Abstract
Mammalian oocytes and eggs are transcriptionally quiescent and depend on post-transcriptional mechanisms for proper maturation. Post-transcriptional mRNA modifications comprise an important regulatory mechanism that can alter protein and miRNA recognition sites, splicing, stability, secondary structure, and protein coding. We discovered that fully grown mouse germinal vesicle oocytes and metaphase II eggs display abundant inosine mRNA modifications compared to growing oocytes from postnatal day 12 oocytes. These inosines were enriched in mRNA protein coding regions (CDS) and specifically located at the third codon base, or wobble position. Inosines, observed at lower frequencies in CDS of somatic tissues, were similarly enriched at the codon wobble position. In oocytes and eggs, inosine modifications lead primarily to synonymous changes in mRNA transcripts. Inosines may ultimately affect maternal mRNA stability by changing codon usage, thereby altering translational efficiency and translationally coupled mRNA degradation. These important observations advance our understanding of post-transcriptional mechanisms contributing to mammalian oocyte maturation.
Keywords: oocyte, inosine, RNA modifications, ADAR RNA editing, post-transcriptional regulation
Summary sentence: Inosine mRNA modifications in germinal vesicle oocytes and MII eggs alter codon composition and may affect translational efficiency and translationally coupled mRNA degradation.
Introduction
The mammalian oocyte has a unique physiological environment because it exhibits high transcriptional activity during growth, followed by transcriptional quiescence when it is stimulated to resume meiosis [1,2]. To maintain cellular processes throughout meiotic maturation, fertilization, and activation of the embryonic genome, the oocyte relies on previously transcribed and stored RNA. Thus, post-transcriptional gene regulation and translational control have increased importance in oocytes and are essential to generate high-quality female gametes [3].
RNA modifications are a common form of post-transcriptional regulation, with more than 50 described in mammals [4]. Of particular interest are RNA modifications that can result in the recoding of mRNA sequence. Currently, the RNA modification most efficient at recoding is inosine. Inosines are formed by deamination of adenosine and can recode RNA to affect codons, splice sites, RNA secondary structure, and RNA recognition motifs [5]. Inosines within mRNA can impact transcript abundance and translational efficiency, increase proteome diversity, and prevent detection of cellular RNA as foreign, thus limiting an interferon response [6–12]. Recent studies demonstrated that both human and mouse oocyte RNA contain inosine modifications [13,14], but the landscape of RNA modifications within the stored RNA pool has not been critically examined.
Inosines arise in RNA by the deamination of adenosine, which is catalyzed by a family of highly conserved double-stranded RNA (dsRNA)-binding proteins, adenosine deaminases acting on RNA (ADARs) [15–17]. Mammals have three ADAR genes: adenosine deaminase, RNA-specific (Adar); adenosine deaminase, RNA-specific, B1 (Adarb1); and adenosine deaminase, RNA-specific, B2 (Adarb2), encoding the proteins ADAR, ADARB1, and ADARB2, respectively [6]. ADAR and ADARB1 are enzymatically active and deaminate dsRNA, whereas ADARB2 has no reported enzymatic activity [18]. Adar is ubiquitously expressed in tissues, while Adarb1 is expressed predominantly in the brain [19,20]. Targeted deletion of Adar in mice results in embryonic lethality due to defective organ development coupled with a systemic interferon response induced by a transcriptome-wide reduction in inosine-containing RNA [21–23]. Consistent with its expression profile, knockout of Adarb1 resulted in loss of a site-specific inosine within Gria2 mRNA in neurons, resulting in seizures and death before postnatal day 20 [24]. While inosine residues within oocyte RNA indicate the presence of an adenosine deaminase enzymes, the abundance of Adar, Adarb1, and Adarb2 within oocytes has not been described.
Due to the unique attributes of transcriptional quiescence in the oocyte, understanding the complete landscape of RNA modifications within the stored RNA pool will be critical to our understanding of oocyte biology. By assessing inosine modifications in the transcriptionally active and transcriptionally inactive oocyte and egg, we have begun to unravel how this singular RNA modification changes during oocyte maturation and how it may potentially affect oocyte mRNA stability and translation during oocyte maturation.
Methods
Oocyte and egg collections
Three biological replicates of postnatal day 12 (PND12) oocytes (6 oocytes per mouse, n = 3) were isolated from the ovaries of wild-type C57BL/6J female animals. PND12 ovaries were incubated with collagenase I (0.1%, v/v, Worthington Biochemical Corporation, Lakewood, NJ) for 30 min to dissociate oocytes. During incubation with collagenase, the ovaries were pipetted up and down every 10 min to facilitate dissociation and then washed through several droplets of culture medium without collagenase. Oocyte RNA was extracted using the PicoPure RNA Isolation Kit (Thermo Fisher Scientific, KIT0204). Germinal vesicle oocytes were collected from 21-day-old wild-type C57Bl/6J females stimulated with pregnant mare’s serum gonadotropin (PMSG). Germinal vesicle (GV) oocytes were maintained in FHM HEPES Buffered medium (Gibco) containing 2.5 μM milrinone to block spontaneous maturation. GV stage oocytes were manually stripped clean of cumulus cells and the zona pellucida removed by brief exposure to acid tyrode’s, prior to extraction of total RNA with TRI Reagent (Ambion). To collect MII eggs, groups of GV oocytes were in vitro matured in the absence of milrinone for 16 h. GV oocytes and MII eggs with visible polar bodies were lysed in groups of 30, directly in SDS sample buffer, with 5% β-mercaptoethanol in preparation for western blot analyses. Mice were maintained in environmentally controlled facilities in the University of Kansas Medical Center (KUMC) in a room with a 14-h light, 10-h dark cycle (7 am to 9 pm) with ad libitum access to food and water. All animal procedures were performed according to an approved KUMC Institutional Animal Care and Use Committee protocol.
Library preparation and RNA sequencing of postnatal day 12 oocytes
RNA-seq libraries for the PND12 oocytes were prepared following the Nugen Ovation Ultralow Library Systems Protocol (Nugen®, Ovation® Ultralow Library Systems User Guide, M01219 v6). Briefly, first-strand and second-strand cDNA synthesis was conducted and purified using Agencourt RNAClean XP beads (Beckman Coulter, A63987). cDNA products were then amplified using SPIA amplification and purified using Qiagen MinElute Reaction Cleanup Kit (Cat# 28204). Amplified cDNA was then fragmented using a Covaris S2 Ultrasonicator and digested using S1 Nuclease treatment. DNA was purified, end repaired, ligated to adaptors, and amplified. The resulting libraries were analyzed on a Bioanalyzer DNA Chip 1000. Fragment distribution was between 150 and 200 base pairs. RNA-seq was performed using single-end Illumina HiSeq 4000. These raw data have been deposited in SRA SRP133083 and are available for sharing.
Sources of germinal vesicle and metaphase II RNA-seq datasets
Wild-type GV oocyte and MII egg RNA-seq data were identified by searching the Gene Expression Omnibus and downloaded from the Sequence Read Archive using SRAdb [25]. Wild-type GV oocyte RNA-seq data were from dataset SRP057558 [26]. The authors of this study demonstrated the high quality of isolated GV oocytes by showing that contemporary oocytes were fully grown, meiotically competent and exhibited successful nuclear envelope breakdown and polar body extrusion [26]. Wild-type MII oocyte RNA-seq data were from datasets SRP034543 [27] and SRP065556 [28]. These samples reflect MII eggs isolated from the oviduct following in vivo maturation. PCA of transcriptionally active PND12 oocytes, and fully grown transcriptionally quiescent GV oocytes and MII eggs demonstrated that the samples were transcriptionally distinct (Supplementary Figure S2E). Control somatic tissue RNA-seq data were obtained from the ENCODE database [29]. Wild-type and AdarE861A/E861A mutant brain RNA-seq data were obtained from SRP098703 (adult SRP098702) [30].
Western blotting
Lysed oocyte and egg samples were subjected to electrophoresis using 10% SDS-PAGE in running buffer at a constant 120 V for 1 h. Proteins were electro-transferred onto PVDF membranes 350mA for 1 h at 4 °C and blocked with 5% (w/v) skim milk in Tris-buffered saline with 0.05% (v/v) Tween-20 (TBST) for 1 h at room temperature. Membranes were then probed with primary anti-ADAR1 (1:1000; sc-73408, Santa Cruz, Dallas, TX) or anti-Actin (1:5000; sc-1616, Santa Cruz, Dallas, TX) antibody overnight at 4 °C in TBST/BSA (50 mM Tris, 150 mM NaCl, 0.05% Tween20, 1.5% BSA) followed by incubation with secondary antibodies (mouse IgG HRP clone NA931V, GE Healthcare Life Sciences, Pittsburgh, PA; or goat IgG HRP, clone sc-2020 Santa Cruz) for 2 h at room temperature in blocking buffer. Membranes were washed three times in TBST and detected by enhanced chemiluminescence (Thermo Fisher Scientific, Waltham, MA).
Transcript abundance analysis
Sequence alignment and transcript abundance calculations for PND12 oocytes, GV oocytes, MII eggs, and mouse somatic tissues were performed by aligning raw RNA-seq reads against mus musculus (mm10) transcripts curated from RefSeq using Kallisto with the following settings: kalisto quant -t 31 -b 100. Differential transcript abundance was calculated using Sleuth in R [31–33].
Validation of identified inosine modification sites
Total RNA was treated with DNase I (Thermo Fisher Scientific, AM2222) followed by cDNA generation using Superscript III (Thermo Fisher Scientific, 18080044) and oligo dT20 primers (Thermo Fisher Scientific, 18418020). Genomic DNA was isolated from tail clips of wild-type C57Bl/6J mice using REDExtract-N-Amp™ Tissue PCR Kit (SigmaAldrich, XNAT-100RXN). Genomic DNA and cDNA were amplified using Phusion® High-Fidelity DNA Polymerase (NEB, M0530). PCR reactions were purified using AMPure XP (Beckman Coulter, A63987) bead purification and Sanger sequenced or cloned using the Zero Blunt™ TOPO™ PCR Cloning Kit (Thermo Fisher Scientific, K286020). Transformations were grown on LB + Kanamycin at 37 °C overnight. Individual colonies were picked and grown in liquid culture, LB + Kanamycin, overnight at 37 °C. Plasmid DNA was extracted from liquid culture using QIAprep Spin Miniprep Kit (Qiagen, 27104). Individual clones were Sanger sequenced using M13 Fwd and M13 Rev primers at GeneWiz (Boston, MA). Primer sequences used in validation Sanger sequencing: Rpa1-F:AAGTACCACTGGTGGCCAGA, Rpa1-R:GGCTAATTAATTGCTTTCCAGTT; Mdc1-F:CCTTCTCAGACCATCGAACAG; Mdc1-R:TGAAGTGAAATTCATAAAGCAAAAA; Wdr37-F:TCACTGATGTTCCTTAGCTCCA, Wdr37-R:AAATGCACTTTTCCCTCAAAAA; Mad21l-F:AGACGGTGAGGAAAAGGATG, Mad21l-R:GCATCAACTGCTTTGTGAGC; Cep85-F:GGAGTGCCAGGGAGGACTTA, Cep85-R:TGTAATCAAAATAGGCAATACAAACC.
Identification and consequence analysis of inosine RNA modifications
We identified putative inosine RNA modifications utilizing a combination of the HISAT2 aligner, the Genome Analysis Tool Kit (GATK), and Ensembl Variant Effect Predictor (VEP) [34–36]. Inosines appear as A-to-G substitutions when comparing RNA-seq data to a reference genome [37]. HISAT2 was used to map raw RNA-seq reads to a mus musculus index built from GRCm38 containing common SNP annotation from dbSNP. Default settings for HISAT2 were used for each type of library aligned (i.e., single-end, paired-end, or stranded, Supplementary Table S2) [35]. Prior to alignment, fastq files were checked for adaptors and trimmed if necessary, using Trimmomatic [38]. The default alignment settings for HISAT2 will report at most 10 valid primary alignments, and we did not increase the amount of multimapping allowed. HISAT2 reports valid alignments that are at or above the minimum alignment score determined by the function 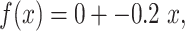 where
where 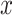 is the length of the read. Mismatches in the alignment were subtracted from the alignment score, reads that fell below the minimum alignment score were not reported. Overall alignment rates with HISAT2 were between 79 and 98% (Supplementary Table S2). RNA/DNA differences were called using the Genome Analysis Toolkit (GATK) RNA-seq variant pipeline with modifications [39]. The program elprep was used to sort, mark duplicates, and index RNA-seq reads (Supplemental Methods).
is the length of the read. Mismatches in the alignment were subtracted from the alignment score, reads that fell below the minimum alignment score were not reported. Overall alignment rates with HISAT2 were between 79 and 98% (Supplementary Table S2). RNA/DNA differences were called using the Genome Analysis Toolkit (GATK) RNA-seq variant pipeline with modifications [39]. The program elprep was used to sort, mark duplicates, and index RNA-seq reads (Supplemental Methods).
Known SNPs were filtered out using Mouse Genomes Project database, Wellcome Trust Sanger Institute mouse strains [40]. Strand information of the variant was inferred from the gene model using Ensembl VEP [34]. VCF files were filtered on allelic depth (AD > 0) and used as input for Ensembl VEP [34]. VEP was used to identify inosine modified transcripts, categorize the location within the transcript, and determine consequence of inosines on coding capacity. Only inosine sites occurring in transcripts with TPM ≥ 1 and having an AD greater than 0 were reported. A transcript was considered to be inosine modified if it contained at least one inosine site. R statistical computation software with the following packages was used to parse VEP output: sleuth, biomaRt, dplyr, plyr, AnnotationFuncs, org. Mm.eg.db, ggplot2 [33,41–43].
Statistics
Statistical analyses were performed using R and Prism 7.0. Results of multiple repeats (number of “n” is presented in figure legends or text) are presented as means ± SEM. Bartlett’s tests were done to ensure equal variance among treatment groups. If data were normally distributed, parametric tests were used. If data showed a variance outside of normal, a non-parametric test was used to determine if statistical differences existed. In cases where only two treatment groups existed, differences were determined by a parametric, unpaired t-test or a non-parametric, unpaired t-test (Mann-Whitney U test). To determine statistical differences between groups with more than two treatment groups, one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests were used, or non-parametric Kruskal-Wallis test with Dunn’s multiple comparisons test were performed. In samples with two variables, a two-way ANOVA followed by Tukey’s multiple comparison tests was performed. χ2 tests were used where appropriate to determine observed versus expected significance and to test differences in populations. Values of P < 0.05 were considered significant.
Detailed line commands for elprep and GATK, equations for the inosine RNA modification false-positive rate, and equations for inosine RNA modification efficiency are provided in the Supplemental Methods.
Results
ADAR is the predominant adenosine deaminase detected in mouse oocytes
To establish which adenosine deaminases were present in oocytes, we first assessed transcript abundance of the adenosine deaminase family members in transcriptionally active PND12 oocytes and fully grown transcriptionally quiescent GV oocytes and MII eggs. In mice, Adar can generate three mRNA isoforms: transcript variants 1 and 3 encode ADAR p150 and transcript variant 2 encodes ADAR p110 (Supplementary Figure S1). Adar transcript variant 2 was the only adenosine deaminase transcript to exhibit significant differential abundance in oocytes and eggs (P < 0.05, one-way ANOVA; Figure 1A). Transcript abundance of the remaining adenosine deaminases, Adarb1, Adarb2, and two related but enzymatically deficient enzymes, adenosine deaminase domain containing 1 (testis specific) (Adad1), and adenosine deaminase domain containing 2 (Adad2) were undetectable or low and not significantly distinct, respectively (Supplementary Figure S2). In contrast, somatic tissues (colon, heart, large intestine, and stomach) expressed all of the Adar family genes (Supplementary Figure S2). Consistent with transcript abundance, ADAR p110 protein was observed in GV oocytes and MII eggs (Figure 1B), with minimal detection in PND12 oocytes. We were unable to detect ADAR p150 protein (Figure 1B). These results indicate that Adar/ADAR is most abundant in transcriptionally inactive, fully grown GV oocytes and MII eggs.
Figure 1.
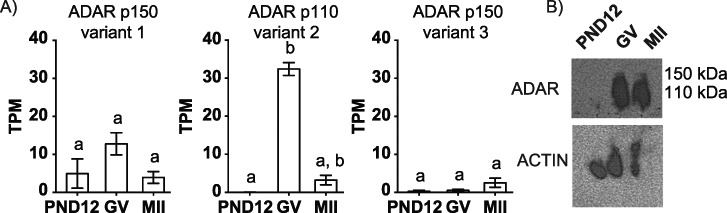
ADAR is predominantly expressed in mouse GV oocytes and MII eggs. (A) Isoform transcript abundance of Adar variants 1, 2, and 3 in PND12 oocytes, GV oocytes, and MII eggs; TPM: transcripts per million. (B) Micro-western dot blot of ADAR and ACTIN in PND12 oocytes, GV oocytes, and MII eggs; n = 30 cells per lane, in 2 μL. a,bMeans ± SEM within a panel that have different superscripts were different (P < 0.05); Kruskal-Wallis test followed by Dunn’s multiple comparison tests.
Identification of inosine sites within mRNA in mouse oocytes and eggs
In order to validate our computational approach to identify inosine sites, we used RNA-seq data from catalytically inactive AdarE861A/E861A brain tissue, which show global reductions of inosine within the transcriptome [30,44]. Total inosine RNA modification sites in brain tissue of the AdarE861A/E861A mice (14058 ± 1313; mean ± SEM) was half of control Adar+/+ mice (25801 ± 1068; P < 0.05, two-way ANOVA). Note that this reduction in inosine sites was not expected to be a complete loss, as brain tissue contains significant ADARB1 mediated inosine RNA modifications (Supplementary Figure S3A) and [45]. Consistent with previous studies, the estimated error rate for the inosine detection pipeline was 0.97% ± 0.05% and 4.05% ± 0.02% for AdarE861A/E861A and control Adar+/+ brain tissue, respectively [37,46]. Overall, these analyses support our approach for identifying inosine within RNA-seq data.
Analysis of oocyte and egg RNA-seq data revealed an abundance of inosine-containing transcripts (Figure 2A). Similar to our observation in brain tissue, the estimated error rate for inosine site detection was 5.94% ± 0.18% for PND12 oocytes, 4.42% ± 0.08% for GV oocyte, and 4.76% ± 0.42% for MII eggs. Notably, fewer inosine-containing transcripts were observed in PND12 oocytes (363 ± 24; mean ± SEM), consistent with the minimal ADAR protein in these oocytes compared to GV oocytes (2540 ± 59) and MII eggs (1468 ± 194; P < 0.05, one-way; Figure 2A). Similar results were observed when data was depicted as inosine sites per gene, rather than per transcript (Supplementary Figure S4A and B). Additionally, we tested whether the increase in inosine-containing transcripts in GV oocytes and MII eggs represented a population of transcripts not present in PND12 oocytes. We observed that the total number of unique transcripts, and genes, detected in GV oocytes and MII eggs was greater than in PND12 oocytes (Figure 2B, and Supplementary Figure S4C). Common transcripts in all three oocyte stages show a similar trend (Figure 2C, and Supplementary Figure S4D). Thus, the increase in inosine RNA modification events observed in GV oocyte and MII egg mRNA is not due to an overall increase in transcripts present but strongly correlates with ADAR abundance within these oocyte stages (Figure 1B). To validate the presence of inosine within mRNA, Sanger sequencing on five identified genes, replication protein A1 (Rpa1), mediator of DNA damage checkpoint 1 (Mdc1), WD repeat domain 37 (Wdr37), MAD2 mitotic arrest deficient-like 1 (Mad2l1), and centrosomal protein 85 (Cep85), was completed in GV oocytes (Figure 2D and E, using two complementary approaches, SI Appendix). In summary, inosine sites were identified within oocyte and egg mRNA, were correlated with ADAR activity, and were confirmed using Sanger sequencing.
Figure 2.
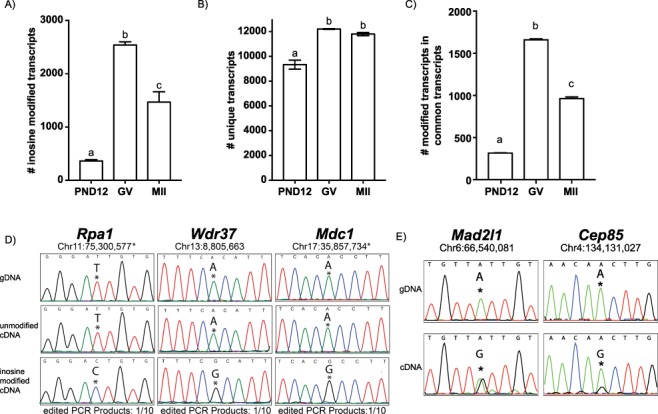
Inosine modifications predominate in fully grown GV oocytes and MII eggs. (A) Number of unique inosine-modified transcripts identified in PND12 oocytes, GV oocytes, and MII eggs. (B) Total number of unique mRNA transcripts per sample type. (C) Number of inosine-modified transcripts among commonly detected transcripts. (D and E) Inosine modifications were validated using Sanger sequencing of TOPO-cloned PCR products (D) or total PCR populations (E) of five genes. Chromosome location is indicated, and the * denotes minus strand of the DNA. a,bMeans ± SEM within a panel that have different superscripts were different (P < 0.05); significance was determined using one-way ANOVA, followed by Tukey’s multiple comparison tests. Only transcripts with TPM ≥ 1 were analyzed.
Oocyte-specific pattern of inosine RNA modifications
In addition to having the fewest inosine-containing transcripts, PND12 oocytes had the lowest proportion of the transcriptome that contained inosine (3.9% ± 0.2%; mean ± SEM), compared to GV oocytes (20.8% ± 0.5%) and MII eggs (11.3% ± 0.7%; P < 0.05, χ2; Figure 3A). Overall, the proportion of inosine-containing transcripts in fully grown oocytes was similar to mouse somatic tissues (Supplementary Figure S5A). Consistent with higher levels of ADAR, GV oocytes and MII eggs have an approximately 2-fold increase in the number of inosine sites per transcript compared to PND12 oocytes (Figure 3B). We did not observe a difference in the number of inosines per transcript among somatic tissues, which express similar numbers of transcripts (Supplementary Figure S5B and C). In addition, the average number of transcripts detected per gene was 1.18 and 1.17 for GV oocytes and MII eggs, respectively (Supplementary Figure S4E), but when only the inosine RNA-modified genes were considered, the number of transcripts significantly increased to 4.3 and 2.5 per gene for the GV oocytes and MII eggs, respectively (P < 0.05, one-way ANOVA; Supplementary Figure S4F).
Figure 3.
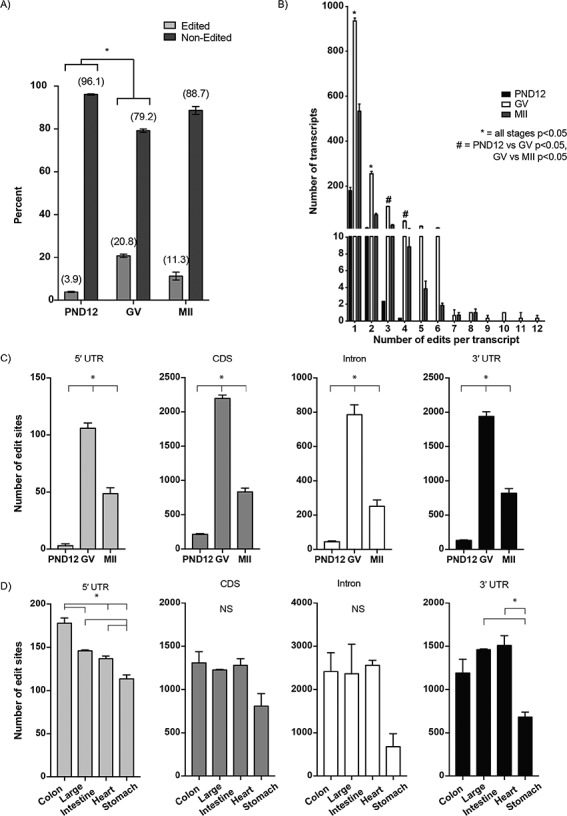
Distinct pattern of inosine modifications in oocytes compared to somatic cells. (A) Proportion of the transcriptome (percentage) that contains inosine modification within PND12 oocytes, GV oocytes, and MII eggs. (B) Number of inosine modified transcripts exhibiting one or multiple inosines per transcript in PND12 oocytes, GV oocytes, and MII eggs. Numbers above line indicate the number of inosines/transcript. (C) Number of inosines within specific regions (5′ UTR, CDS, intron, and 3′ UTR) in mRNA of oocytes and eggs, and (D) somatic tissues of C57BL/6 wild-type mice. *Means ± SEM within panel A are different (P < 0.05); significance was determined using χ2 tests. Only transcripts with TPM ≥ 1 were analyzed.
To further characterize the nature of inosine RNA modifications in oocytes, the location (5′ UTR, coding, intron, and 3′ UTR) of inosines within protein-coding genes was annotated. The highest amount of inosine sites occurred in the CDS and 3′ UTR regions, and these proportions were similar at all oocyte stages (Figure 3C, Supplementary Figure S6A and B). In contrast, somatic tissues exhibited the highest number of inosine sites in intronic regions (Figure 3D, Supplementary Figure S6C). Thus, mouse oocytes display a unique distribution pattern of inosine RNA modifications within transcripts.
Consequences of coding region inosine RNA modifications in mouse oocytes and eggs
Inosines can be recoded by the ribosomal machinery as guansosines, thus potentially changing the protein sequence [47]. Therefore, to understand the consequences of inosine RNA modifications, if any, on the protein coding capacity of GV oocyte and MII egg mRNA, we calculated the frequency of synonymous and nonsynonymous changes. Among the nonsynonymous changes, altered stop codons (stop loss, stop gain, or stop retained) made up less than 0.3% of coding sequence changes. Therefore, inosine modifications generating stop codons were excluded from all further analyses. Coinciding with the increased Adar/ADAR expression in GV oocytes and MII eggs, synonymous RNA changes also increased (P < 0.05, χ2; Figure 4A). Interestingly, somatic tissues displayed synonymous RNA changes similar to that of GV oocytes and MII eggs (Figure 4B). To confirm that this increase in synonymous changes was a result of increased inosine RNA modifications, we analyzed RNA-seq data from AdarE861A/E861A brain tissue. Synonymous RNA changes dramatically decreased in AdarE861A/E861A brain tissue, similar to the ADAR-deficient PND12 oocytes (P < 0.05, χ2; Supplementary Figure S3A and C).
Figure 4.
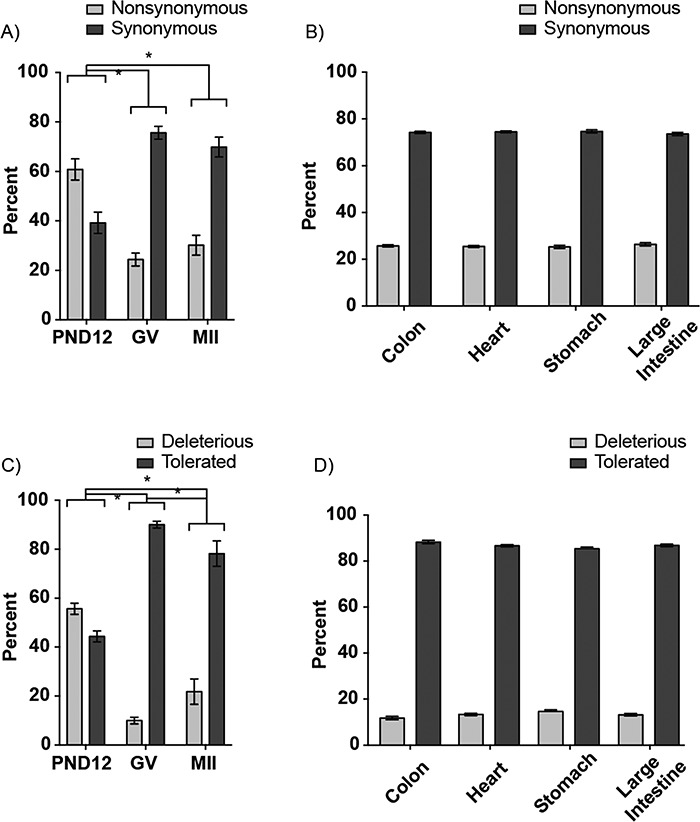
The consequence of inosines in CDS of mouse oocyte transcripts. (A) Proportion of inosine modifications (percentage) in PND12 and GV oocytes, and MII eggs that result in nonsynonymous or synonymous changes was determined for all inosine-modified mRNA transcripts. (B) Percent of inosines in a variety of somatic tissues. (C and D) Proportion of tolerated and deleterious transcripts following Sorting Intolerant From Tolerant (SIFT) analysis of the inosine-modified mRNA transcripts from oocytes and somatic tissues. *Means ± SEM within a panel are different (P < 0.05); significance was determined using χ2 tests. Only transcripts with TPM ≥ 1 were analyzed.
To predict the consequences of the amino acid changes on protein function, the computational tool, Sorting Intolerant From Tolerant (SIFT), was used [48]. Inosine mRNA modifications observed in GV oocytes and MII eggs had increased tolerated amino acid changes when compared to PND12 oocytes (P < 0.05, χ2; Figure 4D). Similarly, somatic tissues exhibited greater tolerated (>80%) versus deleterious changes (Figure 4E). Comparison of AdarE861A/E861A and control Adar+/+ brain tissue indicated that loss of inosine RNA modifications increased the percentage of deleterious changes (P < 0.05, χ2; Figure 4F). In summary, inosine mRNA modifications were associated with increased tolerated, synonymous RNA changes in oocytes, eggs, and somatic tissues.
Enrichment of inosine RNA modifications at the codon wobble position
The abundance of synonymous changes caused by inosine RNA modifications led us to investigate transcriptome codon usage. GV oocytes and MII eggs had 22 and 11 codons, respectively, that were inosine modified more frequently than in PND12 oocytes (Figure 5A; P < 0.05, two-way ANOVA). The difference inprevalence of inosine-modified codons across oocyte stages was not due to changes in abundance of codons (codon bias) within the transcriptome (Supplementary Figure S7). Inosine modifications were enriched at the wobble position of codons in GV oocytes and MII eggs, while PND12 oocytes showed no codon position enrichment (Figure 5B; P < 0.05, one-way ANOVA). Somatic tissues also displayed enrichment of inosine RNA modifications at the wobble position (Figure 5C; P < 0.05, one-way ANOVA). To test if the observed enrichment of inosine at the wobble position was a bonafide inosine signature, we compared brain tissue from control Adar+/+ and AdarE861A/E861A mice. The AdarE861A/E861A brain tissue lacked enrichment of inosines at the wobble position as observed in control Adar+/+ brain tissue (Supplementary Figure S3D; P < 0.05, one-way ANOVA).
Figure 5.
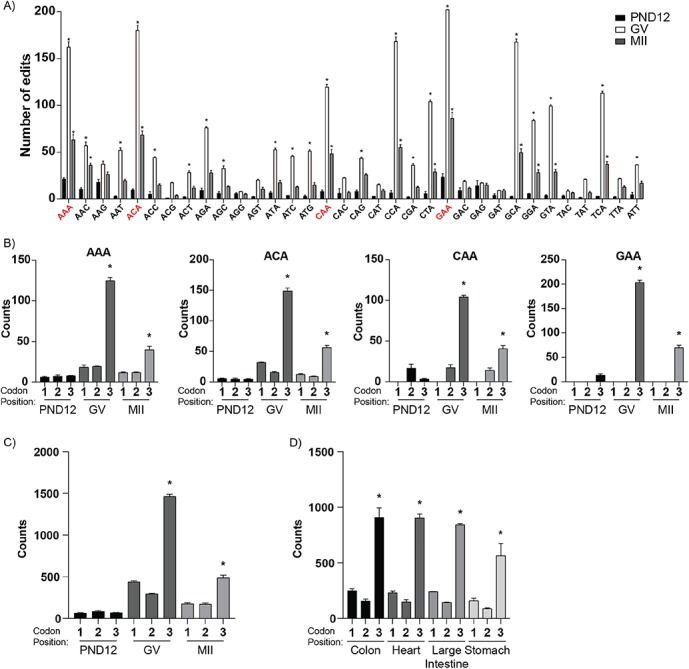
Inosines are enriched at the wobble position of codons. (A) Number of inosines in adenosine-containing codons in PND12, GV, and MII samples. (B) Number of inosines occurring at specific sites among the codons with multiple adenosines that were statistically different in PND12 versus GV and MII samples. Total number of inosine mRNA modifications occurring at the first, second, or third codon position in: (C) PND12, GV, and MII samples, (D) somatic tissues, and (E) brain tissue of Adar+/+ and AdarE861A/E861A mice. *Means ± SEM within a panel were different (P < 0.05); significance was determined using two-way ANOVA tests. Only codons from transcripts with TPM ≥ 1 were analyzed.
Wobble position inosines were enriched in codons with multiple adenosines such as the AAA, ACA, CAA, and GAA codons (Figure 5E; P < 0.05, one-way ANOVA). Among the codons with adenosines at the first and second position, such as AAC, AAG, and AAT, we did not find a site-specific enrichment of inosine modifications (Supplementary Figure S8A). Moreover, evidence for the wobble position inosine RNA modifications in GV oocytes and MII eggs was not dependent upon read depth (Supplementary Figure S8B). We further analyzed previously validated inosine RNA modifications curated by the RADAR database [49], which yielded eight synonymous changes that occur in mice, all of which have been validated in the literature [50–52]. All eight of these substitutions occurred at the codon wobble position (Supplementary Table S1).
Transcriptome-wide inosine RNA modification efficiency at the codon wobble position was highest in GV oocytes (0.481 ± 0.002; mean ± SEM) and MII eggs (0.488 ± 0.004) compared to PND12 oocytes (0.377 ± 0.014; P < 0.05, one-way ANOVA; Supplementary Figure S6D). The inosine RNA modification efficiency at the wobble position of AAA, CAA, and GAA codons was also highest in GV oocytes and MII eggs compared to PND12 oocytes (P < 0.05, two-way ANOVA; Supplementary Figure S6E). Conversely, there was no decrease in inosine RNA modification efficiency at the wobble position of the codon ACA (Supplementary Figure S6E). Overall, fully grown GV oocytes and MII eggs, as well as somatic tissue, showed an inosine RNA modification pattern with enrichment at the codon wobble position that was dependent on ADAR deamination activity.
Discussion
In our studies of mouse oocyte growth and maturation, we identified an enrichment of inosine modifications at the wobble position. Codon-specific inosine RNA modifications were not limited to oocytes but were found in a variety of somatic tissues as well. This observation of an enrichment of inosines at the wobble position has likely not yet been reported because most cells or tissues examined do not have as high a proportion of CDS modifications as mouse oocytes. Furthermore, many studies examining A-to-I RNA editing utilize microfluidics-based multiplex PCR and deep sequencing (mmPCR–seq) [45,53]. mmPCR-seq interrogates inosines at 11103 exonic sites across 557 different loci and will not discover any new inosine sites. Another common method of examining A-to-I RNA editing focuses on hyper-modified sites, where detection of multi-mapped reads from repetitive elements is allowed [54,55]. Because of the difficulties in cataloging multi-mapped reads, we avoided this approach. Our strategy, in combination with the frequent CDS inosine RNA modifications of mouse oocytes, is what led to our novel discovery of wobble position inosine RNA modifications.
During the growth phase of oocytes, RNA accumulates and is stored to support meiotic maturation, fertilization, and early embryonic growth prior to embryonic genome activation [1,2,56,57]. We observed low Adar expression and no detectable protein in transcriptionally active PND12 oocytes. This correlated with a reduced overall number of inosine containing transcripts, a reduced number of inosine sites per transcript, and a decrease in overall inosine modification efficiency. In contrast, transcriptionally quiescent fully grown GV oocytes and MII eggs contained abundant ADAR protein and increased levels of inosine RNA modifications.
The abundance of inosine RNA modifications occurring within the CDS and 3′ UTR regions of oocyte and egg mRNA was unique compared to mouse somatic cell types; however, it is not unprecedented [58]. Cephalopods have a high proportion of inosine modifications occurring in the CDS region of mRNA, and this has been hypothesized to increase proteome diversity and allow for increased adaptability [59]. Porath et al. also documented increased inosine RNA modifications in the gametes of stony coral, Acropora millepora [60], indicating that this post-transcriptional mechanism could be important during early embryogenesis. A study of human oocytes identified that the majority of inosine modifications occurred in the 3′ UTR regions (47.12%), followed by intronic (33.77%), non-coding RNA (17.12%), and 5′ UTR regions (1.01%), with only 0.98% occurring in coding regions [13]. Conversely, a recent study of mouse oocytes using an analysis pipeline different than ours also identified a high proportion of inosine modifications within the CDS region (48.69%), followed by intronic (35.70%) and 3′ UTR (15.61%; [14]). It is unclear if the increased CDS inosine RNA modifications we and Wang et al. observed in mice oocytes, that were not observed in human oocytes, are due to species differences or due to the vitrification process and culture of human oocytes prior to RNA-seq [61]. A recent cross-species analysis of inosine modifications in somatic tissues revealed that species, rather than tissue type, was a greater source of inosine modification differences [45]. Male mouse germ cells also display a unique inosine modification environment, with the majority of inosines occurring as a result of ADARB1 activity, rather than ADAR [62].
In an analysis of high confidence inosine modifications occurring in human cells, Xu et al. calculated that inosine sites resulting in synonymous changes were significantly more common than those resulting in nonsynonymous changes [11]. Our data in mouse oocytes are consistent with this observation. Synonymous changes are known to affect RNA transcript stability as well as translational efficiency, because of tRNA availability and codon usage [63,64]. Of the RNA codons with adenosines, we observed that GV oocytes and MII eggs contained an increased enrichment of inosines at the codon wobble position compared to PND12 oocytes (Figure 5). Of the codons with multiple adenosines, three of four codons showed a decrease in inosine RNA modification efficiency in PND12 oocytes (Supplementary Figure S6E), consistent with the absence of ADAR in these oocytes. Lastly, brain tissue from AdarE861A/E861A mice displayed reduced inosine RNA modifications at the codon wobble position (Supplementary Figure S3D). Therefore, wobble position inosine modifications are not an oocyte-specific phenomenon and depend on ADAR deamination activity.
The molecular mechanism leading to enrichment of inosines at the codon wobble position remains to be explored. Codon-specific RNA modifications are not unprecedented. The RNA modification at cytidine, N4-acetylcytidine, is enriched at the codon wobble position and can affect translational efficiency [65]. Whether inosines at the wobble position have a similar role in translational efficiency in vivo remains to be examined. A recent report by Licht et al. demonstrated that the presence of inosine within mRNA can affect translational efficiency in an in vitro system [47]. In vitro models using purified ADAR and synthetic RNA have demonstrated that ADAR has a trinucleotide preference and specifically modifies the second adenosine nucleotide of the triplet [66]. However, our analysis of CDS inosine RNA modifications using in vivo genetic models of catalytically inactive ADAR demonstrate that within the CDS, inosine frequently occurs at the codon wobble position. Most studies of inosine do not focus on the CDS, and therefore, the wobble position inosine modification has gone unreported. It is possible that mRNA from transcriptionally quiescent oocytes are inosine modified at the first and second position of a codon, but degraded rapidly, resulting in an increase of mRNA with inosine modifications at the codon wobble position. However, even in transcriptionally active somatic cells, we observed inosine more frequently at the codon wobble position. Further experiments are needed to understand the molecular mechanisms leading to inosine modifications at the codon wobble position.
We hypothesize that inosine modifications at the wobble position can affect translational efficiency and subsequently mRNA stability in oocytes through codon usage. In fully grown GV oocytes and MII eggs, previously synthesized maternal mRNA are recruited for translation [67,68]. Maternal transcripts are subjected to translationally coupled degradation, and in the absence of degradation, maternal transcripts accumulate, resulting in embryonic arrest [68]. Thus, the regulatory roles of post-transcriptional and translational control of oocytes are essential to ensure successful embryonic development. Our data suggest a novel mechanism whereby inosine modifications could alter codon usage, which may affect translational efficiency, and ultimately translationally coupled mRNA degradation. Further experimentation will reveal the effects of inosine codon modifications on translation in oocytes.
Supplemental methods
Detailed line commands for elprep and genome analysis tool kit
The program elprep was used to sort, mark duplicates, and index RNA-seq reads with the following commands; elprep --reference-T mm10.fasta --filter-unmapped-reads strict --replace-reference-sequences mm10.dict --replace-read-group “ID:id LB:library PL:illumina PU:machine SM:Or” --mark-duplicates --sorting-order coordinate --nr-of-threads 30 (S7) . GATK RNA-seq variant pipeline was run on the output of elprep with the following sequentially executed commands; GenomeAnalysisTK.jar -T SplitNCigar Reads -R mm10.fa -I infile -o outfile -rf ReassignOneMappingQuality -U ALLOW_N_CIRGAR_READS, GenomeAnalysisTK.jar -T RealignerTargetCreator -R mm10.fa -I infile -known snp.vcf -o outfile -U ALLOW_N_CIGAR_READS, GenomeAnalysisTK.jar -T IndelRealigner -R mm10.fa -I infile -targetIntervals targetfile -know snp.vcf -o outfile -U ALLOW_N_CIGAR_READS, GenomeAnalysisTK.jar -T BaseReaclibrator -I infile -R mm10.fa -knownSites snp.vcf -o outfile, GenomeAnalysisTK.jar -T PrintReads -R mm10.fa -I infile -BQSR infile -o outfile, GenomeAnalysisTK.jar -T HaplotypeCaller -R mm10.fa -I infile -dontUseSoftClippedBases -stand_call_conf 20.0 -o outfile, GenomeAnalysisTK.jar -T VariantFiltration -R mm10.fa -V infile -window 50 -cluster 3 filterName FS -filter “FS > 30” -filterName QD “QD < 2.0” -o outfile.vcf. VEP command vep -i infiole --species “mus_musculus” --cache mus_musculus/ --refseq --all_refseq --tab –everything.
Inosine RNA modification false-positive rate
To determine the false-positive rate, 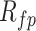 , inherent to our analysis, we used a defined approach [46,69], with minor modifications. Briefly, we assumed that A-to-G / T-to-C and C-to-T / G-to-A variants represent true RNA modifications with known enzymatic sources [70,71]. The remaining eight single nucleotide substitutions were considered background due to the lack of any known enzymatic mechanisms and the observed equal frequency of occurrence within an oocyte stage. The error rate was computed with the following equation:
, inherent to our analysis, we used a defined approach [46,69], with minor modifications. Briefly, we assumed that A-to-G / T-to-C and C-to-T / G-to-A variants represent true RNA modifications with known enzymatic sources [70,71]. The remaining eight single nucleotide substitutions were considered background due to the lack of any known enzymatic mechanisms and the observed equal frequency of occurrence within an oocyte stage. The error rate was computed with the following equation:
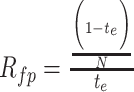
where N is the number of single nucleotide substitution types and te is a proportion calculated with the following equation:
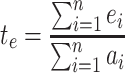 |
where ei is the summation of the A-to-G, T-to-C, C-to-T, and G-to-A RNA modification events occurring within genes, and ai is the summation of all other single nucleotide substitutions within genes.
Inosine RNA modification efficiency
To determine inosine RNA modification efficiency, 0 < 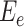 ≤ 1, the inosine RNA modification events were modeled with the following equation:
≤ 1, the inosine RNA modification events were modeled with the following equation:
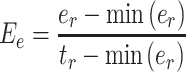 |
where 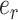 is the total number of reads containing the inosine,
is the total number of reads containing the inosine, 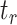 is the total number of reads at the inosine site, and
is the total number of reads at the inosine site, and 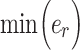 is the minimum number of reads that can be inosine modified at the inosine site, which was set to 1 for this analysis.
is the minimum number of reads that can be inosine modified at the inosine site, which was set to 1 for this analysis.
Conflict of interest
N.S.A. is owner of De Novo Genomics. All other authors declare that they have no conflicting financial interests.
Supplementary Material
Author contributions: P.B., N.S.A., and L.K.C. designed the research; P.B., N.S.A., X.H., S.G., and K.A.V. performed the research; P.B., N.S.A., K.E.L., and L.K.C. analyzed the data; P.B., N.S.A., and L.K.C. wrote the manuscript.
References
- [1]. Piko L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol 1982; 89:362–378. [DOI] [PubMed] [Google Scholar]
- [2]. De La Fuente R, Eppig JJ. Transcriptional activity of the mouse oocyte genome: companion granulosa cells modulate transcription and chromatin remodeling. Dev Biol 2001; 229:224–236. [DOI] [PubMed] [Google Scholar]
- [3]. Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev 2007; 21:682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. O’Connell M. RNA modification and the epitranscriptome; the next frontier. RNA 2015; 21:703–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 2016; 17:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Nishikura K, Sakurai M, Ariyoshi K, Ota H. Antagonistic and stimulative roles of ADAR1 in RNA silencing. RNA Biol 2013; 10:1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Solomon O, Oren S, Safran M, Deshet-Unger N, Akiva P, Jacob-Hirsch J, Cesarkas K, Kabesa R, Amariglio N, Unger R, Rechavi G, Eyal E. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR). RNA 2013; 19:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. McLaughlin PJ, Keegan LP. Conflict RNA modification, host-parasite co-evolution, and the origins of DNA and DNA-binding proteins1. Biochem Soc Trans 2014; 42:1159–1167. [DOI] [PubMed] [Google Scholar]
- [9]. Liddicoat BJ, Chalk AM, Walkley CR. ADAR1, inosine and the immune sensing system: distinguishing self from non-self. WIRES RNA 2016; 7:157–172. [DOI] [PubMed] [Google Scholar]
- [10]. Rosenthal JJ, Seeburg PH. A-to-I RNA editing: effects on proteins key to neural excitability. Neuron 2012; 74:432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Xu G, Zhang J. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A 2014; 111:3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann H-H, Rosenberg BR, Rice CM. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 2018; 172:811–824.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13]. Qiu S, Li W, Xiong H, Liu D, Bai Y, Wu K, Zhang X, Yang H, Ma K, Hou Y, Li B. Single-cell RNA sequencing reveals dynamic changes in A-to-I RNA editome during early human embryogenesis. BMC Genomics 2016; 17:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Wang LY, Guo J, Cao W, Zhang M, He J, Li Z. Integrated sequencing of exome and mRNA of large-sized single cells. Sci Rep 2018; 8:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Nishikura K, Yoo C, Kim U, Murray JM, Estes PA, Cash FE, Liebhaber SA. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J 1991; 10:3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988; 55:1089–1098. [DOI] [PubMed] [Google Scholar]
- [17]. Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A 1989; 86:2647–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18]. Cho D-SC, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem 2003; 278:17093–17102. [DOI] [PubMed] [Google Scholar]
- [19]. Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A 1994; 91:11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature 1996; 379:460–464. [DOI] [PubMed] [Google Scholar]
- [21]. Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 2004; 279:4894–4902. [DOI] [PubMed] [Google Scholar]
- [22]. Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 2004; 279:4952–4961. [DOI] [PubMed] [Google Scholar]
- [23]. Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellaker C, Vesely C, Ponting CP, McLaughlin PJ, Jantsch MF, Dorin J et al. . The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 2014; 9:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000; 406:78–81. [DOI] [PubMed] [Google Scholar]
- [25]. Zhu Y, Stephens RM, Meltzer PS, Davis SR. SRAdb: query and use public next-generation sequencing data from within R. BMC Bioinform 2013; 14:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Pfender S, Kuznetsov V, Pasternak M, Tischer T, Santhanam B, Schuh M. Live imaging RNAi screen reveals genes essential for meiosis in mammalian oocytes. Nature 2015; 524:239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 2015; 16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S-I, Smallwood SA, Chen T, Kelsey G. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev 2015; 29:2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Heraud-Farlow JE, Chalk AM, Linder SE, Li Q, Taylor S, White JM, Pang L, Liddicoat BJ, Gupte A, Li JB, Walkley CR. Protein recoding by ADAR1-mediated RNA editing is not essential for normal development and homeostasis. Genome Biol 2017; 18:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016; 34:525–527. [DOI] [PubMed] [Google Scholar]
- [32]. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A et al. . Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 2017; 14:687–690. [DOI] [PubMed] [Google Scholar]
- [34]. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol 2016; 17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 2013; 10:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Yalcin B, Wong K, Agam A, Goodson M, Keane TM, Gan X, Nellåker C, Goodstadt L, Nicod J, Bhomra A, Hernandez-Pliego P, Whitley H et al. . Sequence-based characterization of structural variation in the mouse genome. Nature 2011; 477:326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. R Core Team. R: A Language and Environment for Statistical Computing; 2013. http://www.R-project.org/. Accessed date: 1/2019–5/2019
- [42]. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 1st ed. New York: Springer-Verlag; 2009. [Google Scholar]
- [43]. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009; 4:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44]. Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 2015; 43:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, Gupte A, Keegan LP et al. . Dynamic landscape and regulation of RNA editing in mammals. Nature 2017; 550:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Yu Y, Zhou H, Kong Y, Pan B, Chen L, Wang H, Hao P, Li X. The Landscape of A-to-I RNA Editome Is Shaped by Both Positive and Purifying Selection. PLoS Genet 2016; 12:e1006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Licht K, Hartl M, Amman F, Anrather D, Janisiw MP, Jantsch MF. Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res 2019; 47: 3–14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48]. Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31:3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Gu T, Buaas FW, Simons AK, Ackert-Bicknell CL, Braun RE, Hibbs MA Canonical A-to-I and C-to-U RNA editing is enriched at 3′UTRs and microRNA target sites in multiple mouse tissues. PLoS One 2012; 7:e33720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Danecek P, Nellåker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, Flint J, Durbin R, Keane TM, Adams DJ. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol 2012; 13:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Cattenoz PB, Taft RJ, Westhof E, Mattick JS. Transcriptome-wide identification of A > I RNA editing sites by inosine specific cleavage. RNA 2013; 19:257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Zhang R, Li X, Ramaswami G, Smith KS, Turecki G, Montgomery SB, Li JB. Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nat Methods 2014; 11:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015; 349:1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Porath HT, Carmi S, Levanon EY. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun 2014; 5:4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Bachvarova R, De Leon V, Johnson A, Kaplan G, Paynton BV. Changes in total RNA, polyadenylated RNA, and actin mRNA during meiotic maturation of mouse oocytes. Dev Biol 1985; 108:325–331. [DOI] [PubMed] [Google Scholar]
- [57]. De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, Eppig JJ. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol 2004; 275:447–458. [DOI] [PubMed] [Google Scholar]
- [58]. Huntley MA, Lou M, Goldstein LD, Lawrence M, Dijkgraaf GJ, Kaminker JS, Gentleman R. Complex regulation of ADAR-mediated RNA-editing across tissues. BMC Genomics 2016; 17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59]. Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJC, Eisenberg E. Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods. Cell 2017; 169:191–202.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Porath HT, Schaffer AA, Kaniewska P, Alon S, Eisenberg E, Rosenthal J, Levanon EY, Levy O. A-to-I RNA Editing in the Earliest-Diverging Eumetazoan Phyla. Mol Biol Evol 2017; 34:1890–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Xue Z, Huang K, Cai C, Cai L, Jiang C-Y, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu J-Y, Horvath S et al. . Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013; 500:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62]. Snyder EM, Licht K, Braun RE. Testicular adenosine to inosine RNA editing in the mouse is mediated by ADARB1. Biol Reprod 2017; 96:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Presnyak V, Alhusaini N, Chen Y-H, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J. Codon optimality is a major determinant of mRNA stability. Cell 2015; 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64]. Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 2016; 35:2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, Fox SD, Zengeya TT et al. . Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 2018; 175:1872–1886.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry 2000; 39:12875–12884. [DOI] [PubMed] [Google Scholar]
- [67]. Romasko EJ, Amarnath D, Midic U, Latham KE. Association of maternal mRNA and phosphorylated EIF4EBP1 variants with the spindle in mouse oocytes: localized translational control supporting female meiosis in mammals. Genetics 2013; 195:349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Yu C, Ji S-Y, Sha Q-Q, Dang Y, Zhou J-J, Zhang Y-L, Liu Y, Wang Z-W, Hu B, Sun Q-Y, Sun S-C, Tang F et al. . BTG4 is a meiotic cell cycle-coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol 2016; 23:387–394. [DOI] [PubMed] [Google Scholar]
- [69]. Ouyang Z, Ren C, Liu F, An G, Bo X, Shu W. The landscape of the A-to-I RNA editome from 462 human genomes. Sci Rep 2018; 8:12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70]. Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, Baysal BE. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun 2015; 6:6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71]. Harjanto D, Papamarkou T, Oates CJ, Rayon-Estrada V, Papavasiliou FN, Papavasiliou A. RNA editing generates cellular subsets with diverse sequence within populations. Nat Commun 2016; 7:12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


