Abstract
Purpose:
To develop and evaluate an automated machine learning (ML) algorithm for segmenting the paraspinous muscles on chest computed tomography (CT) scans to evaluate for presence of sarcopenia.
Methods:
A convolutional neural network based on the U-Net architecture was trained to perform muscle segmentation on a dataset of 1,875 single slice CT images and was tested on 209 CT images of participants in the National Lung Screening Trial (NLST). Low-dose, non-contrast CT examinations were obtained at 33 clinical sites, using scanners from 4 manufacturers. The study participants had a mean age of 71.6 years (range, 70-74 years). Ground truth was obtained by manually segmenting the left paraspinous muscle at the level of the T12 vertebra. Muscle cross-sectional area (CSA) and muscle attenuation (MA) were recorded. Comparison between the ML algorithm and ground truth measures of muscle CSA and MA were obtained using Dice similarity coefficients and Pearson correlations.
Results:
Compared with ground truth segmentation, the ML algorithm achieved median (standard deviation) Dice scores of 0.94 (0.04) in the test set. Mean (SD) muscle CSA was 14.3 (3.6) cm2 for ground truth and 13.7 (3.5) cm2 for ML segmentation Mean (SD) MA was 41.6 (7.6) Hounsfeld units (HU) for ground truth and 43.5 (7.9) HU for ML segmentation. There was high correlation between ML algorithm and ground truth for muscle CSA (r2=0.86; p<0.0001) and MA (r2=0.95; p<0.0001).
Conclusion:
The ML algorithm for measurement of paraspinous muscles compared favorably to manual ground truth measurements in the NLST. The algorithm generalized well to a heterogeneous set of low-dose CT images and may be capable of automated quantification of muscle metrics to screen for sarcopenia on routine chest CT examinations.
Keywords: Muscle, Sarcopenia, Myosteatosis, Chest CT, Machine Learning
INTRODUCTION
Sarcopenia, most commonly defined as the age-related loss of muscle mass and muscle function [1,2], is associated with high health care costs [3,4] and many adverse outcomes including fractures [5], frailty [6,7], functional impairment [6,7], cognitive impairment [8], prolonged hospital stay [9], increased post-operative complications [10], poor health-related quality of life [11], and premature death [6,12]. Functional evaluation of sarcopenia is usually based on measuring grip-strength using a dynamometer or gait speed using a short walk (e.g., 4 or 6 meter) [13–16]. Most operational definitions of sarcopenia also include diagnostic cut-points based on muscle mass determined using dual x-ray absorptiometry (DXA) or bioelectrical impedance analysis (BIA) [13–16].
In the past 5 years, there has been a rapid increase in the use of computed tomography (CT) measurements of muscle to evaluate for sarcopenia [17]. Muscle cross-sectional area (CSA) and muscle attenuation (MA), a surrogate marker for myosteotosis, are the most commonly derived CT metrics [1]. These have been associated with adverse health outcomes in a variety of patient populations. While cancer cohorts have been the most commonly studied [18–22], there is increasing evidence on the value of using CT-derived muscle metrics as prognostic markers in acutely traumatized [23,24], critically ill [25,26], and surgical patients [27,28,29]. In a pragmatic trial of 450 consecutive Medicare patients who had abdominal CTs, Lenchik et al [30] reported that CT-derived muscle metrics predicted 1-year mortality, independent of Charlson comorbidity index, a commonly used prognostic tool
Most clinical trials evaluating for sarcopenia on CT examinations measure muscles of the abdomen or thigh [17]. However, recent studies have shown that muscle measurements on chest CTs are also useful in predicting important clinical outcomes, including mortality [31–36]. While approaches to muscle measurements on chest CT vary, Boutin et al. [36] showed that low paraspinous muscle mass and MA predicted poor survival in patients following hip fracture.
Despite the prognostic value of CT measurements, the current approaches to measuring muscle CSA and MA require laborious manual or semi-automated tissue segmentation, which is not practical for large clinical trials or clinical practice [37–39]. Various approaches have been used to automate muscle segmentation, with atlas-based [40–44] and convolutional neural network (CNN)-based [44–48] approaches showing the greatest promise. Popuri et al [40] developed an atlas-based approach for automatic total abdominal muscle segmentation, reporting a similarity coefficient of 0.91 in a test set of 1069 abdominal CT images. For the same muscle groups, Lee et al [45] developed a CNN-based approach for automatic muscle segmentation reporting a similarity coefficient of 0.93 in a test set of 150 abdominal CT images.
To extend the work of prior studies on abdominal CT exams, we developed an automated CNN-based algorithm for paraspinous muscle segmentation on chest CT exams. The objective of this study was to validate our machine learning (ML) algorithm for measuring muscle CSA and MA on CT examinations from community-dwelling older adult participants in the National Lung Screening Trial (NLST). Our hypothesis is that the ML algorithm will be as accurate as manual segmentation, even on low-dose chest CT used for lung cancer screening.
METHODS
Dataset
The dataset included chest CT examinations from community-dwelling older adult participants in the NLST [49]. The NLST enrolled 53,454 participants, aged 55-74 years, who were current or former smokers with a smoking history of 30 pack-years or more, at 33 medical centers in the United States, from 9/2002 to 4/2004 [49]. Subjects were randomly assigned to two arms: low-dose chest CT (n=26,722) or single-view posteroanterior chest radiography (n=26,732) [49]. The study concluded that three annual low-dose screening CTs reduced mortality from lung cancer by 20% in comparison with three annual single-view chest radiographs in this high-risk cohort [50].
For the current study, a subgroup of oldest participants (age 70-74 years at enrollment) in the CT arm was examined in order to maximize the prevalence of sarcopenia. The study cohort had a mean body mass index of 27.2 kg/m2 and had 64.1% male participants.
CT Acquisition
CT scans were acquired using unenhanced, ungated, low-dose protocols on various models of scanners from 4 major manufacturers (General Electric, Siemens, Philips, Toshiba). CT acquisition parameters of scans are listed in Table 1. CT parameters for each manufacturer are included in the Appendix. Scans with asymmetry between paraspinous muscles (e.g., scoliosis, degenerative diseases, or prior surgery) as well as scans with internal or external artifacts were excluded (n=107), resulting in 2084 CT examinations included in the current study.
Table 1:
Acquisition parameters and reconstruction characteristics of CT images.
| Training n=1875 |
Testing n=209 |
|||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Peak kilovoltage (kVp) | 122.0 | 6.0 | 121.6 | 5.5 |
| X-Ray Tube Current (mA) | 107.1 | 50.9 | 116.0 | 58.0 |
| Slice Thickness (mm) | 1.9 | 0.5 | 1.9 | 0.5 |
| Pixel Size (mm) | 0.7 | 0.1 | 0.7 | 0.1 |
| Reconstruction Diameter (mm) | 337.1 | 36.1 | 335.6 | 34.8 |
| Field of View (mm) | 313.6 | 29.3 | 311.9 | 28.1 |
| Image Size (pixels) | 512 x 512 | |||
Ground Truth Segmentation
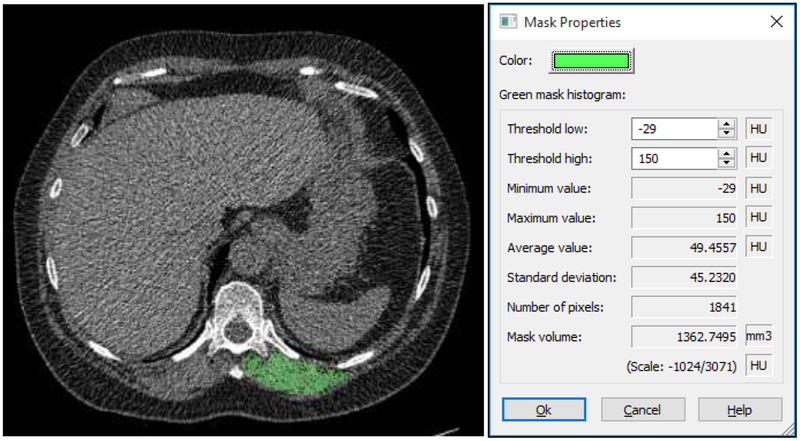
Ground truth was obtained by manually segmenting the left paraspinous muscle on a single CT image at the level of the T12 pedicle using Mimics software (Materialise, Leuven, Belgium) (Figure 1). Muscle thresholds were set at −29 to +150 Hounsfield units (HU) and muscle CSA and MA were recorded. All manual segmentations were performed by an experienced operator and supervised by the principal investigator with 14 years of experience in CT image segmentation.
Figure 1.
Representative low-dose CT image shows paraspinous muscle cross sectional area and attenuation manually measured at the level of T12 vertebra.
Pre-processing
Prior to training in our CNN architecture, our model ground truth images were pre-processed. Training requires a representation of the segmentation in which all pixels corresponding to muscle are explicitly identified. Since the software used for ground truth segmentation did not allow for export of isolated regions of interest, a thresholding method was used to separate the segmentation mask from underlying pixels. This method is described in detail in the Appendix.
Convolutional Neural Network Model
Our method for automatic muscle segmentation adopted the U-Net convolutional neural network architecture [51], with two 50% dropout layers added. There were 24 convolutional layers. In the downsampling path, these layers consisted of adjacent pairs of convolutional layers followed by a pooling layer. The ten downsampling convolutional layers - five pairs - were configured with 64, 128, 256, 512, and 1024 features. The upsampling path consisted of four blocks, with each block containing an upsampling layer, a convolutional layer, a concatenation of the convolutional layer, and the skip connection from the last convolutional layer of the same resolution in the downsampling path, and an additional pair of convolutional layers. The twelve upsampling convolutional layers - four triplets - were configured with 512, 256, 128, and 64 features. Finally, the last convolutional layer was connected to a two-feature convolutional layer, which connected to a final one-feature convolutional layer with a 1x1 kernel that produced the mapping of each pixel to the two classes: tissue present or tissue absent. The downsampling and upsampling paths were mirror images of one another, and consisted of 2x2 max-pooling and 2x2 upsampling layers, resulting in 5 levels with the following resolutions: 512x512, 256x256, 128x128, 64x64, and 32x32.
Two dropout layers, both configured to drop 50% of inputs, were used to control overfitting. These layers surrounded the deepest downsampling level. The first dropout layer followed the second 512-feature convolutional layer at the 64x64 level. The second dropout layer followed the concatenation of the first dropout layer and the output of the second 1024-feature convolutional layer at the 32x32 level
Our ground truth dataset was randomly divided into training and validation subsets, using ninety percent of the data for training and ten percent of the data for validation. For both the training and validation phases, the raw image inputs were split into three separate channels: the raw pixel values (“raw”), the raw values clipped to the interval [−1024, 1024] and rescaled to [−1, 1] (“clipped”), and the clipped values equalized using the CLAHE adaptive histogram equalization algorithm (“CLAHE”) [52].
Training
To improve the generalizability of the trained model, we used image augmentation techniques, which expand the size of the training dataset by creating artificial training images via manipulations of the existing training data. In particular, we used simple affine transformations, including image rotation (−1° to 1°), translation (up to 0.5% of the image dimensions horizontally and vertically), and scaling (99% to 101%). Augmented mask images were binarized by setting all pixels above 0.5 to 1, and all others 0, to avoid aliasing at the mask edges.
Because the muscle mask pixels were only a small portion of the image pixels, commonly-used loss functions including mean squared error and binary cross-entropy did not achieve high accuracy. Instead, we used an adaptation of a smoothed Dice similarity coefficient algorithm [53] coupled with class weighting, with the class weights calculated to be balanced based on the training masks. This loss function ensured that the quality of the segmentation appropriately influenced the model’s training. Class weights were computed using scikit-learn’s [76] compute_class_weight function in “balanced” mode, which calculates class weights based on the data in the training mask images. The weight of each class was determined by the ratio of the number of pixels in the training data to the product of the total number of classes and the number of samples in each class: n_samples / (n_classes * np.bincount(y)). Applied to our dataset, the weight of the non-tissue class was 0.51 and the weight of the tissue class was 39.3. Network weights were initialized using the He normal initializer [54]. Training itself used the stochastic gradient descent optimizer Adam [55], with a learning rate of 1x10−5.
Testing
Following training, the images in the test set were passed through the resulting model Like the training images, these images were separated into 3 input channels: raw, clipped, and CLAHE.
The output layer of the U-Net network was a 1 x 1 convolutional layer with a sigmoid activation function, which returned values in the interval [0,1]. Thus, the output of the model was an image with the same dimensions as the original input image, where each pixel was assigned a continuous value, indicating the probability of that pixel representing the target paraspinous muscle tissue.
All predictions were normalized by dividing each pixel’s probability by the maximum probability observed across all pixels in the image and pixels with resulting values below 0.5 were excluded. Additionally, because we had a priori knowledge that the muscle tissue had HU values in the interval [−29,150], all pixels with HU values outside that range were excluded from the predicted segmentation.
Evaluation of the performance of the ML algorithm consisted of two-parts: 1) mask-oriented comparison of the spatial overlap between the ML and ground truth segmentations and 2) tissue-oriented comparison of the muscle CSA and MA. Spatial overlap of the mask images was evaluated using the Dice similarity coefficient (DSC), where X represents the set of pixels in the ground truth mask and Y represents the set of pixels in the predicted mask.
Muscle CSA and MA similarity was evaluated using the Pearson correlation coefficient.
Implementation
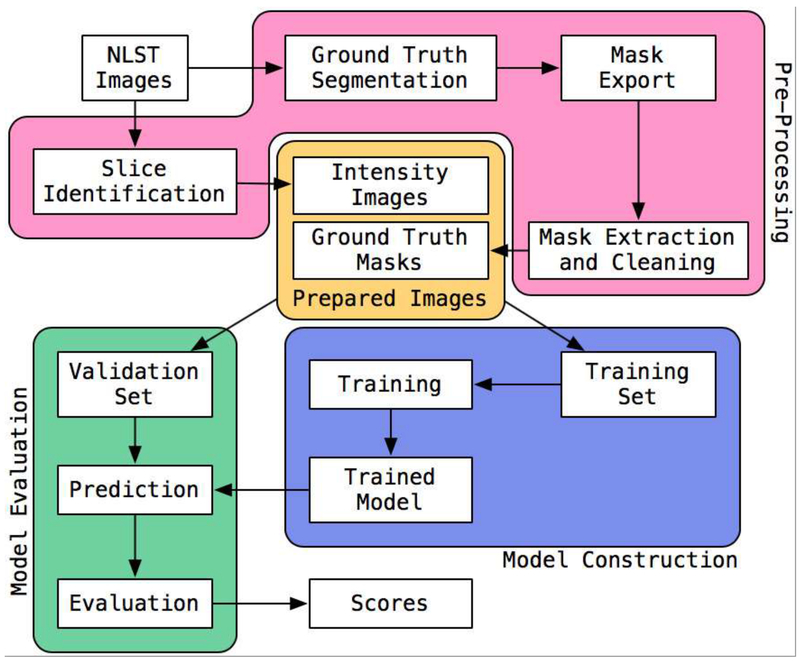
Our ML algorithm was implemented using open source tools. We constructed an initial data preparation pipeline and the main training, evaluation, and reporting pipeline (Figure 2). These were implemented using the Snakemake [56] tool To provide images of segmentations for quality control purposes, additional scripts built on scikit-image and ImageMagick were implemented (Figure 3).
Figure 2.
Diagram of the machine learning pipeline used for deriving muscle metrics from CT images.
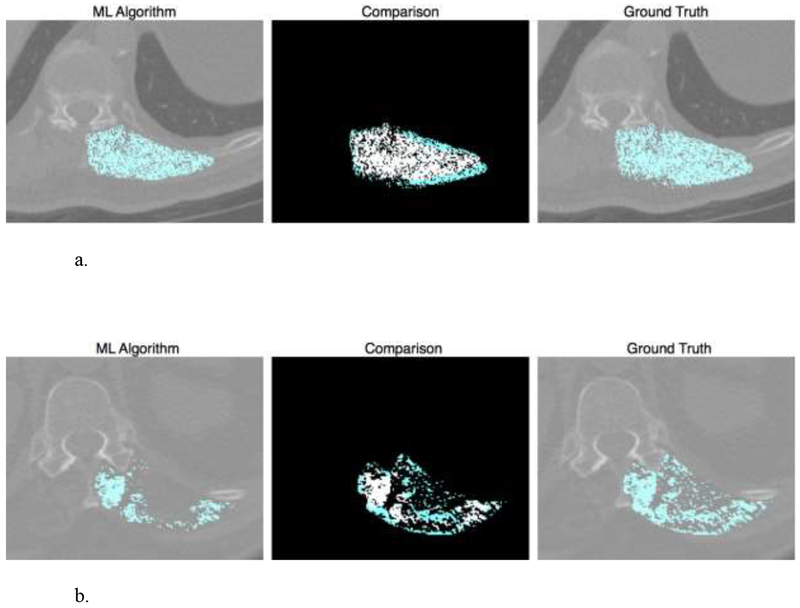
Figure 3.
Representative quality control image shows the probability map of our machine learning algorithm compared to ground truth segmentation.
The input pipeline provided functionality to extract and clean the ground truth mask and prepare the input image and metadata files for automated processing in the main pipeline. The training pipeline used the Keras-based [57] implementation of U-net provided by [58] with the TensorLlow [59] backend. Image augmentation was incorporated using Kerns’ ImageDataGenerator class operating in generator mode with a fixed initial seed to ensure that the generated training data was identical between invocations. Lurther implementation details are provided in the Appendix.
The input data was prepared on a Mid 2010 Mac Pro (Intel Xeon W3530, 32GB RAM). The main pipeline was performed using our university Distributed Environment for Academic Computing (DEAC) high-performance computing cluster on nodes with 44 CPU cores, 496 GB of RAM, and dual Tesla P100 16 GB GPUs.
The model was trained for 200 epochs using a mini-batch size of 5 with 375 mini-batch steps per epoch. Training of the model took 15 hours, 28 minutes of wall-clock time; inference of each subject took 15 seconds of wall-clock time.
RESULTS
The ML algorithm was evaluated using 209 CT images, not included in the training set. Mean (SD) muscle CSA was 14.3 (3.6) cm2 for manual ground truth and 13.7 (3.5) cm2 for automatic ML segmentation. Mean (SD) muscle attenuation was 41.6 (7.6) HU for ground truth and 43.5 (7.9) HU for ML segmentation.
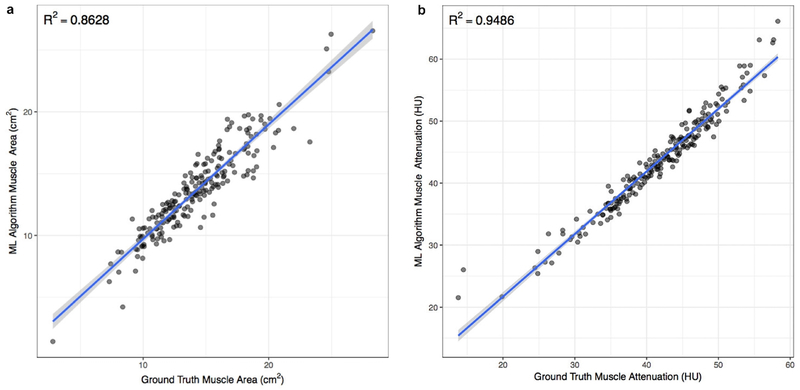
There was high correlation in muscle CSA (r2=0.86; p<0.0001) and MA (r2=0.95; p<0.0001) between ground truth and machine learning segmentation (Figure 4).
Figure 4.
(a) Correlation plot of muscle cross sectional area determined with machine learning algorithm and ground truth. (b) Correlation plot of muscle attenuation determined with machine learning algorithm and ground truth.
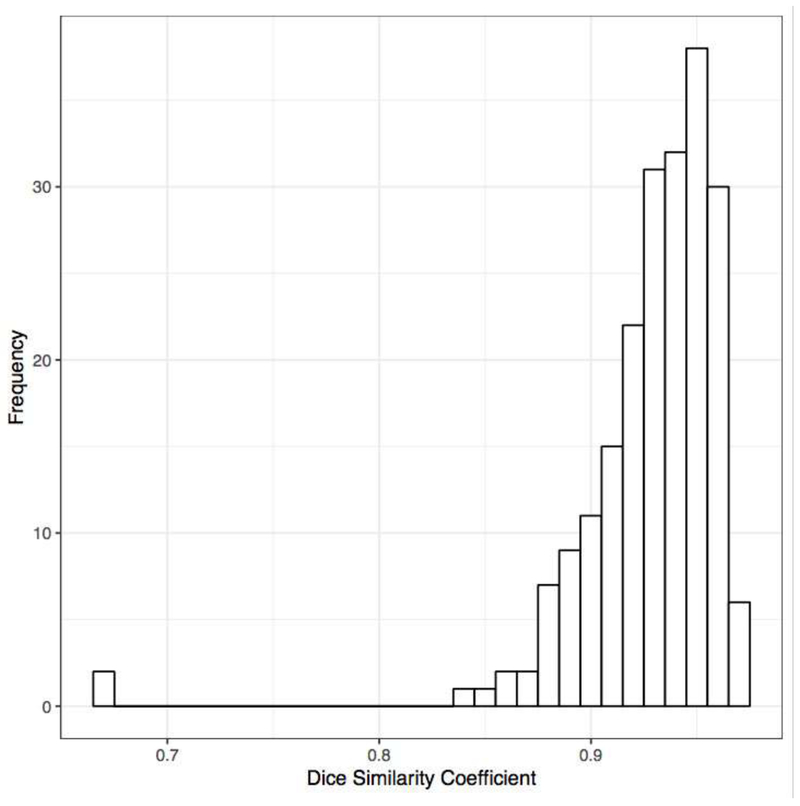
Compared to ground truth segmentation, the ML algorithm had a median Dice similarity coefficient of 0.94 (SD=0.04) (Figure 5).
Figure 5.
Histogram of Dice similarity coefficient for the test set of CT images.
The ML algorithm produced some poor predictions, usually due to under-segmentation of tissue. Most of these appear to be caused by low tissue contrast (Figure 6).
Figure 6.
a. Low contrast CT image resulting in poor machine learning segmentation.
b. Low contrast CT image resulting in poor machine learning segmentation
DISCUSSION
ML algorithms for tissue segmentation are becoming increasingly common in neuroradiology and cardiovascular imaging [60,61]. Similar pipelines for organ and tissue segmentation are being developed for abdominal CT scans [62–65]. Our study helps extend ML approaches to tissue segmentation on chest CTs. Our newly developed fully-automated ML algorithm for paraspinous muscle segmentation is highly accurate when validated against a heterogeneous set of chest CT images in older adults in the NLST.
Our algorithm performed better for MA than for muscle CSA. Larger errors in CSA measurement were expected because muscle boundaries may be difficult to distinguish from adjacent tissues, especially the subcutaneous fat which follows the contour of paraspinous muscle and, when edematous, may have tissue attenuation similar to muscle.
Our results compare favorably to previous studies reporting on automated CNN-based approaches to muscle segmentation. Weston et al. [46] used a U-net architecture to train on 2340 abdominal images at the level of L3. In a test set of 270 images, they reported a Dice similarity coefficient for total abdominal muscles of 0.96. They did not compare performance of MA versus muscle CSA. Because the abdominal muscle groups measured in their study are much larger than the paraspinous muscles measured in our study, somewhat better performance of their algorithm was not unexpected. In addition to differences in study subjects and CT doses between the two studies, specific differences in their ML algorithm that may explain results include: an additional downsampling layer, somewhat different convolution parameters, initializing weights using a Glorot normal initializer, using tanh (instead of ReLU) as their main activation function, and using categorical cross-entropy for loss.
Our results also compare favorably to Lee et al. [45] who used VGG-Net to segment the total abdominal muscles at the level of L3. They trained on 250 cases, and reported a Dice similarity index of 0.93 in a test set of 150 cases. They also reported a mean 3.7% error for muscle CSA, when compared to ground truth. Like our study, their cohort included all 4 CT scanner manufacturers. Unlike our study, these were not low-dose CTs and the acquisition parameters were more standardized.
While Lee H, at al [45] and Weston AD, et al [46] used deep learning methods for muscle segmentation, they segmented all visualized muscles on abdominal CT images, rather than a single muscle group on chest CT images. A recent systematic review by Amini B, et al [17] shows that there are many valid approaches for evaluating muscle mass on CT images and that there are specific reasons for different muscle groups to be evaluated, either alone, or in combination. The overarching goal for sarcopenia screening is to select a muscle group that is most clinically relevant. On chest CT images, the paraspinous muscle group at the level of T12 is more clinically relevant than other muscle groups since it has been shown to predict mortality in older adults [36].” Importantly, because T12 level is frequently visualized not only on chest CTs but also on abdominal CTs, our algorithm can be used on both types of scans, further increasing clinical utility. To our knowledge there are no prior studies using CNNsto automate muscle segmentation on chest CTs. However, Popuri et al [40] used an atlas-based approach to automatically segment muscles on chest CTs at the level of T4. Their algorithm uses a finite element method and a deformable shape model based on ideal template-based segmentation and muscle shape. After training on 60 thoracic images, they reported a Jaccard similarity index of 0.91 in a test set of 530 images. Their slightly poorer performance than ours may be explained in part by greater variability of muscle morphology at the level of T4, depending on the position of the arms during CT acquisition
The potential clinical impact of measuring muscle area and attenuation on chest CTs cannot be overstated. Rosman DA, et al. [66] examined utilization of imaging over two decades using Medicare claims. They reported utilization rates of 134 per 1000 Medicare beneficiaries for chest CTs compared to 191 per 1000 for abdomen/pelvis CTs. Thus, the increased screening potential of using chest as well as abdominal CTs is considerable. More importantly, compared to 1994, the 2013 utilization rates for chest CTs increased by 350% while the utilization for abdominal/pelvis CT increased 93%. If the trends continue, there will be even greater clinical utility of screening for sarcopenia using chest CTs in the future.
Limitations
Our study has several limitations and strengths. The main limitation is that the CT slice cannot be automatically selected. Recent studies [67,68] have used CNN-based approaches to automatically select the L3 slice on CT images. Similarly, we are working on automatically selecting the T12 slice, which will enable us to develop a more efficient pipeline for muscle segmentation on chest CTs. Another limitation is that only low dose CT scans were used. However, our use of low dose images was intentional for both technical and clinical reasons. The technical reason for using low dose images is that our algorithm should work even better on regular dose CT scans that have less image noise. The clinical reasons are that lung cancer screenings are becoming increasingly more common and that value of these screening may be enhanced by opportunistically measuring other prognostic phenotypes, including muscle area and attenuation. The major strength of our study is that the dataset included CT images that are heterogeneous in field of view, voxel spacing, convolution kernel, reconstruction algorithm, image quality, and scanner manufacturer and model For this reason, our model should be more generalizable than previously published models.
Future Directions
Our ML algorithm for measuring muscle mass and myosteatosis on chest CTs could be adapted to other body regions such as the abdomen and thigh, facilitating research on muscle metrics as biomarkers of aging and cachexia. On abdominal CT examinations, muscle measurements are usually obtained as part of a body composition analysis that includes visceral adipose tissue and subcutaneous adipose tissue measurements. Similarly, our ML algorithm can be adapted to measure other tissues in the chest, including pericardial adipose tissue and bone tissue.
CONCLUSION
With automated secondary analysis of clinical CT examinations already being obtained for routine indications, the ML algorithm promises to help identify patients at risk for sarcopenia, investigate the need for follow-up and therapeutic interventions, develop better tools for prognosis, and ultimately improve patient outcomes.
Supplementary Material
ACKNOWLEDGEMENT
Computations were performed on the Wake Forest University’s DEAC Cluster, a centrally managed resource with support provided in part by the University.
FUNDING
National Institute of Health Grants: P30 AG021332 (Pepper Center) and UL1TR001420 (CTSI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lenchik L, Boutin RD. Sarcopenia: Beyond Muscle Atrophy and into the New Frontiers of Opportunistic Imaging, Precision Medicine, and Machine Learning. Semin Musculoskelet Radiol. 2018;22(3):307–322. [DOI] [PubMed] [Google Scholar]
- 2.Shaw SC, Dennison EM, Cooper C. Epidemiology of Sarcopenia: Determinants Throughout the Lifecourse. Calcif Tissue Int. 2017. September;101(3):229–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruyère O, Beaudart C, Ethgen O, Reginster JY, Locquet M. The health economics burden of sarcopenia: a systematic review. Maturitas. 2019. January;119:61–69. [DOI] [PubMed] [Google Scholar]
- 4.Norman K, Otten L. Financial impact of sarcopenia or low muscle mass - A short review. Clin Nutr. 2018. September 27 pii: S0261-5614(18)32464-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Hao Q, Ge M, Dong B. Association of sarcopenia and fractures in community-dwelling older adults: a systematic review and meta-analysis of cohort studies. Osteoporos Int. 2018. June;29(6):1253–1262. [DOI] [PubMed] [Google Scholar]
- 6.Kelley GA, Kelley KS. Is sarcopenia associated with an increased risk of all-cause mortality and functional disability? Exp Gerontol. 2017. October1;96:100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaudart C, Zaaria M, Pasleau F, Reginster JY, Bruyere O. Health Outcomes of Sarcopenia: A Systematic Review and Meta-Analysis. PLoS One. 2017. January 17;12(1):e0169548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang KV, Hsu TH, Wu WT, Huang KC, Han DS. Association Between Sarcopenia and Cognitive Impairment: A Systematic Review and Meta-Analysis. J Am Med Dir Assoc. 2016. December 1;17(12):1164.e7–1164.e15. [DOI] [PubMed] [Google Scholar]
- 9.DeAndrade J, Pedersen M, Garcia L, Nau P. Sarcopenia is a risk factor for complications and an independent predictor of hospital length of stay in trauma patients. J Surg Res. 2018. January;221:161–166. [DOI] [PubMed] [Google Scholar]
- 10.Tsekoura M, Kastrinis A, Katsoulaki M, Billis E, Gliatis J. Sarcopenia and Its Impact on Quality of Life. Adv Exp Med Biol 2017;987:213–218. [DOI] [PubMed] [Google Scholar]
- 11.Joglekar S, Nau PN, Mezhir JJ. The impact of sarcopenia on survival and complications in surgical oncology: A review of the current literature. J Surg Oncol. 2015. October;112(5):503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batsis JA, Mackenzie TA, Barre LK, Lopez-Jimenez F, Bartels SJ. Sarcopenia, sarcopenic obesity and mortality in older adults: results from the National Health and Nutrition Examination Survey III. Eur J Clin Nutr. 2014. September;68(9):1001–7. [DOI] [PubMed] [Google Scholar]
- 13.Cruz-Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyere O, Cederholm T, Cooper C, Landi F, Rolland Y, Sayer AA, Schneider SM, Sieber CC, Topinkova E, Vandewoude M, Visser M, Zamboni M; Writing Group for the European Working Group on Sarcopenia in Older People 2 (EWGSOP2), and the Extended Group for EWGSOP2 . Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing. 2018. October 12. [Google Scholar]
- 14.Dent E, Morley JE, Cruz-Jentoft AJ, Arai H, Kritchevsky SB, Guralnik J, Bauer JM, Pahor M, Clark BC, Cesari M, Ruiz J, Sieber CC, Aubertin-Leheudre M, Waters DL, Visvanathan R, Landi F, Villareal DT, Fielding R, Won CW, Theou O, Martin FC, Dong B, Woo J, Flicker L, Ferrucci L, Merchant RA, Cao L, Cederholm T, Ribeiro SML, Rodríguez-Mañas L, Anker SD, Lundy J, Gutiérrez Robledo LM, Bautmans I, Aprahamian I, Schols JMGA, Izquierdo M, Vellas B. International Clinical Practice Guidelines for Sarcopenia (ICFSR): Screening, Diagnosis and Management. J Nutr Health Aging. 2018;22(10):1148–1161. [DOI] [PubMed] [Google Scholar]
- 15.Cawthon PM. Recent Progress in Sarcopenia Research: a Focus on Operationalizing a Definition of Sarcopenia. Curr Osteoporos Rep. 2018. December;16(6):730–737. [DOI] [PubMed] [Google Scholar]
- 16.Han A, Bokshan SL, Marcaccio SE, DePasse JM, Daniels AH. Diagnostic Criteria and Clinical Outcomes in Sarcopenia Research: A Literature Review. J Clin Med. 2018. April 8;7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amini B, Boyle SP, Boutin RD, Lenchik L. Approaches to Assessment of Muscle Mass and Myosteatosis on Computed Tomography (CT): A Systematic Review. J Gerontol A Biol Sci Med Scr 2019. February 6 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daly LE, Prado CM, Ryan AM. A window beneath the skin: how computed tomography assessment of body composition can assist in the identification of hidden wasting conditions in oncology that profoundly impact outcomes. The Proceedings of the Nutrition Society. 2018;77:135–151. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Moynagh MR, Multinu F, Cliby WA, McGree ME, Weaver AL, et al. Muscle composition measured by CT scan is a measurable predictor of overall survival in advanced ovarian cancer. Gynecologic oncology. 2016;142:311–316. [DOI] [PubMed] [Google Scholar]
- 20.Deng HY, Hou L, Zha P, Huang KL, Peng L. Sarcopenia is an independent unfavorable prognostic factor of non-small cell lung cancer after surgical resection: A comprehensive systematic review and meta-analysis. Eur J Surg Oncol 2018. October 15 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Mintziras I, Miligkos M, Wächter S, Manoharan J, Maurer E, Bartsch DK. Sarcopenia and sarcopenic obesity are significantly associated with poorer overall survival in patients with pancreatic cancer: Systematic review and meta-analysis. Int J Surg. 2018. November;59:19–26. [DOI] [PubMed] [Google Scholar]
- 22.Shachar SS, Williams GR, Muss HB, Nishijima TF. Prognostic value of sarcopenia in adults with solid tumours: A meta-analysis and systematic review. Eur J Cancer. 2016. April;57:58–67. [DOI] [PubMed] [Google Scholar]
- 23.Fairchild B, Webb TP, Xiang Q, Tarima S, Brasel KJ. Sarcopenia and frailty in elderly trauma patients. World J Surg. 2015. February;39(2):373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan SJ, Pham TN, Arbabi S, Gross JA, Damodarasamy M, Bentov I, Taitsman LA, Mitchell SH, Reed MJ. Association of Radiologic Indicators of Frailty With 1-Year Mortality in Older Trauma Patients: Opportunistic Screening for Sarcopenia and Osteopenia. JAMA Surg. 2017. February 15;152(2):e164604. [DOI] [PubMed] [Google Scholar]
- 25.Moisey LL, Mourtzakis M, Cotton BA, Premji T, Heyland DK, Wade CE, Bulger E, Kozar RA; Nutrition and Rehabilitation Investigators Consortium (NUTRIC). Skeletal muscle predicts ventilator-free days, ICU-free days, and mortality in elderly ICU patients. Crit Care. 2013. September 19;17(5):R206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welch CK Hassan-Smith Z, A Greig C, M Lord J, A Jackson T. Acute Sarcopenia Secondary to Hospitalisation - An Emerging Condition Affecting Older Adults. Aging Dis. 2018. February 1;9(1):151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell PM, Collinge CA, O’Neill DE, Bible JE, Mir HR Sarcopenia Is Predictive of 1-Year Mortality After Acetabular Fractures in Elderly Patients. J Orthop Trauma. 2018. June;32(6):278–282. [DOI] [PubMed] [Google Scholar]
- 28.Bokshan SL, Han AL, DePasse JM, Eltorai AE, Marcaccio SE, Palumbo MA, Daniels AH. Effect of Sarcopenia on Postoperative Morbidity and Mortality After Thoracolumbar Spine Surgery. Orthopedics. 2016. November 1;39(6):e1159–e1164. [DOI] [PubMed] [Google Scholar]
- 29.Hale AL, Twomey K, Ewing JA, Langan EM 3rd, Cull DL, Gray BH. Impact of sarcopenia on long-term mortality following endovascular aneurysm repair. Vase Med. 2016. June;21(3):217–22. [DOI] [PubMed] [Google Scholar]
- 30.Lenchik L, Lenoir KM, Tan J, et al. Opportunistic Measurement of Skeletal Muscle Size and Muscle Attenuation on Computed Tomography Predicts One-year Mortality in Medicare Patients. J Gerontol A Biol Sci Med Sci 2018. August 16 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recio-Boiles A, Galeas JN, Goldwasser B, Sanchez K, Man LMW, Gentzler RD, Gilders lee ve J, Hollen PJ, Gralla RJ. Enhancing evaluation of sarcopenia in patients with non-small cell lung cancer (NSCLC) by assessing skeletal muscle index (SMI) at the first lumbar (LI) level on routine chest computed tomography (CT). Support Care Cancer. 2018. July;26(7):2353–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim EY, Kim YS, Park I, Ahn HK, Cho EK, Jeong YM, Kim JH. Evaluation of sarcopenia in small-cell lung cancer patients by routine chest CT. Support Care Cancer. 2016. November;24(11):4721–6. [DOI] [PubMed] [Google Scholar]
- 33.Nemec U, Heidinger B, Sokas C, Chu L, Eisenberg RL. Diagnosing Sarcopenia on Thoracic Computed Tomography: Quantitative Assessment of Skeletal Muscle Mass in Patients Undergoing Transcatheter Aortic Valve Replacement. Acad Radiol. 2017. September;24(9):1154–1161. [DOI] [PubMed] [Google Scholar]
- 34.Troschel FM, Kuklinski MW, Knoll SJ, Best TD, Muniappan A, Gaissert HA, Fintelmann FJ. Preoperative thoracic muscle area on computed tomography predicts long-term survival following pneumonectomy for lung cancer. Interact Cardiovasc Thorac Surg. 2018. October 10. [DOI] [PubMed] [Google Scholar]
- 35.Fuchs G, Thevathasan T, Chretien YR, Mario J, Piriyapatsom A, Schmidt U, Eikermann M, Fintelmann FJ. Lumbar skeletal muscle index derived from routine computed tomography exams predict adverse post-extubation outcomes in critically ill patients. J Crit Care. 2018. April;44:117–123. [DOI] [PubMed] [Google Scholar]
- 36.Boutin RD, Bamrungchart S, Bateni CP, Beavers DP, Beavers KM, Meehan JP, Lenchik L. CT of Patients with Hip Fracture: Muscle Size and Attenuation Help Predict Mortality. AIR Am J Roentgenol 2017;208(6):W208–W215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engelke K, Museyko O, Wang L, Laredo JD. Quantitative analysis of skeletal muscle by computed tomography imaging—State of the art. Journal of orthopaedic translation 2018. October 1 ;15:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murea M, Lenchik L, Register TC, et al. Psoas and Paraspinous Muscle Index as a Predictor of Mortality in African American Men with Type 2 Diabetes Mellitus. J Diabetes Complications. 2018;32(6):558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson DE, Quinn E, Parker E, Allaire BT, Muir JW, Rubin CT, Magaziner J, Hannan MT, Bouxsein ML, Kiel DP. Associations of Computed Tomography-Based Trunk Muscle Size and Density With Balance and Falls in Older Adults. J Gerontol A Biol Sci Med Sci 2016. June;71(6):811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popuri K, Cobzas D, Esfandiari N, et al. Body Composition Assessment in Axial CT Images Using FEM-Based Automatic Segmentation of Skeletal Muscle. IEEE Trans Med Imaging. 2016;35(2):512–20. [DOI] [PubMed] [Google Scholar]
- 41.Yokota F, Otake Y, Takao M, Ogawa T, Okada T, Sugano N, Sato Y. Automated muscle segmentation from CT images of the hip and thigh using a hierarchical multi-atlas method. Int J Comput Assist Radiol Surg. 2018. July;13(7):977–986. [DOI] [PubMed] [Google Scholar]
- 42.Pérez-Carrasco JA, Acha B, Suárez-Mejías C, López-Guerra JL, Serrano C. Joint segmentation of bones and muscles using an intensity and histogram-based energy minimization approach. Comput Methods Programs Biomed. 2018. March;156:85–95. [DOI] [PubMed] [Google Scholar]
- 43.Hu P, Huo Y, Kong D, Carr JJ, Abramson RG, Hartley KG, Landman BA. Automated Characterization of Body Composition and Frailty with Clinically Acquired CT. Comput Methods Clin Appl Musculoskelet Imaging 2018;10734:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kullberg J, Hedström A, Brandberg J, Strand R, Johansson L, Bergström G, Ahlström H. Automated analysis of liver fat, muscle and adipose tissue distribution from CT suitable for large-scale studies. Sci Rep. 2017. September 5;7(1):10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H, Troschel FM, Tajmir S, et al. Pixel-Level Deep Segmentation: Artificial Intelligence Quantifies Muscle on Computed Tomography for Body Morphometric Analysis. J Digit Imaging. 2017;30(4):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weston AD, Korfatis P, Kline TL, Philbrick KA, Kostandy P, Sakinis T, Sugimoto M, Takahashi N, Erickson BJ. Automated Abdominal Segmentation of CT Scans for Body Composition Analysis Using Deep Learning. Radiology. 2018. December 11:181432. [DOI] [PubMed] [Google Scholar]
- 47.Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys Med Biol. 2016. September 7;61(17):6553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somasundaram E, Deaton J, Kaufman R, Brady S. Fully automated tissue classifier for contrast-enhanced CT scans of adult and pediatric patients. Phys Med Biol. 2018. June 27;63(13):135009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.National Lung Screening Trial Research Team, Aberle DR, Berg CD, et al. The National Lung Screening Trial: Overview and Study Design. Radiology 2011;258:243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England Journal of Medicine 2011;365:395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention 2015 Oct 5 (pp. 234–241). Springer, Cham. [Google Scholar]
- 52.Zuiderveld K Contrast limited adaptive histogram equalization. Graphics gems. 1994:474–85. [Google Scholar]
- 53.Jocic M Deep learning tutorial for Kaggle ultrasound nerve segmentation competition, using Keras, https://github.com/jocicmarko/ultrasound-nerve-segmentation, 2017.
- 54.He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. InProceedings of the IEEE international conference on computer vision 2015. (pp. 1026–1034). [Google Scholar]
- 55.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014. December 22.
- 56.Köster J, Rahmann S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012. August 20;28(19):2520–2. [DOI] [PubMed] [Google Scholar]
- 57.Chollet F, et al. Keras, https://keras.io, 2015.
- 58.zhixuhao, Implementation of deep learning framework – U-Net, using Keras, https://github.com/zhixuhao/unet, 2018.
- 59.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-scale machine learning on heterogeneous systems, Software available from tensorflow.org, 2015. [Online], Available: https://www.tensorflow.org/.
- 60.Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Isgum I. Automatic Segmentation of MR Brain Images With a Convolutional Neural Network. IEEE Trans Med Imaging. 2016. May;35(5):1252–1261. [DOI] [PubMed] [Google Scholar]
- 61.Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016. May;30:108–119. [DOI] [PubMed] [Google Scholar]
- 62.Summers RM. Progress in Fully Automated Abdominal CT Interpretation. AIR Am J Roentgenol 2016. July;207(l):67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koyuncu H, Ceylan R, Sivri M, Erdogan H. An Efficient Pipeline for Abdomen Segmentation in CT Images. Journal of digital imaging. 2018. April 1;31(2):262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sahiner B, Pezeshk A, Hadjiiski LM, Wang X, Drukker K, Cha KH, Summers RM, Giger ML. Deep learning in medical imaging and radiation therapy. Medical physics. 2018. October 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Belharbi S, Chatelain C, Herault R, Adam S, Thureau S, Chastan M, Modzelewski R Spotting L3 slice in CT scans using deep convolutional network and transfer learning. Comput Biol Med. 2017. August 1;87:95–103. [DOI] [PubMed] [Google Scholar]
- 66.Rosman DA, Duszak R Jr, Wang W, Hughes DR, Rosenkrantz AB. Changing Utilization of Noninvasive Diagnostic Imaging Over 2 Decades: An Examination Family–Focused Analysis of Medicare Claims Using the Neiman Imaging Types of Service Categorization System. American Journal of Roentgenology. 2018. February;210(2):364–8. [DOI] [PubMed] [Google Scholar]
- 67.Kanavati F, Islam S, Aboagye EO, Rockall A. Automatic L3 slice detection in 3D CT images using folly-convolutional networks. arXiv preprint arXiv:l811.09244. 2018. November 22.
- 68.Perthen JE, Ali T, McCulloch D, Navidi M, Phillips AW, Sinclair RCF, Griffin SM, Greystoke A, Petrides G. Intra- and interobserver variability in skeletal muscle measurements using computed tomography images. Eur J Radiol 2018. December;109:142–146. [DOI] [PubMed] [Google Scholar]
- 69.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2018. [ROnline]. Available: https://www.R-project.org/. [Google Scholar]
- 70.Whitcher B, Schmid VJ, and Thornton A, “Working with the DICOM and NIfTI data standards in R,” Journal of Statistical Software, vol 44, no. 6, pp. 1–28, 2011. [ROnline}. Available: http://www.jstatsoft.org/v44/i06/. [Google Scholar]
- 71.Brett M, Hanke M, Markiewicz C, et al. Nipy/nibabel: 2.3.1, Oct. 2018. doi: 10.5281/zenodo.1464282 [Online]. Available: 10.5281/zenodo.1464282. [DOI]
- 72.Jones E, Oliphant T, Peterson P. {SciPy}: open source scientific tools for {Python}. [Online; accessed 2018-12-13], Available: http://www.scipy.org/.
- 73.Oliphant TE. A guide to NumPy. USA: Trelgol Publishing; 2006. December 7. [Google Scholar]
- 74.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011. February 1;54(3):2033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012; 62(2): 782–790. [DOI] [PubMed] [Google Scholar]
- 76.Van der Walt S van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T, and the scikit-image contributors, PeerJ 2, e453 (2014). PeerJ. 2014;2:e453. [Online]. Available: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit-learn: Machine learning in Python. Journal of machine learning research 2011;12(Oct):2825–30. [Google Scholar]
- 78.McKinney W Data structures for statistical computing in python. InProceedings of the 9th Python in Science Conference 2010 Jun 28 (Vol. 445, pp. 51–56). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.