half-maximum lysis and TNF-α release. Similar observations were also made with IFN-γ release (Supplementary Fig. 3).
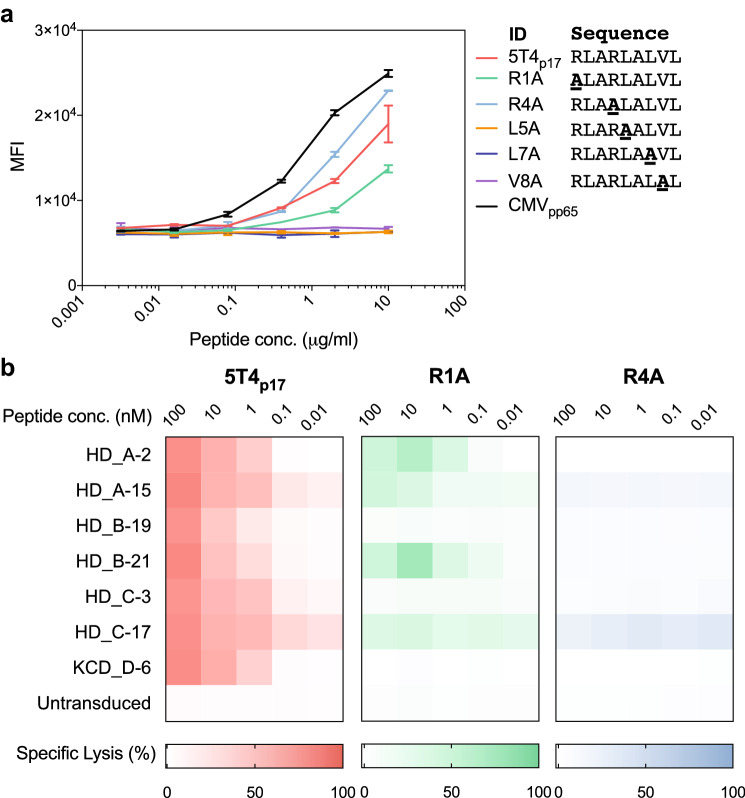
To closely examine the TCR-peptide/major histocompatibility complex (MHC) binding specificity for 5T4p17-specific TCRs, we synthesized five 5T4p17 peptide variants containing non-alanine residues substituted each to the nonpolar, aliphatic alanine residue, except for the two conserved anchor residues essential for HLA-A2 binding (RLARLALVL; lysine at position 2 and position 9) [38]. MHC binding affinities of alanine-substituted and proband 5T4p17 peptides were measured by cell surface HLA-A2 stabilization on T2 cells using flow cytometry (Fig. 3a). The 5T4p17, R1A, and R4A peptides stabilized HLA-A2 on the surface of T2 cells with a measurable increase of HLA-A2 staining. The alanine-substituted peptides L5A, L7A, and V8A showed no capacity to bind to HLA-A2 suggesting side chains at these positions may contribute to the peptide–MHC binding interaction.
Fig. 3.
Cytotoxicity of 5T4p17-specific TCR transduced CD8+ T-cells against T2 pulsed with alanine-substituted 5T4p17 peptides. a Binding of alanine-substituted 5T4p17 peptides to HLA-A2 was determined by stabilization of HLA-A2 on the surface of peptide-pulsed T2 cells. CMVpp65 is an HLA-A2+ T-cell epitope from CMV and serves as a positive control. Cell surface HLA-A2 was assessed by immunostaining and flow cytometry. b CD8+ T-cells expressing 5T4p17-specific TCRs were tested for recognition of T2 cells pulsed with the progenitor 5T4p17 peptide or HLA-A2 binding alanine-substituted variants in a 4-h cytotoxicity assay with a 10:1 E:T
We then performed a cytotoxicity assay with the seven 5T4p17-specific TCR expressing T-cell lines and T2 targets pulsed with the 5T4p17, R1A, or R4A peptides. Switching the positively charged arginine at position 4 to nonpolar alanine (R4A) disrupts T-cell recognition for this sequence by all 7 of the 5T4p17-specific TCRs (Fig. 3b). The R1A, the arginine to alanine changes at position 1 (R1A) differentially preserved 5T4p17-specific TCR recognition. Three of the seven 5T4p17 specific TCRs (HD_A-2, HD_A-15, and HD_B-21) killed T2 targets pulsed with R1A at high peptide concentration (Fig. 3b).
5T4 p17-specific TCR transduced CD8+ T-cells lyse 5T4+/HLA-A2+ tumor targets
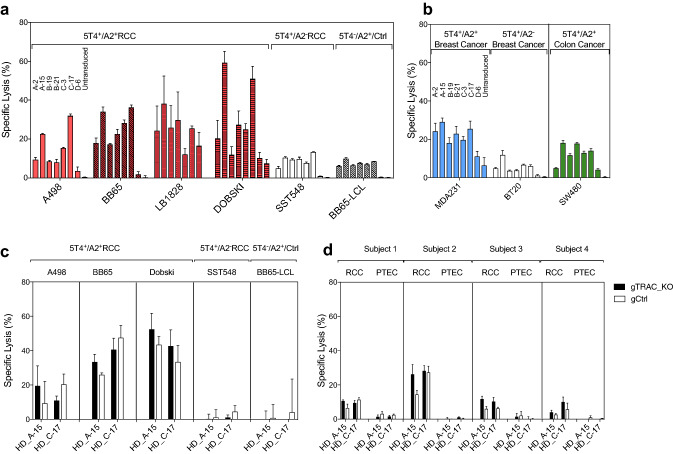
We tested the cytotoxicity of 5T4p17-specific TCR-expressing T-cells against 5T4+/HLA-A2+ RCC (A498, BB65, LB1828, DOBSKI), breast cancer (MDA231) and a colon cancer-cell line (SW480) versus HLA-mismatched (SST548, BT20) or 5T4-negative target-cells (BB65-LCL). T-cells expressing each of the seven 5T4p17-specific TCRs demonstrated specific lysis above background levels with control targets for at least one 5T4+/HLA-A2+ tumor line (Fig. 4a, b). Among the seven TCRs, T-cells expressing HD_A-15 and HD_C-17 consistently had the highest lytic potency against all 5T4+/HLA-A2+ targets tested including RCC, breast cancer, and colon cancer-cell lines.
Fig. 4.
Cytotoxicity of 5T4p17-specific TCR-transduced CD8+ T-cells against 5T4-expressing tumor targets. CD8+ T-cells expressing 5T417–25-specific TCRs were tested for recognition of tumor target lines in a 4-h cytotoxicity assay with 10:1 E:T. For each target cell-line, 5T417–25-specific TCR transduced effector CD8+ T-cells were plotted in the following order: HD_A-2, HD_A-15, HD_B-19, HD_B-21, HD_C-3, HD_C-17, KCD_D-6, and untransduced. a Targets are 5T4+/HLA-A2+ RCC cell lines (A498, BB65, TREP, DOBSKI; red bars), the 5T4+/HLA-A2− RCC line (SST548; open bars) and the 5T4−/HLA-A2+ LCL (BB65-LCL; gray shading). b Targets are 5T4+/HLA-A2+ breast cancer line (MDA231; blue bars), a 5T4+/HLA-A2− breast cancer line (BT20; open bars) and 5T4+/HLA-A2+ colon cancer line (SW480, green bars). CD8+ T-cells with (gTRAC_KO) or without (gCtrl) native TRA disruption expressing 5T4 p17-specific TCRs were tested for recognition of c 5T4+/HLA-A2+ RCC cell-lines (A498, BB65, DOBSKI), the 5T4+/HLA-A2− RCC line (SST548), the 5T4−/HLA-A2+ BB65-LCL, and d paired primary 5T4+/HLA-A2+ RCC and autologous PTEC cell-lines from four RCC patients
In some experiments, low-level lysis by 5T4 TCR-transduced T-cells was observed with HLA-mismatched (SST548, BT20) or 5T4-negative target-cells (BB65-LCL) versus control-target cell lysis seen with the originating T-cell clones. We speculated that low-level cytolytic activity for control-target cells could represent alloreactivity of the heterogeneous endogenous TCR repertoire expressed on the activated, CD8+ effector population transduced with 5T4-specific TCRs. To test this hypothesis, we transiently introduced CRISPR/Cas9 guide-RNA targeting of endogenous TRAC by electroporation into healthy donor CD8+ T-cells. CRISPR/Cas9 mediated TRAC targeting disrupted native TCR expression in 97.8% of electroporated T-cells indicated by loss of surface CD3 protein expression (Supplementary Fig. 4a, d). These CD3-negative T-cells were then transduced with lentiviral vectors directing 5T4-specific TCR expression. Residual CRISPR/Cas9 activity was not anticipated to interfere with 5T4-TCR expression due to nucleotide changes in TRAC resulting from codon optimization of the 5T4-TCR sequences. The efficiency of lentiviral transduction and surface level of 5T4-specific TCRs remained unchanged compared with control electroporated T-cells (Supplementary Fig. 4b, c, e, f).
Transduced CD8+ T-cells with or without native TRAC disruption expressing HD_A-15 or HD_C-17 TCRs were re-tested for recognition of 5T4+/HLA-A2+ RCC (A498, BB65, DOBSKI) versus control targets demonstrating preserved lytic potency for the 5T4+/HLA-A2+ RCC lines and absent recognition of the control targets, including the HLA-mismatched SST548 RCC or 5T4-negative targets BB65-LCL and HLA-A2+/5T4− fibroblast lines (Fig. 4c and Supplementary Fig. 5). The effector activity profile for 5T4-TCR transduced T-cells with native TRAC disruption was indistinguishable from the originating T-cell clones [10].
Transduced CD8+ T-cells with or without native TRAC disruption expressing HD_A-15 or HD_C-17 TCRs were then tested for recognition of primary 5T4+/HLA-A2+ RCC tumor cells versus autologous PTEC isolated from 4 donors (Supplementary Fig. 6). 5T4-TCR transduced T-cells with native TRAC disruption demonstrated specific tumor lysis for all 4 RCC tumors (5–30% specific lysis) with no lytic activity for the patient-matched syngeneic 5T4− PTEC targets (Fig. 4d and Supplementary Fig. 6).
5T4p17-specific TCR transduced CD8+ T-cells detect transporter associated with antigen processing (TAP)-independent processing of 5T4p17 in T2 cells
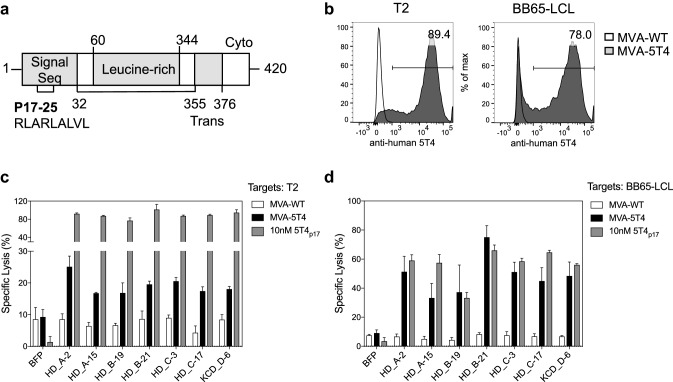
One unique characteristic of 5T4p17 is its location within the signal sequence of 5T4 (Fig. 5a). Prior studies have shown that some T-cell epitopes within leader domains can load MHC-class I via a TAP-independent pathway as an alternative to the classic antigen presentation pathway mediated by proteasome degradation and TAP transport [39]. The T2 cell-line has bi-allelic deletions of chromosome 6 spanning the TAP1/2 genes [40]. Therefore, we tested the cytotoxicity of 5T4p17-specific TCR expressing T-cells against the T2 cell-line (HLA-A2+) versus a wild-type LCL target-line (BB65-LCL, HLA-A2+). Both T2 cells and BB65-LCLs express comparable 5T4 protein on their surface when infected with a recombinant vaccinia virus encoding the full-length 5T4 gene (MVA-5T4, Fig. 5b). 5T4p17-specific T-cells consistently recognized and lysed MVA-5T4 infected T2 cells above the background killing of MVA-WT infected-T2 targets (Fig. 5c), suggesting the presence of TAP1/2-independent presentation of the 5T4p17 epitope in T2 cells. The specific lysis for 5T4-expressing T2 targets was always less than for 5T4-expressing BB65-LCL (Fig. 5d). These results suggest that in the antigen processing-competent target-cells, 5T4p17-processing and MHC-class I presentation may result from both the classical processing pathway as well as by a TAP-independent mechanism.
Fig. 5.
Cytotoxicity of 5T4p17-specific TCR-transduced CD8+ T-cells against MVA-5T4 infected targets. a Schematic of human 5T4 protein, p17–25 is in the signal sequence region. Trans: transmembrane region, Cyto: cytoplasmic region. b T2 cells and BB65-LCL were infected with MVA-WT or MVA-5T4, and cell surface expression of 5T4 was analyzed by immunostaining and flow cytometry 24 h after infection. The fraction of cells positive for cell surface 5T4 after MVA-5T4 infection is indicated. CD8+ T-cells expressing 5T4p17-specific TCRs were tested for recognition of c T2 and d BB65-LCL cells infected by MVA-WT, MVA-5T4, or uninfected cells pulsed with 10 nM 5T4p17 peptide in a 6-h cytotoxicity assay at a 10:1 E:T
Discussion
In this report, we have identified 7 unique TCRs with specificity for the 5T4p17 epitope presented by HLA-A2. Our analysis of the TRA and TRB gene segment usage, as well as TRA-CDR3 amino acid sequences, identified a high degree of homology between TCRs. We identified clones with shared V-and J-gene segments isolated from unrelated donors for both TRA and TRB. Three similar α-chains from two donors (KCD_D-6, HD_C-3, and HD_C-17) utilized the same TRAV38-2/TRAJ45 and TRBV6-3, differing by only one amino acid within TRA-CDR3. It has been shown that TCRs against the same tumor-associated epitope use biased sets of variable gene segments. For example, CD8+ T-cells in patients and healthy donors specific for an HLA-A2 associated Wilms’ tumor 1 epitope (WT1126–134) shared the usage of TRBV3, 6, 7, 20, 27 [41]; and CD8+ T-cell clones specific for the HLA-A2 associated Melanoma antigen recognized by T-cells 1 (MART-126–35) epitope used TRAV12-2 [42]. These similarities reflect the convergent nature of peptide/MHC-TCR binding in different individuals likely as a result of physical constraints on TCR sequence to generate high avidity binding to the peptide/MHC complex. We speculate that interactions between a negatively-charged CDR3 region with the positively charged 5T4p17 is a key contributor to the TCR recognition of the 5T4p17 epitope sequence.
To evaluate the suitability of 5T4p17-specific TCRs for T-cell therapy, we assessed the expression profile, antigen specificity, lytic potency, and off-target reactivity for the seven TCRs. Each TCR efficiently assembles in transduced-healthy CD8+ T-cells, is readily detectable at the cell surface, and redirects potent T-cell effector functions including cytotoxicity and cytokine release for HLA-A2+-target cells pulsed with 5T4p17. Differences in avidity between these TCRs were distinguished when 5T4p17-specific TCR-transduced CD8+ T-cells were tested against limiting concentration of 5T4p17 revealing 10-fold lower avidity for the HD_B-19 TCR versus the other sequences. The specificity of the TCR’s also differed for recognition of an alanine-substituted version of 5T4p17 at position 1 (R1A) recognizable by only three of the seven TCRs (HD_A-15, HD_C-17, and HD_B-21). Most importantly, the cytolytic activity of 5T4p17-specific TCR transduced CD8+ T-cells was demonstrated with a panel of 5T4+/HLA-A2+ tumor targets including RCC, breast, and colon cancer tumor lines and primary RCC tumor in short-term culture. T-cells transduced with HD_A-15 or HD_C-17 TCRs demonstrated the highest and most consistent specific lysis for the tumor lines tested.
An NCBI peptide blast search reveals that the 9-mer 5T4p17–25 sequence is unique to 5T4 in the human peptidome. The 8-mer 5T4p18–25 sequence is also unique to 5T4, indicating a potential cross-reactivity of R1A 5T4p17–25 specific TCRs for a heterologous protein containing a shared 5T4p18–25 sequence is not expected. In performing functional testing of 5T4p17-specific TCR transduced CD8+ T-cells, we noted low-level cytolytic activity for 5T4− or HLA-A2− target cells that had not been observed with the originating T-cell clones. Using CRISPR/Cas9 mediated TRAC targeting and disruption of native TCR expression, we provide direct evidence that allo-reactivity of the heterogenous endogenous TCR repertoire expressed on the activated, CD8+ effector population transduced with 5T4-specific TCRs accounts for 5T4 independent-target reactivity. 5T4-TCR transduced T-cells with native TRAC disruption showed no lytic activity for 5T4-negative cells representing healthy tissues that included LCL, fibroblast, and PTEC targets. TRAC disruption may also prevent the risk of TCR chain mispairing that could result in unexpected auto-reactivity [43]. Whether disruption of native TCR expression in transduced T-cells could augment specific antigen recognition of transduced TCRs by eliminating competition for TCR signaling co-factors is an area of ongoing investigation.
We searched a database of all publicly available TRB-CDR3 sequences curated by our research group (currently > 320 million unique TRB-CDR3s) for 5T4p17-associated TRB-CDR3 sequences. All seven of the TRB-CDR3s sequences from 5T4p17 specific TCRs were identified in non-RCC bearing individuals. In one public dataset of TRB-CDR3 sequencing from 678 healthy individuals [44], identical TRB-CDR3 sequences to HD_A-15 and HD_C-17 were found in 44 (12 HLA-A2+) and 5 (1 HLA-A2+) healthy bone marrow samples, respectively. The natural occurrence of these TRB-CDR3s in HLA-A2+ healthy donors suggests the possibility for 5T4-reactive TCRs to circulate in the repertoire of a subset of healthy donors in addition to the individuals providing research samples for our original T-cell culture experiments.
Our sequence analyses have demonstrated that naturally occurring 5T4p17-specific T-cells are present at very low-frequency. Our studies set an estimate of precursor frequency for these T-cells as low as 1 in 107 CD8+ T-cells in responding donors [10]. These data are consistent with immune monitoring studies performed with MVA-5T4 vaccination that have also revealed a heterogenous response phenotype for induced 5T4-specific T-cell immunity and low titer of measured T-cell responses [13]. Inadequate T-cell priming and expansion likely accounts for the failure of the tumor antigen-specific cancer vaccines MVA-5T4 and IMA901 to show improved survival for RCC patients in phase III clinical trials [45, 46].
In contrast to 5T4-specific vaccination, engineered autologous T-cells expressing CARs or TCRs specific for 5T4 represents an emerging therapy platform that, in principle, will facilitate a massive expansion of tumor-reactive T-cells in treated subjects. 5T4 is a transmembrane antigen and can, therefore, be targeted by either CAR or TCR-based strategies. Preclinical assessment of 5T4-CARs has demonstrated recognition of human nasopharyngeal carcinoma in vitro [8] and ovarian cancer in a murine xenograft model [47]. A potential advantage for TCR targeting of the 5T4p17 epitope is its location within the signal sequence of the 5T4 protein. It has been well established that a subset of T-cell epitopes within leader domain sequences can load class I MHC by a TAP-independent processing pathway [39]. Deficient TAP expression is a common tumor-associated phenotype thought to contribute to tumor escape from immune surveillance [48, 49]. We observed specific lysis of MVA-5T4 infected, TAP-deficient T2-target cells above background levels with 5T4p17-specific TCR-transduced CD8+ T-cells indicating the 5T4p17 epitope can be processed by a TAP-independent pathway. The robust specific lysis of 5T4p17-specific TCR-transduced CD8+ T-cells for MVA-5T4 infected LCL targets also suggests the intriguing clinical scenario that subjects treated with TCR-engineered 5T4p17-specific T-cells could be subsequently vaccinated with MVA-5T4 to re-stimulate the transduced effector T-cells against the 5T4 target in vivo.
In summary, TCR-engineered 5T4p17-specific CD8+ T-cells are of interest for future testing as a cellular-immunotherapy product in subjects with 5T4+ tumors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Oxford BioMedica (Oxford, UK) for providing MVA-WT and MVA-5T4; Dr. B. Van den Eynde (Ludwig Institute for Cancer Research, Brussels, Belgium) for providing RCC- and LCL cell-lines; Dr. Phillip D. Greenberg and Dr. Tom B. Schmitt (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing the lentivirus packaging plasmids (pRSV-REV, pMD2-G, pMDLg/pRRE, pRRLSIN) and TM-LCL line.
Abbreviations
- 5T4
Trophoblast glycoprotein, TPBG
- ADCs
Antibody–drug conjugates
- CDR3
Complementarity-determining region 3
- cPPT
Central polypurine tract
- CTLA4
Cytotoxic T-lymphocyte associated protein 4
- IMGT
International Immunogenetics Information System
- MART-1
Melanoma antigen recognized by T-cells 1
- MSCV
Murine stem cell virus
- mTOR
Mammalian target of rapamycin
- MVA
Modified vaccinia virus Ankara
- PTEC
Proximal tubule endothelial cell
- RCC
Renal cell carcinoma
- TAP
Transporter associated with antigen processing
- TET
Tetramer
- TKIs
Tyrosine kinase inhibitors
- TNF-α
Tumor necrosis factor-α
- TRA
T-cell receptor α chain
- TRA-CDR3
CDR3 region from TCR-α gene
- TRAC
Constant region of TCR-α gene
- TRB
T-cell receptor β chain
- TRB-CDR3
CDR3 regions from the TCR-β gene
- TRBC
Constant region of TCR-β gene
- WT1
Wilms’ tumor 1
- WPRE
Woodchuck posttranscriptional regulatory element
Author contributions
SST, EHW, and YX designed the study; YX, AJM and MJC performed the experiments; AMHT performed the TCRB sequencing; YX and DGC performed the single-cell sequencing analysis; YX and SST wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by a generous gift from Dave Jackson.
Compliance with ethical standards
Conflict of interest
Aprovisional patent application was filed by Yuexin Xu, Edus H. Warren and Scott S. Tykodi regarding TCRs described in this study. The other authors declare that they have no conflict of interest.
Ethical approval and ethical standard
All study protocols and donor consent documents were approved by the Institutional Review Board of Fred Hutchinson Cancer Research Center, Seattle, Washington, USA and included protocols 1495 (skin biopsy), 999.209 (skin biopsy), 868 (healthy donor leukapheresis), 1246 (cancer patient leukapheresis), and 1810 (RCC tumor, normal kidney, peripheral blood).
Informed consent
Written informed consent was obtained from all individual donors of biological samples for research use included in this study. Kidney cancer donor D consent for the collection of RCC tumor tissue for study 1810 also authorized collection of clinical data from the donor’s electronic medical record, including treatment history and disease status.
Cell line authentication
The cell lines T2, A498, MDA-231, BT-20, and SW480 were obtained from American Type Culture Collection (Manassas, VA). The cell line lenti-X 293T virus packaging cells was obtained from Clontech Laboratories (Mountain View, CA). The RCC tumor lines LB1828, BB65, DOBSKI, and the BB65-LCL line were a generous gift from Dr. Benoît Van den Eynde (Ludwig Institute for Cancer Research, Brussels, Belgium). The TM-LCL line was a generous gift from Dr. Phillip D. Greenberg and Dr. Tom B. Schmitt (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). The phenotypes of these lines were verified by the source before arriving at our facility. The phenotypic characterization of RCC tumor line SST548 was previously verified [10]. Paired RCC tumor and PTEC cultures from four clear-cell RCC tumors and adjacent normal kidney cortex, as well as three fibroblast lines derived from skin biopsies of three additional donors were established following specific cell culture protocols to ensure propagation of the targeted cell types [50–52]. Expression of the 5T4 tumor marker on the four primary RCC lines was verified by flow cytometry, as shown in Supplementary Fig. 6. No 5T4 expression was detected on any fibroblast or PTEC lines [10]. PCR-based HLA-typing of genomic DNA was performed for all cell lines to select for HLA-A2 expression [10]. Mycoplasma was routinely tested. Phenotypes for cell morphology and growth rate were monitored regularly and did not change over time. All the experiments were performed with cells at a low passage number (≤ 10).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edus H. Warren and Scott S. Tykodi contributed equally to this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Fyfe G, Fisher RI, Rosenberg SA, Sznol M, Parkinson DR, Louie AC. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. J Clin Oncol. 1995;13(3):688–696. doi: 10.1200/JCO.1995.13.3.688. [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthelemy P, Porta C, George S, Powles T, Donskov F, Neiman V, Kollmannsberger CK, Salman P, Gurney H, Hawkins R, Ravaud A, Grimm MO, Bracarda S, Barrios CH, Tomita Y, Castellano D, Rini BI, Chen AC, Mekan S, McHenry MB, Wind-Rotolo M, Doan J, Sharma P, Hammers HJ, Escudier B, CheckMate I. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378(14):1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern PL, Harrop R. 5T4 oncofoetal antigen: an attractive target for immune intervention in cancer. Cancer Immunol Immunother. 2017;66(4):415–426. doi: 10.1007/s00262-016-1917-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Southall PJ, Boxer GM, Bagshawe KD, Hole N, Bromley M, Stern PL. Immunohistological distribution of 5T4 antigen in normal and malignant tissues. Br J Cancer. 1990;61(1):89–95. doi: 10.1038/bjc.1990.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffiths RW, Gilham DE, Dangoor A, Ramani V, Clarke NW, Stern PL, Hawkins RE. Expression of the 5T4 oncofoetal antigen in renal cell carcinoma: a potential target for T-cell-based immunotherapy. Br J Cancer. 2005;93(6):670–677. doi: 10.1038/sj.bjc.6602776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borghaei H, Alpaugh K, Hedlund G, Forsberg G, Langer C, Rogatko A, Hawkins R, Dueland S, Lassen U, Cohen RB. Phase I dose escalation, pharmacokinetic and pharmacodynamic study of naptumomab estafenatox alone in patients with advanced cancer and with docetaxel in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2009;27(25):4116–4123. doi: 10.1200/JCO.2008.20.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X, Zheng H, Luo W, Zhang Q, Liu J, Yao K. 5T4-specific chimeric antigen receptor modification promotes the immune efficacy of cytokine-induced killer cells against nasopharyngeal carcinoma stem cell-like cells. Sci Rep. 2017;7(1):4859. doi: 10.1038/s41598-017-04756-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damelin M, Geles KG, Follettie MT, Yuan P, Baxter M, Golas J, DiJoseph JF, Karnoub M, Huang S, Diesl V, Behrens C, Choe SE, Rios C, Gruzas J, Sridharan L, Dougher M, Kunz A, Hamann PR, Evans D, Armellino D, Khandke K, Marquette K, Tchistiakova L, Boghaert ER, Abraham RT, Wistuba II, Zhou BB. Delineation of a cellular hierarchy in lung cancer reveals an oncofetal antigen expressed on tumor-initiating cells. Cancer Res. 2011;71(12):4236–4246. doi: 10.1158/0008-5472.CAN-10-3919. [DOI] [PubMed] [Google Scholar]
- 10.Tykodi SS, Satoh S, Deming JD, Chou J, Harrop R, Warren EH. CD8 + T-cell clones specific for the 5T4 antigen target renal cell carcinoma tumor-initiating cells in a murine xenograft model. J Immunother. 2012;35(7):523–533. doi: 10.1097/CJI.0b013e318261d630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins RE, Gore M, Shparyk Y, Bondar V, Gladkov O, Ganev T, Harza M, Polenkov S, Bondarenko I, Karlov P, Karyakin O, Khasanov R, Hedlund G, Forsberg G, Nordle O, Eisen T. A randomized phase II/III study of naptumomab estafenatox + IFN alpha versus IFN alpha in renal cell carcinoma: final analysis with baseline biomarker subgroup and trend analysis. Clin Cancer Res. 2016;22(13):3172–3181. doi: 10.1158/1078-0432.CCR-15-0580. [DOI] [PubMed] [Google Scholar]
- 12.Sapra P, Damelin M, Dijoseph J, Marquette K, Geles KG, Golas J, Dougher M, Narayanan B, Giannakou A, Khandke K, Dushin R, Ernstoff E, Lucas J, Leal M, Hu G, O’Donnell CJ, Tchistiakova L, Abraham RT, Gerber HP. Long-term tumor regression induced by an antibody-drug conjugate that targets 5T4, an oncofetal antigen expressed on tumor-initiating cells. Mol Cancer Ther. 2013;12(1):38–47. doi: 10.1158/1535-7163.MCT-12-0603. [DOI] [PubMed] [Google Scholar]
- 13.Amato RJ, Shingler W, Goonewardena M, de Belin J, Naylor S, Jac J, Willis J, Saxena S, Hernandez-McClain J, Harrop R. Vaccination of renal cell cancer patients with modified vaccinia Ankara delivering the tumor antigen 5T4 (TroVax) alone or administered in combination with interferon-alpha (IFN-alpha): a phase 2 trial. J Immunother. 2009;32(7):765–772. doi: 10.1097/CJI.0b013e3181ace876. [DOI] [PubMed] [Google Scholar]
- 14.Grupp SA, Kalos M, Barrett D, Aplenc R, Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, Milone MC, Levine BL, June CH. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turtle CJ, Hanafi LA, Berger C, Hudecek M, Pender B, Robinson E, Hawkins R, Chaney C, Cherian S, Chen X, Soma L, Wood B, Li D, Heimfeld S, Riddell SR, Maloney DG. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8 + and CD4 + CD19-specific chimeric antigen receptor-modified T cells. Sci Transl Med. 2016;8(355):355ra116. doi: 10.1126/scitranslmed.aaf8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins PF, Kassim SH, Tran TL, Crystal JS, Morgan RA, Feldman SA, Yang JC, Dudley ME, Wunderlich JR, Sherry RM, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee CC, Li YF, El-Gamil M, Rosenberg SA. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21(5):1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Angelo SP, Melchiori L, Merchant MS, Bernstein D, Glod J, Kaplan R, Grupp S, Tap WD, Chagin K, Binder GK, Basu S, Lowther DE, Wang R, Bath N, Tipping A, Betts G, Ramachandran I, Navenot JM, Zhang H, Wells DK, Van Winkle E, Kari G, Trivedi T, Holdich T, Pandite L, Amado R, Mackall CL. Antitumor activity associated with prolonged persistence of adoptively transferred NY-ESO-1 (c259)T cells in synovial sarcoma. Cancer Discov. 2018;8(8):944–957. doi: 10.1158/2159-8290.CD-17-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doran SL, Stevanovic S, Adhikary S, Gartner JJ, Jia L, Kwong MLM, Faquin WC, Feldman S, Somerville R, Sherry RM, Yang JC, Rosenberg SA, Hinrichs CS. Genetically engineered T-cell therapy for HPV-associated epithelial cancers: a first in human, phase I/II clinical trial. J Clin Oncol. 2018;36(15_suppl):3019. doi: 10.1200/jco.2018.36.15_suppl.3019. [DOI] [Google Scholar]
- 19.Nagarsheth N, Norberg S, Doran SL, Kanakry JA, Adhikary S, Schweitzer C, Astrow SH, Faquin WC, Gkitsas N, Highfill S, Stroncek D, Hinrichs C. Regression of epithelial cancers in humans following t-cell receptor gene therapy targeting human papillomavirus-16 E7. J Clin Oncol. 2018;36(15_suppl):3043. doi: 10.1200/jco.2018.36.15_suppl.3043. [DOI] [Google Scholar]
- 20.Sherwood AM, Desmarais C, Livingston RJ, Andriesen J, Haussler M, Carlson CS, Robins H. Deep sequencing of the human TCRgamma and TCRbeta repertoires suggests that TCRbeta rearranges after alphabeta and gammadelta T cell commitment. Sci Transl Med. 2011;3(90):90ra61. doi: 10.1126/scitranslmed.3002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32(7):684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12(5):380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 23.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 24.Scaviner D, Lefranc MP. The human T cell receptor alpha variable (TRAV) genes. Exp Clin Immunogenet. 2000;17(2):83–96. doi: 10.1159/000019128. [DOI] [PubMed] [Google Scholar]
- 25.Folch G, Lefranc MP. The human T cell receptor beta variable (TRBV) genes. Exp Clin Immunogenet. 2000;17(1):42–54. doi: 10.1159/000019123. [DOI] [PubMed] [Google Scholar]
- 26.Jones S, Peng PD, Yang S, Hsu C, Cohen CJ, Zhao Y, Abad J, Zheng Z, Rosenberg SA, Morgan RA. Lentiviral vector design for optimal T cell receptor gene expression in the transduction of peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Hum Gene Ther. 2009;20(6):630–640. doi: 10.1089/hum.2008.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren EH, Greenberg PD, Riddell SR. Cytotoxic T-lymphocyte-defined human minor histocompatibility antigens with a restricted tissue distribution. Blood. 1998;91(6):2197–2207. doi: 10.1182/blood.V91.6.2197. [DOI] [PubMed] [Google Scholar]
- 28.Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23(9):2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodie SJ, Lewinsohn DA, Patterson BK, Jiyamapa D, Krieger J, Corey L, Greenberg PD, Riddell SR. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat Med. 1999;5(1):34–41. doi: 10.1038/4716. [DOI] [PubMed] [Google Scholar]
- 30.Tykodi SS, Warren EH, Thompson JA, Riddell SR, Childs RW, Otterud BE, Leppert MF, Storb R, Sandmaier BM. Allogeneic hematopoietic cell transplantation for metastatic renal cell carcinoma after nonmyeloablative conditioning: toxicity, clinical response, and immunological response to minor histocompatibility antigens. Clin Cancer Res. 2004;10(23):7799–7811. doi: 10.1158/1078-0432.CCR-04-0072. [DOI] [PubMed] [Google Scholar]
- 31.Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2003;31(1):307–310. doi: 10.1093/nar/gkg085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blevins SJ, Pierce BG, Singh NK, Riley TP, Wang Y, Spear TT, Nishimura MI, Weng Z, Baker BM. How structural adaptability exists alongside HLA-A2 bias in the human alphabeta TCR repertoire. Proc Natl Acad Sci USA. 2016;113(9):E1276–E1285. doi: 10.1073/pnas.1522069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borrman T, Cimons J, Cosiano M, Purcaro M, Pierce BG, Baker BM, Weng Z. ATLAS: a database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes. Proteins. 2017;85(5):908–916. doi: 10.1002/prot.25260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt TM, Aggen DH, Stromnes IM, Dossett ML, Richman SA, Kranz DM, Greenberg PD. Enhanced-affinity murine T-cell receptors for tumor/self-antigens can be safe in gene therapy despite surpassing the threshold for thymic selection. Blood. 2013;122(3):348–356. doi: 10.1182/blood-2013-01-478164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banu N, Chia A, Ho ZZ, Garcia AT, Paravasivam K, Grotenbreg GM, Bertoletti A, Gehring AJ. Building and optimizing a virus-specific T cell receptor library for targeted immunotherapy in viral infections. Sci Rep. 2014;4:4166. doi: 10.1038/srep04166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ofran Y, Kim HT, Brusic V, Blake L, Mandrell M, Wu CJ, Sarantopoulos S, Bellucci R, Keskin DB, Soiffer RJ, Antin JH, Ritz J. Diverse patterns of T-cell response against multiple newly identified human Y chromosome-encoded minor histocompatibility epitopes. Clin Cancer Res. 2010;16(5):1642–1651. doi: 10.1158/1078-0432.CCR-09-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 39.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salter RD, Howell DN, Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21(3):235–246. doi: 10.1007/BF00375376. [DOI] [PubMed] [Google Scholar]
- 41.Morimoto S, Oka Y, Tsuboi A, Tanaka Y, Fujiki F, Nakajima H, Hosen N, Nishida S, Nakata J, Nakae Y, Maruno M, Myoui A, Enomoto T, Izumoto S, Sekimoto M, Kagawa N, Hashimoto N, Yoshimine T, Oji Y, Kumanogoh A, Sugiyama H. Biased usage of T cell receptor beta-chain variable region genes of Wilms’ tumor gene (WT1)-specific CD8 + T cells in patients with solid tumors and healthy donors. Cancer Sci. 2012;103(3):408–414. doi: 10.1111/j.1349-7006.2011.02163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinto S, Sommermeyer D, Michel C, Wilde S, Schendel D, Uckert W, Blankenstein T, Kyewski B. Misinitiation of intrathymic MART-1 transcription and biased TCR usage explain the high frequency of MART-1-specific T cells. Eur J Immunol. 2014;44(9):2811–2821. doi: 10.1002/eji.201444499. [DOI] [PubMed] [Google Scholar]
- 43.Mastaglio S, Genovese P, Magnani Z, Ruggiero E, Landoni E, Camisa B, Schiroli G, Provasi E, Lombardo A, Reik A, Cieri N, Rocchi M, Oliveira G, Escobar G, Casucci M, Gentner B, Spinelli A, Mondino A, Bondanza A, Vago L, Ponzoni M, Ciceri F, Holmes MC, Naldini L, Bonini C. NY-ESO-1 TCR single edited stem and central memory T cells to treat multiple myeloma without graft-versus-host disease. Blood. 2017;130(5):606–618. doi: 10.1182/blood-2016-08-732636. [DOI] [PubMed] [Google Scholar]
- 44.Dean J, Emerson RO, Vignali M, Sherwood AM, Rieder MJ, Carlson CS, Robins HS. Annotation of pseudogenic gene segments by massively parallel sequencing of rearranged lymphocyte receptor loci. Genome Med. 2015;7:123. doi: 10.1186/s13073-015-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rini BI, Stenzl A, Zdrojowy R, Kogan M, Shkolnik M, Oudard S, Weikert S, Bracarda S, Crabb SJ, Bedke J, Ludwig J, Maurer D, Mendrzyk R, Wagner C, Mahr A, Fritsche J, Weinschenk T, Walter S, Kirner A, Singh-Jasuja H, Reinhardt C, Eisen T. IMA901, a multipeptide cancer vaccine, plus sunitinib versus sunitinib alone, as first-line therapy for advanced or metastatic renal cell carcinoma (IMPRINT): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17(11):1599–1611. doi: 10.1016/S1470-2045(16)30408-9. [DOI] [PubMed] [Google Scholar]
- 46.Amato RJ, Hawkins RE, Kaufman HL, Thompson JA, Tomczak P, Szczylik C, McDonald M, Eastty S, Shingler WH, de Belin J, Goonewardena M, Naylor S, Harrop R. Vaccination of metastatic renal cancer patients with MVA-5T4: a randomized, double-blind, placebo-controlled phase III study. Clin Cancer Res. 2010;16(22):5539–5547. doi: 10.1158/1078-0432.CCR-10-2082. [DOI] [PubMed] [Google Scholar]
- 47.Owens GL, Sheard VE, Kalaitsidou M, Blount D, Lad Y, Cheadle EJ, Edmondson RJ, Kooner G, Gilham DE, Harrop R. Preclinical assessment of CAR T-cell therapy targeting the tumor antigen 5T4 in ovarian cancer. J Immunother. 2018;41(3):130–140. doi: 10.1097/CJI.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seliger B, Jasinski-Bergner S, Quandt D, Stoehr C, Bukur J, Wach S, Legal W, Taubert H, Wullich B, Hartmann A. HLA-E expression and its clinical relevance in human renal cell carcinoma. Oncotarget. 2016;7(41):67360–67372. doi: 10.18632/oncotarget.11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst. 2013;105(16):1172–1187. doi: 10.1093/jnci/djt184. [DOI] [PubMed] [Google Scholar]
- 50.Detrisac CJ, Sens MA, Garvin AJ, Spicer SS, Sens DA. Tissue culture of human kidney epithelial cells of proximal tubule origin. Kidney Int. 1984;25(2):383–390. doi: 10.1038/ki.1984.28. [DOI] [PubMed] [Google Scholar]
- 51.Lahn M, Kohler G, Schmoor C, Dengler W, Veelken H, Brennscheidt U, Mackensen A, Kulmburg P, Hentrich I, Jesuiter H, Rosenthal FM, Fiebig HH, Sommerkamp H, Farthmann EH, Hasse J, Mertelsmann R, Lindemann A. Processing of tumor tissues for vaccination with autologous tumor cells. Eur Surg Res. 1997;29(4):292–302. doi: 10.1159/000129536. [DOI] [PubMed] [Google Scholar]
- 52.Riddell SR, Rabin M, Geballe AP, Britt WJ, Greenberg PD. Class I MHC-restricted cytotoxic T lymphocyte recognition of cells infected with human cytomegalovirus does not require endogenous viral gene expression. J Immunol. 1991;146(8):2795–2804. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.