A long-sought calmodulin- and Ca2+-dependent NAD kinase that is conserved in the plant lineage is the missing link between Ca2+ signaling, metabolism, and the oxidative burst.
Abstract
NADP(H) is an essential cofactor of multiple metabolic processes in all living organisms, and in plants, NADP(H) is required as the substrate of Ca2+-dependent NADPH oxidases, which catalyze a reactive oxygen species burst in response to various stimuli. While NADP+ production in plants has long been known to involve a calmodulin (CaM)/Ca2+-dependent NAD+ kinase, the nature of the enzyme catalyzing this activity has remained enigmatic, as has its role in plant physiology. Here, we used proteomic, biochemical, molecular, and in vivo analyses to identify an Arabidopsis (Arabidopsis thaliana) protein that catalyzes NADP+ production exclusively in the presence of CaM/Ca2+. This enzyme, which we named NAD kinase-CaM dependent (NADKc), has a CaM-binding peptide located in its N-terminal region and displays peculiar biochemical properties as well as different domain organization compared with known plant NAD+ kinases. In response to a pathogen elicitor, the activity of NADKc, which is associated with the mitochondrial periphery, contributes to an increase in the cellular NADP+ concentration and to the amplification of the elicitor-induced oxidative burst. Based on a phylogenetic analysis and enzymatic assays, we propose that the CaM/Ca2+-dependent NAD+ kinase activity found in photosynthetic organisms is carried out by NADKc-related proteins. Thus, NADKc represents the missing link between Ca2+ signaling, metabolism, and the oxidative burst.
As sessile organisms, plants have evolved mechanisms to react quickly to stress conditions, such as changes in temperature, salinity, or pathogen attacks. A common response to stress is a cytosolic calcium (Ca2+) influx followed by an apoplastic burst of reactive oxygen species (ROS; Grant et al., 2000). This ROS burst is generated by plasma membrane NADPH oxidases known as respiratory burst oxidase homologs (RBOHs; Torres and Dangl, 2005) and in turn regulates adaptation mechanisms such as gene expression, epigenetic changes, and long-distance signal transduction (Choi et al., 2017; Liebthal and Dietz, 2017; Chapman et al., 2019). RBOH oxidase activity is dependent on Ca2+ binding to their EF-hand domains and is stimulated by phosphorylation by Ca2+-dependent protein kinases (Dubiella et al., 2013) as well as CIPK/CBL complexes (Calcineurin B-Like Protein/CBL-Interacting Protein Kinase; Drerup et al., 2013).
A rapid increase in the NADP(H) pool size is observed in response to plant treatment with a pathogen elicitor (Harding et al., 1997; Pugin et al., 1997) and may be required to sustain the ROS burst by fueling RBOH proteins. Since most (∼70%–90%) of plant NAD+ kinase activity is dependent on binding calmodulin (CaM) in its Ca2+-loaded conformation (Anderson and Cormier, 1978), it was proposed (Harding et al., 1997) that the protein responsible for this activity may also be stimulated by the elicitor-induced Ca2+ influx. NADP+ produced by this enzyme may then be converted to NADPH (the substrate of RBOH proteins) by NADP-isocitrate dehydrogenase (Mhamdi et al., 2010) or by the reducing branch of the oxidative pentose phosphate pathway (Pugin et al., 1997; Scharte et al., 2009).
Several studies have described the CaM/Ca2+-dependent NAD+ kinase activity in plants using partially purified enzymatic preparations from plant tissues. These studies allowed finding this protein activity in a wide variety of plant species (Dieter and Marmé, 1984; Delumeau et al., 2000; Turner et al., 2004) and characterizing its kinetic parameters (Delumeau et al., 2000; Turner et al., 2004) as well as its preferences for specific CaM and CaM-like isoforms (Turner et al., 2004). However, the protein responsible for CaM/Ca2+-dependent NAD+ kinase activity has not been identified. In particular, among the three Arabidopsis (Arabidopsis thaliana) NAD+ kinases identified to date, the plastidial NADK2 binds CaM in vitro in a Ca2+-dependent way (Turner et al., 2004; Dell’Aglio et al., 2013), but its activity does not require CaM binding (Turner et al., 2004). Thus, this lack of knowledge of the identity of the plant CaM-dependent NAD+ kinase has prevented a thorough characterization of its role in plant physiology, and in particular in the production of the stress-induced ROS burst.
Here, we report the characterization of an Arabidopsis CaM/Ca2+-dependent NAD+ kinase that displays all the properties of the elusive enzyme. We show that this NAD+ kinase, which we named NADKc (for NAD kinase-CaM dependent), is associated with the mitochondrial periphery and is involved in sustaining the ROS burst induced by the bacterial elicitor flagellin22.
RESULTS AND DISCUSSION
NADKc Is a CaM/Ca2+-Dependent NAD+ Kinase
We obtained an Arabidopsis protein extract enriched in CaM/Ca2+-dependent NAD+ kinase activity by a four-step purification procedure described in Supplemental Materials and Methods. The last step consisted of binding the protein on a CaM-charged matrix in the presence of Ca2+ and its subsequent release with an excess of the Ca2+ chelator EGTA (Supplemental Fig. S1). We then used mass spectrometry-based proteomics to identify proteins enriched in the EGTA elution compared with the Ca2+-containing washing steps (Supplemental Table S1). We reasoned that putative CaM/Ca2+-dependent NAD+ kinases should display the following characteristics: (1) have a molecular mass between 50 and 65 kD, to respect the size range previously calculated by Delumeau et al. [2000]; (2) be annotated as ATP-binding proteins (but not as a protein kinase), since plant CaM-activated NAD kinase uses ATP as a substrate (Anderson and Cormier, 1978); (3) contain a predicted CaM-binding site (following the guidelines of Rhoads and Friedberg [1997]); and (4) have no previously assigned enzymatic activity. Our analysis revealed only one protein, encoded by the At1g04280 gene, that fulfilled all these criteria.
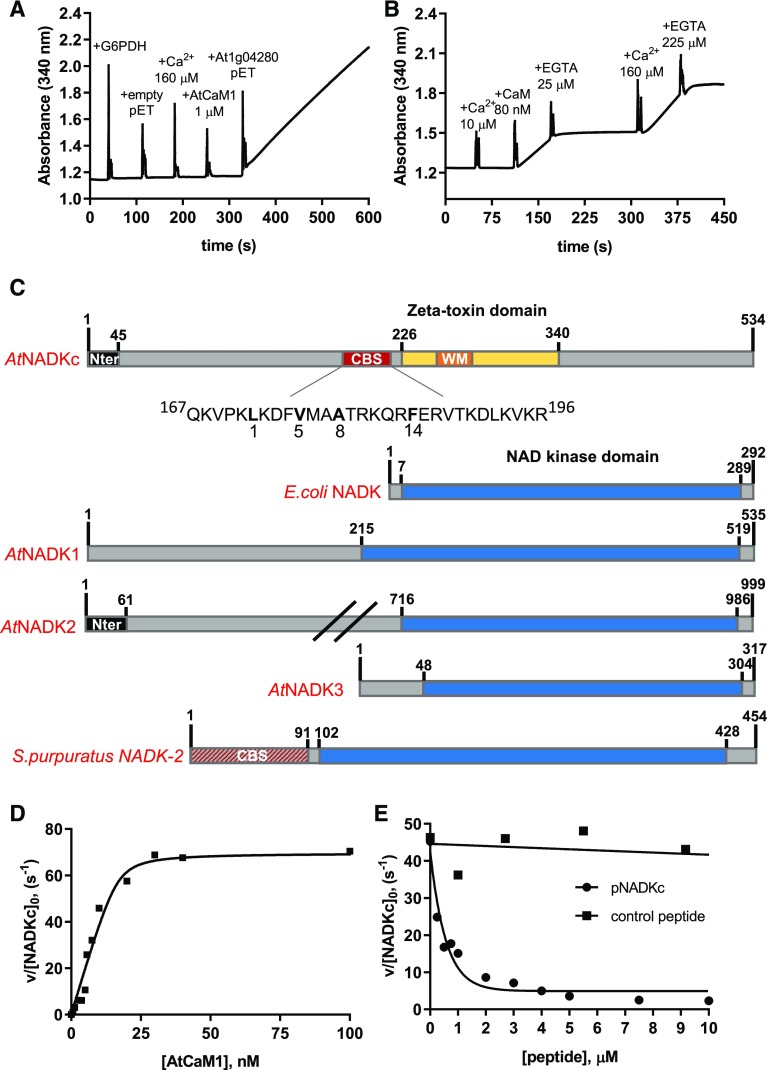
To confirm its CaM/Ca2+-dependent NAD+ kinase activity, we expressed the full-length recombinant protein coded by At1g04280 in Escherichia coli with an N-terminal His tag. We compared the NAD+ kinase activity of two E. coli extracts: one obtained from an At1g04280-expressing strain and the second from a strain containing an empty vector. As shown in Figure 1A, no NAD+ kinase activity (which in our test was detected as an increase of A340) was detected in the E. coli strain containing the empty vector, not even after the addition of an excess of Ca2+ and of Arabidopsis Calmodulin1 (AtCaM1). In contrast, addition of the At1g04280-expressing E. coli extract to the same reaction mixture immediately revealed NAD+ kinase activity.
Figure 1.
Biochemical properties of a CaM-dependent NAD+ kinase identified in Arabidopsis. A, NAD+ kinase activity measured in an E. coli extract expressing an empty pET28b(+) and an E. coli extract expressing At1g04280. Spikes correspond to the moments of addition of Glc-6-P dehydrogenase (G6PDH), Ca2+, AtCaM1, and E. coli extracts (10 μg). B, NAD+ kinase activity in an E. coli bacterial extract expressing At1g04280. Ca2+, AtCaM1, and EGTA were added at different times, as indicated in the graph. C, Schematic representation of the NADKc primary sequence and comparison with previously known NAD+ kinases. Yellow, ζ-Toxin domain (InterPro homologous superfamily: IPR010488); black, N-terminal region with putative organelle target sequence; red, putative conserved type A 1-8-14 CaM-binding site (detailed below the scheme); orange, Walker A motif (WM; ATP-binding site); blue, NAD+ kinase domain (InterPro homologous superfamily: IPR016064); red/gray, N-terminal sequence expected to contain a CaM-binding site according to Love et al. (2015). Sequences used for comparison (UniProt) are as follows: E. coli NAD+ kinase, P0A7B3; Arabidopsis NAD+ kinases, AtNADK1, Q56YN3; AtNADK2, Q9C5W3; AtNADK3, Q500Y9; S. purpuratus NAD+ kinase-2 (sea urchin CaM-dependent NAD+ kinase; Love et al., 2015), C3RSF7. D, Affinity of NADKc recombinant protein for AtCaM1. Activity of the purified NADKc recombinant protein after denaturation in urea and subsequent refolding was measured in the presence of 50 μm Ca2+ and as a function of [AtCaM1]. Experiments were performed in triplicate, and data shown are from one representative experiment. Binding data were analyzed assuming tight binding. The Kd value for AtCaM1 binding varied from 0.6 to 1 nm. E, Inhibition of NADKc activity by competition with the putative CaM-binding site (black circles). Black squares correspond to results obtained with a negative control peptide, which does not bind AtCaM1.
Activity measurements with a partially purified At1g04280 enzyme confirmed the lack of NAD+ kinase activity in the absence of AtCaM1/Ca2+ and its appearance, within seconds, upon the addition of both AtCaM1 and Ca2+. This NAD+ kinase activity was suppressed by EGTA and restored by the addition of an excess of Ca2+, showing that the CaM/Ca2+-dependent enzyme activation is an all-or-none, reversible process (Fig. 1B). In contrast, while CaM/Ca2+-dependent NAD+ kinases have also been described in invertebrates, animal NAD+ kinase activity is only slightly increased by CaM/Ca2+ addition. For example, CaM induces a 3.5-fold increase of the NADK-2 activity of the sea urchin Strongylocentrotus purpuratus (Love et al., 2015).
Based on these results, we identified the At1g04280 gene product as the long-sought CaM/Ca2+-dependent NAD+ kinase enzyme previously found in several plant species (Anderson and Cormier, 1978; Delumeau et al., 1998; Turner et al., 2004) and named it Arabidopsis NADKc for NAD kinase-CaM dependent.
AtNADKc Peculiar Features in Primary Sequence and Enzyme Activity
The primary sequence of NADKc (Fig. 1C) contains (1) an N-terminal region predicted to contain a transmembrane helix (amino acids 1–45); (2) a domain of unknown function (amino acids 46–225) that includes a conserved putative CaM-binding site (CBS; Supplemental Fig. S2A); and (3) a C-terminal kinase domain (amino acids 226–340) similar to bacterial type II ζ-toxin domains (Khoo et al., 2007), which is predicted to contain a conserved P-loop for ATP binding (Walker A motif, amino acids 236–250; Supplemental Fig. S2B). Interestingly, these features are not shared with all other NAD+ kinases known to date, from bacteria, plants, and animals (Fig. 1C; Kawai et al., 2001; Turner et al., 2004; Chai et al., 2006; Love et al., 2015).
To optimize NADKc expression levels in E. coli and improve the solubility of the protein, we removed the first 38 residues constituting the predicted transmembrane helix. The shorter version, 6HIS-Δ38NADKc, was partially purified by Ni-NTA affinity chromatography (Supplemental Fig. S3, lane 3). Activity assays with saturating AtCaM1/Ca2+ concentrations revealed that the NAD+ kinase activity of NADKc was specific toward NAD+, as no activity could be detected with NADH or deamido-NAD+ (NAAD; Table 1). Like most P-loop-containing kinases (Das et al., 2013), the enzyme displayed broad specificity for the phosphoryl donor, as ATP, CTP, GTP, and UTP could be used interchangeably and produced similar efficiencies (Table 1). The enzyme catalytic constant with CTP or ATP was close to 40 s−1 in the presence of Ca2+ and AtCaM1 (i.e. about 10-fold higher than that reported for plant CaM/Ca2+-independent NAD+ kinases and other NAD+ kinases from bacteria and animals [0.5–7 s−1]; Kawai et al., 2001; Turner et al., 2004; Chai et al., 2006; Love et al., 2015).
Table 1. NADKc kinetic parameters.
n.d., Not detected.
| Substrate Varied | Constant Substrate | Km | kcat | kcat/Km |
|---|---|---|---|---|
| µm | s−1 | µm−1 s−1 | ||
| ATP | NAD+ (10 mm) | 203 ± 30 | 41 ± 2 | 0.2 |
| CTP | NAD+ (10 mm) | 283 ± 70 | 42 ± 1 | 0.15 |
| GTP | NAD+ (10 mm) | 522 ± 135 | 26 ± 1 | 0.05 |
| UTP | NAD+ (10 mm) | 207 ± 26 | 29.5 ± 2 | 0.14 |
| NAD+ | ATP (8 mm) | 147 ± 17 | 42 ± 2 | 0.28 |
| NADH+ | ATP (8 mm) | n.d. | n.d. | n.d. |
| NAAD+ | ATP (8 mm) | n.d. | n.d. | n.d. |
To characterize the interaction of NADKc with CaM, we further purified the recombinant enzyme by urea denaturation and rapid dilution (Supplemental Materials and Methods). The refolded protein (Supplemental Fig. S3, lanes 4 and 5) produced a single band on an SDS-PAGE gel and had an increased catalytic constant (70 s−1) compared with the partially purified enzyme (40 s−1; Table 1) and high affinity for CaM (Kd = 0.6–1 nm; Fig. 1D), similar to the value of 0.4 nm reported for the tomato (Solanum lycopersicum) CaM-dependent NAD+ kinase (Delumeau et al., 2000).
In conclusion, compared with all other NAD+ kinases known to date, NADKc displays unique structural as well as catalytic features that make it particularly suitable for rapid NADP+ production following Ca2+ signals.
Identification of a CaM-Binding Peptide in the NADKc N-Terminal Domain
To verify that the NADKc N-terminal domain is involved in CaM binding, we measured NADKc activity in the presence of a synthetic peptide containing the putative type A 1-8-14 CaM-binding sequence (amino acids 167–196; Fig. 1C; Rhoads and Friedberg, 1997) in a competitive assay. As shown in Figure 1E, the presence of the putative NADKc CaM-binding peptide decreased the stimulation of NADKc by AtCaM1, as expected if AtCaM1, trapped by the peptide in excess, was no longer available for NADKc activation. The reduction in reaction rate was hyperbolically related to the peptide concentration (50% inhibition of initial activity = 0.5 μm). In contrast, another unrelated peptide from the short-chain dehydrogenase TIC32 protein (At4g23430), which cannot specifically bind AtCaM1 (Dell’Aglio, 2013), was also tested. An excess of this control peptide had no effect on NAD+ kinase activity (Fig. 1E).
These data suggest that the NADKc peptide identified plays a major role in the AtCaM1/Ca2+-dependent activation of the NADKc enzyme. We hypothesize that it could be an anchoring point for CaM in the full-length protein, facilitating activation of the kinase domain by an as yet unknown mechanism.
NADKc Is Located at the Mitochondrial Periphery
To assess the NADKc localization in vivo, we produced strains containing several YFP-tagged NADKc versions: (1) the NADKc full-length protein fused to YFP at its C-terminal region (construct NADKc-YFP); (2) the NADKc N-terminal region (amino acids 1–45) fused to YFP (construct NADKcNter-YFP); and (3) YFP fused at the N terminus of the whole NADKc protein sequence (construct YFP-NADKc). All fusion proteins were inserted into expression plasmids under the control of the cauliflower mosaic virus (CaMV) 35S promoter.
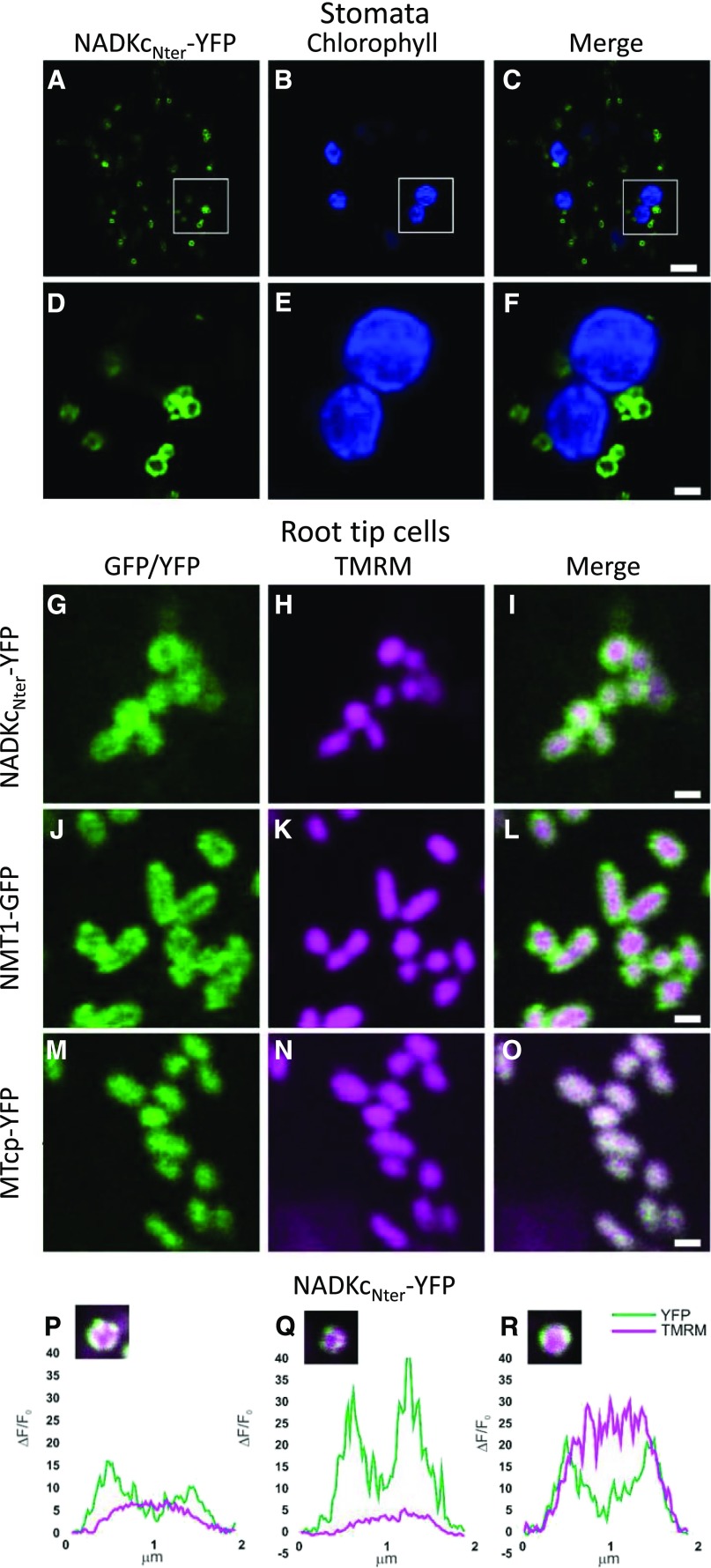
Arabidopsis lines and Nicotiana benthamiana leaves containing the NADKc:YFP construct showed protein clusters that likely constitute nonspecific aggregates caused by high expression of a membrane construct (Supplemental Fig. S4, bottom row). However, in N. benthamiana leaves (Supplemental Fig. S4, first and second rows) and Arabidopsis seedlings, the NADKcNter-YFP was targeted to ring-like structures in both stomata (Fig. 2, A–F) and root tip cells (Fig. 2, G–O). In the root tips, the NADKcNter-YFP protein colocalized with the mitochondrial matrix marker tetramethylrhodamine, methyl ester (TMRM), but the fluorescence signal of NADKcNter-YFP was more peripheral than the TMRM signal (Fig. 2, G–I). As a comparison, we observed Arabidopsis root tip cells expressing NMT1-GFP, an outer mitochondrial membrane protein (Fig. 2, J–L; Wagner et al., 2015), as well as MT-cpYFP (Fig. 2, M–O), a pH biosensor located in the mitochondrial matrix (Schwarzländer et al., 2011; Behera et al., 2018). This comparison clearly showed a higher resemblance of the NADKcNter-YFP signal profile to the NMT1-GFP profile than to the MT-cpYFP profile, suggesting a localization at the outer mitochondrial membrane.
Figure 2.
Analysis of submitochondrial localization of NADKcNter-YFP. A to C, Confocal laser scanning microscopy images from stomata guard cells of a representative Arabidopsis seedling stably expressing NADKcNter-YFP. Bar = 5 µm. D to F, Higher magnification of the regions of interest shown in A to C (white squares). Bar = 1 µm. A and D, YFP fluorescence in green. B and E, Chlorophyll fluorescence in blue. C and F, Merged YFP and chlorophyll fluorescences. G to I, Confocal laser scanning microscopy images from root tip cells of a representative Arabidopsis seedling stably expressing NADKcNter-YFP and stained with the mitochondrial matrix marker TMRM. Bar = 1 µm. J to L, Confocal laser scanning microscopy images from root tip cells of a representative Arabidopsis seedling stably expressing NMT-GFP and stained with the mitochondrial matrix marker TMRM. Bar = 1 µm. M to O, Confocal laser scanning microscopy images from root tip cells of a representative Arabidopsis seedling stably expressing MT-cpYFP and stained with the mitochondrial matrix marker TMRM. Bar = 1 µm. G and M, YFP fluorescence in green. J, GFP fluorescence in green. H, K, and N, TMRM fluorescence in magenta. I, L, and O, Merged YFP/GFP and TMRM fluorescences. NMT1-GFP and MT-cpYFP were used as markers for the outer mitochondrial membrane and matrix, respectively. P to R, Normalized pixel intensity distributions in the YFP and TMRM fluorescence channels plotted centrally across three individual mitochondria of a seedling expressing the NADKcNter-YFP.
To corroborate this conclusion, we measured the pixel intensity distribution of various TMRM-stained mitochondria from NADKcNter-YFP-transformed plants. While the TMRM fluorescence intensity peak was located at the center of the mitochondria, NADKcNter-YFP fluorescence intensity formed two distinctive peaks at opposite sides from the center where fluorescence intensity was at its minimum (Fig. 2, P–R). This pattern matches the one previously measured for the outer membrane-localized NMT1-GFP protein (Wagner et al., 2015).
We successfully achieved the expression of YFP-NADKc only in transiently transformed N. benthamiana leaves, where fluorescence was dispersed inside the cytosol (Supplemental Fig. S4, third row). This result was probably due to the NADKc N terminus being hidden in the middle of the sequence and is consistent with the hypothesis that the N-terminal region of NADKc is important for the protein to be correctly addressed to the mitochondria.
Protein overexpression by a strong promoter such as CaMV 35S is prone to promote protein aggregates and overexpression artifacts. However, using cell fractionation, early works on the CaM/Ca2+-dependent NAD+ kinase activity in plants located this enzyme at the mitochondrial periphery (either inner or outer mitochondrial membrane) in both maize (Zea mays; Dieter and Marmé, 1984; Sauer and Robinson, 1985) and oat (Avena sativa; Pou de Crescenzo et al., 2001). More recently, NADKc was detected in the mitochondria by two proteomic studies (Klodmann et al., 2011; Wagner et al., 2015) and at the plasma membrane by one study (Mitra et al., 2007). Our results therefore corroborate and extend previous findings obtained using different approaches.
NADKc Enhances flg22 Response in Arabidopsis Seedlings
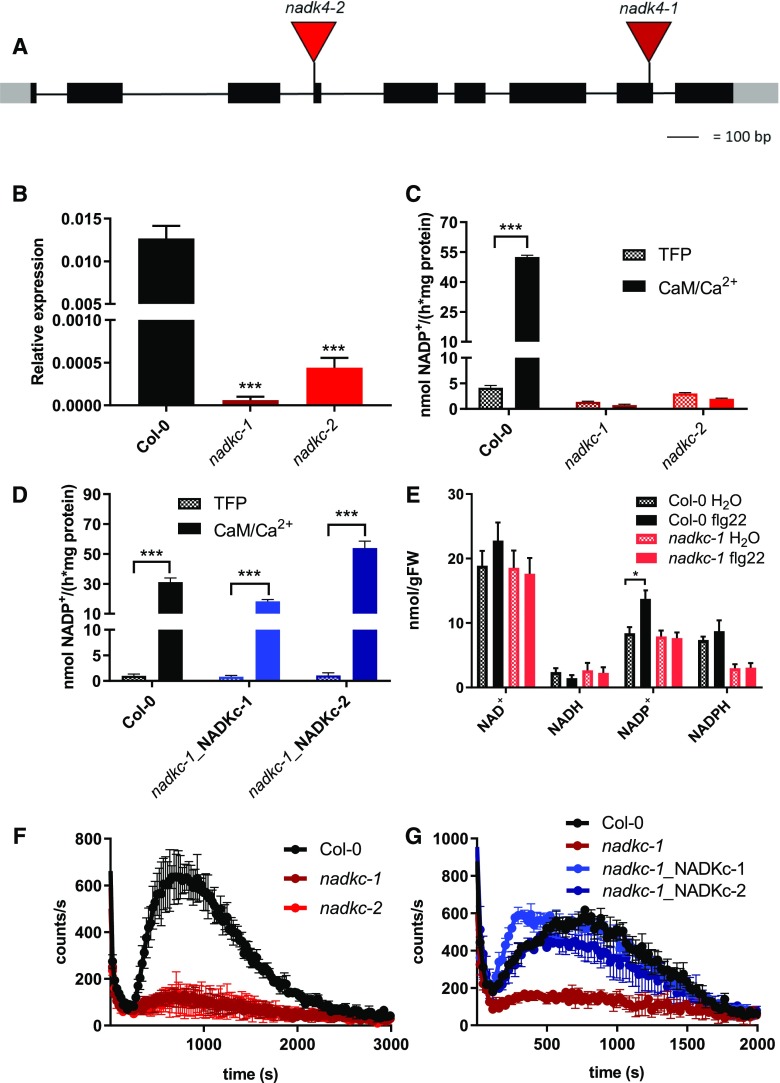
To investigate the physiological role of NADKc, we analyzed the two Arabidopsis T-DNA insertion lines, SALK_006202 and GABI-KAT_311H11, hereafter called nadkc-1 and nadkc-2 (Fig. 3A). NADKc transcripts were reduced by more than 95% in both lines (Fig. 3B).
Figure 3.
The CaM/Ca2+-dependent NAD+ kinase activity of Arabidopsis seedlings is absent in nadkc mutants. A, Schematic representation of the NADKc gene and positions of the T-DNA insertions in the nadkc-1 and nadkc-2 mutant lines. B, NADKc transcript levels in Col-0 and nadkc mutant seedlings. Levels are expressed relative to those of GAPDH. Data shown correspond to means ± sd, n = 3. C, NAD+ kinase activity measured in Col-0 and nadkc mutant plants (7-d-old whole plantlets) in the presence of the CaM inhibitor TFP (40 μm) or AtCaM1 (250 nm) and Ca2+ (0.5 mm). Data shown correspond to means ± sd, n = 4. D, NAD+ kinase activity measured in 7-d-old Col-0 and mutant whole plantlets complemented with NADKc (nadkc-1_NADKc-1 and nadkc-1_NADKc-2) in the presence of the CaM inhibitor TFP (40 μm) or AtCaM1 (250 nm) and Ca2+ (0.5 mm). E, NAD(P)+ and NAD(P)H concentrations in 7-d-old seedlings exposed (flg22; 1 μm) or unexposed (H2O) for 12 min to the bacterial elicitor flg22. Eighty to 100 mg of tissue was used per measurement. Data shown correspond to means ± sd for three biological replicates. FW, Fresh weight. F, Flg22 (1 μm)-induced oxidative burst in 7-d-old Col-0 and nadkc mutant seedlings (30 plantlets per well). Data shown correspond to means ± sd for four wells. G, Flg22 (1 μm)-induced oxidative burst in Col-0, nadkc-1 mutant, and mutant plants complemented with NADKc (nadkc-1_NADKc-1 and nadkc-1_NADKc-2) measured in 7-d-old seedlings, 30 plantlets per well. Data shown correspond to means ± sd for four wells. Asterisks indicate significant differences between two conditions based on Welch’s t test (*, P < 0.05 and ***, P < 0.001).
To confirm the unique role of NADKc for CaM/Ca2+-dependent NADP+ production in Arabidopsis seedlings, NAD+ kinase activity was measured in protein extracts from Columbia-0 (Col-0) and mutant seedlings in the presence of both trifluoroperazine (TFP), a CaM inhibitor, and AtCaM1/Ca2+ (Fig. 3C). In Col-0 plants, the activity measured in the presence of AtCaM1/Ca2+ was more than 10-fold higher than in the presence of TFP, while in nadkc-1/2 mutants, no difference was observed between the two conditions. In both mutants, activity levels were similar to those measured in Col-0 plants in the presence of TFP, confirming the absence of NADKc activity in these mutants. Consistent with these results, two complemented lines obtained by stably transforming nadkc-1 with full-length NADKc under the control of the CaMV 35S promoter (lines nadkc-1_NADKc-1 and nadkc-1_NADKc-2) had NADKc activity levels similar to Col-0 (Fig. 3D).
Neither nadkc mutant showed any visible growth impairment when grown under short-day or long-day photoperiods (Supplemental Fig. S5,A and B), and photosynthetic parameters (variable fluorescence over maximum fluorescence, electron transfer rate, and non-photochemical quenching; Maxwell and Johnson, 2000) were the same in all genotypes (Supplemental Fig. S5, C–E). This suggests that NADKc is not involved in photosynthesis-driven growth.
As CaM-dependent NAD+ kinase activity was previously associated with the generation of the oxidative burst triggered by plant response to elicitors (Harding et al., 1997; Grant et al., 2000), we expected to observe a decrease in a pathogen elicitor-induced extracellular ROS burst in nadkc-1/2 mutants coupled with lower NADP pools with respect to Col-0 seedlings.
We therefore exposed 7-d-old Arabidopsis seedlings to the bacterial elicitor flg22, followed by measurements of NAD(P)+ and NAD(P)H concentrations and ROS production. As shown in Figure 3E, no statistically significant differences were observed between Col-0 and nadkc-1 in NAD(P)+ and NAD(P)H concentrations before the flg22 treatment. However, the flg22 challenge induced an increase in the Col-0 NADP+ cellular concentration, which was absent in the nadkc seedlings. Moreover, dramatic reduction in ROS accumulation (more than 90%) was observed in the nadkc-1/2 mutants (Fig. 3F), but complementation with the NADKc full-length protein restored ROS accumulation up to wild-type levels (Fig. 3G).
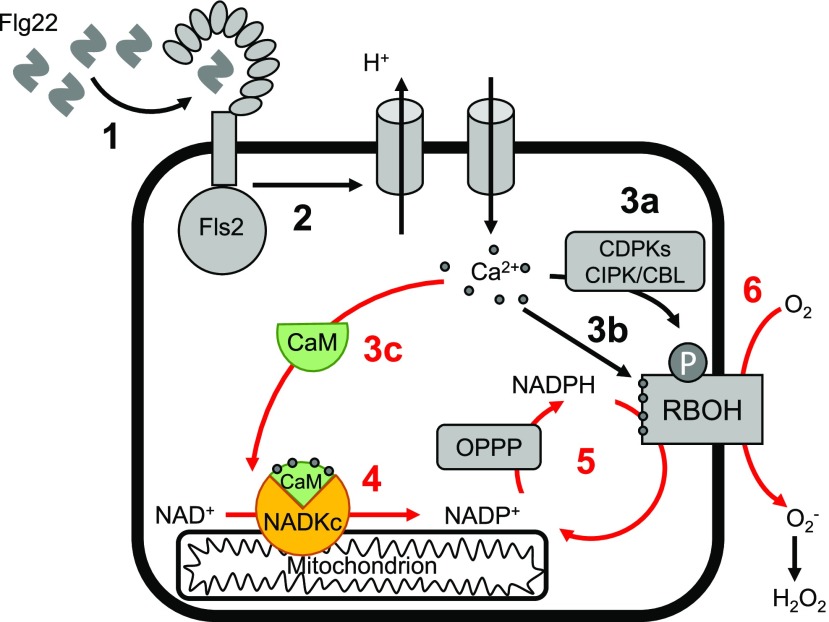
Based on these results, we propose a role for NADKc in producing NADP(H) needed to sustain the elicitor-induced ROS burst in Arabidopsis seedlings (Fig. 4).
Figure 4.
Hypothetical model of the role of CaM/Ca2+-dependent NADKc in sustaining the flg22-induced oxidative burst in Arabidopsis seedlings. Numbers refer to known sequential events; red numbers highlight events related to NADKc activation: 1, binding of flg22 elicitor to the Fls2 receptor (Sun et al., 2013); 2, activation of proton efflux and Ca2+ influx; 3a, Ca2+-dependent activation of CDPKs and CIPK/CBLs that phosphorylate RBOH proteins; 3b, Ca2+ binding to RBOH proteins; 3c, Ca2+ binding to CaM, leading to CaM structural modification and formation of the CaM/NADKc complex; 4, activation of NADP+ production by NADKc; 5, increased flux in the oxidative pentose phosphate pathway (OPPP), leading to a higher availability of NADPH; 6, production of the extracellular oxidative burst by NADPH oxidases (RBOH proteins).
Distribution of CaM-Dependent NAD+ Kinase Activity in the Green Lineage
The domain organization of NADKc (i.e. an approximately 200-amino acid domain of unknown function at the N terminus with a putative CBS followed by a kinase domain annotated ζ-toxin domain; Fig. 1C) was only found in angiosperms, bryophytes, and some algae.
To better trace the evolution of the plant CaM-dependent NAD+ kinase, we compared gene sequences and NADK activity in several plants and algae, and we built a maximum likelihood phylogenetic tree with representative putative NADKc proteins (Supplemental Fig. S6). The phylogenetic tree showed that plant NADKc-like proteins form four major clusters that correspond to the main plant phylogenetic groups, with the exception of gymnosperms and pteridophytes. Many plants, especially dicots, contain several genes encoding this protein, suggesting duplication events across evolution. Interestingly, the two other NADKc homologs present in the Arabidopsis genome (At1g06750 and At2g30630) seem to have a pollen-specific expression pattern (Krishnakumar et al., 2015).
Among algae, while the genomes of Chlamydomonas reinhardtii, Ostreococcus taurii, and Chara braunii appeared devoid of NADKc-like sequences, the genomes of Coccomyxa subellipsoidea, Ulva mutabilis, Klebsormidium flaccidum, Spyrotaenia minuta, Entransia fimbriata, Mougeotia spp., and Spirogyra spp. all harbor one NADKc-like sequence. In particular, NAD+ kinase genes of K. flaccidum, E. fimbriata, Mougeotia spp., and Spirogyra spp. contain a clear CBS, while in C. subellipsoidea, U. mutabilis, and S. minuta, this region is less conserved (Supplemental Fig. S2A). In agreement with the genomic survey, CaM-dependent NAD+ kinase activity could be successfully measured in the moss Marchantia polymorpha and the filamentous alga K. flaccidum but not in the unicellular alga C. reinhardtii (Table 2).
Table 2. Comparison of CaM-dependent NADK activity in different photosynthetic organisms.
NADK activity (nmol h−1 mg−1 protein) was measured in soluble protein extracts from Arabidopsis, M. polymorpha, K. flaccidum, and C. reinhardtii as detailed in “Materials and Methods.” n.d., Not detected.
| Parameter | Arabidopsis | M. polymorpha | K. flaccidum | C. reinhardtii |
|---|---|---|---|---|
| Total activitya | 31.2 ± 2.8 | 6.9 ± 0.7 | 5.8 ± 0.3 | 24.6 ± 1.4 |
| CaM/Ca2+-independent activityb | 1.0 ± 0.3 | 1.0 ± 0.1 | 1.8 ± 0.3 | 29.7 ± 2.7 |
| CaM/Ca2+-dependent activityc | 30.2 | 5.9 | 4.0 | n.d. |
NADK activity measured in the presence of CaM/Ca2+ represents CaM-independent plus CaM-dependent activity.
NADK activity measured in the presence of TFP (CaM inhibitor) represents CaM-independent NADK activity.
CaM/Ca2+-dependent activity is the difference between total NADK activity and CaM/Ca2+-independent activity. NADK activity in C. reinhardtii is independent of CaM/Ca2+ (the difference is within experimental error).
Interestingly, both the total CaM-dependent NAD+ kinase activity and the percentage of CaM-dependent NADK activity in the total NADK activity increase from K. flaccidum (4 nmol h−1 mg−1; 66.7%) to M. polymorpha (5.9 nmol h−1 mg−1; 85.7%) and to Arabidopsis (30.2 nmol h−1 mg−1; 96.8%). It is therefore possible that the importance of the CaM control on NADKc-like proteins increased during the evolution of plant lineages and became a key element of the ROS response to elicitors in angiosperms and, quite likely, other abiotic/biotic stress conditions that trigger Ca2+ fluxes.
CONCLUSION
Overall, we have identified unambiguously NADKc as the CaM/Ca2+-dependent NAD+ kinase of Arabidopsis seedlings. Its identification allows answering earlier questions concerning its physiological role: consistent with its localization at the mitochondrial periphery, this enzyme has no role in photosynthesis but can regulate the ROS burst by sustaining the activity of RBOH proteins. Besides being essential for the elicitor-induced oxidative burst of Arabidopsis, this enzyme may participate in other plant developmental and stress responses involving Ca2+ fluxes. This would stem from the evolutionary recruitment of a distinctive combination of a CaM-binding region and a type II ζ-toxin domain, which would provide it with regulatory properties different from its animal counterpart.
MATERIALS AND METHODS
Chemicals
All chemicals were from Sigma-Aldrich.
Plant Growth and Isolation of Homozygous NADKc Lines
Arabidopsis (Arabidopsis thaliana) Col-0 ecotype was used in this study. Plants were grown under 65% humidity and either long-day (16 h of light [85 μmol photons m−2 s−1]/8 h of dark) or short-day (8 h of light [90 μmol photons m−2 s−1]/16 h of dark) conditions. Daytime temperature was set to 20°C and nighttime temperature to 18°C.
The two T-DNA insertion lines, nadkc-1 (SALK_006202) and nadkc-2 (GABI_311H11), were obtained from the Nottingham Arabidopsis Stock Centre/Arabidopsis Biological Resource Center (Alonso et al., 2003; Kleinboelting et al., 2012). Lines were selected in the appropriate antibiotic (kanamycin for nadkc-1 and sulfadiazine for nadkc-2) and genotyped by PCR using left border primer LBb1.3 (nadkc-1) or LB GABI-KAT (nadkc-2) and the appropriate specific primers listed in Supplemental Table S2. PCR products were sequenced to confirm the precise position of each insertion.
Klebsormidium flaccidum (Hori et al., 2014; SAG335-2b curated as Klebsormidium nitens) was obtained from the Department of Experimental Phycology and Culture Collection of Algae (Göttingen Universität). The alga was grown on agar plates under continuous light (60 μmol photons m−2 s−1) in the Modified Bolds 3N Medium (https://utex.org/products/modified-bolds-3n-medium) without vitamins.
Marchantia polymorpha was collected in the forest (GPS coordinate 45.335088, 5.632257), and Chlamydomonas reinhardtii (C137 strain) was grown in Tris-acetate phosphate medium at 24°C under continuous low white light (40 μmol photons m−2 s−1) exposure. Protein extracts were prepared as described in Supplemental Materials and Methods for Arabidopsis.
Additional procedures are described in Supplemental Materials and Methods.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At1g04280 and under UniProt accession number Q0WUY1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. CaM-affinity purification of native CaM-dependent NAD+ kinase from Arabidopsis plantlets.
Supplemental Figure S2. Features of NADKc primary sequence and its homologs in other plant and algae species.
Supplemental Figure S3. Purification of recombinant NADKc produced in E. coli.
Supplemental Figure S4. NADKcNter-YFP associates with mitochondria in N. benthamiana leaves.
Supplemental Figure S5. Phenotype of nadkc mutants.
Supplemental Figure S6. Phylogeny of NADKc.
Supplemental Table S1. List of Arabidopsis proteins bound on a CaM-affinity column in the presence of Ca2+ and eluted by EGTA identified by mass spectrometry.
Supplemental Table S2. Primers used in this study.
Supplemental Materials and Methods. Protein purification procedures, biochemical and physiological analyses, imaging procedure, and phylogenetic analyses.
ACKNOWLEDGMENTS
We thank Markus Schwarzländer (University of Münster) for providing the NMT1:GFP and MT-cpYFP Arabidopsis lines and Elsa Clavel-Coibrié (Laboratoire de Physiologie cellulaire Végétale) for help with analysis of the complemented mutant plants. Part of this work was carried out at NOLIMITS, an advanced imaging facility established by the University of Milan. We thank the live microscopy facility MuLife of IRIG/DBSCI, funded by CEA Nanobio and labex Gral for equipment access and use.
Footnotes
This work was supported by the French National Research Agency (grant no. ANR-10-13 LABEX-04 GRAL Labex, Grenoble Alliance for Integrated Structural Cell Biology). G.F. acknowledges support from Human Frontier Science Program grant RGP0052/2015 (Photosynthetic Light Utilization and Ion Fluxes: Making the Link), and A.C. acknowledges support from Unimi Piano di Sviluppo di Ateneo (transition grant 2015/2017-Horizon 2020 Linea 1A). The proteomic analyses were partially supported by the French National Research Agency ProFI Grant (ANR-16 10-INBS-08-01).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson JM, Cormier MJ (1978) Calcium-dependent regulation of NAD kinase. Biochem Biophys Res Commun 84: 595–602 [DOI] [PubMed] [Google Scholar]
- Behera S, Zhaolong X, Luoni L, Bonza MC, Doccula FG, De Michelis MI, Morris RJ, Schwarzländer M, Costa A (2018) Cellular Ca2+ signals generate defined pH signatures in plants. Plant Cell 30: 2704–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai MF, Wei PC, Chen QJ, An R, Chen J, Yang S, Wang XC (2006) NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J 47: 665–674 [DOI] [PubMed] [Google Scholar]
- Chapman JM, Muhlemann JK, Gayomba SR, Muday GK (2019) RBOH-dependent ROS synthesis and ROS scavenging by plant specialized metabolites to modulate plant development and stress responses. Chem Res Toxicol 32: 370–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WG, Miller G, Wallace I, Harper J, Mittler R, Gilroy S (2017) Orchestrating rapid long-distance signaling in plants with Ca2+, ROS and electrical signals. Plant J 90: 698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Aglio E (2013) The regulation of plastidial proteins by calmodulins. PhD thesis. Université de Grenoble, Grenoble, France.
- Das U, Wang LK, Smith P, Shuman S (2013) Structural and biochemical analysis of the phosphate donor specificity of the polynucleotide kinase component of the bacterial pnkp⋅hen1 RNA repair system. Biochemistry 52: 4734–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Aglio E, Giustini C, Salvi D, Brugière S, Delpierre F, Moyet L, Baudet M, Seigneurin-Berny D, Matringe M, Ferro M, et al. (2013) Complementary biochemical approaches applied to the identification of plastidial calmodulin-binding proteins. Mol Biosyst 9: 1234–1248 [DOI] [PubMed] [Google Scholar]
- Delumeau O, Montrichard F, Laval-Martin DL (1998) NAD+ kinase activity, calmodulin levels during the growth of isolated cells from Lycopersicon pimpinellifolium and kinetic constants of the calmodulin-dependent NAD+ kinase. Plant Sci 138: 436–52 [Google Scholar]
- Delumeau O, Renard M, Montrichard F (2000) Characterization and possible redox regulation of the purified calmodulin-dependent NAD+ kinase from Lycopersicon pimpinellifolium. Plant Cell Environ 23: 1267–1273 [Google Scholar]
- Dieter P, Marmé D (1984) A Ca2+, calmodulin-dependent NAD kinase from corn is located in the outer mitochondrial membrane. J Biol Chem 259: 184–189 [PubMed] [Google Scholar]
- Drerup MM, Schlücking K, Hashimoto K, Manishankar P, Steinhorst L, Kuchitsu K, Kudla J (2013) The calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol Plant 6: 559–569 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Harding SA, Oh SH, Roberts DM (1997) Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J 16: 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5: 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Mori S, Mukai T, Hashimoto W, Murata K (2001) Molecular characterization of Escherichia coli NAD kinase. Eur J Biochem 268: 4359–4365 [DOI] [PubMed] [Google Scholar]
- Khoo SK, Loll B, Chan WT, Shoeman RL, Ngoo L, Yeo CC, Meinhart A (2007) Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J Biol Chem 282: 19606–19618 [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V, Hanlon MR, Contrino S, Ferlanti ES, Karamycheva S, Kim M, Rosen BD, Cheng CY, Moreira W, Mock SA, et al. (2015) Araport: The Arabidopsis information portal. Nucleic Acids Res 43: D1003–D1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love NR, Pollak N, Dölle C, Niere M, Chen Y, Oliveri P, Amaya E, Patel S, Ziegler M (2015) NAD kinase controls animal NADP biosynthesis and is modulated via evolutionarily divergent calmodulin-dependent mechanisms. Proc Natl Acad Sci USA 112: 1386–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebthal M, Dietz KJ (2017) The fundamental role of reactive oxygen species in plant stress response In Sunkar R, ed, Plant Stress Tolerance. Humana Press, New York, pp 23–39 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: A practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Mauve C, Gouia H, Saindrenan P, Hodges M, Noctor G (2010) Cytosolic NADP-dependent isocitrate dehydrogenase contributes to redox homeostasis and the regulation of pathogen responses in Arabidopsis leaves. Plant Cell Environ 33: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Mitra SK, Gantt JA, Ruby JF, Clouse SD, Goshe MB (2007) Membrane proteomic analysis of Arabidopsis thaliana using alternative solubilization techniques. J Proteome Res 6: 1933–1950 [DOI] [PubMed] [Google Scholar]
- Pou de Crescenzo MA, Gallais S, Léon A, Laval-Martin DL (2001) Tween-20 activates and solubilizes the mitochondrial membrane-bound, calmodulin dependent NAD+ kinase of Avena sativa L. J Membr Biol 182: 135–146 [DOI] [PubMed] [Google Scholar]
- Pugin A, Frachisse JM, Tavernier E, Bligny R, Gout E, Douce R, Guern J (1997) Early events induced by the elicitor cryptogein in tobacco cells: Involvement of a plasma membrane NADPH oxidase and activation of glycolysis and the pentose phosphate pathway. Plant Cell 9: 2077–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F (1997) Sequence motifs for calmodulin recognition. FASEB J 11: 331–340 [DOI] [PubMed] [Google Scholar]
- Sauer A, Robinson DG (1985) Calmodulin dependent NAD-kinase is associated with both the outer and inner mitochondrial membranes in maize roots. Planta 166: 227–233 [DOI] [PubMed] [Google Scholar]
- Scharte J, Schön H, Tjaden Z, Weis E, von Schaewen A (2009) Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc Natl Acad Sci USA 106: 8061–8066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, Logan DC, Fricker MD, Sweetlove LJ (2011) The circularly permuted yellow fluorescent protein cpYFP that has been used as a superoxide probe is highly responsive to pH but not superoxide in mitochondria: Implications for the existence of superoxide ‘flashes.’. Biochem J 437: 381–387 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou J-M, Chai J (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Vanderbeld B, Snedden WA (2004) Cloning and characterization of two NAD kinases from Arabidopsis: Identification of a calmodulin binding isoform. Plant Physiol 135: 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Behera S, De Bortoli S, Logan DC, Fuchs P, Carraretto L, Teardo E, Cendron L, Nietzel T, Füßl M, et al. (2015) The EF-hand Ca2+ binding protein MICU choreographs mitochondrial Ca2+ dynamics in Arabidopsis. Plant Cell 27: 3190–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]