Abstract
Dynamic mRNA modification in the form of N6-methyladenosine (m6A) adds considerable richness and sophistication to gene regulation. The m6A mark is asymmetrically distributed along mature mRNAs, with approximately 35% of m6A residues located within the coding region (CDS). It has been suggested that methylation in CDS slows down translation elongation. However, neither the decoding feature of endogenous mRNAs nor the physiological significance of CDS m6A has been clearly defined. Here, we found that CDS m6A leads to ribosome pausing in a codon-specific manner. Unexpectedly, removing CDS m6A from these transcripts results in a further decrease of translation. A systemic analysis of RNA structural datasets revealed that CDS m6A positively regulates translation by resolving mRNA secondary structures. We further demonstrate that the elongation-promoting effect of CDS methylation requires the RNA helicase-containing m6A reader YTHDC2. Our findings established the physiological significance of CDS methylation and uncovered non-overlapping function of m6A reader proteins.
Subject terms: Biochemistry, Chemical biology, Genetics
Several functions have been proposed for m6A in RNA metabolism, yet little is known regarding the specific functions of individual m6A readers. Here, the authors observe that the m6A reader YTHDC2 — which contains an RNA helicase domain — acts on the coding region to promotes mRNA translation by resolving secondary structures.
Introduction
A grand challenge in the postgenomic era is to elucidate complex layers of regulatory elements beyond the nucleotide sequence. mRNAs carry the genetic information that is translated by ribosomes. Both the 5′ and 3′ untranslated regions (UTRs) bear many cis-acting elements that are intricately linked to the regulation of translation initiation. The importance of the coding region (CDS) is apparent because the elongation speed directly controls the translational output. Recent findings from ribosome profiling studies show that the translation machinery proceeds not at a constant rate but rather in a stop-and-go traffic manner1,2. Frequent ribosomal pausing decreases the overall translation efficiency (TE) by reducing the elongation speed and limiting the amount of free ribosomes available for other protein synthesis. Factors contributing to ribosomal pausing are likely to be multifaceted. Besides the nucleotide sequence, the flexible nature of mRNA molecules implies that particular shape can also encode regulatory information guiding translational control3. One fundamental question is how cells fine-tune the TE for individual transcripts by integrating parallel codes embedded within the nucleotide sequence.
One such parallel code is the chemical modification of nucleotides within mRNAs4. To date, more than 150 distinct modifications have been identified on RNA species5. N6-methyladenosine (m6A) is the most abundant internal base modification occurring on eukaryotic mRNAs. The m6A content varies substantially across various species, tissues, and cellular environments6, suggesting an extensive regulation of methylation dynamics. The m6A topology is achieved by two opposing enzyme systems: the methyltransferase complex comprising a core heterodimer of METTL3–METTL14 (refs 7,8), and m6A demethylases FTO and ALKBH5 (refs 9,10). The biological effect of m6A largely depends on m6A reader proteins, such as YTH domain-containing proteins11. A recent study reported that m6A also repels certain RNA-binding proteins12, forming an additional layer in controlling dynamic RNA–protein interaction. By affecting nearly all the aspects of mRNA metabolism, m6A marks an ever growing list of cellular and physiological functions.
Despite the tremendous progress in the functional characterization of m6A modification, the regional effects of mRNA methylation remain obscure. For mature mRNAs, there is a strong enrichment of m6A around the stop codon and 3′ UTR13,14. The asymmetric m6A deposition suggests that regional methylation may have distinct functional consequences. Previous studies reported that the cytosolic m6A readers YTHDF1 and YTHDF3 promote cap-dependent mRNA translation presumably via 3′ UTR methylation15,16. Recent m6A-seq studies also revealed m6A peaks in 5′ UTR and start codons when plotting the peak density along the transcriptome17. Intriguingly, m6A in the 5′ UTR could facilitate cap-independent translation through a process involving eIF3 (ref. 18), although the exact nature of this process remains unclear. Besides m6A in the untranslated regions, approximately 35% of m6A residues are located within the CDS. Using an elegant single molecule-based in vitro translation system, it has been demonstrated that m6A interferes with the decoding process by affecting tRNA accommodation, thereby slowing down translation elongation19. However, neither the decoding feature of methylated codons within endogenous mRNAs nor the physiological significance of CDS methylation has been clearly defined.
Given the forward movement of ribosome during elongation, the CDS methylation could impede the translation by directly affecting the decoding process or indirectly blocking elongation via m6A-binding proteins. Acting as brakes and roadblocks, these mechanisms are expected to result in prominent ribosome pausing. Such persistent ribosome stalling is expected to trigger the mRNA surveillance system and subsequent mRNA degradation. However, the logical and mechanistic relationships between CDS methylation and ribosome dynamics are poorly understood, if such relationships exist. Here we set out to determine the relationship between CDS m6A modification and ribosome behavior. Unexpectedly, we found that CDS methylation positively regulates translation by resolving mRNA secondary structures. Intriguingly, the translation-promoting effect of m6A modification requires YTHDC2, the only RNA helicase-containing m6A reader. Our findings establish the physiological significance of m6A methylation in CDS and the unique role of YTHDC2 in translation elongation suggests nonoverlapping functions of m6A reader proteins.
Results
CDS m6A occurs on mRNAs with low TE
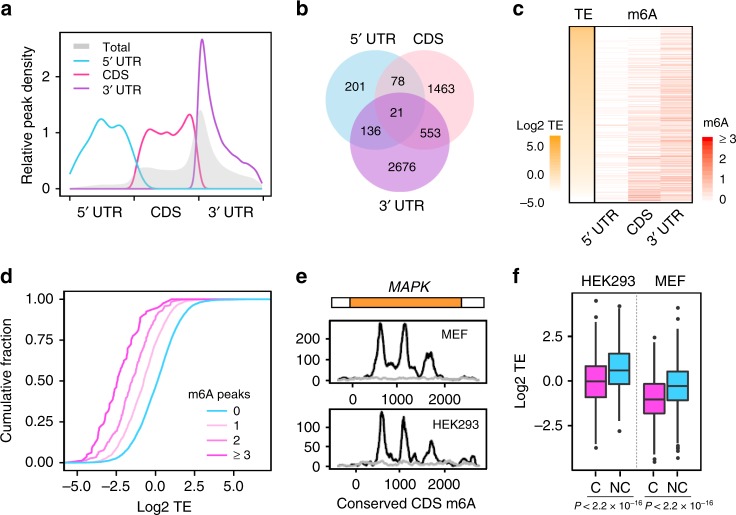
We began our analyses by identifying m6A peaks from mouse embryonic fibroblasts (MEF) using a method described before20. To avoid false positives due to background noise and possible bias of peak calling, only METTL3 or WTAP sensitive m6A peaks are used in the following analysis. From a total of 15,646 m6A peaks identified from MEF cells, only 8352 peaks show >50% decrease of methylation upon depletion of METTL3 or WTAP. As expected, these m6A peaks are mostly enriched near the stop codon (Fig. 1a, gray line). Based on their positions within mRNA, we classified these m6A sites into three regions: 5′ UTR (449 peaks from 436 genes), CDS (2825 peaks from 2115 genes) and 3′ UTR (4044 peaks from 3386 genes). Notably, more than half of the transcripts contain one regional m6A (Fig. 1b). Only a few messengers harbor m6A in all three regions. To reveal possible relationships between regional methylation and gene functions, we searched for common biological themes among transcripts bearing m6A in different regions. Interestingly, genes involved in transcriptional regulation are overrepresented among mRNAs with CDS methylation (Supplementary Fig. 1).
Fig. 1.
CDS m6A is enriched in transcripts with inactive translation. a Distribution of m6A sites along transcripts with different regional methylation. Transcripts are grouped into 5′ UTR, CDS, and 3′ UTR methylation based on the identified m6A sites. Only METTL3 or WTAP sensitive m6A sites are considered. The distribution of total m6A sites was shown in gray. UTR: untranslated region, CDS: protein coding region. b A Venn diagram shows the overlapping of transcripts containing m6A sites in different regions. c Translation efficiency (TE, left column) and m6A density across the transcriptome are presented as parallel heat-maps. Note the inverse correlation between CDS m6A methylation and TE. d Translation efficiency (TE) is plotted as accumulative fractions for mRNAs with different amount of m6A peaks in CDS. e A representative example (MAPK) of evolutionary conserved m6A sites between MEF and HEK293 cells. The coverage of m6A is shown as black lines and the input as gray. f A box plot shows the translation efficiency (TE) of mRNAs containing conserved (C) and non-conserved (NC) m6A sites in MEF and HEK293 cells (Wilcox test, all P values < 2.2 × 10−16). The median of TE in each group is indicated by a center line, the box shows the upper and lower quantiles, whiskers shows the 1.5× interquartile range, and the outliers are indicated by points. Source data are provided as a Source Data file.
Given the crucial role of CDS in translation elongation, we next sought to determine the correlation between CDS methylation and TE. Using ribosome profiling (Ribo-seq) data sets obtained from MEF cells, we computed TE by normalizing ribosome density with the corresponding mRNA levels. Notably, neither 5′ UTR nor 3′ UTR methylation correlates with TE in a significant manner (Fig. 1c). Only when CDS methylation is considered, does an inverse correlation become evident between m6A levels and TE. In comparison to the non-methylated mRNAs, transcripts with CDS methylation show significantly reduced ribosome occupancy (Wilcox-test, P < 2.2 × 10−16, Supplementary Fig. 2a). Remarkably, as the number of CDS m6A peaks increases, the ribosome occupancy decreases correspondingly with mRNAs bearing >3 m6A peaks exhibiting the lowest TE (Fig. 1d). The similar finding was observed by analyzing CDS m6A coverage (Supplementary Fig. 2b), which is further corroborated by the distribution of TE for transcripts with or without CDS m6A (Supplementary Fig. 2c). The negative correlation between CDS methylation and ribosome occupancy also holds true in a human cell line HEK293 (Supplementary Fig. 2d), as well as mouse embryonic stem cells (ESCs) and embryonic bodies (EBs)21 (Supplementary Fig. 2e). Since the calculated TE is normalized by mRNA levels, the inverse correlation between TE and CDS methylation cannot be explained by m6A-mediated mRNA degradation. It is rather consistent with the notion that CDS methylation occurs on transcripts with relatively inactive translation.
We noticed that certain m6A peaks in CDS are highly reproducible in different cell lines, suggesting a functional conservation. One typical example is MAPK7 that bears three prominent CDS m6A peaks (Fig. 1e). Using m6A-seq data sets obtained from human (HEK293) and mouse (MEF) cells, we identified approximately 10% of m6A peaks (429 peaks of 316 mRNAs) as the conserved methylation sites in CDS. Notably, transcripts harboring the conserved m6A sites exhibit significantly lower ribosome occupancy than ones containing the non-conserved sites (Fig. 1f). Therefore, CDS methylation could be evolved and retained on certain transcripts with functional significance.
CDS m6A methylation leads to ribosome pausing
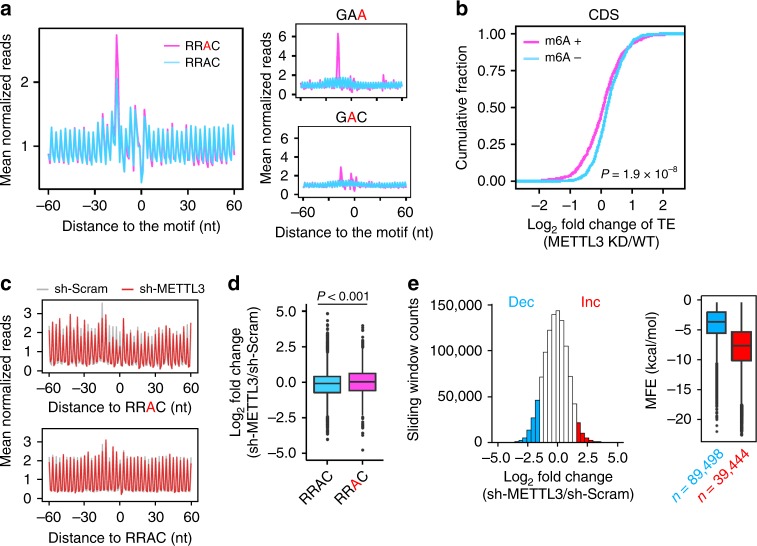
A previous study demonstrated that presence of m6A interferes with the decoding process of ribosomes by affecting tRNA accommodation at the A site, at least in the in vitro translation system reconstituted from Escherichia coli19. We then examined whether CDS m6A methylation leads to ribosome pausing at specific mRNA positions in mammalian cells. We took advantage of single nucleotide resolution m6A sites identified from HEK293 cells22. In addition, we used Ribo-seq data sets obtained in HEK293 without cycloheximide pretreatment to avoid technical artifacts23. When transcripts are aligned to the identified m6A site, we observed an elevated ribosome density at the −15 nt position when the 5′ end of reads are counted (Fig. 2a). This position corresponds to the methylated codon at the ribosomal A site. The approximately threefold higher ribosome density when the A site codon is methylated suggests a delayed codon:anti-codon interaction in the presence of m6A. The similar result was also seen in MEFs (Supplementary Fig. 3a), although the low resolution m6A mapping possibly underestimates the m6A-induced ribosome pausing in these cells. Our analysis provides an in vivo evidence that CDS m6A methylation affects the decoding process of endogenous transcripts.
Fig. 2.
Characterize the role of CDS m6A in translation efficiency. a Aggregation plots show the mean ribosome densities along mRNA regions aligned to the RRAC motif with (pink line) or without (blue line) m6A modification. Right panels show the mean ribosome densities along mRNA regions aligned to GAA or GAC codons with (pink line) or without (blue line) m6A modification. A minus position value indicates upstream of m6A sites, whereas a positive value indicates downstream of m6A sites. The m6A site in RRAC motif or codons is highlighted by red. b The fold change of translation efficiency upon METTL3 knockdown is plotted as accumulative fractions (Wilcox test, P = 1.9 × 10−8) for mRNAs bearing CDS methylation (m6A+) or not (m6A−). Both groups have similar levels of basal TE. c Aggregation plots show the mean ribosome densities along mRNA regions aligned to the RRAC motif with (top panel) or without (bottom panel) m6A modification. Plots from cells with or without METTL3 knockdown are color coded. The m6A site in RRAC motif is highlighted by red. d From the same data sets as c, the fold change of ribosome accumulation upstream of the motif in response to METTL3 knockdown is calculated for transcripts with or without m6A modification (Wilcox test, P < 0.001). For the boxplot in d and e, the median value in each group is indicated by a center line, the box shows the upper and lower quantiles, whiskers shows the 1.5× interquartile range, and the outliers are indicated by points. e A histogram shows the distribution of changes of regional ribosome density in response to METTL3 knockdown. A sliding window of 30 nt in length with a step of 3 nt are used to calculate the local ribosome density. Regions with <1/3-fold (Dec, blue) and >3-fold (Inc., red) changes are highlighted by color coding. The right box plot shows the predicted minimum folding free energy (MFE) for regions with <1/3-fold (Dec) and >3-fold (Inc.) changes (Wilcox test, P < 2.2 × 10−16). Source data are provided as a Source Data file.
When the methylated codon enters the A site, the m6A could be in any positions of the triplet. Based on the consensus sequence of m6A (Supplementary Fig. 3b), we calculated the ribosome occupancy for different types of methylated codons. Notably, GAC (A as the methylated adenosine) accounts for the majority of methylated codons (41%). However, only a modest increase (approximately twofold) of ribosome pausing was observed when the methylated GAC triplet occupies the A site (Fig. 2a, right panel). Intriguingly, the less abundant GAA codon (10%) exhibited the strongest ribosome pausing among all the methylated codons. This is not due to the wobble position because the same m6A position in both AGA and GGA codons is only associated with a slight increase of ribosome density (Supplementary Fig. 3c). This finding suggests a strong codon-specific effect of m6A on ribosome dynamics.
CDS m6A methylation promotes TE
The m6A-caused delay in the decoding process potentially explains the negative correlation between CDS methylation and the TE. However, correlation does not imply causation. If reduced TE is a direct consequence of CDS methylation, removal of m6A modification from CDS is expected to increase the translational output. To test this possibility, we knocked down the core m6A methyltransferase METTL3 from MEF cells using shRNA. As reported previously24, METTL3 knockdown resulted in nearly 30% decrease of global protein synthesis as determined by [35S] metabolic labeling (Supplementary Fig. 4a). Since METTL3 knockdown reduces mRNA methylation in a non-specific manner, it is possible that the effect of CDS methylation is masked by m6A in other regions. We therefore stratified mRNAs based on regional methylation followed by comparison of TE fold changes before and after METTL3 knockdown. While altering 3′ UTR methylation has modest effects on TE, reducing 5′ UTR m6A levels decreased ribosome occupancy (Supplementary Fig. 4b). This is consistent with previous findings about the potential role of 5′ UTR m6A methylation in non-canonical mRNA translation18,24. To our surprise, depletion of CDS methylation as a result of METTL3 knockdown also led to reduced TE (Fig. 2b). The same feature holds true in a human cell line HeLa (Supplementary Fig. 4c). In addition, we conducted the same analysis using published data sets obtained from EBs and ESCs. It is clear that transcripts bearing CDS methylation were more sensitive to METTL3 depletion than the m6A negative control by decreasing TE (Supplementary Fig. 4d).
The positive role of CDS methylation in translation is seemingly contradictory to the m6A-induced ribosomal pausing effect. We next examined whether the removal of m6A from CDS would eliminate ribosomal pausing. While MEF cells with scramble shRNA control showed a prominent ribosome pausing at the methylated A site (Fig. 2c, gray line), the elevated peak at the same site was largely diminished in MEFs lacking METTL3 (Fig. 2c, red line). Despite the lack of prominent pausing peaks, we observed an increased ribosome density upstream of the m6A site relative to the downstream region. This was not seen at the RRAC site without methylation (Fig. 2c, bottom panel). To achieve a more quantitative analysis, we computed the ratio of ribosome density in regions before and after the CDS m6A sites. Upon METTL3 knockdown, there was a significant increase of ribosome density in regions upstream of the m6A site (Fig. 2d). Using sliding window analysis (Supplementary Fig. 5a), we identified regional ribosome pausing induced by METTL3 knockdown across the whole transcriptome. Intriguingly, the regions with pausing sites caused by the lack of CDS methylation tend to form stable secondary structures (low minimum folding energy, MFE) (Fig. 2e). To substantiate this finding further, we repeated the same analysis using cells lacking ALKBH5, a m6A demethylase. Under the elevated m6A levels, we observed fewer pausing regions, and less structural features in pausing regions. (Supplementary Fig. 5b). Collectively, these results suggest that CDS methylation promotes TE by reducing ribosomal pausing, probably involving m6A-mediated structural switches.
CDS m6A are associated with relaxed mRNA structures
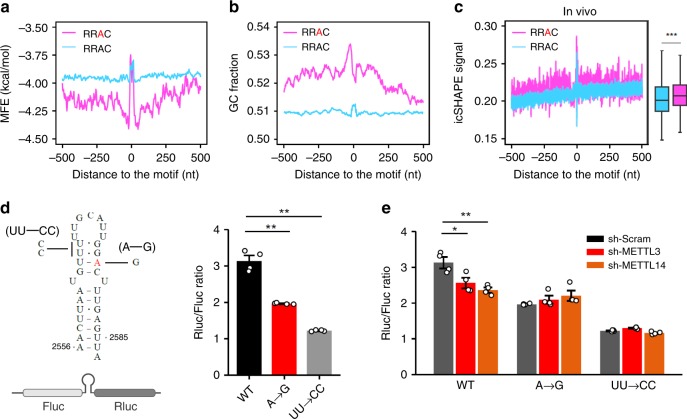
Previous studies demonstrated that m6A influences mRNA folding by acting as a structural switch25. In particular, m6A installation destabilizes RNA secondary structures26. To explore the mechanism explaining the positive role of CDS m6A in translation, we first analyzed the structural potential predicted by ViennaRNA in regions with or without m6A modification. From the mouse transcriptome, the methylated region displays a greater potential for stable secondary structures than the nonmethylated counterpart (Fig. 3a and Supplementary Fig. 6a). Consistently, analysis of the sequence context in methylated regions revealed a significant increase of the G/C content (Fig. 3b, Wilcox test, P < 2.2 × 10–16). These sequence-based structural prediction clearly indicates that m6A methylation sites are preferentially deposited to the regions tending to form stable structures. To probe the mRNA folding status in vivo, we took advantage of the icSHAPE data sets derived from MEF cells27. Unexpectedly, the methylated region shows higher icSHAPE signals than the non-methylated counterpart (Wilcox test, P < 0.001) (Fig. 3c), an indication of more single-stranded signals. The discrepancy between in vitro and in vivo structures was further confirmed by comparison of Gini index of methylated and non-methylated regions (Supplementary Fig. 6b, c). These results suggest that m6A modification in CDS likely eliminates local structures despite the relatively high GC content.
Fig. 3.
CDS m6A methylation resolves mRNA secondary structures. a The predicted minimum folding free energy (MFE) is plotted along mRNA regions surrounding the CDS RRAC motif with (pink line) or without (blue line) m6A modification. A sliding window with 30 nt in length and a step of 3 nt was used to calculate MFE. For each window, the central position is used for alignment. A minus position value indicates upstream of m6A sites, whereas a positive value indicates downstream of m6A sites. The m6A site in RRAC motif is highlighted by red. Notably, a lower MFE value indicates a higher potential for RNA secondary structures. b The GC content is plotted along mRNA regions surrounding the CDS RRAC motif with (pink line) or without (blue line) m6A modification. c The in vivo icSHAPE signal is plotted along mRNA regions surrounding the CDS RRAC motif with (pink) or without (blue) m6A modification. Notably, a higher in vivo icSHAPE signal indicates a less structured region. The right boxplot shows the average of icSHAPE signals across mRNA regions from –500 nt to 500 nt relative to the RRAC motif with (pink) or without (blue) m6A modification (Wilcox test, ***P < 0.001). The median of icSHAPE signals in each group is indicated by a center line, the box shows the upper and lower quantiles, whiskers shows the 1.5× interquartile range. The outliers are not shown. d The left panel shows the schematic of a dual luciferase reporter with a sandwiched secondary structure derived from MALAT1 (2556–2587). Both UU → CC and A → G mutants are also shown. The m6A site is highlighted by red. The right panel shows the ratio of Rluc/Fluc in transfected cells expressing wild type or indicated mutants. Error bars, mean ± s.e.m.; Single-tailed t test, n = 4, *P < 0.05, **P < 0.01. e The ratio of Rluc/Fluc in transfected cells expressing wild type or mutant reporters, with either METTL3 or METTL14 knockdown. Error bars, mean ± s.e.m.; single-tailed t test, n = 4, *P < 0.05, **P < 0.01. Source data are provided as a Source Data file.
As an independent validation, we repeated our analyses using the available structural and m6A data sets derived from HepG2 cells. Once again, methylated CDS regions in human transcriptome have a stronger tendency of forming stable secondary structures as evidenced by lower MFE and higher GC content (Supplementary Fig. 6d, e). However, there was a clear decrease of PARS scores in mRNA regions containing CDS m6A modification (Supplementary Fig. 6f), an indication of increased single-strand RNA signals. Therefore, CDS methylation in the form of m6A acts to prevent the formation of stable secondary structures.
CDS m6A modification resolves mRNA secondary structures
As ribosome occupancy can be influenced by both initiation and elongation, potential pausing sites in CDS could increase the ribosome density with reduced translational output. To directly demonstrate the critical role of m6A in CDS structures and subsequent translational outcomes, we constructed a fusion reporter by inserting a structural motif between firefly luciferase (Fluc) and renilla luciferase (Rluc) (Fig. 3d). The structural motif is derived from the non-coding RNA MALAT1 ranging from 2556 to 2586 nt (ENST00000534336.1), which includes a well-characterized m6A site at position 2577. The resultant fusion protein contains an extra 20 amino acids between Fluc and Rluc (see Supplementary Methods). It has been demonstrated that m6A modification at the site of A2577 destabilizes RNA folding25, which enables us to investigate the effect of RNA structural changes in translation. To evaluate the effect of m6A-induced structural switch, we first created a mutant by replacing the nucleotide A at position 2577 with G (A → G), which abolishes methylation at this site. Consistent with the more stable stem loop structure in the absence of m6A, we observed a significant decrease (~35%) of downstream Rluc translation when compared to the wild type (Fig. 3d). Next, we mutated the dinucleotides UU at positon 2566 and 2567 to CC (UU → CC), which makes the structure more stable than wildtype by changing the two G•U base pairs to G•C. Indeed, we observed a further decrease of Rluc translation from this mutant (~63%) (Fig. 3d). Therefore, the secondary structure in CDS acts as a roadblock for elongating ribosomes.
The decreased Rluc translation in the A → G mutant supports the positive role for m6A in translation. To test the possibility that CDS m6A methylation promotes translation by resolving mRNA secondary structures, we examined the Rluc/Fluc ratio in cells lacking m6A methyltransferases. In cells with METTL3 or METTL14 knockdown, we observed 15% and 18% reduction of Rluc translation, respectively (Fig. 3e). This was not due to the pleiotropic effects of m6A writers because the same cells exhibited little decrease of Rluc translation for the A → G mutant. The methylation status of these reporters was confirmed using SELECT, a site-specific m6A detection method28 (Supplementary Fig. 7a). The UU → CC mutant largely diminishes the effect of m6A-dependent structural rearrangement25. As a result, this mutant is resistant to METTL3 or METTL14 knockdown (Fig. 3e).
To substantiate this finding further, we knocked down m6A demethylases ALKBH5 or FTO. Although both A → G and UU → CC mutants maintained the similar Rluc/Fluc ratio, we observed modest, but significant, increase of Rluc translation (Supplementary Fig. 7b). The modest effect is presumably due to the high basal levels of methylation (60–80%) at the position of 2577 (ref. 6), leaving limited room for further increase of m6A. Collectively, we conclude that m6A modification in the CDS, in spite of causing codon-specific ribosome pausing, promotes the overall TE by resolving stable secondary structures.
YTHDC2 promotes TE
mRNA structures in CDS are commonly believed to be unwound by elongating ribosomes29. In line with this notion, transcriptome-wide structural mapping revealed that RNAs tend to assume more secondary structures in vitro than in vivo27,30. Since CDS methylation is enriched in transcripts with relatively inactive translation, one interesting question is whether m6A modification itself is able to resolve mRNA secondary structures. If so, a similar folding status of methylated regions is expected between in vitro and in vivo. However, this is not the case. Although the methylated regions showed increased in vivo icSHAPE signals (more single strand of RNA, Fig. 3c), the same region exhibited little difference of icSHAPE signals obtained in vitro when compared to the nonmethylated counterpart (Supplementary Fig. 7c). In addition, mRNAs with CDS methylation showed decreased in vivo icSHAPE signals upon METTL3 depletion (Supplementary Fig. 7d). This result argues that m6A modification relies on other factors inside cells in order to effectively unfold stable CDS structures.
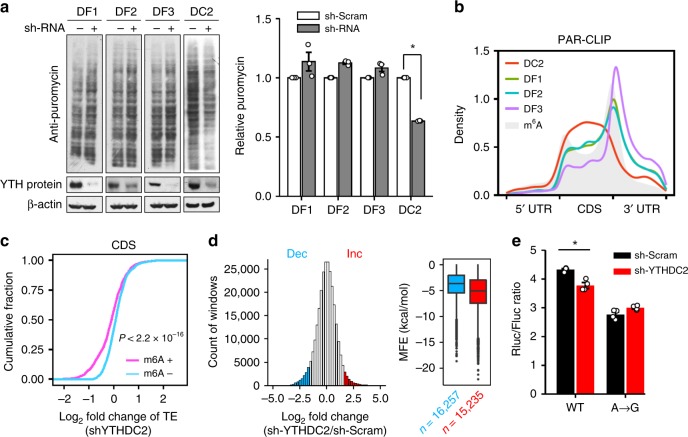
To search for potential m6A-interacting proteins that are capable of resolving CDS structures, we examined all the cytoplasmic YTH domain-containing m6A reader proteins. YTHDF1, YTHDF2, and YTHDF3 have been suggested to function cooperatively in the cytoplasm to promote efficient translation and degradation of specific m6A-containing mRNAs15,16,31. The multi-domain m6A reader YTHDC2 has been demonstrated to have an ATP-dependent RNA helicase activity32,33. If any of these m6A readers are actively participated in CDS methylation-promoted translation, silencing the corresponding genes would result in reduced translational output. We knocked down each m6A readers from HEK293 cells followed by measurement of global protein synthesis. Puromycin labeling of nascent polypeptides revealed that knocking down YTHDF1-3 had minimal effect on the global scale of mRNA translation (Fig. 4a). Remarkably, knocking down YTHDC2 led to a 40% reduction of protein synthesis. This is consistent with the observation that silencing YTHDC2, but not other m6A readers, significantly decreased the cell growth rate (Supplementary Fig. 8a). The translational effect of YTHDC2 is further corroborated by the polysome profiling in cells lacking YTHDC2 (Supplementary Fig. 8b). The sustained polysome in the absence of YTHDC2 is an indication of elongation pausing. Although YTHDC2 is highly expressed in testes cells (Supplementary Fig. 8c), it is clear that depleting this m6A reader readily affects protein synthesis even in cultured cells.
Fig. 4.
YTHDC2 promotes translation efficiency by acting on CDS m6A. a Puromycin labeling assay shows the global protein synthesis in HEK293 cells lacking each individual cytoplasmic m6A reader proteins. The right panel shows the quantitative results based on puromycin signals normalized with β-actin. Error bars, mean ± s.e.m.; t est, n = 3, *P < 0.05. DF1: YTHDF1, DF2: YTHDF2, DF3: YTHDF3, DC2: YTHDC2. b Distribution of the binding sites of cytoplasmic m6A reader proteins across the human transcriptome. All binding sites are identified from PAR-CLIP data sets obtained from HeLa cells. The distribution of m6A sites is shown as gray. c The fold change of translation efficiency upon YTHDC2 knockdown is plotted as accumulative fractions (Wilcox test, P < 2.2 × 10−16) for transcripts bearing CDS methylation (m6A+) or not (m6A−). Both groups have similar levels of basal TE. d A histogram shows the distribution of changes of regional ribosome density in response to YTHDC2 knockdown. A sliding window of 30 nt in length with a step of 3 nt are used to calculate the local ribosome density. Regions with <1/3-fold (Dec, blue) and >3-fold (Inc., red) changes are highlighted by color coding. The right box plot shows the predicted minimum folding free energy (MFE) for regions with <1/3-fold (Dec) and >3-fold (Inc.) changes (Wilcox test, P < 2.2 × 10−16). The median of MFE in each group is indicated by a center line, the box shows the upper and lower quantiles, whiskers shows the 1.5× interquartile range, and the outliers are indicated by points. e The ratio of Rluc/Fluc in transfected cells expressing wild type or mutant reporters, with or without YTHDC2 knockdown. Error bars, mean ± s.e.m.; Single-tailed t test, n = 4, *P < 0.05. Source data are provided as a Source Data file.
YTHDC2 resolves mRNA secondary structures
YTHDC2 contains multiple domains that bind to RNAs with distinct nucleotide preferences34. By reanalyzing the PAR-CLIP data sets obtained from HeLa cells, we found a strong enrichment of YTHDC2 binding sites in the CDS, rather than the 3′ UTR where the other cytoplasmic m6A readers are enriched (Fig. 4b). In addition, these binding sites have increased m6A coverage relative to the random sequences (Supplementary Fig. 8d). To substantiate this finding further, we conducted PAR-CLIP using MCF7 cells transfected with Flag-tagged YTHDC2 (Supplementary Fig. 8e). It is clear that the majority of YTHDC2 binding sites are enriched in CDS. Recent studies reported that YTHDC2 directly interacts with the small ribosomal subunit35, which suggests the involvement of YTHDC2 in translation of certain transcripts36–38. Notably, YTHDC2 has several motifs that are characteristic of the DEAH/RNA helicase A family, implying a functional connection with RNA structures. To investigate whether YTHDC2 helps resolve mRNA structures marked by m6A, we conducted Ribo-seq in HEK293 cells with or without YTHDC2 knockdown. By comparing the TE of transcripts with or without CDS methylation, we found that mRNAs bearing methylated CDS are more sensitive to YTHDC2 depletion than ones with 3′ UTR methylation (Fig. 4c). Notably, YTHDC2 knockdown has little effect on the translation of mRNAs with 5′ UTR methylation (Supplementary Fig. 8f), confirming the regional effect of YTHDC2.
We next examined whether YTHDC2 depletion reduces translation by inducing ribosomal pausing. Indeed, regions with increased ribosome density in the absence of YTHDC2 exhibit stronger structural features (lower MFE, Fig. 4d). Finally, we assessed the role of YTHDC2 in the translation of structural reporters. Much like METTL3 knockdown (Fig. 3e), silencing YTHDC2 decreases the ratio of Rluc/Fluc (Fig. 4e). Importantly, the A → G mutant maintained the Rluc/Fluc ratio, suggesting that YTHDC2 facilitates ribosome movement over the structural hurdle in an m6A-dependent manner. To confirm the critical role of helicase activity, we conducted a rescue experiment by introducing an E332Q mutation to inactivate the helicase of YTHDC2 (ref. 33) (Supplementary Fig. 9a). Luciferase reporter assay showed that only the wild type YTHDC2, but not the helicase-dead mutant, was able to restore the Rluc/Fluc ratio (Supplementary Fig. 9b). Taken together, our results indicate that m6A modification in CDS promotes translation of structured mRNA by recruiting the RNA helicase-containing YTHDC2.
Discussion
Recent studies have uncovered multiple roles of m6A in regulating translation. However, both positive and negative influences of this epitranscriptomic mark on protein production has been reported. Since methylation at different mRNA regions may have distinct functions, it is important to dissect regional effects of m6A on translation. In this regard, it is intriguing that approximately 35% of m6A residues are located within the CDS. Besides the embedded coding information, modified nucleotides like m6A could confer additional layer of regulation for the decoding process. Indeed, single molecule analysis of ribosome dynamics using an in vitro translation system derived from E. coli revealed that the presence of m6A in the CDS delays translation elongation19. Using synthetic mRNAs, random incorporation of m6A in general reduces translational output in transfected cells17. Consistently, for endogenous transcripts, we found that the greater the amount of m6A in CDS, the lower the TE. In addition, we demonstrated that methylated codons lead to pausing of elongating ribosomes, although the m6A is clearly not the sole determinant. The seemingly negative role of CDS methylation in the form of m6A in translation raises an intriguing question: what is the evolutionary benefit by creating potential roadblocks on endogenous transcripts?
To our surprise, removing CDS m6A from methylated transcripts did not result in enhanced translation. In fact, it further reduces the TE at least for some transcripts. This counter-intuitive result serves as a warning against attaching direct causative effects to what are merely suggestive correlations. Since the negative correlation is based on a pool of mRNAs with differential methylation, validating the causative effect requires examination of individual transcripts. Notably, the majority of methylated mRNAs only contain one reliable m6A peak and the density of CDS methylation on average is lower than that in 3′ UTR. Intriguingly, CDS m6A residues are usually located in regions that tend to form relatively stable secondary structures. This feature provides a plausible explanation why CDS m6A is highly enriched in mRNAs with less active translation. However, in the absence of METTL3, we observed newly formed ribosome pausing in mRNAs with methylated CDS. These results suggest that m6A installation is favorable for translation of structured mRNAs. For unstructured mRNAs with active translation, m6A modification likely reduces the TE by acting as a resistor. For structured mRNAs with poor TE, additional m6A marks could serve as a transconductor, facilitating translation elongation by resolving stable structures. How the structural region is selected for methylation is not known. It is possible that typical RNA structures may provide additional signals for recognition by m6A methyltransferases.
A major mechanism through which m6A regulates the fate of mRNAs is by recruiting m6A reader proteins4. Both YTHDF1 and YTHDF3 have been shown to mediate translational effects of m6A, mainly at the UTR regions15,16. It remains unclear how these reader proteins preferentially recognize the m6A mark in 3′ UTR, but not CDS. YTHDC2 is the largest YTH domain-containing protein and the only member of the family to contain helicase domains. YTHDC2 is highly expressed in testis and several recent studies demonstrated its crucial role in spermatogenesis32,33,36,37. It will be interesting to determine whether the transcriptome in germ cells tend to have more stable secondary structures. Regardless, many other human tissues express YTHDC2 at substantial levels, suggesting that the function of YTHDC2 is not limited to germ cells. Akin to the tissue-specific expression of YTHDC2, different cell types have varied subcellular localizations of m6A readers (Supplementary Fig. 10). Therefore, the functionality of m6A reader is likely context-dependent.
Unlike other m6A readers, silencing YTHDC2 from cells in culture resulted in a severe growth defect, arguing for a crucial role for YTHDC2 in cell physiology. Recent studies indicate that YTHDC2 increases the TE of a small subset of mRNAs with highly structured 5′ UTR38. Other studies have suggested the ability of YTHDC2 to promote TE33,36, albeit the underlying mechanism remained elusive. We found that YTHDC2 promotes translation of structured mRNAs by resolving secondary structures. It is possible that YTHDC2 gained this unique feature by possessing motifs that are characteristic of the DEAH/RNA helicase A family. Intriguingly, YTHDC2 has also been shown to accelerate mRNA decay. In particular, YTHDC2 interacts with the 5′ → 3′ exonuclease XRN1 (ref. 35). It is conceivable that YTHDC2 relies on CDS methylation signals to coordinate translation and decay of structured mRNAs. Since many of these transcripts encode transcriptional regulators, this coordination offers a means to effectively fine-tune the protein levels in a needed basis. Broadly, CDS methylation forms an important layer of translational regulation by acting as a controllable switch via a specialized m6A reader YTHDC2.
Methods
Cell lines and reagents
293T and MEF cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum. Antibodies used in this study are listed as follows: anti-YTHDF1 (Abcam ab99080, 1:1,000 WB), anti-YTHDF2 (Proteintech 24744-1-AP, 1:1000 WB), anti-YTHDF3 (Santa Cruz sc-377119, 1:1000 WB), anti-YTHDC2 (Abcam ab176846, 1:1000 WB), anti-METTL3 (Abnova H00056339-B01P, 1:1000 WB), anti-METTL14 (Sigma HPA038002, 1:1000 WB), anti-FTO (Phosphosolutions 597-FTO, 1:1000 WB), anti-ALKBH5 ((Proteintech 16837-1-AP, 1:1000 WB), anti-puromycin (Developmental Studies Hybridoma Bank-PMY-2A4, 1:100 WB), anti-Flag (M2) antibody (F1804), anti-m6A (Millipore ABE572) and anti-β-actin (Sigma-A5441, 1:2,000 WB). [35S]-methionine was purchased from PerkinElmer (#NEG772007MC).
Plasmids
Human YTHDC2 was cloned into pcDNA3 vector. To create the E332Q mutant, site-directed mutagenesis was performed using Q5 Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer manual. Primers: 5′-TGAAGTGCATCAAAGGGATCG-3′ and 5′-CAAGACATCTCGCAATAGG-3′. Mutation was identified by Sanger DNA sequencing.
Lentiviral shRNAs
shRNA targeting sequences based on RNAi consortium at Broad Institute (http://www.broad.mit.edu/rnai/trc) are listed as below:
YTHDC2 (human): 5′-GCCCACAGATTGGCTTATTTA-3′;
YTHDF1 (human): 5′-CCGCGTCTAGTTGTTCATGAA-3′;
YTHDF2 (human): 5′-CCACAGGCAAGGCCCAATAAT-3′;
YTHDF3 (human): 5′-GATAAGTGGAAGGGCAAATTT-3′;
METTL3 (mouse): 5′-GCTACAGGATGACGGCTTTCT-3′;
METTL14 (mouse): 5′-GGATCAAAGGAA CCGTGAAGC-3′;
FTO (mouse): 5′-GCTGAGGCAGTTCTGGTTTCA-3′;
ALKBH5 (mouse): 5′-GCCTCAGGACATT AAGGAACG-3′;
Scramble control sequence: 5′-AACAGTCGCGTTTGCGACTGG-3′.
shRNA targeting sequences were cloned into DECIPHER™ pRSI9-U6-(sh)-UbiC-TagRFP-2A-Puro (Cellecta, CA). Lentiviral particles were packaged using Lenti-X 293T cells (Clontech). Virus-containing supernatants were collected at 48 h after transfection and filtered to eliminate cells. Cells were infected by the lentivirus for 48 h before selection with 1–2 μg ml−1 puromycin.
Immunoblotting
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (50 mM Tris (pH 6.8), 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol), lysates were heated at 95 °C for 5 min, and then centrifuged at 16,000g for 5 min at 4 °C, supernatants were collected and subjected to immunoblotting. Proteins were separated on a SDS-PAGE and transferred to Immobilon-P membranes (Millipore). Membranes were blocked for 1 h in TBS containing 5% nonfat milk and 0.1 % Tween-20, followed by incubation with primary antibodies overnight at 4 °C. After incubation with horseradish peroxidase-coupled secondary antibodies at room temperature for 1 h, immunoblots were visualized using enhanced chemiluminescence (ECLPlus, GE Healthcare).
Cell proliferation assay
A total of 2000 cells (293T cells with sh-Scramble, sh-YTHDC2, sh-YTHDF1, sh-YTHDF2, sh-YTHDF3, and sh-YTHDC1) per well were seeded into 96-well plates, followed by cell culture for 96 h. The cell viability was detected by adding 10 μl of Cell Counting Kit-8 solution (Dojindo) to each well and the absorbance was detected by TECAN Spak10TM at the wavelength of 450 nm after incubation at 37 °C for 1–2 h.
Puromycin labeling
Cells at 80–90% confluence were changed with fresh medium 2 h before harvesting, and then were treated with 10 µg ml−1 puromycin for 10 min. After washing twice with cold Dulbecco's PBS (DPBS), cells were lysed with SDS-PAGE sample buffer, and proteins were separated on SDS-PAGE and transferred to Immobilon-P membranes. Membranes were blocked for 1 h in TBS containing 5% nonfat milk and 0.1% Tween-20, followed by incubation with puromycin antibodies (1:100 dilution) overnight at 4 °C. After incubation with horseradish peroxidase-conjugated anti-mouse IgG (1:10000 dilution) for 1 h at room temperature, the membrane was visualized using enhanced chemiluminescence.
[35S] Radiolabeling
MEF cells were briefly incubated in methionine- and cysteine-free media before addition of 50 mCi of [35S]-methionine. Labeling was stopped by ice-cold DMEM containing 100 mM of cycloheximide. Cells were washed with PBS containing 100 mM of cycloheximide, lysed with polysome lysis buffer. For the quantitation of [35S]-Met-labeled proteins, cell lysates were resolved on a 10% Tris-Glycine SDS-PAGE and radiography captured by Typhoon 9400. Quantification of [35S] methionine incorporation was done using ImageJ software.
Dual-luciferase reporter assay
The partial DNA sequence of Malat1 encoding the typical RNA secondary structure (AACUUAAUGUUUUUGCAUUGGACUUUGAGUU) along with the coding region of Renilla luciferase were cloned into pcDNA-Fluc vector using XbaI and AgeI sites. The stop codon of Fluc and the start codon of Rluc no longer exist. The resulted fusion protein contains an extra 20 amino acids between Fluc and Rluc. The cloning primers were listed as follows: Forward, 5′-GCTCTAGAGTAATTACCAACTTAATGTCCTTGCATTGGACTTTGAGTTATGATTATTTTTTAACTTCGAAAGTT-3′; Reverse, 5′-GCACCGGTTTATTGTTCATTTTTG-3′. The mutant plasmids (A → G and UU → CC) for the pcDNA-Fluc-Malat1SS-Rluc reporter were generated using the Q5® Site-Directed Mutagenesis Kit (New England Biolabs) and primers listed below:
Q5-Mat1SS-(A-G)-F, TTTGCATTGGGCTTTGAGTTATG;
Q5-Mat1SS-(A-G)-R, AACATTAAGTTGGTAATTACTC;
Q5-Mat1SS-(CC-TT)-F, AACTTAATGTTTTTGCATTGGACTTTG;
Q5-Mat1SS-(CC-TT)-R, GGTAATTACTCTAGACACG.
pcDNA-Fluc-Malat1SS-Rluc and its mutant reporters were transfected into control or indicated knockdown MEF cells. After 24 h, Firefly and Renilla luciferase activities in individual cell lysate were measured using Dual-Luciferase® Reporter Assay System (Promega). At least three biological replicates were performed for each cell line and reporter.
SELECT for detection of m6A
Five microgram total RNA was incubated with 40 nM Up Primer, 40 nM Down Primer and 5 μM dNTP in 17 μl 1 × CutSmart buffer (50 mM KAc, 20 mM Tris-HAc, 10 mM MgAc2, 100 μg/ml BSA) and annealed in the programs below: 90 °C (1 min), 80 °C (1 min), 70 °C (1 min), 60 °C (1 min), 50 °C (1 min), and 40 °C (6 min). Subsequently, the 17 μl annealing products were incubated with a 3 μl of enzyme mixture containing 0.01 U Bst 2.0 DNA polymerase, 0.5 U SplintR ligase and 10 nmol ATP. The final 20 μl reaction mixture was incubated at 40 °C for 20 min, denatured at 80 °C for 20 min and kept at 4 °C. Quantitative PCR was run at the following condition: 95 °C, 5 min; (95 °C, 10 s; 60 °C, 45 s) × 40 cycles. The SELECT products of indicated site were normalized to the RNA abundance of indicated transcript bearing this site.
Ribosome profiling
Cells at 80–90% confluence were changed with fresh medium to remove the dead cells 3–4 h before harvesting. Four 10 cm dishes of cells were harvested in 450 µl lysis buffer (10 mM HEPES, pH 7.4, 100 mM KCl, 5 mM MgCl2, 1% Triton X-100) containing CHX (100 µg ml−1), then centrifuged at 12,000g, 4 °C for 10 min. The supernatant was collected and subjected to sucrose gradient sedimentation. Sucrose solutions were prepared in polysome buffer (10 mM HEPES, pH 7.4, 100 mM KCl, 5 mM MgCl2). 15–45% (w/v) sucrose density gradients were freshly made in SW41 ultracentrifuge tubes (Backman) using Gradient Master (BioComp Instruments). Totally, 500 µl of the cell lysates was loaded onto sucrose gradients followed by centrifugation for 2.5 h at 32,000 rpm, 4 °C in a SW41 rotor. Separated samples were fractionated at 1.5 ml min−1 through an automated fractionation system (Isco) that continually monitors OD254 values.
RNA-seq and m6A-seq
For RNA-seq, 12 µg Trizol extracted total RNAs were fragmentated by using freshly prepared RNA fragmentation buffer (10 mM Tris-HCl, pH 7.0, 10 mM ZnCl2) and heating at 94 °C for 5 min, followed by adding 1 µl 0.5 M EDTA to terminate. For m6A immunoprecipitation, 1 mg fragmented RNA was incubated with 15 μg anti-m6A antibody in 1 × IP buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Igepal CA-630) for 2 h at 4 °C. The m6A-IP mixture was then incubated with Protein A beads for additional 2 h at 4 °C on a rotating wheel. After washing 3 times with IP buffer, bound RNA was eluted using 100 μl elution buffer (6.7 mM N6-Methyladenosine 5′-monophosphate sodium salt in 1 × IP buffer), followed by ethanol precipitation. Precipitated RNA was used for cDNA library construction and high-throughput sequencing described below.
cDNA library construction
For Ribo-seq, E. coli RNase I (Ambion) was added into the pooled fractions from ribosome profiling (100 U per 100 µl) and incubated at 4 °C for 1 h to convert the polysome into monosome. Total RNAs were extracted using Trizol LS reagent (Invitrogen).
RNase I digested RNA extracts (Ribo-seq) and fragmented RNAs (RNA-seq and m6A-seq) were dephosphorylated for 1 h at 37 °C in a 15 µl reaction (1 × T4 polynucleotide kinase buffer, 10 U SUPERase_In and 20 U T4 polynucleotide kinase). The products were separated on a 15% polyacrylamide TBE-urea gel (Invitrogen) and visualized using SYBR Gold (Invitrogen). Selected regions in the gel corresponding to 40–60 nt (for RNA-seq) or 25–35 nt (for Ribo-seq) were excised. RNA fragments were dissolved by soaking overnight in 400 μl RNA elution buffer (300 mM NaOAc, pH 5.5, 1 mM EDTA, 0.1 U ml−1 SUPERase_In). The gel debris was removed using a Spin-X column (Corning), followed by ethanol precipitation. Purified RNA fragments were resuspended in nuclease-free water.
Totally, 0.15 μg linker (rApp/NNNNCTGTAGGCACCATCAAT/3ddC) was added to the RNA fragments, heated at 70 °C for 90 s and then cooled to room temperature, followed by ligation for 3 h at 22 °C in a 20 µl reaction (1 × T4 Rnl2 reaction buffer, 10 U SUPERase_In, 15% PEG8000 and 20 U T4 RNA ligase 2 truncated). The reaction was heat inactivated at 80 °C for 10 min and the products were separated on a 10% polyacrylamide TBE-urea gel and selected regions in the gel corresponding to 65–85 nt (for RNA-seq) or 50–70 nt (for Ribo-seq) were excised. RNA fragments were dissolved by soaking overnight in 400 μl RNA elution buffer, purified RNA fragments were re-suspended in nuclease-free water.
For reverse transcription, the following oligos containing barcodes were used:
(Phos)CTANNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC(SpC18)CACTCA(SpC18)TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG
(Phos)AGCNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC(SpC18)CACTCA(SpC18)TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG
(Phos)ATTNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC(SpC18)CACTCA(SpC18)TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG
(Phos)CCGNNNAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC(SpC18)CACTCA(SpC18)TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG
where Phos represents phosphorylation, NNN represents random sequence, SpC18 represents Hexa-ethyleneglycol spacer.
The linker ligated RNA sample was mixed with 0.5 mM dNTP and 2.5 mM synthesized primer and incubated at 75 °C for 5 min, followed by incubation on ice for 3 min. The reaction mix was then added with 20 mM Tris (pH 8.4), 50 mM KCl, 5 mM MgCl2, 10 mM DTT, 40 U RNaseOUT and 200 U SuperScript III. Reverse transcription reaction was performed according to the manufacturer’s instruction. Reverse transcription products were separated on a 10% polyacrylamide TBE-urea gel. The extended first-strand product band was expected to be approximately 200 nt, and the corresponding region was excised. The cDNA was recovered by using DNA gel elution buffer (300 mM NaCl, 1 mM EDTA). First-strand cDNA was circularized in 20 μl of reaction containing 1 × CircLigase buffer, 2.5 mM MnCl2, 1 M Betaine and 100 U CircLigase II (Epicenter). Circularization was performed at 60 °C for 1 h, and the reaction was heat inactivated at 80 °C for 10 min, then was precipitated by ethanol.
Deep sequencing
Circular template was amplified by PCR by using the Phusion high-fidelity (HF) enzyme (NEB) according to the manufacturer’s instructions. The PCR forward primer: 5′-AATGATACGGCGACCACCGAGATCTACAC-3′ and reverse primer: 5′-CAAGCAGAAGACGGCATACGAGATGTGACTGGAGTTCAGACGTGTGCT CTTCCG-3′
were used to create DNA suitable for sequencing. The PCR contains 1 × HF buffer, 0.2 mM dNTP, 0.5 µM oligonucleotide primers, and 0.5 U Phusion polymerase. PCR was carried out with an initial 30 s denaturation at 98 °C, followed by 12 cycles of 10 s denaturation at 98 °C, 20 s annealing at 65 °C, and 20 s extension at 72 °C. PCR products were separated on a nondenaturing 8% polyacrylamide TBE gel. Expected DNA at 180 bp was excised and recovered. After quantification by Agilent BioAnalyzer DNA 1000 assay, equal amounts of barcoded samples were pooled into one sample. Approximately, 5 pM mixed DNA samples were used for cluster generation followed by sequencing by using sequencing primer 5′-CGACAGGTTCAGAGTTCTACAGTCCGACGATC-3′ (Illumina HiSeq).
Alignment of sequencing reads
The 3′ adapters and low quality bases were trimmed by Cutadapt39. The trimmed reads with length <15 nucleotides were excluded. The remaining reads were mapped to the mouse transcriptome using Bowtie40 with parameters: -a --best -m1 --strata. To construct the transcriptome, the annotation file from ENSEMBL database (GRCm38) was used. For each gene, the mRNA with longest CDS was selected. In the case of equal CDS length, the longest transcript was used. For read alignment, a maximum of two mismatches were permitted. To avoid ambiguous, the reads that were mapped to multiple positions were excluded.
Prediction of m6A peak
We used a similar method reported previously to identify m6A peaks in the immunoprecipitation sample as compared to the input sample. In brief, a sliding window of 50 nucleotides with a step of 25 nucleotides was employed to scan each transcript. For each window with maximum read coverage higher than 10, a peak-over-median score (POM) was derived by calculating the ratio of the mean read coverage in the window to the median read coverage of corresponding transcript. The windows with POM higher than three in IP sample were obtained. The same processes were performed in input sample. The windows found in input sample were eliminated from following analyses. The windows that overlapped at least single nucleotide were merged into one cluster. Finally, a peak over input (POI) score was assigned to each cluster by calculating the ratio of POM in the IP sample to that in the input sample. The cluster with POI score higher than three were retrieved, and defined as m6A-enriched cluster. The peak position with maximum coverage in each m6A-enriched cluster was defined as the position of m6A peak. The adenosine site of the nearest RRAC motif was defined as m6A residue. To reduce noises for background reads and possible bias from peak calling method, only the m6A peaks that were found in all biological replicates were used. m6A peaks in METTL3 and WTAP knockdown samples were predicted using the same method. The m6A peaks with a decrease of POI score up to 50% after METTL3 or WTAP knockdown were defined as WTAP or METTL3 sensitive m6A peaks. All m6A sites were classified into different mRNA segments: 5′ UTR, CDS and 3′ UTR, according to the positions of m6A. In current study, we mainly focused on CDS m6A, therefore m6A peaks around the start codon (−15 nt, +100 nt) and the stop codon (−100 nt, +15 nt) were not included in analyses.
Single nucleotide m6A
We used single nucleotide m6A to investigate the effect of m6A on ribosome movement. The data were obtained from the supplementary file of the study of Linder et al22.
m6A-positive mRNAs and negative controls
m6A-positive mRNAs refer to the mRNAs that contain at least one m6A peak. Nonmethylated mRNAs were defined as the mRNAs that none of the conserved methylation site was found. We found CDS m6A tends to be enriched in the transcripts with inactive translation, therefore, to exclude possible biases caused by the difference in basal translational levels, we did not use a full set of non-methylated mRNAs as negative control. Instead, for each m6A-positive mRNA, k-nearest neighbors algorithm was used to define a negative control. In brief, we used three non-methylated mRNAs (K = 3), which show a similar ribosome density (±10%) to m6A positive mRNA in wild type, as negative control. The mean value of nonmethylated mRNAs was used.
When analyzing sequence context and ribosome densities around m6A, the nonmethylated adenosine sites of RRAC motifs in the same transcript were defined as negative control. The methylated regions refer to the interval of −400 to +100 nucleotides relative to the methylated codons.
Ribosome density of transcript
For each transcript, RPKM was used to estimate the ribosome density of transcript. To exclude the effect of RNA level, ribosome density was normalized by corresponding RNA level. mRNAs with RPKM < 1 were excluded.
Ribosome densities around m6A residues
To investigate the effect of m6A on ribosome movement, ribosome densities along mRNA regions in the interval of −60 to +60 nucleotides relative to methylated codons were calculated. First, for each methylated codon, footprint reads at each position in the interval of −60 to +60 nucleotides were normalized by the total reads of such region. The regions with total reads <20 were excluded. Second, mean ribosome densities were obtained by averaging the normalized footprint reads at the same positions.
Identification of regions with ribosomal pausing
To identify structure related ribosomal pausing, we firstly used a sliding window with 30 in length and a step of 3 nt to scan the transcript. Ribosome densities, normalized by the averaged density of CDS, between scramble cells and knockdown cells in the same window were compared. The regions with fold change of knockdown over scramble higher than three were defined as pausing regions. By contrast, the regions with fold change < 1/3 were used as negative controls.
mRNA secondary structure analysis
For each transcript, a sliding window of 30 nucleotides with a step of 3 nucleotides was used to calculate RNA minimum fold free energy (MFE) along transcript. For each window, MFE was calculated by ViennaRNA41, using default parameters. To investigate whether an evolutionally conserved structure is formed in the flanking regions of m6A methylation sites, 30 random sequences were generated by shuffling nucleotides of native sliding sequence while controlling for the dinucleotides content42. A z-score value for each native sequence was calculated as reported before43. For aggregation plot, a mean MFE in each window was calculated by averaging MFE values of the windows in the same position. icSHAPE27 and PARS44, both can capture RNA secondary structure at a transcriptome-wide level, were used to estimate the effect of m6A on mRNA folding.
m6A conservation analysis
For m6A between HEK293 and MEF, we retrieved orthologous sequences from ENSEMBL, only one-to-one orthologs were used. The orthologs were then aligned by Muscle45 using default parameters. The positions of m6A peaks were adjusted according to the positions of insertions and deletions between orthologs. The adjusted m6A peaks that were found at the same position of the aligns were defined as conserved m6A. m6A peaks only found in human or mouse were defined non-conserved m6A.
Analysis of PAR-CLIP
After filtering out the low-quality reads, the remaining PAR-CLIP reads were aligned to human genome using bowtie, with parameters: −v 2 −m 10 --best --strata. PAR-CLIP peaks were predicted by PARalyzer using default parameters. The genome coordinates of PAR-CLIP peaks were mapped to coordinates of the longest transcript using custom Perl script.
Motif analysis
Motif analyses were performed by MEME46.
Gene ontology (GO) analysis
GO analyses were performed by DAVID GO.
tAI calculation
The tRNA adaptation index (tAI) was calculated using the method reported before47. tRNA copy number used for tAI calculation was downloaded from the GtRNAdb database.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We’d like to thank Qian lab members for critical reading and comments. We are grateful to Cornell University Life Sciences Core Laboratory Center for sequencing support. This work was supported in part by US National Institutes of Health (R21CA227917, R01GM1222814) and HHMI Faculty Scholar (55108556) to S.-B.Q.
Source Data
Author contributions
Y.M. and S.-B.Q. conceived the project, designed the experiments, and wrote the paper. Y.M. conducted all the data analysis. L.D. performed the shRNA knockdown experiments and functional characterization. X.-M.L. conducted structural reporter assay. J.G. constructed Flag-tagged YTHDC2 cell lines. H.M. and B.S. contributed to PAR-CLIP of YTHDC2. All authors discussed the results and edited the paper.
Data availability
All new sequencing data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE129194. All other published sequencing data have been cited in main text, the GEO Series accession numbers of published sequencing data are listed in Supplementary Data 1. All other relevant data are available from the corresponding author on request. The source data underlying Fig. 4a and Supplementary Fig. 9a are provided as Source Data files.
Code availability
All the procedures but sequencing mapping were completed using custom Perl scripts, which are available upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-13317-9.
References
- 1.Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015;16:651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 3.Leppek K, Das R, Barna M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–1856. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 12.Edupuganti RR, et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017;24:870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 14.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer KD, et al. 5’ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J, et al. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 2016;23:110–115. doi: 10.1038/nsmb.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz S, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geula S, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 22.Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreev DE, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coots, R. A., et al. m6A facilitates eIF4F-independent mRNA translation. Mol. Cell68, 504–514.e7 (2017). [DOI] [PMC free article] [PubMed]
- 25.Liu N, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kierzek E, Kierzek R. The synthesis of oligoribonucleotides containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines via post-synthetic modification of precursor oligomers. Nucleic Acids Res. 2003;31:4461–4471. doi: 10.1093/nar/gkg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Y, et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N6 -methyladenosine modification. Angew. Chem. Int. Ed. Engl. 2018;57:15995–16000. doi: 10.1002/anie.201807942. [DOI] [PubMed] [Google Scholar]
- 29.Qu X, et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustoe AM, et al. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell. 2018;173:181–195. e118. doi: 10.1016/j.cell.2018.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain D, et al. ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife. 2018;7:e30919. doi: 10.7554/eLife.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojtas MN, et al. Regulation of m6A transcripts by the 3ʹ→ 5ʹ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell. 2017;68:374–387. e312. doi: 10.1016/j.molcel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 34.Patil DP, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretschmer J, et al. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. RNA. 2018;24:1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu PJ, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey, A. S., et al. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife6, e26116 (2017). [DOI] [PMC free article] [PubMed]
- 38.Tanabe A, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016;376:34–42. doi: 10.1016/j.canlet.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 39.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Erickson BW. Significance of nucleotide sequence alignments: a method for random sequence permutation that preserves dinucleotide and codon usage. Mol. Biol. Evol. 1985;2:526–538. doi: 10.1093/oxfordjournals.molbev.a040370. [DOI] [PubMed] [Google Scholar]
- 43.Clote P, Ferre F, Kranakis E, Krizanc D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominissini D, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey, T. L., et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res.37, W202–W208 (2009). [DOI] [PMC free article] [PubMed]
- 47.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All new sequencing data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession number GSE129194. All other published sequencing data have been cited in main text, the GEO Series accession numbers of published sequencing data are listed in Supplementary Data 1. All other relevant data are available from the corresponding author on request. The source data underlying Fig. 4a and Supplementary Fig. 9a are provided as Source Data files.
All the procedures but sequencing mapping were completed using custom Perl scripts, which are available upon request.