Abstract
Aims:
Acinic cell carcinoma of the breast (ACC) is a rare histologic form of triple-negative breast cancer (TNBC). Despite its unique histology, targeted sequencing analysis has failed to identify recurrent genetic alterations other than those found in common forms of TNBC. Here, we subjected three breast ACCs to whole-exome and RNA-sequencing, seeking to define whether they would harbor a pathognomonic genetic alteration.
Methods and Results:
DNA and RNA samples from three breast ACCs were subjected to whole-exome sequencing and RNA-sequencing, respectively. Somatic mutations, copy number alterations, mutational signatures and fusion genes were determined using state-of-the-art bioinformatics methods. Our analyses revealed TP53 hotspot mutations associated with loss of heterozygosity of the wild-type allele in two cases. Mutations affecting homologous recombination (HR) DNA repair-related genes were found in two cases, and an MLH1 pathogenic germline variant was detected in one case. In addition, copy number analysis revealed the presence of a somatic BRCA1 homozygous deletion and focal amplification of 12q14.3–12q21.1, encompassing MDM2, HMGA2, FRS2 and PTPRB. No oncogenic in-frame fusion transcript was identified in the three breast ACCs analyzed.
Conclusions:
No pathognomonic genetic alterations were detected in the ACCs analyzed. These tumors have somatic genetic alterations similar to those of common forms of TNBC and may display HR deficiency or microsatellite instability. These findings provide further insights as to why ACCs which are usually clinically indolent may evolve into or in parallel with high-grade TNBC.
Keywords: breast cancer, massively parallel sequencing, acinic cell carcinoma, DNA damage repair
INTRODUCTION
Acinic cell carcinoma of the breast (ACC) is an exceedingly rare special histologic type of breast cancer.1 ACCs of the breast are morphologically similar to their salivary gland conterparts, and are characterized by infiltrative microglandular or solid-nest structures composed of cells with diffuse serous differentiation with abundant eosinophilic to amphophilic cytoplasm and coarse or fine granules resembling Paneth cells.2 Areas composed of clear cells with hypernephroid appearance or non-specific glandular cells may be present.2, 3 Despite their triple negative phenotype, pure ACCs of the breast are low-grade carcinomas that usually follow an indolent clinical behavior,2 but may, nonetheless, be associated with or progress to high-grade triple negative breast cancer (TNBC).3, 4
Previous studies from our group3, 5 and others4, 6 revealed that breast ACCs, microglandular adenosis (MGA) and atypical MGA, which show marked phenotypic overlap, display genetic alterations characteristic of common forms of TNBC, including complex patterns of copy number alterations (CNAs) and highly recurrent TP53 mutations. These observations suggest that these entities may represent a low-grade triple-negative breast neoplasia family with no or minimal metastatic potential even when not associated with high-grade TNBC.5, 7
Notwithstanding their unique phenotype and previous efforts to characterize them by copy number and targeted sequencing analyses, no pathognomonic genetic alterations have been identified underpinning ACCs of the breast. Therefore, we sought to investigate the repertoire of somatic genetic alterations of pure ACCs of the breast by using whole-exome sequencing (WES) and RNA-sequencing. WES allowed for the detection of genetic alterations in genes not surveyed in previous studies of breast ACCs employing targeted sequencing panels. Additionally, RNA-sequencing analysis was performed to define whether pure ACC of the breast would harbor a highly recurrent fusion gene.
METHODS
Subjects and samples
Following Institutional Review Boards approval, formalin-fixed paraffin-embedded tissue blocks of breast pure ACCs were retrieved from the archives of the Department of Pathology of the Nottingham University City Hospital (Nottingham, UK). Samples were anonymized and reviewed by three pathologists (F.P., E.G-R. and J.S.R.-F). Of the two pure ACCs previously subjected to targeted capture sequencing as reported in Guerini-Rocco et al.3, only case ACC12 was included in the present study due to tissue and nucleic acids availability. Cases ACC1 and ACC18 have not been previously reported and are unique to this study.
WES and RNA-Sequencing analysis
DNA was extracted from microdissected representative tumor and normal breast tissue, as previously described,8 and subjected to WES at the Integrated Genomics Operations (IGO) of Memorial Sloan Kettering Cancer Center (MSKCC), as previously described.9–11 Tumor RNA samples were subjected to RNA-sequencing at the MSKCC’s IGO, as previously described.10, 11 Detailed analysis methods are described in the Supplementary Methods.
RESULTS
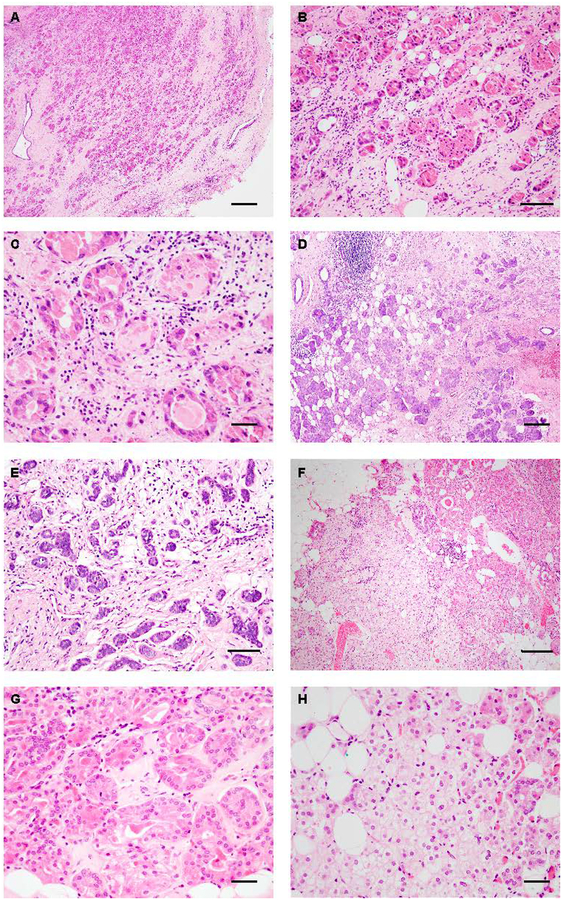
All breast ACCs studied here featured an infiltrative microglandular growth pattern and cytoplasmic eosinophilic Paneth cell-like granules (Figure 1A–1H). In addition, ACC18 focally displayed areas composed of clear cells with hypernephroid appearance (Figure 1H). All ACCs were of histologic grade 1, not associated with high-grade TNBC or any other lesions, including microglandular adenosis and lacked estrogen receptor (ER) and HER2 expression (Supplementary Table 1).
Figure 1. Histologic characteristics of acinic cell carcinomas of the breast.
(A-B) Representative photomicrographs of hematoxylin and eosin (H&E)-stained acinic cell carcinomas of the breast (ACCs) from this study. (A-C) ACC1 displays an infiltrative growth (A) with microglandular features (B) and is composed of Paneth-like cells with coarse intracytoplasmic granules (C). (D-E) ACC12 displays a microglandular growth pattern (D) with cells featuring an amphophilic cytoplasm with fine granules (E). (F-H) ACC18 displays microglandular areas (F) composed of eosinophilic cells with coarse granules (G) and hypernephroid areas composed of clear cells (H). Scale bar, 200 μm (A, D), 50 μm (B, and E), 20 μm (C, G and H) and 100 μm (F).
To determine whether breast ACCs would be underpinned by a pathognomonic genetic alteration, these cases were subjected to RNA-sequencing and WES (Supplementary Tables 2 and 3). RNA-sequencing analysis identified potential fusion transcripts only in ACC1, all of which were out-of-frame and likely constitute passenger events (Supplementary Table 2). One of these fusion genes, an out-of-frame TC2N-FBLN5 intra-chromosomal fusion gene, displayed a low oncogenic potential as defined by OncoFuse (Supplementary Table 3) and was also found to be present in two ER-positive invasive breast carcinomas of no special type (IDC-NST) from The Cancer Genome Atlas (TCGA) dataset.12
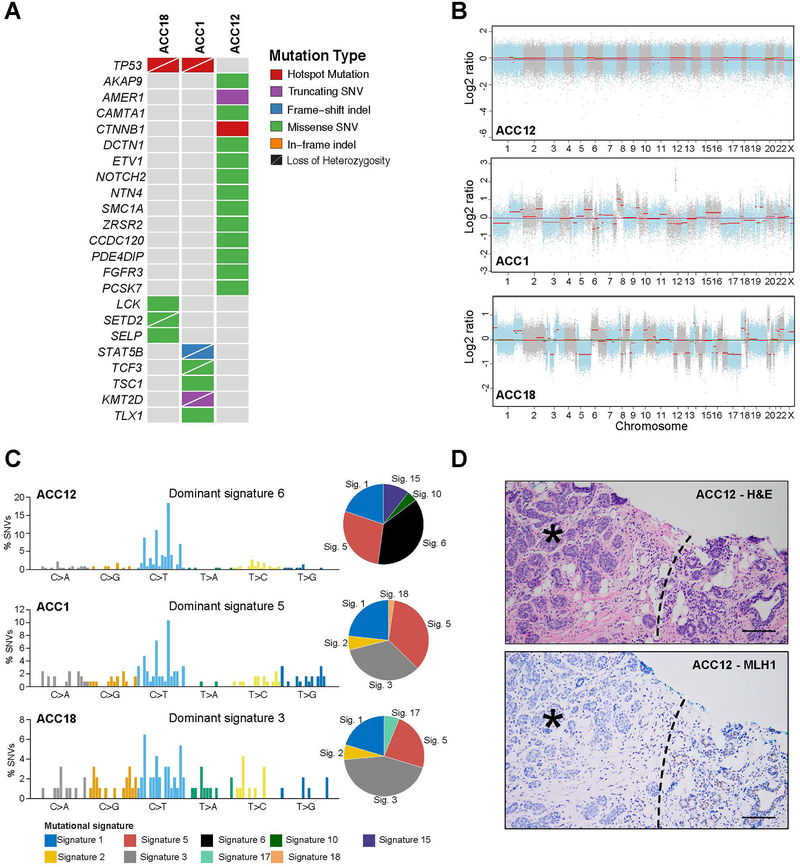
ACCs displayed a median of 173 (range, 92–230) non-synonymous somatic mutations as defined by WES (Supplementary Table 4), several affecting cancer genes (Figure 2A). Our analysis revealed clonal TP53 hotspot mutations associated with loss of heterozygosity (LOH) of the wild-type allele in two ACCs. In contrast, the TP53 wild-type ACC (ACC12) harbored a pathogenic MLH1 germline mutation (c.790+2dupT) and a clonal hotspot mutation in CTNNB1 (c.1004A>T, Figure 2A, Supplementary Figure 1 and Supplementary Table 4). Other likely pathogenic mutations included a truncating mutation in KMT2D (MLL2), a histone methyltranferase whose inactivation results in genome instability13 and is frequently mutated in common forms of TNBC14, associated with LOH in case ACC1 (Figure 2A, Supplementary Figure 1 and Supplementary Table 4). No cancer gene other than TP53 was found to be mutated in >1 of the ACCs studied here.
Figure 2. Repertoire of somatic mutations and mutational signatures of the acinic cell carcinomas of the breast.
(A) Non-synonymous somatic mutations affecting cancer-related genes13–15 and mutations shared among cases identified in the acinic cell carcinomas of the breast (ACCs; n=3) subjected to whole-exome sequencing (WES). Cases are shown in columns and genes in rows. (B) Copy number plots depicting segmented Log2 ratios (y-axis) plotted according to genomic position (x-axis). Chromosomes are demarcated by alternating blue and gray colors (C) Mutational signatures of all somatic SNVs in breast ACCs (n=3). Pie charts indicate the proportion of the different mutational signatures identified in each case. (D) Representative hematoxylin and eosin micrograph of ACC12 arising in a patient with an MLH1 germline mutation (top) and micrograph depicting loss of MLH1 expression in the tumor cells (*). Normal breast (right lower corner) shows retention of MLH1 expression. Dashed line Scale bar, 50 μm. SNV, single nucleotide variant. Sig, signature; SNV, single nucleotide variant.
Copy number analysis revealed that ACC1 and ACC18 displayed complex copy number profiles, with multiple gains and losses and focal high-level amplifications, in contrast, ACC12 showed a rather quiet copy number profile (Figure 2B). Copy number alterations (CNAs) present in both ACC1 and ACC18 included gains of 1q, 2q and 8q and losses of 3p, 12p, 12q, 14q, 17p and 17q. In line with previous studies reporting ACCs arising in BRCA1 germline mutation carriers, we identified a somatic homozygous deletion in 17q21.31 encompassing BRCA1 in ACC18 (Figure 1B). In addition, we observed that ACC18 harbored a focal amplification in 20p12.3 encompassing PCNA, which encodes for Proliferating Cell Nuclear Antigen, a key promoter of processive DNA synthesis.15 ACC1 was found to harbor a high-level amplification of 12q14.3–12q21.1, which encompasses several cancer genes, such as MDM2, HMGA2, WIF1, FRS2 and PTPRB. In contrast, and consistent with its DNA mismatch repair deficiency, ACC12 displayed a simple genome without detectable copy number alterations.
We next sought to determine whether breast ACCs would display genomic features suggestive of HRD or other biological processes that would confer genomic instability. ACC1 was found to display a dominant signature 5, ascribed to aging (Figure 2C). ACC18, which harbored a BRCA1 homozygous deletion, displayed genomic features suggestive of HRD, including a dominant signature 3 (HRD-related),16 along with a high large-scale state transition (LST) score (24)17, a high telomeric allelic imbalance (NtAI) score (23)18 and a high number of ‘small deletions’ >5bps (Figure 2C and Supplementary Table 5). Although we did not identify somatic mutations in MLH1 or in other core MMR genes, ACC12, which harbored a pathogenic germline splice site mutation in MLH1 (c.790+2dupT), a key tumor suppressor of the mismatch repair (MMR) system (Supplementary Table 4), displayed a dominant mutational signature 6, ascribed to defective MMR19 (Figure 2C), as well as high levels of microsatellite instability (MSI-H) as determined by MSIsensor20 (Supplementary Table 5). Consistent with these findings, this case additionally showed loss of MLH1 protein expression in the tumor by immunohistochemistry (Figure 2D).
DISCUSSION
Previous studies from our group and others have suggested that ACCs of the breast and MGA, entities with overlapping histologic characteristics, are part of the spectrum of low-grade triple negative disease, and harbor genomic features indistinguishable from those of common forms of TNBC3, 5, 7, 21. Further supporting this notion, our study revealed few recurrently mutated genes, such as TP53, and complex copy number profiles.
Most importantly, our findings provide further support to the association between breast ACC and HRD through BRCA1 inactivation. Our results demonstrate that ACC18 harbors a BRCA1 homozygous deletion (Figure 2C). In conjunction with previous reports by our group and others3, 22, loss-of-function alterations affecting BRCA1 concurrent with TP53 somatic mutations seem not to be uncommon in breast ACCs, even in those lacking a high-grade TNBC component. Our findings, however, suggest the tantalizing possibility that both BRCA1 and TP53 loss of function may not be sufficient for a TNBC to display high-grade features and that inactivation of these two genes may not sufficient for the development of high-grade TNBC.3, 5
Here we also described an ACC (ACC12) that lacked mutations affecting TP53, displayed a simple copy number profile, high MSI levels and a dominant signature 6 (MSI-related). This case arose in a patient carrier of a germline MLH1 splice-site mutation. Although we did not identify a somatic genetic alteration in MLH1, we cannot rule out that the second MLH1 allele could have been inactivated by epigenetic silencing via promoter hypermethylation, as described in colorectal and endometrial carcinoma.23, 24
Our study has important limitations, including the small sample size, due to the rarity of this entity and the fact that only archival samples were available for analysis. We were unable to perform any methylation analyses to interrogate epigenetic silencing as a mechanism of inactivation of the second MLH1 allele in ACC12 owing to the lack of residual DNA from this case. Despite these limitations, our data lend further support to the notion that ACCs of the breast are genetically heterogeneous and display genomic features overlapping with those of common forms of TNBCs. These tumors appear not to be driven by a highly recurrent mutation or oncogenic fusion gene. Most importantly, our findings suggest that at least some ACCs of the breast may arise in the setting of HRD or MSI through distinct molecular mechanisms. Even though we could not establish a definitive causal link between BRCA1 mutations or MLH1 germline mutations and breast ACCs, our study demonstrates that HDR deficiency and MSI-H happen in ACCs, and that the ACCs analyzed displayed high levels of genetic instability (either HDR defects or MSI-H). Additional studies on the genetic or epigenetic basis of breast ACCs are warranted.
Supplementary Material
Supplementary Methods
Supplementary Figure 1. Cancer cell fractions of non-synonymous somatic mutations affecting cancer-related genes identified in the acinic cell carcinomas (ACCs) of the breast by whole-exome sequencing.
Supplementary Table 1. Clinicopathologic characteristics of the acinic cell carcinomas of the breast included in this study.
Supplementary Table 2. Fusion genes identified by RNA-sequencing analysis of acinic cell carcinomas of the breast
Supplementary Table 3. Whole-exome Sequencing statistics.
Supplementary Table 4. Non-synonymous somatic mutations identified in the acinic cell carcinomas of the breast by whole-exome sequencing.
Supplementary Table 5. Genomic features of homologous recombination deficiency and microsatellite instability in the acinic cell carcinomas of the breast included in this study
ACKNOWLEDGEMENTS
This study was funded by the Breast Cancer Research Foundation. FB is supported by the Department of Pathology of the Stanford University School of Medicine Pathology Trainee Mentored Award in Precision Health and Earlier.org - Friends for An Earlier Breast Cancer Test® Medical Research Grant. BW is funded by a Cycle for survival grant. Research reported in this paper was supported in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant No P30CA008748).
Footnotes
Conflict of interest: JSR-F reports personal/consultancy fees from Goldman Sachs, REPARE Therapeutics, VolitionRx, Page.AI, Roche Tissue Diagnostics, Ventana Medical Systems, Novartis and Genentech, outside the scope of the submitted work. E.G.R. received honoraria from Thermo Fisher Scientific, Biocartis, Roche, MSD, AstraZeneca, Novartis, outside the scope of the submitted work. RGM reports personal/consultancy fees from Oxford Genetics, outside the scope of this work. All other authors declare no conflicts of interest.
REFERENCES
- 1.Roncaroli F, Lamovec J, Zidar A, Eusebi V. Acinic cell-like carcinoma of the breast. Virchows Arch 1996;429;69–74. [DOI] [PubMed] [Google Scholar]
- 2.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. Who classification of breast tumors. IARC: Lyon, 2012. [Google Scholar]
- 3.Guerini-Rocco E, Hodi Z, Piscuoglio S et al. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: An exploratory, hypothesis-generating study. J Pathol 2015;237;166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conlon N, Sadri N, Corben AD, Tan LK. Acinic cell carcinoma of breast: Morphologic and immunohistochemical review of a rare breast cancer subtype. Hum Pathol 2016;51;16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geyer FC, Berman SH, Marchio C et al. Genetic analysis of microglandular adenosis and acinic cell carcinomas of the breast provides evidence for the existence of a low-grade triple-negative breast neoplasia family. Mod Pathol 2017;30;69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kahn R, Holtveg H, Nissen F, Holck S. Are acinic cell carcinoma and microglandular carcinoma of the breast related lesions? Histopathology 2003;42;195–196. [DOI] [PubMed] [Google Scholar]
- 7.Geyer FC, Pareja F, Weigelt B et al. The spectrum of triple-negative breast disease: High- and low-grade lesions. Am J Pathol 2017;187;2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martelotto LG, De Filippo MR, Ng CK et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol 2015;237;179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geyer FC, Li A, Papanastasiou AD et al. Recurrent hotspot mutations in hras q61 and pi3k-akt pathway genes as drivers of breast adenomyoepitheliomas. Nat Commun 2018;9;1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pareja F, Brandes AH, Basili T et al. Loss-of-function mutations in atp6ap1 and atp6ap2 in granular cell tumors. Nat Commun 2018;9;3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pareja F, Lee JY, Brown DN et al. The genomic landscape of mucinous breast cancer. J Natl Cancer Inst 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Wang Q, Tang M et al. Tumorfusions: An integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 2018;46;D1144–D1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantidakis T, Saponaro M, Mitter R et al. Mutation of cancer driver mll2 results in transcription stress and genome instability. Genes Dev 2016;30;408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weisman PS, Ng CK, Brogi E et al. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod Pathol 2016;29;476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe KN, Moldovan GL. Forging ahead through darkness: Pcna, still the principal conductor at the replication fork. Mol Cell 2017;65;380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nik-Zainal S, Davies H, Staaf J et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534;47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popova T, Manie E, Rieunier G et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with brca1/2 inactivation. Cancer Res 2012;72;5454–5462. [DOI] [PubMed] [Google Scholar]
- 18.Birkbak NJ, Wang ZC, Kim JY et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012;2;366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nik-Zainal S, Morganella S. Mutational signatures in breast cancer: The problem at the DNA level. Clin Cancer Res 2017;23;2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niu B, Ye K, Zhang Q et al. Msisensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30;1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen PP. So-called acinic cell carcinoma of the breast arises from microgladular adenosis and is not a distinct entity. Mod Pathol 2017;30;1504. [DOI] [PubMed] [Google Scholar]
- 22.Ripamonti CB, Colombo M, Mondini P et al. First description of an acinic cell carcinoma of the breast in a brca1 mutation carrier: A case report. BMC Cancer 2013;13;46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine AJ, Phipps AI, Baron JA et al. Clinicopathologic risk factor distributions for mlh1 promoter region methylation in cimp-positive tumors. Cancer Epidemiol Biomarkers Prev 2016;25;68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varley KE, Mutch DG, Edmonston TB, Goodfellow PJ, Mitra RD. Intra-tumor heterogeneity of mlh1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res 2009;37;4603–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Supplementary Figure 1. Cancer cell fractions of non-synonymous somatic mutations affecting cancer-related genes identified in the acinic cell carcinomas (ACCs) of the breast by whole-exome sequencing.
Supplementary Table 1. Clinicopathologic characteristics of the acinic cell carcinomas of the breast included in this study.
Supplementary Table 2. Fusion genes identified by RNA-sequencing analysis of acinic cell carcinomas of the breast
Supplementary Table 3. Whole-exome Sequencing statistics.
Supplementary Table 4. Non-synonymous somatic mutations identified in the acinic cell carcinomas of the breast by whole-exome sequencing.
Supplementary Table 5. Genomic features of homologous recombination deficiency and microsatellite instability in the acinic cell carcinomas of the breast included in this study