Analysis of natural variation in Arabidopsis provides insight into adaptive constraints and underlying gene functions influencing the composition and properties of outer seed mucilage.
Abstract
On imbibition, Arabidopsis (Arabidopsis thaliana) seeds release polysaccharides from their epidermal cells that form a two-layered hydrogel, termed mucilage. Analysis of a publicly available data set of outer seed mucilage traits of over 300 accessions showed little natural variation in composition. This mucilage is almost exclusively made up of rhamnogalacturonan I (RGI), highlighting the importance of this pectin for outer mucilage function. In a genome-wide association study, observed variations in polymer amount and macromolecular characteristics were linked to several genome polymorphisms, indicating the complexity of their genetic regulation. Natural variants with high molar mass were associated with a gene encoding a putative glycosyltransferase called MUCILAGE-RELATED70 (MUCI70). muci70 insertion mutants produced many short RGI polymers that were highly substituted with xylan, confirming that polymorphism in this gene can affect RGI polymer size. A second gene encoding a putative copper amine oxidase of clade 1a (CuAOα1) was associated with natural variation in the amount of RGI present in the outer mucilage layer; cuaoα1 mutants validated its role in pectin production. As the mutant phenotype is unique, with RGI production only impaired for outer mucilage, this indicates that CuAOα1 contributes to a further mechanism controlling mucilage synthesis.
Myxospermous seeds accumulate polysaccharides in the epidermal cells of the seed coat during seed development, and these are released on imbibition to form a sticky hydrogel that encapsulates the seed. This seed coat specialization is found in a large number of plant families (Yang et al., 2012b), with mucilage composition varying between species (Western, 2012; Phan and Burton, 2018). The model plant Arabidopsis (Arabidopsis thaliana) is myxospermic, and the composition and structure of its mucilage are the best characterized to date, largely because of the facility of molecular genetic studies in this species. Moreover, seeds without mucilage are viable in glasshouse or laboratory conditions, and as diverse cell wall polysaccharides and structural proteins are found as constituents, it has proved to be an excellent system for studying the properties and interactions between plant cell wall polymers.
Arabidopsis mucilage consists of two layers: a water-extractable outer layer and an adherent inner layer (Western et al., 2000; Macquet et al., 2007a). In the reference accession Col-0, the major constituent of both layers is the pectin domain rhamnogalacturonan I (RGI; Macquet et al., 2007a), which is formed of a repeating disaccharide, [→2)-α-l-Rha-(1→4)-α-d-GalA-(1→]n. The outer mucilage layer is essentially pure, unbranched RGI, in contrast to a more complex composition and structure for the inner layer. This is also a remarkable attribute when compared with cell wall pectin, which is a complex mixture of several different domain types whose relative amounts vary depending on the plant source. This makes Arabidopsis outer mucilage an excellent substrate for the study of structural modifications (Deng et al., 2006).
Mutant studies have shown that these RGI domains are initially synthesized with a few galactan and arabinan side chains that are then hydrolyzed in the later stages of seed coat differentiation (Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009a). The removal of these side chains increases the hydrophilic properties of RGI and contributes to mucilage release from the epidermal cells. More recently, it has been shown that mucilage RGI also has xylan substitutions that are present in a tight proportionality to the number of constituent disaccharides (Ralet et al., 2016). The inner mucilage layer contains cellulose, which is an essential structural component that provides a scaffold for RGI (Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011). The RGI polymers appear to be attached to this scaffold through the high binding affinity between the xylan side chains and cellulose (Ralet et al., 2016). Small amounts of the pectin domain homogalacturonan (HG) also seem to be present in the inner layer of mucilage, as treatment with the calcium chelator EDTA or polygalacturonase makes it appear less compact (Western et al., 2001; Macquet et al., 2007a), which suggests that calcium cross links between HG contribute to increasing interactions between polymers. Another minor mucilage component, galactoglucomannan, has also been shown to contribute to the organization of inner mucilage, as its absence results in more densely packed polymers (Yu et al., 2014; Voiniciuc et al., 2015b). Finally, a small amount of protein is present in mucilage (Tsai et al., 2017), and the arabinogalactan protein SALT-OVERLY SENSITIVE5 (SOS5) has been shown to play a role in the adherence of RGI polymers to the seed (Harpaz-Saad et al., 2011; Griffiths et al., 2014, 2016).
The identification of mutants with altered mucilage characteristics has been essential for determining the contribution and interactions described above. They have also enabled the identification of glycosyltransferases (GTs) involved in key steps of pectin synthesis, for example the recent characterization of a novel GT, RHAMNOGALACTURONAN I RHAMNOTRANSFERASE1 (RRT1), and GALACTURONOSYLTRANSFERASE11 (GAUT11; Takenaka et al., 2018; Voiniciuc et al., 2018). Both induced and naturally occurring mucilage mutants have been obtained, and in addition to the identification of the genes containing causal mutations, the latter have provided important information about the genetic basis for natural variation (Macquet et al., 2007b; Saez-Aguayo et al., 2013, 2014; Voiniciuc et al., 2016). Functional studies of mucilage production have mainly involved large-effect mutants, identified in forward genetic screens. Quantitative traits are, however, often produced by the combined contribution of small-effect genes, and the application of genome-wide association studies (GWAS) provides an alternative means of identifying such loci. This approach has previously been applied to inner mucilage phenotypes, and while no causal polymorphisms were identified, the reduced expression of a gene involved in galactoglucomannan synthesis was linked to more compact inner mucilage for a group of accessions (Voiniciuc et al., 2016).
The production of mucilage polysaccharides depletes a significant amount of carbon fixed by the mother plant, and while mucilage is nonessential in the laboratory, it must make an important contribution to fitness in the natural environment. This role, however, remains equivocal, and no unifying function has yet been demonstrated (North et al., 2014). Diverse ecophysiological roles have been suggested, including that seed mucilage aids germination, dispersion, seedling growth, or interaction with soil microorganisms (for review, see Yang et al., 2012b). Furthermore, the reason for the two layers observed in Arabidopsis mucilage is unknown, although it has been postulated that they each have a particular function, because of their different composition and properties (Macquet et al., 2007a). In order to optimize growth and reproduction in varying and suboptimal environmental conditions, plants adapt, and this is observed as natural variation within a species. As Arabidopsis is highly selfing, natural populations have little heterozygosity, making this species particularly suitable for the study of natural variation (Long et al., 2013). Many natural variants have been collected for Arabidopsis, and although a link was previously found between seed flotation caused by nonrelease of mucilage in some of these Arabidopsis accessions and their site of collection (Saez-Aguayo et al., 2014), why seeds of most Arabidopsis populations accumulate and release mucilage is unknown.
To address this, analysis of natural variability in seed mucilage structure, composition, and amount in relation to habitat would be necessary. A study of inner mucilage in around 280 Arabidopsis accessions previously found variation in the composition and structure of this layer (Voiniciuc et al., 2016). No association was found, however, between this variation and the geolocalization of their collection sites. Outer mucilage composition had only been analyzed for a handful of Arabidopsis accessions used routinely in laboratories until recently, when advances in analytical techniques used for examining polysaccharide characteristics were exploited in a high-throughput analysis to generate a data set for six outer mucilage characteristics (Poulain et al., 2019). Here, we exploit this to describe the variation in composition, amount and properties of polysaccharides in the outer mucilage layer. In addition to identifying accessions with atypical properties and geolocalizing a group with similar properties to a mountain range in central Asia, the data were exploited in GWAS. The characterization of two gene candidates identified confirmed their roles in modulating the properties and/or synthesis of mucilage RGI.
RESULTS
Outer Mucilage Traits Show Significant Variation between Arabidopsis Accessions
To determine the extent of natural variation in outer mucilage characteristics, we used a data set for six outer mucilage traits obtained for outer mucilage extracted from a panel of 306 accessions by gentle shaking in water (Poulain et al., 2019). These traits were GalA and neutral sugar (NS) contents, indicating the quantity and composition of outer mucilage polysaccharides, while the molar mass at peak maximum (Mp), intrinsic viscosity (IV), hydrodynamic radius (Rh), and radius of gyration (Rg) reflect the macromolecular characteristics of the major population of polymers separated by size-exclusion chromatography (Macquet et al., 2007a; Sullivan et al., 2011; Griffiths et al., 2016). Together, these characteristics provide information about the conformation, size, and volume the polymer occupies when hydrated. Data for five accessions were excluded from analyses: these correspond to Shahdara, Neo-3, Neo-6, and Sus-1, where no values were obtained because of their defect in mucilage release (Macquet et al., 2007b; Saez-Aguayo et al., 2014), and Dja-5, which has the same haplotype as Dja-1 (Simon et al., 2012), which shows delayed and incomplete mucilage release in water (Saez-Aguayo et al., 2013). The low values observed for Dja-5 for GalA and NS contents (Poulain et al., 2019) indicate that it shares the same mucilage phenotype, so that values were likely to be unrepresentative.
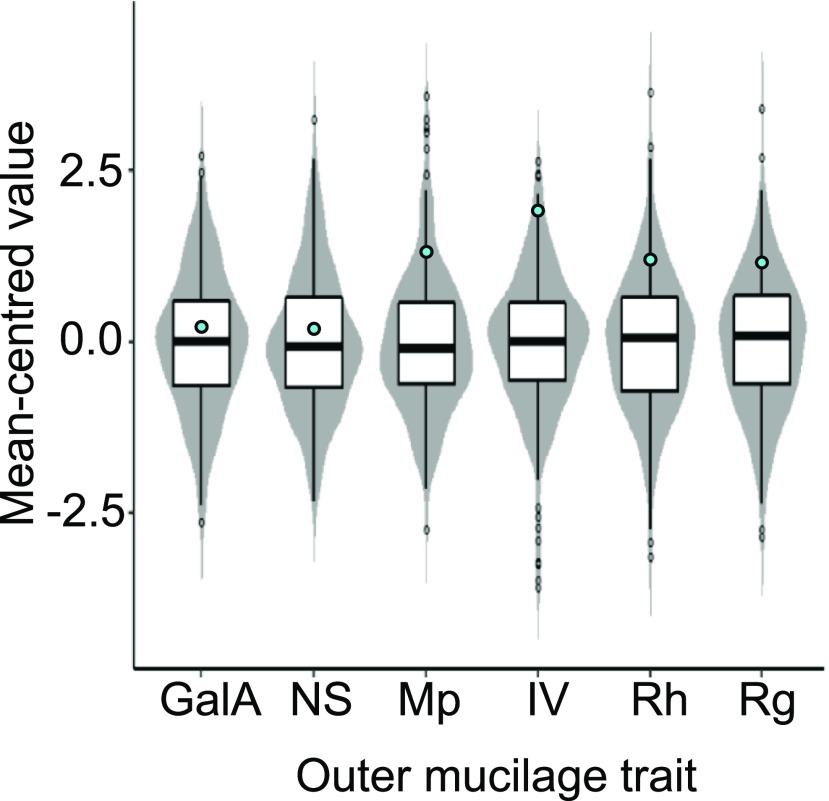
Each outer mucilage trait examined showed extensive variation between accessions, which only appeared to follow a normal distribution for GalA and Rg (Shapiro-Wilk normality test, P > 0.05). While similar levels of variation were observed for the two traits related to mucilage quantity, the traits that exhibited the largest and smallest range of variability were both related to polymer size and conformation, being Rh and IV, respectively (Fig. 1; Table 1). Outlier accessions were identified for each trait, notably 62 for NS content compared with only three for Rg (Fig. 1; Supplemental Table S1). Values for the reference accession Col-0 were within the interquartile range for mucilage amount but were higher than the upper quartile for all the macromolecular parameters, even being among the outliers with high IV. This variability in outer mucilage traits suggests that the natural habitat in which the accessions grow shapes these characteristics and that they may be adaptive.
Figure 1.
Natural variation observed between 301 Arabidopsis accessions for seed outer mucilage traits. Values have been transformed to mean-centered values to enable comparison of variation for each trait displayed as violin plots overlaid with the corresponding box plot, where outliers are indicated as circles. Turquoise circles indicate the positions of values for the reference accession Col-0.
Table 1. Summary of variation observed between Arabidopsis accessions for seed outer mucilage traits in absolute values.
| Value | GalUA | NS | Mp | IV | Rh | Rg |
|---|---|---|---|---|---|---|
| mg g−1 seed | mg g−1 seed | kD | mL g−1 | nm | nm | |
| Maximum | 15.7 | 15.1 | 829 | 654 | 64 | 110 |
| Mean | 10.5 | 9.6 | 550 | 499 | 46 | 81 |
| Minimum | 5.5 | 5.6 | 339 | 290 | 30 | 57 |
RGI Is the Major Component of Outer Mucilage in Arabidopsis
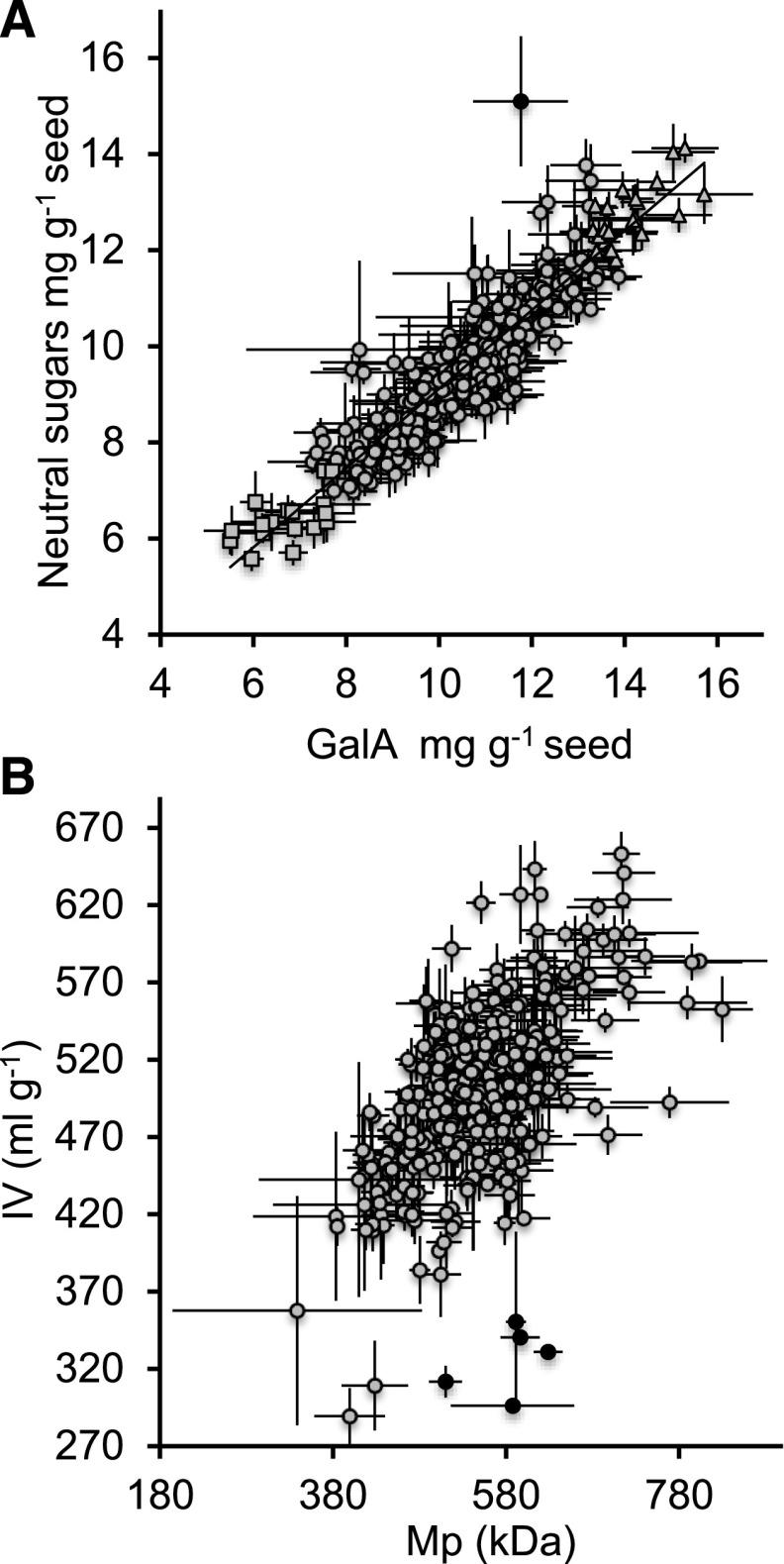
The composition of Arabidopsis outer mucilage has been extensively characterized for a small number of accessions, including the reference Col-0, and found to be composed almost exclusively of unbranched RGI (Goto, 1985; Western et al., 2000, 2001; Penfield et al., 2001; Macquet et al., 2007a). As RGI is made up of repeating [→2)-α-l-Rha-(1→4)-α-d-GalA-(1→] disaccharides, GalA contents for each accession were plotted against their NS contents to determine whether this was the case for the much larger panel of accessions studied here (Fig. 2A). As the latter includes Gal, Ara, Man, Xyl, and Fuc, in addition to Rha, deviation of the composition from unbranched RGI would be expected to shift the proportion of GalA to NS from a 1:1 ratio. This molar proportionality appeared to be conserved for most accessions, with a highly significant Pearson correlation coefficient between these traits (P < 10−9) and the linear regression intersecting the NS axis at 0.86, which is close to the 0.84:1 difference in MW for rhamnosyl:galacturonosyl residues; only one accession showed a clear shift toward more NS, Blh-1 (AV180). This indicates that the composition of outer mucilage over the range of Arabidopsis populations is stable. Furthermore, among outliers with a significant difference from the global mean for a given trait (P < 0.0001), 21 out of 24 high GalA outliers were common to high NS outliers, while 18 out of 23 low GalA outliers were common to the low NS outliers (Supplemental Table S1). These common outliers were termed high outer mucilage (HOM) and low outer mucilage (LOM) accessions, respectively (Tables 2 and 3). While this large number of common accessions is in accord with the stable composition of outer mucilage, it should be noted that more NS outliers were identified than GalA, which meant that a larger proportion was not common to GalA outliers, 12 and nine specific to high or low NS, respectively. This suggests that while the major component is RGI, a small degree of variation might exist for outer mucilage composition, toward a limited amount of either RGI branching or HG domains. In contrast to the stability of outer mucilage composition, very large differences were observed in the quantity of outer mucilage, with nearly threefold differences in total polysaccharide sugars between the lowest and highest values (Fig. 2A; Table 1).
Figure 2.
Relationship between seed outer mucilage traits for 301 Arabidopsis accessions. A, Sugar contents. B, Macromolecular properties. Atypical accessions are shown as black circles, high outer mucilage (HOM) accessions as triangles, and low outer mucilage (LOM) accessions as squares. Values are means of four biological repeats ± se.
Table 2. Site and country from which HOM accessions were collected.
Country of origin is indicated by ISO 3166 code (https://www.iso.org/iso-3166-country-codes.html). Symbols indicate coordinate reliability: *, reliable location; §, estimated location (often based on nearby town/city); #, no reliable location within country (corresponds to coordinates for capital city of country of origin). –, Not applicable.
| Versailles Identification Number | Accession Name | Country of Origin | Collection Site Coordinates (Latitude/Longitude) |
|---|---|---|---|
| 14 | Rom-5 | FRA | * 45.53793/4.858646 |
| 68 | Te-0 | FIN | § 60.059383/23.298025 |
| 114 | Is-0 | GER | § 50.477205/7.588778 |
| 133 | Ep-0 | GER | § 50.180479/8.516972 |
| 203 | Hodja-Obi-Garm | TJK | # 38.550475/68.781166 |
| 211 | Ksk-1 | UK | § 54.601734/−3.136282 |
| 235 | Sav-0 | CZE | § 50.086863/12.381974 |
| 255 | Niigata | JPN | § 37.988803/139.02008 |
| 298 | Js-0 | Unknown | – |
| 330 | Kly-4 | RUS | * 51.3166/82.5666 |
| 346 | Rak-3 | RUS | * 51.8666/80.05 |
| 351 | Chab-1 | RUS | * 53.6/79.3833 |
| 355 | Bij | RUS | * 52.5166/85.2666 |
| 356 | Sij-1 | UZB | * 41.75/70.0833 |
| 472 | Fl-3 | FIN | # 60.184755/24.937363 |
| 513 | Sv-0 | DEN | § 55.652764/11.282516 |
| 515 | Ts-1 | ESP | § 41.725795/2.930889 |
| 517 | Ts-5 | ESP | § 41.725795/2.930889 |
| 532 | Kar-2 | KGZ | * 42.3002/74.3691 |
| 537 | Zal-3 | KGZ | § 42.7964/76.3496 |
| 547 | Altaï-2 | CHN | * 47.756944/88.3975 |
Table 3. Site and country from which LOM accessions were collected.
Country of origin is indicated by ISO 3166 code (https://www.iso.org/iso-3166-country-codes.html). Symbols indicate coordinate reliability: *, reliable location; §, estimated location (often based on nearby town/city); #, no reliable location within country (corresponds to coordinates for capital city of country of origin).
| Versailles Identification Number | Accession Name | Country of Origin | Collection Site Coordinates (Latitude/Longitude) |
|---|---|---|---|
| 8 | Pyl-1 | FRA | * 47.194192/4.039306 |
| 10 | Par-9 | FRA | * 46.646643/−0.248251 |
| 13 | Hag-2 | UK | * 49.675771/−1.800928 |
| 18 | Con-1 | FRA | * 49.200371/3.204345 |
| 35 | Wc-1 | GER | § 52.60506/10.078897 |
| 65 | Gy-0 | FRA | § 48.77475/2.084824 |
| 90 | Rsch-4 | RUS | § 56.266741/34.327612 |
| 128 | Fr-2 | GER | § 50.129647/8.681717 |
| 145 | Hn-0 | GER | § 51.338183/8.266811 |
| 148 | Mr-0 | ITA | § 37.174762/14.770203 |
| 171 | Lc-0 | UK | § 57.338604/−4.423599 |
| 197 | Enkheim-T | GER | § 50.156517/8.753686 |
| 257 | Sakata | JPN | § 38.942641/139.831009 |
| 265 | N10 | RUS | § 54.7073/20.564125 |
| 287 | N16 | RUS | # 55.794025/37.611237 |
| 397 | Sq-8 | UK | * 51.25/0.41 |
| 399 | CIBC-17 | UK | * 51.25/0.41 |
| 544 | Had-1b | LBN | * 33.923056/35.695333 |
Variation in Outer Mucilage Macromolecular Properties Is Independent of the Quantity of Mucilage Produced
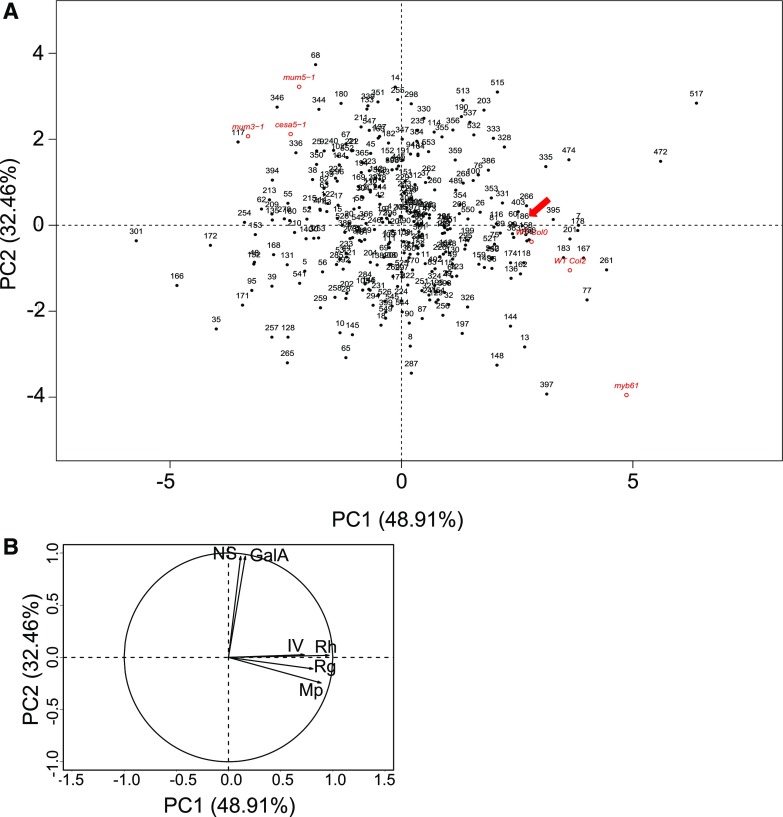
To examine in more detail the variation and relationships between outer mucilage traits, principal component analysis (PCA) was carried out. The first two components described over 80% of the variance observed in outer mucilage traits (48.91% and 32.46%, respectively). Accessions were relatively evenly scattered over the central area of the PCA (Fig. 3A). The corresponding variables factor map showed that the four macromolecular characteristics correlated positively with factor loading for PC1, while the two sugar content traits loaded positively with PC2 (Fig. 3B). The orthogonal factor loadings for sugar contents with respect to macromolecular properties demonstrated that these characteristics were completely independent for outer mucilage. As previous studies have shown that RGI branches change the volume of the polymer (Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009a), the independence of these traits is in agreement with outer mucilage being mainly unbranched RGI for the majority of Arabidopsis accessions and indicates that the properties of polymers are unrelated to the quantity of polymer produced. The relationships between accessions were examined using hierarchical clustering to class accessions into groups based on their outer mucilage traits. This indicated that accessions could be classified into five groups, with group 2 containing the four accessions that were previously termed floating mucilage releasing (FMR; Supplemental Fig. S1; Supplemental Table S2). The five groups contained similar numbers of accessions: 61, 66, 55, 77, and 42 accessions.
Figure 3.
PCA of six outer mucilage traits for 301 Arabidopsis accessions. A, Plot of principal components 1 and 2 (PC1 and PC2). Numbers correspond to Versailles accession numbers. The positions of control genotypes in red were projected onto the plot based on values obtained for the same six traits. The position of the reference accession Col-0 is indicated by the red arrow. B, Variables factor map showing loadings for each trait contributing to PC1 and PC2.
The relationship between outer mucilage macromolecular traits was investigated further in bivariate analyses. Highly significant Pearson correlation coefficients were observed between all four traits, although that for IV versus Rg was slightly lower (P < 10−9 versus P < 9.259 × 10−9; Supplemental Fig. S2). As Rh is calculated using the IV values, this linear correlation was expected. Furthermore, IV is indicative of the volume occupied by a polymer in solution, and when a polymer varies uniquely in length, as would be the case for outer mucilage composed of unbranched RGI, a close relationship is expected between molar mass and IV. Nevertheless, a set of accessions appeared to deviate from this linear correlation (Fig. 2B). These accessions comprised AV254 (Hiroshima), AV257 (Sakata), AV258 (Tokushima), AV259 (Yamagata), and AV549 (Qar8a). The observed shift in the relationship between Mp and IV for these accessions suggests that their outer mucilage polymers are present in a more compact form, which could be associated with either increased branching or other factors influencing polymer folding, such as structural proteins, like SOS5, previously observed in Arabidopsis mucilage (Harpaz-Saad et al., 2011; Griffiths et al., 2014, 2016).
Chemotype Grouping of Accessions with Known Mutants Indicates Potential Mechanisms Underlying Trait Variation
The six outer mucilage traits had also been analyzed for seeds of six control genotypes comprising four previously characterized induced mutants and their corresponding wild-type backgrounds (Poulain et al., 2019). The mutants used, cesa5-1, cesa5mum3-1, mum5-1, and myb61, have modified outer mucilage properties and/or amounts (Penfield et al., 2001; Sullivan et al., 2011; Ralet et al., 2016). These mutants are affected in genes for a cellulose synthase catalytic subunit, a putative xylosyltransferase, and a MYB transcription factor, respectively (Penfield et al., 2001; Desprez et al., 2007; Sullivan et al., 2011; Ralet et al., 2016). These data were projected onto the PCA to see whether the induced mutant outer mucilage chemotypes colocalized with those of accessions (Fig. 3A). The two wild-type controls, wild-type Col-0 and wild-type Col-2, were located in a similar region of the PCA, close to the AV186 Col-0 reference accession, confirming the reproducibility of the analyses for these six traits; Col-2 is closely related to Col-0, being a fifth-generation single-seed descent from Col-0 (Voiniciuc et al., 2013). Interestingly, all four mutant controls were localized on the edge of the PCA, indicating that some of their outer mucilage traits had extreme properties compared with natural variants. The values obtained for each trait for the induced mutants would place them as an outlier for two or more traits, with more extreme values observed for cesa5-1, cesa5mum3-1, and mum5-1 for the radius of gyration and myb61 for Mp and IV (Fig. 3A; Poulain et al., 2019).
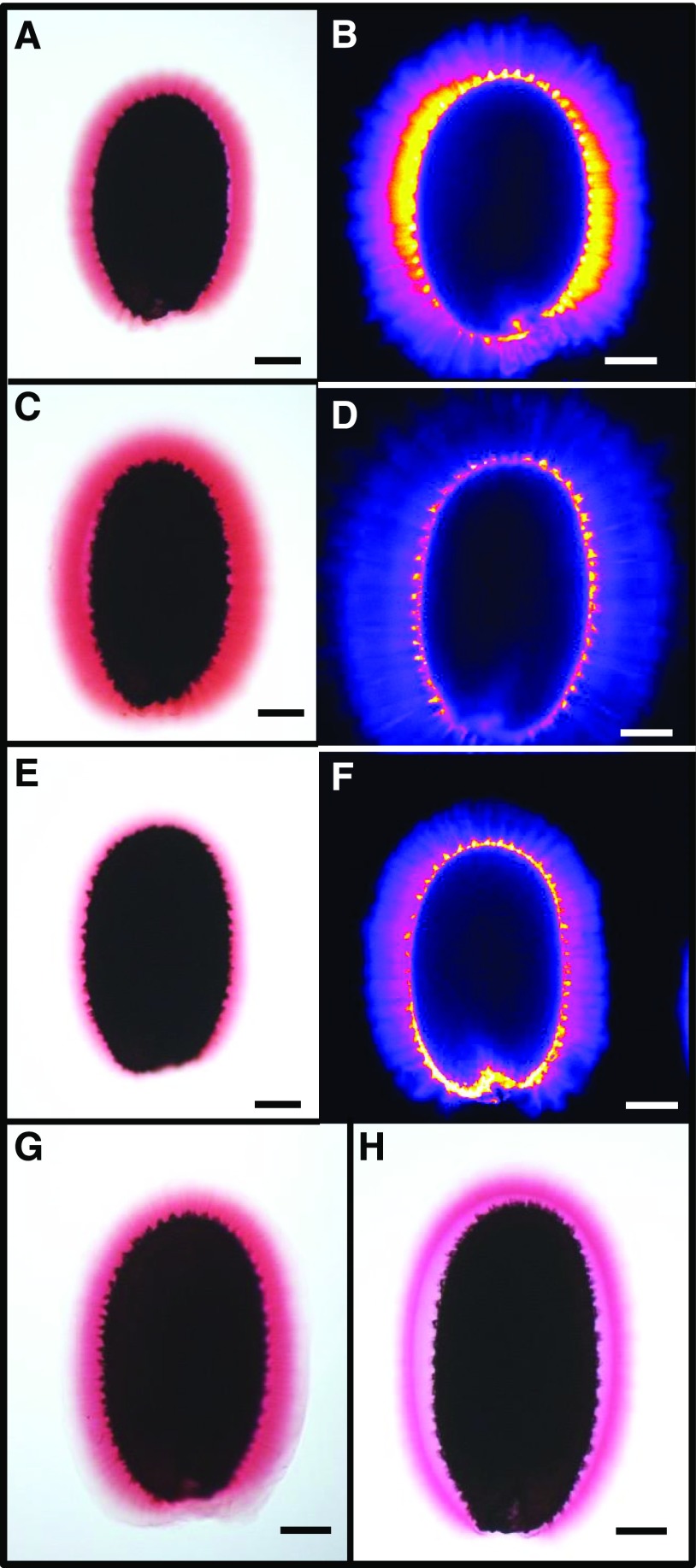
The three mutants affected in the formation of the inner mucilage layer because of defective attachment of RGI to cellulose were grouped together in the top left-hand quarter of the PCA, near the AV336, AV344, AV346, and AV350 accessions. These four accessions were previously classified as FMR and had HOM sugar contents and reduced inner mucilage cellulose staining, similar to cesa5-1 (Saez-Aguayo et al., 2014). This suggested that two other nearby accessions, Te-0 (AV68) and Ga-0 (AV117), might have similar redistribution of inner mucilage polysaccharides to outer mucilage. The pectin and cellulose in the inner mucilage layer of these two accessions were examined using Ruthenium Red staining and Direct Red 23, respectively. While Te-0 formed a large halo of mucilage, the Ga-0 inner mucilage layer appeared thinner than that of the Col-0 reference accession (Fig. 4, A–F). To determine if the Ga-0 accession was a natural mutant allele for the CESA5 or MUM5 gene, crosses were performed between Ga-0 and cesa5-2 and mum5-1, both recessive mutant alleles, and mucilage released from F1 seed coats on F2 seeds was examined. In both cases, the inner mucilage layer appeared as wide as the reference Col-0, indicating that Ga-0 was not affected in these genes (Fig. 4, G and H).
Figure 4.
Comparison of inner mucilage released from imbibed seeds of Te-0 and Ga-0 accessions with that of the Col-0 reference. Ruthenium red staining of pectin (A, C, E, G, and H) and Calcofluor staining of β-glycans (B, D, and F) are shown. A and B, Col-0 seeds. C and D, Te-0 seeds. E and F, Ga-0 seeds. G, Ga-0 × cesa5-2 F2 seeds. H, Ga-0 × mum5-1 F2 seeds. Images B, D, and F are shown using the Fire look-up table. Bars = 100 µm.
Adaptation of Outer Mucilage Traits to Environmental Conditions
The outer mucilage data set had been generated using seeds produced from two independent series of plants grown and harvested together; two seed lots from each series were used for subsequent mucilage analyses (Poulain et al., 2019). Environmental variations could have occurred during seed production, despite the plants being grown in controlled light and temperature conditions. For example, a randomized planting scheme was used, which might have varied the extent of shading from neighboring plants or exposure to air currents. Analysis of the data for any effects associated with the series in which the plant was produced was carried out and found a shift in values for all six traits in certain accessions between the first and second series. Nonetheless, different subsets of accessions were affected for each trait, and more than 250 accessions were unaffected for each. This suggests that different environmental conditions influence each trait and that genetic variation exists between accessions concerning their response to external factors, which might be related to adaptive responses in their natural habitats.
To go further toward linking genetic variation with adaptive responses, we examined in more detail the geographic localization of collection sites for outlier accessions classed as HOM or LOM. This attribution, based on two associated traits, involved a larger number of outliers than identified for the other outer mucilage traits (Tables 2 and 3). One of the HOM group of accessions corresponds to AV347, which was previously characterized as an FMR, originating from close to the Russian border with Kazakhstan (Saez-Aguayo et al., 2014). Interestingly, of the 18 HOM accessions having reliable or estimated coordinates for their point of collection, another eight came from central Asia and were collected near a mountainous region extending from the Altai in Russia through the Tian Sian, running through China and Kyrgyzstan down to Tadjakistan (Supplemental Fig. S3). This contrasted with the two Russian accessions with relatively accurate coordinates in the LOM group, which were from sites close to Europe.
GWA Highlights Distinct Genetic Factors That Influence Different Macromolecular Characteristics for Outer Mucilage Polysaccharides
Data from a 250K single-nucleotide polymorphism (SNP) chip are available from the 1001 Genomes project for 199 of the 301 accessions studied above (Cao et al., 2011; Gan et al., 2011; Long et al., 2013). This was used to perform a GWAS to identify genomic regions associated with mucilage quantity (GalA/NS) or measured values for macromolecular characteristics (IV/Mp/Rg). Traits related to mucilage quantity or properties were analyzed independently, and two sets of accessions were obtained after filtering to remove those with high variability between replicates (Supplemental Tables S3 and S4); the GalA/NS set comprised 194 accessions, and the IV/Mp/Rg set comprised 193 accessions. GWAS was performed for all five traits after data filtering, using the GWA-portal (https://gwas.gmi.oeaw.ac.at/) and both nonparametrical Wilcoxon rank-sum test (KW) and accelerated mixed-model (AMM) methods. Manhattan plots were obtained after filtering with a minor allele count (mac) of greater than 15 (Fig. 5; Supplemental Fig. S4).
Figure 5.
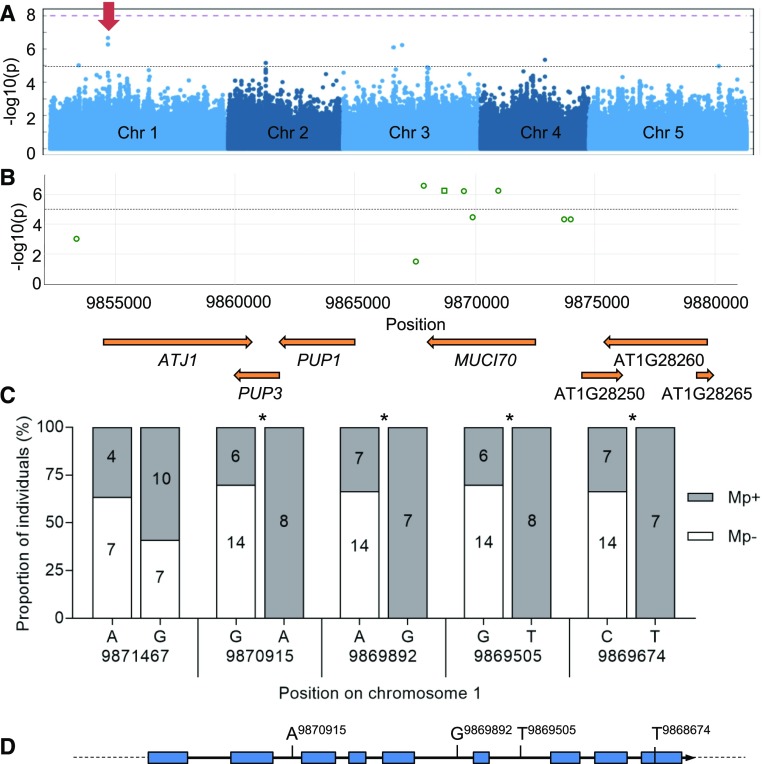
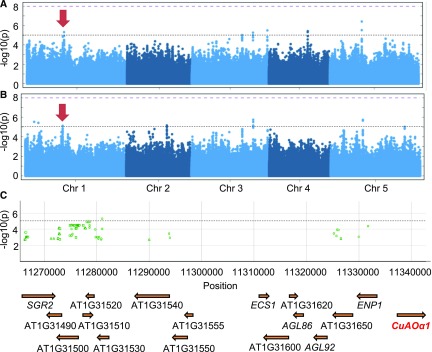
GWAS of Arabidopsis seed outer mucilage links MUCI70 to variations in polysaccharide molar mass. A, Manhattan plot showing the individual SNPs associated with the outer mucilage polysaccharide trait Mp using a panel of 193 natural accessions and the accelerated mixed-model GWA method. The log-transformed P values from the test of association are plotted as a function of chromosomal position. The genome-wide significance threshold (purple dashed line) was corrected for multiple testings by the Bonferroni method. The black dashed line represents the –log(P) = 5 threshold for significance used in this study. The red arrow indicates the peak on chromosome 1 presented in the closeup in B. Manhattan plots were obtained from GWA-Portal (https://gwas.gmi.oeaw.ac.at/). B, Closeup view of the significant peak on chromosome 1. The most significant SNPs, indicated with green outlines, are found inside the MUCI70 (At1g28240) sequence (mac > 15). Squares indicate synonymous coding polymorphisms, and circles indicate SNPs located in introns or intergenic regions. C, Frequency of SNPs in MUCI70 for 28 accessions classed as outliers for the Mp trait. Histograms indicate the proportions of SNPs for five genome positions; only the nucleotide variants in Supplemental Table S6 that are observed for at least six of the outlier accessions are shown. Asterisks indicate alleles present with significantly different proportions from that expected with a random distribution (χ2 test, P < 0.05). D, Polymorphisms relative to Col-0 found within MUCI70 DNA sequence from the Arabidopsis 1001 Genome Browser (http://signal.salk.edu/atg1001/3.0/gebrowser.php) enriched in high Mp outlier accessions.
The GWAS analysis did not highlight SNPs significantly associated with the variation of any trait using both GWA methods and the stringent threshold [−log10(P) > 8] corrected for multiple testing by the Bonferroni method. This method is designed to minimize false-positive associations, but true-positive associations that do not meet statistical significance can be missed, especially for complex traits influenced by many loci with small effects (Yang et al., 2010; Vinkhuyzen et al., 2013). Consequently, an arbitrary and less stringent threshold corresponding to a –log(P) of 5 was used in order to identify regions of interest. Given the lower threshold used, only the peaks that were colocalized using the AMM and KW methods were considered. The nine SNPs that reached this threshold for traits related to mucilage macromolecular characteristics are listed in Table 4, of which one is associated with variations in IV, seven with Mp, and two with Rg. The presence of only one colocalized SNP, between Mp and Rg (Fig. 5A; Supplemental Fig. S4), suggests that different genes modulate these traits, which was unexpected considering the strong correlations observed between all three traits (Fig. 2B; Supplemental Fig. S2).
Table 4. List of SNPs associated with outer mucilage physicochemical properties in GWA analysis and their nearest annotated genes.
The positions of SNPs with an association above the −log(P) = 5 threshold on Manhattan plots for both GalUA (GalA) and NS contents or for IV, Mp, or Rg are indicated with their P value scores. Only the SNPs colocalizing using both nonparametric Wilcoxon rank-sum test and accelerated mixed-model GWA methods were retained. The predicted effect of the SNP change compared with the Col-0 reference sequence is indicated, as well as the nearest gene, its annotation, and putative function obtained from Araport (Krishnakumar et al., 2015). Seed coat expression level corresponds to the maximum value in the seed coat data set, obtained from the Bio-Array Resource eFP browser (Winter et al., 2007), based on the data set generated by Le et al. (2010) for RNA extracted from laser-capture microdissected seed tissues.
| Chromosome | Position | Score | Effect | Nearest Gene | Araport 11 Annotated Function | Expression in Seed Coat |
|---|---|---|---|---|---|---|
| 1 | 4849215 | Mp 5.02 | Intergenic | AT1G14180 | RING/U-box superfamily protein | 65.3 ± 9.1 |
| 9867819 | Mp 6.67 | Intergenic | AT1G28240 | MUCILAGE-RELATED70 (MUCI70) | 187.8 ± 31.9 | |
| 9868674 | Mp 6.28 | Synonymous coding | ||||
| 9869505 | Intron | |||||
| 9870915 | ||||||
| 11281107 | GalA 5.33 | Intergenic | AT1G31530 | DNase I-like superfamily | 15.3 ± 9.9 | |
| NS 5.13 | ||||||
| 3 | 10050704 | Mp 6.23 | Intergenic | AT3G27220 | Galactose oxidase | 55.6 ± 13.9 |
| Rg 6.16 | ||||||
| 4 | 11011073 | Mp 5.35 | Intron | AT4G20400 | JUMONJI14 | 108.0 ± 17.0 |
| 17699116 | Rg 5.25 | Intergenic | AT4G37670 | N-ACETYL-l-GLUTAMATE SYNTHASE2 | 39.2 ± 35.0 | |
| NAGS2 | ||||||
| 5 | 9699695 | GalA 6.43 | Intergenic | AT5G27470 | Seryl-tRNA synthetase/ligase | 244.9 ± 116.0 |
| NS 5.74 | ||||||
| 16205167 | IV 5.1 | Intergenic | AT5G40460 | Cyclin-dependent kinase inhibitor (SMR6) | 37.8 ± 3.0 |
A Putative GT, MUCILAGE-RELATED70, Modulates the Macromolecular Characteristics of RGI Polymers in Outer Mucilage
The gene regions containing the different SNPs associated with the variation in the natural accessions were examined in more detail (Table 4). A region of chromosome 1 contained four SNPs above the threshold of significance for association with outer mucilage polysaccharide Mp (Fig. 5A), and these were located within the MUCILAGE-RELATED70 (MUCI70) sequence (Fig. 5B). This gene was recently shown to play a critical role in the synthesis of seed mucilage pectin (Voiniciuc et al., 2018). MUCI70 was found to contribute to the formation of RGI domains in mucilage and was proposed to be a member of a novel GT family. In agreement with this study, two independent mutant lines, muci70-1 and muci70-2, produced less total mucilage sugars than the wild type (Supplemental Fig. S5A; Supplemental Table S5). Furthermore, production of RGI was reduced in both mucilage layers (Supplemental Fig. S5B), and although Rha contents were decreased in the outer layer of mucilage, Xyl contents increased (Supplemental Fig. S5C).
Variation in the nucleotide sequence of MUCI70 was examined in more detail for accessions showing significant differences (P < 0.05) from the global mean value for Mp for which sequence data were available (Supplemental Table S6). The frequency of the different variants observed among these accessions was determined, and four were only present in a subset with high Mp: Alc-0, Ak-1, C24, Pn-0, Se-0, Sq-8, Ts-1, and Ts-5 (Fig. 5C). This indicated linkage disequilibrium (LD) for their coinheritance with the high Mp trait, supporting MUCI70 as a strong candidate for one of the genes modulating this complex trait. These polymorphisms include a synonymous coding substitution and three SNPs located in introns (Fig. 5D). Although synonymous coding mutations are not expected to affect protein function, it is now recognized that they can impact RNA processing, protein folding, or translation (Hurst, 2011; Plotkin and Kudla, 2011). Moreover, the MUCI70 protein contains a predicted transmembrane domain near its N terminus as well as a Domain of Unknown Function 616, and this synonymous coding mutation is located within the latter.
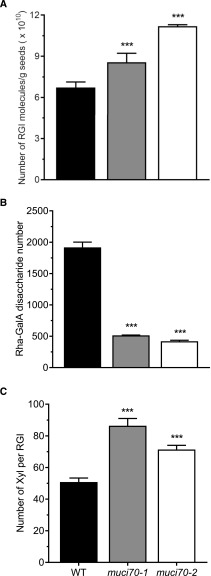
In order to determine whether modified MUCI70 activity could be responsible for the observed modifications in the macromolecular characteristics of outer mucilage polysaccharides, water-extracted mucilage from muci70 seeds was analyzed by high-performance size-exclusion chromatography (HP-SEC) combined with multiple-angle laser light scattering and viscometer (Supplemental Fig. S6; Supplemental Table S7). Values for all three measured macromolecular characteristics, Mp, IV, and Rg, were significantly lower for outer mucilage polysaccharides from muci70 seeds than those of the wild type. As MUCI70 expression is lower in these mutants (Voiniciuc et al., 2018), this demonstrates that altering MUCI70 activity affects polymer macromolecular characteristics, in accordance with the observed polymorphisms being potentially causal for the natural variation in these traits. Nevertheless, the polydispersity index showed that the population of RGI polymers produced had a similar level of size variation in both wild-type and mutant outer mucilage (Supplemental Table S7). As stated above, unbranched RGI is the major component of outer mucilage for the Col-0 reference accession, which allows the average number and length of RGI polymers to be estimated from their Mp. In this way, the number of RGI molecules produced in muci70 mutants was calculated from Mp and sugar content values and found to be significantly higher than that of the wild type (Fig. 6A; Supplemental Table S7), despite total RGI production being reduced (Supplemental Fig. S5; Supplemental Table S5; Voiniciuc et al., 2018). This was because the length of RGI polymers was greatly reduced in mutants (Fig. 6B) and highlighted that a larger number of short RGI polymers were synthesized when MUCI70 expression was reduced. Moreover, the proportion of Xyl molecules per RGI polymer was higher in mutants (Fig. 6C) and confirmed the increase in Xyl linkages previously observed in muci70 mutant lines (Voiniciuc et al., 2018). Taken together, these results indicate that RGI polymers in muci70 mutants are shorter and have more xylan substitutions or longer xylan side chains.
Figure 6.
Seed outer mucilage from muci70 mutants has a large number of short RGI polymers with more Xyl substitutions. A, A larger number of polymer molecules are produced in muci70 mutants compared with the wild type (WT), despite total RGI production being smaller. B, RGI polymer length, expressed as the total number of repeating Rha and GalA disaccharides, is shorter in outer mucilage from muci70 mutant seeds compared with the wild type. C, The proportion of Xyl molecules per RGI polymer is higher in muci70 mutants. Values are means and error bars are se of 2 < n < 3 biological replicates. Asterisks indicate Dunnett pairwise comparison with the wild type: ***, P < 0.001.
The Properties of Polymers in the Inner Mucilage Layer Are Modified by MUCI70
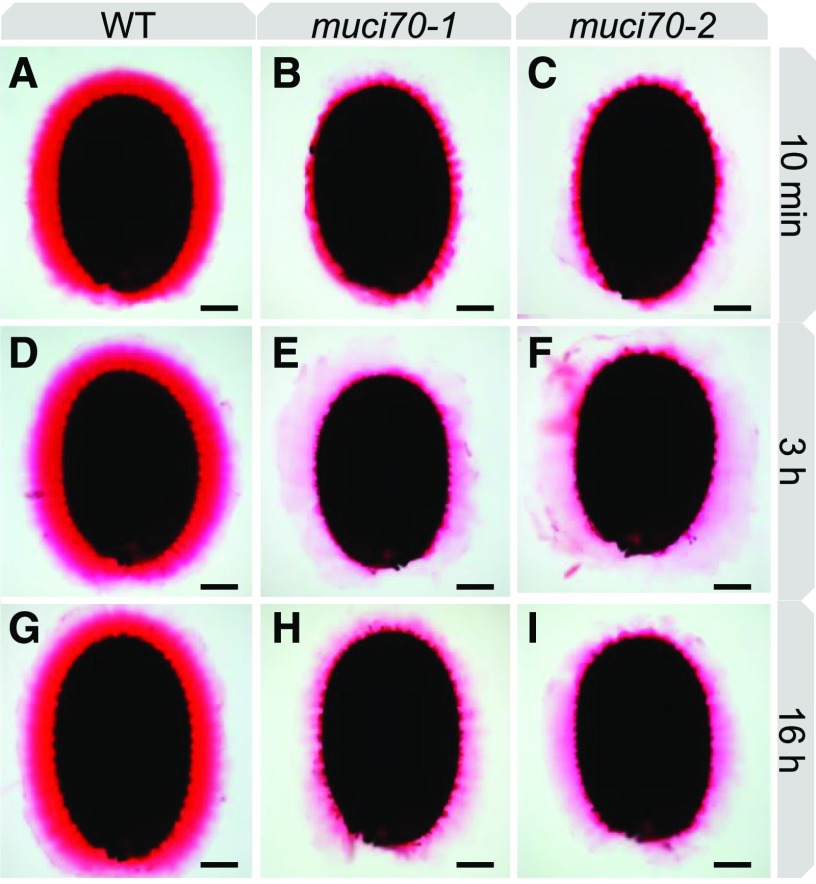
Impaired mucilage production in muci70 mutants was previously associated with seed flotation because mucilage was not released (Voiniciuc et al., 2018). Similar to other mutants with defective mucilage release, this phenotype was rescued by imbibition in the calcium chelator EDTA. In certain mutants, the mucilage release defect can also be complemented in water by extending imbibition time (Saez-Aguayo et al., 2013). Mucilage release from seeds of muci70 mutants was, therefore, observed by ruthenium red staining after imbibition in water for increasing lengths of time. After a short imbibition of 10 min, around 20% of muci70 seeds floated, and inner mucilage was observed as a thin layer as previously described (Fig. 7, A–C; Voiniciuc et al., 2018). This contrasted with the halo observed after 3 h of imbibition, which was similar in width to that of wild-type seeds, but was only stained the same intense red close to the seed and was surrounded by weakly stained mucilage that had an uneven outline (Fig. 7, C–E). Remarkably, following a more prolonged imbibition of 16 h, the weaker stained mucilage was greatly reduced in the majority of mutant seeds, in contrast to the wild type, where the mucilage layer appeared unaltered (Fig. 7, E–G). This suggested that muci70 inner mucilage includes a population of polymers that become detached from the seed over time.
Figure 7.
Comparison of the inner layer of mucilage released from seeds imbibed for different periods of time in water. Ruthenium red staining is shown for mucilage pectin from wild-type Col-0 (WT; A, D, and G), muci70-1 (B, E, and H), and muci70-2 (C, F, and I). Bars = 100 µm.
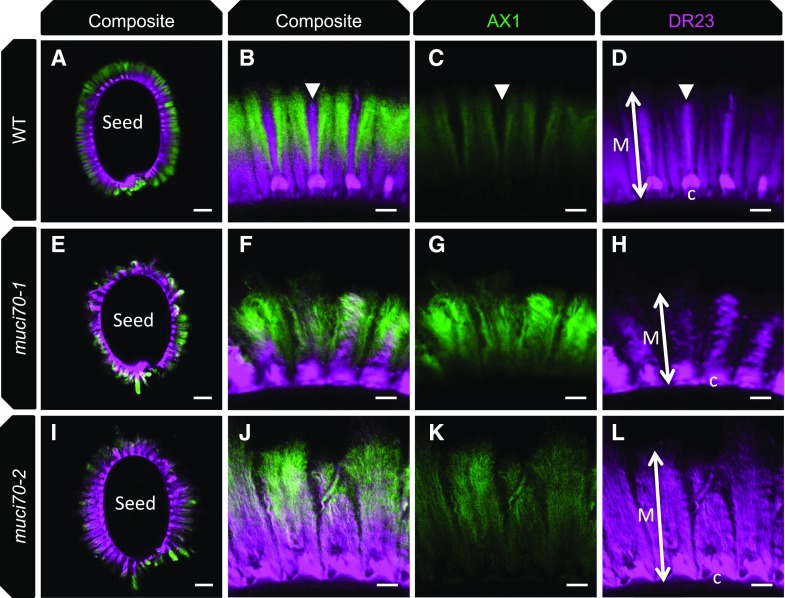
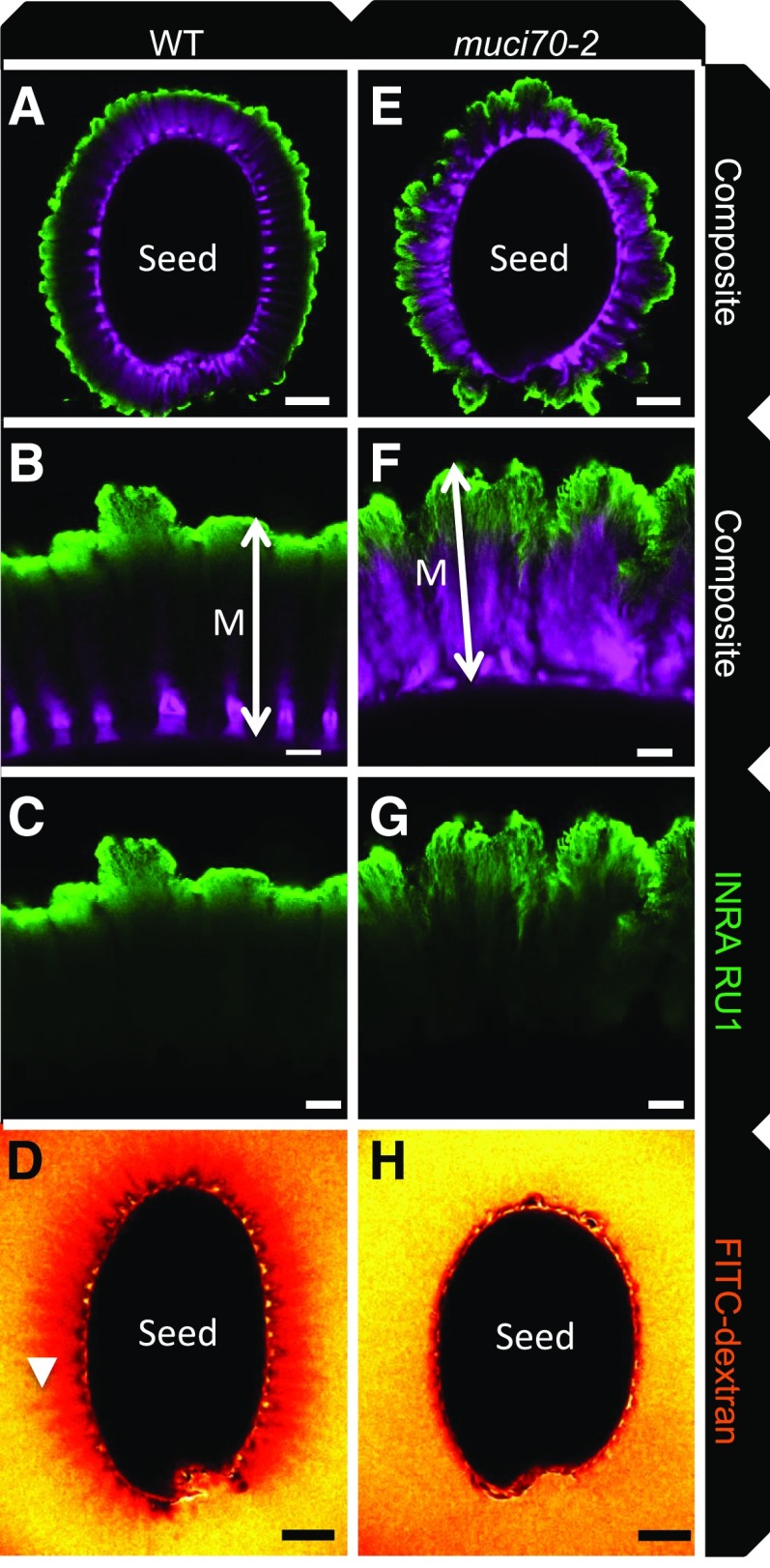
To determine whether the weakly stained region of muci70 inner mucilage was caused by a modification in the distribution of the polymers, notably xylan and RGI, whose amounts differed in biochemical analyses (Supplemental Fig. S5), immunolabeling was carried out using antibodies that bind specifically to these polysaccharide epitopes, AX1 and INRA-RU1, respectively (Guillon et al., 2004; Ralet et al., 2010). In mutant seeds, the intensity of the signal observed with the AX1 antibody was stronger than that observed in the wild type (Fig. 8, C, G, and K), in agreement with the higher Xyl contents of mutant mucilage extracts (Supplemental Fig. S5; Voiniciuc et al., 2018). Nevertheless, in composite images where AX1 labeling was combined with that of cellulose staining, the difference was less apparent, independent of whether the mutant seed had a large or small halo of inner mucilage (Fig. 8, A, B, E, F, I, and J). This was because of the overlap of signal from labeling of the two polysaccharides in the mutant, which contrasted with the wild type, where the xylan labeling was in zones with little cellulose staining and separated from the rays of cellulose radiating out from the top of a columella. The INRA-RU1 antibody is an IgM that binds to RGI and only labels the periphery of wild-type Col-0 mucilage (Fig. 9, A–C; Voiniciuc et al., 2015b). In the muci70 mutant, the labeling appeared to penetrate more deeply into the mucilage layer, indicating that the mucilage polymers were less densely packed. Mucilage porosity was, therefore, examined further using 140-kD dextran labeled with fluorescein isothiocyanate (FITC). The exclusion of these fluorescent molecules from the inner mucilage creates a dark region around the seed in the wild type, in particular from the rays of cellulose (Fig. 9D), whereas in muci70, the inner mucilage appeared to fluoresce homogenously (Fig. 9H). This increased porosity of muci70 inner mucilage and the loss of polymers on prolonged imbibition suggested that, similar to the outer layer, the length of RGI polymers was reduced. Together, these results indicated that the organization of inner mucilage polymers was altered in muci70 mutants.
Figure 8.
Labeling of xylan and cellulose in the inner mucilage released from wild-type (WT) and muci70 seeds. Confocal microscopy optical sections of seeds and mucilage are from mature seeds following 16 h of imbibition showing xylan epitopes labeled with the AX1 antibody (green) and cellulose stained with Direct Red 23 (DR23; magenta). A to D, Wild-type Col-0. E to H, muci70-1. I to L, muci70-2. A, E, and I show whole seeds, with higher magnifications of mucilage from the same seeds in B, C, and D, F, G, and H, or J, K, and L, respectively. A, B, E, F, I, and J show composite images of double labeling with AX1 and DR23. Arrowheads indicate the zone where a cellulose ray projected from the top of a columella (C), and double-headed arrows indicate the width of the inner mucilage layer (M). Bars = 100 µm (A, E, and I) or 20 µm (B, C, D, F, G, H, J, K, and L).
Figure 9.
Polymers in muci70 inner mucilage are less densely packed than those of the wild type (WT). Confocal microscopy optical sections of seeds and mucilage are from mature seeds following 16 h of imbibition showing RGI epitopes labeled with the INRA-RU1 antibody (green) and cellulose stained with Direct Red 23 (magenta; A, B, C, E, F, and G) or FITC-dextran fluorescence (D and H). A to D, Wild-type Col-0. E to H, muci70-2. A, D, E, and H show whole seeds. Higher magnifications of mucilage from the seeds in A and E are shown in B and C or F and G, respectively. A, B, E, and F show composite images of double labeling with INRA-RU1 and DR23. The arrowhead indicates a cellulose ray projecting from the top of a columella, and double headed arrows indicate the width of the inner mucilage layer (M). Bars = 100 µm (A, D, E, and H) or 20 µm (B, C, F, and G).
GWA Implicates a Copper Amine Oxidase Family Member in the Production of Mucilage Pectin
Only two SNPs were associated with the variation of both GalA and NS contents above the threshold of significance (Fig. 10; Table 4). As GWAS is based on the principle that a particular phenotype shared by a subset of individuals will be highly linked to neighboring genetic variations because of LD (Kim et al., 2007), we also examined the annotated functions and transcription profiles of genes located near the SNP peaks. In the same way as a previous GWAS using natural variability in Arabidopsis seed mucilage structure (Voiniciuc et al., 2016), we considered neighboring genes within 1 million bases of the SNP peaks, which represents 100 times the average LD decays found in Arabidopsis populations. A promising candidate, At1g31670, was identified on chromosome 1 near the SNP at position 11281107 (Fig. 10C; Table 4). This candidate gene was strongly expressed in the seed coat, with highest abundance at the maturation green stage of embryogenesis (Supplemental Fig. S7), when mucilage polysaccharide accumulation occurs, while its expression in other plant tissues was extremely low, indicating that it was seed specific. Moreover, the subcellular location database for Arabidopsis proteins, SUBA, predicts that the protein is secreted to the apoplast, in agreement with a role in mucilage metabolism.
Figure 10.
GWAS of Arabidopsis seed outer mucilage links a copper amine oxidase family member to mucilage quantity. A and B, Manhattan plots showing the SNPs associated with outer mucilage polysaccharide traits using a panel of 194 natural accessions and the accelerated mixed-model GWA method: GalA content (A) and NS content (B). The log-transformed P values from the test of association are plotted as a function of chromosomal position. The genome-wide significance threshold (purple dashed lines) was corrected for multiple testing by the Bonferroni method. Black dashed lines represent the –log(P) = 5 threshold for significance used in this study. Red arrows indicate a colocalized SNP peak on chromosome 1 that is presented in the closeup in C. Manhattan plots were obtained from GWA-Portal (https://gwas.gmi.oeaw.ac.at/). C, Closeup view of the significant peak on chromosome 1 for GalA content. CuAOα1 (At1g31670) is located near the highest SNPs (mac > 15), highlighted by green outlines. Triangles indicate nonsynonymous and squares indicate synonymous coding polymorphisms, and circles indicate SNPs located in introns or intergenic regions.
This gene is annotated as a member of the copper amine oxidase family that contains 10 members in Arabidopsis, which are divided into three clades, and as a putative copper amine oxidase of clade 1a, it has been named CuAOα1 (Tavladoraki et al., 2016). This superfamily of enzymes catabolizes plant polyamines that are involved in several physiological processes, such as cell proliferation, differentiation, and defense responses, and they are found at high levels in the cell walls of many species (for review, see Tavladoraki et al., 2016). As no function has yet been assigned to the CuAOα1 family member that is specifically and highly expressed in the seed coat when mucilage production is ongoing, it was considered a promising candidate for further experiments.
CuAOα1 May Contribute to Natural Variability in Mucilage Production
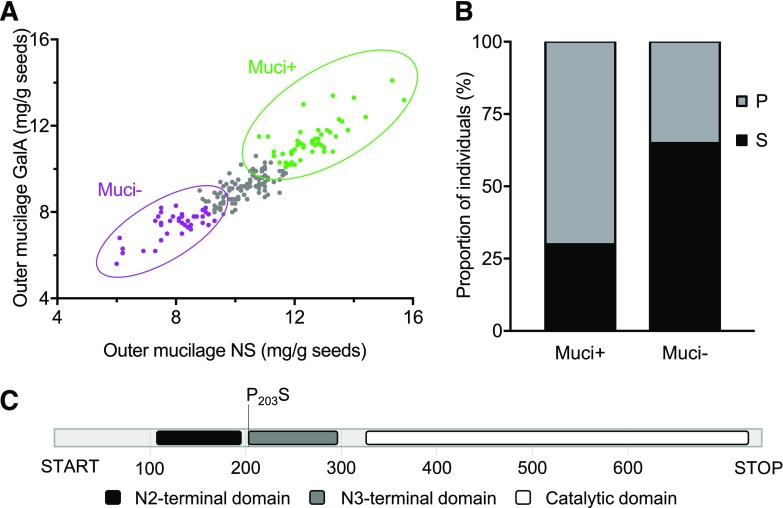
To examine further the potential role of CuAOα1 in mucilage production, the predicted amino acid substitutions occurring in the protein in different accessions were analyzed. An agglomerative hierarchical clustering of the 194 accessions used for GWAS indicated that they could be classed into three groups (Supplemental Table S8). The first comprises 41 accessions with low mucilage contents (muci−), the second 51 accessions with high mucilage contents (muci+), and 102 accessions have intermediate phenotypes (Fig. 11A). Sequence data were available on the 1001 Genomes browser for 68 accessions from muci− and muci+ groups, and a number of amino acid substitutions were observed within their protein sequences. In order to determine whether any were enriched in either group, χ2 tests of independence were carried out (Supplemental Table S9). Only one substitution, Pro-203 to Ser, appeared to be preferentially associated with reduced mucilage pectin production, with a χ2 test P of 0.042 (Fig. 11B; Supplemental Table S9). While 65% of individuals in the muci− group had this substitution, 70% of individuals from the muci+ group have Pro-203. A schematic representation of CuAOα1 protein domains was obtained from the European Bioinformatics Institute (https://www.ebi.ac.uk/). This polymorphism affects the first amino acid of the N3-terminal structural domain (Fig. 11C) and is a substitution between amino acids with different chemical properties; notably, Pro has unique properties and often is associated with significant protein structural features such as folding (Betts and Russell, 2007). This amino acid polymorphism is likely, therefore, to have a strong effect on protein function.
Figure 11.
Amino acid substitutions in the N3-terminal domain of CuAOα1 protein are associated with reductions in the amount of outer mucilage. A, Schematic representation of CuAOα1 protein and locations of functional domains annotated by the European Bioinformatics Institute (https://www.ebi.ac.uk/). Changes in amino acid sequence that are likely to have an effect on mucilage production and their positions are indicated. B, Hierarchical clustering of natural accessions for GalA and NS contents identified three clusters. Individuals producing less mucilage (muci−) are presented in magenta, and those producing a high amount of mucilage (muci+) are presented in green. C, A higher proportion of accessions has the Pro-203-to-Ser substitution in the muci− group. The observed proportion of both alleles between muci+ and muci− groups differed from a random distribution as determined by a χ2 test (P = 0.042).
CuAOα1 Is Required for the Synthesis of Outer Mucilage RGI
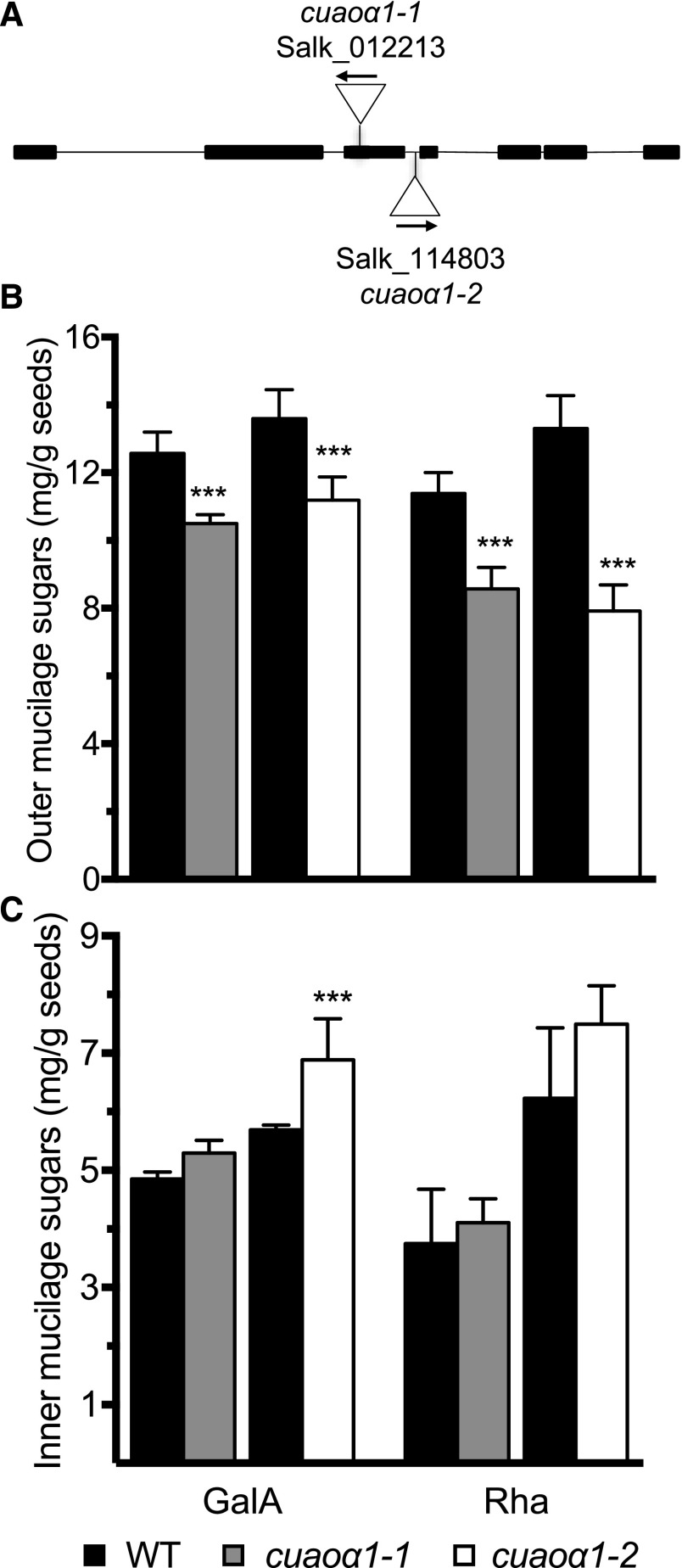
To confirm the role of the CuAOα1 gene in mucilage polysaccharide metabolism, two T-DNA insertion mutant lines were obtained and homozygous individuals selected. The mutant lines cuaoα1-1 and cuaoα1-2 have insertions in the third exon or third intron, respectively (Fig. 12A). Outer and inner mucilage were extracted sequentially from mutant seeds and their wild-type Col-0, followed by GalA and NS quantification (Fig. 12, B and C; Supplemental Table S5). A significant reduction of both GalA and Rha contents was observed in outer mucilage from seeds of both mutant lines compared with the wild type, confirming the involvement of CuAOα1 in RGI production. Rha contents were reduced by 24.5% and 40.7% for cuaoα1-1 and cuaoα1-2, respectively, while reductions in GalA contents were more modest, 16.4% and 17.7% for cuaoα1-1 and cuaoα1-2, respectively (Fig. 12B). The greater impact on Rha contents suggests that mutations in the CuAOα1 gene may specifically impact RGI production without affecting other pectin domains such as HG. In contrast to muci70 mutants, RGI polymer length was not impacted in cuaoα1 mutants, as no recurrent differences in macromolecular characteristics were observed (Supplemental Figs. S6 and S8A). The number of RGI polymers was, however, reduced in both cuaoα1-1 and cuaoα1-2 and confirmed the implication of this candidate gene in RGI production (Supplemental Fig. S8B).
Figure 12.
Mutants in the CuAOα1 gene have reduced outer mucilage RGI contents compared with the wild type (WT). A, Schematic representation of the exon and intron structure of CuAOα1 as annotated by The Arabidopsis Information Resource (http://www.arabidopsis.org/). The positions and orientations of cuaoα1 T-DNA insertions are indicated. B, GalA contents of water-extracted outer mucilage. C, GalA contents of RGase hydrolysates of inner mucilage. Values are means and error bars are se of 2 < n < 3 biological replicates. Asterisks indicate Dunnett pairwise comparison with the wild type: ***, P < 0.001.
The effect of this enzyme on mucilage production appeared to be specific to the outer mucilage layer, as no consistent differences in inner mucilage sugar contents were observed with both mutant alleles compared with the wild type (Fig. 12C). Furthermore, the inner mucilage of cuaoα1 mutants did not exhibit any obvious visual phenotype when stained with ruthenium red (Supplemental Fig. S9). This indicated that the effect on the outer mucilage layer was not because of a redistribution of RGI to the inner layer.
DISCUSSION
RGI Is Central to the Ecophysiological Function of Arabidopsis Outer Mucilage
Natural variation occurs spontaneously and is maintained if polymorphisms acquired provide advantageous or neutral effects on survival in a given environment. In Arabidopsis, a large number of natural accessions have been identified, and for many of these their site of location is known with a certain level of precision. Associating particularities of these environments with genetic polymorphisms and adaptive phenotypes can be carried out based on LD, and this is greatly facilitated in Arabidopsis through the availability of genomic data for 1,001 accessions (Cao et al., 2011). Furthermore, these genomic data allow potential causal mutations to be identified and new functions to be assigned to genes.
In the study presented here, natural variation in polysaccharide traits has been described for polymers extracted from the outer layer of seed mucilage. For the majority of accessions, these polymers are mainly composed of the pectin domain RGI as the quantity of GalA and NS was proportional, in agreement with this polymer being a repeating disaccharide of [→2)-α-l-Rha-(1→4)-α-d-GalA-(1→]n (Fig. 2A). The polysaccharide composition of water-extractable mucilage varies in other species, such as flaxseed (Linum usitatissimum) and psyllium (Plantago spp.), which contain large proportions of heteroxylan (for review, see Phan and Burton, 2018). This suggests that attributes of the RGI polymer are important for the biological function of outer mucilage in Arabidopsis. Nevertheless, all six characteristics examined for outer mucilage polymers showed extensive variation (Fig. 1; Table 1), so while the composition varied little, the amount produced could vary nearly 3-fold between accessions and average polymer length by around 2-fold. This is similar to the approximately 4-fold variation in contents observed previously for galactoglucomannan, a low-abundance polysaccharide in the inner layer of Arabidopsis mucilage (Voiniciuc et al., 2016). The lowest amounts of outer mucilage observed in accessions were, however, still higher than reductions detected for certain induced mutants in transcription factors regulating the differentiation of seed coat epidermal cells, such as ttg1 (greater than 17-fold) or gl2-9 (greater than 5-fold; Arsovski et al., 2009b; Ralet et al., 2016), indicating that the complete elimination of mucilage production would be disadvantageous. This is in agreement with the previous observation that all Arabidopsis accessions with floating seed phenotypes still produced mucilage (Saez-Aguayo et al., 2014). Nevertheless, it is possible that mutation of mucilage genes could also impact polysaccharide production in other plant tissues and negatively affect fitness, so that maintenance of mucilage production is not the causative factor.
Variations were observed in the macromolecular characteristics of the polymers (Table 1), which could suggest that conservation of polymer properties per se is not an absolute requirement for outer mucilage function. As outer mucilage is mainly unbranched RGI, the observed variations in macromolecular characteristics are principally related to changes in polymer length, and the strong proportionality observed between Mp and the three other characteristics determined is in agreement with this (Fig. 2B; Supplemental Fig. S2). Together, the analyses of outer mucilage variation indicate that its function requires a conserved composition, while polymer length and amount may be adapted to particular environments.
Arabidopsis Accessions with Distinctive Outer Mucilage Traits May Have Common Geographic Origins
Following its release, it is likely that outer mucilage would be gradually leached into the surrounding media, as it is not attached to the seed. In this context, among diverse functions proposed for mucilage in other species, two might be coherent for Arabidopsis outer mucilage. It may enhance plant growth through improved soil rheology, similar to chia (Salvia hispanica) or shepherd’s purse (Capsella bursa-pastoris) mucilage, whose adhesive properties can aggregate soil particles and thereby change soil water retention and airflow (Deng et al., 2015; Di Marsico et al., 2018). Alternatively, it could benefit seedling establishment, similar to Artemisia sphaerocephala mucilage, which can act as a carbon source, promoting seedling and soil microbiome growth (Yang et al., 2012a). The properties of RGI polymers might provide a particular advantage to Arabidopsis seeds for one or both of these functions, but while the adaptation of outer mucilage quantity could control the extent of microbiome growth, differences in polymer length are unlikely to have an impact. The modulation of polymer size might, however, have an impact on soil rheology, and this could be tested using outer mucilage extracts from outlier accessions.
A set of five accessions produced outer mucilage that did not respect the linear relationship between outer mucilage macromolecular characteristics, because of a shift to lower IV (Fig. 2B; Supplemental Fig. S2C). This means that the outer mucilage polymers in these accessions are folded differently and occupy a smaller volume. As four of these accessions were from Japan, this might represent a local adaptation, which could be studied further to gain clues concerning the role of RGI polymer conformation with function. Studying the effect of mucilage from these accessions on soil rheology and microbial growth might also provide pointers. Interestingly, the sites of collection for a group of HOM accessions were geolocalized to a mountainous region of central Asia (Table 2; Supplemental Fig. S3), contrasting with the absence of LOM accessions in this area. Although this region includes the collection sites of FMR accessions, with one FMR figuring among the LOM accessions, the HOM phenotype would not appear to be linked to seed flotation, as four LOM accessions had previously been found to have seeds that sink (Saez-Aguayo et al., 2014).
Overall, accessions could be classed into five groups, based on their outer mucilage characteristics (Supplemental Fig. S1; Supplemental Table S2). Accessions with atypical inner mucilage traits have previously also been classed into five groups (Voiniciuc et al., 2016), but there was no link between accessions attributed to these with any of the outer mucilage groups. This suggests that the inner mucilage phenotypes examined are controlled by independent genetic factors and reinforces the proposal that the two mucilage layers have different functions in natural habitats.
MUCI70 Influences RGI Polymer Length and Xylan Substitution
A large number of genes have been catalogued that contribute to mucilage production in Arabidopsis (Francoz et al., 2015), but only seven could potentially play a direct role involved in RGI polymer synthesis, despite it being the main component of mucilage. Notably, an RGI rhamnosyltransferase (RRT1) was recently identified whose mutation reduced mucilage RGI production by 19% (Takenaka et al., 2018). As outer mucilage is almost exclusively RGI, exploiting natural variation for the identification of genes is more likely to uncover direct contributors in its production. Accessions with atypical phenotypes for mucilage release have previously proved to be useful tools for the identification of genes involved in mucilage production (Macquet et al., 2007b; Saez-Aguayo et al., 2013, 2014), and the outliers uncovered here will be important tools for the discovery of further loci. These, however, involve mapping approaches requiring the generation of progeny from crosses between accessions. An alternative, more rapid strategy is to use GWA to provide a list of candidates, and this approach was successfully used here to demonstrate that two genes, MUCI70 and CuAOα1, are involved in the modulation of outer mucilage macromolecular characteristics and/or synthesis, respectively. This indicated that our GWAS represented biologically relevant associations rather than noise, even when applying a lower significance threshold than that commonly calculated with the Bonferroni method.
MUCI70 was recently identified as a putative GT from a previously unclassified CAZy family, and its mutation, together with that of GAUT11, reduced the amount of mucilage RGI produced, with muci70 gaut11 mutants being more strongly impaired than each single mutant (Voiniciuc et al., 2018). Here, GWAS identified MUCI70 as a locus that could modulate the natural variation in RGI macromolecular characteristics (Fig. 5). We found that in addition to defects in RGI synthesis, muci70 mutants produce a larger number of short polymers and that the proportion of Xyl to Rha was increased compared with the wild type (Fig. 6; Supplemental Table S7). Additionally, the inner mucilage of muci70 mutants was more strongly labeled by an antibody that binds xylan (Fig. 8). Mucilage RGI polymers are substituted with xylan (Ralet et al., 2016), and the increase in Xyl per polymer could be caused by either more xylan substitutions or longer xylan side chains. Xylan branches are formed from a 4-linkage of xylosyl onto rhamnosyl in the RGI polymer, followed by extension with 1,4-linked xylosyls, and finishing with a terminal xylosyl. An increase in xylan polymer length would, therefore, lead to an increased proportion of 1,4-Xyl linkages. Based on previous linkage analyses for muci70-1 (Voiniciuc et al., 2018), the ratios of 2,4-Xyl:1,4-Xyl:t-Xyl are similar to those observed in the wild type: 1.1:3.2:1 versus 1.5:3.7:1, respectively. This indicates that the increased Xyl contents in muci70 mucilage is caused by the RGI polymer being more substituted with xylan.
Recently two other enzymes, MUM5/MUCI21 and IRX14, were proposed as the GTs involved in adding or elongating RGI xylan side chains, respectively (Voiniciuc et al., 2015a; Ralet et al., 2016). As irx14 muci70 seeds were more severely impaired in mucilage RGI production than the muci70 single mutant, Voiniciuc et al. (2018) proposed that xylan is essential for the elongation of RGI, which would also explain the increased Xyl contents in muci70. Nevertheless, total mucilage Rha and GalA contents are similar to the wild type in irx14 or mum5 mutants (Voiniciuc et al., 2015a; Ralet et al., 2016), showing that RGI synthesis is not affected when xylan production is limited. Furthermore, we show here that many short, 135- to 165-kD, RGI polymers are synthesized in muci70 (Supplemental Table S7), which is a more drastic effect than previously measured for mum5-1, where only around half the outer mucilage polymers exhibited a modest size reduction to 500 kD (Macquet et al., 2007a). This demonstrates that there is not a linear relationship between polymer length and Xyl contents.
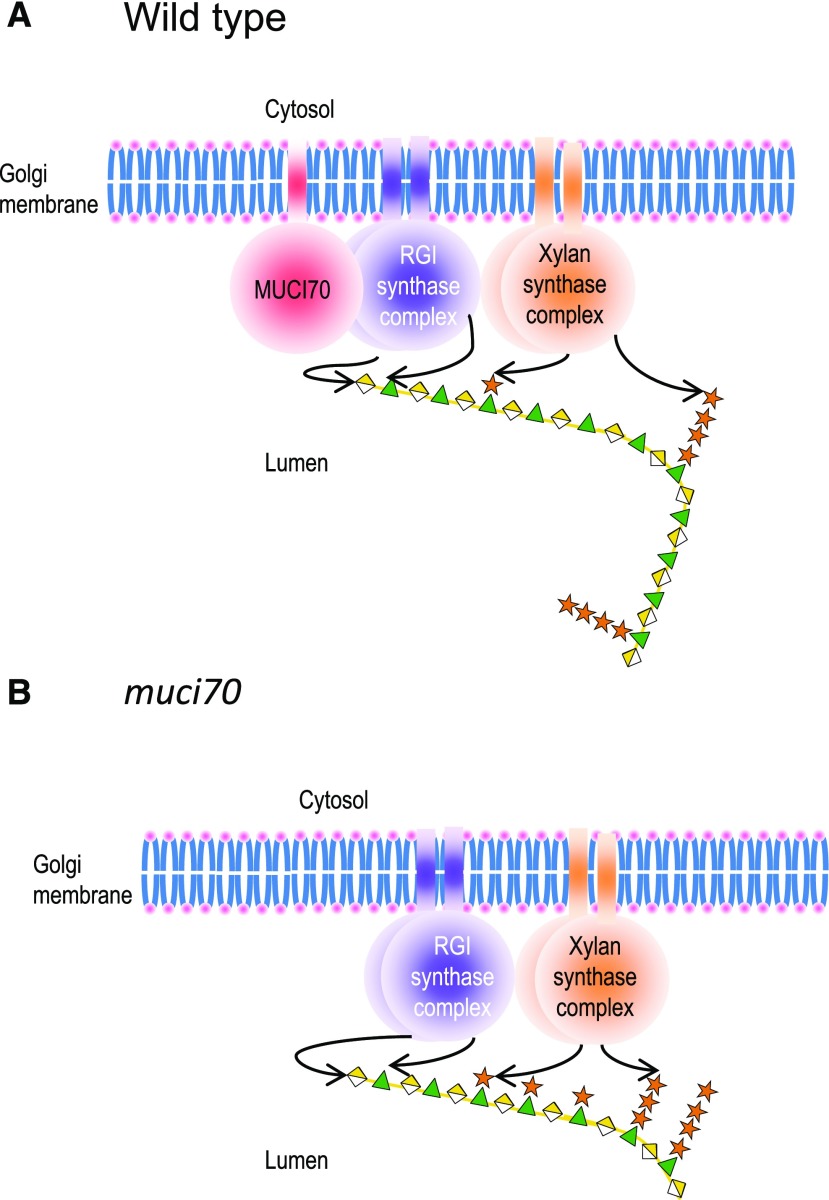
Pectin is synthesized in the Golgi lumen by GTs that transfer glycosyl residues from nucleotide sugars to specific polysaccharide acceptors, and accordingly, MUCI70 is localized to the Golgi (Voiniciuc et al., 2018). It has been proposed that pectin synthesis involves multi-GT enzyme complexes that produce different pectin domains and side chains, as previously observed for HG synthesis with GAUT1/GAUT7 (Atmodjo et al., 2011, 2013). Furthermore, a putative glucuronoarabinoxylan synthase complex from wheat (Triticum aestivum) was found to have xylosyltransferase (XylT), arabinosyltransferase, and glucuronyltransferase activities (Zeng et al., 2010). In this case, mucilage RGI production would, therefore, involve a complex with at least three GTs: a RhaT, a GalAT, and XylT. We propose a model for mucilage RGI synthesis (Fig. 13) involving a multi-GT complex with an RGI synthase complex of RhaT and GalAT associated with a XylT complex that substitutes a Rha unit with a Xyl residue followed by elongation of the xylan chain. As RGI production is not completely impaired in muci70 (Supplemental Fig. S5), this suggests that the presence of MUCI70 promotes RGI backbone synthesis from a basal level. No enzyme activity has yet been demonstrated for MUCI70, and while it may directly contribute as a GT to RGI synthesis, it may only play a structural role, in a similar manner to that previously described for GAUT7 in the HG:GalAT complex (Atmodjo et al., 2011). As the activity of the XylT complex is independent of the rate of RGI backbone production, a slower elongation rate would allow more rhamnosyls to receive xylosyl branches, explaining the higher number of substitutions observed in muci70. To confirm this hypothesis, further genetic and biochemical studies will be required using proteins already identified as potential components of this complex: GALACTURONOSYL TRANSFERASE-LIKE5 (GATL5), GAUT11, IRX14, MUCI70, MUM5, and RRT1. As more and shorter polymers were produced in muci70 mutants, this indicates that synthesis is terminated earlier, perhaps because of crowding by the high level of substitutions. Interestingly, outer mucilage extracts of mum4/rhm2 mutants, defective for the production of the UDP-Rha substrate for RGI synthesis, also contain a greater number of small polymers (Usadel et al., 2004). Conversely, mutation of RRT1 and GATL5 increased mucilage polymer size, despite overall production of mucilage RGI being reduced (Kong et al., 2013; Takenaka et al., 2018). This suggests that RGI polymer termination is not directly related to the elongation rate of the RGI backbone in seed coat epidermal cells.
Figure 13.
Proposed model for the synthesis of seed mucilage RGI by a multi-glycosyltransferase complex located on the Golgi membrane. A, In the wild type, elongation of the RGI backbone by the RGI synthase complex comprises proteins with rhamnosyltransferase and galacturonosyltransferase activities. The presence of MUCI70 in this complex ensures efficient polymerization. B, In the absence of MUCI70, the synthesis of the RGI backbone is slower, which allows more xylan side chains to be added and elongated, resulting in the high number of substitutions observed in muci70. Arrows indicate the transfer of glycosyl residues onto the polymer. Note that the frequency and pattern of xylan branching on RGI remain to be determined and so are not representative, only serving to illustrate the model.
Interestingly, while MUCI70 was potentially associated with natural variation in macromolecular characteristics, it was not linked to differences in mucilage amount, or other macromolecular characteristics, despite insertion mutants having dramatic decreases in RGI production for both mucilage layers and modified IV and Rg (Supplemental Fig. S5; Supplemental Table S7). This suggests that the polymorphisms identified may specifically alter Mp and is in agreement with a cell’s capacity for RGI synthesis being independent of the mechanism controlling RGI polymer size. The absence of associations between MUCI70 polymorphisms and NS contents because of modified Xyl amounts would not be observed, as the latter represent only a small fraction of NS, making changes undetectable. The four SNPs associated with natural accessions having relatively high Mp are predicted to cause a synonymous codon substitution in MUCI70 or are located in introns. Further work is required to determine whether these are indeed causal mutations and if they all contribute to modifying the length of RGI polymers. Such changes might affect the level of protein expression, for example, through codon bias or via regulatory sequences within introns, but understanding how this would impact polymer mass requires further information concerning the RGI synthesis module.
Immunolabeling and FITC-dextran staining indicated that the polymers forming the inner mucilage layer are less densely packed in muci70 than in the wild type (Fig. 9). Furthermore, mucilage polymers appeared to be lost from the inner layer of mutant mucilage on prolonged imbibition in water (Fig. 7). In the wild type, extraction of the inner layer of mucilage requires aggressive procedures involving vigorous shaking, sonication, and strong alkali or enzymatic digestion (Macquet et al., 2007b; North et al., 2014; Voiniciuc et al., 2015b; Zhao et al., 2017), demonstrating that polymers are anchored to the seed coat through strong molecular interactions. Mutant studies and cellulose staining phenotypes have shown that the formation of the inner mucilage layer involves at least two distinct pathways (Griffiths et al., 2016): first, through the adsorption of RGI polymers to cellulose through xylan, and a second, less well understood mechanism involving the arabinogalactan protein SOS5 and the receptor-like kinase FEI2 (Harpaz-Saad et al., 2011; Griffiths et al., 2014, 2016; Ralet et al., 2016). Moreover, macromolecular entanglement of polymers is expected to contribute to polymer attachment (Macquet et al., 2007a), and this will be influenced by polymer size. The observed reduction of the inner mucilage layer over time is, therefore, coherent with shorter muci70 RGI being less enmeshed with other polymers and shedding into the surrounding water. The difference in amount and length of RGI polymers also affected the structure of inner mucilage, as observed by immunolabeling and staining (Figs. 8 and 9). Notably, the central ray of cellulose projecting from the top of a columella was not observed, which is a distinctive phenotype of fei2 and sos5 mutants and suggests that MUCI70 contributes to cellulose ray formation. Nevertheless, the proportion of polymers in outer and inner mucilage layers was similar to the wild type (Supplemental Fig. S5B), unlike fei2 and sos5, where partitioning to outer mucilage is increased (Harpaz-Saad et al., 2011; Griffiths et al., 2016).
Copper Amine Oxidase, a New Player in RGI Production
Our GWAS linked an amino acid substitution in CuAOα1 to natural variation in outer mucilage amount (Figs. 10 and 11). This polymorphism of either Pro or Ser at amino acid 203 is predicted to have a major effect on protein conformation at the beginning of the N3 structural domain (Fig. 11). As insertion mutants in CuAOα1 produced lower amounts of RGI and the muci− group of accessions were preferentially associated with Ser-203, this indicates that this protein form may be less active (Figs. 11C and 12B). It will be important to study the effect of this amino acid substitution on mucilage production to confirm that this polymorphism underlies, at least partly, the natural variation observed between muci+ and muci− accessions.
Among eight Arabidopsis genes considered to encode for putative functional CuAOs, catalytic activity has only been demonstrated for five (Møller and McPherson, 1998; Planas-Portell et al., 2013; Groß et al., 2017). These homodimeric enzymes control polyamine metabolism by catabolizing the oxidation of primary amino groups of polyamines with the simultaneous reduction of oxygen to hydrogen peroxide. CuAOs can also generate another reactive oxygen species (ROS), nitric oxide (Groß et al., 2017). Different family members have different cellular and tissue localizations and have been proposed to have distinct biological roles (Planas-Portell et al., 2013). Apoplastic CuAOs have previously been shown to contribute to cell wall loosening (Tavladoraki et al., 2016), and CuAOα1 is predicted to be localized to the apoplast. Similarly, cell wall peroxidases are involved in apoplastic ROS production, and PEROXIDASE36 (PER36) modulates mucilage release from seed coat epidermal cells (Kunieda et al., 2013). PER36 was required for the fragmentation of the outer primary cell wall and did not have a direct effect on mucilage constituents. As no delay or absence of mucilage release was observed in cuao mutants (Supplemental Fig. S9), CuAOα1 and PER36 would, therefore, have different targets. ROS can induce the oxidation of polysaccharides and proteins, so it is possible that CuAOα1 could act either directly to modify polysaccharides or indirectly on a protein that influences polysaccharide production. Identification of the CuAOα1 target is likely to be challenging, as relatively small polysaccharide modifications can have strong effects, as previously observed for RGI branching (Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009a). Interestingly, CuAOδ (At4g12290) and CuAOε2 (At4g12280) also show strong seed coat expression, and the possibility that the reduction in RGI production in cuao is tempered by functional redundancy could be tested using double or triple mutants. The phenotype of cuao is, however, remarkable as the first example of a modification to outer mucilage production without a concomitant change in inner mucilage amounts (Fig. 12).
In conclusion, our study of natural variation in Arabidopsis outer mucilage has emphasized the importance of RGI as its major component. The characteristics of this polymer varied for the amount and length of polymer produced, and potential links between specific mucilage properties and natural habitat were highlighted for two groups of accessions. A GWAS found LD at eight genome locations, indicating that the inheritance of outer mucilage traits involves a complex genetic regulation. Two associated loci, MUCI70 and CuAOα1, may contribute to the natural variation observed in the amount and/or length of RGI polymer produced, and investigation of the remaining six should lead to the identification of further genes affecting outer mucilage production. Among these, a putative Gal oxidase associated with RGI polymer properties is particularly noteworthy in the light of the recent demonstration of a role for another member of this enzyme family in modulating RGI properties (Šola et al., 2019). Together, our results demonstrate the value of natural variation as a resource for decrypting the underlying genetic basis for traits and their biological context.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The cuaoα1-1 (Salk_012213), cuaoα1-2 (Salk_114803), muci70-1 (Salk_129524), and muci70-2 (Salk_145602) T-DNA insertion mutants in the Arabidopsis (Arabidopsis thaliana) Col-0 background were identified in the SIGnAL database (Alonso et al., 2003; http://signal.salk.edu) and obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info). Homozygous lines were identified by PCR using the primers listed in Supplemental Table S10. Seed production was performed in a glasshouse (18°C–28°C) with a minimum photoperiod of 13 h provided by supplementary lighting from plants grown in nonenriched compost (Stender Substrates). In both growth chambers and the glasshouse, plants were watered with Plan-Prod nutritive solution (Fertil).
Mucilage Extraction and Analysis
A sequential extraction of outer followed by inner mucilage was performed and analyzed for monosaccharide composition, essentially as previously described (Sullivan et al., 2011). First, 150 mg of intact seeds was mixed head-over-tail in 3 mL of distilled water for 3 h at room temperature (24°C). The suspension was centrifuged (8,000g, 3 min), and supernatants filtered through a disposable glass microfiber filter (13 mm diameter, 2.7 μm pore size; Whatman) were analyzed as outer mucilage extracts. Seeds were then rinsed twice in 5 mL of water and once in 5 mL of 50 mm sodium acetate buffer, pH 4.5, for 5 min each, before adding rhamnogalacturonan hydrolase (0.1 nkat; Swiss-Prot Q00018) provided by Novozymes, in 3 mL of 50 mm sodium acetate buffer, pH 4.5, and incubating for 1.5 h at 40°C. Samples were centrifuged (8,000g, 3 min), and the supernatants were collected and filtered as above and analyzed as inner mucilage extracts.
The uronic acid and NS contents were determined by the automated m-hydroxybiphenyl and orcinol methods, respectively (Thibault, 1979; Tollier and Robin, 1979). Mucilage extracts were hydrolyzed with 2 m trifluoroacetic acid at 121°C for 2.5 h, then individual NS were derivatized to alditol acetates (Blakeney et al., 1983) and analyzed by gas-liquid chromatography (Perkin-Elmer gas chromatograph).
For HP-SEC, the outer mucilage extracts were boiled for 5 min and filtered through a polyvinylidene difluoride filter (13 mm diameter, 0.45 μm pore size; Whatman). HP-SEC was performed at room temperature on a system comprising a Shodex OH SB-G precolumn followed by a Shodex OH-Pack SB-805 HQ column. Polymers were eluted out with 50 mm sodium nitrate buffer at a constant flow rate of 60 mL h−1. Measurements were performed using a differential refractometer (VE 3580 RI Detector) and a Viscotek 270 Dual Detector (dual-laser light scattering, λ = 670 nm, 90° and 7°, combined with a differential pressure viscometer; Malvern Instruments). All detectors were calibrated with a pullulan standard having narrow molecular mass distribution (weight-average molar mass = 145,618 D, number-average molar mass = 139,180 D, IV = 54 mL g−1 at 30°C in 0.1 m sodium nitrate, refractive index increment [dn/dc] = 0.147 mL g−1; Malvern Instruments). Samples were injected automatically through a 50-μL loop. Data analyses were carried out using OmniSec version 4.5 software (Malvern Instruments). Calculation of the number of RGI macromolecules was carried out using the following equation:
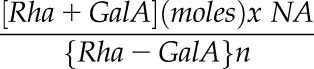 |
where 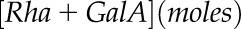 corresponds to outer mucilage Rha + GalA contents expressed in mol per g of seeds,
corresponds to outer mucilage Rha + GalA contents expressed in mol per g of seeds, 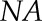 is the Avogadro constant (6.022 × 1023 mol−1), and
is the Avogadro constant (6.022 × 1023 mol−1), and 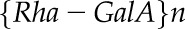 refers to the number of {Rha-GalA} dimers calculated as Mp/322 (molar mass Rha = 164 g mol−1; molar mass GalA = 194 g mol−1 − 36 [i.e. 2H2O for glycosidic linkages]).
refers to the number of {Rha-GalA} dimers calculated as Mp/322 (molar mass Rha = 164 g mol−1; molar mass GalA = 194 g mol−1 − 36 [i.e. 2H2O for glycosidic linkages]).
Statistical Analyses
Data analysis and graphic production were performed using R version 3.5.1 (R Core Team; https://www.R-project.org/). Contrasting global means was performed with the least-squares means (lsmeans) package, version 2.30.0 (Lenth). Normality of trait distributions among accessions was tested using the Shapiro-Wilk method in the stats package, version 3.5.1. PCAs were fit with FactoMineR, version 1.41, and plotted with the factoextra package, version 1.0.5. The violin plot with overlaid boxplot was generated using the ggplot2 R packages, version 3.0.0. For correlations analysis, the basic cor function from the stats package was used, and the associated P values were calculated with the rcorr function from Hmisc, version 4.1.1.
GWAS
Sequence data were available from the 1001 Genomes project (Cao et al., 2011; Horton et al., 2012) for 199 of the Arabidopsis accessions analyzed for outer mucilage traits by Poulain et al. (2019). Data for mucilage macromolecular properties (IV/Mp/Rg) or quantity (GalA/NS) were analyzed separately. The coefficient of variation was calculated as follows:
where s is the sd of the sample and  is the mean of the sample. Accessions showing a coefficient of variation greater than 25% for a given trait were removed from the analysis of that trait, resulting in a set of 193 accessions for IV/Mp/Rg and 194 for GalA/NS. The lsmeans package on R software was used to calculate adjusted means for culture effect from linear models for each trait. GWA was then performed using the GWA-portal (Seren, 2018; https://gwas.gmi.oeaw.ac.at/) and the imputed full sequence data set, applying a Box-Cox transformation for all traits and using the nonparametric KW and AMM GWA methods.
is the mean of the sample. Accessions showing a coefficient of variation greater than 25% for a given trait were removed from the analysis of that trait, resulting in a set of 193 accessions for IV/Mp/Rg and 194 for GalA/NS. The lsmeans package on R software was used to calculate adjusted means for culture effect from linear models for each trait. GWA was then performed using the GWA-portal (Seren, 2018; https://gwas.gmi.oeaw.ac.at/) and the imputed full sequence data set, applying a Box-Cox transformation for all traits and using the nonparametric KW and AMM GWA methods.
Clustering of Accessions and Analysis of Genomic Sequences
An agglomerative hierarchical clustering of the 194 accessions was performed on GalA and NS contents applying Ward’s method for agglomeration using the Euclidean distances on XLSTAT software (Addinsoft 2017, XLSTAT 2017: Data Analysis and Statistical Software for Microsoft Excel). Available CuAOα1 amino acid sequences for accessions used for GWAS were exported from the Arabidopsis 1001 Genome Browser (http://signal.salk.edu/atg1001/3.0/gebrowser.php). Amino acid substitutions were only used for subsequent analysis when the sequence quality indicated that they were reliable. The association of these substitutions with their corresponding phenotypes was tested using a χ2 test of independence at a threshold of 5%.
Cytochemical Staining and Immunolabeling of Adherent Mucilage
Seeds were imbibed in water for 10 min, 3 h, or 16 h prior to staining with 200 µg mL−1 ruthenium red and observation with a light microscope (DRMB; Leica) or staining with Calcofluor at 25 mg mL−1. Following imbibition in water for 16 h, inner mucilage was immunolabeled as described previously with the xylan-binding antibody AX1 or the RGI-binding antibody INRA-RU1 and counterstaining cellulose with 100 µg mL−1 Direct Red 23 (Voiniciuc et al., 2015b; Ralet et al., 2016). Optical sections were obtained using a Leica TCS-SP5-AOBS or Zeiss LSM70 confocal microscope using a 405-, 561-, or 488-nm laser to excite Calcofluor, Direct Red 23, or Alexa Fluor 488 labeling on secondary antibodies, respectively, and detecting fluorescence emission between 412 and 490 nm, 580 and 660 nm, or 500 and 550 nm. For comparison of signal intensity between genotypes in a given experiment, laser gain values were fixed. Similarly, seeds were stained with 150-kD FITC-dextran as described previously (Voiniciuc et al., 2015b) following 16 h of imbibition.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org) database using the loci identifiers At1g28240 (MUCI70) and At1g31670 (CuAOα1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Hierarchical clustering of Arabidopsis accessions based on outer mucilage traits classes them into five groups.
Supplemental Figure S2. Relationships between different macromolecular characteristics of seed outer mucilage polysaccharides for 301 Arabidopsis accessions.
Supplemental Figure S3. Locations of eight HOM amount accessions in central Asia.
Supplemental Figure S4. Manhattan plots showing the SNPs associated with variations in outer mucilage macromolecular characteristics.
Supplemental Figure S5. Two mutant alleles of MUCI70 produce seed mucilage with less pectin, which has a higher level of substitution with Xyl.
Supplemental Figure S6. The outer mucilage from MUCI70 mutant seeds exhibits different macromolecular properties than the wild type on size-exclusion chromatography combined with multiple-angle laser light scattering.
Supplemental Figure S7. CuAOα-1 and MUCI70 are highly expressed in the seed coat during seed development.
Supplemental Figure S8. Seed outer mucilage from CuAOα1 mutants has reduced RGI production.
Supplemental Figure S9. Ruthenium red staining of the inner mucilage layer released from seeds of the cuaoα1-1 mutant compared with the wild type.
Supplemental Table S1. List of outlier accessions for each outer mucilage trait examined determined by significant difference from global mean absolute value.
Supplemental Table S2. Classification of Arabidopsis accessions into five groups based on hierarchical clustering using outer mucilage traits.
Supplemental Table S3. List of accessions and corresponding mucilage quantity values used for GWAS analysis.
Supplemental Table S4. List of accessions and corresponding values for outer mucilage macromolecular characteristics used for GWAS analysis.
Supplemental Table S5. Monosaccharide composition of inner and outer mucilage from seeds of muci70 or cuaoα1 mutants compared with the wild type.
Supplemental Table S6. Details of the nucleotide sequence variation observed between outlier accessions in the transcribed region of the MUCI70 gene.
Supplemental Table S7. Macromolecular characteristics of polymers in outer mucilage extracts from seeds of muci70 or cuaoα1 mutants compared with the wild type determined by HP-SEC combined with multiple-angle laser light scattering.
Supplemental Table S8. Arabidopsis natural accessions can be classed into three groups based on hierarchical clustering analysis of their outer mucilage GalA and NS contents.
Supplemental Table S9. Amino acid substitutions found in Arabidopsis accessions for the CuAOα1protein.
Supplemental Table S10. List of primers used for genotyping.
ACKNOWLEDGMENTS
We thank Susana Saez-Aguayo (University Andés-Bello, Santiago, Chile) for help in developing the calculation for RGI polymer length and Marie-Jeanne Crépeau (Biopolymers, Interactions, Assemblies, Nantes, France) for excellent technical assistance for biochemical analyses. We also thank Dr. Olivier Loudet (Institut Jean-Pierre Bourgin, Versailles, France) for helpful advice for the selection of Arabidopsis accessions from the Versailles collection. We thank the reviewers for their constructive suggestions for improving this article.
Footnotes
This work was funded by the ANR program (grants ANR-08-BLAN-0061 and ANR-14-CE19-0001-01) and the Institut National de la Recherche Agronomique Plant Biology and Breeding Division (grant AAP BAP2013_22 ExMu). The Institut Jean-Pierre Bourgin benefits from the support of the Labex Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
References
- Alonso J M, Stepanova A N, Leisse T J, Kim C J, Chen H, Shinn P, Stevenson D K, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL (2009a) AtBXL1 encodes a bifunctional β-d-xylosidase/α-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Villota MM, Rowland O, Subramaniam R, Western TL (2009b) MUM ENHANCERS are important for seed coat mucilage production and mucilage secretory cell differentiation in Arabidopsis thaliana. J Exp Bot 60: 2601–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, Mohnen D (2013) Evolving views of pectin biosynthesis. Annu Rev Plant Biol 64: 747–779 [DOI] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA III, Orlando R, Scheller HV, Mohnen D (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts M J, Russell R B (2007) Amino-acid properties and consequences of substitutions In Barnes MR, and Gray IC, eds, Bioinformatics for Geneticists. John Wiley and Sons, Ltd, Chichester, United Kingdom, pp 311–342 [Google Scholar]
- Blakeney AB, Harris PJ, Henry RJ, Stone BA (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113: 291–299 [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a β-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, O’Neill MA, York WS (2006) Selective chemical depolymerization of rhamnogalacturonans. Carbohydr Res 341: 474–484 [DOI] [PubMed] [Google Scholar]
- Deng W, Hallett PD, Jeng DS, Squire GR, Toorop PE, Iannetta PPM (2015) The effect of natural seed coatings of Capsella bursa-pastoris L. Medik. (shepherd’s purse) on soil-water retention, stability and hydraulic conductivity. Plant Soil 387: 167–176 [Google Scholar]
- Desprez T, Juraniec M, Crowell EF, Jouy H, Pochylova Z, Parcy F, Höfte H, Gonneau M, Vernhettes S (2007) Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marsico A, Scrano L, Amato M, Gàmiz B, Real M, Cox L (2018) Mucilage from seeds of chia (Salvia hispanica L.) used as soil conditioner: Effects on the sorption-desorption of four herbicides in three different soils. Sci Total Environ 625: 531–538 [DOI] [PubMed] [Google Scholar]
- Francoz E, Ranocha P, Burlat V, Dunand C (2015) Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Gan X, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N. (1985) A mucilage polysaccharide secreted from testa of Arabidopsis thaliana. Arabidopsis Inf Serv 22: 143–145 [Google Scholar]
- Griffiths JS, Crepeau MJ, Ralet MC, Seifert GJ, North HM (2016) Dissecting seed mucilage adherence mediated by FEI2 and SOS5. Front Plant Sci 7: 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JS, Tsai AY, Xue H, Voiniciuc C, Sola K, Seifert GJ, Mansfield SD, Haughn GW (2014) SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol 165: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß F, Rudolf EE, Thiele B, Durner J, Astier J (2017) Copper amine oxidase 8 regulates arginine-dependent nitric oxide production in Arabidopsis thaliana. J Exp Bot 68: 2149–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon F, Tranquet O, Quillien L, Utille JP, Ordaz Ortiz JJ, Saulnier L (2004) Generation of polyclonal and monoclonal antibodies against arabinoxylans and their use for immunocytochemical location of arabinoxylans in cell walls of endosperm of wheat. J Cereal Sci 40: 167–182 [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ (2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. (2011) Molecular genetics: The sound of silence. Nature 471: 582–583 [DOI] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhou G, Abdeen AA, Schafhauser J, Richardson B, Atmodjo MA, Jung J, Wicker L, Mohnen D, Western T, et al. (2013) GALACTURONOSYLTRANSFERASE-LIKE5 is involved in the production of Arabidopsis seed coat mucilage. Plant Physiol 163: 1203–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V, Hanlon M R, Contrino S, Ferlanti E S, Karamycheva S, Kim M, Rosen B D, Cheng C Y, Moreira W, Mock S A, et al. (2015) Araport: The Arabidopsis information portal. Nucleic Acids Res 43: D1003–D1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunieda T, Shimada T, Kondo M, Nishimura M, Nishitani K, Hara-Nishimura I (2013) Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell 25: 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le B H, Cheng C, Bui A Q, Wagmaister J A, Henry K F, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, Nizhynska V, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-d-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S (2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller SG, McPherson MJ (1998) Developmental expression and biochemical analysis of the Arabidopsis atao1 gene encoding an H2O2-generating diamine oxidase. Plant J 13: 781–791 [DOI] [PubMed] [Google Scholar]
- North HM, Berger A, Saez-Aguayo S, Ralet MC (2014) Understanding polysaccharide production and properties using seed coat mutants: Future perspectives for the exploitation of natural variants. Ann Bot 114: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Meissner RC, Shoue DA, Carpita NC, Bevan MW (2001) MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 13: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan JL, Burton RA (2018) New insights into the composition and structure of seed mucilage In Roberts JA, ed, Annual Plant Reviews Online. John Wiley & Sons, New York, doi:10.1002/9781119312994.apr0606 [Google Scholar]
- Planas-Portell J, Gallart M, Tiburcio AF, Altabella T (2013) Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol 13: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G (2011) Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet 12: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain D, Botran L, North HM, Ralet MC (2019) Composition and physicochemical properties of outer mucilage from seeds of Arabidopsis natural accessions. AoB Plants 11: plz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Crépeau MJ, Vigouroux J, Tran J, Berger A, Sallé C, Granier F, Botran L, North HM (2016) Xylans provide the structural driving force for mucilage adhesion to the Arabidopsis seed coat. Plant Physiol 171: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Tranquet O, Poulain D, Moïse A, Guillon F (2010) Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM (2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S, Rondeau-Mouro C, Macquet A, Kronholm I, Ralet MC, Berger A, Sallé C, Poulain D, Granier F, Botran L, et al. (2014) Local evolution of seed flotation in Arabidopsis. PLoS Genet 10: e1004221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü. (2018) GWA-Portal: Genome-wide association studies made easy. Methods Mol Biol 1761: 303–319 [DOI] [PubMed] [Google Scholar]
- Simon M, Simon A, Martins F, Botran L, Tisné S, Granier F, Loudet O, Camilleri C (2012) DNA fingerprinting and new tools for fine-scale discrimination of Arabidopsis thaliana accessions. Plant J 69: 1094–1101 [DOI] [PubMed] [Google Scholar]
- Šola K, Gilchrist EJ, Ropartz D, Wang L, Feussner I, Mansfield SD, Ralet MC, Haughn GW (2019) RUBY, a putative galactose oxidase, influences pectin properties and promotes cell-to-cell adhesion in the seed coat epidermis of Arabidopsis. Plant Cell 31: 809–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM (2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka Y, Kato K, Ogawa-Ohnishi M, Tsuruhama K, Kajiura H, Yagyu K, Takeda A, Takeda Y, Kunieda T, Hara-Nishimura I, et al. (2018) Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat Plants 4: 669–676 [DOI] [PubMed] [Google Scholar]
- Tavladoraki P, Cona A, Angelini R (2016) Copper-containing amine oxidases and FAD-dependent polyamine oxidases are key players in plant tissue differentiation and organ development. Front Plant Sci 7: 824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault JF. (1979) Automatisation du dosage des substances pectiques par la méthode au métahydroxydiphényle. Lebensm Wiss Technol 12: 247–251 [Google Scholar]
- Tollier MT, Robin JP (1979) Adaptation de la méthode à l’orcinol sulfurique au dosage automatique des glucides neutres totaux: Conditions d’application aux extraits d’origine végétale. Ann Technol Agric 28: 1–15 [Google Scholar]
- Tsai AY, Kunieda T, Rogalski J, Foster LJ, Ellis BE, Haughn GW (2017) Identification and characterization of Arabidopsis seed coat mucilage proteins. Plant Physiol 173: 1059–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M (2004) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM (2013) Estimation and partition of heritability in human populations using whole-genome analysis methods. Annu Rev Genet 47: 75–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Engle KA, Günl M, Dieluweit S, Schmidt MH, Yang JY, Moremen KW, Mohnen D, Usadel B (2018) Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol 178: 1045–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Günl M, Schmidt MH, Usadel B (2015a) Highly branched xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol 169: 2481–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Schmidt MH, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Günl M (2015b) MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol 169: 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Zimmermann E, Schmidt MH, Günl M, Fu L, North HM, Usadel B (2016) Extensive natural variation in Arabidopsis seed mucilage structure. Front Plant Sci 7: 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL. (2012) The sticky tale of seed coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Sci Res 22: 1–25 [Google Scholar]
- Western TL, Burn J, Tan WL, Skinner DJ, Martin-McCaffrey L, Moffatt BA, Haughn GW (2001) Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW (2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson G V, Provart N J (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Baskin CC, Baskin JM, Zhang W, Huang Z (2012a) Degradation of seed mucilage by soil microflora promotes early seedling growth of a desert sand dune plant. Plant Cell Environ 35: 872–883 [DOI] [PubMed] [Google Scholar]
- Yang X, Baskin J, Baskin C, Huang Z (2012b) More than just a coating: Ecological importance, taxonomic occurrence and phylogenetic relationships of seed coat mucilage. Perspect Plant Ecol Syst 14: 434–442 [Google Scholar]
- Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G (2014) CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol 164: 1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A (2010) A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol 154: 78–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Qiao L, Wu AM (2017) Effective extraction of Arabidopsis adherent seed mucilage by ultrasonic treatment. Sci Rep 7: 40672. [DOI] [PMC free article] [PubMed] [Google Scholar]