Phosphorylation dynamics of CURVATURE THYLAKOID 1B, PSII core, and Light-harvesting II complex proteins cooperate in regulation of thylakoid lateral heterogeneity under fluctuating light.
Abstract
Thylakoid membranes in land plant chloroplasts are organized into appressed and nonappressed membranes, which contribute to the control of energy distribution between the two photosystems (PSI and PSII) from the associated light-harvesting complexes (LHCs). Under fluctuating light conditions, fast reversible phosphorylation of the N-terminal thylakoid protein domains and changes in electrostatic forces induce modifications in thylakoid organization. To gain insight into the role and dynamics of thylakoid protein phosphorylation, we used targeted proteomics to quantify amounts of the structural proteins CURVATURE THYLAKOID1 (CURT1), including the levels of CURT1B N terminus phosphorylation and acetylation, after short-term fluctuating light treatments of Arabidopsis (Arabidopsis thaliana). The CURT1B protein was localized to a specific curvature domain separated from the margin domain, and specifically depleted of chlorophyll-binding protein complexes. The acetylation and phosphorylation of the CURT1B N terminus were mutually exclusive. The level of CURT1B phosphorylation, but not of acetylation, increased upon light shifts that also led to an increase in PSII core protein phosphorylation. These dynamics were largely absent in the knockout mutant of PSII core protein kinase SER/THR PROTEIN KINASE8 (STN8). Moreover, in mutants impaired in interaction between phosphorylated LHCII and PSI, the phosphorylation dynamics of CURT1B and the amount of the other CURT1 proteins were misregulated, indicating a functional interaction between CURT1B and PSI-LHCII complexes in grana margins. The complex relationships between phosphorylation of PSII, LHCII, and CURT1B support the dynamics of thylakoid protein complexes that are crucial in the optimization of photosynthesis under fluctuating light intensities.
The photosynthetic conversion of solar energy into reducing equivalents and chemical bonds is achieved in plant chloroplasts by the orchestrated action of two thylakoid membrane-integral pigment-binding protein complexes, PSI and PSII, together with cytochrome b6f (Cyt b6f) complex, ATP synthase, light harvesting complexes (LHCs) I and II, and several auxiliary proteins. The regulation of the photosynthetic apparatus is tightly connected to the structural arrangement of the thylakoid membrane with defined appressed and nonappressed regions. The two photosystems are spatially segregated due to defined lateral heterogeneity of the thylakoid membrane system, with PSII predominantly present in the appressed membranes (grana), while PSI is confined to the stroma-exposed nonappressed thylakoid membranes (lamellae; Andersson and Anderson, 1980). How exactly the thylakoid membrane is organized into grana and how the continuity with the nonappressed regions is guaranteed is still a matter of debate (Chow et al., 2005; Mullineaux, 2005; Anderson et al., 2012; Kirchhoff, 2013; Pribil et al., 2014; Järvi et al., 2017). Studies with PSII- or PSI-deficient mutants demonstrate that grana stacking is mainly due to the interaction of the N-terminal domains of the LHCII proteins (Mullet and Arntzen, 1980; Simpson, 1982; Simpson et al., 1989; Ruban et al., 2003; Kim et al., 2009; Belgio et al., 2015).
The N-termini of LIGHT-HARVESTING CHLOROPHYLL A/B-BINDING 1 (LHCB1) and LHCB2, as well as of several other thylakoid proteins, including the PSII core subunits PsbA (D1), PsbC (CP43), PsbD (D2), and PsbH, undergo N-terminal acetylation and concurrent phosphorylation of the next available Thr residues (Vener et al., 2001; Hansson and Vener, 2003). The kinase/phosphatase couples SER/THR PROTEIN KINASE7 (STN7)/THYLAKOID ASSOCIATED PHOSPHATASE 38 (TAP38) and STN8/PSII CORE PHOSPHATASE (PBCP) are responsible for reversible phosphorylation of LHCII and PSII core proteins, respectively (Pesaresi et al., 2011; Rochaix et al., 2012). The dynamics of PSII core and LHCII phosphorylation also affect the interaction between appressed membranes in grana stacks, altering the attractive and repulsive forces in the partition gap (for review, see Chow et al., 2005). The effect of different light qualities versus different white light intensities on the phosphorylation dynamics of LHCII and PSII has been extensively studied. Both LHCII and PSII core are maximally phosphorylated under light treatments favoring PSII excitation, such as red or blue light, while almost complete dephosphorylation is achieved by preferentially exciting PSI with far red light (Tikkanen et al., 2006; Trotta et al., 2016). Changes in white light intensities, reflecting natural growth conditions, instead cause opposite phosphorylation dynamics of the PSII core with respect to LHCII phosphorylation (Rintamäki et al., 1997; Tikkanen et al., 2006, 2010). However, the specific contribution of phosphorylation of either the PSII core or LHCII to the size of the partition gap between thylakoid layers in grana stacks is still an unsolved question (Fristedt et al., 2009; Puthiyaveetil et al., 2017).
Both in vivo and in vitro studies have indicated a decrease in grana diameter and the total area of appressed membranes under red or blue light, i.e. when both the PSII core and LHCII become phosphorylated (Chuartzman et al., 2008; Iwai et al., 2018). However, a similar decrease in appressed thylakoid area was observed in dark-acclimated plants exposed directly to high light (HL; Khatoon et al., 2009; Herbstová et al., 2012) when there is high PSII core phosphorylation but complete dephosphorylation of LHCII (Tikkanen et al., 2008a). Intriguingly, under red light the grana membranes appear loosely appressed in wild-type but not in stn7 plants (Dietzel et al., 2011), in which, as under HL, only the PSII core proteins are highly phosphorylated (Tikkanen et al., 2006). Finally, an increase in the lumen volume and grana margin area has been observed in thylakoids of plants exposed to high photoinhibitory illumination directly after dark acclimation (Herbstová et al., 2012; Yoshioka-Nishimura et al., 2014), indicating that other factors besides PSII/LHCII phosphorylation must be involved in the dynamics of thylakoid heterogeneity.
Recently, members of a family of membrane-integral thylakoid proteins named CURVATURE THYLAKOID1 (CURT1) have been proposed as new key components in shaping grana dimensions in response to changes in light quality and quantity (Armbruster et al., 2013; Pribil et al., 2018). One of them, CURT1B, formerly known as TMP14 or PSAP (At2g4682), was originally described as a PSI subunit interacting with PsaL/PsaH/PsaO (Khrouchtchova et al., 2005; Yu et al., 2008). CURT1B is a relatively abundant protein that is prone to acetylation and phosphorylation at the N terminus (Hansson and Vener, 2003; Bienvenut et al., 2012), but the role and dynamics of these posttranslational modifications (PTMs) of CURT1 proteins have not been clearly demonstrated, besides the fact that the phosphorylation of CURT1B is below the detection limit in stn7 stn8 thylakoids (Ingelsson and Vener, 2012). Since CURT1 proteins are critical for mediating curvature within the grana margins, it has been postulated that CURT1 might be responsible for inducing a local unstacking of grana edges to help the operation of the PSII repair cycle (Yamamoto et al., 2013).
To gain further insights into the role and dynamics of CURT1 proteins, we quantified the amounts of proteins CURT1A–CURT1D and the levels of CURT1A and CURT1B PTMs in Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0; hereafter Arabidopsis) after short-term fluctuating light treatments. The CURT1B N terminus is strictly either phosphorylated, acetylated, or not modified at all, and the shift to light conditions leading to increased PSII core protein phosphorylation is accompanied by increased CURT1B phosphorylation. The response of the phosphorylation of CURT1B, LHCII, and PSII core proteins to changes in light intensity was investigated in several mutants impaired in either LHCII or PSII core phosphorylation, providing a comprehensive picture of the interplay between phosphorylation of these thylakoid proteins in dynamic modulations of the appressed membranes. Such dynamics allow optimal excitation of both photosystems and efficient photosynthetic electron flow under constantly changing natural environments.
RESULTS
Thr-65 Phosphorylation and Ala-64 Acetylation of the CURT1B N Terminus Are Mutually Exclusive
Our first goal was the detection and identification by nanoscale liquid chromatographic tandem mass spectrometry (MS/MS) of the so-called proteotypic peptides, i.e. unique peptides suitable for quantification in selected reaction monitoring (SRM), from all four Arabidopsis CURT1 proteins (CURT1A–curt1d) and their possible PTMs without using enrichment, thus making it possible to contemporaneously quantify both the PTMs and the protein levels (Trotta et al., 2016). Arabidopsis plants (32 d old) grown under an 8-h photoperiod (120 µmol photons m−2 s−1) were used, and thylakoids were isolated at the end of a 16 h dark period (Da1) after subsequent treatment for 2 h at low light (LL; 20 µmol photons m−2 s−1) and after an additional 2 h of HL treatment (1,000 µmol photons m−2 s−1). Thereafter, thylakoids were digested with trypsin and the obtained peptides were identified using a Q-exactive mass spectrometer (Thermo Fisher Scientific; Supplemental Table S1). Besides the proteotypic peptides from all four CURT1 proteins, we identified the previously described CURT1A and CURT1B N-terminal peptides acetylated on Ala-63 (63Ac-ASSEETSSIDTNELITDLK81) and Ala-64 (64Ac-ATTEVGEAPATTTEAETTELPEIVK88), respectively, as well as the CURT1B N-terminal single phosphorylated peptide (64ApTTEVGEAPATTTEAETTELPEIVK88; Supplemental Table S2; Hansson and Vener, 2003; Bienvenut et al., 2012). For the latter peptide, the analysis of generated spectra by phosphoRS (Taus et al., 2011) indicated that the phosphorylation site is most likely at Thr-66 (Supplemental Fig. S2), although this assignment was not evident in all recorded spectra (Supplemental Table S2). Interestingly, the CURT1B N terminus was never phosphorylated and acetylated at the same time, as observed for many membrane-spanning thylakoid proteins, including LHCB1, LHCB2, and PSII core subunits (Supplemental Table S2; Vener et al., 2001; Hansson and Vener, 2003).
SRM was next used for accurate quantification of proteins by monitoring multiple fragments (transitions) generated from selected precursor proteotypic peptides during their entire chromatographic elution (Supplemental Figs. S2 and S3). In this way, it is possible to determine the phosphorylated residue by monitoring the coelution of fragments (y and b ions), which must be generated in case of the presence of a phospho-group (Trotta et al., 2016). Thus, we first tested which transitions corresponding to phosphorylation on Thr-65 or Thr-66 were coeluting with the rest of the transitions from the phosphopeptide 64ApTTEVGEAPATTTEAETTELPEIVK88 (Supplemental Fig. S2). Unlike observations in the MS/MS spectra of the N terminus of CURT1B (Supplemental Fig. S1), we could monitor the transition corresponding to Ala-64 plus phosphorylated Thr-65 (ion b2; Supplemental Fig. S2A, left), indicating that the actual site of reversible CURT1B phosphorylation was Thr-65, as previously suggested (Hansson and Vener, 2003). Moreover, to confirm that the correct peptide was monitored, we analyzed the coeluting transitions of the respective CURT1A and CURT1B peptides extracted from thylakoids of curt1a and curt1b knockout plants. No signals of CURT1A or CURT1B peptides were detected in the preparations of the corresponding knockout plants. In the case of transitions used for quantification of CURT1B phosphorylation (Supplemental Fig. S2B, right), weak signals were detected due to interfering transitions (Supplemental Fig. S2B, left), and these signals were excluded during quantification (Supplemental Table S4).
Phosphorylation, But Not Acetylation, of CURT1B Is Affected by Shifts in Light Intensity
Arabidopsis plants were illuminated under changing white light intensities (illustrated in Fig. 1A), and the levels of phosphorylation of CURT1B (Fig. 1B) and acetylation of CURT1A and CURT1B (Fig. 2), as well as the amount of CURT1 proteins (Supplemental Fig. S3), were quantified by applying SRM. As a control for standardization between the measurements of thylakoid replicates, we also measured the level of the PSI core subunits PsaA and PsaB in parallel with CURT1 proteins (Supplemental Fig. S3). Moreover, since CURT1B and CURT1C interact with small subunits of PSI (Yu et al., 2008), the levels of PSAL and PSAH, which form the LHCII-interacting domain of the PSI complex (Lunde et al., 2003; Rantala et al., 2016), and that of PSAK as a control peripheral subunit, were likewise quantified (Supplemental Tables S3 and S4). Finally, the levels of the six LHCI subunits were monitored in all measurements as a control.
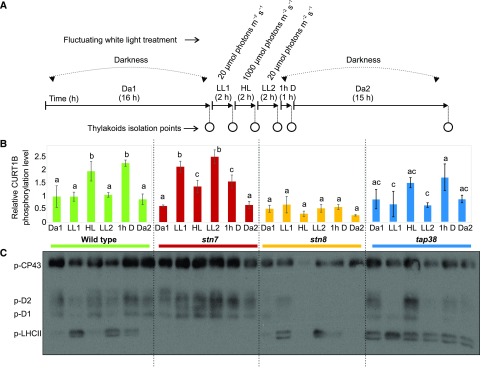
Figure 1.
Phosphorylation dynamics of CURT1B, PSII core, and LHCII proteins in wild-type, stn7, stn8, and tap38 plants. A, Experimental design of the fluctuating white light treatment used to assess the dynamics of protein expression and posttranslational modifications. Plants grown under 120 μmol photons m−2 s−1 with a photoperiod of 8 h light and 16 h dark were subjected to 2 h of LL1 (20 μmol photons m−2 s−1), 2 h of HL (1,000 μmol photons m−2 s−1), 2 h of LL2 (20 μmol photons m−2 s−1), and finally 16 h of dark. Thylakoids were isolated at the end of Da1, at the end of each light shift (LL1, Hl, and LL2), then after 1h D, and finally after Da2. B, Changes in the phosphorylation level of the N-terminal CURT1B peptide 64ATTEVGEAPATTTEAETTELPEIVK88. The level of phosphorylation was calculated as the percentage of the intensity of the phosphorylated peptide with respect to the sum of the intensities of both the phosphorylated peptide and the nonphosphorylated/nonacetylated form of the same peptide. For clarification, the y axis indicates the level of CURT1B phosphorylation with respect to that in the Da1 fraction in the wild type, which was set to 1. The dynamics upon shifts in light intensity are shown for wild-type (green), stn7 (red), stn8 (orange), and tap38 (blue) plants. Lowercase letters indicate statistically different levels of phosphorylation in each genotype (P < 0.05 according to the all-pairwise multiple-comparison one-way ANOVA) and refer to the ANOVA test run within light treatments in the same genotype. No difference was found in the starting Da1 time point among the genotypes. Error bars indicate the SD (n = 3). C, Representative immunoblot (n = 3) based on chl loading (0.5 µg) and immunodecorated with anti-P-Thr antibody, showing changes in phosphorylation levels of PSII core subunits CP43, D1, and D2, and of LHCII. The same sample preparations were analyzed as with SRM (B; Supplemental Fig. S3).
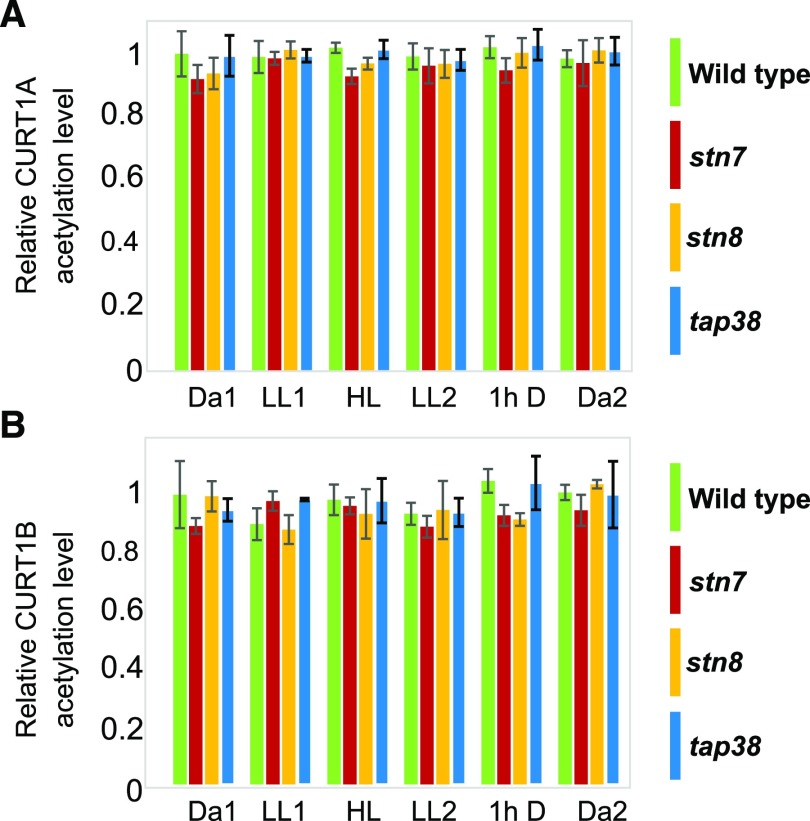
Figure 2.
Acetylation dynamics of CURT1A and CURT1B upon light shifts in wild-type, stn7, stn8, and tap38 CURT1A and CURT1B N-terminal peptides acetylated on Ala-63 (63Ac-ASSEETSSIDTNELITDLK81) and Ala-64 (64Ac-ATTEVGEAPATTTEAETTELPEIVK88), respectively, upon changes in illumination conditions in the wild type (green) and stn7 (red), stn8 (orange), and tap38 mutant lines (blue). A, Relative percentage of acetylation of the CURT1A N terminus. B, Relative percentage of acetylation of the CURT1B N terminus. The level of acetylation was calculated as a percentage of the intensity of the acetylated peptide with respect to the sum of the intensities of all three detected forms of the same peptide (acetylated, phosphorylated, and nonphosphorylated/nonacetylated). For clarity, the y axis indicates the level of acetylation with respect to the wild-type Da1 level, which was set to 1. In contrast to the phosphorylated form of the same peptide, no changes were observed in the relative percentage of the acetylated peptide with respect to the sum of the acetylated, phosphorylated and nonmodified peptides. Error bars indicate the SD (n = 3, all-pairwise multiple-comparison one way ANOVA).
Wild-type Arabidopsis was first assessed for the dynamics of expression of the above-mentioned proteins and of CURT1A and CURT1B PTMs. To this end, plants were treated with a fluctuating white light cycle, and thylakoids were isolated at six time points (Fig. 1A): at the end of the 16 h dark period (Da1), after subsequent 2 h at LL (LL1), then after 2 h of HL, again after 2 h of LL (LL2), and finally after transfer for 1 h to darkness (1h D), followed by 15 h of darkness (Da2; Fig. 1A). Since the applied fluctuations in white light intensity are known to cause opposite PSII core and LHCII protein phosphorylation dynamics (Tikkanen et al., 2006; Trotta et al., 2016), relative Thr phosphorylation levels of PSII core and LHCII were recorded by immunoblotting (Fig. 1C) using the same thylakoid protein samples as for SRM analyses of CURT1B phosphorylation (Fig. 1B).
In wild-type plants (Fig. 1B, green bars), the level of phosphorylation of CURT1B was stable in the shift from Da1 to LL1, while the shift to HL resulted in a 2-fold increase, which reverted back to basal level (Da1) at the end of LL2. The shift to darkness (1h D) again caused a 2-fold increase in the level of CURT1B phosphorylation, which decreased back to the basal level by the end of the subsequent 15-h dark period (Da2). The levels of acetylation of CURT1A (Fig. 2A) and CURT1B (Fig. 2B) remained largely stable upon changing light intensities, indicating that the two PTMs did not influence or interfere with each other. It is noteworthy that none of the proteins quantified (i.e. CURT1A–CURT1D and the PSI and LHCI subunits) varied in abundance during the periods of applied light fluctuation (Supplemental Fig. S3).
STN8 and PSII Core Phosphorylation Are the Main Determinants of Increased CURT1B Phosphorylation
To understand the relationships between CURT1B phosphorylation and that of the PSII core and LHCII proteins, we next monitored the phosphorylation dynamics in light shifts (Fig. 1A), comparing the wild type to stn7, stn8, and tap38 plants. In the wild type (Fig. 1B, green bars), the increases in CURT1B phosphorylation (Fig. 1B, transitions to HL and 1h D) followed an increase in PSII core and a decrease in LHCII protein phosphorylation (Fig. 1C). Lack of STN8 completely abolished the dynamics of CURT1B phosphorylation upon any change in light intensity (Fig. 1B, orange bars), although a basal phosphorylation level was still detectable, consistent with earlier reports on the presence of CURT1B phosphopeptides in stn8 plants (Fristedt and Vener, 2011; Reiland et al., 2011; Ingelsson and Vener, 2012). On the other hand, the dynamics of LHCII phosphorylation in stn8 was similar to that of the wild type (Fig. 1C), indicating that the amount of CURT1B phosphorylation was particularly linked to the function of the STN8 kinase.
Lack of STN7, similar to stn8, did not affect the basal level of CURT1B phosphorylation in darkness (Da1; Fig. 1B, red bars). Instead, the shift of plants from darkness to LL1 caused an increase in CURT1B phosphorylation to levels similar to those observed in the wild type after HL and 1h D treatments. Intriguingly, the shift of stn7 plants to HL decreased CURT1B phosphorylation to an intermediate level between those observed in Da1 and LL1. Levels similar to those seen in LL1 and HL were reached in subsequent shifts of stn7 from HL to LL2 and from LL2 to 1h D. At the end of the Da2 period, the level of phosphorylation was again at the basal level. In general, enhanced CURT1B phosphorylation in stn7 occurred in parallel with an increase in PSII core protein phosphorylation (Fig. 1C), although the dynamics were not identical, especially in the HL phase (see “Discussion”). However, acetylation of CURT1A and CURT1B, and the amounts of PSI subunits, LHCI subunits, and CURT1 proteins assessed by SRM, were not affected by the lack of STN7 or STN8 (Fig. 2; Supplemental Fig. S3).
The tap38 plants, characterized by hyperphosphorylation of LHCII (Rantala et al., 2016), were next used to further assess the phosphorylation dynamics between LHCII and CURT1B. While the basal level of phosphorylation of CURT1B was similar in the wild type and tap38 at the end of Da1, the dynamics of phosphorylation in tap38 appeared attenuated in amplitude, being significantly affected only after the shift from LL2 to 1h D, not after the other shifts in light intensity. Interestingly, in tap38 there was only a weak correlation between CURT1B and PSII core protein phosphorylation dynamics (Fig. 1, B and C), indicating that the effect of hyperphosphorylated LHCII or increased grana stacking (Armbruster et al., 2013; Iwai et al., 2018) overrides other possible determinants of the phosphorylation status of CURT1B. Acetylation of CURT1A and CURT1B and protein levels of the four CURT1 isoforms, as well as the PSI and LHCI subunits, were all unaffected by the lack of TAP38 phosphatase (Fig. 2; Supplemental Fig. S3).
PSI-LHCII Interaction Is Critical for Protein and Phosphorylation Levels of CURT1
The experiments with both the stn7 kinase and tap38 phosphatase mutants indicated a putative interlink between LHCII and CURT1B phosphorylation dynamics besides the link observed with the PSII core protein phosphorylation (Fig. 1). Thus, we tested two other genotypes with impaired LHCII phosphorylation dynamics, i.e. the psb33 plants, which harbor functional STN7 kinase and TAP38 phosphatase (Fristedt et al., 2017), and the psal mutant, which lacks the PSI proteins PSAL and PSAH that form the docking site for p-LHCII (Lunde et al., 2003). In particular, psb33 is unable to phosphorylate LHCII after shifts to LL (Fig. 3; Fristedt et al., 2017), and psal shows constitutive hyperphosphorylated LHCII under all tested light conditions (Fig. 3; Lunde et al., 2003; Rantala et al., 2016). Importantly, the interaction between PSI and the “state-transition” LHCII trimer is compromised in both psb33 and psal.
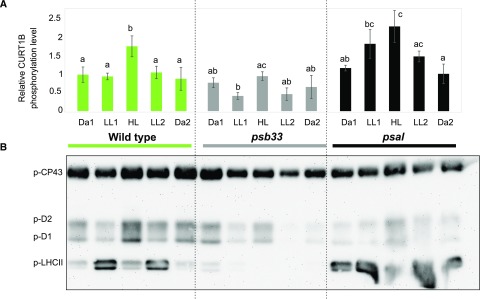
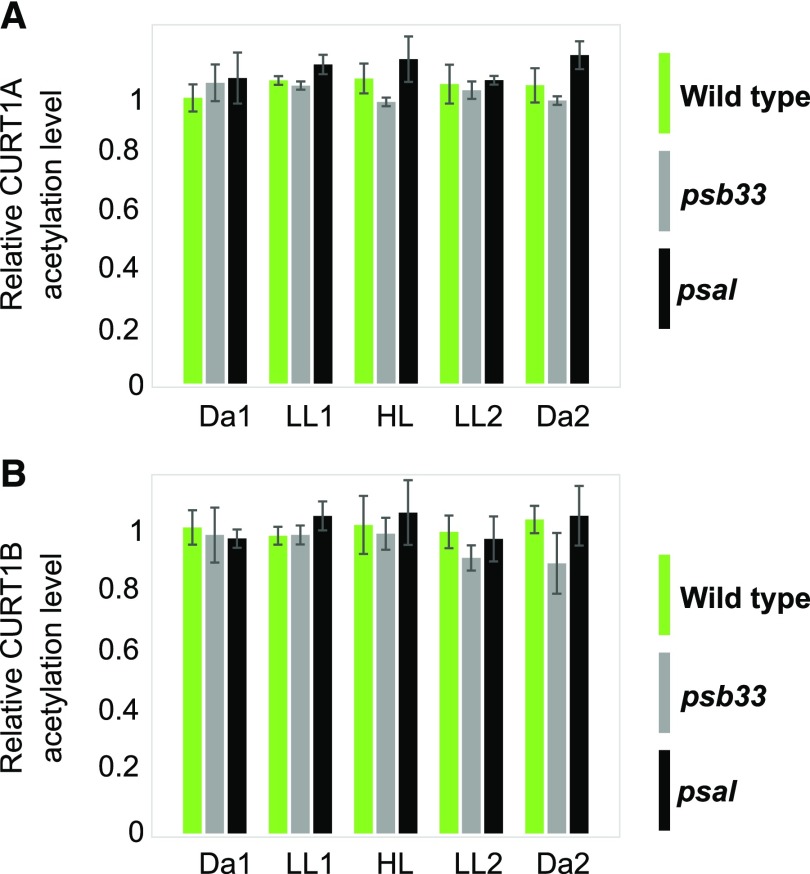
Figure 3.
Phosphorylation dynamics of CURT1B, PSII core, and LHCII N-terminal peptides in wild-type, psb33, and psal plants. A, Changes in the phosphorylation level of CURT1B 64ATTEVGEAPATTTEAETTELPEIVK88 phosphopeptide upon shifts in light intensity, as in Figure 1. The dynamics of CURT1B phosphorylation upon shifting light intensities are shown for wild-type (green), psb33 (gray), and psal (black) plants. In this experiment, the 1h D point (see Figs. 1 and 2) was omitted. The letters indicate statistically different levels of phosphorylation in each genotype (P < 0.05 according to all-pairwise multiple-comparison one-way ANOVA). Error bars indicate the SD (n = 3). B, Representative immunoblot immunodecorated with antip-Thr antibody and showing the changes in levels of phosphorylation of the N terminus of PSII core subunits CP43, D1, and D2, and of LHCII, in the same samples analyzed using SRM (A; Supplemental Fig. S4). Samples corresponding to 0.5 µg of chl were loaded in each well.
When the psb33 and psal plants were subjected to changing light intensities as described above, the basal level of phosphorylation of CURT1B after the long dark period (Da1) was similar in the wild type, psb33, and psal (Fig. 3A, green, gray, and black bars respectively). Thus, the basal level of phosphorylation of CURT1B is independent of the constitutive LHCII phosphorylation observed in psal (Fig. 3B) and tap38 (Fig. 1; Lunde et al., 2003; Rantala et al., 2016) and of the lack of LHCII phosphorylation, despite active STN7 kinase, in psb33 (Fristedt et al., 2017).
The shift of psb33 to LL resulted in a significant decrease of CURT1B phosphorylation to half of the basal level observed in Da1 (Fig. 3A, gray bars). It is thus conceivable that the role of PSB33 is hierarchically above the thylakoid protein phosphorylation-related regulatory events that control the interaction between the state-transition LHCII trimer and PSI, as well as the phosphorylation of CURT1B protein (see “Discussion”). However, it should be noted that in the case of psb33, the associated phosphorylation dynamics of CURT1B and PSII core were similar but attenuated compared to the wild type (Fig. 3B). In contrast, psal demonstrated a strong increase in CURT1B phosphorylation upon transfer to LL1 (Fig. 3A, black bars), similar to the effect described for stn7 (Fig. 1A, red bars). It would follow that in stn7 and psal the increase in CURT1B phosphorylation in LL took place because of the lack of interaction between state-transition LHCII trimer and PSI, irrespective of whether this lack of interaction was due to lack of p-LHCII in stn7 and/or lack of p-LHCII binding to PSI in psal.
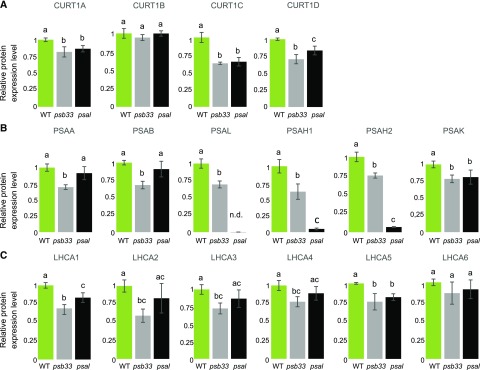
Interestingly, the protein levels of CURT1 isoforms other than CURT1B were strongly affected in both psb33 and psal thylakoids (Fig. 4A), while stn7, stn8, and tap38 had wild-type levels of CURT1 proteins (Supplemental Fig. S3). In particular, the CURT1A, CURT1C, and CURT1D proteins were reduced to almost the same level in both psb33 and psal plants (Fig. 4, gray and black bars, respectively), i.e. to about 80%, 60%, and 70%, respectively, of wild-type protein amounts (Fig. 4A). This suggests a possible effect of the lack of PSB33 or PSAL on the thylakoid ultrastructure. Besides the near absence of the PSAH isoforms in psal (Lunde et al., 2000), the other peripheral subunit, PSAK, also showed lower amounts compared to the wild type (Fig. 4 B, black bars). In contrast to the psal plants, the levels of core and peripheral PSI subunits, as well as the PSI antenna subunits (LHCAs), were lower in psb33 with respect to the wild type (Fig. 4, B and C). The abundance of all the proteins measured remained nearly unchanged at the following time points (Supplemental Fig. S4). Thus, it appears that lack of either the PSB33 or the PSAL protein affects PSI protein stoichiometry in different ways, although it causes a very similar effect on CURT1A, CURT1C, and CURT1D isoforms at the protein level. Finally, as in all genotypes investigated so far, the level of acetylation of CURT1A and CURT1B was not affected in the psb33 and psal plants, suggesting that the two PTMs are acting on different pools of CURT1 proteins (Fig. 5).
Figure 4.
Quantification of CURT1 isoforms, PSI subunits, and LHCA isoforms in wild-type, psb33, and psal plants by SRM. A, Protein level of CURTl isoforms at the end of the Da1 period in thylakoids of wild-type (WT; green), psb33 (gray), and psal (black) plants. B, Protein levels of PSI core subunits PSAA and PSAB, of the PSI subunits responsible for interaction with LHCII (PSAL, PSAHl, and PSAH2), and of the PSI peripheral subunit PSAK at the end of the Da1 period. C, Protein levels of the six isoforms of the PSI antenna LHCA at the end of the Da1 period. Lowercase letters indicate statistically different levels of phosphorylation within genotypes (P < 0.05 according to all-pairwise multiple-comparison one-way ANOVA). Error bars indicate the SD (n = 3). In all cases, the y axis indicates the level of expression of the protein with respect to that in the wild type, which was set to 1.
Figure 5.
Dynamics of acetylation of CURT1A and CURT1B in wild-type (green), psb33 (gray), and psal (black) CURT1A and CURT1B N-terminal peptides on Ala-63 (63Ac-ASSEETSSIDTNELITDLK81) and Ala-64 (64Ac-ATTEVGEAPATTTEAETTELPEIVK88), respectively, upon exposure of the peptides to changing light intensities. A, Relative percentage of acetylation of the CURT1A N terminus. B, Relative percentage of acetylation of the CURT1B N terminus. No significant changes were observed in the relative percentage of acetylated peptide with respect to either the nonacetylated or the nonphosphorylated peptides, as also shown for the stn7, stn8, and tap38 mutants (Fig. 2). Error bars indicate the SD (n = 3, all-pairwise multiple-comparison one-way ANOVA).
Grana Margins and Curvature Domains of Thylakoids Can Be Isolated
To better understand the role of CURT1A and CURT1B proteins, we attempted to localize them more precisely in the heterogeneous thylakoid membrane network. To this end, the thylakoid membranes were isolated from dark-acclimated wild-type plants and subjected to thorough subfractionation to properly separate the grana, margin, stroma, and curvature subfractions.
Wild-type thylakoids at the end of the Da1 dark phase were solubilized with 0.4% digitonin (DIG) according to the protocol of (Kyle et al., 1984), and the fractions corresponding to 10,000g (grana), 40,000g (margins), and 144,000g (stroma membranes) were collected. Besides the pelleted nonappressed stroma membranes at 144,000g, a separate fraction corresponding to the “loose pellet” described by Puthiyaveetil et al. (2014) was collected (Fig. 6A). As shown in Figure 6B (light gray bars), the chlorophyll (chl) a/b ratios of the subfractions, with respect to the starting thylakoids, indicated clear enrichment in chl a in the margins fraction and even more in the stroma membrane fraction, demonstrating an enrichment in PSI and depletion of LHCII, as expected. The loose-pellet fraction, here called the curvature fraction (Fig. 6B), had a chl a/b ratio similar to that of the thylakoid margin subfraction. To analyze the difference between the margin and the curvature fractions, the samples of both intact thylakoids and isolated grana, margin, stroma, and curvature subfractions were loaded on a chl basis on an SDS-PAGE, transferred onto a polyvinylidine fluoride (PVDF) membrane, and probed with CURT1B- and CURT1A-specific antibodies (Fig. 6C). As expected, CURT1B and CURT1A were present mostly in the curvature fraction, where the signal was several-fold higher than in the thylakoids used as a starting material. Since all subfractions were loaded on a chl basis, the high concentrations of CURT1A and CURT1B indicate that the chl-binding protein complexes were present in the curvature subfraction in much lower amounts with respect to the CURT1A and CURT1B proteins.
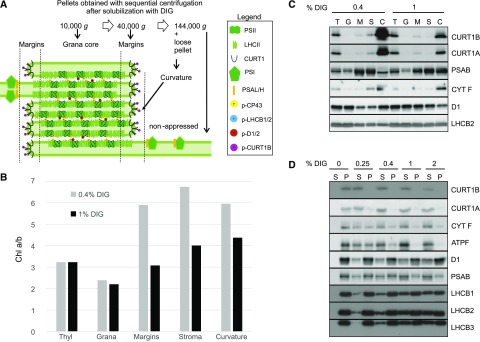
Figure 6.
Biochemical separation of thylakoid membranes into grana core, grana margins, stroma lamellae, and curvature subfractions. A, A scheme of the classical fractionation of the thylakoid domains obtained with sequential centrifugation at the indicated g values. The curvature region in the grana margin domains is obtained as a loose pellet, distinct from the tight pellet containing the nonappressed stroma lamellae, in the 144,000g step (Puthiyaveetil et al., 2014). B, Representative chl a/b ratio in thylakoid (Thyl) subfractions obtained after solubilization with 0.4% (light gray) or 1% (dark gray) DIG (n = 3). Plants were dark adapted (16 h) before thylakoid isolation. C, Representative immunoblots of the fractions shown in B immunodecorated with α-CURT1B, α-CURT1A, α-PSAB, α-CYT F, and α-D1 antibodies. T, Thylakoids; G, grana core; M, margins; S, stroma lamellae; C, curvature fraction. D, Immunoblot of the supernatant and pellet obtained from dark-acclimated thylakoid membranes with increasing concentrations of DIG and immunodecorated with α-CURT1B, α-CURT1A, α-CYT F (for cyt b6f), α-ATPF (for ATPase), α-D1 (for PSII), α-PSAB (for PSI), and α-LHCB1, α-LHCB2, and α-LHCB3 (for LHCII) antibodies. S, supernatant; P, pellet
To verify the existence of a specific curvature subfraction, the same thylakoid starting material was solubilized with 1% DIG, which results in more extensive solubilization of the thylakoids (Rantala and Tikkanen, 2018). As expected, the solubilization with 1% DIG resulted in significantly lower chl a/b ratios in both the grana margin and stroma thylakoid subfractions, as compared to solubilization with 0.4% DIG (Fig. 6B). Likewise, the CURT1B and CURT1A protein signals detected in the curvature subfraction obtained with 0.4% DIG were much more intense than with 1% DIG, indicative of a higher purity of the CURT1-containing subfraction obtained with 0.4% DIG (Fig. 6C). Indeed, the curvature subfraction obtained with 0.4% DIG was much depleted in PSAB and, to a minor extent, also in LHCB2, representing PSI and LHCII complexes, respectively, while the PSII D1 protein was instead enriched (Fig. 6C). Notably, most of the CYT F protein, representing Cyt b6f, was found in the curvature subfraction with both DIG concentrations tested. Moreover, PSII and LHCB2 were mostly confined to the grana fraction of the 0.4%-DIG-solubilized thylakoids, while an even distribution among grana, margins, and stroma subfractions was visible with 1% DIG, indicating that part of the grana was solubilized and present within the 40,000g and 144,000g fractions when 1% DIG was used.
To further test the accessibility of the thylakoid curvature domain to DIG, we next tested the solubility of CURT1A and CURT1B from the thylakoid membrane with increasing concentrations of DIG and compared that with the solubility of the thylakoid embedded protein complexes PSI, PSII, Cyt b6f, ATP synthase, and LHCII (Rantala and Tikkanen, 2018). Wild-type thylakoids isolated at the end of the Da1 period were divided into five different pools and separately solubilized with the different DIG concentrations. Solubilized (supernatant) and insolubilized (pellet) proteins/protein complexes were separated by centrifuging at 18,620g. The resulting pellet was resuspended to the same volume as the supernatant and both fractions were probed by immunoblotting with CURT1A and CURT1B antibodies (Fig. 6D). Already at 0.25% DIG, almost all CURT1A and CURT1B proteins were released from the thylakoid membrane to the supernatant (Fig. 6D). Of the other thylakoid proteins tested, CYT F and ATPF, representing Cyt b6f and ATP synthase, respectively, showed partial release from thylakoids at 0.4% DIG, while PSII, PSI, and especially LHCII were the least prone to becoming solubilized at low concentrations of DIG (Fig. 6D). The different degree of solubilization of the CURT1 proteins with respect to PSI, PSII, and LHCII indicated that the curvature fraction, which is largely devoid of chl-binding protein complexes, represents a separate fraction with respect to the margin domain where the PSII-LHCII-PSI interaction takes place.
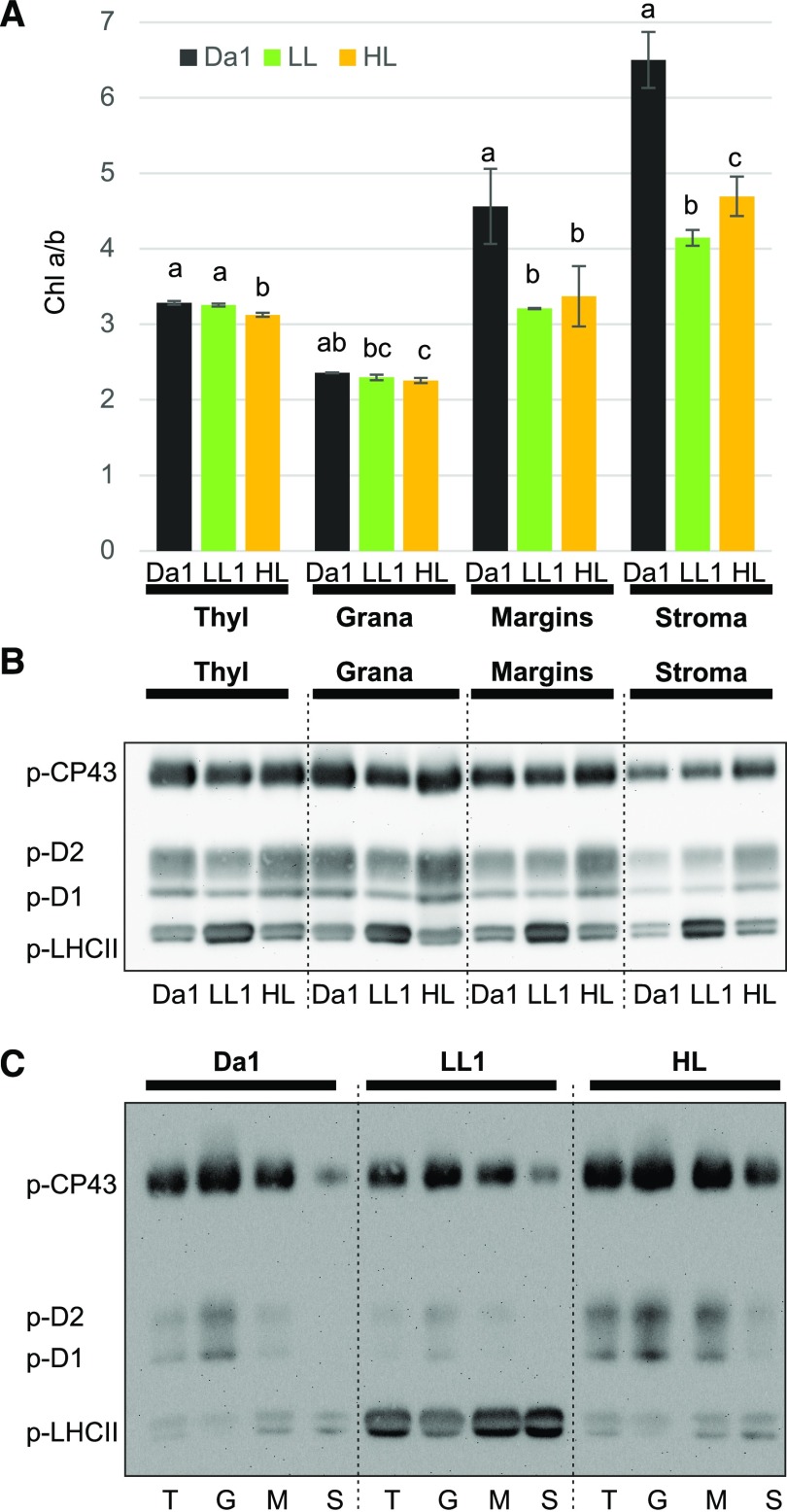
We also attempted to gain insight into the dynamics of the thylakoids fractions, particularly the nonappressed domains where the light-intensity-dependent PSII-LHCII-PSI interaction is likely to occur (Grieco et al., 2015; Suorsa et al., 2015). Illumination of plants is thus expected to result in a lower chl a/b ratio not only in the stroma thylakoids but also in the grana margin fraction, as demonstrated already in the classical fractionation procedure (Kyle et al., 1983, 1984). To analyze the relationship between changes of chl a/b ratio and the phosphorylation pattern of thylakoid proteins, we fractionated thylakoids with 0.4% DIG from Arabidopsis wild-type plants subjected to fluctuating light treatments comprising the Da1, LL1, and HL phases, as indicated in Figures 3 and 5 (Fig. 7A). As expected, the chl a/b ratio of the margin and stroma fractions in LL1 (Fig. 7A, green bars) was significantly lower than in Da1 (Fig. 7A, black bars), and it remained lower than in Da1 also after the subsequent HL phase (Fig. 7A, orange bars). The lower chl a/b ratio in the margin and stroma membrane subfractions in thylakoids isolated from light-exposed plants, as compared to dark-acclimated plants (Fig. 7A), indicates that these are the domains where the lateral heterogeneity of protein complexes is most dynamic (Kyle et al., 1983, 1984; Grieco et al., 2015). To test whether the different chl a/b ratio was accompanied by different phosphorylation patterns of PSII core and LHCII, the proteins from thylakoids, grana, margin, and stroma membrane subfractions were loaded on a chl basis on a SDS-PAGE, separated, transferred onto a PVDF membrane, and finally probed with anti-p-Thr antibody (Fig. 7B). The strongest p-LHCII signal was detected in the LL phase throughout the subfractions. However, the distribution of the p-LHCII signal across all subfractions was rather similar, with a small increase in the margin fraction in all light treatments (Fig. 7C). While the dynamics of PSII core phosphorylation in the grana were similar to those observed in thylakoids, i.e. a decrease after the LL period followed by an increase after the HL period (Fig. 7B) in the margin and stroma thylakoid fractions, the enrichment of PSII core phosphorylation in the HL phases was particularly evident, providing further evidence that the margin domain is the place where the phosphorylated and damaged PSII migrates in light to be subsequently dephosphorylated and degraded to initiate the repair cycle of PSII (Fig. 7B; Järvi et al., 2015, 2017).
Figure 7.
Effect of light intensity shifts on chl a/b ratios and protein phosphorylation in different thylakoid subfractions. A, Chl a/b ratio in thylakoid fractions obtained with 0.4% digitonin (DIG) from 16-h dark-adapted plants (dark bars) subsequently treated with LL for 2 h (green bars) followed by 2 h HL (orange bars). The letters indicate a statistical difference among light treatments in each fraction (P < 0.05 according to all-pairwise multiple-comparison one-way ANOVA). Error bars indicate the SD (n = 3). B, Representative immunoblot of the fractions in A loaded on a chl basis (0.5 µg) and immunodecorated with antip-Thr antibody. C, Representative immunoblot, as in B, where the fractions were loaded to facilitate comparison between fractions within the same light treatment. T, Thylakoids; G, grana core; M, margins; S, stroma lamellae.
DISCUSSION
The lateral heterogeneity of the thylakoid membrane in plant chloroplasts has been investigated for several decades to understand the advantages and dynamics of the segregation of PSII and PSI into appressed grana and nonappressed stroma-exposed membranes, respectively (Andersson and Anderson, 1980; Anderson et al., 2012). Thylakoid heterogeneity plays an important role in short-term acclimation and long-term adaptation of plants to sessile life form on land. Short-term dynamics are based on rapid biophysical and biochemical mechanisms that allow reversible interactions between thylakoid protein complexes to cope with ever changing environmental conditions, particularly fluctuations in the intensity of sunlight. Because grana stacking largely depends on electrostatic forces, the dynamics of phosphorylation and acetylation of the N termini of several thylakoid proteins have been particularly investigated, given the importance of the negatively charged phosphate groups in exerting repulsive and attractive forces as a basis for dynamics of the grana stacks (Chow et al., 2005; Tikkanen and Aro, 2012; Puthiyaveetil et al., 2017). In this work, we shed light on the still elusive localization and dynamics of phosphorylation of the thylakoid protein CURT1B upon shifts in light intensity. The results indicate a strong connection of CURT1B phosphorylation with the dynamics of phosphorylation of the PSII core and LHCII, as well as with the interaction between PSI and LHCII, while also providing more details on the different roles of these three components (PSII, PSI, and LHCII) in shaping the dynamics of the lateral heterogeneity of the thylakoid membrane.
CURT1B Is Localized in a Subfraction of the Grana Margin Domain
One major question in disclosing the underlying mechanisms of the dynamics of thylakoid heterogeneity concerns the isolation and composition of the grana margin regions, which are mostly affected by changing light intensities. The accessibility of this dynamic thylakoid domain to DIG is dependent on the composition of the chl-binding protein complexes and of the phosphorylation status of LHCII (Kyle et al., 1984). At any DIG concentration used for solubilization of thylakoids, the subsequent differential centrifugation steps will gradually pellet the different thylakoid subfractions (domains), as revealed by clearly different chl a/b ratios, with the lowest in the grana core (10,000g fraction), the highest in the stroma membranes (144,000g fraction), and an intermediate chl a/b ratio in the 40,000g subfraction defined as margins (Fig. 6, A and B). A further subcompartment of the margin domain, which contains CURT1B (Fig. 6C), can be assigned as the curvature of the grana edges (Puthiyaveetil et al., 2014; Yoshioka-Nishimura et al., 2014), and the “purity” of this subfraction with respect to CURT1 proteins is negatively affected by increased DIG concentrations (DIG/Chl ratios; Fig. 6, C and D). Notably, Cyt b6f is coenriched with CURT1 proteins in the curvature of the grana edges (Fig. 6C), which deserves further investigation.
CURT1B Phosphorylation Is Enhanced during Specific Light Shifts
While phosphorylation of the PSII core linearly increases upon shifts of plants both from darkness to light and from LL to HL (Figs. 1 and 3; Tikkanen et al., 2006; Trotta et al., 2016), the phosphorylation level of CURT1B is not affected in wild-type thylakoids by a shift from darkness to LL (Fig. 1B, green bars). Instead, two distinct levels of CURT1B phosphorylation can be distinguished in wild-type Arabidopsis. First, a low basal level occurs before turning the lights on (Da1) and after the shift to LL (LL1 and LL2); and second, a 2-fold increase in phosphorylation is observed after the shift from LL1 to HL and the shift from LL2 to darkness for 1 h (1h D), without an apparent effect on the CURT1 protein levels or on the level of CURT1A and CURT1B acetylation (Figs. 1B and 2; Supplemental Fig. S1). These two conditions of different CURT1B phosphorylation levels might be linked to different configurations of the lateral heterogeneity of the thylakoid membrane and the distribution of the chl-protein complexes to allow fluent acclimation of photosynthesis to changing light environments.
The fact that a contemporaneous increase of CURT1B and PSII core protein phosphorylation in wild-type thylakoids occurs both in HL and 1h D (Fig. 1, B and C) might indicate a connection with the PSII repair cycle, which takes place in the grana margins comprising the curvature domain, where CURT1 proteins are located (Fig. 6; Armbruster et al., 2013; Yamamoto et al., 2013; Puthiyaveetil et al., 2014). Indeed, the repulsive forces exerted by increased CURT1B phosphorylation in the edges of grana margins might locally increase the grana partition gaps and thus facilitate the de-stacking of grana edges necessary to accommodate the PSII repair machinery (Yamamoto et al., 2013; Tikkanen and Aro, 2014; Yoshioka-Nishimura et al., 2014). However, the fact that a certain percentage of CURT1B is acetylated, but not phosphorylated, and that this subpopulation of CURT1B is apparently not responsive to light changes, suggests that CURT1 proteins might have additional functions in conditions other than those studied so far (Armbruster et al., 2013; Pribil et al., 2018).
Both PSII Core and LHCII Phosphorylation Affect CURT1B Phosphorylation Dynamics
The fact that the light-dependent CURT1B phosphorylation dynamics observed in wild-type Arabidopsis are absent in stn8 (Fig. 1B), but CURT1B phosphorylation is still detectable, is in line with the idea that the two distinct pools of CURT1B phosphorylation are connected with the repair of PSII. Indeed, in stn8 thylakoids, phosphorylation of the PSII core is almost absent after HL illumination and 1h D (Fig. 1B, orange bars), which are the two light shifts in which CURT1B is maximally phosphorylated in the wild type (Fig. 1B, green bars). However, in the mutants with perturbed LHCII phosphorylation (stn7 and tap38) the relationship between PSII core phosphorylation and CURT1B phosphorylation is altered. Thus, it is conceivable that not only the PSII core protein phosphorylation but also LHCII phosphorylation plays an essential role in the shifts between the two phosphorylation stages of CURT1B (low basal and high) observed in the wild type.
Taking into account the localization of CURT1 proteins in the curvature domain of the grana margins (Fig. 6), the role of LHCII phosphorylation should be contextualized with the dynamics of CURT1B phosphorylation in the same domain. In this respect, the high CURT1B phosphorylation in stn7 at LL with the lack of LHCII phosphorylation and constitutive phosphorylation of LHCII in tap38 at HL are of particular interest. They indicate that the role of LHCII phosphorylation in shaping the thylakoid lateral heterogeneity in the wild type under LL has at least an indirect effect on CURT1B phosphorylation.
How exactly does LHCII phosphorylation contribute to the dynamics of the grana margins? Modulation of the thylakoid lateral heterogeneity by increasing LHCII phosphorylation under low white light has been generally associated with either changes in overall appressed area (Kyle et al., 1983; Rozak et al., 2002; Khatoon et al., 2009; Anderson et al., 2012) or a decrease in the grana width, as in the case of tap38 plants with constitutively phosphorylated LHCII (Armbruster et al., 2013; Iwai et al., 2018). However, in psal and psah plants with constitutively phosphorylated LHCII but a lack of the PSI domain responsible for interaction with LHCII, the grana are much wider and comprise only a few stacks (Lunde et al., 2003; Wood et al., 2019). Thus, the PSI-LHCII interaction is more important than LHCII phosphorylation per se in modulation of thylakoid lateral heterogeneity or decrease in grana diameter. The same conclusions can be drawn from the double mutant stn7 stn8, which is also characterized by wider grana with fewer stacks (Fristedt et al., 2009; Herbstová et al., 2012; Iwai et al., 2018), as well as a complete lack of PSII core and LHCII phosphorylation. It is interesting to note here that CURT1B phosphorylation has previously been reported as absent in stn7 stn8, but not in the stn8 single mutant (Ingelsson and Vener, 2012), which further supports the idea that regulation of CURT1B phosphorylation is affected by both PSII core and LHCII phosphorylation. Moreover, this may indicate that the basal level of phosphorylation of CURT1B results from an overlap of STN7 and STN8 kinase activities, as is the case for PSII core (Fig. 1C), or from involvement of an as yet unknown third kinase.
PSI-LHCII Interaction Involves CURT1 Proteins
In both psb33 and psal mutants the connection between PSI and state-transition LHCII trimer is lost (Lunde et al., 2003; Fristedt et al., 2017; Kato et al., 2017). Moreover, at the molecular level, psb33 plants, like stn7 plants, lack phosphorylation of LHCII under LL, while psal plants, like tap38 plants, have hyperphosphorylated LHCII. However, the LL-induced dynamics of CURT1B phosphorylation observed in stn7 and tap38 (Fig. 1B) showed the opposite behavior in psb33 and psal (Fig. 3A), respectively. Elevated phosphorylation of CURT1B under LL in psal and in stn7 with respect to the wild type suggests that PSI-LHCII interaction is a key factor in the regulation of CURT1B phosphorylation. Recent research has established that the interaction between PSI and LHCII is much wider than previously considered (Grieco et al., 2012; Wientjes et al., 2013; Grieco et al., 2015; Bos et al., 2017, 2019; Schwarz et al., 2018), which suggests a major role for the PSI-LHCII and PSI-LHCII-PSII megacomplexes in shaping thylakoid lateral heterogeneity (Suorsa et al., 2015; Schwarz et al., 2018). In this respect, the previous finding that PSAL and CURT1B might interact (Yu et al., 2008) allows us to speculate that the phosphorylation status of CURT1B possibly has a role in the dynamic interaction of LHCII with PSI in grana margins, which are topologically very close to the curvature domains where CURT1 has been localized (Fig. 6; Armbruster et al., 2013).
The phosphorylation of LHCII, which influences the grana width in tap38 (Armbruster et al., 2013; Iwai et al., 2018) and the loosening of appression in blue light-treated wild-type plants (Dietzel et al., 2011), exerts this effect only if state-transition LHCII trimer is functionally connected to PSI, which is not the case in psal. In psb33, CURT1B phosphorylation has the opposite dynamics with respect to psal, which indicates that a still unknown component, missing in psb33, is hierarchically more important than LHCII and PSII core phosphorylation in determining the phosphorylation status of CURT1B and, thus, the organization of grana upon shifting light intensities.
The striking differences in the phosphorylation level of CURT1B in wild-type, stn7, stn8, and tap38 plants are not accompanied by any changes at the CURT1B protein level, nor that of the other CURT1 proteins or the PSI and LHCI subunits measured here (Supplemental Fig. S1). Instead, both psb33, where the light-induced dynamics of LHCII phosphorylation is absent, and psal, where the LHCII phosphorylation is constitutive, show almost identically lower content of the CURT1A, CURT1C, and CURT1D proteins (Fig. 4). Strikingly, only CURT1B is not affected at the protein level, which is of particular interest since CURT1B is expressed differentially to CURT1A and CURT1C in plants grown in natural light (Pribil et al., 2018). This result suggests that the phosphorylation of the CURT1B N terminus might confer a specific role to this isoform with respect to the other CURT1 proteins and their N-terminal phosphorylations in the conditions tested. In psal plants, the different protein levels of CURT1A, CURT1C, and CURT1D might be explained by the highly increased grana width and reduced number of grana stacks (Lunde et al., 2003; Armbruster et al., 2013; Wood et al., 2019), while a different explanation is required for the results obtained with psb33 plants (Fig. 4). Further analysis of the ultrastructure in psb33 plants will help to clarify this point.
Interactions between PSII Core, LHCII, and CURT1B Protein Phosphorylation in Modulation of Thylakoid Dynamics
Based on the results presented in this work and in the literature, the interactive dynamics between PSII core, LHCII, and CURT1B phosphorylation are summarized in Figure 8. During the dark period, grana stacking is mainly facilitated by the interaction between nonphosphorylated N-terminal domains of LHCII and by phosphorylated CP43 (Figs. 1, 3, and 7, summarized in Fig. 8; Chow et al., 2005; Fristedt et al., 2010; Anderson et al., 2012; Wood et al., 2019). The shift from darkness to LL induces high phosphorylation of LHCII in all thylakoid domains (Fig. 7; Tikkanen et al., 2008b), which results in loosening of the appressed grana stacks and thus widening of the partition gaps (Dietzel et al., 2011; Johnson and Wientjes, 2019). Loosening of the grana stacks is further reinforced by a decrease in CP43 phosphorylation (Figs. 1, 3, and 7; Fristedt et al., 2010). The loosely appressed membranes favor the interaction of phosphorylated LHCII with PSI, resulting in increased area of the margin domain (Fig. 7, summarized in Fig. 8; Grieco et al., 2015). The shift from LL to HL results in dephosphorylation of LHCII and increased phosphorylation of PSII core proteins (Figs. 1, 3, and 7; Tikkanen et al., 2008a; summarized in Tikkanen and Aro, 2014), both of which contribute to a decrease in the size of partition gaps between appressed grana thylakoids (Johnson and Wientjes, 2019). At the same time, increased CURT1B phosphorylation promotes outward bending of the edges of grana margins (curvature domain) to accommodate the PSII repair cycle (Figs. 1, 3, and 7, summarized in Fig. 8; Armbruster et al., 2013; Yamamoto et al., 2013; Yoshioka-Nishimura et al., 2014; Puthiyaveetil et al., 2017). The PSI-LHCII-PSII interaction still takes place in the margins despite dephosphorylation of LHCB2 in HL (Grieco et al., 2012, 2015; Wientjes et al., 2013; Suorsa et al., 2015; Yokono et al., 2015, 2019; Bos et al., 2017; Schwarz et al., 2018), possibly allowing excess energy dissipation via PSI. Thus, the dynamics of phosphorylation of LHCII and CURT1B are perfectly integrated to respond to the fluctuating light intensities (Fig. 8; Grieco et al., 2015; Suorsa et al., 2015; Yokono et al., 2015; Schwarz et al., 2018). The role of CURT1 proteins in the dynamics of thylakoid heterogeneity is still emerging, and considerable work still needs to be done to reveal the possible role of CURT1A and CURT1B acetylation, as well as phosphorylation of other CURT1 proteins, whose cooperation seems to be complex (Armbruster et al., 2013). Our work provides an essential contribution to the understanding of the regulation of photosynthesis in fluctuating environmental conditions and serves in building a comprehensive view of interactive regulation mechanisms by thylakoid protein phosphorylation.
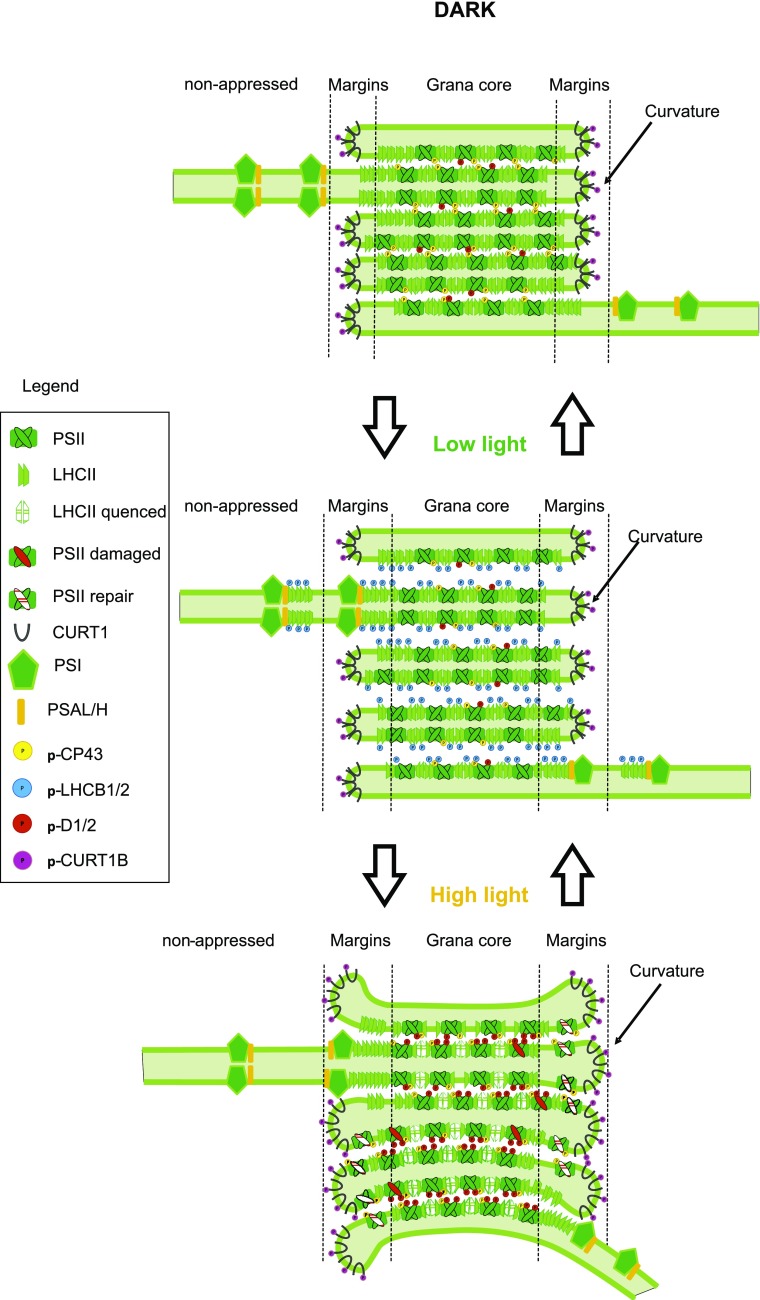
Figure 8.
Scheme of the interaction of the phosphorylation of PSII core, LHCII, and CURT1B. The transition from dark to LL leads to an increase in LHCII phosphorylation and a decrease in CP43 phosphorylation. Consequently, loosening of the appressed grana stacks, and thus widening of the size of the partition gaps, occurs. The increased interaction of phosphorylated LHCII with PSI in the loosely appressed membranes results in increased area of the margin domain. The shift from LL to HL results in a decrease of the size of the partition gaps between grana core thylakoid membranes due to both dephosphorylation of LHCII and increased phosphorylation of PSII core proteins. The increased CURT1B phosphorylation, instead, promotes outward bending of the edges of grana margins (curvature domain), which accommodates the PSII repair cycle. The scheme is based on the results presented in this paper and in pervious literature (Chow et al., 2005; Tikkanen et al., 2008a, 2008b; Fristedt et al., 2010; Dietzel et al., 2011; Anderson et al., 2012; Grieco et al., 2012, 2015; Armbruster et al., 2013; Wientjes et al., 2013; Yamamoto et al., 2013; Tikkanen and Aro, 2014; Yoshioka-Nishimura et al., 2014; Suorsa et al., 2015; Yokono et al., 2015, 2019; Bos et al., 2017; Puthiyaveetil et al., 2017; Schwarz et al., 2018; Johnson and Wientjes, 2019; Wood et al., 2019).
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Light Treatments
Plants of wild-type Arabidopsis (Arabidopsis thaliana) ecotype Columbia, as well as of mutants of STN7 kinase (Bellafiore et al., 2005), STN8 kinase (Vainonen et al., 2005), TAP38 phosphatase (Pribil et al., 2010), PSI subunit PSAL (Lunde et al., 2000), and auxiliary protein PSB33 (Fristedt et al., 2015) were used for experiments. The plants were grown at 23°C under 120 μmol photons m−2 s−1 of light using OSRAM PowerStar HQIT 400/D Metal Halide lamps as the light source, with a photoperiod of 8 h light and 16 h dark in a mixture of soil and vermiculite (1:1). All plants were 32 d old at the time of the experiments. The light treatments were performed with entire intact plants. Besides the 16 h dark period (Da1), the plants were treated for 2 h with LL (20 μmol photons m−2 s−1) or 2 h with HL (1,000 μmol photons m−2 s−1) in the order specified in the Results section. The spectra showing light quality and quantity under the light conditions used are presented in Supplemental Figure S5. The ratios between red, blue, and far-red regions are also presented, demonstrating that there is no enrichment in far red in HL.
Isolation of the Thylakoid Proteins and Subfractionation
Thylakoids membranes were isolated as in Suorsa et al. (2015) in dim light at 4°C from fresh leaves ground in ice-cold grinding buffer (50 mm HEPES/KOH [pH 7.5], 330 mm sorbitol, 5 mm MgCl2, 2.5 mm ascorbate, 0.05% [w/v] BSA, and 10 mm NaF), filtered through two layers of Miracloth, and centrifuged at 5,000g at 4°C for 4 min. The pellet was resuspended in a shock buffer (50 mm HEPES/KOH [pH 7.5], 5 mm sorbitol, 5 mm MgCl2, and 10 mm NaF), followed by centrifugation at 5,000g at 4°C for 4 min. The shock buffer was removed and the pellet resuspended in storage buffer (50 mm HEPES/KOH [pH 7.5], 100 mm sorbitol, 5 mm MgCl2, and 10 mm NaF), followed by centrifugation at 5,000g at 4°C for 4 min and resuspension in small aliquots of storage buffer. The subfractionation of thylakoid membrane with DIG was performed essentially as described in Kyle et al. (1984). Thylakoids resuspended in 15 mm Tricine (pH 7.8), 100 mm sorbitol, 10 mm NaCl, 5 mm MgC12, and 10 mm NaF were solubilized for 8 min at room temperature with a final concentration of 0.4% or 1% (w/v) DIG at 0.5 mg chl/mL, followed by 3 min centrifugation at 1,000g at 4°C to remove the unsolubilized material. The supernatant was subsequently centrifuged at 4°C for 30 min at 10,000g to collect the grana core membranes, followed by 30 min at 40,000g to collect the margins, and finally 1 h at 144,000g to collect the stroma lamellae (tight pellet) and the curvature fraction (loose pellet). The chl content was measured according to Porra et al. (1989). Thylakoids for solubilization with increasing DIG concentrations were resuspended in the same buffer as for subfractionation. Details about the procedure are reported in the text and in Rantala and Tikkanen (2018).
Immunoblotting
Thylakoid proteins were solubilized in Laemmli buffer (Laemmli, 1970) and separated by 12% (w/v) SDS‐PAGE supplemented with 6 m urea. The gels were loaded based on an equal chl concentration. After electrophoresis, the proteins were electroblotted to a PVDF membrane. To assess the level of phosphorylation of D1, D2, CP43 (PSII core), and LHCII, the membranes were probed with antip‐Thr (New England Biolabs), or with antibodies raised against CURT1A or CURT1B (Armbruster et al., 2013), D1, PSAB, CYTF, ATPF, LHCB1, LHCB2, and LHCB3 (Agrisera; www.agrisera.com; catalog numbers AS10704, AS10695, AS08306, AS10 1604, AS01004, AS01003, and AS01002, respectively). Horseradish peroxidase‐linked secondary antibody and ECL reagents (Amersham, GE Healthcare) were used for detection.
Trypsin Digestion and Analysis by Liquid Chromatography Electrospray Ionization MS/MS
Thylakoids were digested as in Fristedt et al, (2017) and Trotta et al. (2016). Samples corresponding to 5 g of chl were solubilized in Laemmli buffer and denatured and purified by running for 0.5 cm in 6% (w/v) SDS-PAGE in the presence of 6 m urea. The corresponding area of the gel was cut in 1-mm3 pieces and protein reduction, alkylation, and trypsin digestion were performed as in Gerotto et al. (2019), with a sequencing grade-modified trypsin (Promega V5111). The ratio of trypsin to chl was 1:10 (w/w). The peptides were extracted thrice with 50% (v/v) acetonitrile (ACN)/5% (v/v) formic acid (FA), dried in a speed vacuum, and resuspended in 2% (v/v) FA. The peptide mixture spiked with iRT peptides according to the manufacturer's instructions (Biognosys) was loaded in a nanoflow HPLC system (EasyNanoLC 1000, Thermo Fisher Scientific) equipped with a 20 × 0.1 mm (inner diameter) precolumn in conjunction with a 150 mm × 75 µm (inner diameter) analytical column, both packed with 5 µm Reprosil C18-bonded silica (Dr Maisch GmbH), and coupled to a Q‐Exactive electrospray ionization-hybrid quadrupole‐orbitrap mass spectrometer (Thermo Fisher Scientific), and analyzed in data-dependent acquisition (DDA). The mobile phase consisted of water/ACN (98:2 [v/v]) with 0.2% (v/v) FA (solvent A) or ACN/water (80:20 [v/v]) with 0.2% (v/v) FA (solvent B) at a flow rate of 300 nL min−1. The peptides were separated by a two‐step gradient elution from 2% (or 5%) to 35% (or 43%) solvent B over 60 min, then from 35% to 100% over 5 min, and subsequently at 100% solvent B for 10 min. The type of fragmentation used was higher‐energy collisional dissociation, and the scan range was set from 300 to 2,000 m/z, with up to 10 data‐dependent MS/MS spectra acquired in each scan and dynamic exclusion set for 10 s
Data Analysis and MS/MS Spectra Interpretation
The acquired MS/MS spectra were analyzed against a nonredundant Arabidopsis proteome (TAIR10; www.arabidopsis.org) as a database supplemented with most common contaminants (35,502 entries in total), using Proteome Discoverer (version 2.2) Software (Thermo Fisher Scientific) with an in‐house Mascot (version 2.4; Matrix Science) search engine. The search parameters were the following: monoisotopic mass, two missed cleavages allowed, precursor mass tolerance 10 ppm, fragment mass tolerance 0.02 D, m/z ≥ 2 + . The variable modifications oxidation of Met (mono Δ = 15.9949) and phosphorylation of Ser, Thr, and Tyr (mono Δ = 79.9663) were allowed, as was carbamidomethylation of Cys (mono Δ = 57.021464) for static modification. Identifications of phosphopeptides and peptides were validated through a PhosphoRS filter (version 3.1; Taus et al., 2011) and a decoy database search, with target false discovery rates of <0.01 (strict) and <0.05 (relaxed). For phosphopeptides the confidence threshold was set to P < 0.05.
SRM Analysis
The MS/MS library generated in DDA was used to generate SRM transitions to the peptides listed in Supplemental Tables S3 and S4 with Skyline software (version 4.2; MacLean et al., 2010). The transitions were analyzed with a triple-stage quadrupole TSQ Vantage QQQ mass spectrometer (Thermo Fisher Scientific), in line with a nanoflow HPLC system similar to the one used with Q-Exactive, and equipped with the same nano‐electrospray ion source used for Q‐Exactive. In this case, the gradient used was 3% to 43% solvent B (80:20 [v/v]) over 25 min, followed by 35% to 100% over 5 min and 5 min of 100% solvent B with a flow rate of 300 nL min−. Measurements in scheduled mode were performed with a cycle time of 2.5 s and a dwell time of ∼20 ms, using a window of 2.5 min to allow a maximum of 80 concomitant transitions.
The intensity of every peptide has been calculated as the sum of the integrated peak area of the three most intense transitions (Supplemental Table S4), or the four most intense for the CURT1B N terminus. The relative level of phosphorylation of CURT1B has been calculated as the percentage of the intensity of the phosphopeptide with respect to the sum of the intensities of the nonphosphorylated peptide plus the phosphopeptide, with the following formula:
| (1) |
where P is the relative level of phosphorylation, t1, t2… are the most intense transitions of the nonphosphorylated form, and pt1, pt2… are the most intense transitions of the phosphorylated form. This kind of calculation allowed comparison of the percentage of phosphorylation through all the mutants in all conditions. In this way, the phosphorylation in Da1 in the wild type was considered as 1 and the rest of the measurements are relative to Da1. For the level of acetylation, the formula was
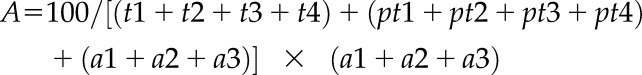 |
(2) |
where A is the relative level of acetylation, t1, t2… and pt1, pt2… are the most intense transitions of the nonphosphorylated and phosphorylated forms, respectively, and a1, a2… are the most intense transitions of the acetylated form. In the case of CURT1A, only the three most intense transitions from the nonphosphorylated and acetylated forms were used (Supplemental Table S4). For protein level, the sum of the intensities of three proteotypic peptides was used (Supplemental Table S4), except in the case of PSAK, for which only two peptides were available, and PSAH1, which has only one unique peptide with respect to PSAH2. For all peptides considered, the missed cleaved form was included if relevant, as was the form with oxidized methionine (Supplemental Tables S3 and S4).
Statistical Analysis
Significance of the differences in phosphorylation, acetylation, and protein level in the time points considered within every genotype were evaluated using an all-pairwise multiple-comparison procedures one-way ANOVA (the Holm-Sidak method) with the Shapiro-Wilk method for normality testing and the Brown-Forsythe method for equal variance testing. The significance of the differences for the basal level of phosphorylation among the genotypes was evaluated in the same way. The letters in the figures refer to the differences within the genotype where P is at least <0.05. In Figures 2 and 4 and Supplemental Figure S3 the letters were not reported because there were no differences. All the statistics were calculated with Sigmaplot (version 13.0) software.
Accession Numbers
The refined SRM dataset, as well as the raw files for both DDA and SRM, can be accessed via https://panoramaweb.org/CURT1B_phosphorylation_dynamics.url. Gene accession numbers are AT1G68830 (STN7), AT5G01920 (STN8), AT4G27800 (TAP38), AT1G71500 (PSB33), AT4G12800 (PSAL), AT3G16140 (PSAH1), AT1G52230 (PSAH2), AT1G30380 (PSAK), ATCG00350 (PSAA), ATCG00340 (PSAB), AT3G54890 (LHCA1), AT3G61470 (LHCA2), AT1G61520 (LHCA3), AT3G47470 (LHCA4), AT1G45474 (LHCA5), AT1G19150 (LHCA6).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Representative MS/MS spectrum of the CURT1B phosphopeptide 64ATpTEVGEAPATTTEAETTELPEIVK88.
Supplemental Figure S2. Identification of the correct phosphorylation site of the CURT1B phosphopeptide 64ATTEVGEAPATTTEAETTELPEIVK88 by monitoring the coeluting fragments (transitions) in SRM.
Supplemental Figure S3. Quantification by SRM of CURT1 isoforms, PSI subunits, and LHCA isoforms in thylakoids of wild-type (green), stn7 (red), stn8 (orange), and tap38 (blue) plants in the six time points indicated in Figures 1 and 2.
Supplemental Figure S4. Quantification by SRM of CURT1 isoforms, PSI subunits, and LHCA isoforms in thylakoids of wild-type (green), psb33 (gray), and psal (black) plants in the remaining four time points indicated in Figures 3 and 4.
Supplemental Figure S5. Wavelength spectra and ratios between blue, red, and far-red regions.
Supplemental Table S1. List of the proteins identified in DDA in thylakoids from Da1, LL1, HL, and LL2 time points.
Supplemental Table S2. List of the peptides identified for CURT1A–CURT1D and of the PSII core subunits with N-terminal acetylation.
Supplemental Table S3. List of transitions used for detection in SRM
Supplemental Table S4. List of transitions used for quantification in SRM.
ACKNOWLEDGMENTS
Mass spectrometry analysis was performed at the Turku Proteomics Facility, University of Turku, and at Åbo Akademi University. The facility is supported by Biocenter Finland. We thank Mikko Tikkanen, Marjaana Rantala, and Steffen Grebe for valuable discussions.
Footnotes
This work was supported by the Academy of Finland (project nos. 307335 and 303757 to E.-M.A.).
Articles can be viewed without a subscription.
References
- Anderson JM, Horton P, Kim E-H, Chow WS (2012) Towards elucidation of dynamic structural changes of plant thylakoid architecture. Philos Trans R Soc Lond B Biol Sci 367: 3515–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Anderson JM (1980) Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim Biophys Acta 593: 427–440 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Labs M, Pribil M, Viola S, Xu W, Scharfenberg M, Hertle AP, Rojahn U, Jensen PE, Rappaport F, et al. (2013) Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25: 2661–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgio E, Ungerer P, Ruban AV (2015) Light-harvesting superstructures of green plant chloroplasts lacking photosystems. Plant Cell Environ 38: 2035–2047 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix J-D (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, Meinnel T, Giglione C (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol Cell Proteomics 11: M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos I, Bland KM, Tian L, Croce R, Frankel LK, van Amerongen H, Bricker TM, Wientjes E (2017) Multiple LHCII antennae can transfer energy efficiently to a single Photosystem I. Biochim Biophys Acta 1858: 371–378 [DOI] [PubMed] [Google Scholar]
- Bos P, Oosterwijk A, Koehorst R, Bader A, Philippi J, van Amerongen H, Wientjes E (2019) Digitonin-sensitive LHCII enlarges the antenna of photosystem I in stroma lamellae of Arabidopsis thaliana after far-red and blue-light treatment. Biochim Biophys Acta 1860: 651–658 [DOI] [PubMed] [Google Scholar]
- Chow WS, Kim E-HH, Horton P, Anderson JM (2005) Granal stacking of thylakoid membranes in higher plant chloroplasts: The physicochemical forces at work and the functional consequences that ensue. Photochem Photobiol Sci 4: 1081–1090 [DOI] [PubMed] [Google Scholar]
- Chuartzman SG, Nevo R, Shimoni E, Charuvi D, Kiss V, Ohad I, Brumfeld V, Reich Z (2008) Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell 20: 1029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel L, Bräutigam K, Steiner S, Schüffler K, Lepetit B, Grimm B, Schöttler MA, Pfannschmidt T (2011) Photosystem II supercomplex remodeling serves as an entry mechanism for state transitions in Arabidopsis. Plant Cell 23: 2964–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Granath P, Vener AV (2010) A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS One 5: e10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Herdean A, Blaby-Haas CE, Mamedov F, Merchant SS, Last RL, Lundin B (2015) PHOTOSYSTEM II PROTEIN33, a protein conserved in the plastid lineage, is associated with the chloroplast thylakoid membrane and provides stability to photosystem II supercomplexes in Arabidopsis. Plant Physiol 167: 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Trotta A, Suorsa M, Nilsson AK, Croce R, Aro E-M, Lundin B (2017) PSB33 sustains photosystem II D1 protein under fluctuating light conditions. J Exp Bot 68: 4281–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Vener AV (2011) High light induced disassembly of photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS One 6: e24565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Willig A, Granath P, Crèvecoeur M, Rochaix J-D, Vener AV (2009) Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21: 3950–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerotto C, Trotta A, Bajwa AA, Mancini I, Morosinotto T, Aro E-M (2019) Thylakoid protein phosphorylation dynamics in a moss mutant lacking SERINE/THREONINE PROTEIN KINASE STN8. Plant Physiol 180: 1582–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieco M, Suorsa M, Jajoo A, Tikkanen M, Aro EM (2015) Light-harvesting II antenna trimers connect energetically the entire photosynthetic machinery—including both photosystems II and I. Biochim Biophys Acta 1847: 607–619 [DOI] [PubMed] [Google Scholar]
- Grieco M, Tikkanen M, Paakkarinen V, Kangasjärvi S, Aro E-M (2012) Steady-state phosphorylation of light-harvesting complex II proteins preserves photosystem I under fluctuating white light. Plant Physiol 160: 1896–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson M, Vener AV (2003) Identification of three previously unknown in vivo protein phosphorylation sites in thylakoid membranes of Arabidopsis thaliana. Mol Cell Proteomics 2: 550–559 [DOI] [PubMed] [Google Scholar]
- Herbstová M, Tietz S, Kinzel C, Turkina MV, Kirchhoff H (2012) Architectural switch in plant photosynthetic membranes induced by light stress. Proc Natl Acad Sci USA 109: 20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelsson B, Vener AV (2012) Phosphoproteomics of Arabidopsis chloroplasts reveals involvement of the STN7 kinase in phosphorylation of nucleoid protein pTAC16. FEBS Lett 586: 1265–1271 [DOI] [PubMed] [Google Scholar]
- Iwai M, Roth MS, Niyogi KK (2018) Subdiffraction-resolution live-cell imaging for visualizing thylakoid membranes. Plant J 96: 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Rantala M, Aro E-M (2017) Oxygenic Photosynthesis—Light Reactions within the Frame of Thylakoid Architecture and Evolution In Barber J, and Ruban AV, eds, Photosynthesis and Bioenergetics. World Scientific, Singapore, pp 243–263 [Google Scholar]
- Järvi S, Suorsa M, Aro EM (2015) Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim Biophys Acta 1847: 900–909 [DOI] [PubMed] [Google Scholar]
- Johnson MP, Wientjes E (2019) The relevance of dynamic thylakoid organisation to photosynthetic regulation. Biochim Biophys Acta doi:10.1016/j.bbabio.2019.06.011 [DOI] [PubMed] [Google Scholar]
- Kato Y, Yokono M, Akimoto S, Takabayashi A, Tanaka A, Tanaka R (2017) Deficiency of the stroma-lamellar protein LIL8/PSB33 affects energy transfer around PSI in Arabidopsis. Plant Cell Physiol 58: 2026–2039 [DOI] [PubMed] [Google Scholar]
- Khatoon M, Inagawa K, Pospísil P, Yamashita A, Yoshioka M, Lundin B, Horie J, Morita N, Jajoo A, Yamamoto Y, et al. (2009) Quality control of photosystem II: Thylakoid unstacking is necessary to avoid further damage to the D1 protein and to facilitate D1 degradation under light stress in spinach thylakoids. J Biol Chem 284: 25343–25352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrouchtchova A, Hansson M, Paakkarinen V, Vainonen JP, Zhang S, Jensen PE, Scheller HV, Vener AV, Aro EM, Haldrup A (2005) A previously found thylakoid membrane protein of 14 kDa (TMP14) is a novel subunit of plant photosystem I and is designated PSI-P. FEBS Lett 579: 4808–4812 [DOI] [PubMed] [Google Scholar]
- Kim E-H, Li X-P, Razeghifard R, Anderson JM, Niyogi KK, Pogson BJ, Chow WS (2009) The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: A study using two chlorophyll b-less mutants. Biochim Biophys Acta 1787: 973–984 [DOI] [PubMed] [Google Scholar]
- Kirchhoff H. (2013) Architectural switches in plant thylakoid membranes. Photosynth Res 116: 481–487 [DOI] [PubMed] [Google Scholar]
- Kyle DJ, Kuang T, Watson JL, Arntzen CJ (1984) Movement of a sub-population of the light harvesting complex (LHCII) from grana to stroma lamellae as a consequence of its phosphorylation. Biochim Biophys Acta 765: 89–96 [Google Scholar]
- Kyle DJ, Staehelin LA, Arntzen CJ (1983) Lateral mobility of the light-harvesting complex in chloroplast membranes controls excitation energy distribution in higher plants. Arch Biochem Biophys 222: 527–541 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lunde C, Jensen PE, Haldrup A, Knoetzel J, Scheller HV (2000) The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408: 613–615 [DOI] [PubMed] [Google Scholar]
- Lunde C, Jensen PE, Rosgaard L, Haldrup A, Gilpin MJ, Scheller HV (2003) Plants impaired in state transitions can to a large degree compensate for their defect. Plant Cell Physiol 44: 44–54 [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010) Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet JE, Arntzen CJ (1980) Simulation of grana stacking in a model membrane system. Mediation by a purified light-harvesting pigment-protein complex from chloroplasts. Biochim Biophys Acta 589: 100–117 [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. (2005) Function and evolution of grana. Trends Plant Sci 10: 521–525 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Pribil M, Wunder T, Leister D (2011) Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: The roles of STN7, STN8 and TAP38. Biochim Biophys Acta 1807: 887–896 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Pribil M, Labs M, Leister D (2014) Structure and dynamics of thylakoids in land plants. J Exp Bot 65: 1955–1972 [DOI] [PubMed] [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D (2010) Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol 8: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Sandoval-Ibáñez O, Xu W, Sharma A, Labs M, Liu Q, Galgenmüller C, Schneider T, Wessels M, Matsubara S, et al. (2018) Fine-tuning of photosynthesis requires CURVATURE THYLAKOID1-mediated thylakoid plasticity. Plant Physiol 176: 2351–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, van Oort B, Kirchhoff H (2017) Surface charge dynamics in photosynthetic membranes and the structural consequences. Nat Plants 3: 17020. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Tsabari O, Lowry T, Lenhert S, Lewis RR, Reich Z, Kirchhoff H (2014) Compartmentalization of the protein repair machinery in photosynthetic membranes. Proc Natl Acad Sci USA 111: 15839–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala M, Lehtimäki N, Aro EM, Suorsa M (2016) Downregulation of TAP38/PPH1 enables LHCII hyperphosphorylation in Arabidopsis mutant lacking LHCII docking site in PSI. FEBS Lett 590: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantala S, Tikkanen M (2018) Phosphorylation-induced lateral rearrangements of thylakoid protein complexes upon light acclimation. Plant Direct 2: e00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, Gerrits B, Rutishauser D, Gruissem W, Rochaix J-D, et al. (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamäki E, Salonen M, Suoranta UM, Carlberg I, Andersson B, Aro EM (1997) Phosphorylation of light-harvesting complex II and photosystem II core proteins shows different irradiance-dependent regulation in vivo. Application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. J Biol Chem 272: 30476–30482 [DOI] [PubMed] [Google Scholar]
- Rochaix J-D, Lemeille S, Shapiguzov A, Samol I, Fucile G, Willig A, Goldschmidt-Clermont M (2012) Protein kinases and phosphatases involved in the acclimation of the photosynthetic apparatus to a changing light environment. Philos Trans R Soc Lond B Biol Sci 367: 3466–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozak PR, Seiser RM, Wacholtz WF, Wise RR (2002) Rapid, reversible alterations in spinach thylakoid appression upon changes in light intensity. Plant Cell Environ 25: 421–429 [Google Scholar]
- Ruban AV, Wentworth M, Yakushevska AE, Andersson J, Lee PJ, Keegstra W, Dekker JP, Boekema EJ, Jansson S, Horton P (2003) Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature 421: 648–652 [DOI] [PubMed] [Google Scholar]
- Schwarz EM, Tietz S, Froehlich JE (2018) Photosystem I-LHCII megacomplexes respond to high light and aging in plants. Photosynth Res 136: 107–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DJ. (1982) Freeze-fracture studies on barley plastid membranes V.viridis-n 34, a photosystem I mutant. Carlsberg Res Commun 47: 215–225 [Google Scholar]
- Simpson DJ, Vallon O, von Wettstein D (1989) Freeze-fracture studies on barley plastid membranes: VIII. In viridis-115, a mutant completely lacking Photosystem II, oxygen evolution enhancer 1 (OEE1) and the α-subunit of cytochrome b-559 accumulate in appressed thylakoids. Biochim Biophys Acta 975: 164–174 [Google Scholar]
- Suorsa M, Rantala M, Mamedov F, Lespinasse M, Trotta A, Grieco M, Vuorio E, Tikkanen M, Järvi S, Aro EM (2015) Light acclimation involves dynamic re-organization of the pigment-protein megacomplexes in non-appressed thylakoid domains. Plant J 84: 360–373 [DOI] [PubMed] [Google Scholar]
- Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K (2011) Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res 10: 5354–5362 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Aro E-M (2014) Integrative regulatory network of plant thylakoid energy transduction. Trends Plant Sci 19: 10–17 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM (2012) Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim Biophys Acta 1817: 232–238 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Kangasjärvi S, Aro E-M (2010) Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol 152: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Nurmi M, Kangasjärvi S, Aro E-M (2008a) Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim Biophys Acta 1777: 1432–1437 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Nurmi M, Suorsa M, Danielsson R, Mamedov F, Styring S, Aro EM (2008b) Phosphorylation-dependent regulation of excitation energy distribution between the two photosystems in higher plants. Biochim Biophys Acta 1777: 425–432 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Piippo M, Suorsa M, Sirpiö S, Mulo P, Vainonen J, Vener AV, Allahverdiyeva Y, Aro E-M (2006) State transitions revisited—a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol Biol 62: 779–793 [DOI] [PubMed] [Google Scholar]
- Trotta A, Suorsa M, Rantala M, Lundin B, Aro EM (2016) Serine and threonine residues of plant STN7 kinase are differentially phosphorylated upon changing light conditions and specifically influence the activity and stability of the kinase. Plant J 87: 484–494 [DOI] [PubMed] [Google Scholar]
- Vainonen JP, Hansson M, Vener AV (2005) STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J Biol Chem 280: 33679–33686 [DOI] [PubMed] [Google Scholar]
- Vener AV, Harms A, Sussman MR, Vierstra RD (2001) Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J Biol Chem 276: 6959–6966 [DOI] [PubMed] [Google Scholar]
- Wientjes E, Van Amerongen H, Croce R (2013) LHCII is an antenna of both photosystems after long-term acclimation. Biochim Biophys Acta 1827: 420–426 [DOI] [PubMed] [Google Scholar]
- Wood WHJ, Barnett SFH, Flannery S, Hunter CN, Johnson MP (2019) Dynamic thylakoid stacking is regulated by LHCII phosphorylation but not its interaction with PSI. Plant Physiol 180: 2152–216631186333 [Google Scholar]
- Yamamoto Y, Hori H, Kai S, Ishikawa T, Ohnishi A, Tsumura N, Morita N (2013) Quality control of Photosystem II: Reversible and irreversible protein aggregation decides the fate of Photosystem II under excessive illumination. Front Plant Sci 4: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokono M, Takabayashi A, Akimoto S, Tanaka A (2015) A megacomplex composed of both photosystem reaction centres in higher plants. Nat Commun 6: 6675. [DOI] [PubMed] [Google Scholar]
- Yokono M, Takabayashi A, Kishimoto J, Fujita T, Iwai M, Murakami A, Akimoto S, Tanaka A (2019) The PSI-PSII megacomplex in green plants. Plant Cell Physiol 60: 1098–1108 [DOI] [PubMed] [Google Scholar]
- Yoshioka-Nishimura M, Nanba D, Takaki T, Ohba C, Tsumura N, Morita N, Sakamoto H, Murata K, Yamamoto Y (2014) Quality control of photosystem II: Direct imaging of the changes in the thylakoid structure and distribution of FtsH proteases in spinach chloroplasts under light stress. Plant Cell Physiol 55: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Yu Q-B, Li G, Wang G, Sun J-C, Wang P-C, Wang C, Mi H-L, Ma W-M, Cui J, Cui Y-L, et al. (2008) Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana. Cell Res 18: 1007–1019 [DOI] [PubMed] [Google Scholar]