A transcription factor-cysteine protease regulatory module is associated with programmed cell death and reactive oxygen species accumulation in nodule senescence.
Abstract
Legume-rhizobia symbiosis is a time-limited process due to the onset of senescence, which results in the degradation of host plant cells and symbiosomes. A number of transcription factors, proteases, and functional genes have been associated with nodule senescence; however, whether other proteases or transcription factors are involved in nodule senescence remains poorly understood. In this study, we identified an early nodule senescence mutant in Medicago truncatula, denoted basic helix-loop-helix transcription factor2 (bhlh2), that exhibits decreased nitrogenase activity, acceleration of plant programmed cell death (PCD), and accumulation of reactive oxygen species (ROS). The results suggest that MtbHLH2 plays a negative role in nodule senescence. Nodules of wild-type and bhlh2-TALEN mutant plants at 28 d postinoculation were used for transcriptome sequencing. The transcriptome data analysis identified a papain-like Cys protease gene, denoted MtCP77, that could serve as a potential target of MtbHLH2. Electrophoretic mobility shift assays and chromatin immunoprecipitation analysis demonstrated that MtbHLH2 directly binds to the promoter of MtCP77 to inhibit its expression. MtCP77 positively regulates nodule senescence by accelerating plant PCD and ROS accumulation. In addition, the expression of MtbHLH2 in the nodules gradually decreased from the meristematic zone to the nitrogen fixation zone, whereas the expression of MtCP77 showed enhancement. These results indicate that MtbHLH2 and MtCP77 have opposite functions in the regulation of nodule senescence. These results reveal significant roles for MtbHLH2 and MtCP77 in plant PCD, ROS accumulation, and nodule senescence, and improve our understanding of the regulation of the nodule senescence process.
Legumes play an important role in sustainable agriculture by increasing crop yields due to their symbiotic association with rhizobia (Fox et al., 2007). Symbiotic nitrogen fixation occurs in special organs, named nodules, and these organs provide a suitable microenvironment for supplying the carbon obtained from photosynthesis to rhizobia. Rhizobia can fix nitrogen by nitrogenase to provide ammonium to legumes (Stonoha-Arther and Wang, 2018). The late stage in the development of nodules, including determinate and indeterminate nodules, regulates their fate, namely senescence. Determinate nodules have nonpersistent meristems and result from the development of radial gradients, as has been observed in Glycine max, Lotus japonicus, Phaseolus vulgaris, and Vigna unguiculata. The senescence of determinate nodules begins at the nodule center and gradually extends to the periphery (Puppo et al., 2005). Indeterminate nodules, such as those in Medicago truncatula, Pisum sativum, Vicia faba, and Trifolium repens, contain five different zones: zone I (meristematic zone), zone II (infection zone), zone III (nitrogen fixation zone), zone IV (senescence zone), and zone V. Zone IV does not exist during the early stages of nodule differentiation but appears with nodule development. In addition, zone IV increases in size with increased age of the host plant and triggers the degradation of symbiosomes in response to aging or environmental stress. Zone V is proximal to zone IV and shows ultra-structural features of free-living bacteria (Timmers et al., 2000; Dupont et al., 2012). During nodule senescence, the color of the nitrogen fixation zone is converted from pink, which is associated with functional leghemoglobin, to green, which is related to nitration of the heme group of leghemoglobin (Navascués et al., 2012). At the ultrastructural level, the following changes in cell organelles and symbiosomes have been observed: the electron density of the cytoplasm becomes progressively lower, the loose plasma membrane separates from the cell wall, the cell wall becomes damaged, bacteroids undergo lysis, and the peribacteroid membrane is degraded (Timmers et al., 2000; Van de Velde et al., 2006). Another study reported the existence of a conical front in the senescence zone of indeterminate nodules (Pérez Guerra et al., 2010). The early signs of senescence occur in some infected cells in the center of the nitrogen fixation zone and then gradually extended to the proximal cell layers of the nodule periphery, leading to the formation of a conical front during nodule growth (Pérez Guerra et al., 2010).
At the senescence stage, several kinases, nodule-specific Cys-rich (NCR) peptides, membrane proteins, and other functional genes in L. japonicus or M. truncatula are involved in nodule senescence. MtSymCRK encodes a Cys-rich receptor-like kinase of the non-Arg Asp (nonRD) family and is required to prevent nodule senescence and to avoid defense-like reactions (Berrabah et al., 2014). DEFECTIVE NITROGEN FIXATION7 (MtDNF7) encodes NCR169, which contributes to bacteroid differentiation and is associated with early senescence of the symbiotic cells (Horváth et al., 2015). Two mutants of MtNCR211, early senescent nodule1 (esn1) and dnf4, both present spherically shaped nodules, ineffective nitrogen fixation, and premature senescence (Xi et al., 2013; Kim et al., 2015). NITROGEN FIXATION SPECIFICITY1 (MtNFS1) and MtNFS2 encode NCR peptides and negatively regulate nitrogen fixation symbiosis in a rhizobial strain-specific manner (Wang et al., 2017; Yang et al., 2017; Wang et al., 2018). SYMBIOTIC SULFATE TRANSPORTER1 (LjSST1) is a nodule-specific sulfate transporter that is localized on the symbiosome membrane (Krusell et al., 2005). INEFFECTIVE GREENISH NODULES1 (LjIGN1) encodes an ankyrin-repeat membrane protein that is essential for the differentiation or persistence of bacteroids and symbiosomes (Kumagai et al., 2007). STATIONARY ENDOSYMBIONT NODULE1 (LjSEN1) is an integral membrane protein required for nitrogen fixation activity and the differentiation of bacteroids or symbiosomes (Hakoyama et al., 2012). REGULATOR OF SYMBIOSOME DIFFERENTIATION (MtRSD) encodes a Cys-2/histone-2 (C2H2) transcription factor that inhibits the expression of VESICLE-ASSOCIATED MEMBRANE PROTEIN721A (MtVAMP721a), which indicates that it influences nitrogen fixation and the differentiation of symbiosomes and bacteroids (Sinharoy et al., 2013). NODULES WITH ACTIVATED DEFENSE1 (MtNAD1) is a small uncharacterized protein localized in the endoplasmic reticulum, which indicates that it plays a positive role in the maintenance of rhizobial endosymbiosis (Wang et al., 2016; Domonkos et al., 2017). MtDNF2 encodes a phosphatidylinositol-specific phospholipase C X domain (PI-PLC-XD) protein, and its mutants exhibit fewer cell layers in the infection zone, where bacteria do not differentiate, and rapid senescence (Bourcy et al., 2013). ASPARTIC PEPTIDASE NODULE-INDUCED1 (LjAPN1) is a nepenthesin-type aspartic peptidase that plays a necessary role in nitrogen fixation symbiosis in a rhizobial strain-dependent manner (Yamaya-Ito et al., 2018). M. truncatula has five nodule-specific PLAT (polycystin-1, lipoxygenase, alpha toxin) domain proteins (NPDs), and lines with different mutation combinations presented smaller nodules, earlier nodule senescence, or ineffective nodules (Trujillo et al., 2019).
In addition to various mutations in symbiotic genes, nodule senescence is also affected by diverse environmental stresses, such as drought, salt, cold, continuous darkness, and nitrate (Dupont et al., 2012), and previous studies have indicated that different stresses accelerate nodule senescence (Pérez Guerra et al., 2010; Dhanushkodi et al., 2018). Moreover, phytohormones, such as abscisic acid, ethylene, and jasmonic acid (JA), might positively regulate nodule senescence (Puppo et al., 2005; Van de Velde et al., 2006; Karmarkar, 2014; Guinel, 2015). The positive role of abscisic acid induces nodule senescence (González et al., 2001; Puppo et al., 2005) and the expression of some senescence-associated genes (SAGs; Fukudome et al., 2019). The positive roles of ethylene and JA have been suggested by the induction of ETHYLENE RESPONSE FACTOR (ERF) transcription factors and genes related to ethylene and JA biosynthesis (Van de Velde et al., 2006; Serova et al., 2017). Ethylene can also trigger plant defense responses and results in necrosis that colocalizes with infected cells by rhizobia (Berrabah et al., 2018). GAs play a negative role in nodule senescence (Van de Velde et al., 2006; Hayashi et al., 2014; Serova et al., 2019), and nitric oxide positively regulates nodule senescence (Cam et al., 2012; Berger et al., 2019).
A pivotal characteristic of the nodule senescence process is the involvement of proteolytic activity, which can act in the degradation of misfolded or modified proteins (Pladys and Vance, 1993). Cys protease (CP; EC3.4.22) uses catalytic Cys as a nucleophile during proteolysis (van der Hoorn, 2008). Most plant Cys proteases belong to the papain (C1) and legumain (C13) families, and metacaspases (family C14) and calpains (family C2, calcium-dependent proteases) have been detected in plants (Grudkowska and Zagdańska, 2004). The plant papain-like Cys proteases (PLCPs) are the most researched of all Cys proteases. PLCP genes are significantly induced in senescent nodules of G. max, Astragalus sinicus, P. sativum, and M. truncatula (Fedorova et al., 2002), such as GmCYSP1 (Alesandrini et al., 2003; Oh et al., 2004), AsNodf32 (Naito et al., 2000), PsCYP1, and PsCYP15A (Kardailsky and Brewin, 1996; Vincent and Brewin, 2000). MsCYP15A, AsNodf32, and MtCP6 belong to the PLCP family and positively regulate nodule senescence (Sheokand et al., 2005; Li et al., 2008; Pierre et al., 2014). The senescence of root nodules is modulated by transcription factors. To date, three transcription factors, namely MtATB2, MtNAC969, and MtNAC920, have been found to be involved in nodule senescence. MtATB2, which encodes a bZIP transcription factor, is more highly expressed in senescent nodules (D’haeseleer et al., 2010). The knockdown of MtNAC969 results in a higher number of amyloplasts in the nitrogen fixation zone and thereby triggers premature senescence (de Zélicourt et al., 2012). MtNAC920, which is induced by nitrate and ethylene, serves as a positive regulator of nodule senescence and directly targets MtCP2, which was previously reported to be a marker gene for nodule senescence (Karmarkar, 2014). However, whether other proteases or transcription factors are involved in nodule senescence remains unclear.
In this study, we demonstrate that a basic helix-loop-helix (bHLH) transcription factor in M. truncatula, named MtbHLH2, is a key regulatory component that negatively regulates nodule senescence. Moreover, we show that MtbHLH2 binds to the promoter of the Cys protease MtCP77 to repress its expression and reveal that MtCP77 plays a positive role in nodule senescence.
RESULTS
MtbHLH2 Is a Negative Regulator of Nodule Senescence
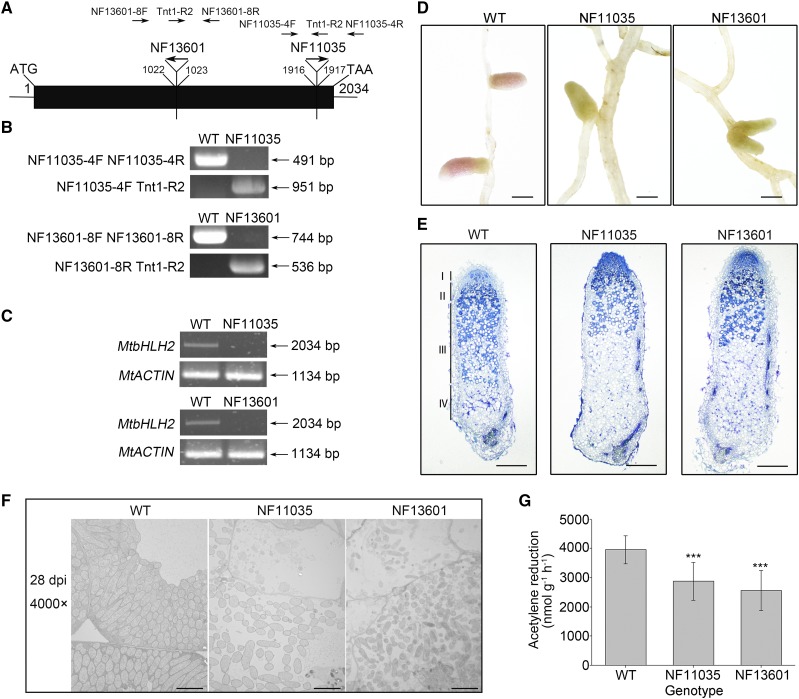
The M. truncatula mutant line NF11035 with early nodule senescence (Fig. 1D) was isolated from the M. truncatula Tnt1 insertion mutant database by forward genetic screening. To identify which gene was involved in nodule senescence, 16 flanking sequence tags (FSTs) available in the M. truncatula Mutant Database at the Noble foundation for this mutant line were analyzed. Only one Tnt1 FST inserted between 1916 and 1917 bp of Medtr5g030430 was linked to the phenotype, and this gene was denoted basic helix-loop-helix transcription factor2 (MtbHLH2; Fig. 1, A and B). We then selected another Tnt1 insertion mutant (NF13601) of MtbHLH2, in which the Tnt1 FST was located between 1022 and 1023 bp of MtbHLH2 (Fig. 1, A and B). The phenotype of NF13601 was consistent with that of NF11035 and was associated with prematurely senescent nodules (Fig. 1D). The MtbHLH2 full-length transcripts in NF11035 and NF13601 were not detected by RT-PCR, which suggested that the expression of MtbHLH2 was impaired (Fig. 1C).
Figure 1.
The mutation of MtbHLH2 results in early-nodule senescence and decreased nitrogen fixation. A, Schematic diagram of the Tnt1 insertion site in the MtbHLH2 gene. The box shows the exon, and this gene has no intron. The orientations of Tnt1 are indicated. B, Identification of homozygous lines of Tnt1-insertion mutants (NF11035 and NF13601). The insertion site analysis was performed at the DNA level. C, RT-PCR analysis of MtbHLH2 transcripts in the wild type (WT) and bhlh2 Tnt1-insertion mutants. MtACTIN was used as the loading control. D, Light micrographs of wild-type and bhlh2 Tnt1-insertion mutant nodules at 28 dpi. Bars = 1 mm. E, Longitudinal sections of toluidine blue-stained wild-type and bhlh2 Tnt1-insertion mutant nodules at 28 dpi. The section thickness is 8 μm. The different zones of the nodules (I–IV) are shown. I, Meristematic zone; II, infection zone; III, nitrogen fixation zone; IV, senescence zone. Bars = 200 μm. F, Electron micrographs of longitudinal ultrathin sections of wild-type and bhlh2 Tnt1-insertion mutant nodule cells from the nitrogen fixation zone at 28 dpi (4000× magnification). Bars = 5 μm. G, The nitrogenase activity of wild type and bhlh2 Tnt1-insertion mutant nodules (NF11035 and NF13601) at 28 dpi was determined by an ARA. The error bars represent the ± sd of the means, and asterisks indicate a significant difference between the wild-type and bhlh2 Tnt1-insertion mutant nodules (one-way ANOVA followed by Tukey’s post-hoc test, ***P < 0.001, n = 19). Similar results were obtained from three independent experiments.
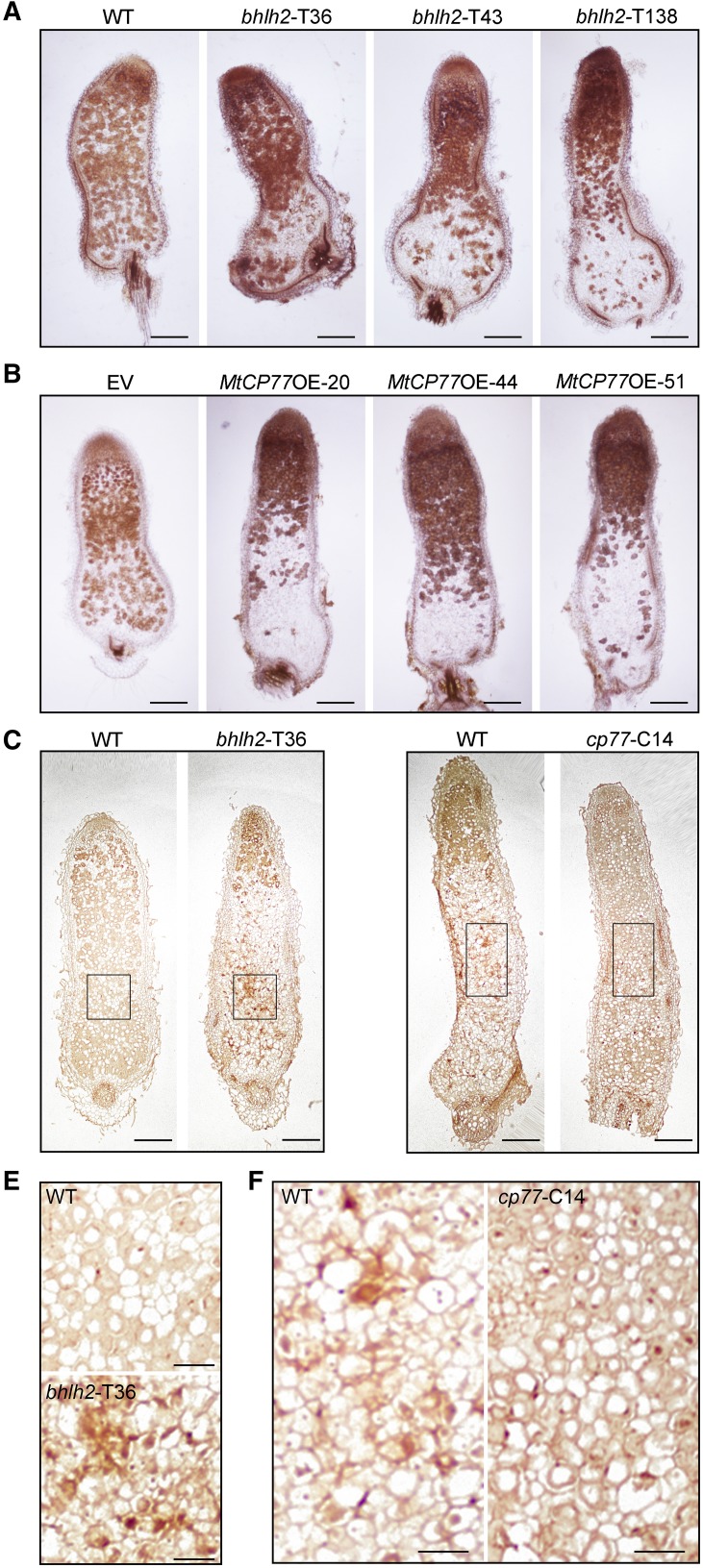
To elucidate the influences of the MtbHLH2 mutation on nodule senescence, the nodules of the wild-type and bhlh2 mutant were harvested at 28 d postinoculation (dpi), sectioned, stained with toluidine blue, and observed. Compared with the pattern observed in the wild type, most of the senescence zone and a small part of the nitrogen fixation zone were found in the bhlh2 nodules (Fig. 1E). To investigate the effects of the MtbHLH2 mutation at the ultrastructural level, the nitrogen fixation zone of the wild type and bhlh2 nodules was observed at 28 dpi with a transmission electron microscope (TEM). In agreement with the light microscopy observations, the nitrogen fixation zone was substantially modified in the bhlh2 mutants. In the wild-type nodules, symbiosomes completely filled the infected cells, whereas in the mutant nodules, symbiosomes appeared to be loosely placed within the infected cells, the host cells died, and the symbiosomes decayed (Fig. 1F). This result suggested that the mutation of MtbHLH2 might promote host cell death and symbiosome decay. The nitrogenase activity of the wild type and bhlh2 mutant nodules at 28 dpi was measured through an acetylene reduction assay (ARA). The nitrogenase activity of the bhlh2 mutants was significantly lower than that of the wild type, which suggested that MtbHLH2 affects the efficiency of symbiotic nitrogen fixation (Fig. 1G).
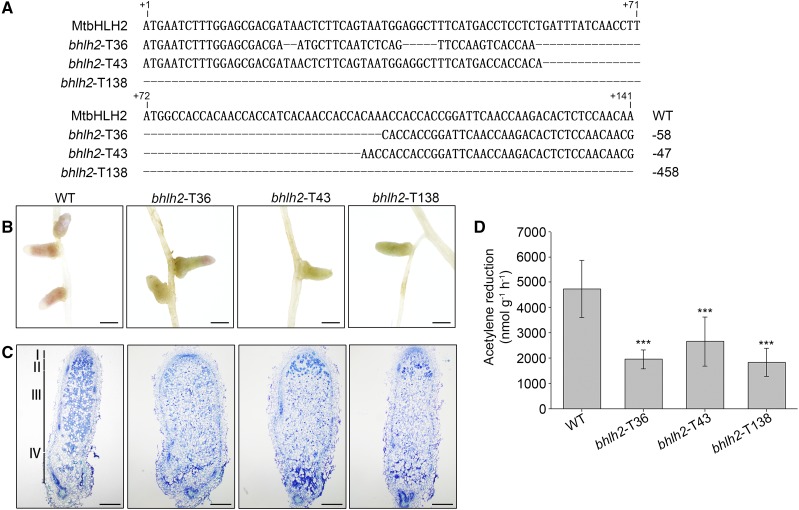
To further determine the function of MtbHLH2 in nodule senescence, MtbHLH2-knockout mutants were obtained using transcription activator-like effector nuclease (TALEN) technology. Stable transgenic lines were generated by Agrobacterium tumefaciens-mediated transformation and identified by sequencing. The bhlh2-T36- and bhlh2-T43-knockout mutants containing 58- and 47-bp deletions, respectively, triggered the frameshift mutation (Fig. 2A). The bhlh2-T138-knockout mutant caused the deletion of a large fragment, which included the transcriptional start site (ATG; Fig. 2A). The phenotype of the bhlh2-TALEN mutants was similar to that of the Tnt1-insertion mutants (Fig. 2, B–D), which further confirmed that the MtbHLH2 single-gene mutation affected nodule senescence.
Figure 2.
Identification and phenotype of the bhlh2-TALEN mutant nodules. A, The bhlh2-TALEN mutants bhlh2-T36, bhlh2-T43, and bhlh2-T138 mutants had deletions of 58, 47, and 458 bp, respectively. The first base of the transcriptional start site (ATG) is shown at position +1. B, Light micrographs of wild-type (WT) and bhlh2-TALEN mutant nodules at 28 dpi. Bars = 1 mm. C, Longitudinal sections of toluidine blue-stained wild-type and bhlh2-TALEN mutant nodules at 28 dpi. The section thickness is 8 μm. The different zones of the nodules (I–IV) are shown. I, Meristematic zone; II, infection zone; III, nitrogen fixation zone; IV, senescence zone. Bars = 200 μm. D, The nitrogenase activity of wild-type and bhlh2-TALEN mutant nodules at 28 dpi was determined by an ARA. The error bars represent the ± sd of the means, and asterisks indicate a significant difference between the wild-type and bhlh2-TALEN mutant nodules (one-way ANOVA followed by Tukey’s post-hoc test, ***P < 0.001, n = 19). Similar results were obtained from three independent experiments.
MtbHLH2 Is Localized in the Nucleus and Encodes a Transcriptional Repressor
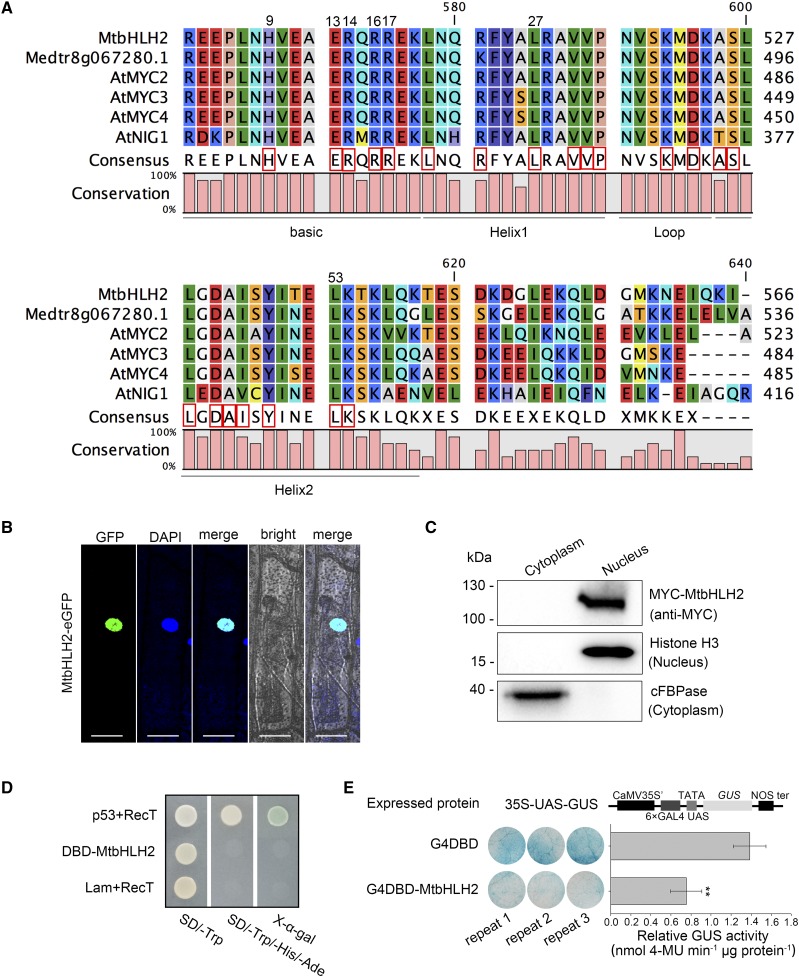
The cDNA fragment of MtbHLH2 was 2034 bp in length, and this gene did not contain introns (Fig. 1A). This transcript was predicted to encode a 677-amino-acid protein (Supplemental Fig. S1A). The typical bHLH motif was located in the C terminus of MtbHLH2, in which the basic region contained five basic amino acids for DNA binding and highly conserved hydrophobic residues in helices 1 and 2 for protein-protein interactions (Fig. 3A). The phylogenetic analysis showed that MtbHLH2 belongs to subfamily III (d+e; Supplemental Fig. S1B; Supplemental Notes S1). These results suggest that MtbHLH2 could be a functional transcription factor.
Figure 3.
MtbHLH2 is localized in the nucleus and serves as a transcriptional repressor. A, bHLH domain structures of some III (d+e) subfamily members in M. truncatula and Arabidopsis. These III (d+e) subfamily sequence alignments were constructed using CLC Sequence Viewer 7.0 software. The conserved residues for DNA binding or protein-protein interaction are shown by red boxes. B, Subcellular localization of MtbHLH2 in onion epidermal cells. The MtbHLH2-eGFP fusion protein driven by the Cauliflower mosaic virus (CaMV) 35S promoter was transiently expressed in onion epidermal cells via particle bombardment. The fluorescence signals, namely GFP and DAPI, were detected by a confocal laser scanning microscope (CLSM) using excitation wavelengths of 488 and 385 nm, respectively. Bars = 50 μm. C, Cellular fractionation and immunoblot analysis of transiently expressed MYC-MtbHLH2 in N. benthamiana leaves. Anti-histone H3 and anti-cFBPase were used as nucleus and cytoplasm markers, respectively. D, MtbHLH2 has no transcriptional activation activity in yeast. The GAL4 DBD-MtbHLH2 construct was transformed into the yeast strain AH109. The transformed yeast solutions were spotted in SD medium lacking Trp, His, and adenine. The plates were incubated for 3 d and examined through an X-α-gal assay. p53+RecT (positive control); Lam+RecT (negative control). E, MtbHLH2 is a transcriptional repressor. The transcriptional activity of MtbHLH2 in N. benthamiana leaves was detected using a GAL4/UAS-based system. CaMV 35S′, 35S promoter without a TATA box; 6×GAL4 UAS, six copies of the GAL4-binding site (UAS); NOS ter, terminator of the nopaline synthase gene; G4DBD, GAL4 DNA-binding domain; G4DBD-MtbHLH2, G4DBD fused with MtbHLH2. Measurements obtained through transient GUS activity assays are shown on the right. The error bars represent the ± sd of the means, and asterisks indicate a significant difference between G4DBD and G4DBD-MtbHLH2 (Student’s t test, **P < 0.01, n = 3).
To address the subcellular localization of MtbHLH2, MtbHLH2-eGFP was transiently expressed in onion (Allium cepa) epidermal cells. Costaining with 4′,6-diamidino-2-phenylindole (DAPI), a dye that is used for nuclear quantitation, showed that MtbHLH2 was localized in the nucleus (Fig. 3B) with an intensity correlation quotient (ICQ) of 0.386 (Supplemental Fig. S2). A protein immunoblot analysis confirmed that MtbHLH2 was detected only in the nucleus (Fig. 3C), which is consistent with the fluorescence data. These findings verified that MtbHLH2 was localized to the nucleus.
To test whether MtbHLH2 serves as a transcriptional activator or repressor, a transient expression assay was performed using yeast cells with the GAL4 DNA-binding domain (DBD) reporter system. Only the positive control grew well on synthetic dropout minimal base medium lacking Trp, His, and adenine and displayed X-α-gal activity. The DBD-MtbHLH2 and negative control did not grow (Fig. 3D), which implied that MtbHLH2 did not act as a transcriptional activator in yeast. The reporter construct 35S-UAS-GUS (Tao et al., 2013) was then cotransformed with G4DBD or G4DBD-MtbHLH2 into Nicotiana benthamiana leaves. Strong GUS staining was obtained with the combination of 35S-UAS-GUS with G4DBD, whereas considerably weakened GUS staining was obtained with the combination of 35S-UAS-GUS with G4DBD-MtbHLH2 in N. benthamiana leaves (Fig. 3E). Transient GUS quantitative experiments were performed, and the results revealed that MtbHLH2 significantly repressed the expression of GUS. These results demonstrated that MtbHLH2 was a transcriptional repressor.
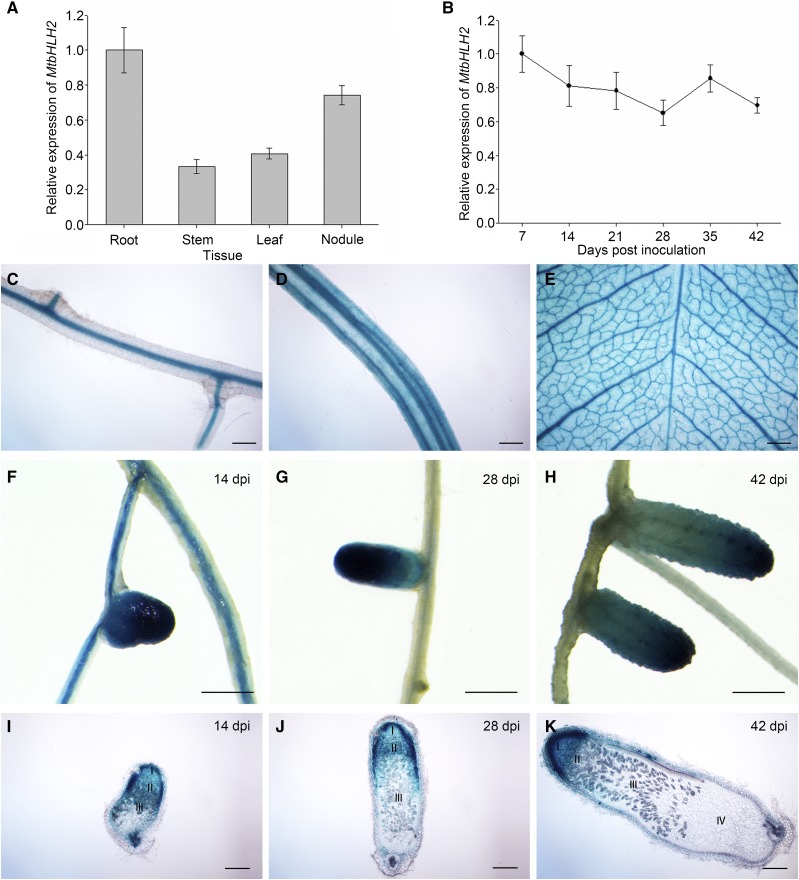
To determine the tissue expression pattern of MtbHLH2, the transcript level of MtbHLH2 in different tissues of M. truncatula R108 at 28 dpi with Sinorhizobium meliloti 1021 (Sm1021) was analyzed by RT-qPCR, and MtACTIN (Medtr3g095530) was selected as the reference gene because the expression of MtACTIN was found to be stable at the lowest M value (M = 0.047 < 0.5). The results showed that MtbHLH2 was expressed in the roots, stems, leaves, and nodules (Fig. 4A). The expression of MtbHLH2 during nodule organogenesis was also analyzed, and the RT-qPCR results indicated that the expression of MtbHLH2 first showed a slow decrease until the lowest point was reached at 28 dpi and then exhibited a slow increase (Fig. 4B). These results were consistent with the data of the M. truncatula Gene Expression Atlas (Supplemental Fig. S3A). The putative promoter-GUS-transgenic lines were obtained from M. truncatula by A. tumefaciens-mediated transformation, and a histochemical analysis of GUS staining in stable transgenic lines was performed at 14, 28, and 42 dpi with Sm1021. The results indicated that MtbHLH2 was expressed in the vascular bundles of roots, stems and nodules as well as in leaf veins (Fig. 4, C–H). The expression of MtbHLH2 decreased gradually from the meristematic zone to the nitrogen fixation zone (Fig. 4, I–K), and this expression pattern was consistent with the data in the Symbimics website (https://iant.toulouse.inra.fr/symbimics; Roux et al., 2014; Supplemental Fig. S3B). To gain insight into the relationship between the MtbHLH2 expression pattern in nodules and its negative regulation of nodule senescence, we performed high-throughput RNA-sequencing (RNA-seq) studies.
Figure 4.
Expression pattern of MtbHLH2. A, The relative expression of MtbHLH2 in the roots, stems, leaves, and nodules of wild-type plants at 28 dpi with Sinorhizobium meliloti 1021 was measured by RT-qPCR using MtACTIN as the reference gene. The error bars represent the ± sd of the means of two biological replicates. B, Relative expression of MtbHLH2 in the nodules of wild-type plants at different time points after inoculation (7, 14, 21, 28, 35, and 42 dpi) with S. meliloti 1021. MtACTIN was used as the reference gene. The error bars represent the ± sd of the means of two biological replicates. C to K, GUS staining of MtbHLH2 expressed in M. truncatula. The expression pattern of the 2.3-kb promoter region of MtbHLH2 was detected. Expression of MtbHLH2 in the roots (C), stems (D), and leaves (E) at 14 dpi with S. meliloti 1021. F to H, GUS staining of nodules on stable transgenic plants at 14 (F), 28 (G), and 42 dpi (H). I to K, Sections of nodules at 14 (I), 28 (J), and 42 dpi (K). The section thickness is 80 μm. The different zones of the nodules (I–IV) are shown. I, Meristematic zone; II, infection zone; III, nitrogen fixation zone; IV, senescence zone. Bars = 200 μm (C–E and I–K) and 1 mm (F–H).
Transcriptome Analysis of the Wild-Type and bhlh2-TALEN Mutant Nodules
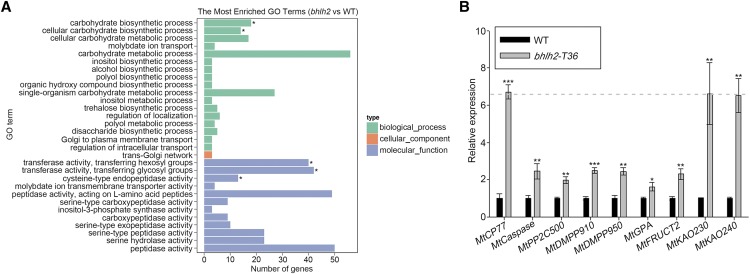
Because the Tnt1 mutants contained many insertion sites, MtbHLH2-TALEN mutants were subjected to transcriptome sequencing. To investigate the target genes and pathways regulated by MtbHLH2, RNA-seq digital gene expression profiling was performed using wild-type and bhlh2-T36 nodules at 28 dpi. The total number of clean reads in the wild-type and bhlh2-T36 nodules was 72,311,379 and 77,666,037, respectively. Among the 929 differentially expressed genes, 433 and 496 genes were upregulated and downregulated (|log2fold change| ≥ 0.6, P ≤ 0.05) in the bhlh2-T36 nodules compared with the wild-type nodules, respectively. The enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways showed that the upregulated genes were related to purine and pyrimidine metabolism, vitamin B6 metabolism, starch and Suc metabolism, and diterpenoid biosynthesis (Supplemental Fig. S4A). The enrichment of KEGG pathways revealed that the downregulated genes were involved in starch and Suc metabolism and the degradation of various branched amino acids, glycosaminoglycan, and amino sugar (Supplemental Fig. S4B). The analysis of the Gene Ontology (GO) terms of the differentially expressed genes showed that these genes were associated with carbohydrate biosynthetic process in the biological process category and in the transfer of hexosyl groups, the transfer of glycosyl groups, and Cys-type endopeptidase activity in the molecular function category (Fig. 5A).
Figure 5.
MtbHLH2 regulates various genes involved in nodule senescence. A, GO analysis of differentially expressed genes at the biological process, cellular component, and molecular function levels. The 30 most significantly enriched GO terms were analyzed using the GOseq R package. GO terms with an adjusted P < 0.05 were considered significantly enriched. B, The relative expression of senescence-, programmed cell death (PCD)-, vitamin B6 metabolism-, starch and Suc metabolism-, and GA biosynthesis-related genes in wild-type (WT) and bhlh2 mutant nodules at 28 dpi was quantified by RT-qPCR using MtACTIN as the reference gene. The error bars represent the ± sd of the means of two biological replicates, and asterisks indicate a significant difference between the wild-type and bhlh2-T36 nodules (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001).
Due to the transcriptional repression activity of MtbHLH2, we subsequently focused on upregulated genes. These genes included MtDMPP950 and MtDMPP910, which influence the homeostasis of the active form of coenzyme vitamin B6 (ShuoHao et al., 2019); MtKAO230 and MtKAO240, which are associated with GA biosynthesis (Sun, 2008); MtGPA, which promotes the production of ADP-Glc, the precursor of amylose (Hädrich et al., 2012); MtFRUCT2, which is related to Suc degradation and might be involved in leaf senescence (Buchanan-Wollaston et al., 2005; Gómez et al., 2006); MtCaspase, which is involved in programmed cell death (PCD) and the hypersensitive response (Coll et al., 2011); and senescence-associated genes (Lohman et al., 1994; Zhang et al., 2012b), such as Cys protease and PP2C-type protein phosphatase. To examine whether these genes were modulated by MtbHLH2, the relative expression of these candidate genes was detected by reverse transcription quantitative PCR (RT-qPCR). The findings revealed that the genes were upregulated in the bhlh2 mutant nodules at 28 dpi compared with those in the wild-type nodules, and the most highly upregulated gene was a PLCP involved in Cys-type endopeptidase activity, which is denoted MtCP77 (Fig. 5B; Supplemental Table S1). The results indicated that MtbHLH2 might negatively regulate the expression of candidate genes that respond to the nodule senescence process.
MtbHLH2 Directly Binds to the Promoter of MtCP77 In Vitro and In Vivo
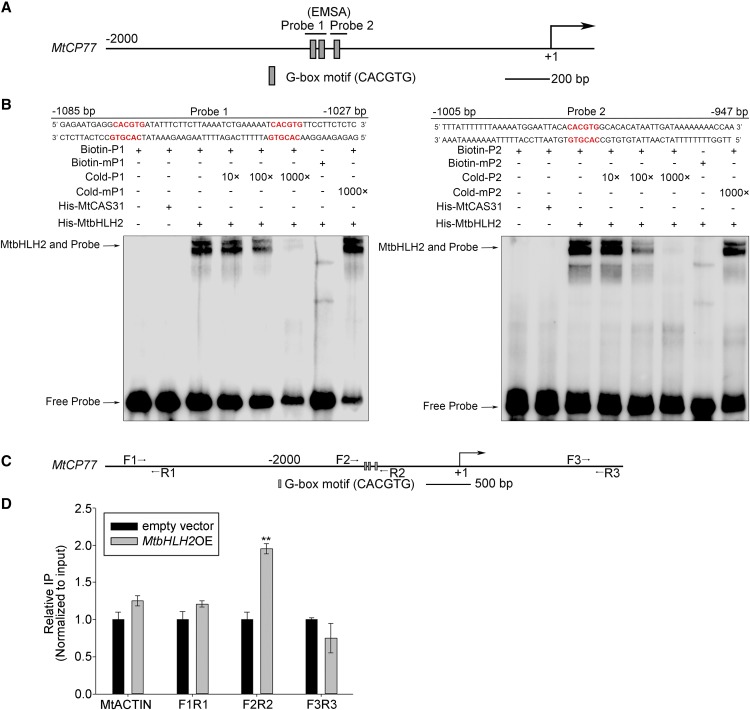
An EMSA was performed to determine whether MtbHLH2 can bind to the promoter of MtCP77. Both probes contained three G-boxes in the MtCP77 promoter (Fig. 6A). The MtbHLH2 protein can bind to both wild-type probes (P1 and P2), and this binding effectively competed with the respective unlabeled probe. MtbHLH2 did not bind to the respective mutant probe (the 5′-AAAAAA-3′ motif instead of the 5′-CACGTG-3′ motif), and neither unlabeled mutant probe competed with the binding of MtbHLH2 to the wild-type probe (Fig. 6B). These results demonstrated that MtbHLH2 could bind to G-box cis-elements in the promoter region of MtCP77 in vitro.
Figure 6.
MtbHLH2 directly binds to the promoter of MtCP77. A, Schematic diagram of G-box cis-elements in the promoter of MtCP77 for the EMSA assay. The sequence 2000 bp upstream of the transcriptional start site (ATG) and part of the coding sequence of MtCP77 are shown. The transcriptional start site is shown at position +1. B, EMSA for the analysis of MtbHLH2 binding to G-box cis-elements in the promoter of MtCP77. A purified His-MtbHLH2 protein (200 ng) was incubated with biotin-labeled probes (Biotin-P1 and Biotin-P2, 20 fM) or biotin-labeled mutant probes (Biotin-mP1 and Biotin-mP2, 20 fM). For the competition assay, unlabeled probes with different concentrations (from 10 to 1000 times, Cold-P1 and Cold-P2) or unlabeled mutant probes (1000 times, Cold-mP1 and Cold-mP2) were added. mP, Mutant probe with the 5′-AAAAAA-3′ motif instead of 5′-CACGTG-3′. The G-box cis-elements are presented in bold red text. MtCAS31, a dehydrin, was used as a negative control. The numbers of bp upstream of the transcriptional start site (ATG) are indicated on the primer sequence. C, Schematic diagram of G-box cis-elements in the promoter of MtCP77 for the ChIP assay. The approximate sequence 4000 bp upstream of the transcriptional start site (ATG) and part of the coding sequence of MtCP77 are shown. The transcriptional start site is shown at position +1. Three pairs of primers were used for RT-qPCR: the F2/R2 pair containing the G-box cis-elements in the MtCP77 promoter region (−1080 to −771 bp), the F1/R1 pair located upstream of the G-box cis-elements (−3622 to −3493 bp), and the F3/R3 pair located downstream of the G-box cis-elements (+1451 to +1623 bp). D, In vivo ChIP assay showing the binding of MtbHLH2 to the promoter of MtCP77. The ChIP assay was performed using EV-transgenic plants and MtbHLH2OE-transgenic plants at 28 dpi with S. meliloti 1021. EV-transgenic plants were used as the negative control. MtACTIN was used as the reference gene. The error bars represent the ± sd of the means of two independent experiments (Student’s t test, **P < 0.01). Three G-box cis-elements are shown by gray boxes.
Because MtbHLH2 could bind to the promoter of MtCP77 in vitro, whether MtbHLH2 could also bind to the promoter of MtCP77 in vivo was then investigated. A chromatin immunoprecipitation (ChIP) experiment was performed using empty vector (EV)-transgenic lines and MtbHLH2-FLAG (MtbHLH2OE)-transgenic lines (Supplemental Fig. S5). The anti-FLAG antibody was used for immunoprecipitation. In the nodules and roots collected from the materials at 28 dpi, the MtCP77 promoter fragment F2R2 (primers MtCP77-ChIP-qF2 and MtCP77-ChIP-qR2) containing the G-box cis-element was enriched in MtbHLH2-FLAG-transgenic lines compared with the EV-transgenic lines used as a control, whereas promoter fragments lacking the G-box cis-elements, namely F1R1 (primers MtCP77-ChIP-qF1 and MtCP77-ChIP-qR1) and F3R3 (primers MtCP77-ChIP-qF3 and MtCP77-ChIP-qR3), which were upstream and downstream of F2R2, respectively, were not enriched (Fig. 6, C and D). These results indicated that the MtbHLH2 protein could bind to the MtCP77 promoter in vivo, which suggests that MtbHLH2 could directly regulate MtCP77 in M. truncatula.
MtCP77 and MtbHLH2 Show Opposite Expression Patterns from the Meristematic Zone to the Nitrogen Fixation Zone
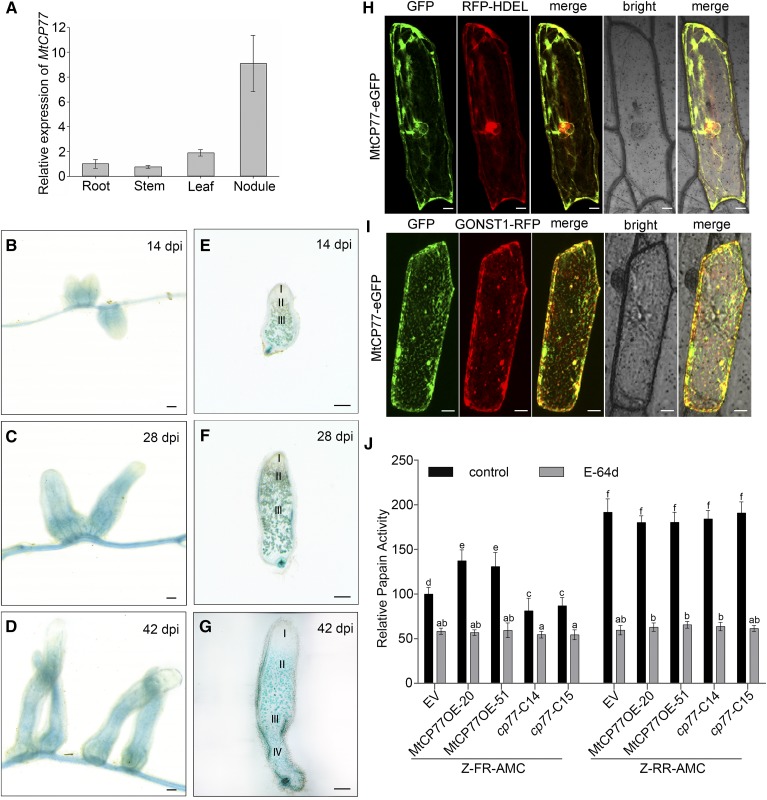
To explore the tissue expression pattern of MtCP77, the transcript levels of MtCP77 in different tissues of M. truncatula R108 at 28 dpi with S. meliloti 1021 were analyzed by RT-qPCR. The results showed that the expression of MtCP77 in the nodules was higher than that in the roots, stems, and leaves (Fig. 7A), which suggested that MtCP77 might primarily function in the nodules. Putative M. truncatula promoter-GUS-transgenic lines were obtained by A. tumefaciens-mediated transformation, and a histochemical analysis of GUS staining in stable transgenic lines at 14 dpi, 28 dpi, and 42 dpi with Sm1021 was performed. The tissue expression pattern of MtCP77 in nodules was analyzed, and the results showed that MtCP77 was mainly expressed in the nitrogen fixation zone and vascular bundles (Fig. 7, B–D). The expression of MtCP77 was increasingly enhanced from the meristematic zone to the nitrogen fixation zone (Fig. 7, E–G), which was consistent with the data in the Symbimics website (https://iant.toulouse.inra.fr/symbimics; Roux et al., 2014; Supplemental Fig. S3C) and opposite to the MtbHLH2 expression pattern. These results imply that MtCP77 may be involved in nodule senescence and is inhibited by MtbHLH2.
Figure 7.
Expression pattern and Cys protease activity of MtCP77. A, The relative expression of MtCP77 in the roots, stems, leaves, and nodules of wild-type plants at 28 dpi with S. meliloti 1021 was quantified by RT-qPCR using MtACTIN as the reference gene. The error bars represent the ± sd of the means of two biological replicates. B to G, GUS staining of MtCP77 expressed in M. truncatula nodules. The expression pattern of the 2-kb promoter region of MtCP77 was detected. B to D, GUS staining of nodules on stable transgenic plants at 14 (B), 28 (C), and 42 dpi (D). E to G, Sections of nodules at 14 (E), 28 (F), and 42 dpi (G). The section thickness is 80 μm. The different zones of the nodules (I–IV) are shown. I, Meristematic zone; II, infection zone; III, nitrogen fixation zone; IV, senescence zone. Bars = 1 mm. H and I, Subcellular localization of MtCP77 in onion epidermal cells. MtCP77-eGFP fusion protein driven by the CaMV35S promoter was transiently coexpressed with RFP-HDEL (ER marker, H) and GONST1-RFP (Golgi marker, I) in onion epidermal cells via particle bombardment. The fluorescence signals, i.e. GFP and RFP, were detected by CLSM using excitation wavelengths of 488 nm and 546 nm, respectively. Bars = 20 μm. J, The Cys protease activity of MtCP77 in EV, MtCP77OE, and Mtcp77. The protein extracts of roots of EV, MtCP77OE, and Mtcp77 at 14 dpi were used to characterize their proteinase activity. The Cys activity of MtCP77 was assayed for Z-FR-AMC (cathepsin-l) and Z-RR-AMC (cathepsin-B) proteolytic activity, and the papain inhibitor E-64d was used to inhibit the Cys protease activity. The data are represented as a percentage of the Z-FR-AMC proteolytic activity of the EV. The error bars represent the means ± sd of three biological replicates, and significant differences were determined with ANOVA with a post-hoc Tukey test and are indicated by letters (P < 0.05).
To address the subcellular localization of MtCP77, MtCP77-eGFP alongside RFP-HDEL (endoplasmic reticulum [ER] localization marker; Zhang et al., 2015) or GONST1-RFP (Golgi localization marker; Tse et al., 2006) were transiently coexpressed in onion epidermal cells. MtCP77 colocalized with HDEL (Fig. 7H) with an ICQ of 0.402 (Supplemental Fig. S6A) and partially colocalized with GONST1 (Fig. 7I) with an ICQ of 0.316 (Supplemental Fig. S6B). Therefore, MtCP77 was found to be localized in the ER and Golgi apparatus.
MtCP77 Is a Functional Cys Protease That Plays a Positive Role in Nodule Senescence
The genomic fragment of MtCP77 is 1672 bp in length, and this gene contains three introns (Supplemental Fig. S7A). The cDNA fragment of MtCP77 is 1092 bp in length, and this transcript is predicted to encode a 363-amino -acid protein (Supplemental Fig. S7B). MtCP77 belongs to the PLCP family, which is also referred to as the C1A family. The PLCP subfamilies 1 to 6 and 8 have an inconsecutive ERFNIN (ExxxRxxxFxxNxxx[I/V]xxxN) motif in the prodomain that contains an autoinhibitory domain. The PLCPs contain a catalytic triad (Cys-25-His-159-Asn-175), which implies that they are functional Cys proteases (Richau et al., 2012). Phylogenetic analysis showed that MtCP77 belongs to the CathL-like subfamily (Supplemental Fig. S7C; Supplemental Notes S2). MtCP77 contains the catalytic triad Cys-22-His-25-Asn-179, which is not a typical catalytic triad and shows a change in the His position; however, this Cys-22-His-25-Asn-179 triad was mutually adjacent according to the predicted protein structure (Supplemental Fig. S7D), suggesting that MtCP77 is likely a functional Cys protease.
To elucidate the biological functions of MtCP77 in nodule senescence, transgenic M. truncatula lines overexpressing MtCP77 and Empty Vector (EV) were obtained by A. tumefaciens-mediated transformation. Three stable transgenic lines (MtCP77OE-20, MtCP77OE-44, and MtCP77OE-51) were selected following RT-qPCR and western blotting analysis (Supplemental Fig. S8). Furthermore, MtCP77-knockout mutants were generated using clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) technology via A. tumefaciens-mediated transformation. The transgenic plants were identified by sequencing. The cp77-C14-knockout mutant containing a 4-bp deletion triggered the frameshift mutation (Supplemental Fig. S9). The cp77-C15- and cp77-C30-knockout mutants contained a guanine (G) insertion and an adenine (A) insertion, respectively, which led to an insertion mutation (Supplemental Fig. S9). Overall, these overexpression lines and mutants were suitable to analyze the biological function of MtCP77.
The Cys protease activity of MtCP77 in EV, MtCP77OE, and Mtcp77 was assayed with Z-FR-AMC (cathepsin-l activity) and Z-RR-AMC (cathepsin-B activity), two papain substrates. Compared that in the EV root lines, Z-FR-AMC proteolytic activity was significantly increased with the overexpression of MtCP77 and decreased with the mutation of MtCP77, whereas no significant difference in Z-RR-AMC proteolytic activity was observed (Fig. 7J). Furthermore, both Z-FR-AMC and Z-RR-AMC proteolytic activity were inhibited in the presence of the papain inhibitor E-64d (Fig. 7J). These results indicated that MtCP77 is a functional CathL-like Cys protease and imply that MtCP6 is involved in the nodule senescence process.
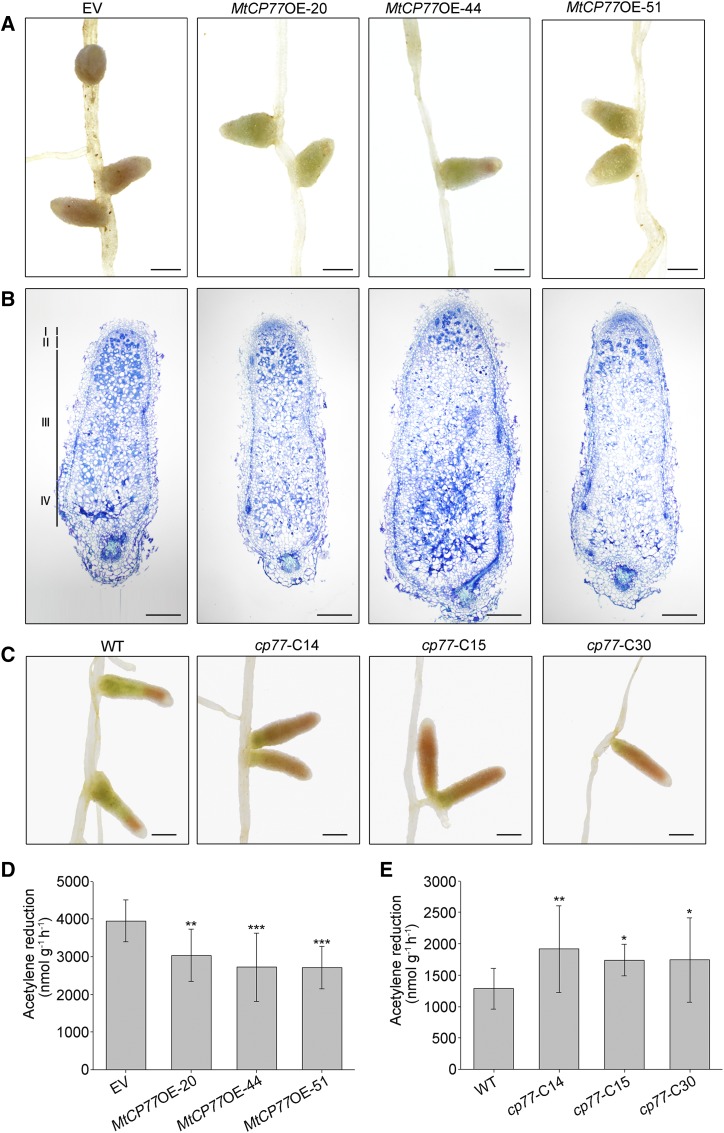
The nodule developmental phenotype and nitrogenase activity assays showed that the overexpression of MtCP77 led to an acceleration of nodule senescence and a decline in nitrogenase activity (Fig. 8, A, B, and E), which is consistent with the findings obtained for bhlh2 mutants. At 49 dpi, the wild-type nodules were green, which suggested that the nodules had entered the senescence stage, and the cp77 nodules were pink (Fig. 8D). The nitrogenase activity of cp77 nodules was significantly higher than that of the wild-type nodules, as demonstrated by the ARA (Fig. 8F). In summary, the mutation of MtCP77 delays nodule senescence.
Figure 8.
MtCP77 positively regulates nodule senescence. A, Light micrographs of nodules in EV-transgenic plants and MtCP77OE-transgenic plants at 28 dpi. Bars = 1 mm. B, Longitudinal sections of toluidine blue-stained nodules in EV-transgenic plants and MtCP77OE-transgenic plants at 28 dpi. The section thickness is 8 μm. The different zones of the nodules (I–IV) are shown. I, Meristematic zone; II, infection zone; III, nitrogen fixation zone; IV, senescence zone. Bars = 200 μm. C, Light micrographs of wild-type (WT) and cp77 mutant nodules at 49 dpi. Bars = 1 mm. D, The nitrogenase activity of EV-transgenic and MtCP77OE-transgenic plant nodules at 28 dpi was determined by an ARA. The error bars represent the ± sd of the means, and asterisks indicate a significant difference between the EV and MtCP77OE-transgenic plants (one-way ANOVA followed by Tukey’s post-hoc test, **P < 0.01, ***P < 0.001, n = 19). E, The nitrogenase activity of the wild-type and cp77 nodules at 49 dpi was determined by an ARA. The error bars represent the ± sd of the means, and asterisks indicate a significant difference between the wild-type and cp77 mutant nodules (one-way ANOVA followed by Tukey’s post-hoc test, *P < 0.05, **P < 0.01, n ≥ 15).
MtbHLH2 and MtCP77 Play Opposite Roles in Plant PCD and ROS Accumulation during Nodule Senescence
To determine whether MtbHLH2 and MtCP77 can affect plant PCD and reactive oxygen species (ROS) accumulation, a terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) assay and 3,3′-diaminobenzidine (DAB) staining were performed. Wild type, bhlh2 mutant, EV, and MtCP77OE nodules were used for DAB staining at 28 dpi. Compared with the wild-type or EV nodules, the bhlh2 and MtCP77OE nodules presented higher concentrations of hydrogen peroxide (H2O2; Fig. 9, A and B). Additionally, a TUNEL assay, which was used to detect PCD events in plant tissues with cell death signals, showed that the bhlh2 nodules exhibited strong DNA fragmentation at 28 dpi compared with the wild-type nodules (Fig. 9, C and E), and weaker staining of DNA fragmentation was observed in the cp77 nodules than in the wild-type nodules at 49 dpi (Fig. 9, D and F). Therefore, MtbHLH2 and MtCP77 play opposite roles in plant PCD and ROS accumulation during nodule senescence.
Figure 9.
MtbHLH2 and MtCP77 play opposing roles in plant PCD and ROS accumulation. A and B, The accumulation of H2O2 in longitudinal sections of bhlh2 (A) or MtCP77OE nodules (B) at 28 dpi was detected. A higher concentration of H2O2 was found in the bhlh2 (A) or MtCP77OE nodules (B). Bars = 200 μm. C and D, DNA fragmentation in longitudinal sections of wild-type (WT) and bhlh2 nodules at 28 dpi (C) or wild-type and cp77 nodules at 49 dpi (D) was detected by TUNEL assay. DNA fragmentation resulting from the cell death signaling cascade is shown as brown signals in both plant nuclei and rhizobia of bhlh2 nodules at 28 dpi (C) or wild-type nodules at 49 dpi (D). Bars = 200 μm. E and F, Magnification of the boxed areas in (C) and (D). Bars = 50 μm.
MtbHLH2 Represses the Expression of MtCP77 to Prevent Early Nodule Senescence in M. truncatula
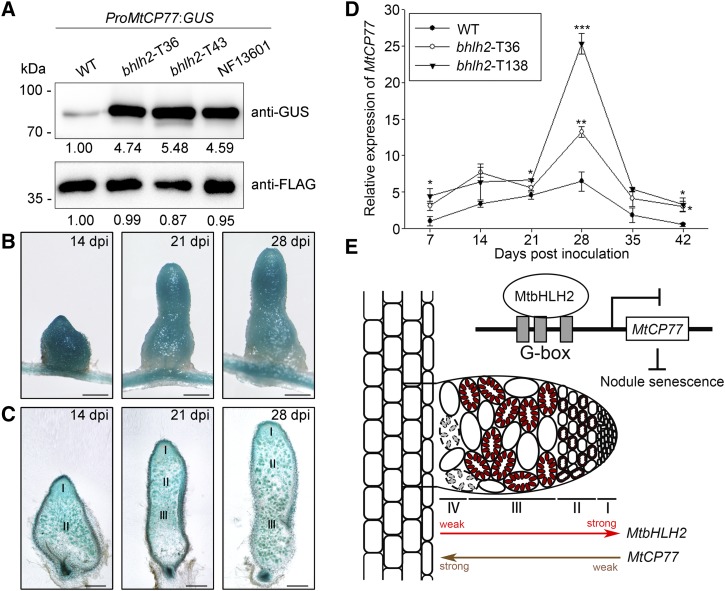
To confirm that endogenous MtbHLH2 can repress the expression of MtCP77, ProMtCP77:GUS was inserted by Agrobacterium rhizogenes-mediated hairy root transformation (Supplemental Fig. S10). The construct was expressed in the wild-type and bhlh2 nodules, and an immunoblot analysis was performed to test the protein level of GUS using GFP-FLAG as the loading control. GUS was barely detectable in ProMtCP77:GUS/wild type and strongly expressed in ProMtCP77:GUS/bhlh2-T36, ProMtCP77:GUS/bhlh2-T43, and ProMtCP77:GUS/NF13601 (Fig. 10A). In addition, GUS staining of ProMtCP77:GUS/bhlh2-T36 showed that, apart from nitrogen fixation and senescence zone, GUS was expressed in the meristematic zone (Fig. 10, B and C), similar to ProMtCP77:GUS/wild type. Thus, endogenous MtbHLH2 suppresses the expression of MtCP77 with respect to both expression level and expression tissue area in nodules. We further compared the expression pattern of MtCP77 in wild type and bhlh2-TALEN mutants at different stages of nodule development. RT-qPCR analysis confirmed that during nodule development, the relative expression of MtCP77 progressively increased until it reached a peak at 28 dpi and then gradually decreased. The expression of MtCP77 in the bhlh2 nodules was always higher than that in the wild-type nodules and significantly higher than that in the wild-type nodules at 28 dpi (Fig. 10D). These results imply that MtbHLH2 can repress the expression of MtCP77 at different stages of nodule development.
Figure 10.
MtbHLH2 represses the expression of MtCP77. A, MtbHLH2 represses the expression of MtCP77 in M. truncatula. The 2-kb promoter region of MtCP77 was used for this assay. ProMtCP77:GUS, which carried an expression box of GFP-FLAG under the control of the CaMV35S promoter, was transformed into the wild type (WT) and bhlh2 mutants (bhlh2-T36, bhlh2-T43, and NF13601) by A. rhizogenes-mediated hairy root transformation. Composite transgenic plants were harvested, and proteins were extracted from ProMtCP77:GUS/wild type, ProMtCP77:GUS/bhlh2-T36, ProMtCP77:GUS/bhlh2-T43, and ProMtCP77:GUS/ NF13601 for immunoblot analysis. GFP-FLAG was used as a loading control. ImageJ software was used to quantify the immunoblot bands. B and C, GUS staining of MtCP77 expression in the nodules of ProMtCP77:GUS/bhlh2-T36. B, GUS staining of nodules on ProMtCP77:GUS/bhlh2-T36 hairy roots at 14, 21, and 28 dpi. C, Sections of nodules at 14, 21, and 28 dpi. The section thickness is 80 μm. The different zones of the nodules (I–III) are shown. Bars = 1 mm. D, The relative expression of MtCP77 in wild-type and bhlh2 mutant nodules (bhlh2-T36 and bhlh2-T138) at different time points after inoculation (7, 14, 21, 28, 35, and 42 dpi) with S. meliloti 1021 was quantified by RT-qPCR using MtACTIN as the reference gene. The error bars represent the ± sd of the means of two biological replicates, and asterisks indicate a significant difference between the wild-type and bhlh2 mutant nodules (one-way ANOVA followed by Tukey’s post-hoc test, *P < 0.05, **P < 0.01, ***P < 0.001). E, A proposed model for the regulation of MtbHLH2 during nodule senescence in M. truncatula. In the nodules, the expression of MtbHLH2 gradually decreases from the meristematic zone to the nitrogen fixation zone, whereas the expression of MtCP77 gradually increases. The binding of MtbHLH2 to the promoter of MtCP77 inhibits MtCP77 expression to delay nodule senescence. The different zones of the nodules (I–IV) are shown. I, Meristematic zone; II, infection zone; III, nitrogen fixation zone; IV, senescence zone.
In this study, we found a repressive transcription factor, MtbHLH2, that negatively regulates nodule senescence, and mutants defective in MtbHLH2 show premature senescence and decreased nitrogenase activity. In the nodules, the expression of MtbHLH2 and MtCP77 gradually decreased and increased, respectively, from the meristematic zone to the nitrogen fixation zone. The binding of MtbHLH2 to the promoter of MtCP77 inhibited MtCP77 expression to delay nodule senescence, which suggests that MtbHLH2 can prevent early nodule senescence to ensure effective nitrogen fixation by inhibiting the expression of MtCP77 (Fig. 10E). Taken together, the results show that MtbHLH2 is a negative regulator of nodule senescence and inhibits the expression of MtCP77, which positively modulates plant PCD, ROS accumulation, and nodule senescence in M. truncatula.
DISCUSSION
Approximately 100 bHLH genes exist in the M. truncatula genome (Young et al., 2011), and to date, only two legume bHLHs related to nodulation have been reported. GmSAT1, which is also called GmbHLHm1, encodes a symbiotic ammonium transporter and is associated with nodule development and NH4+ transport (Kaiser et al., 1998; Marini et al., 2000; Chiasson et al., 2014). MtbHLH1, which is particularly expressed in the roots and nodules, exerts a critical influence on nodule vascular patterning and nodule-to-plant nutrient exchange (Godiard et al., 2011). However, bHLH transcription factors involved in nodule senescence have not yet been identified in M. truncatula. In this study, mutants showing early nodule senescence were identified, and the gene responsible for this process was MtbHLH2 (Figs. 1 and 2).
The analysis of transcriptome data indicated that MtbHLH2 regulates vitamin B6 metabolism (Supplemental Fig. S4A). As an important cofactor, vitamin B6 is involved in many biochemical reactions, including decarboxylation, transamination, elimination, racemization, and transsulfuration reactions, which are mainly related to the synthesis of amino acids (Drewke and Leistner, 2001). The active forms of vitamin B6 are pyridoxal-5-phosphate and pyridoxamine-5-phosphate, which act as cofactors of many enzymes involved in the metabolism of amino acids. In this study, MtDMPP950 and MtDMPP910, which encode 2,3-diketo-5-methylthio-1-phosphopentane phosphatases, were upregulated in the bhlh2 mutant nodules at 28 dpi compared with that in the wild-type nodules (Fig. 5B). These enzymes convert pyridoxal-5-phosphate and pyridoxamine-5-phosphate into pyridoxal and pyridoxamine, respectively, and thereby hinder amino acid synthesis. These results suggest that MtbHLH2 might also be involved in nodule senescence by affecting the synthesis of amino acids.
The analysis of transcriptome data also showed that MtbHLH2 modulates diterpenoid biosynthesis, which was found to be associated with GA biosynthesis (Supplemental Fig. S4A). Ent-kaurene synthase, cytochrome P450 family ent-kaurenoic acid oxidase (KAO), and GA 2-beta-dioxygenase (also called GA 2-oxidase, MtGA2ox) are the key enzymes in the GA biosynthetic pathway (Sun, 2008), and the transcriptome data obtained in this study showed that these enzymes were upregulated in the bhlh2 mutant nodules. The RT-qPCR results also showed that MtKAO230 and MtKAO240 were significantly upregulated in the bhlh2 mutant nodules (Fig. 5B), and these enzymes are associated with the first steps of GA biosynthesis and promote the generation of GA12, the common precursor of all types of GAs in plants (Hedden and Phillips, 2000). Previous studies have reported that GA might inhibit nodule senescence (Van de Velde et al., 2006; Serova et al., 2019), so the up-regulation of MtKAO230 and MtKAO240 appeared to contradict the phenotype of bhlh2. However, the transcriptome data obtained in this study also indicated that MtGA2ox was upregulated in bhlh2 (Supplemental Fig. S11), and this up-regulation might result in conversion of the bioactive form of GA into the inactive form to maintain GA homeostasis in bhlh2. These results suggest that the regulation of GA biosynthesis might also be involved in the nodule senescence processes regulated by bHLH2.
The target gene MtCP77, which is modulated by MtbHLH2, was identified by transcriptome sequencing. The expression of MtCP77 in nodules was higher than that in roots, stems, and leaves (Fig. 7A), which suggested that MtCP77 might play a crucial role in root nodules. This study showed that the overexpression of MtCP77 decreased the nitrogen fixation abilities of nodules and promoted nodule senescence (Fig. 8, A, B, and D), and the cp77 mutants exhibited the opposite phenotype (Fig. 8, C and E). In agreement with previous studies, the knockdown of AsNodf32, a PLCP, delayed nodule senescence (Li et al., 2008), and the down-regulation of MtCP6 increased the nitrogen fixation abilities of nodules and delayed nodule senescence; moreover, the overexpression of MtCP6 accelerated nodule senescence (Pierre et al., 2014). Additionally, the overexpression of MtCP77 triggered a decrease in nitrogenase activity and high concentrations of hydrogen peroxide (H2O2; Figs. 8D and 9B), and the mutation of MtCP77 resulted in an increase in nitrogenase activity and a reduction in DNA fragmentation (Figs. 8E and 9, D and F), which suggested the occurrence of PCD and ROS accumulation during nodule senescence. These results were similar to those obtained in previous studies, which showed that PCD events could be regarded as the initiation of senescence (Alesandrini et al., 2003). The ROS that were accumulated in response to MtCP77 overexpression (Fig. 9B) might produce highly oxidizing conditions to result in nodule senescence, which implies the occurrence of a redox shift in the nodules from a highly reducing status, which is required for nitrogen fixation in nodules, to highly oxidizing conditions, which might lead to senescence (Puppo et al., 2005).
Many endogenous plant Cys proteases are located in the vacuole (Callis, 1995). Many vacuolar proteases are transported from the ER to the vacuole through the Golgi apparatus, and this transport is a Golgi-dependent pathway (Okamoto, 2006; Xiang et al., 2013). In general, PLCPs that do not contain ER retention signal Lys-Asp-Glu-Leu-tail Cys proteases are first transported from the ER to the Golgi apparatus and are then recognized by vacuolar sorting receptor on the Golgi apparatus, leading to the transport of proenzymes to the vacuoles. During or after transport to the vacuoles, the proenzymes are converted into mature enzymes via proteolytic processing (Okamoto, 2006). This study revealed that MtCP77 is localized in the ER and Golgi apparatus (Fig. 7, H and I), which might be due to the presence of the full-length coding sequence of MtCP77 as a proenzyme in the onions and its inability to be transported to the vacuoles without undergoing proteolysis and becoming a mature enzyme. The subcellular localization of MtCP77 in M. truncatula should be further studied.
In conclusion, this study describes an important regulatory module of nodule senescence, namely MtbHLH2-MtCP77, that is associated with plant PCD and ROS accumulation and ultimately leads to nodule senescence.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Medicago truncatula ecotype R108 was used as the wild-type material in all the experiments performed in this study. Both bhlh2 Tnt1-insertion mutants were isolated from the M. truncatula mutant database (primers are listed in Supplemental Table S2). The T1 generation bhlh2 Tnt1-insertion mutants were used for phenotypic experiments.
The full-length coding sequences of MtbHLH2 and MtCP77 were cloned into pCAMBIA1307-FLAG (primers are listed in Supplemental Table S2). Stable transgenic plants were obtained by Agrobacterium tumefaciens-mediated transformation as previously described (Cosson et al., 2006).
Seeds were sterilized for germination as described previously (Duan et al., 2017). For nodule sectioning and the ARA, seedlings were grown on Fåhraeus solid and liquid media (Fåhraeus, 1957) without nitrogen (pH 6.5) in square dishes and containing a vermiculite:perlite (5:2, v/v) mixture, respectively. For all the experiments, the plants were incubated in a growth chamber at 24°C with a 16-h light/8-h dark cycle and 70% relative humidity.
RNA Extraction and RT-qPCR
Total RNA was extracted using the TRIzol reagent (Invitrogen), and 2 μg of total RNA was used for reverse transcription. The RT-qPCR analysis was performed using a CFX-96 Real-Time System (Bio-Rad) with TB Green Premix ExTaq (TaKaRa). To verify the expected stable expression of the reference genes during nodule development, the stability of four candidate reference genes (MtACTIN, MtACTIN11, MtH3L, and MtRBP1) at different time points after inoculation with Sinorhizobium meliloti 1021 was assessed using geNorm software (Vandesompele et al., 2002). The expression of MtACTIN was stable at the lowest M value (M = 0.047 < 0.5). MtACTIN was selected as the reference gene, and the relative expression data were normalized to MtACTIN expression. The mean and sd values were calculated from the results of two biological replicates. The primers used in this study are listed in Supplemental Table S2. MtACTIN11, MtH3L, and MtRBP1 genes were used as references (Plet et al., 2011).
Rhizobial Inoculation and ARA
Seven-day-old seedlings were inoculated with S. meliloti 1021 for the nodulation assay, and the nitrogen fixation ability of rhizobia was measured using an ARA as previously described (Li et al., 2018).
Histochemical GUS Staining
The 2.3-kb promoter of MtbHLH2 and the 2-kb promoter of MtCP77 upstream of the transcriptional start site (ATG) were amplified by PCR from M. truncatula genomic DNA and cloned into pCAMBIA1381 carrying the GUS reporter gene (primers are listed in Supplemental Table S2). The transgenic plants were stained as previously described (Li et al., 2018).
Microscopic Analysis
Nodules were fixed overnight at 4°C with formalin-acetic-alcohol (50% [v/v] ethanol, 5% [v/v] acetic acid, and 3.7% [v/v] formalin) solution. The fixed materials were dehydrated in an ethanol series and subsequently embedded in Paraplast (Sigma). Longitudinal sections (8 μm) were prepared using an RM2235 microtome (Leica) and stained with 0.5% (w/v) toluidine blue for 10 min. The nodules for GUS staining were embedded in 6% (w/v) agarose, and longitudinal sections (80 μm) were cut using a VT1000S vibratome (Leica). The sections were photographed using a light microscope (Olympus BX51).
For electron microscopy analysis, the nodules were fixed in 2.5% (v/v) glutaraldehyde, washed three times with phosphate buffer (pH 7.2), fixed in 1% (v/v) osmic acid, washed three more times, dehydrated in an acetone series, and embedded with a Spurr Embedding Kit. Serial sections were prepared using a UC6 microtome (Leica) equipped with a diamond knife, and the sections were photographed using a JEOL JEM-1230 transmission electron microscope.
Subcellular Localization and Confocal Microscopy
MtbHLH2-GFP, MtCP77-GFP with RFP-HDEL (ER), and GONST1-RFP (Golgi marker) were transiently expressed or coexpressed in onion (Allium cepa) epidermal cells as previously described (Duan et al., 2017). The intensity correlation analysis was described previously (Li et al., 2004). The ICQ is based on nonparametric test analysis of the product of the differences from the mean (PDM) values: −0.5 ≤ ICQ < 0, segregated staining; ICQ ∼ 0, random staining; 0 < ICQ ≤ +0.5, dependent staining. The samples were photographed using a confocal laser scanning microscope (CLSM, Olympus FluoView FV1000).
Nuclear/Cytoplasmic Fraction and Immunoblot Assays
A. tumefaciens EHA105 carrying the MYC-MtbHLH2 plasmid was infiltrated into Nicotiana benthamiana leaves (primers are listed in Supplemental Table S2). Three-day-infiltration samples were extracted as previously described (Du et al., 2015). An immunoblot analysis was performed with anti-MYC (Sigma), antihistone H3 (Agrisera, nuclear fraction marker), and anticFBPase (Agrisera, cytosolic fraction marker).
Analysis of Transcriptional Activation or Repression in Yeast Cells and N. benthamiana Leaves
MtbHLH2 was cloned into the pGBKT7 vector, and yeast transcriptional activation was performed as previously described (Shan et al., 2012). DBD-MtbHLH2 was cloned into pCAMBIA1302, and an assay of transcriptional repression in N. benthamiana leaves was performed as previously described (Tao et al., 2013; primers are listed in Supplemental Table S2).
Design, Assembly, and Identification of TALEN Constructs of MtbHLH2
The target sequence of MtbHLH2 was designed using TAL Effector Nucleotide Targeter 2.0 (https://tale-nt.cac.cornell.edu/) as previously described (Ma et al., 2015). The two target sequences of MtbHLH2 had 16 and 19 repeat-variable diresidues, which were assembled by the three-step golden gate method as previously described (Cermak et al., 2011) and added to the pCAMBIA1300 vector (primers listed in Supplemental Table S2). The T1 generation bhlh2-TALEN mutants are used for phenotypic experiments.
Transcriptome Analysis
Sequencing was carried out by Novogene. At 28 dpi, wild-type and bhlh2-T36 nodules were used for digital high-throughput RNA-seq gene expression profiling, and 3 μg of total RNA per sample was then used to generate nondirectional Illumina sequencing libraries. Each library was generated using the SE50 single-read sequencing method with the Illumina HiSeq 2500 platform. The gene annotation files were referenced against the M. truncatula genome (version 4.0) and downloaded from the J. Craig Venter Institute (http://www.jcvi.org/medicago/). Genes with an adjusted P value ≤ 0.05 were considered differentially expressed. The significantly enriched GO terms were analyzed using the GOseq R package (Young et al., 2010). The statistical analysis of KEGG pathway enrichment was performed using KOBAS software (Mao et al., 2005).
Protein Purification and EMSA
His-MtbHLH2 was expressed in Escherichia coli BL21 and purified by Ni affinity chromatography according to the manufacturer’s instructions. Electrophoretic mobility shift assay (EMSA) experiments were performed using a LightShift Chemiluminescent EMSA kit (Thermo Fisher) as previously described (Duan et al., 2017; primers are listed in Supplemental Table S2).
Hairy Root Transformation and Immunoblot Assays
ProMtCP77:GUS carried an expression box, GFP-FLAG, driven by the CaMV35S promoter, which was used as a loading control (primers listed in Supplemental Table S2). Composite transgenic plants of wild type and bhlh2 mutants were created by Agrobacterium rhizogenes-mediated hairy root transformation as previously described (Boisson-Dernier et al., 2001). Immunoblot analysis was performed with anti-GUS (Sigma) and anti-FLAG (Sigma).
Design and Identification of CRISPR/Cas9 Constructs of MtCP77
The target site of MtCP77 was designed using CRISPR-PLANT (https://www.genome.arizona.edu/crispr/CRISPRsearch.html). The length of each of the two target sites of MtCP77 was 19 bp. The PCR fragment was amplified from pCBC-DT1T2, and the purified PCR fragment (T1T2-PCR), combined with the binary vector pHSE401, was established by restriction-ligation reactions as previously described (Xing et al., 2014; primers are listed in Supplemental Table S2). The T1 generation of cp77-Cas9 mutants are used for phenotypic experiments.
Cys Protease Enzymatic Activities
The Cys protease enzymatic activities were determined as in a previous report (Pierre et al., 2014). The 14-dpi roots were harvested, and the total protein was extracted. The extraction buffer contained 50 mm Tris, pH 8, 5 mm EDTA, 20 mm NaCl, and 10% (v/v) glycerol. A Bradford protein assay (Bio-Rad) was used to quantify the protein concentrations. N-acetyl-FR7-amido-4-methylcoumarin (Sigma, #C9521) and N-acetyl-RR7-amido-4-methylcoumarin (Sigma, #C5429) were used as substrates to assay cathepsin-l and cathepsin-B activity, respectively. The reaction mix contained 100 mm sodium acetate (pH 5.5), 1 mm EDTA, 1 mm dithiothreitol, 10 μg/mL total protein and 50 μm of substrate and was incubated at 28°C for 20 min; the papain inhibitor E-64d (Sigma, #E8640) was used as a negative control. Then, the proteolytic product, coumarin, was assayed with a spectrofluorimeter (Hitachi F-7000) at a 380-nm excitation wavelength and a 460-nm emission wavelength. Activity was measured as cleavage of substrate over time, and its values were expressed as Δ(fluorescence).min−1.
ChIP
ChIP was performed as described previously (Saleh et al., 2008), with some modifications (Zhang et al., 2016; Duan et al., 2017). At 28 dpi, 2.5 g root and nodule samples were cross linked and then sonicated using a Bioruptor Plus (Diagenode; low power, on for 60 s, off for 60 s, four cycles). Anti-FLAG (Sigma) was used to immunoprecipitate the target MtbHLH2-FLAG fusion protein. The enriched DNA fragments that contrasted with the input samples were detected by RT-qPCR (primers are listed in Supplemental Table S2) using MtACTIN as the negative control.
DAB Staining and TUNEL Assay
At 28 dpi, nodules were collected for DAB staining as previously described (Zhou et al., 2015). At 28 or 49 dpi, nodules were fixed in formalin-acetic-alcohol solution, and TUNEL assays were performed using the DeadEnd Colorimetric TUNEL System (Promega) according to the manufacturer’s instructions.
Phylogenetic Analysis
Protein sequences of bHLH transcription factors or PLCPs in M. truncatula and Arabidopsis (Arabidopsis thaliana) were obtained from the JCVI and TAIR databases, respectively. The multiple sequence alignment was analyzed using ClustalW2 software (Larkin et al., 2007) or the MAFFT online service (Katoh et al., 2002, 2017), and a phylogenetic tree was generated using PhyML with the neighbor-joining method or the maximum likelihood method (Guindon et al., 2010). The numbers on the branches show bootstrap values based on 1000 replicates. The phylogenetic analysis was performed using EvolView (Zhang et al., 2012a).
Statistical Analysis
All data analyses and calculations of the means and sd were performed using Sigma Plot 12.0 and Microsoft Excel 2010. The statistical significance of the differences was assessed by Student’s t test or one-way ANOVA followed by Tukey’s post-hoc test using SPSS software (version 16.0). The P values for each statistical test are reported.
Accession Numbers
The sequence data obtained in this study can be found in the GenBank data library under the accession numbers MK798038 (MtbHLH2) and MK798039 (MtCP77). The raw data used for the RNA-seq analysis have been submitted to the NCBI database under the accession number PRJNA383274.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic diagram of the domain structure in MtbHLH2 and phylogenetic analysis of MtbHLH2.
Supplemental Figure S2. Analysis of the intensity of the colocalization of MtbHLH2 with the nuclear dye DAPI.
Supplemental Figure S3. Expression patterns of MtbHLH2 and MtCP77.
Supplemental Figure S4. Analysis of pathways enriched with differentially expressed genes regulated by MtbHLH2.
Supplemental Figure S5. Molecular identification of MtbHLH2OE-transgenic plants.
Supplemental Figure S6. Analysis of the intensity of the colocalization of MtCP77 with an ER marker and a Golgi marker.
Supplemental Figure S7. Gene structure and phylogenetic analysis of MtCP77.
Supplemental Figure S8. Molecular identification of MtCP77OE-transgenic plants.
Supplemental Figure S9. The sequence of MtCP77 in cp77-C after editing by CRISPR/Cas9.
Supplemental Figure S10. Schematic diagram of the ProMtCP77:GUS construct.
Supplemental Figure S11. The expression of GA2ox1 and GA2ox2 in nodules of R108 and bhlh2-1.
Supplemental Table S1. Analysis of upregulated gene expression profiles from RNA-seq data used for RT-qPCR.
Supplemental Table S2. List of primers used in this study.
Supplemental Notes S1. The protein sequences of bHLH transcription factors in M. truncatula and Arabidopsis that were used as an input for the phylogenetic tree analysis.
Supplemental Notes S2. The protein sequences of PLCPs in M. truncatula and Arabidopsis that were used as an input for the phylogenetic tree analysis.
ACKNOWLEDGMENTS
The authors thank Dr. Genji Qin (State Key Laboratory of Protein and Plant Gene Research, College of Life Sciences, Peking University, China) for kindly providing 35S-UAS-GUS vector used in this study. The authors also thank Dr. Qijun Chen (State Key Laboratory of Plant Physiology and Biochemistry, College of Biological Sciences, China Agricultural University, China) for kindly providing CRISPR/Cas9 vector used in this study.
Footnotes
This work was financially supported by grants from the National Natural Science Foundation of China (31772658 and 31571587) and the Project for Extramural Scientists of the State Key Laboratory of Agrobiotechnology (2018SKLAB6-22).
Articles can be viewed without a subscription.
References
- Alesandrini F, Mathis R, Van de Sype G, Herouart D, Puppo A (2003) Possible roles for a cysteine protease and hydrogen peroxide in soybean nodule development and senescence. New Phytol 158: 131–138 [Google Scholar]
- Berger A, Boscari A, Frendo P, Brouquisse R (2019) Nitric oxide signaling, metabolism and toxicity in nitrogen-fixing symbiosis. J Exp Bot 70: 4505–4520 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Balliau T, Aït-Salem EH, George J, Zivy M, Ratet P, Gourion B (2018) Control of the ethylene signaling pathway prevents plant defenses during intracellular accommodation of the rhizobia. New Phytol 219: 310–323 [DOI] [PubMed] [Google Scholar]
- Berrabah F, Bourcy M, Eschstruth A, Cayrel A, Guefrachi I, Mergaert P, Wen J, Jean V, Mysore KS, Gourion B, et al. (2014) A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol 203: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P (2013) Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol 197: 1250–1261 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Callis J. (1995) Regulation of protein degradation. Plant Cell 7: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam Y, Pierre O, Boncompagni E, Hérouart D, Meilhoc E, Bruand C (2012) Nitric oxide (NO): A key player in the senescence of Medicago truncatula root nodules. New Phytol 196: 548–560 [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiasson DM, Loughlin PC, Mazurkiewicz D, Mohammadidehcheshmeh M, Fedorova EE, Okamoto M, McLean E, Glass AD, Smith SE, Bisseling T, et al. (2014) Soybean SAT1 (Symbiotic Ammonium Transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc Natl Acad Sci USA 111: 4814–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson V, Durand P, d’Erfurth I, Kondorosi A, Ratet P (2006) Medicago truncatula transformation using leaf explants. Methods Mol Biol 343: 115–127 [DOI] [PubMed] [Google Scholar]
- D’haeseleer K, De Keyser A, Goormachtig S, Holsters M (2010) Transcription factor MtATB2: About nodulation, sucrose and senescence. Plant Cell Physiol 51: 1416–1424 [DOI] [PubMed] [Google Scholar]
- de Zélicourt A, Diet A, Marion J, Laffont C, Ariel F, Moison M, Zahaf O, Crespi M, Gruber V, Frugier F (2012) Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J 70: 220–230 [DOI] [PubMed] [Google Scholar]
- Dhanushkodi R, Matthew C, McManus MT, Dijkwel PP (2018) Drought-induced senescence of Medicago truncatula nodules involves serpin and ferritin to control proteolytic activity and iron levels. New Phytol 220: 196–208 [DOI] [PubMed] [Google Scholar]
- Domonkos Á, Kovács S, Gombár A, Kiss E, Horváth B, Kováts GZ, Farkas A, Tóth MT, Ayaydin F, Bóka K, et al. (2017) NAD1 controls defense-like responses in Medicago truncatula symbiotic nitrogen fixing nodules Following rhizobial colonization in a bacA-independent manner. Genes (Basel) 8: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewke C, Leistner E (2001) Biosynthesis of vitamin B6 and structurally related derivatives. Vitam Horm 61: 121–155 [DOI] [PubMed] [Google Scholar]
- Du JL, Zhang SW, Huang HW, Cai T, Li L, Chen S, He XJ (2015) The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol Plant 8: 1053–1068 [DOI] [PubMed] [Google Scholar]
- Duan M, Zhang R, Zhu F, Zhang Z, Gou L, Wen J, Dong J, Wang T (2017) A lipid-anchored NAC transcription factor is translocated into the nucleus and activates Glyoxalase I expression during drought stress. Plant Cell 29: 1748–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont L, Alloing G, Pierre O, El Msehli S, Hopkins J, Herouart D, Frendo P (2012) The legume root nodule: From symbiotic nitrogen fixation to senescence In Nagata T, ed, Senescence. InTech, Rijeka, Croatia, pp 137–168 [Google Scholar]
- Fåhraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JE, Gulledge J, Engelhaupt E, Burow ME, McLachlan JA (2007) Pesticides reduce symbiotic efficiency of nitrogen-fixing rhizobia and host plants. Proc Natl Acad Sci USA 104: 10282–10287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukudome M, Watanabe E, Osuki KI, Imaizumi R, Aoki T, Becana M, Uchiumi T (2019) Stably transformed Lotus japonicus plants overexpressing phytoglobin LjGlb1-1 show decreased nitric oxide levels in roots and nodules as well as delayed nodule senescence. Plant Cell Physiol 60: 816–825 [DOI] [PubMed] [Google Scholar]
- Godiard L, Lepage A, Moreau S, Laporte D, Verdenaud M, Timmers T, Gamas P (2011) MtbHLH1, a bHLH transcription factor involved in Medicago truncatula nodule vascular patterning and nodule to plant metabolic exchanges. New Phytol 191: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez LD, Baud S, Gilday A, Li Y, Graham IA (2006) Delayed embryo development in the ARABIDOPSIS TREHALOSE-6-PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. Plant J 46: 69–84 [DOI] [PubMed] [Google Scholar]
- González EM, Gálvez L, Arrese-Igor C (2001) Abscisic acid induces a decline in nitrogen fixation that involves leghaemoglobin, but is independent of sucrose synthase activity. J Exp Bot 52: 285–293 [DOI] [PubMed] [Google Scholar]
- Grudkowska M, Zagdańska B (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51: 609–624 [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guinel FC. (2015) Ethylene, a hormone at the center-stage of nodulation. Front Plant Sci 6: 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hädrich N, Hendriks JH, Kötting O, Arrivault S, Feil R, Zeeman SC, Gibon Y, Schulze WX, Stitt M, Lunn JE (2012) Mutagenesis of cysteine 81 prevents dimerization of the APS1 subunit of ADP-glucose pyrophosphorylase and alters diurnal starch turnover in Arabidopsis thaliana leaves. Plant J 70: 231–242 [DOI] [PubMed] [Google Scholar]
- Hakoyama T, Niimi K, Yamamoto T, Isobe S, Sato S, Nakamura Y, Tabata S, Kumagai H, Umehara Y, Brossuleit K, et al. (2012) The integral membrane protein SEN1 is required for symbiotic nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol 53: 225–236 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Gresshoff PM, Ferguson BJ (2014) Mechanistic action of gibberellins in legume nodulation. J Integr Plant Biol 56: 971–978 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Horváth B, Domonkos Á, Kereszt A, Szűcs A, Ábrahám E, Ayaydin F, Bóka K, Chen Y, Chen R, Murray JD, et al. (2015) Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci USA 112: 15232–15237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Finnegan PM, Tyerman SD, Whitehead LF, Bergersen FJ, Day DA, Udvardi MK (1998) Characterization of an ammonium transport protein from the peribacteroid membrane of soybean nodules. Science 281: 1202–1206 [DOI] [PubMed] [Google Scholar]
- Kardailsky IV, Brewin NJ (1996) Expression of cysteine protease genes in pea nodule development and senescence. Mol Plant Microbe Interact 9: 689–695 [DOI] [PubMed] [Google Scholar]
- Karmarkar V. (2014) Transcriptional regulation of nodule development and senescence in Medicago truncatula. PhD thesis. Wageningen University, Wageningen, The Netherlands [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Chen Y, Xi J, Waters C, Chen R, Wang D (2015) An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci USA 112: 15238–15243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Hakoyama T, Umehara Y, Sato S, Kaneko T, Tabata S, Kouchi H (2007) A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol 143: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF (2004) A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J Neurosci 24: 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Feng H, Wen J, Dong J, Wang T (2018) MtCAS31 aids symbiotic nitrogen fixation by protecting the leghemoglobin MtLb120-1 under drought stress in Medicago truncatula. Front Plant Sci 9: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou L, Li Y, Chen D, Tan X, Lei L, Zhou J (2008) A nodule-specific plant cysteine proteinase, AsNODF32, is involved in nodule senescence and nitrogen fixation activity of the green manure legume Astragalus sinicus. New Phytol 180: 185–192 [DOI] [PubMed] [Google Scholar]
- Lohman KN, Gan S, John MC, Amasino RM (1994) Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant 92: 322–328 [Google Scholar]
- Ma L, Zhu F, Li Z, Zhang J, Li X, Dong J, Wang T (2015) TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds. PLoS One 10: e0143877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793 [DOI] [PubMed] [Google Scholar]
- Marini AM, Springael JY, Frommer WB, André B (2000) Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol Microbiol 35: 378–385 [DOI] [PubMed] [Google Scholar]
- Naito Y, Fujie M, Usami S, Murooka Y, Yamada T (2000) The involvement of a cysteine proteinase in the nodule development in Chinese milk vetch infected with Mesorhizobium huakuii subsp. rengei. Plant Physiol 124: 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navascués J, Pérez-Rontomé C, Gay M, Marcos M, Yang F, Walker FA, Desbois A, Abián J, Becana M (2012) Leghemoglobin green derivatives with nitrated hemes evidence production of highly reactive nitrogen species during aging of legume nodules. Proc Natl Acad Sci USA 109: 2660–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CJ, Lee H, Kim H, An CS (2004) Isolation and characterization of a root nodule-specific cysteine proteinase cDNA from soybean. J Plant Biol 47: 216–220 [Google Scholar]
- Okamoto T. (2006) Transport of proteases to the vacuole: ER export bypassing Golgi? In Robinson DG, ed, The Plant Endoplasmic Reticulum. Springer-Verlag, Heidelberg, Germany, pp 125–139 [Google Scholar]
- Pérez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M (2010) Comparison of developmental and stress-induced nodule senescence in Medicago truncatula. Plant Physiol 152: 1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre O, Hopkins J, Combier M, Baldacci F, Engler G, Brouquisse R, Hérouart D, Boncompagni E (2014) Involvement of papain and legumain proteinase in the senescence process of Medicago truncatula nodules. New Phytol 202: 849–863 [DOI] [PubMed] [Google Scholar]
- Pladys D, Vance CP (1993) Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol 103: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM, de Felipe MR, Harrison J, Vanacker H, Foyer CH (2005) Legume nodule senescence: Roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol 165: 683–701 [DOI] [PubMed] [Google Scholar]
- Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stüber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RA (2012) Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol 158: 1583–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Serova TA, Tikhonovich IA, Tsyganov VE (2017) Analysis of nodule senescence in pea (Pisum sativum L.) using laser microdissection, real-time PCR, and ACC immunolocalization. J Plant Physiol 212: 29–44 [DOI] [PubMed] [Google Scholar]
- Serova TA, Tsyganova AV, Tikhonovich IA, Tsyganov VE (2019) Gibberellins inhibit nodule senescence and stimulate nodule meristem bifurcation in pea (Pisum sativum L.). Front Plant Sci 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, Li XP, Chen WX, He QG, Chen JY, et al. (2012) Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J Exp Bot 63: 5171–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheokand S, Dahiya P, Vincent JL, Brewin NJ (2005) Modified expression of cysteine protease affects seed germination, vegetative growth and nodule development in transgenic lines of Medicago truncatula. Plant Sci 169: 966–975 [Google Scholar]
- ShuoHao H, Jing L, Jie Z, JianYun Z, LongQuan H (2019) Identification and characterization of a pyridoxal 5′-phosphate phosphatase in tobacco plants. Plant Sci 278: 88–95 [DOI] [PubMed] [Google Scholar]
- Sinharoy S, Torres-Jerez I, Bandyopadhyay K, Kereszt A, Pislariu CI, Nakashima J, Benedito VA, Kondorosi E, Udvardi MK (2013) The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25: 3584–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonoha-Arther C, Wang D (2018) Tough love: Accommodating intracellular bacteria through directed secretion of antimicrobial peptides during the nitrogen-fixing symbiosis. Curr Opin Plant Biol 44: 155–163 [DOI] [PubMed] [Google Scholar]
- Sun TP. (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6: e0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Guo D, Wei B, Zhang F, Pang C, Jiang H, Zhang J, Wei T, Gu H, Qu LJ, et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers AC, Soupène E, Auriac MC, de Billy F, Vasse J, Boistard P, Truchet G (2000) Saprophytic intracellular rhizobia in alfalfa nodules. Mol Plant Microbe Interact 13: 1204–1213 [DOI] [PubMed] [Google Scholar]
- Trujillo DI, Silverstein KAT, Young ND (2019) Nodule-specific PLAT domain proteins are expanded in the Medicago lineage and required for nodulation. New Phytol 222: 1538–1550 [DOI] [PubMed] [Google Scholar]
- Tse YC, Lo SW, Hillmer S, Dupree P, Jiang L (2006) Dynamic response of prevacuolar compartments to brefeldin a in plant cells. Plant Physiol 142: 1442–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol 141: 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. (2008) Plant proteases: From phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Brewin NJ (2000) Immunolocalization of a cysteine protease in vacuoles, vesicles, and symbiosomes of pea nodule cells. Plant Physiol 123: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yu H, Luo L, Duan L, Cai L, He X, Wen J, Mysore KS, Li G, Xiao A, et al. (2016) NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol 212: 176–191 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu J, Li H, Yang S, Körmöczi P, Kereszt A, Zhu H (2018) Nodule-specific cysteine-rich peptides negatively regulate nitrogen-fixing symbiosis in a strain-specific manner in Medicago truncatula. Mol Plant Microbe Interact 31: 240–248 [DOI] [PubMed] [Google Scholar]
- Wang Q, Yang S, Liu J, Terecskei K, Ábrahám E, Gombár A, Domonkos Á, Szűcs A, Körmöczi P, Wang T, et al. (2017) Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc Natl Acad Sci USA 114: 6854–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J, Chen Y, Nakashima J, Wang SM, Chen R (2013) Medicago truncatula esn1 defines a genetic locus involved in nodule senescence and symbiotic nitrogen fixation. Mol Plant Microbe Interact 26: 893–902 [DOI] [PubMed] [Google Scholar]
- Xiang L, Etxeberria E, Van den Ende W (2013) Vacuolar protein sorting mechanisms in plants. FEBS J 280: 979–993 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya-Ito H, Shimoda Y, Hakoyama T, Sato S, Kaneko T, Hossain MS, Shibata S, Kawaguchi M, Hayashi M, Kouchi H, et al. (2018) Loss-of-function of ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1) in Lotus japonicus restricts efficient nitrogen-fixing symbiosis with specific Mesorhizobium loti strains. Plant J 93: 5–16 [DOI] [PubMed] [Google Scholar]
- Yang S, Wang Q, Fedorova E, Liu J, Qin Q, Zheng Q, Price PA, Pan H, Wang D, Griffitts JS, et al. (2017) Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc Natl Acad Sci USA 114: 6848–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao S, Lercher MJ, Hu S, Chen WH (2012a) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40: W569–W572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Xia X, Zhang Y, Gan SS (2012b) An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J 69: 667–678 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hu X, Zhang Y, Miao Z, Xie C, Meng X, Deng J, Wen J, Mysore KS, Frugier F, et al. (2016) Opposing control by transcription factors MYB61 and MYB3 increases freezing tolerance by relieving C-repeat binding factor suppression. Plant Physiol 172: 1306–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lu D, Xu G, Finlayson SA, He P, Shan L (2015) The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J Exp Bot 66: 3353–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]