Introduction
H. pylori alters the gastric microbial composition and causes reduced gastric inflammatory disease in children in Chile1–3. These findings suggest that H. pylori-associated gastric microbiota may influence the pathogenesis of H. pylori infection beginning in childhood, at least in Chile. Here we characterized the gastric microbiota in H. pylori-infected children in an entirely different geographic region of South America, namely Venezuela, where H. pylori also is endemic4, and determined the impact of H. pylori eradication on bacterial community structure.
Methods
Sixteen children (≤13 years) with nausea and abdominal discomfort without antibiotic or antacid therapy during the preceding month underwent endoscopic antral biopsy for H. pylori urease testing and histology (both required positive for infection) and microbiota analysis. Eleven subjects were infected and treated with amoxicillin, clarithromycin and omeprazole for 14 days. Two months later, the children were re-biopsied for H. pylori testing and microbiota analysis. Biopsy DNA was isolated, the V4 region of the 16S rRNA bacterial gene was amplified by PCR, and 250 base single end reads were sequenced and analyzed, as previously described3.
Results
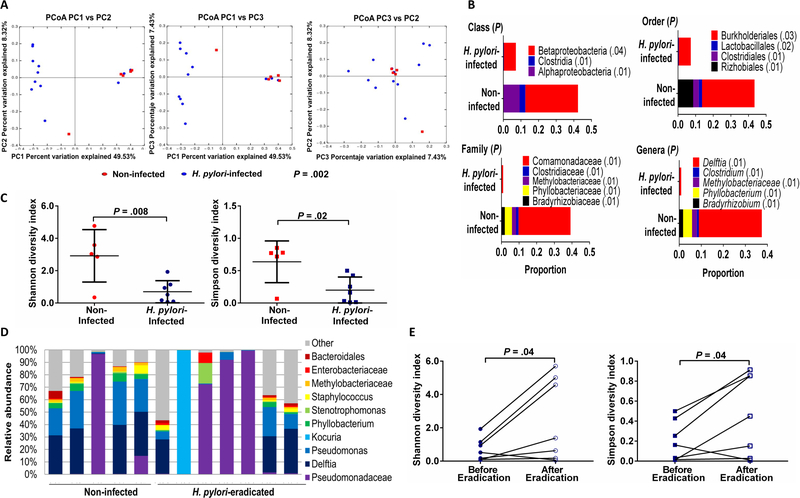
The overall gastric bacterial community structure (β-diversity) in the H. pylori-infected children was different from that of non-infected children using unweighted UniFrac (P=.002) (Figure 1A), weighted UniFrac (P=.005) and Bray-Curtis analyses (P=.006). This finding suggested that H. pylori impacted the gastric commensal bacterial composition in infected children and prompted us to determine whether difference in the abundance of taxa with a prevalence >1% correlated with difference in the bacterial communities between infected and non-infected children. The relative abundance of taxa in the H. pylori-infected children was significantly reduced at the class (3 taxa), order (4 taxa), family (5 taxa) and genera (5 taxa) levels compared with taxa harbored by non-infected children (P=.01-.04), as determined by the Kruskal-Wallis test (Figure 1B). Since reduced gut microbial diversity is associated with certain disease processes, we determined the relative phylotype abundances (α-diversity) for the gastric microbiota. H. pylori-infected children harbored significantly less diverse gastric bacterial communities compared with non-infected children by both Shannon (P=.008) and Simpson (P=.02) indices (Figure 1C). These findings suggested that H. pylori impacted the gastric microbial community structure of children residing in this northern region of South America.
Figure 1.
Gastric microbiota in Venezuelan children is modified by H. pylori infection and reverts to that of non-infected children after H. pylori eradication. (A) β-diversity of the gastric microbiota in H. pylori-infected (n=11) and non-infected children (n=5) determined by permutational multivariate analysis of variance (PERMANOVA) and illustrated by unweighted UniFrac distances with each dot representing one subject in the principal coordinate analysis (PCoA) presented in 2-dimensional plots. P value determined by PERMANOVA test in QIIME. (B) Taxa abundances (when >1% of total bacterial DNA) at the class, order, family and genera levels among H. pylori-infected and non-infected children. P values determined by Kruskal-Wallis test with multiple comparison correction and false discovery rate analysis. When a genus could not be assigned, family is listed. (C) α-diversity of the gastric microbiota in H. pylori-infected (n=11) and non-infected children (n=5) using Shannon and Simpson indices. P values determined by Student’s t-test. (D) Taxa abundances of the 10 most abundant gastric bacteria (when >1% of total bacterial DNA) in children without H. pylori (5) and children in whom H. pylori was eradicated (n=7). P>.05, Kruskal-Wallis test. (E) α-diversity of the microbiota in H. pylori-infected children before (n=11) and after successful H. pylori eradication (n=7) using Shannon and Simpson indices. P values determined by Student’s t-test.
To substantiate these findings, we next analyzed the gastric microbial composition in H. pylori-infected children whose H. pylori was successfully eradicated by antibiotic treatment (7 of the 11 treated children). The bacterial community structure after H. pylori clearance was not significantly different from that of non-infected children (β-diversity) based on unweighted UniFrac analysis (P=.107), as well as weighted UniFrac analysis (P=.439) and Bray-Curtis analysis (P=.260). Overall, the relative proportion of the 10 most abundant bacterial taxa after H. pylori eradication did not differ significantly from that of non-infected children (Kruskal-Wallis test; P>0.5) (Figure 1D). Kocuria, an environmental Gram-positive bacteria that may inhabit skin and mucus membranes, is typically amoxicillin-resistant, and may cause systemic infection in immunocompromised hosts, dominated the microbiota in a single child. Finally, gastric bacterial diversity increased significantly after clearance of H. pylori, based on comparisons of Shannon and Simpson indices (both P=.04) (Figure 1E), reflecting the more diverse bacterial composition after H. pylori eradication. Thus, eradication of H. pylori in infected Venezuelan children was associated with restoration of the gastric microbiota to the community structure of non-infected children.
Discussion
This first characterization of the gastric microbiota in Venezuelan children is important because the microbial composition associated with H. pylori infection may impact the risk for gastric disease sequelae, as shown for adults in mountainous Pacific South America5. Further, in countries with a high prevalence of H. pylori, gastric cancer in adults is strongly related to acquiring H. pylori in childhood. However, children have reduced levels of H. pylori-associated gastric inflammation, the key risk factor for gastric cancer, compared with infected adults1,2. Indeed, gastric cancer in children is exceptionally rare, although H. pylori is typically acquired in early childhood. That H. pylori infection in children is associated with an altered gastric microbiome, as we previously reported for Chilean children3 and confirmed here, raises the possibility that the gastric microbiota in infected children may contribute to mucosal changes such as increased regulatory T-cell responses in infected children1–3. In a mouse model, H. pylori accelerates the pathogenesis of gastric cancer 6 and has more impact on the abundance of specific bacterial taxa in younger than older mice7. In addition, H. pylori infection in neonatal, but not adult, mice promotes tolerance to H. pylori and protects against preneoplastic lesions8.
The complex consortia of generally non-cultivatable bacteria colonizing the stomach requires a molecular approach to identify these microbial communities. Using 16S rDNA sequencing, we show that H. pylori clearance restores the gastric microbial composition to the community structure of non-infected children, suggesting H. pylori eradication impacts the associated microbiota. Thus, should the gastric microbial composition altered by H. pylori contribute to H. pylori-associated disease, eradication of H. pylori with restoration of the microbial community structure to that of non-infected children may modify infection sequelae. Notably, the class, order and family of bacterial taxa impacted by the presence of H. pylori in the Venezuelan children is different from the taxa reported to be impacted by H. pylori in Chilean children3, except for reduced abundance of the acid tolerant order Lactobacillales in both populations. The more prevalent bacterial genera (>1% abundance) associated with H. pylori also did not overlap in the Venezuelan and Chilean children. The lack of concordance in the gastric microbial community structure among children in these two populations may reflect the impact of different diets, nutritional status and geographic location. However, both studies demonstrate that the presence of H. pylori substantially alters the gastric microbiome in children. While acknowledging the small size of our study population, we provide a starting point for future studies on the clinical impact of restoring the microbiota to that of non-infected children. Thus, characterization of the gastric microbiota in pediatric H. pylori infection in endemic regions of the world may provide insight into the role of the microbiome in disease pathogenesis.
Acknowledgements
The authors are grateful for the assistance of the Venezuelan pediatricians, who under difficult social and economic conditions, have contributed to this project and by M. Sandoval in obtaining the specimens. CAS, RP, PDS, PRH conceived and designed the study. RP recruited the patients and procured the specimens. CDM performed the sequencing. CAS, WJVDP, CDM, PDS analyzed and interpreted the data. CAS, PDS, PRH wrote the manuscript.
Funding. This work was supported by Conicyt-PIA Anillo grant ACT172097; The UAB Microbiome Center; Center for Clinical and Translational Science grant UL1TR001417 (NCATS, NIH); UAB Microbiome Resource supported by the School of Medicine Comprehensive Cancer Center grant P30CA013148; NIH grants HD088954 and RR20136; Research Service of the Veterans Administration; and The DeGregorio Family Foundation.
Abbreviations
- rDNA
ribosomal DNA
- PCoA
principal coordinate analysis
- QIIME
quantitative insight into microbial ecology
- OTU
operational taxonomic unit
- PERMANOVA
permutational multivariate analysis of variance
Footnotes
Conflict of interest. The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Harris PR, et al. Gastroenterology 2008; 134:491–9. [DOI] [PubMed] [Google Scholar]
- 2.Serrano C, et al. Mucosal Immunol 2013;6:950–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brawner KM, et al. Mucosal Immunol 2017;10:1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos IS, et al. Public Health Nutrition 2009;12:1862–1870. [DOI] [PubMed] [Google Scholar]
- 5.Yang I, et al. Sci. Rep. 2016;6:18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox JG, et al. Nat Med 2000;6:536–542. [DOI] [PubMed] [Google Scholar]
- 7.Kienesberger S, et al. Cell Rep. 2016;14:1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold IC, et al. Gastroenterology 2011;140:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]