Abstract
The gut microbiota, including pathogenic microorganisms and probiotics, has been involved in tumor initiation and progression by regulating the components of intestinal flora. Canmei formula (CMF), a traditional Chinese medicine, chronicled in the Chuang Yang Jing Yan Quan Shu, has been clinically used as an adjuvant therapy to treat patients with colorectal carcinoma (CRC) in China. In this study, we investigate the treatment effect of CMF in the azoxymethane (AOM) and dextran sodium sulfate (DSS) induced and high-fat diet augmented colitis-associated colorectal cancer in vivo, and explore its mechanism of action. We found that CMF treatment relieved the inflammation and alteration of the gut microbiota and significantly inhibited the development of intestinal adenoma. Linear discriminant analysis showed that the flora diversity in the normal mice, model mice and CMF treatment mice was different. At the family level, the relative abundance of Desulfovibrionaceae decreased in CMF groups. The relative abundance of Desulfovibrionaceae were lower in the CMF groups than in model group, whereas Rikenellaceae and Alistipes were increased. Altogether our results indicate that CMF treatment ameliorate colitis-associated colorectal carcinogenesis by modulating the composition of the gut microbiota in vivo.
Keywords: Canmei formula (CMF), traditional Chinese medicine, gut microbiota, AOM/DSS, colorectal carcinogenesis
Introduction
Colorectal carcinoma (CRC) is the second leading cause of cancer-related death worldwide (1). There are about 1.4 million new CRC patients and 700,000 deaths each year (2). Colorectal adenoma (CRA) refers to any lesion that originates from the surface of colorectal mucosa and protrudes into the intestinal cavity. It is a precancerous lesion of CRC and has the characteristics of high recurrence and high incidence of cancerous change (3). Microscopic removal is currently the main treatment for CRA, which can effectively reduce the incidence of CRC and reduce mortality by 67% (4). However, microscopic resection did not reduce the recurrence rate of CRA. Aspirin, celecoxib, and cyclooxygenase-2 inhibitors are used clinically to treat inflammatory bowel disease, but the efficacy is limited according to the large sample clinical data (5). To date, there is no therapeutic agent specific for this disease. It is critical to explore drugs that can safely and effectively prevent and treat the recurrence of CRA.
In China, traditional Chinese medicine (TCM) has a long history dating back several thousands of years ago. Results from both pre-clinical laboratory and human studies have indicated that TCM has been widely used in anticancer therapy. Previous studies have indicated that PHY906, a modified formulation derived from Huang-Qin-Tang, could ameliorate chemotherapy-induced GI tract toxicity (6, 7). Qingjie Fuzheng granules (QFGs) has been proven to be able to inhibit proliferation and induce apoptosis of CRC cells (8). These studies indicate that TCM play important roles in cancer treatment and could be expected to become a promising therapeutic candidate for tumor.
Over the past decade, CRC have emerged as one of the most studied human conditions link to gut microbiota. Studies indicate that changes in the composition of gut microbiota, one of the most important internal environmental factors, are closely associated with the progression of CRC (9, 10). At present, the study on the secretion of toxin-activated inflammation by the intestinal flora is the most in-depth study of CRA carcinogenesis (3). The changes of intestinal flora are positively correlated with colorectal cancer. The secretion of toxins by the bacteria is a key factor for activating inflammation and oxidative stress pathways (11, 12). A number of studies have confirmed that Enterococcus faecalis, Escherichia coli, Enterotoxin-producing Bacteroides fragilis (ETBF), Streptococcus bovis, Fusobacterium nucleatum, and Helicobacter pylori are the main species that can induce cancer (3, 12, 13). Among them, Fusarium nucleatum induces uncontrolled intestinal epithelial cell growth by activating β-catenin; E. coli produces colibactin with genotoxicity (3).
Canmei formula (CMF), a classical traditional Chinese herbal formulation with Titanium and Marci Hieronymi, is widely used to treat colorectal diseases. The active constituents in CMF, including Citric acid, DL-TYROSINE, L(-)-Carnitine, L-Tyrosine, Ambrosic acid and others, have been reported (14, 15). However, there is no study on the effect of CMF in modulating gut microbiota. In our previous studies, 16sDNA sequencing technique was used to compare the change of gut microbiota induced by CMF treatment in AOM/DSS and high-fat diet-induced CRC mice (16, 17). This study provides evidence of the modulations of gut microbiota through CMF treatment, which will help understanding the host-microbe interactions during the treatment of CRC.
Materials and Methods
Preparation of the Extracts for CMF
Mume Sieb and Marci Hieronymi were purchased from Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine. Mume Sieb was the fruit of Prunus mume. The Marci Hieronymi is a dry body of the larvae of the camphor family, Bombyx mori linnaeus, 4 ~ 5 years old, infected with Beauveria bassiana. They were formally identified by the department of pharmacy, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China.
CMF was formulated with Titanium and Marci Hieronymi in a ratio of 1:1. All the herbs were purchased from Yueyang hospital herbal pharmacy. Formulation was performed as described below. Briefly, 980 g mixture was extracted twice for 1.5 and 1 h for each time. The filtrates were concentrated in a vacuum at 60°C and dried to obtain the CMF extract powder of 335.2 g, the rate of the extract was 32.1%.The extract obtained in this part is CMF-L/H. The same process was repeated twice for the CMF extract. Then the liquid was poured through the column of the macroporous resin, and then the resin column was rinsed with pure water, 20% ethanol and 90% ethanol. After alcohol precipitation, the filtrateswere concentrated in a vacuum at 60°C and dried to obtain the CMF extract powder of 60.3 g, the rate of the purified product was 18%. The extract obtained in this part is used as CMF-A. The quality of preparations was controlled according to the guidelines from Chinese food and drug administration (CFDA).
Liquid Chromatograph and Mass Spectrometry
The solution obtained by the above preparation method was stored at 4°C. For HPLC analysis, the solution was centrifuged at 12,000 rpm for 10 min, then the supernatant was filtered through a 0.22 μm membrane before injection into the HPLC system. Analysis were performed by using Dionex UltiMate 3000 HPLC system (ThermoFisherScientific, USA) with a diode array detector. The C18 column (150 × 2.1 mm, 2.5 μm) was used with a flow rate of 0.3 ml·min−1. The injection volume was 5 μl, and the column temperature was maintained at 40°C. The mobile phase was composed of (A) aqueous formic acid (0.1%, v/v) and (B) acetonitrile (0.1%, v/v) under following gradient elution: 95% A from 0 to 0.5 min, 95-2% A from 0.5 to 15 min, 2% A from 15 to 17 min, 2-95% A from 17 to 17.5 min, 95% A from 17.5 to 25 min. Mass spectrometry was performed on a Q Exactive high-resolution benchtop quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific, USA) using a heated electrospray ionization (HESI) source for ionization of the target compounds in positive and negative ion modes. The key parameters were as follows: ionization voltage, +3.5 kV/−3.2 Kv; sheath gas pressure, 40 arbitrary units; auxiliary gas, 10 arbitrary units; heat temperature, 350°C. For the compounds of interest, a scan range of m/z 120–1,500 was chosen. Resolution for higher energy collisional dissociation cell (HCD) spectra was set to 17,500 at m/z 120 on the QExactive.
Animal Experiments and Grouping
The animal procedures and care in this study was approved by the IRB of Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine. Seven-week-old female C57/BL6 mice were purchased from Shanghai Jihui Experimental Animal Breeding Co., Ltd. After arrival, the animals were quarantined for the first 7 days. Under controlled conditions: humidity 50 + 10%, 12/12 h light/dark cycle, and temperature ~23°C, all animals were housed in plastic cages (6 mice/cage) and provided with free drinking water and basic diet. AOM, the colonic carcinogen, was purchased from Sigma-Aldrich, USA. DSS with a molecular weight of 36,000–50,000 (No.216011080) was purchased from MPBIO Company, USA. For the induction of tumorigenesis, mice were injected with AOM (12 mg/kg body weight) intraperitoneally. Starting 5 day after the injection, animals received 2.5% DSS in the drinking water for 5 days. At the same time, the mice were given a high-fat diet. Then mice were given normal drinking water and normal diet for 2 weeks and subjected to two more DSS treatment cycles. Subsequently, mice were randomized into experimental and control groups. Group 1, NC, n = 8. Experimental groups included 5 groups: group 2: treatment with AOM and DSS (MC, n = 8); group 3: treatment with AOM, DSS, and Aspirin (Aspirin, n = 8); group 4: treatment with AOM, DSS, and CMF ethanol extract (CMF-A, n = 8); group 5: treatment with AOM, DSS, and CMF aqueous extract (CMF-L, n = 8); group 6: treatment with AOM, DSS, and CMF water extract high dose group(CMF-H, n = 8). Aspirin was given every day at the dose of 1.4 mg/kg/d. CMF-L and CMF-H were given at the doses of 3.65 and 7.3 g/kg/d, respectively. CMF-A was given at the doses of 0.657 g/kg/d. In the clinical practice of Chinese herbal medicine, the prescription dose of CMF is usually 60 g of herbal materials per day. When this human dose was converted into an animal dose (a person of 60 kg, and a conversion factor of 9.1 between human and mouse), it was equivalent to the low dose (3.65 g/kg/d) used in this study. All animals were sacrificed at the 18th week after the corresponding drug was administrated.
Colon Tissue Collection and Processing
After mice were sacrificed, the colorectal adenoma tissues were rapidly isolated, freed of adherent tissues and rinsed with phosphate buffer saline. Subsequently, tumor biopsy specimens of 1 cm-length were fixed with 4% paraformaldehyde for 24 h, embedded in paraffin and sectioned into 4-μm-thick slices. Slices were hydrated and stained with hematoxylin and eosin (H&E) followed by microscopic examination. Then portions of adenoma tissues were homogenized (10% W/V) in (20 mM Tris-HCI containing 1 mM EDTA, PH 7.4) by a glass homogenizer. The homogenates were centrifuged at 3,000 × g for 20 min at 4°C and supernatants were stored at −80°C.
Real Time Quantitative Reverse Transcription PCR
According to the modified methods as described, the RNA extracts of the colorectal tissues were prepared (8). By normalizing target mRNA Ct values to those for GAPDH (Ct), the relative amount of target mRNA was determined by the comparative threshold (Ct) method. The primer sequences used were as follows:
MUS-NF-κB-F (bp1297): (ATGCACCGTAACAGCAGGAC);
MUS-NF-κB-R (bp1405C): (TGTCATCCGTGCTTCCAGTG);
MUS-IL-17C-F (bp269) (AGGAGGTGCTGGAAGCTGAC);
MUS-IL-17C-R (bp391C) (TGCATCCACGACACAAGCAT);
GAPDH-F (5′-GTGAGGCCGGTGCTGAGTAT-3′);
GAPDH-R (5′-GTGCAGGATGCATTGCTGAC-3′);
ELISA Assay
Concentration of IL-17C in serum and NF-κB in supernatants of colonic tissues were quantified by IL-17C assay kit and NF-κB assay kit (Shanghai Pusheng Biological Technology Co., Ltd. China) according to manufacturer's instruction.
Fecal DNA Extraction and Illumina Miseq Sequencing
Genomic DNA was extracted from every stool sample using the FastDNA Spin Kit for Soil (MP Biomedical, LLC, catalog 116560-200) following the manufacturer's instructions.V3 and V4 region of the 16s rDNA sequencing was amplified by Primer F (5′-AACGGGAAGACAACGTACGG-3′) and Primer R (5′-CAGATGCAGGAGGACATGTC-3′) with barcode sequence. Library was constructed following the manufacturer's instructions of the Ion Plus Fragment Library Kit and sequenced by Ion S5 Sequencer.
Bioinformatics Analysis
Reads was filtered and chimera sequence was removed.
The read pairs were de-multiplexed based on their unique molecular barcodes and overlapping reads were merged using USEARCH v7.0.1001 software. Operational taxonomic unit (OTU) picking was conducted using the QIIME (Quantitative Insights Into Microbial Ecology) software package. By using UCLUST, 16S rRNA gene sequences were clustered at a similarity cutoff value of 97%. By the SILVA reference database (https://www.arb-silva.de/download/), matching of OTUs to bacteria was conducted. By mapping the de-multiplexed reads to the identified OTUs, abundances were recovered. Alpha andbeta diversity plots were also generated using QIIME. The non-weighted UniFrac approach was used to measure the beta diversity between five bacterial cave communities. The relative abundance of transporter genes was predicted and performed by picrust analysis. Independent of the taxonomic analysis, aclosed-reference OTU picking protocol (QIIME) and the Greengenes database (http://greengenes.lbl.gov) were used to pre-cluster at 97% identity, and 97% of the OTUs were picked. The obtained OTU table was normalized by 16S rRNA copy number, and the functional genes were predicted according to the Kyoto Encyclopedia ofGenes and Genomes (KEGG) catalog. By using SPSS version 15.0 statistical software (IBM, Armonk, NY, USA), Fisher's test and the Mann-Whitney U testwere used to analyze, respectively, the gender distribution of subjects and intergroup differences at the genus level in each subcluster. The linear discriminant analysis (LDA) effectsize (LEfSe) method with default settings was used to analyze the differences among groups in phyla, class, order, family, and genera levels.
Statistical Analysis
SPSS software version 15.0 was used for statistical analysis. The data were presented as mean ± SD. Unpaired students' t-test was used to compare the means of the two groups. One-way analysis of variance and Adonis were used to compare the means of more than two groups. A level of P < 0.05 was considered as statistically significant.
Results
Identification of the Main Components in the Extract of CMF
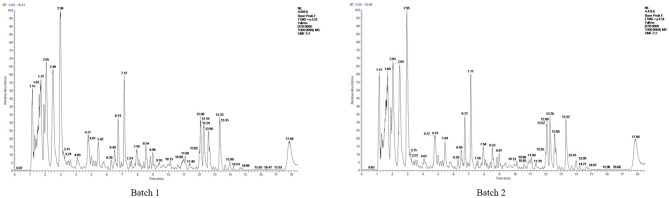
To identify the components of Mume Sieb and Marci Hieronymi extract, Q Exactive high-performance benchtop quadrupole-Orbitrap LC-MS/MS was performed. The main components in the extract of CMF were identified according to the elemental composition data determined from accurate mass measurements and comparison with the literature data. Forty-one compounds were identified in the CMF extract and demonstrated in Table 1. In addition, to exclude the possibility of manufacturing problems relating to procession, extraction, handling, and storage, two batches of CMF were evaluated by HPLC. We found that the magnitude, number, and retention time of the peaks were similar between the two batches (Figure 1). These results confirmed the stability of the CMF extraction process.
Table 1.
Major chemical constituents identified in CMF.
| No. | Name | RT (min) | Formula | Molecular weight | Area (max.) |
|---|---|---|---|---|---|
| 1 | DL-Arginine | 1.28 | C6 H14 N4 O2 | 174.1117 | 1300061.86 |
| 2 | L(-)-Carnitine | 1.37 | C7 H15 N O3 | 161.10527 | 9750518.42 |
| 3 | DL-Stachydrine | 1.40 | C7 H13 N O2 | 143.09471 | 1508022.19 |
| 4 | Adenine | 1.42 | C5 H5 N5 | 135.05457 | 12469462.57 |
| 5 | Guanine | 1.43 | C5 H5 N5 O | 151.04946 | 7435505.89 |
| 6 | Hypoxanthine | 1.57 | C5 H4 N4 O | 136.03858 | 12616671.18 |
| 7 | Nicotinic acid | 1.58 | C6 H5 N O2 | 123.03221 | 1086991.17 |
| 8 | L-Tyrosine | 1.64 | C9 H11 N O3 | 181.07401 | 6502680.25 |
| 9 | Adenosine | 1.69 | C10 H13 N5 O4 | 267.09676 | 26955456.82 |
| 10 | Adenine | 1.69 | C5 H5 N5 | 135.05457 | 3750819.71 |
| 11 | 2-Hydroxycinnamic acid | 1.72 | C9 H8 O3 | 164.04745 | 37877896.83 |
| 12 | Hypoxanthine | 1.76 | C5 H4 N4 O | 136.03858 | 2774356.88 |
| 13 | L-Norleucine | 2.07 | C6 H13 N O2 | 131.09474 | 55189905.01 |
| 14 | Alanyltyrosine | 2.81 | C12 H16 N2 O4 | 230.12691 | 433071.84 |
| 15 | Nornicotine | 2.88 | C9 H12 N2 | 148.10023 | 477190.87 |
| 16 | 2-Oxindole | 3.52 | C8 H7 N O | 151.06347 | 420066.93 |
| 17 | 2′-Deoxyadenosine | 3.54 | C10 H13 N5 O3 | 251.10184 | 148660.32 |
| 18 | Glycyl-L-leucine | 4.17 | C8 H16 N2 O3 | 188.11627 | 4092286.76 |
| 19 | Chlorogenic acid | 4.80 | C16 H18 O9 | 354.09487 | 36388021.14 |
| 20 | tert-Butyl N-[1-(aminocarbonyl)-3-methylbutyl]carbamate | 5.41 | C11 H22 N2 O3 | 230.16338 | 2831504.35 |
| 21 | D-(+)-Tryptophan | 5.45 | C11 H12 N2 O2 | 204.08996 | 21112273.16 |
| 22 | 3,4-Dihydroxybenzaldehyde | 5.90 | C7 H6 O3 | 138.03188 | 196915.08 |
| 23 | Leucylproline | 5.94 | C11 H20 N2 O3 | 228.14765 | 2951420.35 |
| 24 | 7-Hydroxycoumarine | 6.51 | C9 H6 O3 | 162.03159 | 3160911.99 |
| 25 | 6-Methylquinoline | 6.93 | C10 H9 N | 143.07372 | 1669486.73 |
| 26 | 3-Phenylpropanoic acid | 7.18 | C9 H10 O2 | 150.06808 | 50678.13 |
| 27 | 3-O-feruloyl-D-quinic acid | 7.63 | C17 H20 O9 | 368.11073 | 285397.82 |
| 28 | 4-Coumaric acid | 7.90 | C9 H8 O3 | 164.04747 | 616864.76 |
| 29 | Rutin | 7.92 | C27 H30 O16 | 610.15385 | 383860.64 |
| 30 | N-Acetyl-L-leucine | 7.95 | C8 H15 N O3 | 173.10535 | 172744.27 |
| 31 | Quercetin-3β-D-glucoside | 8.13 | C21 H20 O12 | 464.09641 | 532784.34 |
| 32 | 3,5-Dimethoxybenzoic acid | 8.22 | C9 H10 O4 | 182.05817 | 712514.01 |
| 33 | Quercetin | 8.52 | C15 H10 O7 | 302.0428 | 150731.57 |
| 34 | Kaempferol-7-O-glucoside | 8.53 | C21 H20 O11 | 448.10099 | 407020.68 |
| 35 | (3R,4S)-4,6,8-Trihydroxy-7-methoxy-3-methyl-3,4-dihydro-1H-isochromen-1-one | 8.89 | C11 H12 O6 | 222.05297 | 117315.38 |
| 36 | (2E)-3-(3,4-Dimethoxyphenyl)acrylic acid | 9.07 | C11 H12 O4 | 208.07356 | 2293865.83 |
| 37 | Ambrosic acid | 10.00 | C15 H20 O4 | 264.13609 | 678788.38 |
| 38 | Andrographolide | 10.34 | C20 H30 O5 | 350.20662 | 1261013.91 |
| 39 | Ipratropium | 11.70 | C20 H29 N O3 | 331.21202 | 224430.08 |
| 40 | Pinolenic acid | 13.35 | C18 H30 O2 | 278.22456 | 293978.53 |
| 41 | α-Eleostearic acid | 14.23 | C18 H30 O2 | 278.22457 | 1744840.46 |
Figure 1.
HPLCprofile of two batches of CMF. The chemical fingerprint of CMF batch #1 and batch #2 was measured by HPLC analysis.
CMF Attenuated AOM/DSS-Induced Colitis-Associated Tumorigenesis
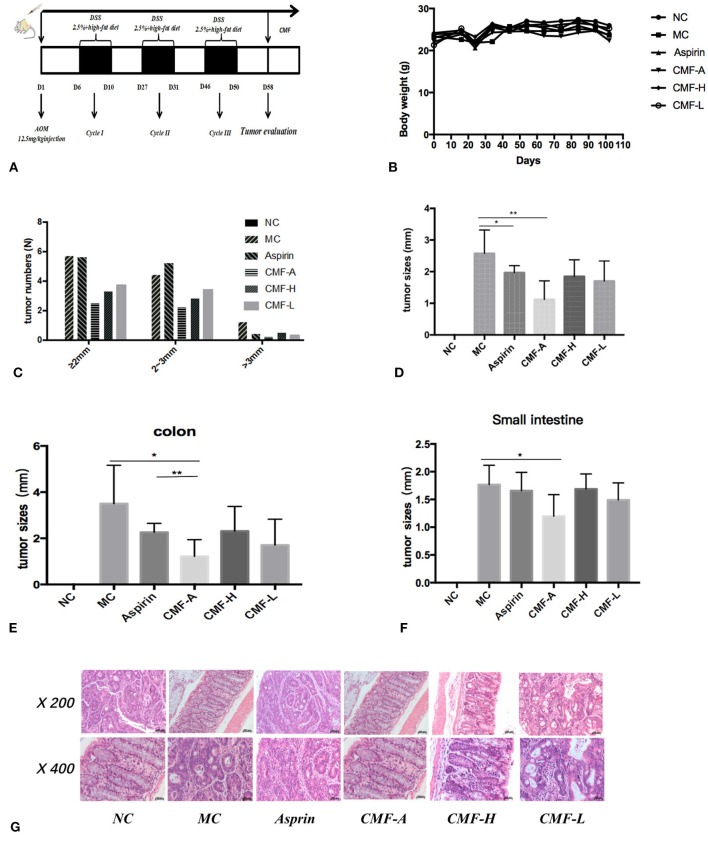
To evaluate the protective effect of CMF on the development of colitis-associated cancer, the well-established AOM/DSS and high-fat diet-induced colitis-associated colorectal tumor model in C57BL/6 mice were used (Figure 2A). Initially, the toxicity of CMF was tested in vivo. We did not find weight loss or behavioral changes in all mice. These results indicate that each of those extracts of CMF were well-tolerated (Figure 2B). AOM/DSS colorectal cancer mice were induced as described. All mice were treated with CMF once a day. After CMF treatment, tumor development and growth was inhibited in AOM/DSS colorectal cancer mice (Figures 2C–F). As a control, aspirin treatment effectively inhibited the occurrence of intestinal adenomas of 3 mm and above, but had no inhibitory effect on the occurrence of adenomas smaller than 3 mm (Figure 2C). Different extracts of CMF can significantly inhibit the growth of intestinal glands. Figures 2D–F shows that the ethanol extract of CMF significantly inhibited the occurrence of intestinal adenoma, and there is no difference in the occurrence of adenoma in the colon and small intestine. Histological examination revealed that mucosal inflammation, aberrant crypt foci adenoma, ulcer, or dysplasia in the colon tissues of the model group. Histological studies indicated that there was a large adenocarcinoma inside mucosa with several abnormal cells exhibiting cylindrical shape, large nuclei, increasing nuclear/cytoplasmic (N/C) ratio and cellular cleavagein AOM/DSS group (Figure 2G). CMF remarkably relieved the condition. These results suggest that CMF treatment inhibits inflammation-associated carcinogenesis in the AOM/DSS and high-fat diet mice.
Figure 2.
CMF treatment reduced the incidence of CRA in mice. C57BL/6 mice were subjected to an AOM-based CRC induction protocol using three cycles of 2.5% DSS in drinking water. (A) Diagram shows the experimental course of AOM/DSS mouse model. (B) Body weights of AOM/DSS group and AOM/DSS + CMF group (1, 3, 4, 5, 6). (C) Histogram showing the size distribution of tumors. (D–F) Tumor sizes in different parts determined by Spot software for microscopic tumors or a caliper for macroscopic tumors. Average tumor size ± S.D. is shown; (G) H&E stains of serial sections of colons. *P < 0.05; **P < 0.01. Data are presented as mean ± SD of mice in each group.
CMF Treatment Repressed NF-κB and IL-17C Signaling in AOM/DSS Induced Mice
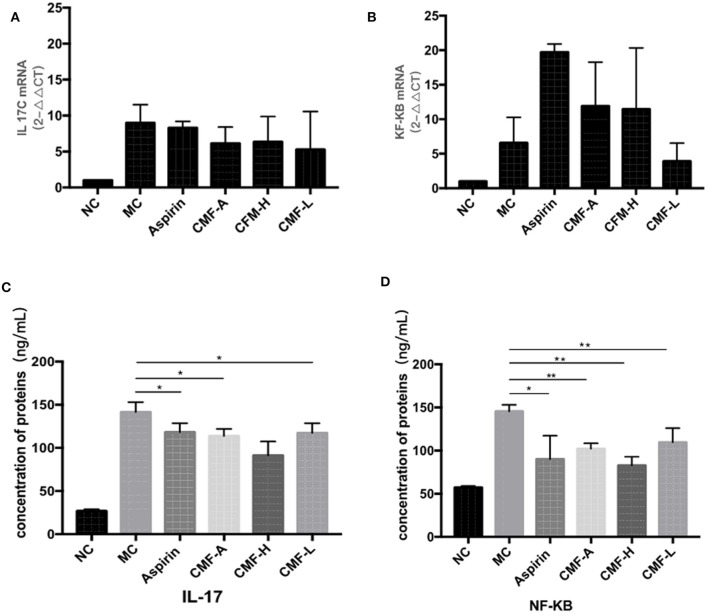
As NF-κB and IL-17C play important roles in the development of CRC, the mRNA expression of IL-17C and NF-κB collected from CRC mice were detected by Quantitative real-time PCR (Figures 3A,B). IL-17C was upregulated in the colorectal tissues of AOM/DSS mice. However, treatment with different extracts of CMF, including water extract and alcohol extract, effectively suppressed the mRNA expression of IL-17C. Drugs treatment, including aspirin, induced the mRNA expression of NF-κB. ELISA assay was performed to detect the protein expressions of inflammatory factors. As it is illustrated in Figure 3C, the protein levels of IL-17C in the serum of AOM/DSS-induced mice were significantly highest among mice with other drug intervention. The level of NF-κB significantly reduced in the tumor tissues of CMF-A, CMF-H, CMF-L, and Aspirin groups when compared to MC group (Figure 3D). All these results indicate that CMF treatment represses the expression of NF-κB and IL-17C.
Figure 3.
CMF changed the inflammatory microenvironment and regulated the expression of IL-17C and NF-κB in vivo. mRNA level of IL-17C (A) and NF-κB (B) in the colonic tissues were measured by real-time RT-PCR. All data are expressed as the mean ± S.D, *P < 0.05, **P < 0.01 compared with AOM/DSS group. The level of IL-17C in serum (C) and the level of NF-κB in tumor tissue homogenate (D) were measured using ELISA kits. All data are expression as the mean ± S.D, *P < 0.05, **P < 0.01 compared with AOM/DSS group.
CMF Treatment Regulated Gut Microbiota
To examine the effect of CMF on the regulation of components of gut microbiota, high-throughput pyrosequencing was performed with an Illumine MiSeq platform to generate 39584 high quality and valid sequences from 16 samples from different groups. We conducted a bar-coded pyrosequencing run to analyze the changes of gut microbiota in the five studied groups. In total, 39,584 available reads and 1461 OUTs were obtained. Shannon diversity and rarefaction curves are shown in Figures 4A,B. The rarefaction curves were consistent with the current sequencing, demonstrating that most of the flora diversity has been captured in all samples. What's more, the overlap of OUTs between groups revealed that 482 OUTs coexisted in both the MC group and CMF-A group, 496 OUTs coexisted in both the NC group and CMF-A group, 514 OUTs coexisted in both the NC group and MC group and 467 OUTs coexisted in both the MC group and Aspirin group (Figures 4C,D). In addition, we examined the flora community richness and diversity in mice. No statistical difference was observed for the ACE, Chao, Shannon, and Simpson indices among these groups (Figures 4E–H), although Simpson index were lower, but other indices are higher in NC group and the different CMF extract groups compared with MC group. Nonmetric multidimensional scaling (NMDS) analysis indicated the changes of overall structure of gut microbiota among different groups (Figure 4I). These results showed that the diversity of bacterial population of the NC group was higher than that of the MC group. There was a statistical difference between the CMF-H group and the MC group. These results suggested that CMF treatment increases the diversity of the intestinal flora.
Figure 4.
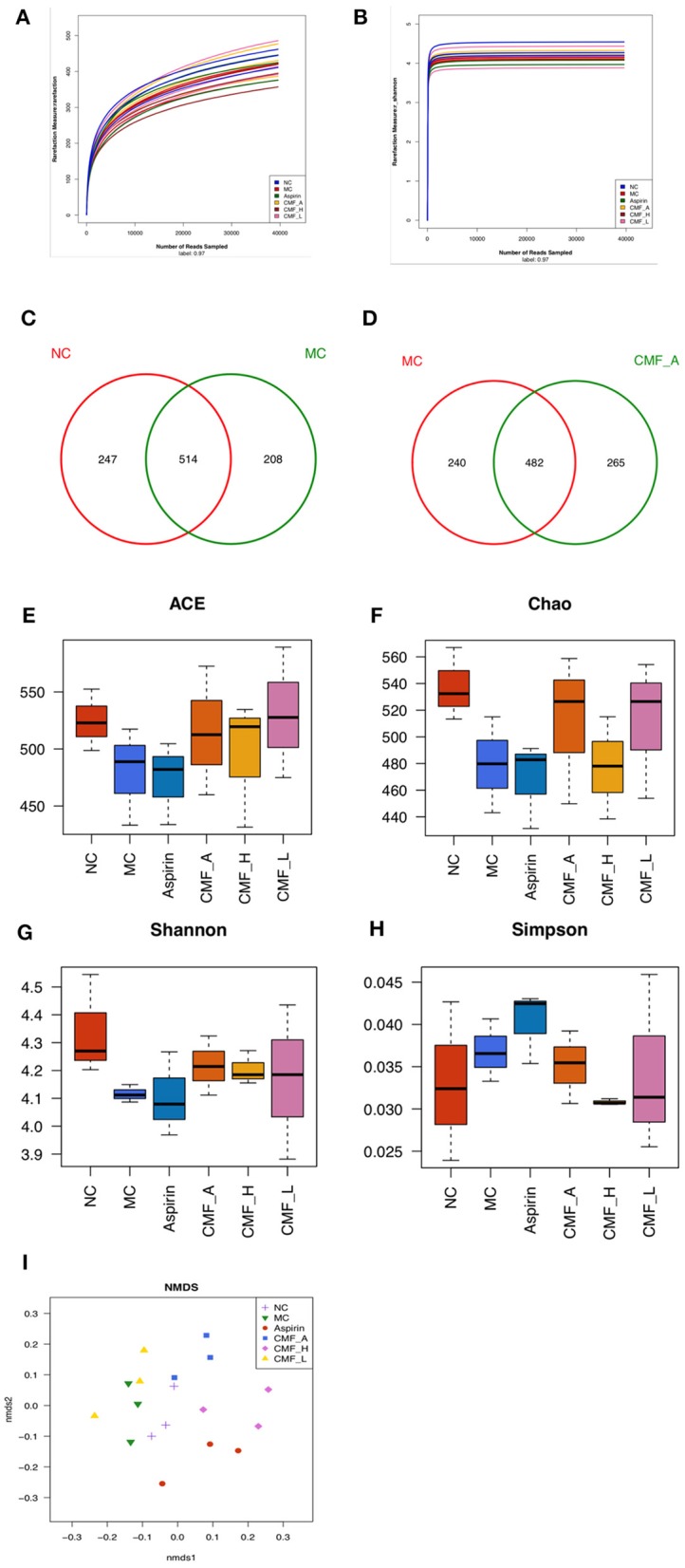
Illumina Miseq sequencing data showed CMF modulates the overall component of gut microbiota. (A) Rarefaction curves determined at the 97% similarity level. (B) Shannon-Wiener curves of samples, calculated using mothur. (C–F) Venn diagram of OUTs in the different groups. (G–I) Community richness and diversity index of the model control group (MC), normal control group (NC), aspirin group, and CMF intervention groups (CMF-A/H/L) on ACE, Chao, Shannon, Simpson analysis.
Analysis of the Relative Abundance of Microbiota at the Family Level and Genus Level
The family muribaculaceae, bacteroidales, and prevotellaceae were the most prevalent taxa in these groups (Figure 5A). It was found that these bacteria are dominant bacteria at the genus level (Figure 5B). Intraindividual changes were detected in these groups. Compared with the model group, the abundance of a small number of bacteria increased or decreased to varying degrees, and most of the bacteria remain unchanged. At the family level, abundance of Desulfovibrionaceae decreased in groups that were treated with CMF. Conversely, a transient CMF-treatment increase several family microbiota, including Rikenellaceae, Erysipelotrichaceae, Lactobacillaceae, Streptococcaceae, and Tannerellaceae. At the genus level, the abundance of genus Alistipes, Faecalibaculum, Lactobacillus, and Parabacterioides, were up-regulated after the treatment of CMF. The genus Parasutterella was found down-regulated in CMF treatment groups. It is worth mentioning that genus Desulfovibrionaceae_uncultured acted to AOM/DSS-mediated gut microbiota dysbiosis had statistical difference (Figure 5F). There arechanges in the gut microbiota but there is no statistical difference in these changes (Figures 5C–E).
Figure 5.
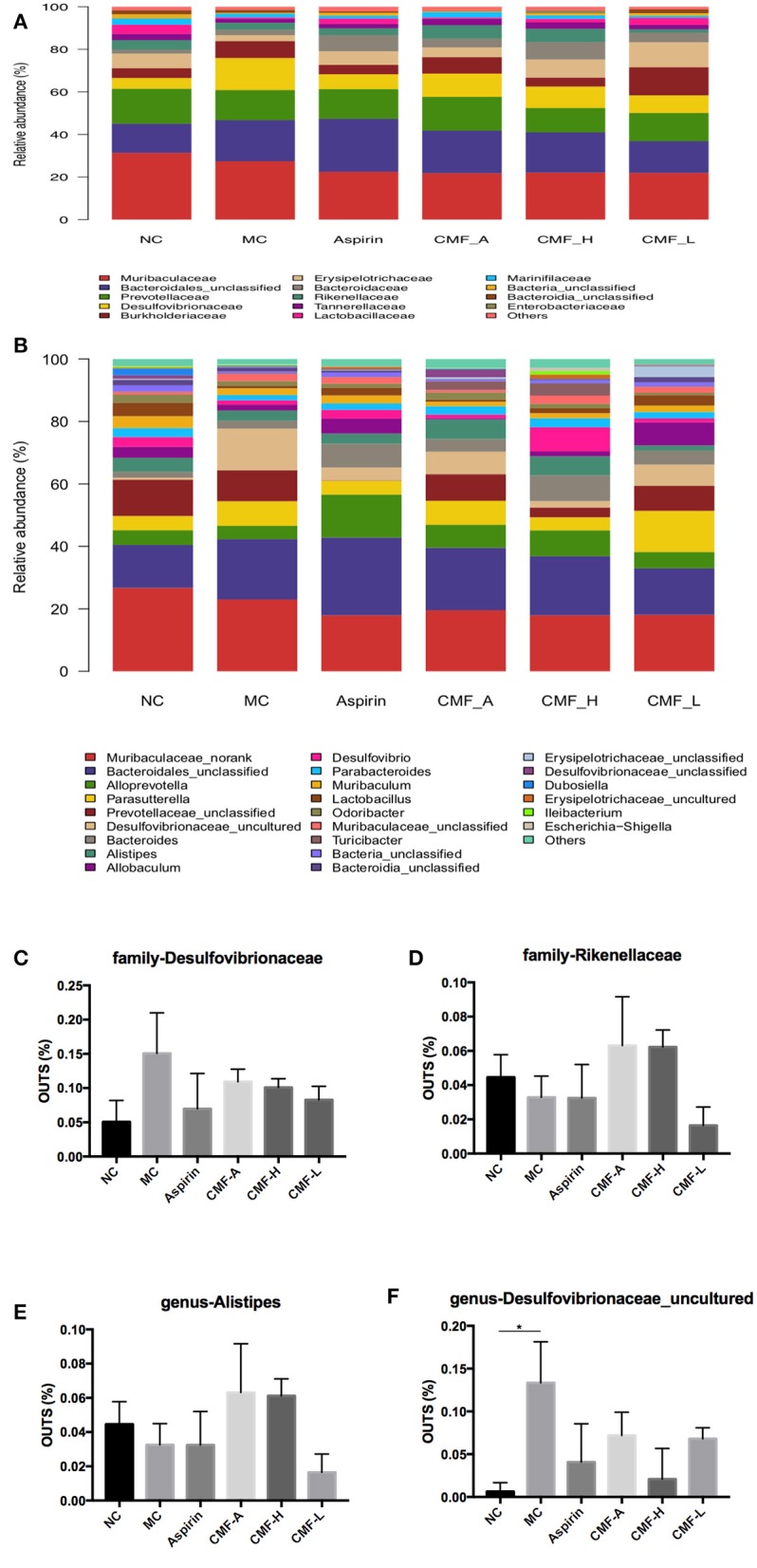
Comparison of gut microbiota among groups. Microbiota distribution at the family level (A) and the genus level (B). Relative abundance measured with qPCR for family-Desulfovibrionaceae, family- Rikenellaceae, and genus-Alistipes, genus- Desulfovibrionaceae_uncultured (C–F). Superscript letters indicate statistical differences (*P < 0.05).
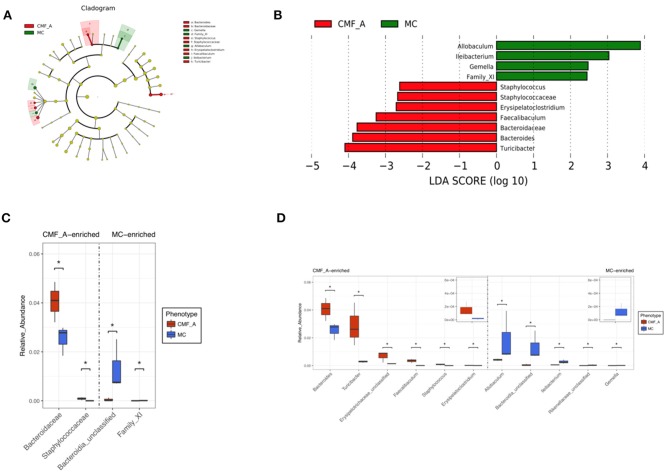
To systematically examine the alteration of taxa abundance according to CMF treatment, we conducted LEfSe (LDA Effect Size). The microbial cladogram indicates that the gut microbiome significantly altered in Bacteroidaceae and Staphylococcaceae, ranging from family to genus level (Figures 6A,C). At the genus level, the relative abundance of Turicibacter, Bacteroides, Bacteroidaceae, Faecalibaculum, Erysipelatoclostridium, and Staphylococcus were enriched in mice treated with the CMF-A diet (Figures 6B,D).
Figure 6.
Comparison of gut microbiota between CMF-A group and MC group. Cladogram of linear discriminant analysis (LDA) coupled with effective size measurement showing differentially abundant genera (A). Histogram of LDA scores for differentially abundant genera (B). qPCR identified the difference in CMF-A group and MC group of mice and t-test for significance testing at family level (C) and genus level (D). Superscript letters indicate statistical differences (*P < 0.05).
Discussion
Traditional Chinese medicine (TCM) have been used for the treatment of diseases and health conditions for several thousands of years. In the treatment of CRC, the application of TCM has received worldwide attention. Many studies have demonstrated the anti-tumor effect of TCM in CRC therapy. For example, Wei et al. showed that JianPi JieDu decoction (JPJD) is able to inhibit tumorigenesis, metastasis, as well as angiogenesis of tumors through the mTOR/HIF-1α/VEGF pathway (18). Wu et al. demonstrated that Gegen Qinlian decoction (GGT) can ameliorate gut-toxicity through anti-inflammatory pathways, inhibition of neutrophil migration, anti-oxidative stress via the Nrf2/Keap-1 pathway in the colon (19). Wing lam et al. reported that PHY906 reduced the CPT-11-induced gastrointestinal toxicity by inhibiting multiple steps of inflammation and the promotion of intestinal progenitor cell repopulation (20). Extensive studies have revealed that TCM antitumor drugs work by inhibiting tumor proliferation, promoting tumor apoptosis, and affecting diverse molecular targets (21–23).
The alterations of cancer genetic and molecular targets in CRC treatment have been extensively reported. Recent studies have reported that the regulation of gut microbiota may affect carcinogenesis (24–28). Intestinal flora is a direct factor of intestinal cancer. In this study, we found that CMF can ameliorate colitis-associated colorectal carcinogenesis in mice by modulating the fecal microbiota. The gut microbiota, including Fusobacterium (F.) nucleatum (29) and Firmicutes, Bacteroidetes Ley et al. (30), plays an important role in human health. In addition, many factors will affect and destroy the balance of intestinal microbiota, including the invasion of antigens, activation of immune cells, and production of cytokines. Therefore, investigating the relationship between CMF and the gut microbiota in AOM/DSS-induced colitis in miceis critical.
In our study, the anti-inflammatory role of CMF in the in vivo model of colitis-associated colorectal carcinogenesis induced by AOM/DSS and high-fat diet in C57BL/6 micewere investigated. Two different solvent extraction components of CMF were used to treat these mice to evaluate its therapeutic effect. The results showed that each of these extracts of CMF were well-tolerated. Histopathology findings were consistent with previous experimental results (31). The CRC mice treated with AOM/DSS and high-fat diet presented more severe condition than CMF-treated mice. In contrast, administration of CMF decreased tumor number and tumor size. Especially the ethanol extract of CMF can effectively inhibit the occurrence of colitis-associated colorectal carcinogenesis in the colon and small intestine.
Previous studies showed that tumor-prone mice colonized with E. coli and B. fragilis and increased IL-17 expression that promotes colon tumorigenesis in AOM mice (3, 32). It was reported that spontaneous activation of NF-κB was detected in tissues of human colorectal cancer (31, 33). We found that CMF could decrease the expression level of IL-17C and inhibit the activity of NF-κB.
High-throughput sequencing determined microbiological composition in mice. Most of the diversity of microorganisms was captured in all samples. The ACE, Chao, Simpson, and Shannon indices reveal that the microbial diversity in control mice and CMF treated mice were greater than in the model mice (16). The microflora analysis show that there are significant differences between the control mice and the model mice. There was a statistical difference between the ethanol extract of CMF treated mice and the model mice. This result indicated that the ethanol extract of CMF could increase the diversity of the intestinal flora. We evaluated phylum, class, order, family, and genus, the gut microbiota community in all samples. From the experiment, Muribaculaceae, Bacteroidales, and Prevotellaceae were the dominant family found in the gut. In accordance with family criteria, these flora occupied a main tier in the genus level. Focus on certain bacteria, statistical results reveal that CMF reversed the gut microbiota dysbiosis in CRC mice, including inhibiting the growth of Desulfovibrionaceae and promoting the Rikenellaceae. Desulfovibrionaceae, a kind of sulfate-reducing and endotoxin-producing bacteria, were mostly enriched in mice with long term high-fat feeding. Previous studies found that the family Desulfovibrionacea is positively associated with obesity (9, 34–36). This phenomenon was also found in our study. Wu et al. found that relative abundances of Rikenellaceae increased in the AOM/DSS mice (32). This result was in consistent with our study. We speculate that Rikenellaceae might be a bacterium that could cause cancer.
Because the results of our study showed that the alcohol extract of CMF is more effective than that of the water extract, we analyzed the difference of flora of the mice treated with the alcohol extract of CMF and the model mice. The results showed that the gut microbiome significantly altered in Bacteroidaceae and Staphylococcaceae, ranging from family to genus level. As shown in our results, at genus level, the relative abundance of Turicibacter, Bacteroides, Faecalibaculum, Erysipelatoclostridium, and Staphylococcus were enriched in mice treated with the CMF-A diet. In a previous study, the abundance of Bacteroidaceae had an important influence in the active progression of colitis in DSS-treated mice (16), which was in contrast to our study. Liu et al. found that the abundance of Turicibacter decreased in obesity mice (37, 38). Similarly, the Turicibacter was also reduced in AOM/DSS and high-fat diet mice in this study. Therefore, we suspect that Turicibacter, Bacteroides, Faecalibaculum, Erysipelatoclostridium, and Staphylococcus were the beneficial bacteria in the gut. But the specific role of these gut microbiota needs to be further investigated.
In summary, we report that CMF, a two-herb formulation, has reduced the incidence of CRC by inhibiting inflammatory factors and regulating gut microbiota. In addition, the mechanism of action of CMF is through the direct action on inflammatory factors or the indirect effect on regulating gut flora. Understanding the role of gut microbiota would be helpful for the prevention of CRC development.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by IRB of Yueyang Hospital Affiliated with Shanghai University of Traditional Chinese Medicine. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
XF, HZ, and GX contributed to study design, data interpretation, and manuscript preparation. YL contributed to animal rearing. DH contributed to data acquisition and analysis.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (81403360). The funders play no role in data collection and analysis, design, decision to publish, or preparation of the manuscript.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. (2016) 66:7–30. 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv. Nutr. (2015) 418–9. 10.3945/an.116.012211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. (2018) 359:592–7. 10.1126/science.aah3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. (2013) 369:1095–105. 10.1056/NEJMoa1301969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan RF, Grainge MJ, Shepherd VC, Armitage NC, Muir KR. ukCAP Trial Group. Aspirin and folic acid for the prevention of recurrent colorectal adenomas. Gastroenterology. (2008) 134:29–38. 10.1053/j.gastro.2007.10.014 [DOI] [PubMed] [Google Scholar]
- 6.Kummar S, Copur MS, Rose M, Wadler S, Stephenson J, O'Rourke M, et al. A phase I study of the chinese herbal medicine PHY906 as a modulator of irinotecan-based chemotherapy in patients with advanced colorectal cancer. Clin Colorectal Cancer. (2011) 10:85–96. 10.1016/j.clcc.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 7.Lam W, Jiang Z, Guan F, Hu R, Liu SH, Chu E, et al. The number of intestinal bacteria is not critical for the enhancement of antitumor activity and reduction of intestinal toxicity of irinotecan by the Chinese herbal medicine PHY906 (KD018). BMC Complement Altern Med. (2014) 14:490 10.1186/1472-6882-14-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Liu JX, Shang HX, Lin S, Zhao JY, Lin JM. Qingjie Fuzheng granules inhibit colorectal cancer cell growth by the PI3K/AKT and ERK pathways. World J Gastrointest Oncol. (2019) 11:377–92. 10.4251/wjgo.v11.i5.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Li Y, Sun M, Xin L, Wang T, Wei L. The Chinese herbal formula Shenzhu Tiaopi Granule results in metabolic improvement in type 2 diabetic rats by modulating the gut microbiota. Evid Based Complement Alternat Med. (2019) 2019:6976394. 10.1155/2019/6976394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, Li J, et al. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients' survival? A pilot study on relevant mechanism. Oncotarget. (2016) 7:46158–72. 10.18632/oncotarget.10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RL, Jemal A, Wender RC, Gansler T, Ma J, Brawley OW. An assessment of progress in cancer control. CA Cancer J Clin. (2018) 68:329–39. 10.3322/caac.21460 [DOI] [PubMed] [Google Scholar]
- 12.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. (2017) 153:1621–33. 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- 13.Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, et al. AMPK-mediated BECN1 phosphorylation promotes ferroptosis by directly blocking system X(c)(-) activity. Curr Biol. (2018) 28:2388–99. 10.1016/j.cub.2018.05.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Li Q, Fu X. Research progress in chemical constituents and pharmacological effects of Wumei. Shanghai J. Tradit. Chin. Med. (2017) 296–300. 10.16305/j.1007-1334.2017.S1.078 [DOI] [Google Scholar]
- 15.Xu C, Shang S, Liu M, Yang M. Research progress on chemical constituents and pharmacological activities of silkworm, Bombyx mori. Chinese Pharm. (2014) 3732–4. 10.6039/j.issn.1001-0408.2014.39.29 [DOI] [Google Scholar]
- 16.Yang Y, Chen G, Yang Q, Ye J, Cai X, Tsering P, et al. Gut microbiota drives the attenuation of dextran sulphate sodium-induced colitis by Huangqin decoction. Oncotarget. (2017) 8:48863–74. 10.18632/oncotarget.16458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DH, Hossain MA, Kang YJ, Jang JY, Lee YJ, Im E. Baicalein, an active component of Scutellaria baicalensis. Georgi, induces apoptosis in human colon cancer cells and prevents AOM/DSS-induced colon cancer in mice. Int J Oncol. (2013) 43:1652–8. 10.3892/ijo.2013.2086 [DOI] [PubMed] [Google Scholar]
- 18.Peng W, Zhang S, Zhang Z, Xu P, Mao D, Huang S, et al. Jianpi Jiedu decoction, a traditional Chinese medicine formula, inhibits tumorigenesis, metastasis, and angiogenesis through the mTOR/HIF-1α/VEGF pathway. J Ethnopharmacol. (2018) 224:140–8. 10.1016/j.jep.2018.05.039 [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Wang D, Yang X, Fu C, Zou L, Zhang J. Traditional Chinese medicine Gegen Qinlian decoction ameliorates irinotecan chemotherapy-induced gut toxicity in mice. Biomed Pharmacother. (2019) 109:2252–61. 10.1016/j.biopha.2018.11.095 [DOI] [PubMed] [Google Scholar]
- 20.Lam W, Bussom S, Guan F, Jiang Z, Zhang W, Gullen EA, et al. The four-herb Chinese medicine PHY906 reduces chemotherapy-induced gastrointestinal toxicity. Sci Trans Med. (2010) 2:45. 10.1126/scitranslmed.3001270 [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Liu H, Zhou Z, Parise RA, Chu E, Schmitz JC. Herbal formula Huang Qin Ge Gen Tang enhances 5-fluorouracil antitumor activity through modulation of the E2F1/TS pathway. Cell Commun Signal. (2018) 16:7. 10.1186/s12964-018-0218-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan L, Zhou M, Wasan HS, Zhang K, Li Z, Guo K, et al. Jiedu Sangen decoction inhibits the invasion and metastasis of colorectal cancer cells by regulating EMT through the Hippo signaling pathway. Evid Based Complement Alternat Med. (2019) 2019:1431726. 10.1155/2019/1431726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Q, Ouyang Y, Meng F, Noolvi MN, Avvaru SP, More UA, et al. A review of pharmacological and clinical studies on the application of Shenling Baizhu San in treatment of Ulcerative colitis. J Ethnopharmacol. (2019):112105. 10.1016/j.jep.2019.112105 [DOI] [PubMed] [Google Scholar]
- 24.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. (2013) 342:967–70. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. (2013) 342:971–6. 10.1126/science.1240537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. (2015) 350:1084–9. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vètizou M, Pitt JM, Daillè re R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. (2015) 350:1079–84. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. (2016) 167:1125–36.e8. 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. (2017) 170:548–63.e16. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. (2006) 444:1022–3. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 31.Lv Z, Wang Z, Luo L, Chen Y, Han G, Wang R, et al. Spliceosome protein Eftud2 promotes colitis-associated tumorigenesis by modulating inflammatory response of macrophage. Mucosal Immunol. (2019) 12:1164–73. 10.1038/s41385-019-0184-y [DOI] [PubMed] [Google Scholar]
- 32.Chan JL, Wu S, Geis AL, Chan GV, Gomes TAM, Beck SE, et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. (2019) 12:164–77. 10.1038/s41385-018-0085-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo M, Li Z, Huang Y, Shi M. Polysaccharides from Nostoc commune Vaucher activate macrophages via NF-κB and AKT/JNK1/2 pathways to suppress colorectal cancer growth in vivo. Food Funct. (2019) 10:4269–79. 10.1039/C9FO00595A [DOI] [PubMed] [Google Scholar]
- 34.Sun J, Qi C, Zhu H, Zhou Q, Xiao H, Le G, et al. IgA-targeted Lactobacillus jensenii modulated gut barrier and microbiota in high-fat diet-fed mice. Front Microbiol. (2019) 10:1179. 10.3389/fmicb.2019.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Lam KL, Hu J, Ge S, Zhou A, Zheng B, et al. Chlorogenic acid alleviates obesity and modulates gut microbiota in high-fat-fed mice. Food Sci Nutr. (2019) 7:579–88. 10.1002/fsn3.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C, Ouyang M, Guo Q, Jia J, Liu R, Jiang Y, et al. Changes in the intestinal microecology induced by bacillus subtilis inhibit the occurrence of ulcerative colitis and associated cancers: a study on the mechanisms. Am J Cancer Res. (2019) 9:872–86. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Crott JW, Lyu L, Pfalzer AC, Li J, Choi SW, et al. Diet- and genetically-induced obesity produces alterations in the microbiome, inflammation and Wnt pathway in the intestine of Apc(+/1638N) mice: comparisons and contrasts. J Cancer. (2016) 7:1780–90. 10.7150/jca.15792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M, Wu Y, Li J, Bao Y, Guo Y, Yang W. The dynamic changes of gut microbiota in Muc2 deficient mice. Int J Mol Sci. (2018) 19:E2809. 10.3390/ijms19092809 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.