High-throughput genome editing is achieved in recalcitrant plant species using a ternary vector system in combination with morphogenic regulators and CRISPR/Cas modules.
Abstract
The lack of efficient delivery methods is a major barrier to clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas)-mediated genome editing in many plant species. Combinations of morphogenic regulator (MR) genes and ternary vector systems are promising solutions to this problem. In this study, we first demonstrated that MR vectors greatly enhance maize (Zea mays) transformation. We then tested a CRISPR/Cas9 MR vector in maize and found that the MR and CRISPR/Cas9 modules have no negative influence on each other. Finally, we developed a novel ternary vector system to integrate the MR and CRISPR/Cas modules. Our ternary vector system is composed of new pGreen-like binary vectors, here named pGreen3, and a pVS1-based virulence helper plasmid, which also functions as a replication helper for the pGreen3 vectors in Agrobacterium tumefaciens. The pGreen3 vectors were derived from the plasmid pRK2 and display advantages over pGreen2 vectors regarding both compatibility and stability. We demonstrated that the union of our ternary vector system with MR gene modules has additive effects in enhancing maize transformation and that this enhancement is especially evident in the transformation of recalcitrant maize inbred lines. Collectively, our ternary vector system-based tools provide a user-friendly solution to the low efficiency of CRISPR/Cas delivery in maize and represent a basic platform for developing efficient delivery tools to use in other plant species recalcitrant to transformation.
Since 2013, a revolutionary genome-editing tool, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas), has enabled plant researchers and crop breeders to create mutants of target plant genes easily and efficiently (Altpeter et al., 2016; Chen et al., 2019). CRISPR/Cas-based de novo domestication of wild tomato (Solanum pimpinellifolium) provided a new paradigm for crop breeding (Birchler, 2017; Rodriguez-Leal et al., 2017; Lemmon et al., 2018; Li et al., 2018; Zsögön et al., 2018; Butt et al., 2019). However, this strategy requires high-throughput gene editing, and the low efficiency of crop transformation has been a bottleneck. Successful applications of CRISPR/Cas in plants depend on efficient delivery of the reagents into cells, and lack of appropriate delivery methods is the major barrier to CRISPR/Cas-mediated gene editing in a variety of plant species (Altpeter et al., 2016). The available delivery systems include indirect methods that use Agrobacterium tumefaciens or a virus as mediators and direct methods that use protoplast transfection or particle bombardment (Svitashev et al., 2016; Liang et al., 2017; Ran et al., 2017; Liu et al., 2019).
Agrobacterium-mediated delivery is the most popular method of CRISPR/Cas reagent delivery and accounts for more than 80% of the reported editing events, covering almost all model plant species, major field crops, and vegetable and fruit crops (Ran et al., 2017). Other delivery methods are suitable for special purposes but are technically complicated, dependent on expensive equipment, or require further development (Ran et al., 2017; Zhao et al., 2017; Demirer et al., 2019; Kwak et al., 2019; Toda et al., 2019; Zhang et al., 2019). New CRISPR tools have almost always been first reported in rice (Oryza sativa) rather than in other crop plants, primarily because of the availability of high-efficiency Agrobacterium-mediated rice transformation (Ran et al., 2017; Chen et al., 2019; Hua et al., 2019; Ren et al., 2019; Zhong et al., 2019). For the same reason, only in rice has high-throughput generation of CRISPR mutants been realized (Lu et al., 2017; Meng et al., 2017). Recently, using pollen of maize (Zea mays) haploid-inducer lines to deliver CRISPR/Cas9 reagents into female gametophytes of different maize lines, two groups have created haploid maize lines with homozygous mutations of target genes (Kelliher et al., 2019; Wang et al., 2019). This strategy provides a rapid route for maize breeding based on genome editing but still depends on transgenic maize lines to produce pollen harboring the CRISPR/Cas9 reagents.
One drawback of Agrobacterium-mediated plant transformation is that current methods are generally suitable only for a limited number of species and depend strongly on the recipient genotypes, especially in monocots (Altpeter et al., 2016). However, methods of Agrobacterium-mediated transformation have been improving recently (Lowe et al., 2016; Mookkan et al., 2017; Anand et al., 2018; Che et al., 2018). Using morphogenic regulator (MR) genes and a ternary vector system, Lowe et al. (2018) achieved callus-free and genotype-independent maize transformation. The integration of these two modules with various CRISPR/Cas tools could allow more efficient genome editing. However, the ability of researchers to combine these modules is restricted by the limited availability of both original and derived tools in public plasmid repository platforms, such as Addgene. In addition, the restrictions are not favorable for iterative updating of the CRISPR/Cas delivery system for use in maize and other plant species.
In this study, we developed a new ternary vector system to unite the MR and CRISPR/Cas9 modules. We demonstrate that these tools greatly enhance Agrobacterium-mediated delivery of CRISPR/Cas9 reagents in maize, thus improving the throughput of genome editing. Our new ternary vector system provides a basic platform to develop efficient CRISPR/Cas tools in other plant species recalcitrant to transformation. In addition, our vectors are designed so that the single-guide RNA (sgRNA) construction is compatible with one-step Golden Gate cloning and Gibson assembly, which provides a user-friendly protocol for the generation of final CRISPR/Cas MR vectors.
RESULTS
Since ternary vector systems and MR genes have additive effects on maize transformation (Anand et al., 2018; Lowe et al., 2018), we first constructed MR vectors and tested their effects on maize transformation. We then developed a novel ternary vector system to integrate MR modules and tested the combinatorial effects of the vector system and MR modules on maize transformation.
Morphogenic Regulators Enhance Agrobacterium-Mediated Delivery of CRISPR-Cas Reagents
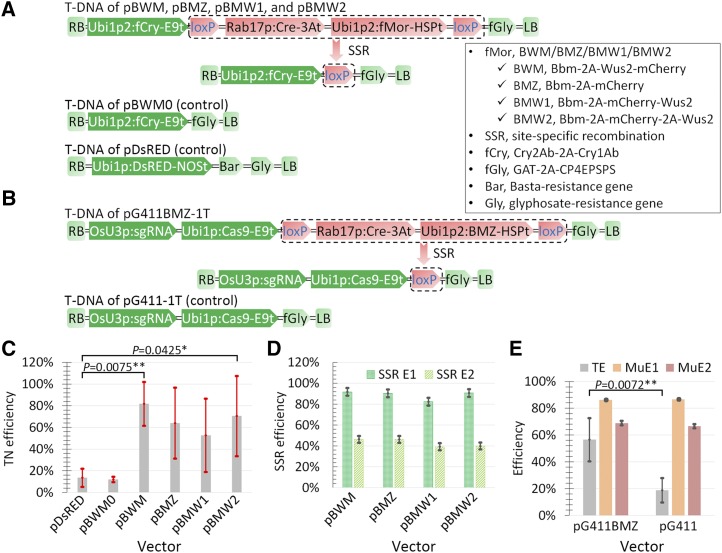
For more efficient delivery of CRISPR/Cas9 components and thus higher throughput of CRISPR/Cas9 genome editing in maize, we first tested an MR-mediated transformation-enhancing system. We fused the maize MR gene Wuschel2 (Wus2) to the fluorescent reporter gene mCherry (Wus2-mCherry or mCherry-Wus2) to attempt to weaken Wus2 activity because a high level of Wus2 has negative effects on transformed cells (Lowe et al., 2016). To lessen the number of expression cassettes, we further fused the maize MR gene Baby boom (Bbm) to these fused genes using 2A as a linker (Zhao et al., 2014). We named these two fusion genes BWM (Bbm-2A-Wus2-mCherry) and BMW1 (Bbm-2A-mCherry-Wus2). For comparison, we also generated two additional fusion genes, BMZ (Bbm-2A-mCherry) and BMW2 (Bbm-2A-mCherry-2A-Wus2; Fig. 1A), encoding fusion polyproteins that are subject to self-cleavage between the last two amino acids of the 19-amino acid 2A linker polypeptide. We fused two glyphosate-resistance genes, GAT and CP4EPSPS, as a selection marker for transgenic lines (fGly). We fused two maize codon-optimized insect-resistance genes, Cry2Ab and Cry1Ab, as a representative transgene (Zhao et al., 2014). We also generated a test MR vector harboring the CRISPR/Cas9 components (pG411BMZ-1T) and a CRISPR/Cas9 control vector (pG411-1T; Fig. 1B). The Cre/loxP module was added to allow the inducible excision of MR genes to produce healthy, fertile T0 transgenic plants, since ectopic Bbm expression results in aberrant phenotypes (Lowe et al., 2016).
Figure 1.
MR genes enhanced CRISPR-Cas delivery. A, T-DNA structures of four MR-testing vectors and two control vectors. The T-DNA region of the MR-testing vectors following Cre/loxP site-specific recombination (SSR) induced by desiccation is indicated. RB and LB, T-DNA right and left borders, respectively; Ubi1p2, maize Ubi1 promoter with the intron shortened. Terminators used were pea (Pisum sativum) rbcS-E9 (E9t), pea 3A (3At), Arabidopsis (Arabidopsis thaliana) HSP18.2 (HSPt), and Agrobacterium NOS (NOSt). Bar, Basta resistance gene; Gly and fGly, glyphosate-resistance genes. Bar, Gly, and fGly were selection marker genes for transgenic lines. B, T-DNA structures of the CRISPR/Cas9 MR vector, pG411BMZ-1T (before and after desiccation-induced SSR), and the corresponding control vector, pG411-1T. C, Maize transformation (TN) efficiencies for vectors in A. The values represent averages of four transformation experiments. D, SSR efficiencies for MR vectors in A. SSR E1 and SSR E2 represent the frequencies of all SSR lines or complete SSR lines, respectively, relative to the total number of transgenic lines detected. E, Efficiencies of maize transformation and mutations for the two CRISPR/Cas9 vectors in B. TE, Transformation efficiency. The values represent averages of five transformation experiments, and the error bars represent sd. A total of 29 pG411BMZ-1T and 15 pG411-1T transgenic lines were used for analysis of mutations, and the mutant lines harboring a wild-type allele were categorized as chimeric mutants. MuE1, Mutation efficiency for all three types of mutations (homozygous, biallelic, or chimeric) of ZmCDPK10; MuE2, mutation efficiency for two types of mutations (homozygous and biallelic). The error bars represent sd. P values were calculated by Student’s t test: *, P < 0.05 and **, P < 0.01.
We transformed maize with the four MR and two control vectors (Fig. 1A). The transformation efficiencies of the MR vectors were greatly increased, specifically 3.9- to 6.8-fold that of the control vectors (Fig. 1C; Supplemental Table S1). Moreover, Bbm alone (vector pBMZ) worked well to improve transformation efficiency.
As described by Lowe et al. (2016), a desiccation step was introduced during transformation to induce the expression of Cre, which is driven by the Rab17 drought-specific promoter; in turn, this activated Cre/loxP SSR, leading to excision of the MR components flanked by 34-bp loxP sites (Lowe et al., 2016). We examined the efficiency of this excision using PCR amplification. We regarded the lines as complete SSR lines when the PCR amplification denoting excision was positive and the two PCR amplifications denoting no excision were negative. We regarded the lines as partial SSR lines when the PCR amplification denoting excision was positive and either of the two PCR amplifications denoting no excision was also positive. The average frequencies of all (partial + complete) SSR lines and complete SSR lines relative to the total number of transformed lines were 88.8% and 43%, respectively (Fig. 1D; Supplemental Table S2). These results indicate that the inducible Cre/loxP SSR components in our MR vectors work well; the relatively low proportion of complete SSR lines could be attributed to insufficient desiccation owing to the high ambient humidity at the time of these experiments.
To test the editing efficiency of the CRISPR/Cas9 MR vector with an sgRNA targeting the maize CDPK10 gene (Fig. 1B), we transformed maize with this MR vector together with a control vector containing the CRISPR/Cas9 elements but without the MR elements. Consistent with the above results, the CRISPR/Cas9 MR vector conferred much higher transformation efficiency than the control vector (Fig. 1E; Supplemental Table S3). Mutation analysis of the target gene indicated that the editing efficiency of the MR vector was comparable to that of the control vector (Fig. 1E; Supplemental Table S4). Excision analysis of the loxP-flanked fragment indicated that the frequencies of SSR lines and complete SSR lines relative to the total number of transformed lines were 93.1% and 69%, respectively (Supplemental Table S4). These results suggest that MR genes do not affect the editing efficiency of CRISPR/Cas and that CRISPR/Cas components do not affect transformation enhancement by MR genes.
The Union of a Novel Ternary Vector System and MR Genes Has Additive Effects on Maize Transformation
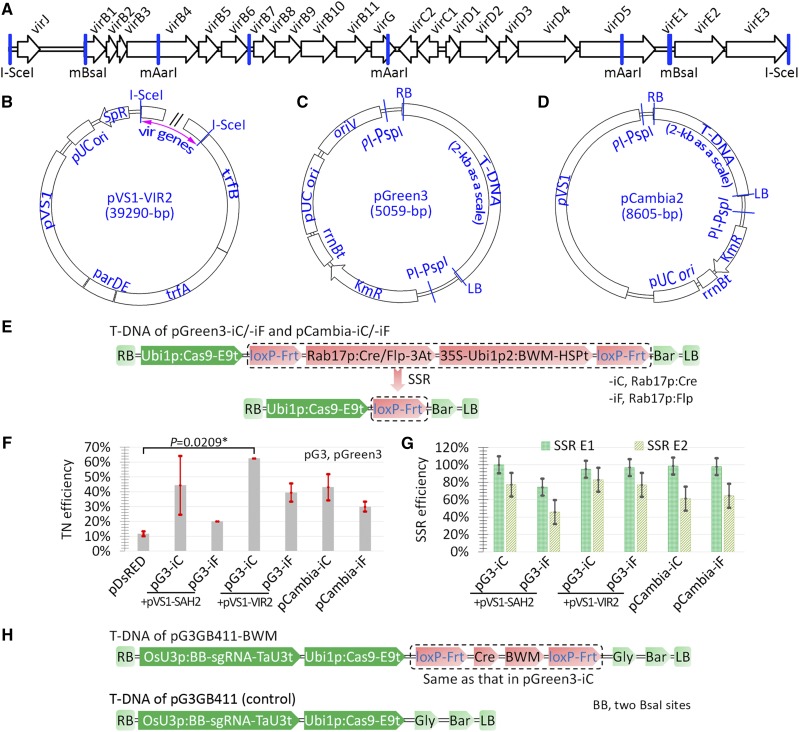
To further enhance the Agrobacterium-mediated delivery of CRISPR-Cas reagents, we developed a novel ternary vector system (Fig. 2). In this system, the new binary vectors were based on a pGreen-like backbone, and the new virulence helper plasmid pVS1-VIR2 also served as a replication helper plasmid for these new binary vectors. The first-generation pGreen vectors were unstable in Escherichia coli and in Agrobacterium due to inefficient transcription termination of the kanamycin-resistance gene (KmR). Inserting the ampicillin-resistance gene terminator behind the KmR open reading frame improved the stability in E. coli of the modified plasmids, named pGreen2 (Hellens et al., 2000; Thole et al., 2007; Murai, 2013). The pGreen and pGreen2 vectors were based on the pSa replication system in Agrobacterium, whereas our new pGreen-like vectors, named pGreen3, were based on the pRK2 replication system. The pGreen3 vectors (Fig. 2C) harbor the replication origin of pRK2 (oriV) and depend on replication helper plasmids (pVS1-VIR2 or pVS1-SAH2) harboring the trfA region of pRK2 for propagation in Agrobacterium (Fig. 2B). As discussed later, the advantages of pGreen3 over pGreen2 vectors include an excellent compatibility with the pVS1 replication system in Agrobacterium and a greater stability in E. coli and Agrobacterium.
Figure 2.
Combinatorial effects of a novel ternary vector system and MR genes on maize transformation. A, Physical map of the virulence gene cluster cloned from the Ti plasmid of Agrobacterium strain EHA105. The two I-SceI sites, the three mutated AarI sites (mAarI), and the two mutated BsaI sites (mBsaI) are indicated. B, Physical map of pVS1-VIR2. The I-SceI-flanked fragment (indicated by the pink double-headed arrow) is the same as that in A. The map of pVS1-SAH2 is similar to that of pVS1-VIR2 except with no I-SceI fragment. The trfA, trfB, and parDE regions from pRK2 function in replication or stability. The pUC ori and pVS1 regions function in replication in E. coli and Agrobacterium, respectively. SpR, Spectinomycin resistance gene. C, Physical map of the pGreen3 binary vector backbone. The T-DNA region is flanked by two PI-PspI sites, and only the 2-kb T-DNA was left for comparison of the sizes of the backbones. oriV, Replication origin from pRK2; rrnBt, E. coli rrnB terminator. D, Physical map of the slightly modified pCambia backbone. E, T-DNA structures of four test vectors. F, Maize transformation efficiencies of the vectors shown in E. The map of pDsRED is shown in Figure 1A. The values represent averages of two transformations, and the error bars represent sd. P values were calculated by Student’s t test: *, P < 0.05. G, SSR efficiencies. H, T-DNA structure of pG3GB411-BWM designed for user-friendly assembly of sgRNA expression cassettes. TaU3t, Wheat (Triticum aestivum) U3 terminator. See Figure 1 for the other annotations.
To test the additive effect on maize transformation of the combination of the ternary vector system and MR genes, we constructed two MR binary vectors based on the pGreen3 backbone and two SSR systems. These two vectors, named pGreen3-iC and pGreen3-iF, harbored the Cre/loxP and Flp/Frt SSR systems, respectively (Fig. 2, C and E). We also constructed two pCambia-based MR binary vectors named pCambia-iC and pCambia-iF for comparison (Fig. 2, D and E). Consistent with the above results, all of the MR vectors conferred much higher transformation efficiencies than the control vector (Fig. 2F; Supplemental Table S5). As predicted, the results also demonstrated that for the MR vectors with the same SSR module, the ternary vector system pGreen3/pVS1-VIR2 conferred much higher transformation efficiencies than the dual binary system pGreen3/pVS1-SAH2 and the pCambia binary vectors (Fig. 2F; Supplemental Table S5). The Flp/Frt vectors conferred much lower transformation efficiencies than the Cre/loxP vectors (Fig. 2F; Supplemental Table S5) for unknown reasons and also seemed to have lower SSR efficiencies in the dual binary system (Fig. 2G; Supplemental Table S6).
Since transgenic components would be unnecessary and could be separated from a mutant line once mutations were induced, we defined single-copy T-DNA integrations as quality events for genome editing. For MR vectors, we also considered complete SSR as a factor of quality events. To evaluate the efficiency of quality events, we analyzed T-DNA copy number in 55 SSR-C and eight SSR-P lines from the combination of pGreen3-iC and pVS1-VIR2 using droplet digital PCR (Collier et al., 2017). The results indicated that approximately 73% of SSR-C lines harbored single T-DNA copies (Supplemental Table S7). Thus, the efficiency of quality events to produce single-copy and completely excised T0 plants was approximately 61% (73% × 83%). The results also indicated that approximately 75% of SSR-P lines harbored single T-DNA copies (Supplemental Table S7), suggesting that most SSR-P lines were formed at two-cell or later stages of somatic embryogenesis and multiple-copy T-DNA integrations were not the main reason for the formation of SSR-P lines. At two-cell or later stages, the reversible integration events other than the excision events, caused by the circular DNA molecules excised at one-cell stage, may also be involved in the formation of SSR-P lines.
Finally, we transformed recalcitrant maize inbred line ND88 using the pGreen3-iC/pVS1-VIR2 ternary vector system. The transformation efficiency of the ternary vector system reached an average of 17.2% ± 3.1% according to four experiments transforming a total of 450 immature embryos. By contrast, the transformation efficiency of the control vector pDsRED was only 0.8% ± 1%, which was similarly based on four experiments transforming 450 immature embryos in total. Thus, our results suggest that our ternary vector system in combination with MR modules may enable high-efficiency transformation of other recalcitrant maize inbred lines. In addition, we constructed a pGreen3-based CRISPR/Cas9 MR cloning vector, named pG3GB411-BWM, to facilitate user-friendly assembly of sgRNA expression cassettes (Fig. 2H). We also constructed a pGreen3-based CRISPR/Cas9 cloning vector named pG3GB411 as a control for use with pG3GB411-BWM (Fig. 2H).
DISCUSSION
Callus-free, genotype-independent maize transformation promises a bright future for high-throughput genome editing in maize (Altpeter et al., 2016; Lowe et al., 2016, 2018). However, CRISPR/Cas tools integrated with MR components are still unavailable from public plasmid repository platforms. This issue led us to develop a new ternary vector system to combine the MR and CRISPR/Cas9 components. The integrated modules facilitate higher-throughput gene editing in maize and build a foundation for iterative updates of CRISPR/Cas tools in maize and other plant species. In addition, since the CRISPR/Cas9 MR vectors pG411BMZ (Fig. 1B) and pG3GB411-BWM (Fig. 2H) are compatible with Golden Gate cloning and Gibson assembly of sgRNA cassettes, the tools are very user friendly in terms of ease of use, efficiency, time, and cost (Xing et al., 2014).
Our new ternary vector system is composed of pGreen3 binary vectors and a virulence helper plasmid, pVS1-VIR2, which also serves as a replication helper plasmid for pGreen3 vectors. As compared with pGreen2, pGreen3 vectors have the advantage of compatibility with the pVS1 replication system in Agrobacterium, which makes them able to constitute a ternary vector system with the pVS1-derived virulence helper plasmid. Before developing the pGreen3 vectors, we had constructed four pVS1-derived helper plasmids of pGreen2 using the native promoter or a synthetic Em7 promoter to drive pSa-RepA and by inserting the two expression cassettes at the same site in two orientations. However, Agrobacterium colonies harboring pGreen2 and the helper plasmids grew slowly and poorly, which suggested that pGreen2 was poorly compatible with pVS1-derived helper plasmids in Agrobacterium. The results also suggested that the compatibility of pGreen-like binary vectors with their replication helper plasmids is complicated and depends not only on incompatibility groups but also on other factors, such as plasmid copy number. Since pVS1 plasmids (approximately 20 copies per cell) have much higher copy number than pSa (approximately two copies per cell) in Agrobacterium (Anand et al., 2018), the presence of the pVS1 helper plasmid may lead to overaccumulation of pSa-RepA, and accordingly to uncontrollable propagation of pGreen2, which may aggravate the instability of pGreen2. This incompatibility problem led us to develop the pGreen3 vectors. Three reasons may account for the much better compatibility of pGreen3 with pVS1-derived helper plasmids in Agrobacterium relative to that of pGreen2. First, pRK2 and pVS1 belong to different incompatibility groups, which means that they can stably coexist in the same cell. In fact, the coexistence of pVS1- and pRK2-derived binary vectors appeared to increase the stability of pRK2 derivatives (Murai, 2013). Second, pRK2 per se is more stable than pSa (Murai, 2013). Third, the copy number difference between pRK2 (approximately 10 copies per cell) and pVS1 (approximately 20 copies per cell) is much smaller than that between pSa (approximately two copies) and pVS1 (Anand et al., 2018).
A further advantage of pGreen3 vectors is that they are more stable than pGreen2 vectors. We frequently encountered a problem with some Agrobacterium colonies harboring the helper pSoup and our pGreen2-like vectors (Xing et al., 2014); these colonies grew slowly in liquid media, although they grew normally on agar plates. This problem suggests that the pSa replication system, which is necessary for pGreen2 vector propagation in Agrobacterium, is much less stable than other replication systems. In fact, pCH30 and pCH32, which harbor the pSa replication system, are not stable in Agrobacterium in the absence of selection (Hamilton, 1997). In comparison with the pGreen2 vectors, our pGreen3 vectors grew normally in solid and liquid media, demonstrating that pGreen3 vectors are more stable than pGreen2.
Since the virulence helper plasmid pVS1-VIR2 harbors trfA, trfB, and parDE fragments from pRK2 (Fig. 2B), the propagation and stability of pGreen3 in Agrobacterium are able to be maintained. In addition, pVS1-derived vectors per se are very stable (Murai, 2013; Anand et al., 2018). Thus, the virulence helper pVS1-VIR2 and the pGreen3 binary vectors constitute a novel and stable ternary vector system. Since the additional virulence genes help to transfer large T-DNAs (Hamilton, 1997), this ternary vector system may be especially useful for the delivery of T-DNAs harboring MR modules (greater than 8 kb). Thus, the combination of ternary vectors and MR-based transformation-enhancing modules may provide a basic platform for the development of high-throughput gene-editing tools in a variety of plant species. The spectinomycin-resistance helper plasmid is also compatible with the kanamycin-resistance pRi binary vectors such as pRi101 (TaKaRa Bio), BIBAC (Hamilton, 1997; Zhang et al., 2017), and TAC (Lin et al., 2003). Thus, the virulence helper plasmid can be combined with other binary vectors to produce additional ternary systems. Since the combination of pVS1 virulence helper plasmids and pRi binary vectors is more efficient than other combinations (Anand et al., 2018), it will be interesting to compare our ternary vector system with this optimal combination. Since the vir gene cluster in the helper plasmid pVS1-VIR2 is flanked by two I-SceI sites (Fig. 2, A and B), the fragment can be easily transferred to other vectors with different Agrobacterium replication systems by digestion with I-SceI. This feature facilitates the generation of more ternary vector systems. In addition, the spectinomycin resistance of the helper plasmid is compatible with the gentamycin resistance of some Agrobacterium strains, such as GV3101/pMP90. This feature can therefore offset the incompatibility problem posed by gentamycin-resistance virulence-helper plasmids, such as pPHP71539 (Anand et al., 2018), and thus provides an important alternative.
Altogether, our ternary vector system-based platform, in combination with appropriate MR genes and promoters, will facilitate the development of high-efficiency tools for Agrobacterium-mediated delivery of CRISPR-Cas reagents and thus enable high-throughput genome editing in a variety of plant species.
CONCLUSION
We developed a novel ternary vector system that, in combination with MRs and CRISPR/Cas modules, may facilitate higher-throughput genome editing in recalcitrant plant species. The compatibility of the sgRNA construction with one-step Golden Gate cloning and Gibson assembly provides a user-friendly protocol for the generation of final CRISPR/Cas binary vectors. The vectors are available from Addgene and/or MolecularCloud (GenScript) for sharing with academic investigators for noncommercial research.
MATERIALS AND METHODS
Vector Construction
Detailed descriptions of the vector construction are provided in Supplemental Materials and Methods S1. All primers used in this study are listed in Supplemental Table S8. Annotated sequences of the vectors described in this study together with their DNA samples are available from Addgene and/or MolecularCloud (GenScript) for sharing with academic investigators for noncommercial research. The synthetic Bbm and Wus2 genes were based on the sequence of B73. The sgRNA expression cassette was generated as previously described (Xing et al., 2014).
Maize Transformation and Analysis of the Transformation Efficiency
We used Agrobacterium tumefaciens strain EHA105 for the pCambia-based binary vectors. For Agrobacterium strains EHA105 and LBA4404 harboring the helper plasmids pVS1-SAH2 and pVS1-VIR2, respectively, we added 150 mg L−1 spectinomycin and 25 mg L−1 rifampicin to the growth medium. Once we introduced the pGreen3 binary vectors into the engineered Agrobacterium strains, we added 100 mg L−1 kanamycin (instead of spectinomycin) and 25 mg L−1 rifampicin to the growth medium. We used maize (Zea mays) ND73, a B73-derived inbred line, for maize transformation unless otherwise specified. Maize transformation followed published protocols with minor modifications (Lowe et al., 2016). For selection of transgenic lines, we supplemented the growth medium with 25 mg L−1 glyphosate for fGly or Gly selection marker genes or 3 mg L−1 bialaphos for the Bar selection gene. We confirmed transgenic lines by PCR using primers fGly-IDF/-IDR, Bar-IDF/-IDR, and Gly-IDF/-IDR for the fGly, Bar, and Gly selection genes, respectively. We regarded PCR-positive plants as transgenic lines. We calculated transformation efficiency as the frequency of regenerated transgenic lines relative to the number of immature embryos used for transformation.
Analysis of SSR-Based Excision
We used four primers (Up-loxP1-F, Dn-loxP1-R, Up-loxP2-F, and Dn-loxP2-R), located upstream or downstream of the first or second loxP site, to detect excisions of the MR modules in pBWM, pBMZ, pBMW1, pBMW2, and pG411BMZ-1T. We used Up-loxP1-F and Dn-loxP2-R for PCR amplification (535 bp) to detect T-DNA undergoing excision. We used two PCR amplifications to detect T-DNA undergoing no excision: one (322 bp) with the primers Up-loxP1-F and Dn-loxP1-R, and the other (748 bp) with the primers Up-loxP2-F and Dn-loxP2-R. To detect excisions of the MR modules in pGreen3-iC/iF and pCambia-iC/iF, we replaced the Dn-loxP2-R primer with Dn-loxP2-R2 and amplified the three PCR fragments using the primer pairs Up-loxP1-F/Dn-loxP2-R2 (519 bp), Up-loxP1-F/Dn-loxP1-R (473 bp), and Up-loxP2-F/Dn-loxP2-R2 (383 bp), respectively. To detect excisions of the MR modules in pG3GB411-BWM-derived vectors, we replaced the Dn-loxP2-R with Dn-loxP2-R3 and amplified the three PCR fragments using the primer pairs Up-loxP1-F/Dn-loxP2-R3 (657 bp), Up-loxP1-F/Dn-loxP1-R (473 bp), and Up-loxP2-F/Dn-loxP2-R3 (521 bp), respectively.
Analysis of Mutations
To analyze mutations of CDPK10, we amplified a fragment surrounding the target site of CDPK10 by PCR using gene-specific primers CDPK10-F/R. We submitted purified PCR products for direct sequencing with primer CDPK10-F. Biallelic, heterozygous, and chimeric mutations with multiple superimposed sequencing chromatograms were analyzed using the Web tool DSDecode (Liu et al., 2015).
Accession Numbers
Sequence data from this article can be found in Supplemental Data Set S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Transformation efficiencies of the MR vectors.
Supplemental Table S2. SSR efficiencies of the MR modules.
Supplemental Table S3. Transformation efficiencies of the two CRISPR/Cas9 vectors.
Supplemental Table S4. Mutation efficiencies of the two CRISPR/Cas9 vectors and SSR efficiency of the MR module.
Supplemental Table S5. Transformation efficiencies of the MR-harboring ternary vectors.
Supplemental Table S6. SSR efficiencies of the MR module in the ternary vectors.
Supplemental Table S7. T-DNA copy number in 55 SSR-C and eight SSR-P lines.
Supplemental Table S8. Primers used in this study.
Supplemental Data Set S1. Annotated sequences of the vectors.
Supplemental Materials and Methods S1. Vector construction.
ACKNOWLEDGMENTS
The seeds were created by the Maize Functional Genomic Platform of China Agricultural University. We thank our colleagues from the platform for help in maize transformation.
Footnotes
This work was supported by grants from the National Crop Breeding Fund (grant no. 2016YFD0101804), the National Natural Science Foundation of China (grant nos. 31872678 and 31670371), and the National Transgenic Research Project (grant no. 2016ZX08009002).
Articles can be viewed without a subscription.
References
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, et al. (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28: 1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Bass SH, Wu E, Wang N, McBride KE, Annaluru N, Miller M, Hua M, Jones TJ (2018) An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol Biol 97: 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA. (2017) Editing the phenotype: A revolution for quantitative genetics. Cell 171: 269–270 [DOI] [PubMed] [Google Scholar]
- Butt H, Eid A, Momin AA, Bazin J, Crespi M, Arold ST, Mahfouz MM (2019) CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol 20: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Anand A, Wu E, Sander JD, Simon MK, Zhu W, Sigmund AL, Zastrow-Hayes G, Miller M, Liu D, et al. (2018) Developing a flexible, high-efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnol J 16: 1388–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70: 667–697 [DOI] [PubMed] [Google Scholar]
- Collier R, Dasgupta K, Xing YP, Hernandez BT, Shao M, Rohozinski D, Kovak E, Lin J, de Oliveira MLP, Stover E, et al. (2017) Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J 90: 1014–1025 [DOI] [PubMed] [Google Scholar]
- Demirer GS, Zhang H, Matos JL, Goh NS, Cunningham FJ, Sung Y, Chang R, Aditham AJ, Chio L, Cho MJ, et al. (2019) High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol 14: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton CM. (1997) A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene 200: 107–116 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hua K, Tao X, Han P, Wang R, Zhu JK (2019) Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant 12: 1003–1014. [DOI] [PubMed]
- Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, et al. (2019) One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37: 287–292 [DOI] [PubMed] [Google Scholar]
- Kwak SY, Lew TTS, Sweeney CJ, Koman VB, Wong MH, Bohmert-Tatarev K, Snell KD, Seo JS, Chua NH, Strano MS (2019) Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat Nanotechnol 14: 447–455 [DOI] [PubMed] [Google Scholar]
- Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, Van Eck J, Lippman ZB (2018) Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plants 4: 766–770 [DOI] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C (2018) Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36: 1160–1163 [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, et al. (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8: 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Liu YG, Xu X, Li B (2003) Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proc Natl Acad Sci USA 100: 5962–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Nannas NJ, Fu FF, Shi J, Aspinwall B, Parrott WA, Dawe RK (2019) Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 31: 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie X, Ma X, Li J, Chen J, Liu YG (2015) DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant 8: 1431–1433 [DOI] [PubMed] [Google Scholar]
- Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W (2018) Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev Biol Plant 54: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, et al. (2016) Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28: 1998–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ye X, Guo R, Huang J, Wang W, Tang J, Tan L, Zhu JK, Chu C, Qian Y (2017) Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol Plant 10: 1242–1245 [DOI] [PubMed] [Google Scholar]
- Meng X, Yu H, Zhang Y, Zhuang F, Song X, Gao S, Gao C, Li J (2017) Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol Plant 10: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Mookkan M, Nelson-Vasilchik K, Hague J, Zhang ZJ, Kausch AP (2017) Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep 36: 1477–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai N. (2013) Review: Plant binary vectors of Ti plasmid in Agrobacterium tumefaciens with a broad host-range replicon of pRK2, pRi, pSa or pVS1. Am J Plant Sci 4: 932–939 [Google Scholar]
- Ran Y, Liang Z, Gao C (2017) Current and future editing reagent delivery systems for plant genome editing. Sci China Life Sci 60: 490–505 [DOI] [PubMed] [Google Scholar]
- Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H (2019) Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant 12: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480.e478 [DOI] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7: 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole V, Worland B, Snape JW, Vain P (2007) The pCLEAN dual binary vector system for Agrobacterium-mediated plant transformation. Plant Physiol 145: 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda E, Koiso N, Takebayashi A, Ichikawa M, Kiba T, Osakabe K, Osakabe Y, Sakakibara H, Kato N, Okamoto T (2019) An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat Plants 5: 363–368 [DOI] [PubMed] [Google Scholar]
- Wang B, Zhu L, Zhao B, Zhao Y, Xie Y, Zheng Z, Li Y, Sun J, Wang H (2019) Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol Plant 12: 597–602 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Demirer GS, Zhang H, Ye T, Goh NS, Aditham AJ, Cunningham FJ, Fan C, Landry MP (2019) DNA nanostructures coordinate gene silencing in mature plants. Proc Natl Acad Sci USA 116: 7543–7548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Wang XH, Dong L, Wang ZP, Liu B, Lv J, Xing HL, Han CY, Wang XC, Chen QJ (2017) MISSA 2.0: An updated synthetic biology toolbox for assembly of orthogonal CRISPR/Cas systems. Sci Rep 7: 41993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Liu M, Tan M, Gao J, Shen Z (2014) Expression of Cry1Ab and Cry2Ab by a polycistronic transgene with a self-cleavage peptide in rice. PLoS ONE 9: e110006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Meng Z, Wang Y, Chen W, Sun C, Cui B, Cui J, Yu M, Zeng Z, Guo S, et al. (2017) Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat Plants 3: 956–964 [DOI] [PubMed] [Google Scholar]
- Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, et al. (2019) Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant 12: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36: 1211–1216 [DOI] [PubMed] [Google Scholar]