Much like the spikes that deter birds from sitting on fences, trichomes (hair-like projections on the leaf surface) are the epidermis’ first line of defense, discouraging insects and other pests (Levin, 1973). In addition to their role protecting the plant, trichomes are an excellent marker to study developmental biology: they arise from the same progenitor cells as stomata and pavement cells, are easily visible, and their patterning is under strict genetic control.
To better understand trichome development, we must understand two core processes: trichome patterning (or the fate determination of progenitor cells) and trichome differentiation, both of which have been most studied in Arabidopsis (Arabidopsis thaliana). The decision to adopt a trichome or other epidermal cell identity is regulated antagonistically by two protein complexes termed the initiator complex and the inhibitor complex (Fig. 1A). The initiator complex consists of GLABRA1 (GL1), GL3 or ENHANCER OF GL3 (EGL3), and TRANSPARENT TESTA GLABRA1 (TTG1; for review, see Pattanaik et al., 2014). This initiator complex activates the expression of GL2, whose transcription factor product initiates trichome morphogenesis (Szymanski et al., 1998). Replacement of GL1 by an inhibitor renders the complex unable to activate GL2 expression, preventing trichome morphogenesis. While the expression of most trichome inhibitors is regulated by the initiator complex, a small number are not, including TRICHOMELESS1 (TCL1) and TCL2. To date, the roles of TCL1 and TCL2 in trichome development in leaves have not been extensively studied, and their upstream regulators in this early stage of development are unknown.
Figure 1.
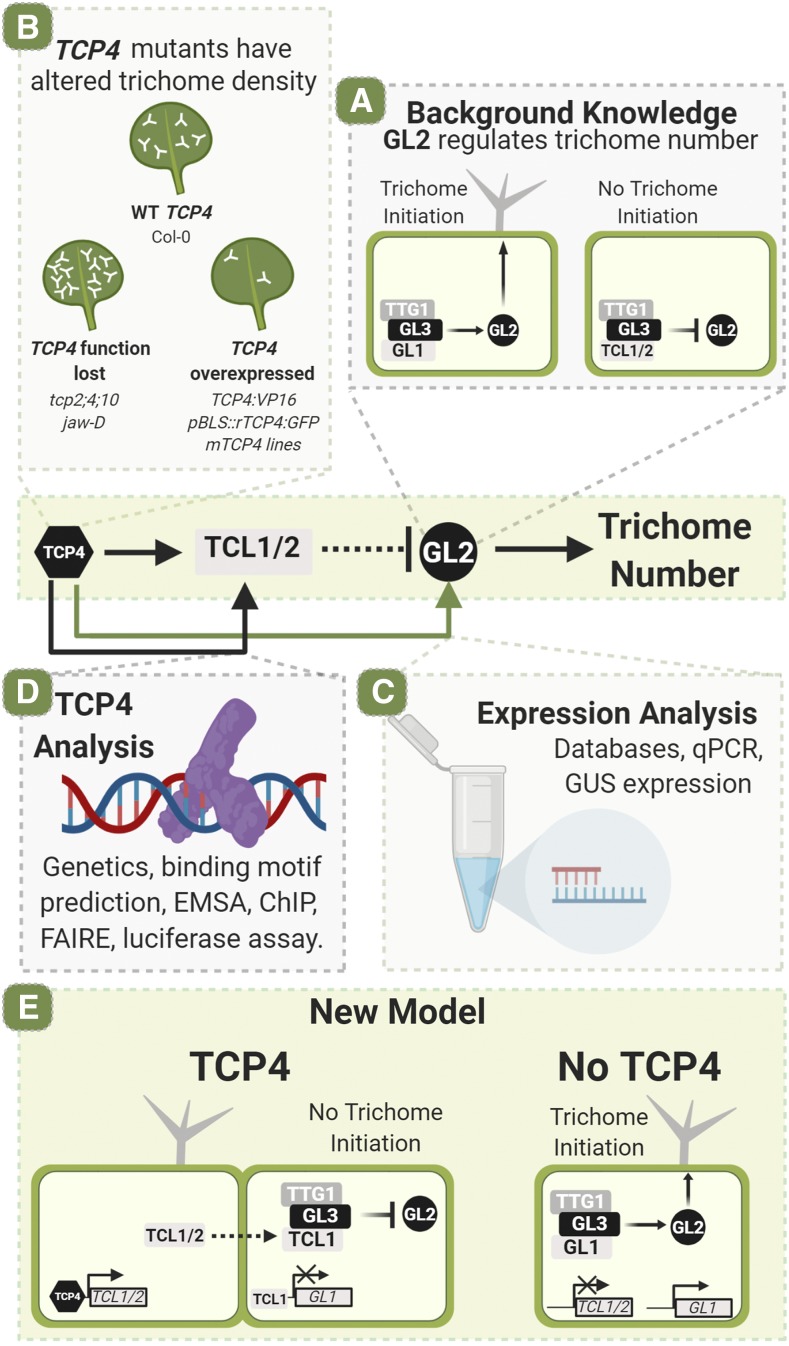
Graphical summary of the article. A, Previous work has shown that GL2 positively regulates trichome initiation. B, This study uses several lines with altered TCP4 expression. Lines where TCP4 function is lost are tcp2;4;10 and jaw-D. Lines where TCP4 is overexpressed are TCP4:VP16, pBLS::rTCP4:GFP, inducible mTCP4 jawD;GR, inducible mTCP4 Col-0;GR, and p35S::mTCP4:GR. The wild-type (WT) Columbia-0 (Col-0) is used as a control. C, Several techniques were used to study the indirect effect of TCP4 on GL2 and thus trichome number, including the use of existing databases, quantitative PCR (qPCR), and GUS expression in stably transformed lines. D, Several techniques were used to analyze TCP4 binding and activity, including genetic screens, binding motif prediction, electrophoretic mobility shift assay (EMSA), chromatin immunoprecipitation (ChIP), formaldehyde-assisted isolation of regulatory element (FAIRE), and transient luciferase assays. E, The authors propose that TCP4 affects trichome initiation through directly binding to the TCL1/2 promoter, which in turn inhibits GL2. Figure created with biorender.com. (Adapted from figure 6E of Vadde et al., 2019.)
In this issue of Plant Physiology, Vadde et al. (2019), from the Indian Institute of Science, present their comprehensive characterization of the role of a class II TCP (a family named after the first four studied members: TEOSINTE BRANCHED1 [TB1; Zea mays], CYCLOIDIA [CYC; Antirrhinum majus], and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1/2 [PCF1/2; Oryza sativa]) in the regulation of TCL1 and TCL2, and therefore its role in trichome development on Arabidopsis leaves. The authors hypothesized that upstream regulators of TCL1 and TCL2 would be transcription factors present for the duration of leaf morphogenesis. The TCPs are a family of plant-specific transcription factors that can be split broadly into two classes, which act antagonistically to promote (class I) or repress (class II) cell proliferation (for review, see Martín-Trillo and Cubas, 2010). TCPs are known to regulate several developmental processes, including leaf shape determination, petal and stamen development, and circadian clock function (Li, 2015). In addition, TCP expression overlaps both temporally and spatially with factors involved in trichome initiation, making them a potential candidate in this process.
Plants with TCP4 (a class II TCP) gain of function have previously been observed to have fewer trichomes on their leaf surfaces compared with wild-type plants (Efroni et al., 2008). Vadde et al. (2019) confirmed this observation through examining two TCP4 gain-of-function mutants: TCP4:VP16, in which TCP4 is fused to a viral activation domain; and pBLS::rTCP4:GFP, which expresses a miR319-resistant form of TCP4 fused to GFP under an early leaf-specific promoter. Both plant lines showed reduced trichome density compared with the wild type. Conversely, two TCP4 loss-of-function lines, the triple mutant tcp2 tcp4 tcp10 and jawD (an overexpressor of miR319, whose targets include class II TCPs TCP2, TCP3, TCP4, TCP10, and TCP24), displayed an increased trichome density compared with the wild type (Fig. 1B).
Publicly available gene expression data show that the positive regulators of trichome initiation GL1 and GL2 are significantly up-regulated in TCP loss-of-function mutants and vice versa in gain-of-function mutants. This was validated by Vadde et al. (2019) by quantitative PCR time-course experiments and GUS assays (Fig. 1C), demonstrating that TCP4 suppresses the expression of these two genes. As the class II TCPs are transcriptional activators, it was hypothesized that this down-regulation of GL1 and GL2 by TCP4 is an indirect effect.
To identify which genes act downstream of TCP4 in GL2 transcriptional regulation, transcript levels of known trichome initiator and trichome inhibitor complex genes were compared between a TCP4 gain-of-function line and the wild type. Four genes, TCL1, TCL2, ENHANCER OF TRY AND CPC2 (ETC2), and ETC3, were up-regulated in the TCP4 gain-of-function line. Comparison with microarray data previously produced in the Nath lab revealed that only TCL1 and TCL2 are activated soon after TCP4 induction (Challa et al., 2016). TCL1 and TCL2 activation is associated with down-regulation of GL1, supporting the model that TCP4 prevents trichome morphogenesis via activation of TCL1 and TCL2, which constitute part of the trichome inhibition complex.
Vadde et al. (2019) go on to show that TCP4 directly targets TCL1 and TCL2. In the absence of protein synthesis, induction of TCP4 increased TCL1 and TCL2 transcript levels by at least twofold. Putative TCP4 binding sites were identified in the regulatory regions of both TCL1 and TCL2. These in silico predictions were verified by in vitro electrophoretic mobility shift assays, which demonstrated that TCP4 binds specifically to these sequence motifs, and chromatin immunoprecipitation showed that TCP4 is recruited to these sites in the genome. Formaldehyde-assisted isolation of regulatory element assays confirmed that the presence of TCP4 increases chromatin accessibility in the promoters of TCP1 and TCP2. Furthermore, an in vivo luciferase assay in isolated protoplasts and an in planta GUS assay both confirmed that TCP4 binds to and activates the TCL1/2 genes, supporting the model that trichome morphogenesis is inhibited by TCP4 through direct activation of TCL1 and TCL2 (Fig. 1D).
Further genetic experiments showed that TCL1 is required for the reduction in trichome number observed in the leaves of TCP4 overexpression lines. Here, examination of trichomes on the inflorescence stems and older leaves could have been included to determine whether TCP4 regulation of TCL1 is restricted to young leaves or is ubiquitous throughout plant development.
The thorough work presented in this article is an important step forward in our understanding of trichome development, summarized in Figure 1E. For the first time, an upstream transcription factor controlling inhibitory regulators of trichome morphogenesis in leaves has been identified. Excitingly, TCP4 is also implicated in the repression of cell proliferation (Schommer et al., 2014) and in fundamental leaf patterning (Koyama et al., 2017), and thus may play an important role in the coordination of development. As there is some degree of functional redundancy within the TCP family (Danisman et al., 2013), whether additional TCPs are involved in trichome initiation and patterning must await further study. TCPs are ubiquitous across plant species; thus, advances in our understanding of their role in coordinating development will also be beneficial when considering economically important crop species.
References
- Challa KR, Aggarwal P, Nath U (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28: 2117–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van Dijk ADJ, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent GC, Immink RGH (2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot 64: 5673–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175: 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. (1973) The role of trichomes in plant defence. Q Rev Biol 48: 3–15 [Google Scholar]
- Li S. (2015) The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal Behav 10: e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: A family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Patra B, Singh SK, Yuan L (2014) An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 5: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Debernardi JM, Bresso EG, Rodriguez RE, Palatnik JF (2014) Repression of cell proliferation by miR319-regulated TCP4. Mol Plant 7: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Vadde BVL, Challa KR, Sunkara P, Hegde AS, Nath U (2019) The TCP4 transcription factor directly activates TRICHOMELESS1 and 2 and suppresses trichome initiation. Plant Physiol 181: 1587–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]