A newly developed maize Mutator insertional library named ChinaMu represents the largest sequence-indexed insertional resource for maize to date.
Abstract
Sequence-indexed insertional libraries are important resources for functional gene study in model plants. However, the maize (Zea mays) UniformMu library covers only 36% of the annotated maize genes. Here, we generated a new sequence-indexed maize Mutator insertional library named ChinaMu through high-throughput sequencing of enriched Mu-tagged sequences. A total of 2,581 Mu F2 lines were analyzed, and 311,924 nonredundant Mu insertion sites were obtained. Based on experimental validation, ChinaMu contains about 97,000 germinal Mu insertions, about twice as many as UniformMu. About two-thirds (66,565) of the insertions are high-quality germinal insertions (positive rate > 90%), 89.6% of which are located in genic regions. Furthermore, 45.7% (20,244) of the 44,300 annotated maize genes are effectively tagged and about two-thirds (13,425) of these genes harbor multiple insertions. We tested the utility of ChinaMu using pentatricopeptide repeat (PPR) genes. For published PPR genes with defective kernel phenotypes, 17 out of 20 were tagged, 11 of which had the previously reported mutant phenotype. For 16 unstudied PPR genes with both Mu insertions and defective kernel phenotypes, 6 contained insertions that cosegregated with the mutant phenotype. Our sequence-indexed Mu insertional library provides an important resource for functional genomics study in maize.
The general approach to identifying gene function is based on the analysis of phenotypic variation between wild-type and mutant organisms. Sequence-indexed insertional libraries provide important resources for functional genomics studies in model plants such as Arabidopsis (Arabidopsis thaliana; Alonso et al., 2003; Kuromori et al., 2004) and rice (Oryza sativa; Jeong et al., 2006; Zhang et al., 2006; Krishnan et al., 2009; Wang et al., 2013). For example, in Arabidopsis, about 74% of the genes have been targeted by more than 88,000 transfer DNA (T-DNA) insertions (Alonso et al., 2003). In rice, about 60% of the genes have been targeted by 246,566 T-DNA, Ds/dSpm, or Tos17 insertions (Wang et al., 2013).
As an important crop and model plant, maize (Zea mays) plays an important role in both global food security and scientific research. The release of the B73 reference genome in 2009 has greatly facilitated the study of functional genes in maize (Schnable et al., 2009). According to a recent report, a total of 44,300 genes were annotated in the newly improved B73 reference genome (Jiao et al., 2017), but the vast majority of them were not functionally characterized. Therefore, there is an urgent need to generate high-quality sequence-indexed insertional resources at the genome-wide level to strengthen a platform for functional genomics in maize.
Several transposon-based genome-wide mutation libraries have been developed in maize, using either the Activator/Dissociation (Ac/Ds; McClintock, 1947) or Robertson’s Mutator (Mu; Robertson, 1978) system. Mutator has a high copy number and ∼100-fold the mutagenizing efficiency of Ac/Ds (Walbot, 2000), without apparent local transposition bias (Bennetzen, 1996), making the Mutator system ideal for the construction of genome-wide mutation libraries in maize. Several Mu-based mutation libraries have been constructed in maize (Bensen et al., 1995; Raizada et al., 2001; May et al., 2003; Stern et al., 2004). However, only the UniformMu library is sequence-indexed and has a high germinal insertion rate (McCarty et al., 2005). The UniformMu library currently contains 39,864 insertions that have tagged 15,950 genes (https://www.maizegdb.org/uniformmu). Although UniformMu is an important resource for maize functional gene study, it covers only about 36% of annotated maize genes. Therefore, tremendous efforts are needed to increase the coverage of the maize genome.
In this study, we created a Mu insertional library named ChinaMu, consisting of about 20,000 F2 Mu lines, by crossing pollen of a Mu-starter line (see “Results”) to the B73 inbred line. The Mu flanking sequences from 2,581 F2 Mu lines were isolated and sequenced using a Mu-tag enrichment approach coupled with high-throughput sequencing. In total, 66,565 high-quality insertion sites were identified, tagging 20,244 (45.7%) of the annotated genes in the maize genome. Together with the UniformMu library, about 52.2% of the annotated maize genes are now tagged. The ChinaMu library is currently the largest sequence-indexed insertional library in maize, providing an important resource for maize functional genomics study.
RESULTS
Construction of the Mutant Library
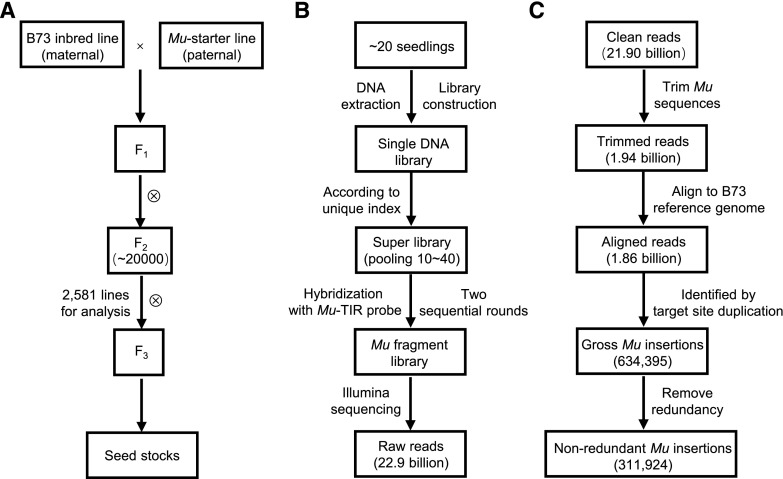
The Mu-starter line contained MuDR and the a1-mum2 gene (Bensen et al., 1995). The spotted aleurone phenotype of a1-mum2 identifies autonomous MuDR activity in the genome (Pooma et al., 2002). The seeds from the Mu-starter line were planted, and the pollen was used to pollinate B73 plants to generate F1 seeds. About 20,000 F2 ears were obtained through self-pollination of the F1 plants; of these, 2,581 F2 ears were randomly chosen for analysis in this study (Fig. 1A). Phenotypic scoring of seed-defective mutants in the 2,581 F2 lines indicated a seed mutation rate of about 9.1%, with 139 defective kernel mutants and 96 empty pericarp mutants. The mutation rate of this library was slightly higher than that of UniformMu (5% to 7%), based on seed mutant frequency (McCarty et al., 2005).
Figure 1.
Crossing scheme and experimental process of ChinaMu. A, Overview of ChinaMu crossing scheme. B, The process of Mu insertions isolation experiment. C, The identification pipeline of Mu insertion sites.
Isolation and Sequencing of Mu-Tagged Sequences
To construct a DNA library for each F2 line, the leaf tissues of about 20 F2 seedlings from a single F2 ear were pooled and then subjected to genomic DNA extraction (Fig. 1B). This pooling strategy could minimize the impact of somatic transpositions in the following analysis, because somatic transpositions would represent a smaller part of the total DNA than would germinal transpositions. The genomic DNA of each F2 line was then mechanically sheared, fractionated, and ligated to an adapter with a unique sequencing index to construct a DNA library. These DNA libraries were accurately quantified using a Bioanalyzer 2100 instrument. A super library was constructed by pooling 10 to 40 DNA libraries with equal DNA molar amounts.
To isolate Mu-containing DNA fragments, the super library was then hybridized to a biotinylated 60-mer oligonucleotide designed according to the end of the Mu terminal inverted repeats (TIRs; Williams-Carrier et al., 2010). Two successive hybridization steps were performed to enrich the DNA fragments containing Mu sequences. The products from hybridization were quality checked by a Mu-enrichment test (see “Materials and Methods”), and then sequenced by Hiseq Xten (Illumina).
In this study, the 2,581 F2 lines were subjected to Mu-tag isolation and sequencing analysis, and 22.9 billion raw reads were obtained. After removing adapter sequences and low-quality reads, 21.9 billion clean reads were retained for further analysis (Fig. 1C).
Identification of Mu Insertion Sites
To obtain sequence information for the Mu insertion sites in each F2 line, reads containing any member of the Mu family were extracted from the 21.9 billion clean reads. This yielded total 6.29 billion Mu-containing reads. These Mu-containing reads were trimmed for Mu sequences, and 1.94 billion Mu flanking sequence tags (FSTs) were obtained. The FSTs were then aligned to the B73 reference genome (v4.0) using Bowtie 2 (Langmead and Salzberg, 2012). About 1.86 billion FSTs with the best alignments were retained (Fig. 1C).
Because the FSTs on both sides of each Mu insertion were sequenced, a reliable Mu insertion site could be identified by searching for overlapping FSTs containing the same target site duplication (TSD; Barker et al., 1984; Supplemental Fig. S1). The FSTs without TSD features were further removed for the following scenarios. Due to the presence of original Mu insertions in the B73 reference genome, the FSTs cannot form TSD overlaps to the flanking sequences of pre-existing Mu sites in the B73 genome. But these FSTs are capable of forming stacks next to ends of these Mu sites. Based on the distance and sequences between the stacks, 30 ancestral insertions (Supplemental Dataset S1) with 0.39 billion FSTs were identified in B73 females and filtered out. Moreover, due to genomic variation between the Mu-starter line genome and the B73 reference genome, and/or the potential chimeric amplification during Mu sequence enrichment and library preparation, about 0.73 billion FSTs that only formed one side of the stacks (no overlaps) were also removed. A total of 634,395 reliable Mu insertion sites containing TSD features remained, with 0.74 billion aligned FSTs (Fig. 1C).
These identified Mu insertions contained redundant Mu insertions existing in multiple F2 lines. The redundant ancestor insertion sites were reduced to one for each site, leaving a total of 311,924 unique Mu insertion sites from the 2,581 F2 lines (Fig. 1C).
Criteria for Identification of Germinal Mu Insertions
The identified unique Mu insertions contained both germinal and somatic insertions; however, only germinal insertions are stably transmitted to the next generation and thus useful for further genetic studies. Theoretically, germinal insertions are present in almost every cell, whereas somatic insertions are present in only a small fraction of cells. The number of sequenced FSTs could reflect the prevalence of each insertion among the pooled seedlings; however, due to the variation in enrichment efficiency and sequencing depth among different samples, the FSTs needed to be properly normalized before making this comparison. The FSTs at each insertion site were normalized to the total aligned FSTs in each F2 line. This normalization resulted in comparable numbers of FST reads between different F2 lines. Normalized FST read counts (NFCs) per insertion site showed an approximately exponential distribution (Supplemental Fig. S2).
To determine the ideal NFC cutoff value for germinal insertion identification, eight F2 lines were subjected to biological replicate analysis. Two batches of seedlings were started from each of the eight lines. Mu germinal insertions were predicted to be those present in both replicate samples. The inheritance of these putative germinal Mu insertions was further tested by looking for their presence in the corresponding F3 genomic DNAs. Primers were designed to amplify DNA flanking putative insertion sites in combination with a Mu-TIR-specific primer. An authentic germinal insertion should be present both in the F2 DNA and in the corresponding F3 DNA.
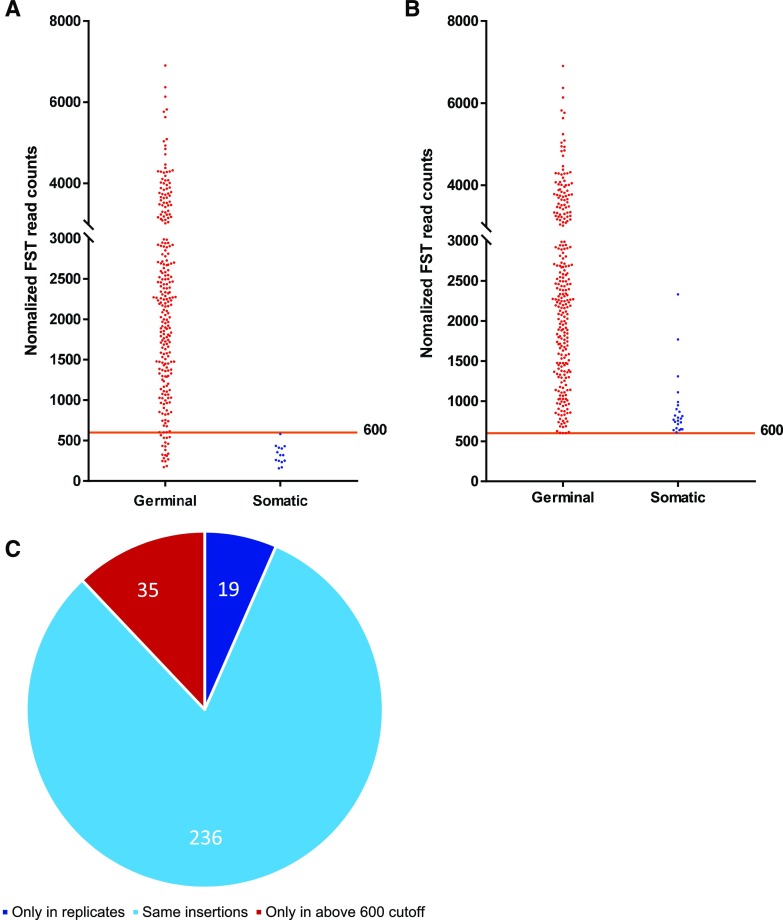
A total of 269 unique insertions present in both replicates were tested for inheritance in the corresponding F3 generation. In this way, 255 insertions (94.8%) were experimentally verified as germinal insertions. There were 14 insertions that were not detected in F3 generation, all with NFCs below 600 (Fig. 2A; Supplemental Dataset S2). When a threshold of 600 NFCs was applied, about 92.5% (236) of the 255 verified germinal insertions were retained. Therefore, 600 NFCs was chosen as a practical cutoff number for identifying germinal insertions.
Figure 2.
Comparison of the two methods in replicate analysis. NFCs denotes Normalized FST (Flanking Sequence Tag) read counts. Red and blue dots represent the germinal and somatic insertions verified by experiment, respectively. The orange line represents the cutoff number to classify the germinal and somatic insertions. A, The NFCs distribution of insertions obtained from replicate method. These 269 unique insertions that presented both in the pair of replicates were tested for inheritance in the corresponding F3 generation; 255 (94.8%) of them were validated as germinal, and 14 insertions with NFCs below 600 were somatic. B, The NFCs distribution of insertions obtained from 600 cutoff method. There were 294 unique insertions with NFCs above 600 in the eight samples. For the same inheritance test, 271 (92.2%) were validated as germinal and 23 were somatic. C, The statistics of germinal insertions validated by the two methods. A total of 290 germinal insertions were validated by the two methods. The recovery rate of 600 cutoff was 93.4% [(236 + 35)/290]. The recovery rate of the replicated method was 87.9% [(236 + 19)/290].
In the data from the eight pairs of replicates, there were 58 insertions with NFCs above 600 that were only present in one of the replicates. These 58 insertions were tested for inheritance, and 35 were found to be germinal insertions. Therefore, there were 290 (255 + 35) experimentally verified germinal insertions among the eight pairs of replicates (Fig. 2C; Supplemental Dataset S2). If the cutoff value of 600 NFCs was applied, a total of 294 unique insertions had an NFC above 600 in the eight pairs (Fig. 2B). Furthermore, 93.4% (271/290) of the germinal insertions could be recovered from the entire dataset, and the positive rate (confirmed germinal insertions out of those initially detected by the method) was 92.2% (271/294; Fig. 2, B and C). When the replicate method was used, 87.9% (255/290) of the germinal insertions could be recovered, and the positive rate was 94.8% (255/269; Fig. 2, A and C). However, the replicate method would double the cost of data production. Based on these results, 600 NFCs was chosen as the cutoff value for germinal insertion identification in the following analysis.
Identification of Germinal Mu Insertions
To test the cutoff of 600 NFCs for germinal insertion identification in other nonreplicated lines, 295 insertion sites with >600 NFCs from 153 F2 lines were randomly selected based on the distribution of NFCs per insertion site (Supplemental Fig. S2; Supplemental Dataset S3), and tested for their inheritance in their corresponding F3 DNAs. The result showed that 90.8% (268) of the tested insertion sites were germinal. When the cutoff of 600 NFCs was applied to the entire dataset of 2,581 F2 lines, 66,565 putative germinal insertion sites were identified (Table 1). Among them, 28 insertions that were present in more than 2,000 F2 lines suggested that they were fixed in the Mu-starter male parent (Supplemental Dataset S1). A set of 10,577 insertions occurred in two or more F2 families, but were not ubiquitous. These insertions are probably inherited by plants with closely related pedigrees. Besides, there were 55,960 new germinal transpositions, giving an average of 22 new germinal transpositions per line.
Table 1. The prediction of putative germinal insertions based on experiment.
| NFCs Section | Proportion (%)a | Total Insertionsb | Putative Germinal Insertionsc |
|---|---|---|---|
| Above 600 | 268/295 (90.8) | 66,565 | 66,565 |
| 100–600 | 24/150 (16.0) | 195,912 | ∼31,000 |
| Below 100 | 0/30 | 49,447 | — |
| Total | — | 311,924 | ∼97,000 |
The proportion of germinal insertions validated by experiment.
The number of the nonredundant insertions in corresponding NFCs section.
In terms of insertions with NFCs above 600, the proportion of germinal insertions is 90.8%; therefore, these insertions were all regarded as putative germinal insertions for further analysis. The number of putative germinal insertions with 100–600 NFCs was calculated based on the experiment. Because the experiment did not obtain the proportion of germinal insertions with NFCs below 100, the number of putative germinal insertions is unavailable in this section.
In the replicate analysis, a substantial but lower percentage of germinal insertions were found among the insertions with NFCs below 600, but there were no germinal insertions below 100 NFCs. Therefore, to test the inheritance of the insertions with NFCs below 600, these insertions were divided into two categories: less than 100 NFCs and 100–600 NFCs. A total of 30 insertions with NFCs below 100 were tested, but none of them was detected in the corresponding F3 generation (Table 1; Supplemental Dataset S3). A total of 150 insertions with 100–600 NFCs from 51 lines were tested (Supplemental Dataset S3), and 16% (24/150) of them were germinal. Based on these analyses, it was predicted that there were about 31,000 additional germinal insertions from the 195,912 nonredundant insertions within the category of 100–600 NFCs (Table 1).
According to these calculations, the entire dataset of 2,581 F2 lines, named ChinaMu, included about 97,000 (66,565 + 31,000) germinal insertions (Table 1), which is about 2.4 times the number in UniformMu. Due to the relatively lower germinal insertion rate (16%) in the sites below 600 NFCs, such sites should be first tested for germinal inheritance before further use. In the following analysis, for data accuracy, only the 66,565 high-quality insertions with NFCs above 600 were used.
Distribution Patterns of Germinal Mu Insertions
To evaluate ChinaMu as a resource for maize functional genomics study, we compared the ChinaMu data to well-established UniformMu data (McCarty et al., 2005). A total of 39,864 UniformMu insertion sites obtained from MaizeGBD were aligned to the B73 v4.0 reference genome for ease of comparison (see “Materials and Methods”).
To examine the distribution of Mu insertions across chromosomes, the Mu insertion site numbers from both libraries were plotted in fixed windows of 500 kb. This analysis revealed a nonuniform distribution on each of the chromosomes (Supplemental Fig. S3). Similar distribution patterns of insertion sites were observed in ChinaMu and UniformMu. Each chromosome exhibited a ‘bowl-shape’ pattern (i.e. greater preference for insertions in the distal portions than in pericentromeric regions) of the frequencies of Mu insertions per 500 kb. Noticeably, some insertion hotspots were observed in certain chromosome regions in both libraries (Supplemental Fig. S3).
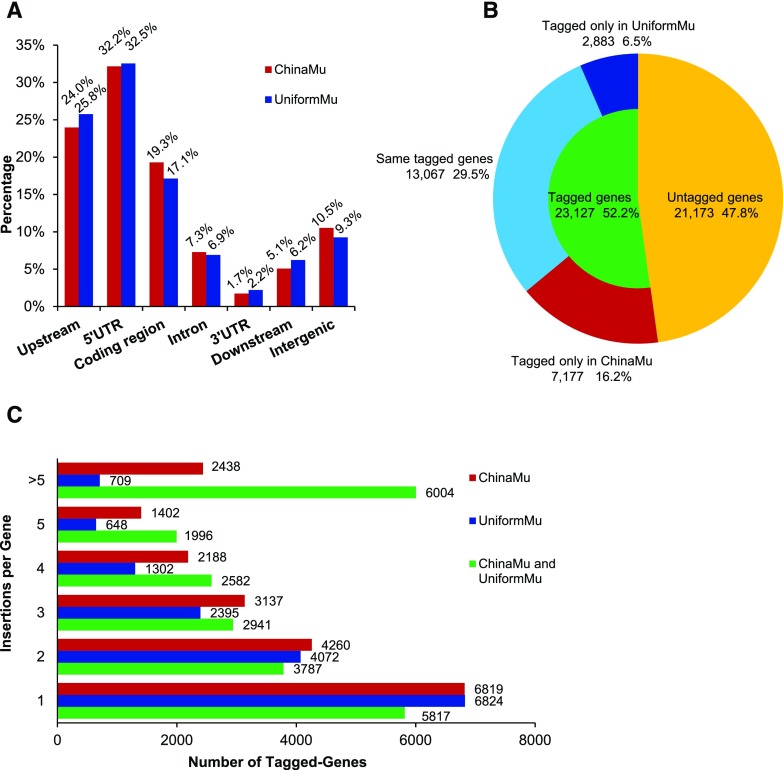
To explore the distribution of Mu insertions relative to gene structure, the insertions were categorized according to insertion position using SnpEff (Cingolani et al., 2012; see “Materials and Methods”; Fig. 3A). All identified insertions were categorized as variations in intergenic or genic regions with the upstream and downstream regions considered to extend 2 kb from the 5′ or 3′ untranslated region (UTR), respectively. Mu exhibits a strong preference for genic regions: the percentages of Mu germinal insertions from ChinaMu and UniformMu located in genic regions were about 89.6% and 90.7%, respectively (Fig. 3A). Similar nonuniform distribution patterns of insertion sites in genic regions were observed in both libraries. Overall, 5′ UTR regions had the highest frequencies of Mu insertions, and Mu insertions occurred at much higher rates in coding regions than in introns. By contrast, the 3′ UTR and downstream regions of genes had relatively low frequencies of Mu insertions. These findings are all consistent with a previous report (Liu et al., 2009).
Figure 3.
Distribution pattern, gene coverage, and insertional alleles of Mu insertions from ChinaMu and UniformMu. A, For each annotation, digits and percentages above the column denote the numbers and percentages of Mu insertions corresponding with this effect. All identified insertions were categorized as variations in intergenic or genic regions with the upstream and downstream regions considered to extend 2 kb from the 5′ or 3′ UTR, respectively. Percentages may not add up to 100% because of rounding. B, When ChinaMu and UniformMu were compared, the proportion of genes that were tagged (green) and untagged (yellow) by the two libraries was determined. Tagged genes contained the same tagged genes (sky blue) for the two libraries; genes tagged only in ChinaMu (red) and genes tagged only in UniformMu (blue). C, Red and blue denote unique tagged-genes from ChinaMu and UniformMu, respectively. Green represents tagged-genes from the combination of two libraries.
Gene Coverage and Insertion Frequency of ChinaMu
The 66,565 high-quality insertions from ChinaMu covered 20,244 genes, representing 45.7% of the 44,300 annotated genes in the B73 v4.0 reference genome (Fig. 3B). Although 39,864 insertions from UniformMu covered 15,950 genes, 13,067 of them were also tagged in ChinaMu. Moreover, 7,177 genes (16.2% of total maize genes) were only covered by insertions from ChinaMu. In the two libraries combined, 52.2% (23,127) of maize genes were covered by Mu insertions (Fig. 3B).
To further analyze the number of Mu insertions per gene, we counted the numbers of insertional alleles (Fig. 3C). In ChinaMu, 13,425 genes (30.3% of total maize genes) have two or more insertional alleles, whereas 6,819 genes have only a single insertion. In UniformMu, 9,126 genes (20.6% of total maize genes) have two or more insertional alleles, and 6,824 genes have a single insertion. When the two libraries are combined, the genes with two or more allelic insertions increases to 17,310 (39.1% of total maize genes), accounting for near three-quarters (74.8%) of all tagged genes in both libraries. Moreover, 6,004 genes have more than 5 allelic insertions (Fig. 3C).
Test of ChinaMu with PPR Genes
To test the utility of ChinaMu for functional gene identification, pentatricopeptide repeat (PPR) genes were used as a test model. PPR genes compose a large plant gene family with biological functions highly related to posttranscriptional regulation in mitochondria and chloroplasts (Barkan and Small, 2014). In this study, 509 putative PPR genes were predicted by a 35-amino acid sequence motif using the HMMER3 program (http://www.ebi.ac.uk/Tools/hmmer/) applying an E-value ≤ 1E-10 (see “Materials and Methods”; Supplemental Dataset S4). In the ChinaMu library, 1,187 high-quality Mu insertions were identified in 382 (75%) of the predicted PPR genes, among which 287 genes harbored more than one insertion per gene (Supplemental Dataset S5).
Among the 20 PPR genes that have been characterized as functioning in mitochondria and producing defective kernel or empty pericarp phenotypes, 17 were tagged in ChinaMu by 76 Mu insertions (Table 2; Supplemental Dataset S6). Among these 76 Mu insertions, 37 of them were located in exonic regions of 11 PPR genes. The corresponding seed stocks of these 37 insertions all had predicted seed mutant phenotypes according to published results. Although three insertions validated as somatic by PCR experiments were located in exonic regions, the corresponding seed stocks had no visible phenotype (Supplemental Dataset S6). In addition, 34 of the 76 insertions were in upstream, downstream, and intron gene regions, and the corresponding 34 seed stocks had no visible mutant phenotype. However, for 2 insertions upstream of dek35 (Chen et al., 2017) and close to the exon (44 and 36 bp upstream of the coding region), the corresponding 2 seed stocks had visible phenotypes (Supplemental Dataset S6).
Table 2. Summary of published and newly identified PPR genes in ChinaMu.
‘Dek’ and ‘emp’ represent defective kernel and empty pericarp, respectively.
| Gene ID | Allelesa | Mutant Phenotypeb | Full Namec | Reference |
|---|---|---|---|---|
| Zm00001d002098 | 1 | emp | emp12 | Sun et al., 2018 |
| Zm00001d003543 | 6 | dek | dek37 | Dai et al., 2018 |
| Zm00001d007100 | 2 | No visible phenotype | smk1 | Li et al., 2014 |
| Zm00001d008298 | 1 | emp | emp7 | Sun et al., 2015 |
| Zm00001d011559 | 10 | No visible phenotype | emp16 | Xiu et al., 2016 |
| Zm00001d013136 | 1 | No visible phenotype | dek36 | Wang et al., 2017 |
| Zm00001d022480 | 8 | emp | emp9 | Yang et al., 2017 |
| Zm00001d033749 | 10 | dek | dek35 | Chen et al., 2017 |
| Zm00001d033869 | 4 | emp | emp4 | Gabotti et al., 2014 |
| Zm00001d033992 | 1 | No visible phenotype | emp10 | Cai et al., 2017 |
| Zm00001d034253 | 8 | emp | emp18 | Li et al., 2019 |
| Zm00001d034428 | 3 | No visible phenotype | ppr78 | Zhang et al., 2017 |
| Zm00001d034882 | 3 | dek | dek2 | Qi et al., 2017 |
| Zm00001d038257 | 5 | dek | dek19 | Dong et al., 2019 |
| Zm00001d042039 | 3 | emp | emp5 | Liu et al., 2013 |
| Zm00001d047013 | 3 | No visible phenotype | dek39 | Li et al., 2018 |
| Zm00001d052450 | 7 | emp | emp11 | Ren et al., 2017 |
| Zm00001d012097 | 2 | dek | dek47 | This study |
| Zm00001d012528 | 2 | dek | dek48 | This study |
| Zm00001d014663 | 4 | dek | dek49 | This study |
| Zm00001d022184 | 1 | dek | dek50 | This study |
| Zm00001d028388 | 1 | dek | dek51 | This study |
| Zm00001d044843 | 4 | dek | dek52 | This study |
Different mutant seed stocks could have the same PPR gene inserted at different sites.
The ‘dek’ or ‘emp’ represents at least one of alleles had corresponding mutant phenotype.
The newly identified PPR genes in this study continued to be named according to the known mutant name.
Among the PPR genes with unknown function, 16 were selected based on having both Mu insertions in the coding region and a visible defective kernel mutant phenotype (Supplemental Table S1). For each gene, 20 individual kernels from an F2 family were scored as mutant or nonmutant, and site-specific primers were designed for genotyping. Gene-specific primers flanking the insertion site were used to detect the wild-type genotype, and the combination of one gene-specific primer and a Mu-TIR-specific primer was used to detect the Mu insertion. All 16 of these insertions were detected in the corresponding seed stocks; moreover, six insertions showed cosegregation with the seed mutant phenotype. Therefore, the insertions in these 6 PPR genes could be the causative mutations for the defective kernel phenotypes (Table 2; Supplemental Fig. S4).
DISCUSSION
Here, we report construction of the ChinaMu library, the largest sequence-indexed insertional library in maize. This insertional library is based on the Mutator transposon: newly transposed Mu elements were recovered by probe hybridization, followed by high-throughput sequencing. A total of 2,581 F2 Mu lines were analyzed, and 311,924 nonredundant Mu insertions were identified. A protocol for germinal insertion identification was developed, and 66,565 high-quality germinal insertions were identified (Table 1). This number is about 1.7-fold of that of the UniformMu library (39,864). The ChinaMu library alone tagged 45.7% (20,244) of maize annotated genes. When compared with UniformMu, 7,177 (16.2%) genes were newly tagged by ChinaMu (Fig. 3B). With this new sequence-indexed resource, the percentage of tagged maize genes increased from 36% to 52.2% (Fig. 3B), and the average number of insertion alleles per gene increased from 2.3 to 4.1 (Fig. 3C). Therefore, this newly developed ChinaMu library markedly increased the availability of tagged genes in the maize genome, and will greatly facilitate the functional genomic study of maize.
Although ChinaMu and UniformMu are both Mu insertional libraries, there are still some differences between them. The entire ChinaMu library was from a single Mu-active plant named Mu-starter. In contrast, UniformMu library has multiple mutagenic parents. Because the ChinaMu library was generated by the crossing between the Mu-starter line and the B73 inbred line, the F2 lines contain 58 (28 from Mu-starter line and 30 from B73 inbred line) ancestral insertions (Supplemental Dataset S1). Due to multiple-round backcrossing between the mutagenic parents and the W22 inbred line, the UniformMu library is in a uniform W22 background with 21 ancestral insertions (McCarty et al., 2005). Although 18 new germinal transpositions occurred in each UniformMu line, only five of them were captured on average (McCarty et al., 2005). However, ChinaMu library captured 22 new germinal insertions per line on average. There are two possible reasons for more new insertions recovered in the ChinaMu compared with those in the UniformMu library. The technologies used in ChinaMu library analysis, including probe hybridization enrichment, high-throughput sequencing, and bioinformatic analysis pipeline, enabled a more efficient and comprehensive identification of new Mu insertions. Different mutagenic parent lines could have different MuDR activities. The Mu-starter line might have higher MuDR activity than the UniformMu lines.
Maize is an excellent model for transposon genetics study, and transposon mutagenesis is frequent in maize (Settles, 2005). Maize contains several highly active endogenous DNA transposons, such as Ac/Ds and Mutator (McClintock, 1947; Robertson, 1978), that can be easily adopted for large-scale mutagenesis. The transposon insertions are usually large enough to be highly effective and genetically stable in causing substantial disruptions of gene function (McCarty and Meeley, 2009). Furthermore, transposon insertions are relatively easy to identify using molecular genetics and high-throughput sequencing technologies. Moreover, the maize genome contains relatively low copy numbers of transposons, and it is easier to correlate genotype to phenotype than with saturated EMS mutagenesis (Lu et al., 2018).
Although a number of Mu-based mutation libraries have been constructed in maize (Bensen et al., 1995; May et al., 2003; Fernandes et al., 2004; Stern et al., 2004), two challenges were encountered: one was the inefficiency of recovering and identifying newly transposed insertions; the other was the lack of an effective way to distinguish germinal insertions from the high background of somatic insertions. In this study, we successfully overcame both challenges. With the improved protocol of Mu-tag enrichment by probe hybridization, coupled with high-throughput next-generation sequencing, Mu insertions in each F2 line were efficiently recovered and sequenced. The strategy of pooling 10 to 40 individual F2 lines further increased the throughput for data production. An optimized bioinformatics pipeline increased the efficiency of data processing and the precision of Mu insertion site prediction.
To effectively distinguish germinal insertions from somatic insertions, two methods were attempted in this study. The first method involved identifying germinal insertions using replicated samples, which was also used for UniformMu analysis by Mu-seq (McCarty et al., 2013). The second involved establishing a practical criterion for germinal insertions (Williams-Carrier et al., 2010). This latter method was suggested in the PML Mu-Illumina study, but the suggested criterion was not experimentally validated. In this study, we first normalized the FST read counts in different samples. We then compared the two methods according to the recovery rate and the positive rate using eight pairs of replicated samples (Fig. 2). Based on this comparison, a 600-NFC cutoff number was experimentally determined. Although the positive rate in the replicate method was higher (94.8% versus 92.2% of insertions detected by the method were germinal), the recovery rate was lower (87.9% versus 93.4% of the total germinal insertions) than that obtained using the NFC cutoff method. Moreover, the NFC cutoff method was cost effective (only 50% that of the replicate method), and more efficient for data processing; therefore, it became the choice in this study. Experimental validation of the 600-NFC cutoff in a large dataset indicated a positive rate of 90.8%, which is similar to the 89% of UniformMu (Settles et al., 2007).
Although ChinaMu represents the largest sequence-indexed insertional library in maize, there is great room for future improvement. So far, the combination of ChinaMu and UniformMu contains a total of 95,736 germinal insertions and has tagged 52.2% of the maize annotated genes. This number is less than those of Arabidopsis (more than 88,000 insertions, 74% of genes tagged) and rice (246,566 insertions, about 60% of genes tagged; Alonso et al., 2003; Wang et al., 2013). Because adequate F2 mutant lines are available, and the pipeline for Mu-tag isolation, sequencing, and analysis has been optimized, we can continue to expand the ChinaMu dataset to archive better tagged gene coverage. The expansion plan for ChinaMu is already being implemented. In the current pipeline, we used the B73 (v4.0) reference genome due to its high assembly and annotation quality. Although B73 was one of the parents used to construct the F2 lines, the Mu-starter line is the one that harbored the initial Mu transpositions. For this reason, a fully sequenced Mu-starter line genome could further improve Mu FSTs alignment and increase the number of high-quality germinal insertions.
As the number of Mu insertions increased, the number of insertional alleles per gene also increased (Fig. 3C), and, therefore, the efficiency of tagging new genes decreased. This was also observed in insertional libraries of rice: up to 246,566 T-DNA, Ds/dSpm, or Tos17 insertions covered only about 60% of the genes (Wang et al., 2013). Previous studies (Kolesnik et al., 2004; Hsing et al., 2007; Zhang, 2007) together with our data strongly suggest that natural insertion mutations would hardly achieve whole-genome saturation. Because Mu insertions concentrate in genomic regions with epigenetic marks of open chromatin near the transcription start site (Liu et al., 2009; Springer et al., 2018), some treatment or measure (Ransom et al., 2009) can be attempted to promote the opening of chromatin, thereby facilitating Mu insertions to occur in otherwise inaccessible regions.
To facilitate functional genomics study in the maize community, the dataset and seed collections generated in this study are publicly available at http://chinamu.jaas.ac.cn/Default.aspx.
MATERIALS AND METHODS
Plant Material
The maize (Zea mays) seed stock 330I (a1-mum2; A2 C1 C2 MuDR R1) containing active Mu was obtained from the Maize Genetics Cooperation Stock Center (https://www.maizegdb.org/data_center/stock?id=96309). The 330I stock was planted and self-pollinated, and a cob with all spotted seeds was selected as the Mu-starter line. The seeds from the Mu-starter line were planted, and the pollen was used to pollinate B73 inbred line plants to generate F1 seeds. All F1 seeds were spotted, and they were planted and selfed to obtain F2 ears. F3 seeds were obtained through self-pollination of F2 plants.
DNA Extraction and Preparation
Twenty seeds from each F2 line were germinated on germination paper at room temperature. At the two-leaf stage, equal amounts of young leaf tissue were collected from each germinated seedling and pooled. Genomic DNA was extracted from the pooled tissue using the phenol–chloroform extraction method (Porebski et al., 1997). The integrity and quantity of genomic DNA were ensured by running agarose gel electrophoresis and Qubit Fluorometer (Invitrogen). With the help of two commercial platforms (Mega Genomics; Annoroad Gene Tech), ∼1 μg of genomic DNA was fragmented into 300- to 600-bp lengths using the Bioruptor Picoruptor Sonicator (Diagenode). Libraries were constructed commercially using the KAPA HTP Library Preparation Kit (Kapa Biosystems) and AnnoLib DNA Library Prep Kit (Annoroad Gene Tech).
Isolation of Mu Insertions
FSTs of Mu insertions were isolated by the modified method of Mu-Illumina (Williams-Carrier et al., 2010). According to the barcodes of different samples, equimolar amounts of samples from 10 to 40 libraries were pooled for hybridization. Hybridization was performed between pooled samples and a biotinylated 60-mer oligonucleotide (Integrated DNA Technologies), which was designed based on the end of the Mu TIR (Williams-Carrier et al., 2010). Two successive hybrid enrichment steps were performed. The hybrids were captured by Dynabeads M-270 Streptavidin (Invitrogen, 653-05). With use of primers that bind to the ends of DNA fragment adapters, 16 and 15 cycles of PCR were used to bulk up the recovered DNA after the first and second selection rounds, respectively. PCR product was cleaned using a Universal DNA Purification Kit (TIANGEN Biotech). The effectiveness of the Mu-tag enrichment was tested by calculating the multiple of ancestral Mu insertions (located within a 47.3-Mb region of maize chromosome 2) using an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific).
The quality of the enrichment product was assessed using the QIAxcel-Advanced instrument (QIAGEN). All libraries were sequenced commercially (Mega Genomics; Annoroad Gene Tech) by Hiseq Xten (Illumina) to generate 150-bp paired-end reads and 8-bp indexed reads.
Identification of Mu Insertion Sites
Raw reads were categorized by their barcode sequences. Reads containing low-quality bases or adapter sequences were removed. Two indexed libraries including 15 Mu family sequences and B73 genome (v4.0; Jiao et al., 2017) were generated by the Bowtie2-build program (Langmead and Salzberg, 2012). Clean data for each sample were aligned to the Mu-indexed library using the Bowtie2 tool in local mode to extract the Mu-containing reads, and then Mu TIR sequences were removed. Trimmed sequences with lengths ≥20 bp were realigned to the maize B73 v4 reference genome (Jiao et al., 2017) using Bowtie2 in local mode with –U, –S options. To facilitate further analysis, the view command in SAMtools program (0.1.19; Li, 2011) was used to transform the alignment outputs from SAM to BAM format. The BAM file was sorted and then transformed to SAM file by sort and view commands. Because the flanking sequences on both sides of Mu insertion can be amplified and sequenced, the featured TSD can be found according to the overlapping sequences (Supplemental Fig. S1). The read number of each site in the genome was counted from the sorted SAM file.
Conversion of UniformMu Insertion Sites
To identify the physical position of the UniformMu insertions on the B73 reference genome (v4.0), an in-house Perl script was used to extract 200-bp flanking sequences of Mu insertion sites (B73 v2; https://www.maizegdb.org/uniformmu). These flanking sequences were aligned to the B73 reference genome (v4.0) via the BLASTN program, using a cutoff E-value ≤1E-20.
Annotation of Mu Insertion Sites
SnpEff (Cingolani et al., 2012) was used to annotate the functional effects of each insertion based on the B73 reference genome version 4.0 (Jiao et al., 2017). All identified insertion sites were categorized as variations in intergenic and gene regions with the upstream and downstream regions considered to extend 2 kb from the 5′ or 3′ UTR, respectively, and all other SnpEff parameters were default settings. Functional annotations for the genes disrupted by likely moderate- and high-effect candidate insertions were obtained from Gramene (Tello-Ruiz et al., 2018).
Site-Specific PCR
The insertion site flanking sequences with 500 bp upstream and 500 bp downstream were extracted using an in-house Perl script. The high-throughput and genome-wide InDel marker development software mInDel was used to discover dimorphic InDel markers (Lv et al., 2016). Primers are listed in Supplemental Datasets S2 and S3. After primer design, the optimal dimorphic InDel markers predicted by mInDel were validated by agarose gel electrophoresis using W22 and B73 genomic DNA. The DNA template was isolated from the F3 generation. It was amplified with a site-specific primer and a degenerate Mu TIR primer (5′-GAAGCCAACGCCAWCGCCTCYATTTCGTCGAAT).
Prediction of PPR Genes
The PPR is a 35-amino acid sequence motif. Raw Hidden Markov model (HMM) data for the PPR gene family motif seed file (PF01535) were downloaded from the Pfam v32.0 database (http://pfam.xfam.org/). The motif file was then used to query the B73 v4 Ensembl-41 (ftp://ftp.ensemblgenomes.org/pub/plants/release-41/fasta/zea_mays/pep/; Goodstein et al., 2011) with the HMMER3 program (http://www.ebi.ac.uk/Tools/hmmer/) applying an E-value ≤ 1E-10 (Finn et al., 2011). The N-terminal signal peptides for all of the PPR protein sequences in B73 were then predicted using the signal peptide sequence prediction program TargetP (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000).
Statistical Analyses
Excel (2016 version) was used to perform the normalization of FST reads based on the total aligned reads. The NFCs stand for reads of insertion site per million mapped reads. All the NFCs were counted by excel. The number of insertion sites in different NFCs (Supplemental Fig. S2) were calculated with a fixed window of 100NFCs by R software.
Resource Distribution
The datasets generated during the current study are available at http://chinamu.jaas.ac.cn/Default.aspx.
Accession Number
Sequence data in this manuscript have been deposited in National Center for Biotechnology InformationnSequence Read Archive under accession number PRJNA547529.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Screen captures showing sequence reads marking the Mu insertion sites in Zm00001d004291.
Supplemental Figure S2. The number of insertion sites in different NFCs.
Supplemental Figure S3. Distribution comparison of Mu germinal insertion sites on 10 Chromosomes between ChinaMu and UniformMu.
Supplemental Figure S4. Mu insertions in predicted PPR genes cosegregated with mutant phenotype.
Supplemental Table S1. Cosegregation analysis of unstudied PPR genes for seed mutants.
Supplemental Dataset S1. Ancestral insertions from mutagenic male and B73 female parent.
Supplemental Dataset S2. Information for validated insertion sites from replicate analysis.
Supplemental Dataset S3. Information for validated insertion sites with different NFCs section.
Supplemental Dataset S4. Detailed information for PPR genes predicted by conserved amino acid motif.
Supplemental Dataset S5. Predicted PPR genes were tagged in ChinaMu.
Supplemental Dataset S6. PPR genes with published functional analysis were tagged in ChinaMu.
Acknowledgments
We appreciate the help of Dabin Xu (Mega Genomics) to this project. We also thank Yanhua Chen, Mingliang Zhang, Yuwei Chang, Yuanliang Bao, and Bingquan Zhang (College of Agronomy and Biotechnology, China Agricultural University) for technical assistance.
Footnotes
This work was supported by National Natural Science Foundation of China (31425019, to R.S.) and the Jiangsu Agriculture Science and Technology Innovation Fund [CX (18) 1001].
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Barker RF, Thompson DV, Talbot DR, Swanson J, Bennetzen JL (1984) Nucleotide sequence of the maize transposable element Mul. Nucleic Acids Res 12: 5955–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. (1996) The Mutator transposable element system of maize. Curr Top Microbiol Immunol 204: 195–229 [DOI] [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F (2017) Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J 91: 132–144 [DOI] [PubMed] [Google Scholar]
- Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R (2017) Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol Plant 10: 427–441 [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang IL, Coon M, Nguyen T, Wang L, Land S, Lu X, Ruden DJ (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Luan S, Chen X, Wang Q, Feng Y, Zhu C, Qi W, Song R (2018) Maize Dek37 encodes a P-type PPR protein that affects cis-splicing of mitochondrial nad2 intron 1 and seed development. Genetics 208: 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Tu M, Feng Y, Zdepski A, Ge F, Kumar D, Slovin JP, Messing J (2019) Candidate gene identification of existing or induced mutations with pipelines applicable to large genomes. Plant J 97: 673–682 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fernandes J, Dong Q, Schneider B, Morrow DJ, Nan G-L, Brendel V, Walbot V (2004) Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol 5: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabotti D, Caporali E, Manzotti P, Persico M, Vigani G, Consonni GJPS (2014) The maize pentatricopeptide repeat gene empty pericarp4 (emp4) is required for proper cellular development in vegetative tissues. Plant Sci 223: 25–35 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokshar DS (2011) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al. (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol 63: 351–364 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S (2004) Establishing an efficient Ac/Ds tagging system in rice: Large-scale analysis of Ds flanking sequences. Plant J 37: 301–314 [DOI] [PubMed] [Google Scholar]
- Krishnan A, Guiderdoni E, An G, Hsing YI, Han CD, Lee MC, Yu SM, Upadhyaya N, Ramachandran S, Zhang Q (2009) Mutant resources in rice for functional genomics of the grasses. Plant Physiol 149: 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K (2004) A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J 37: 897–905 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gu W, Sun S, Chen Z, Chen J, Song W, Zhao H, Lai J (2018) Defective Kernel 39 encodes a PPR protein required for seed development in maize. J Integr Plant Biol 60: 45–64 [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang YF, Hou M, Sun F, Shen Y, Xiu ZH, Wang X, Chen ZL, Sun SS, Small I, Tan BC (2014) Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J 79: 797–809 [DOI] [PubMed] [Google Scholar]
- Li XL, Huang WL, Yang HH, Jiang RC, Sun F, Wang HC, Zhao J, Xu CH, Tan BC (2019) EMP 18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytol 221 896–907 [DOI] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM, Fu Y, Nettleton D, Schnable PS (2009) Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet 5: e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC (2013) Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25: 868–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Liu J, Ren W, Yang Q, Chai Z, Chen R, Wang L, Zhao J, Lang Z, Wang H, et al. (2018) Gene-indexed mutations in maize. Mol Plant 11: 496–504 [DOI] [PubMed] [Google Scholar]
- Lv Y, Liu Y, Zhao HJBG (2016) mInDel: A high-throughput and efficient pipeline for genome-wide InDel marker development. BMC Genomics 17: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May BP, Liu H, Vollbrecht E, Senior L, Rabinowicz PD, Roh D, Pan X, Stein L, Freeling M, Alexander D, Martienssen R (2003) Maize-targeted mutagenesis: A knockout resource for maize. Proc Natl Acad Sci USA 100: 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Latshaw S, Wu S, Suzuki M, Hunter CT, Avigne WT, Koch KE (2013) Mu-seq: Sequence-based mapping and identification of transposon induced mutations. PLoS One 8: e77172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Meeley RB (2009) Transposon resources for forward and reverse genetics in maize In Handbook of Maize. Springer, pp 561–584 [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, et al. (2005) Steady-state transposon mutagenesis in inbred maize. Plant J 44: 52–61 [DOI] [PubMed] [Google Scholar]
- McClintock B. (1947) Cytogenetic studies of maize and Neurospora. Carnegie Inst Washington Year Book 46: 146–152 [Google Scholar]
- Pooma W, Gersos C, Grotewold E (2002) Transposon insertions in the promoter of the Zea mays a1 gene differentially affect transcription by the Myb factors P and C1. Genetics 161: 793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski S, Bailey LG, Baum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15: 8–15 [Google Scholar]
- Qi W, Yang Y, Feng X, Zhang M, Song RJG (2017) Mitochondrial function and maize kernel development requires Dek2, a pentatricopeptide repeat protein involved in nad1 mRNA splicing. Genetics 205: 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada MN, Nan G-L, Walbot V (2001) Somatic and germinal mobility of the RescueMu transposon in transgenic maize. Plant Cell 13: 1587–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, Liu C, Bartholomew B, Tyler JK (2009) FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 284: 23461–23471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F (2017) EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J Exp Bot 68: 4571–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DS. (1978) Characterization of a mutator system in maize. Mutat Res Fundam Mol Mech Mutagen 51: 21–28 [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Settles A. (2005) Maize community resources for forward and reverse genetics. Maydica 50: 405 [Google Scholar]
- Settles AM, Holding DR, Tan BC, Latshaw SP, Liu J, Suzuki M, Li L, O’Brien BA, Fajardo DS, Wroclawska E, et al. (2007) Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Anderson SN, Andorf CM, Ahern KR, Bai F, Barad O, Barbazuk WB, Bass HW, Baruch K, Ben-Zvi G, et al. (2018) The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet 50: 1282–1288 [DOI] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A (2004) Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci 9: 293–301 [DOI] [PubMed] [Google Scholar]
- Sun F, Wang X, Bonnard G, Shen Y, Xiu Z, Li X, Gao D, Zhang Z, Tan BC (2015) Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J 84: 283–295 [DOI] [PubMed] [Google Scholar]
- Sun F, Xiu Z, Jiang R, Liu Y, Zhang X, Yang Y-Z, Li X, Zhang X, Wang Y, Tan BC (2018) The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. J Exp Bot 70: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello-Ruiz M, Naithani S, Stein J, Gupta P, Campbell M, Olson A, Wei S, Preece J, Geniza M, Jiao Y, et al. (2018) Gramene 2018: Unifying comparative genomics and pathway resources for plant research. Nucleic Acids Res 46: D1181–D1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V. (2000) Saturation mutagenesis using maize transposons. Curr Opin Plant Biol 3: 103–107 [DOI] [PubMed] [Google Scholar]
- Wang G, Zhong M, Shuai B, Song J, Zhang J, Han L, Ling H, Tang Y, Wang G, Song R (2017) E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol 214: 1563–1578 [DOI] [PubMed] [Google Scholar]
- Wang N, Long T, Yao W, Xiong L, Zhang Q, Wu C (2013) Mutant resources for the functional analysis of the rice genome. Mol Plant 6: 596–604 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde R-A, Coalter R, Barkan A (2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–77 [DOI] [PubMed] [Google Scholar]
- Xiu Z, Feng S, Yun S, Zhang X, Jiang R, Bonnard G, Zhang J, Tan B (2016) EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis‐splicing, complex I assembly and seed development in maize: For cell and molecular biology. Plant J 85: 507–519 [DOI] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang HC, Sun F, Huang WL, Song S, Xu C, Tan BC (2017) The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol 214: 782–795 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S (2006) RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34: D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. (2007) Strategies for developing green super rice. Proc Natl Acad Sci USA 104: 16402–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-F, Suzuki M, Sun F, Tan BC (2017) The mitochondrion-targeted PENTATRICOPEPTIDE REPEAT78 protein is required for nad5 mature mRNA stability and seed development in maize. Mol Plant 10: 1321–1333 [DOI] [PubMed] [Google Scholar]