Abstract
Formation of the calvaria is a multi-staged process and is regulated by multiple genetic factors. Disruption of normal calvarial development usually causes craniosynostosis, a prevalent birth defect characterized by premature fusion of calvarial bone. Recent studies have identified mutations of KMT2D allele in patients with craniosynostosis, indicating a potential role for Kmt2d in calvarial development. KMT2D mutations have also been implicated in Kabuki syndrome, which features a distinct facial appearance, skeletal abnormality, growth retardation and intellectual disability. However, the expression pattern of Kmt2d has not been fully elucidated. In the present study we examined the expression pattern of Kmt2d at multiple stages of embryo development in mice, with a focus on the craniofacial tissues. Our in situ hybridization results showed that Kmt2d mRNA is expressed in the developing calvarial osteoblasts, epithelia and neural tissues. Such an expression pattern is in line with the phenotype in Kabuki syndrome, suggesting that Kmt2d plays an intrinsic role in normal development and homeostasis of these craniofacial tissues.
Keywords: Kmt2d, in situ hybridization, calvarial bones, mouse
Introduction
Craniosynostosis is a prevalent birth defect with occurrence of 1/2500 worldwide(Johnson and Wilkie, 2011; Senarath-Yapa et al., 2012). It is diagnosed by premature fusion of calvarial bones, which are normally connected by fibrous sutures. Calvarial development is a highly coordinated process from progenitors of two distinct origins. The calvarium is composed of a pair of frontal bones and parietal bones and a single interparietal bone. While the frontal bones and the central portion of the interparietal bone are originated from the neural crest, the parietal bones and the lateral portion of the interparietal bone are derived from the mesoderm cells(Yoshida et al., 2008). In mice, development of the frontal bones and parietal bones begins at E10.5. The progenitor cells undergo extensive proliferation and differentiation when they migrate along the basal-apical axis towards the supraorbital domain(Deckelbaum et al., 2012). These osteoblast progenitors can be well identified by alkaline phosphatase activity and Runx2 expression beginning at E12.5–E13.5. Subsequently, the frontal bone primordia expand rostrally and the parietal bone primordia grow caudally while separated by the sutural cells. In following stages, these primordia undergo further expansion and differentiation, and form the calvarial vault encasing the developing brain(Deckelbaum et al., 2012; Ishii et al., 2015).
The calvarium development is an intricate process regulated by a genetic network composed of multiple factors. Disruption of genetic regulation could result in a variety of calvarial defects such as craniosynostosis (Ishii et al., 2015; Johnson and Wilkie, 2011). While many genes have been implicated in regulation of calvarial development, novel mutations have been identified in recent researches, suggesting that the mechanisms underlying normal calvarial development remain further elucidated. Kmt2d gene encodes a H3K4 methyltransferase, and mutations in KMT2D allele have been identified in Kabuki syndrome, which is characterized by distinguished facial features and craniosynostosis(Ng et al., 2010; Niikawa et al., 1988). Multiple independent reports have shown that KMT2D mutations are correlated to craniosynostosis in human(Armstrong et al., 2005; Topa et al., 2017; Zollino et al., 2017). However, it remains unclear whether KMT2D regulates calvarial development directly or via an indirect manner. To address this question, we have examined expression pattern of Kmt2d mRNA in the developing mouse embryos, with a focus on the craniofacial tissues.
Our in situ hybridization results with Kmt2d antisense probe show that Kmt2d is expressed in multiple cell contexts during craniofacial development, and its expression is detected in the calvarial osteoblasts in multiple stages, indicating Kmt2d plays an autonomous role in calvarial morphogenesis.
2. Results
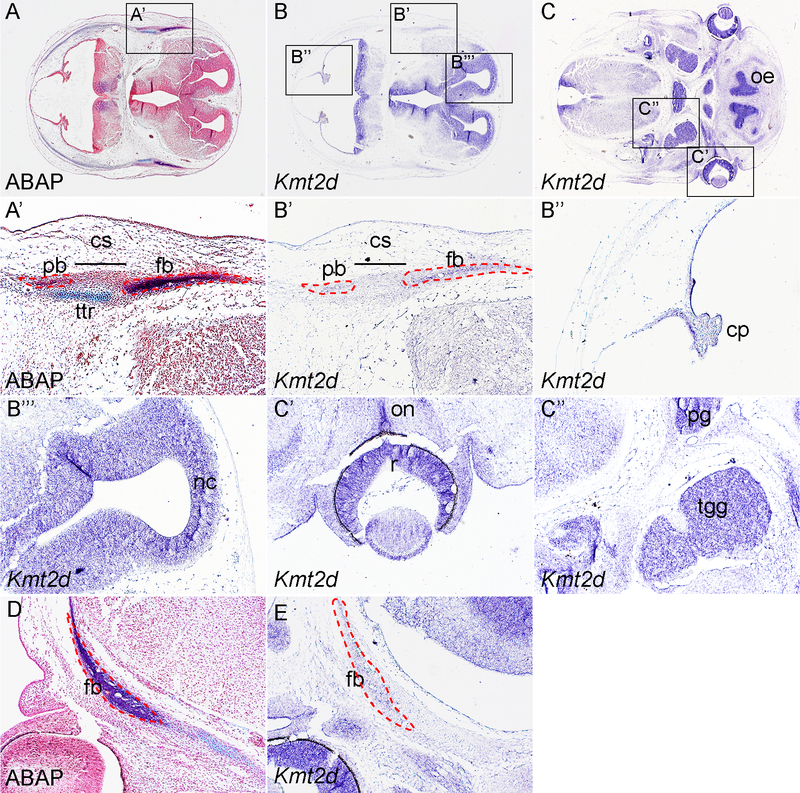
To examine the expression pattern of Kmt2d in the craniofacial tissues, we have carried out in situ hybridization using Kmt2d antisense probe on sections of C57BL6J mice at multiple stages. At E13.5, alcian blue/alkaline phosphatase (ABAP) staining reveals the expression pattern of the osteoprogenitors of the frontal bone and parietal bone separated by the coronal suture (Fig 1A, A’). Our data show that Kmt2d mRNA is expressed in both primordia, indicating Kmt2d is implicated in the early osteogenesis of the calvarial bones(Fig 1B, 1B’). This expression pattern is also confirmed by in situ hybridization data in frontal sections of E13.5 embryos (Fig 1D, E). In addition, Kmt2d transcripts are also detected in other cell types in the craniofacial region, especially in the neuronal tissues. These includes the choroid plexus(Fig 1B”), the roof of neopallial cortex(Fig 1B”#x2019;), the olfactory epithelium, the retina and optic nerve, the trigeminal ganglia and the pituitary gland(Fig 1C – C”).
Fig 1. Expression pattern of Kmt2d in E13.5 mouse embryo.
(A, A’) ABAP staining on cross section of E13.5 wild-type embryonic head at the level of calvarial primordia. A’ is inset of A. (B) In situ hybridization using Kmt2d antisense probe on cross section at the same level of A. B” and B” are insets of B. (C) Expression pattern of Kmt2d on cross section at level of the retina. C’ and C” are insets of C. (D) ABAP staining on frontal section of E13.5 wild-type embryonic head. (E) In situ hybridization using Kmt2d antisense probe on frontal section at comparable level of D. cp, choroid plexus; cs, coronal suture; fb, frontal bone; nc, neopallial cortex; oe, olfactory epithelium; on, optic nerve; pb, parietal bone; pg, pituitary gland; r, retina; tgg, trigeminal ganglion; ttr, tectum transversum.
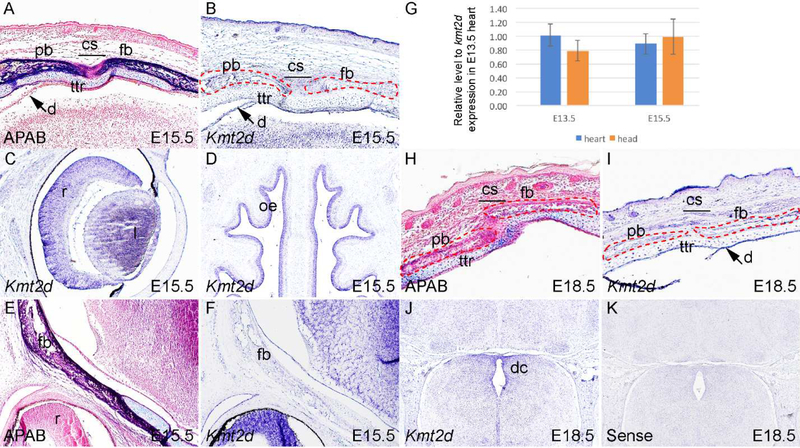
At E15.5, ABAP staining shows that the frontal bone and parietal bone approach approximately, while the tectum transversum, the lateral portion of the developing chondrocranium is located between the calvarium and the dura (Fig 2A). At the same level, Kmt2d is expressed in the ectoderm, the osteoblasts of the frontal bone and parietal bone, and the dura (Fig 2B). Basal level of Kmt2d mRNA is also detected in the ttr chondrocyte and the neurons of the cerebral cortex (Fig 2B). In consistent to its expression pattern at E13.5, Kmt2d transcripts are also identified in the retina, lens, and ectoderm of nasal septum (Fig 2C, D), with many of which enriched with neurons. Kmt2d expression in the osteoblasts, lens and neurons are also confirmed by in situ hybridization results in frontal sections at same stage (Fig 2E, F). It has been documented that kmt2d is expressed in the heart and is required for embryonic heart development(Ang et al., 2016). Comparison of kmt2d expression in the embryonic heart and head at E13.5 and E15.5 by q-PCR reveals that kmt2d exhibits comparable expression level in these tissues (Fig 2G).
Fig 2. Kmt2d expression in mouse embryos at E15.5 and E18.5.
(A) ABAP staining of mouse embryonic head at E15.5 at the level of calvarial primordia. (B–D) In situ hybridization using Kmt2d antisense probe on cross section at the same level with A. (E) ABAP staining in frontal section of mouse embryonic head at E15.5. (F) In situ hybridization using Kmt2d antisense probe on frontal section at the same level with A. (G) Quantitative PCR results showing relative expression level of Kmt2d mRNA in the embryonic heart and head at E13.5 (n=10) and E15.5 (n=9). (H) ABAP staining of mouse embryonic head at E18.5 at the level of coronal suture. (I) Expression pattern of Kmt2d at the same level of H. (J) Expression pattern of Kmt2d in the brain tissues. (K) In situ hybridization using Kmt2d sense probe on cross section at the same level with G, serving a negative control. cs, coronal suture; dc, diencephalon; fb, frontal bone; l, lens; oe, olfactory epithelium; pb, parietal bone; r, retina; ttr, tectum transversum.
At E18.5, the calvarial osteoblasts undergo further differentiation and crystallization is initiated. As a result, AP activity of these cells starts to decline when compared to their E15.5 counterparts in the frontal bone and parietal bone (Fig 2H). At this stage, Kmt2d exhibits an expression pattern similar to that of E15.5 in the ectoderm and osteoblast, but its expression level dramatically increases in the ttr chondrocytes and the dura (Fig 2I). In the developing brain, Kmt2d-expressing cells are dispersed throughout a wide region, but are clustered in the ependymal lining of the third ventricle and the floor of the diencephalon (Fig 2J). On the other hand, the sense probe fails to detect obvious expression, validating the experimental data obtained with Kmt2d anti-sense probe (Fig 2K).
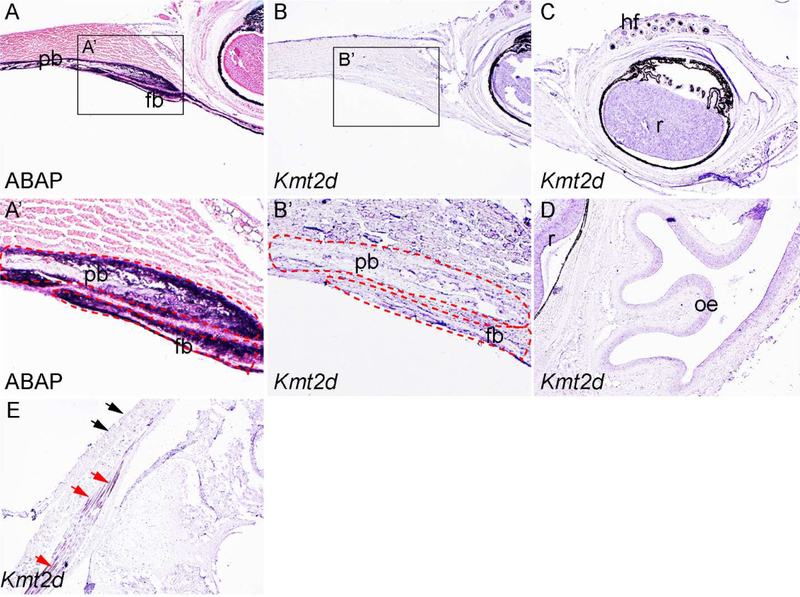
Next we asked whether Kmt2d is expressed in calvarial bones at postnatal stages. To this end we have performed in situ hybridization in postnatal (P) 5 days old samples. At this stage, the boundary of frontal bone and parietal bone can be easily identified with AP staining (Fig 3A, A’). Our in situ hybridization data show that Kmt2d expression persists in the ectoderm, but dramatically decreased in the calvarial osteoblasts (Fig 3B, B’). At this stage, Kmt2d is also expressed in the hair follicle, retina and olfactory epithelium (Fig 3C, D). Kmt2d has been shown important for muscle development (Lee et al., 2013). In line with this report, we have detected Kmt2d expression in muscle cells adjacent to the interparietal bone (red arrows in Fig 3E). Our data also reveal that Kmt2d expression exhibits a restricted pattern in the craniofacial epithelial cells, as we detected high level Kmt2d expression in the epithelium next to the eye (Fig 3B), no obvious signal was identified in the epithelial cells adjacent to the interparietal bone (black arrows in Fig 3E).
Fig 3. Expression pattern of Kmt2d at P5.
(A, A’) ABAP staining on cross section of P5 mouse head at the level of coronal suture. A’ is inset of A. (B, B’) In situ hybridization using Kmt2d antisense probe on cross section at the same level with A. B’ is the inset of B. (C, D) Expression pattern of Kmt2d on the cross section at the level of retina. (E) Kmt2d expression in the craniofacial muscle cells. Red arrows, Kmt2d expressing cells in muscle fibers adjacent to interparietal bone and hindbrain. Black arrows, epithelial cells showing negative results of Kmt2d expression. fb, frontal bone; hf, hair follicle; oe, olfactory epithelium; pb, parietal bone; r, retina.
3. Discussion
In this study, we have investigated the expression pattern of Kmt2d in a systematic fashion at multiple stages in mice. Since frequent mutations in KMT2D allele are found in Kabuki Syndrome, which features distinct facial malformation and craniosynostosis, our study focuses on the craniofacial tissues. Our results show that Kmt2d is expressed in the calvarial osteoblasts at different stages, indicating Kmt2d plays an intrinsic role in calvarial bone development. Our data also discovered Kmt2d expression in the retina, olfactory epithelium, and brain tissues, which are enriched with neurons, suggesting a role for Kmt2d in neuronal development. Our findings are in line with the phenotype of Kabuki Syndrome, which is also featured with intellectual disability and skeletal abnormalities.
Another feature of Kabuki Syndrome is dermatoglyphic abnormalities, indicating that Kmt2d is essential for epidermal development. In support of this notion, our finding confirms that Kmt2d is expressed in a restricted pattern in the epithelial tissues (Fig 2B, 2I, 3B and 3E). Tooth is also regarded as an epidermal appendage. Interestingly, a recent report shows that KMT2D is expressed in the epithelium of tooth germs (Porntaveetus et al., 2018), and provides a plausible explanation of the tooth abnormality in Kabuki Syndrome patients.
Based on the expression pattern of Kmt2d, it is interesting to analyze its role in development of these cell types. Currently multiple genetically engineered mice have been developed to study the role of Kmt2d in development and disease models. However, global inactivation of Kmt2d in mice causes early embryonic lethality at E9–E12, prevent further analysis of this gene in multiple tissues such as the calvarial bones(Bjornsson et al., 2014; Lee et al., 2013). With the availability of the conditional alleles, it has been found that Kmt2d plays essential roles in formation of multiple tissues, including cardiovascular organogenesis, adipogenesis and myogenesis via epigenetic mechanisms(Ang et al., 2016; Lee et al., 2013). It still remains unclear whether Kmt2d is crucial for normal calvarial development, and our data provide a rationale to investigate its role in this process.
4. Experimental procedures
4.1. Animals
All animal experimentation was approved by the Institutional Animal Care and Use Committee of Tulane University. C57BL/6 mice are obtained from Jackson Laboratory (Bar Harbor, ME) and are maintained in Tulane Uptown Vivarium. For embryo collection, vaginal plug was checked daily in the mating females, and noon on the day when vaginal plug was found was defined embryonic day (E) 0.5.
4.2. Histology
Staged embryos were dissected in PBS, and the embryonic heads were separated using Dumont tweezers style 5. The samples were then fixed in 4% paraformaldehyde (PFA) in PBS at 4 degree. Following overnight fixation, the embryonic heads were dehydrated by gradient ethanol washes and were embedded in paraffin. Postnatal samples were decalcified in 10% EDTA for 48 hours before dehydration and embedding. Transverse sections and coronal sections at 10 micrometers were obtained using a HM325 Thermo Fisher microtome and were subject for following analysis.
4.3. Alkaline Phosphatase/Alcian Blue (AP-AB) staining
AP-AB staining was performed as described previously(Yang et al., 2018). Briefly, sections were deparaffinized, hydrated and incubated in 0.03% nitro-blue tetrazolium chloride (NBT) and 0.02% 5-bromo-4-chloro-3’-indolyphosphate p-toluidine salt (BCIP) to detect AP activity. Slides were then rinsed in water and dipped in 1% alcian blue 8GX (A5268, Sigma) in 0.1N HCl to stain cartilage, and counterstained with 0.1% nuclear fast red solution.
4.4. In situ hybridization
A 786 bps of Kmt2d cDNA fragment was amplified using primers 5-CCGAATCAAACAGGGTCGGA −3 and 5-ACATAGGGCTGGCAGGAAAC-3 and cloned into pCR™ 2.1-TOPO® TA vector (Thermo Fisher, K450001). To generate Kmt2d antisense RNA probe, the plasmid was linearized with KpnI and was transcribed with T7 RNA polymerase. Kmt2d sense probe was generated by linearization with EcoRV and was transcribed with Sp6. In situ hybridization was performed on sections using both antisense and sense probes, with the latter serving a negative control. Experimental procedure of In situ hybridization was carried out as described previously (Yang et al., 2018).
4.5. Quantitative PCR
Expression of Kmt2d mRNA in embryonic heart and head at E13.5 and E15.5 were analyzed by real-time quantitative RT-PCR. The embryonic heart and head (with removal of brain) were dissected from staged embryos in ice cold PBS, and total RNA was extracted using the RNeasy Mini Kit (Qiagen 74104). Samples were prepared from 10 individual embryos at E13.5 and 9 at E15.5. First strand cNDA was synthesized using 1 g of total RNA, oligo-dT primers and SuperScript III RT (Invitrogen 18080). Quantitative PCR was performed on Bio-Rad iCycler using SYBR super mix kit (Bio-Rad 170–8880). The following protocol was used: step 1: 95°C for 10 min; step 2: 95°C for 10 s; step 3: 55° for 30 s; repeat to step 2 39× (40 c ycles in total). β-actin expression was used as an internal control. The primers are as follows: mouse Kmt2d, 5′-CCTCTGATGGAGTTACCGCT-3′ and 5′-GGTATGGGGCCGTTTATAGTGT-3′; mouse β-actin, 5′-GCAAGTGCTTCTAGGCGGAC-3′ and 5′-AAGAAAGGGTGTAAAACGCAGC-3’.
5. Acknowledgements
We thank Amy Pierce from Tulane University Uptown Vivarium for taking care of the animals. The authors are grateful to the He lab members for the comments. This work is supported by NIH/NIDCR grant R00DE024617 to F.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Ang SY, Uebersohn A, Spencer CI, Huang Y, Lee JE, Ge K and Bruneau BG (2016). KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development 143, 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Abd El Moneim A, Aleck K, Aughton DJ, Baumann C, Braddock SR, Gillessen-Kaesbach G, Graham JM Jr., Grebe TA, Gripp KW, et al. (2005). Further delineation of Kabuki syndrome in 48 well-defined new individuals. Am. J. Med. Genet. A 132a, 265–272. [DOI] [PubMed] [Google Scholar]
- Bjornsson HT, Benjamin JS, Zhang L, Weissman J, Gerber EE, Chen YC, Vaurio RG, Potter MC, Hansen KD and Dietz HC (2014). Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci. Transl. Med 6, 256ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckelbaum RA, Holmes G, Zhao Z, Tong C, Basilico C and Loomis CA (2012). Regulation of cranial morphogenesis and cell fate at the neural crest-mesoderm boundary by engrailed 1. Development 139, 1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Sun J, Ting MC and Maxson RE (2015). The Development of the Calvarial Bones and Sutures and the Pathophysiology of Craniosynostosis. Curr. Top. Dev. Biol 115, 131–156. [DOI] [PubMed] [Google Scholar]
- Johnson D and Wilkie AO (2011). Craniosynostosis. Eur. J. Hum. Genet 19, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. (2013). H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2, e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. (2010). Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet 42, 790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikawa N, Kuroki Y, Kajii T, Matsuura N, Ishikiriyama S, Tonoki H, Ishikawa N, Yamada Y, Fujita M, Umemoto H, et al. (1988). Kabuki make-up (Niikawa-Kuroki) syndrome: a study of 62 patients. Am. J. Med. Genet 31, 565–589. [DOI] [PubMed] [Google Scholar]
- Porntaveetus T, Abid MF, Theerapanon T, Srichomthong C, Ohazama A, Kawasaki K, Kawasaki M, Suphapeetiporn K, Sharpe PT and Shotelersuk V (2018). Expanding the Oro-Dental and Mutational Spectra of Kabuki Syndrome and Expression of KMT2D and KDM6A in Human Tooth Germs. Int J Biol Sci 14, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senarath-Yapa K, Chung MT, McArdle A, Wong VW, Quarto N, Longaker MT and Wan DC (2012). Craniosynostosis: Molecular pathways and future pharmacologic therapy. Organogenesis 8, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topa A, Samuelsson L, Lovmar L, Stenman G and Kolby L (2017). On the significance of craniosynostosis in a case of Kabuki syndrome with a concomitant KMT2D mutation and 3.2 Mbp de novo 10q22.3q23.1 deletion. Am. J. Med. Genet. A 173, 2219–2225. [DOI] [PubMed] [Google Scholar]
- Yang T, Moore M and He F (2018). Pten regulates neural crest proliferation and differentiation during mouse craniofacial development. Dev. Dyn 247, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y and Iseki S (2008). Cell lineage in mammalian craniofacial mesenchyme. Mech. Dev 125, 797–808. [DOI] [PubMed] [Google Scholar]
- Zollino M, Lattante S, Orteschi D, Frangella S, Doronzio PN, Contaldo I, Mercuri E and Marangi G (2017). Syndromic Craniosynostosis Can Define New Candidate Genes for Suture Development or Result from the Non-specifc Effects of Pleiotropic Genes: Rasopathies and Chromatinopathies as Examples. Front. Neurosci. 11, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]