Abstract
The automated segmentation of organs and tissues throughout the body using computed tomography (CT) and magnetic resonance imaging (MRI) has been rapidly increasing. Research into many medical conditions has benefited greatly from these approaches by allowing the development of more rapid and reproducible quantitative imaging markers. These markers have been used to help diagnose disease, determine prognosis, select patients for therapy, and follow responses to therapy. Because some of these tools are now transitioning from research environments to clinical practice, it is important for radiologists to become familiar with various methods used for automated segmentation. The Radiology Research Alliance of the Association of University Radiologists convened an Automated Segmentation Task Force to conduct a systematic review of the peer-reviewed literature on this topic. The systematic review presented here includes 408 studies and discusses various approaches to automated segmentation using CT and MRI for neurologic, thoracic, abdominal, musculoskeletal, and breast imaging applications. These insights should help prepare radiologists to better evaluate automated segmentation tools and apply them not only to research, but eventually to clinical practice.
Keywords: segmentation, machine learning, quantitative imaging, CT, MRI
Introduction
Various approaches to automated segmentation of CT and MR images are widely used in research environments and promise to transform clinical practice [1–12]. Radiologists involved in interpreting images in patients with cancer, obesity, cardiovascular disease, neurodegeneration, osteoporosis, arthritis, and many other conditions will benefit from these approaches as they help clinicians diagnose disease, determine prognosis, select patients for therapy, and follow responses to therapy. To enable this transition from research to patient care, radiologists should become familiar with various methods used for automated segmentation of CT and MR images.
Segmentation refers to identifying the boundaries of an object in the image. Frequently, the object is an organ, a tissue, a pathologic lesion, or another structure used for diagnosis or management of a particular disease. Traditional approaches to segmentation rely on manual or semi-automated delineation of the object of interest. While these approaches are effective, they are time-consuming and impractical for large scale research studies and even less practical for clinical practice. As a result, many fully automated approaches to tissue segmentation are being developed.
Automated segmentation methods using CT and MRI are generally built on basic image processing of pixel intensities and/or textural features (e.g., relationships between groups of pixels), and may incorporate advanced model-based, atlas-based, or machine learning (ML) techniques [13–16]. Segmentation techniques can be broadly divided into supervised and unsupervised.
Supervised techniques require prior training that is most commonly performed manually. These methods typically include pre-processing such as intensity normalization (e.g., histogram-based, reference tissue), followed by classification (e.g., artificial neural networks, k-nearest neighbors, Bayesian, random decision forests), and feature selection based on intensity, spatial, texture, or contextual information [13–16]. They are considered to be more accurate, but require expert annotation which is both expensive and time consuming.
Unsupervised segmentation techniques do not require any training and are generally considered less accurate than supervised techniques. These methods usually incorporate clustering (e.g., fuzzy c-means, expectation-maximization) and spatial information (e.g., Markov random fields, graph cut, anatomical/topological atlases) to segment the image [13–16]. They also commonly rely on labeled atlases.
The reported performance of supervised and unsupervised segmentation techniques varies greatly, depending in part on the validation metrics used [13–16]. Generally, the validation is based on the assessment by experts. Since evaluation by a single expert may be biased, some studies employ multiple experts using techniques such as Simultaneous Truth and Performance Level Estimation (STAPLE) [17]. In some cases, the segmentation techniques are validated against established pipelines, such as FreeSurfer (https://surfer.nmr.mgh.harvard.edu/), SPM (https://www.fil.ion.ucl.ac.uk/spm/), or FSL (https://www.fmrib.ox.ac.uk/fsl). Another approach uses publically available “challenge databases” for training and validation of automated segmentation techniques.
Many validation metrics have been used for quantitative comparison between the automated segmentation results and ground truth [12–16]. These metrics include: Dice similarity coefficient (DSC), Jaccard index, volume difference, Hausdorff distance, intraclass correlation, and Pearson’s coefficient. The DSC calculates the overlap between two binary segmentation results by accounting for both the intersection and the union of the two results. More familiar metrics are also occasionally used, including: sensitivity, specificity, accuracy, positive predictive value, and negative predictive value.
Previous reviews of segmentation have focused on CT or MRI of a single body region, often combining semi-automated and automated approaches, and rarely using the methodologic rigor of a systematic review. There is a need for a systematic review that focuses on automated segmentation using CT and MRI of the entire body.
We provide such a systematic review of the automated segmentation methods and discuss how these methods have been used in neurologic, thoracic, abdominal, musculoskeletal, and breast imaging. Ultimately, we hope to prepare radiologists for eventual integration of these techniques into their clinical practice.
Methods
Identification of Studies
This systematic review was performed according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement guidelines [18]. A systematic literature search was conducted in the PubMed/MEDLINE, Embase via Ovid, and Cochrane Central Register of Controlled Trials (CENTRAL) via Ovid databases from January 1, 2007 through February 26, 2018 (date of final search execution). A list of Medical Subject Headings (MeSH) and keywords targeting full-text, automated segmentation using CT or MRI was formulated by a joint collaboration between task force members and research librarians (Appendix A1).
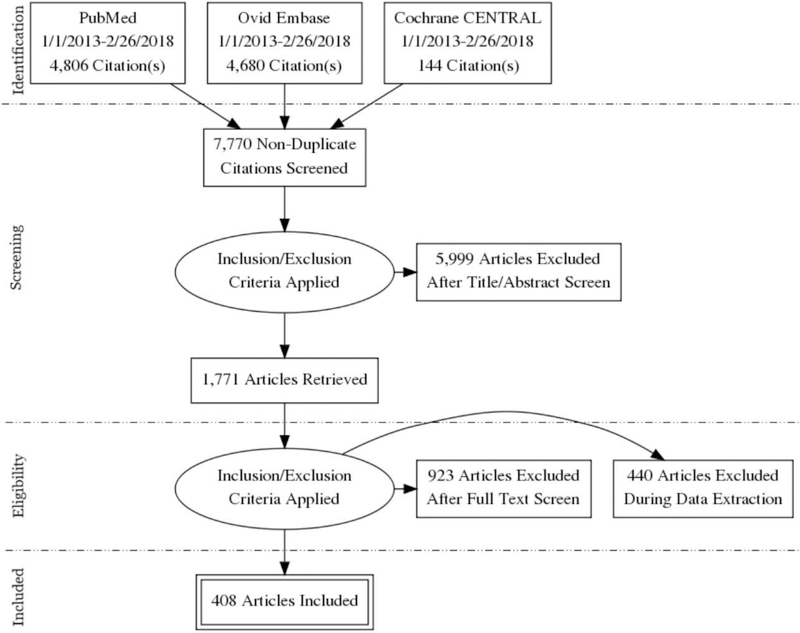
Figure 1 is a PRISMA flow diagram showing identification, screening, eligibility, and inclusion of articles. After initial review of the articles, studies prior to 2013 were excluded as not sufficiently up to date to reflect the current application of automated segmentation methodology. An additional search was performed using a second set of keywords (Appendix A2), resulting in an additional 226 studies. After removal of duplicates, a total of 7,770 citations were identified.
Figure 1:
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram showing identification, screening, eligibility, and inclusion of articles.
Each of the initial 7,770 citations was independently screened at the title/abstract level by two fellowship trained sub-specialty radiologists and/or experienced imaging researchers using predefined exclusion criteria: 1) animal, cadaver, or phantom studies, 2) radiation oncology studies, 3) dental studies, 4) studies using ultrasound, nuclear medicine, functional MRI, magnetic resonance spectroscopy, or diffusion tensor imaging, 5) studies using semi-automated or manual segmentation, 6) reviews and meta-analysis, 7) studies with fewer than 20 subjects.
After title/abstract screening, 1,771 articles were included in the full-text screening. Using the same exclusion criteria, an additional 923 articles were excluded. The remaining articles were divided by sub-specialty based on abstract keywords (neuroimaging, thoracic imaging, abdominal imaging, musculoskeletal imaging, breast, and adipose tissue imaging). At time of sub-specialty data extraction, 440 articles were excluded (Table 1). Studies excluded as outside the scope for this review were: studies of fetal or neonatal neurologic segmentation and studies of pathologic tissues or organs in thoracic and abdominal segmentation. Other exclusions were: free software (i.e., FreeSurfer, SPM, and FSL), commercial software (i.e., Neuroquant), or studies published in ArXiv for neurologic segmentation; non-human subjects, duplicate manuscripts, modality other than CT or MRI, non-English, reviews or meta-analyses, or use of commercial software for thoracic segmentation; duplicate studies for abdominal segmentation; cadaveric study, modality other than CT or MRI for musculoskeletal segmentation; duplicate or breast CT studies for breast segmentation. Following all exclusions, 408 articles were included in the systematic review.
Table 1:
Exclusions during sub-specialty review
| Reason for Exclusion | Neuro n=237 | Thoracic n=67 | Abd n=40 | MSK n=63 | Breast n=20 | Adipose n=13 |
|---|---|---|---|---|---|---|
| Sample size < 20 | 41 | 26 | 20 | 14 | 4 | 7 |
| Not fully automated | 5 | 5 | 6 | 7 | 1 | 3 |
| Classification without segmentation | 0 | 1 | 3 | 7 | 4 | 2 |
| Insufficient detail on segmentation method | 10 | 0 | 1 | 9 | 0 | 0 |
| Application of existing methods | 79 | 0 | 0 | 0 | 3 | 0 |
| Conference abstract | 58 | 8 | 0 | 14 | 5 | 1 |
| Full text not available | 5 | 7 | 1 | 6 | 0 | 0 |
| Outside the scope | 13 | 4 | 7 | 0 | 0 | 0 |
| Other exclusions | 26 | 16 | 2 | 6 | 3 | 0 |
Methodological Quality
The methodological quality of 408 studies was assessed independently by two reviewers (LL and LH) according to a modified National Heart, Lung and Blood Institute (NHLBI) Case Series Quality Assessment Tool [19]. The NHLBI Quality Assessment Tool assesses study objectives, population, outcome measures, statistical method, and provides a Good, Fair, or Poor quality rating. Disagreements were resolved by consensus.
Data Extraction
During full-text review, the following data were extracted: segmented organ or tissue, imaging modality, segmentation technique, sample size, and validation method. Because there is no single accepted classification method to describe segmentation methods, we applied a modified classification scheme, described by Withey and Koles [20]. The segmentation techniques were divided into the following categories: 1) Thresholding, 2) Statistical, 3) Deformable model, 4) Graph search, 5) Multi-resolution, 6) Atlas-based, 7) Texture analysis, 8) Neural network, and 9) Hybrid (i.e., combination of more than one of the above methods) (Table 2).
Table 2:
Classification of segmentation methods
| Method | Segmentation sub-categories |
|---|---|
| Thresholding | Intensity |
| Adaptive | |
| Statistical | Statistical pattern recognition |
| Mixture model, k-nearest neighbor classifiers, Bayesian classifiers | |
| Expectation-maximization, Markov random field, iterative procedure | |
| Prior information modeling | |
| C-means clustering, fuzzy c-means clustering, random forests | |
| Deformable model | Active contour, active surface, active shape, snake, level-set |
| Statistical shape | |
| Appearance model | |
| Graph search | Dynamic programming |
| Graph cut | |
| Watershed | |
| Multi-resolution | Scale reduction |
| Atlas-based | Single atlas |
| Multi-atlas | |
| Texture analysis | Neighboring voxel relationships (intensity, gradient, entropy, etc.) |
| Neural network | Convolutional neural networks (CNN) |
| Multi-class CNN | |
| Multi-scale CNN | |
| Visual geometry group (VGG) | |
| Adversarial network | |
| Deep learning | |
| Deep belief network | |
| Hybrid | More than one method listed above |
Results
408 studies met the inclusion criteria, including 145 (36%) neurologic, 78 (19%) thoracic, 87 (21%) abdominal, 58 (14%) musculoskeletal, 20 (5%) breast, and 20 (5%) adipose tissue studies. Using the NHLBI Quality Assessment Tool, all studies (100%) received a quality rating of Good.
Neurologic Segmentation
145 studies met the inclusion criteria (Appendix B1). MRI was used in 137 (94%) studies, CT in 7 (5%) studies, and CT and MRI in 1 (1%) study. The MR field strengths were: 1.5T (n=31), 3T (n=34), both 1.5T and 3T (n=24), 1T (n=2), 7T (n=2), and not specified (n=45).
The most common pulse sequence for normal brain segmentations was 3D T1-weighted (n=56). For segmenting lesions in multiple sclerosis, gliomas, white matter hyperintense lesions, and stroke, FLAIR was combined with T1-weighted and/or proton-density and/or T2-weighted sequences (n=53).
The brain was segmented in 139 (96%) studies and the spinal cord in 6 (4%) studies. The brain structure most commonly segmented was the hippocampus (n=13). At the tissue level, techniques focused on segmenting lesions, gray matter, white matter, and cerebrospinal fluid. A few studies focused on other structures such as caudate, basal ganglia, thalami, individual gyri within a lobe, or brain stem.
A variety of automated segmentation techniques were used (Table 3). The most commonly used methods were statistical (n=59), atlas-based (n=38), and neural network (n=19). The number of subjects whose images were used for training and validation varied greatly across the studies (mean=218, range = 20–3672).
Table 3:
Automated segmentation methods by anatomic region
| Segmentation Method | Neuro n=145 | Thoracic n=78 | Abd n=87 | MSK n=58 | Breast n=20 | Adipose n=20 |
|---|---|---|---|---|---|---|
| Thresholding | 10 | 19 | 7 | 0 | 1 | 1 |
| Statistical | 59 | 15 | 7 | 4 | 5 | 0 |
| Deformable model | 6 | 24 | 14 | 13 | 2 | 3 |
| Graph search | 3 | 2 | 9 | 1 | 1 | 0 |
| Multi-resolution | 0 | 0 | 0 | 0 | 0 | 0 |
| Atlas-based | 38 | 8 | 19 | 6 | 0 | 3 |
| Texture analysis | 6 | 7 | 0 | 0 | 0 | 0 |
| Neural network | 19 | 5 | 14 | 4 | 3 | 1 |
| Hybrid (combination of more than one method) | 6 | 8 | 17 | 30 | 8 | 12 |
Most studies validated automated segmentation techniques against the ground truth determined by manual segmentation (n=102). Some techniques were validated against other established segmentation techniques (n=28). Finally, some validation used challenge datasets including Medical Image Computing and Computer Assisted Intervention (MICCAI), Brain Tumor Segmentation (BraTS), and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (n=11). While many different validation metrics were used, the most common was the DSC (n=46).
Automated segmentation techniques were used in normal subjects as well as in subjects with various diseases or conditions including: Alzheimer’s disease, multiple sclerosis, stroke, cancer, and epilepsy.
Thoracic Segmentation
78 studies met the inclusion criteria (Appendix B2). CT was used in 30 (38%) of studies, MRI in 27 (35%), CT angiography (CTA) in 14 (18%), MR angiography (MRA) in 2 (3%). Five studies (6%) used a combination of modalities (e.g., CT and CTA).
Segmented organs included the heart (n=39), lungs and airways (n=19), and blood vessels (n=16). Four studies segmented multiple organs. Combinations included the heart and coronary arteries, heart and liver, heart and solid abdominal organs, and segmentation of all intrathoracic organs (i.e., skin, bones, and mediastinal structures).
All but one of the studies that segmented the lungs used CT (n=17). Most of the studies that segmented the blood vessels used CT or CTA (n=13). 62% of the studies that segmented the heart used MRI (n=24).
Cardiac segmentation typically isolated one or more cardiac chambers; most commonly, the left ventricle. Lung segmentation typically focused on isolating the lungs. A few studies segmented the individual lobes or the trachea and bronchial tree. Blood vessel segmentation typically focused on the coronary arteries, with some studies focusing on the great vessels.
A variety of automated segmentation techniques were used (Table 3). The most commonly used methods were deformable models (n=24), thresholding (n=19), and statistical (n=15). Most studies validated automated segmentation techniques against the ground truth determined by manual segmentation. In addition, 30% (n=25) of studies compared their method to previously published segmentation method. A small number of studies relied on visual comparison as their validation method; this technique was almost exclusively used to evaluate vascular segmentations. While many different validation metrics were used, the most common was the DSC (n=39).
The average number of exams used for training and validation varied widely across all studies (mean=151, range = 20–2500). Larger datasets were seen in studies using CT (mean=232) compared to those using MRI (mean=99). Even smaller datasets were seen in studies using CTA (mean=67) and MRA (mean=31).
Only a small number of studies used publicly available datasets, such as COPDgene, Lung Imaging Database, Cardiac Atlas Project Database, or the LIDC-IDRI database. Several studies used data from prior challenges, including the Rotterdam coronary CTA challenge, LOLA11 lung lobe segmentation challenge, and the 2012 MICCAI RV segmentation challenge.
Automated segmentation techniques were used in normal subjects as well as in subjects with various diseases or conditions including : cardiovascular diseases (e.g., coronary artery disease, prior myocardial infarction, ventricular hypertrophy, or congestive heart failure) and pulmonary diseases (e.g., chronic obstructive pulmonary disease or interstitial lung diseases).
Abdominal Segmentation
87 studies met the inclusion criteria (Appendix B3). CT was used in 50 (57%) studies and included non-contrast and contrast-enhanced studies in various phases (i.e., arterial or portal venous). MRI was used in 36 (41%) studies and employed a variety of sequences including multiplanar T2- and T1-weighted pre- and post-contrast images. One study used both CT and MR images for multi-organ segmentation.
The organs segmented were: prostate (n=24), liver (n=20), kidneys (n=10), spleen (n=5), pancreas (n=5), colon (n=2), gallbladder (n=2), esophagus (n=2), bladder (n=1), and female pelvic floor (n=1). The remaining 15 studies segmented more than one organ; ranging from two organs (e.g., liver and spleen) to up to 14 distinct structures including the aorta, inferior vena cava, mesenteric vessels, and uterus.
A variety of automated segmentation techniques were used (Table 3). The most commonly used methods were atlas-based (n=19), deformable models (n=14), and neural networks (n=14). 17 studies used a combination of more than one method. Most studies (n=82) compared performance of the automated segmentation technique to manual segmentation. The average number of exams used for validation was 68 (range = 20–400). While many different validation metrics were used, the most common was the DSC (n=62).
Musculoskeletal Segmentation
58 studies met the inclusion criteria (Appendix B4). CT was used in 20 (34%) studies, MRI in 37 (64%), and both CT and MRI in 1 (2%).
Segmentation of the spine was most common (n=16). Other segmented regions included: thigh/femur (n=13), pelvis/hip (n=10), knee (n=14), wrist (n=3), shoulder (n=2), lower leg (n=1), skull (n=1), and whole body (n=2). Five studies segmented multiple regions (e.g., femur and pelvis, spine and pelvis).
Bone was the most common tissue segmented (n=35). Additional segmented tissues included articular cartilage (n=14), fibrocartilage (n=3), skeletal muscles (n=9), intervertebral discs (n=4), bone-cartilage interface (n=1), spinal canal (n=1), and dural sac (n=1). Eight studies segmented more than one tissue.
A broad range of automated segmentation techniques were used (Table 3). The most commonly used methods were deformable models (n=13). Thirty studies used a combination of methods. Most studies compared performance of the automated segmentation technique to manual segmentation. The average number of exams used for training and validation was 170 (range = 20–2117). While many different validation metrics were used, the most common was the DSC (n=42).
Automated segmentation techniques were used in normal subjects as well as in subjects with various diseases or conditions including: osteoporosis, osteoarthritis, rheumatoid arthritis, avascular necrosis, fractures, meniscal injury, cancer, bone metastases, craniosynostosis, spinal stenosis, disc herniation, and disc degeneration. Some studies focused only on normal subjects.
Breast Segmentation
20 studies met the inclusion criteria (Appendix B5). All studies used MRI. Sequences used for segmentation included axial non-fat-suppressed non-contrast T1-weighted images (n=8), multiple sequences (n=6), post-contrast T1-weighted or subtraction images (n=1), non-contrast T1-weighted fat suppressed (n=3), Dixon-based sequences (n=1), and sagittal non-fat-suppressed non-contrast T1-weighted images (n=1).
Most studies evaluated breast fibroglandular tissue (FGT) segmentation alone (n=11), with a subset extrapolating FGT segmentation to post-contrast images to evaluate background parenchymal enhancement (BPE) (n=3), or to evaluate FGT, BPE and lesion-level segmentation (n=6).
A broad range of automated segmentation techniques were used (Table 3). The most commonly used method was statistical (n=5). Eight studies used more than one method. Most studies compared automated segmentation to manually drawn contours as ground truth. The average number of exams used for training and validation was 90 (range = 20–400). The most common validation metric was the DSC (n=16).
Adipose Tissue Segmentation
20 studies met the inclusion criteria (Appendix B6). CT was used in 12 (60%) and MRI in 8 (40%). The following examinations were used: abdominal CT (n=5), abdominal MRI (n=4), thoracic CT (n=8), thigh CT (n=1), thigh MRI (n=2), calf MRI (n=1), and whole body MRI (n=1). Two studies segmented multiple regions (e.g., abdomen and thorax, thigh and calf). Most studies performed segmentation of tissue volumes (n=17) rather than tissue cross-sectional area (n=3).
Abdominal visceral and subcutaneous adipose tissue were most commonly segmented (n=8), followed by epicardial or pericardial adipose tissue (n=7), thigh intermuscular adipose tissue and subcutaneous adipose tissue (n=2), and brown supraclavicular adipose tissue (n=1).
Atlas-based and deformable model techniques were used equally (Table 3). However, hybrid techniques were most common (n=12). In all studies, manual segmentations were used as the reference standard. The average number of exams used for training and validation was 100 (range = 20–530). The most common validation metric used was the DSC (n=13).
Automated segmentation techniques were used in normal subjects as well as in subjects with various diseases or conditions including: obesity, diabetes, metabolic syndrome, and osteoarthritis. Some studies focused only on normal subjects.
Discussion
This is the first systematic review of automated segmentation that includes CT and MRI across all anatomic regions. The most significant finding of this review is that there is wide variability in approaches to automated segmentation, regardless of anatomic region or image modality. Another finding is that automated segmentation of the brain is far ahead of other organs and tissues. In fact, 79 studies on neuro-segmentation were excluded from our review because they applied existing automated segmentation methods to clinical research, rather than developing new methods. Similarly, there were more studies of thoracic, abdominal, and musculoskeletal segmentation compared to breast and adipose tissue segmentation.
Our results should be interpreted in the context of prior literature on the three generations of medical image segmentation [20]. Fully automated methods often combine multiple segmentation operations and incorporate at least one second- or third-generation approach.
First-generation approaches use pixel intensities or connectedness to apply basic heuristics or one-time operations to segment the image. These include edge tracing (i.e., boundary segmentation) and thresholding and region growing (e.g., volume segmentation) [21].
Second-generation methods incorporate uncertainty models and optimization methods which typically avoid heuristics. These include approaches to segment volumes (i.e., statistical pattern recognition, c-means clustering), boundaries (i.e., deformable models), or other (i.e., graph search, neural networks, multiresolution methods) [20]. They commonly use classifiers for separating clusters, including k-nearest neighbors (k-NN), artificial neural networks (ANN), support vector machines (SVM), and random forest classifiers. Statistical pattern recognition is used to assign pixels a probability of belonging to a tissue class based on pixel intensity and/or texture classifiers (e.g., Bayesian, k-NN, maximum likelihood, expectation-maximization, Markov random field) [22–26]. In c-means clustering (including fuzzy c-means clustering), pixels are grouped into a known number of clusters based on pixel intensity or local texture by minimizing an objective function [22]. Deformable models (e.g., active contours, active surfaces, snakes, level-sets) can conform to image features over time under the influence of internal and external forces to achieve a local optimum [26]. Graph search techniques treat pixels as interconnected nodes in a graph, where graph cuts between the interconnected nodes are defined based on minimization of a cost function to produce a globally optimized segmentation [27]. ANNs learn from training data to classify pixels into predefined classes. In multiresolution segmentation, a high-to-low resolution image stack is created by iteratively blurring and down sampling the original image; pixels with similar features between the stacked images are linked as belonging to the same object to perform the segmentation [28].
Third-generation approaches incorporate higher-level knowledge such as a priori information, expert defined rules, and models of the shape or appearance of the target object, and include methods to segment volumes (e.g., atlas-based segmentation, rule-based segmentation) and/or boundaries (e.g., shape models, appearance models) [20]. Atlases of segmented images can be mapped to an unsegmented image, with the atlas supplying probabilities for statistical pattern recognition based on anatomical, shape, size, and textural features [29]. Segmentation can incorporate automated rule logic based on anatomy, intensity, texture, and/or shape [20]. Statistical shape models, such as the active shape model [30], are an extension of deformable models, where deformation is restricted by statistical bounds of the model. Active appearance models incorporate an “image patch” of shape plus intensity data into the statistical model to segment an object [31].
Despite this hierarchy, there is no consensus on how to categorize various segmentation methods. For example: Withey and Koles [20] categorize based on first-, second-, and third-generation; García-Lorenzo et al. [32] categorize based on supervised and unsupervised approaches; Danelakis et al. [33] categorize into: data-driven, feature-based, atlas-based, statistical, tissue-based, and lesion-based. For the purposes of our systematic review, we divided the segmentation techniques into: thresholding, statistical, deformable model, graph search, multi-resolution, atlas-based, texture, neural network, and hybrid. Our aim was to use a classification system that would be useful for readers who are new to the field.
Our review found a wide variability among automated segmentation methods: 1) statistical, atlas-based, and neural network methods were especially common in neuro-segmentation; 2) statistical, thresholding, and deformable model methods were most common in thoracic segmentation, 3) deformable model, atlas-based, and neural network methods were common in abdominal segmentation, and 4) hybrid models, employing a combination of techniques, were most common for musculoskeletal, breast, and adipose tissue segmentation. Although many different validation metrics were used to evaluate automated segmentation of various organs and tissues, the most common was the Dice similarity coefficient (DSC).
Neuroimaging Segmentation
Segmentation of the brain and spine using MRI poses particular challenges owing to variability in acquisition parameters including slice thickness, resolution, matrix size, TR, and TE. To minimize this variability, preprocessing of images is critical. Preprocessing typically includes registration of images to a common template (i.e., Montreal Neurologic Institute) or co-registration of images, skull stripping, signal intensity normalization, noise reduction, and bias or field inhomogeneity correction [34].
In this systematic review, we did not discuss the advantages or disadvantages of various segmentation techniques as this is addressed in detail in prior narrative reviews [35]. It is difficult to compare different segmentation techniques as there is no consensus regarding optimal evaluation metrics or standardization of databases. This may be mitigated by the use of challenge datasets and development of guidelines for ML researchers [5].
Automated image segmentation relies increasing on ML, including U-net and other convolutional neural networks [36,37]. However ML techniques require a large amount of annotated data for training, validation, and testing. Currently, there are not many large datasets that can be used to develop ML approaches to segmentation. Because there is heterogeneity in MRI protocols as well as differences in tissue contrast that vary with field-strength and MRI vendor, a segmentation technique for one MRI protocol may not be applicable for different protocols.
There have been two prior systematic reviews of segmentation using brain MRI [38,39] and one systematic review on using spine MRI [40]. Dicke et al. [38] performed a systematic review of brain MRI focusing on the creation of atlases for segmentation of normal brains and included 66 studies from October 2010 to August 2016. Cover et al. [39] performed a systematic review focused on segmentation of the corpus-callosum and included 36 studies prior to March 2016. Rak and Tonnies [40] performed a review of spine segmentation and 98 studies but did not follow the PRISMA guidelines. Our systematic review follows the PRISMA guidelines and includes 145 studies using MRI and CT of the brain and spine.
Unlike the systematic reviews that have been few in number, conventional narrative reviews of neuro-segmentation have increased dramatically. Between 2013 and 2018, there were 12 reviews of brain MRI [33,35,37,41–48], 3 of spinal cord MRI [49–51], 3 of brain CT [52–54], and 2 of spine CT [55,56]. While these reviews provide valuable information on neuro-segmentation techniques they do not provide the methodologic rigor of a systematic review and are subject to a selection bias.
Thoracic Segmentation
This systematic review revealed multiple methods for automated segmentation of the organs of the thorax using CT and MRI. As expected, more methods used CT, which allows more accurate evaluation of the lungs and airways. However, more than half the studies of the heart used MRI. While 16 studies focused on vascular segmentation, only 6 evaluated the coronary arteries.
In a prior review of machine learning segmentation Slomka et al. [57] reported that most coronary segmentation methods require some manual initialization. Another review by Moccia et al. [58] reported that vascular segmentation requires a variety of approaches partly because there is no single approach for different types of vessels and that pathologic vessels may require different approaches than normal vessels.
The most commonly used automatic segmentation techniques that were identified in this review are generally the same as those identified in other published reviews. In particular, for segmentation of the heart on MRI, deformable models were the most common techniques [59]. Automated segmentation of the cardiac chambers is particularly challenging owing to normal or pathologic deformation of the heart during the cardiac cycle [60]. The field is very active, with many automated cardiac segmentation methods presented in recent conferences and on preprint websites that have not yet reached peer-reviewed journals [61].
The datasets used to evaluate automated segmentation methods in the thorax are relatively small. This is likely because the ground-truth used to evaluate these methods is resource-intensive and time-consuming to produce. Unlike in brain imaging, there are significantly fewer automated segmentation methods in the thorax that use public datasets.
Surprisingly, only 6% of the studies in our review used neural network approaches to automated segmentation in the thorax. Currently, these techniques are widely used for thoracic classification, but not segmentation. Since there is increasing number of studies using neural networks for pulmonary nodule segmentation [62,63], it is likely that the same methods will eventually be applied to other tissues in the thorax.
Abdominal Segmentation
The development of automated segmentation techniques for abdominal and pelvic organs lags behind other body regions, especially the brain. The major challenges include: 1) variability in the shape, size and position of the anatomical structures of interest, 2) poor contrast between adjacent organs and surrounding tissues (edge detection), 3) motion artifacts that can cause image blurring, and 4) change in organ position relative to other fixed anatomical structures [64].
Despite these challenges, an accurate approach to automated segmentation and measurements of abdominopelvic organs is highly desirable for many clinical indications. For example, accurate preoperative liver volumetry is becoming standard of care prior to major hepatic resection or for partial living donor transplantation [65]. Manual and semi-automated liver segmentation techniques are time-consuming and have high inter- and intra- reader variability. Accurate automated segmentation of kidneys is especially valuable when monitoring patients with autosomal dominant polycystic kidney disease [66]. Accurate segmentation of splenic volume aids in monitoring patients with infection and splenic diseases [67,68]. There are many other current clinical applications, including focal lesion detection in all abdominal organs, bowel segmentation for the detection of obstruction and inflammatory bowel disease, lymph node measurement, radiation treatment planning, and detection of aortoiliac atherosclerosis [12].
Common approaches to segmentation in abdominopelvic organs include detection of features, edges or intensities, strong or weak shape and/or location priors, thresholding, clustering methods, and deformable models with most automated segmentation techniques employing a combination of these techniques. Based on this systematic review, there are many automated segmentation techniques that can be applied to abdominal organs. However most of these have been validated in relatively small studies.
Direct comparison of the different approaches to automated segmentation is problematic as validation techniques vary from study to study. Challenges such as those sponsored by the MICCAI conferences and online publicly available datasets, will certainly foster the development of many more automated segmentation techniques for the abdomen.
Musculoskeletal Segmentation
Studies of automated segmentation of bone commonly use CT, owing to the high image contrast between bone and soft tissue structures [69]. Studies using MRI are increasingly common for the evaluation of joints and soft tissues [70]. Although many studies use a combination of methods to automatically segment bone, the deformable model, atlas-based, statistical, and graph search methods are the typical components of these automated algorithms. Many approaches also incorporate first-generation segmentation methods such as thresholding, region growing, edge detection, and edge tracing into the segmentation framework.
Some studies have compared the performance of different segmentation methods in the same cohort of healthy and diseased subjects from public datasets such as SpineWeb [71]. Recent studies of six automated vertebral segmentation methods reported DSCs of 0.87–0.95 for normal spines and 0.54–0.90 for osteoporotic spines [72,73].
Studies that automatically segmented intervertebral discs most commonly use deformable models and statistical segmentation approaches. Recent segmentation competition challenge reported on eight automated segmentation algorithms with DSC ranging from 0.82–0.92 [74].
Automated segmentation of musculoskeletal tissues has also benefited from ML. Neural networks [75] can now automatically segment joint tissues in approximately 5 seconds, while maintaining high DSCs.
The most common techniques used to segment cartilage and muscle include random forests, nearest neighbors, SVM, and k-mean clustering. When comparing different ML approaches, the results may vary based on study population. For MRI of muscle, Gadermayr et al. [76] reported that a basic thresholding approach was often sufficient, but shape based graph cuts produced the best results in patients with severe fatty infiltration of muscle. Many recent studies incorporate multiple models, such as combining localized classification via 2D and 3D convolutional neural networks (CNNs) with statistical anatomical knowledge via 3D statistical shape models [77].
Many studies have focused on automated articular cartilage segmentation on knee MRI using large publicly available datasets including the Osteoarthritis Initiative (OAI) and the MICCAI grand challenge “Segmentation of Knee Images 2010”. Recent studies used voxel-based relaxometry to obtain fully automated analysis of cartilage composition using T1-rho MRI [78]. Although early studies focused on only one anatomic structure (e.g., cartilage), it is increasingly common to segment multiple structures (e.g., bone, cartilage, meniscus). For example, a fully automated segmentation pipeline has been constructed for evaluation of both morphological and quantitative knee MRI data by combining a deep CNN and three dimensional (3D) simplex deformable modeling [79].
For skeletal muscle segmentation, most studies still use manual or semi-automated techniques, but this is beginning to change [80–84]. With MRI, one proposed strategy is automated quantification of whole-body muscle volumes. Karlsson et al. [81] developed an automated segmentation method based on multiatlas segmentation of intensity corrected water-fat separated image volumes, reporting high accuracy and reproducibility. Yang et al. [82] segmented muscle on a single image at the mid femur level (rather than tissue volumes), reporting shorter scan times and diminished post-processing computational costs.
The use of automated segmentation of muscle using CT is also increasing [11]. Lee et al. [82] used a deep learning system to automatically segment the muscle cross-sectional area of CT slices at the L3 vertebral body level, with an average of less than 3.7% difference between predicted and ground truth muscle cross-sectional area, while reducing segmentation time from 30 minutes to 0.17 seconds.. Current challenges for automated segmentation of muscle using CT include a tendency to underestimate muscle area in general, while overestimating muscle area in subjects with edematous fat [83].
Breast Segmentation
Breast segmentation on MRI consists of three separate challenges: 1) distinguishing breast-chest wall and breast-air boundaries, 2) separating breast FGT from fat, and 3) distinguishing abnormal breast enhancement from normal BPE.
There are unique challenges to breast segmentation that have limited large-scale application to date. Although Dixon-based sequences and other fat-specific sequences offer superior fat and glandular tissue segmentation, these are not routinely incorporated into most clinical breast MRI protocols. In comparison, T1-weighted pre- and post-contrast acquisitions are essential to diagnostic breast MRI but demonstrate B0 and B1 inhomogeneity across the parenchyma at the breast-air boundary and across the coil gradient [84]. Without appropriate correction of the varying enhancement caused by sources of inhomogeneity, image-processing methods can be inaccurate. Finally, the routine use of fat suppression can introduce additional artifact that must be corrected. Although breast MRI can be performed at both 1.5T and 3.0T, there is no evidence that segmentation differs between these field strengths.
There are two prior systematic reviews of breast segmentation. Wang et al. [85] reviewed breast and chest segmentation studies but did not include lesion-level automated segmentation studies or ML studies. Codari et al. [86] reviewed only ML studies. In contrast, our review includes fully automated breast segmentation examining FGT, BPE and lesion segmentation approaches.
In developing approaches for fully automated breast segmentation, hybrid approaches, incorporating both statistical and atlas-based methods are most common. For example, Wu et al. [87] used a fuzzy C means atlas-based method on sagittal T1W fat-suppressed imaging in 60 breast MRIs, achieving a Pearson correlation coefficient of r = 0.92 for FGT% and a DSC of 0.67.. Similar template based approaches using fat-water separation have demonstrated high reliability for FGT separation [88,89]. However, fat-water separation techniques are not commonly used in routine clinical breast imaging. Statistical (particularly fuzzy c-means) and thresholding techniques have also shown promise in FGT segmentation. However, ML approaches will likely predominate in the future. Recent studies of SVM-based approaches and U-net approaches have resulted in higher DSCs and relatively fast processing times compared to more traditional approaches [90,91].
Lesion segmentation on breast MRI is particularly challenging. Deep learning techniques show the most promise in automatic lesion segmentation. Dalmiş et al. [92] used a 2D U-net CNN to evaluate 361 cases using early post-contrast images to create a CADe system with a computation performance metric (CPM) of 0.6429, significantly higher than the CPM value of 0.5325 obtained by a previous CADe system utilizing a full dynamic breast MRI (p=0.008).
Strengths and Limitations
Our systematic review has several strengths and limitations. A major strength is that we used a comprehensive search strategy using three databases (PubMed, Embase, and Cochrane), yielding 7770 citations reviewed by sub-specialty experts using strict exclusion and inclusion criteria, resulting in 408 studies. Another strength is that we included organs and tissues of the entire body, rather than one region, and that we included both CT and MRI. One limitation is that we did not perform a meta-analysis to determine which automated method is most valuable. However, due to wide variability in the segmentation methods as well as the approaches to their validation, such meta-analysis is currently not feasible.
Future Directions
Future studies of automated image segmentation will undoubtedly involve methodological innovations as well as new clinical applications.
Most innovative methods at this time appear to involve deep learning (DL). DL is a subfield of machine learning (ML), which in turn is a subfield of artificial intelligence (AI). DL is based on artificial neural networks. Neural networks are machine learning devices modeled after the human brain. DL employs neural networks, consisting of layers of nonlinear processing units that successively process the numerical input data. A single processing unit (the artificial neuron) typically receives multiple inputs, combines them as a weighted sum, and applies to the results some form of a nonlinear transformation [93, 94]. Through training, the network learns the weights required to achieve its task [93, 94]. Especially well-suited to image analysis are the convolutional neural networks (CNNs). These networks use convolution kernels (i.e., filters) that are shared among all neurons in the same layer, drastically reducing the number of parameters in the network and allowing training with a relatively smaller number of datasets. CNNs have been successfully applied to image processing, segmentation, classification, and prediction [95]. U-net is a CNN that consists of encoding and decoding stages and is particularly well suited for image segmentation [96]. Recently, generative adversarial networks have also been gaining popularity [97, 98]. One major hurdle for further development of DL methods is the need for large datasets of annotated images for training. However, increasing availability of publicly available image databases such as ADNI, LONI, OASIS, should improve access to training images. Importantly, all DL methods require significant computational power. Increased availability of graphical processing units as well as open source frameworks such as Apache MXNet, PyTorch, Caffe, and Chainer has provided the needed computational resources to allow for increased application of DL methods to CT and MR image segmentation.
Future clinical applications of DL methods may include triaging patients with potentially life-threatening conditions or reducing common interpretive errors [12]. Future research applications will include large-scale integration of clinical, genomic, proteomic, metabolomic, and radiomic data as part of epidemiological studies as well as pragmatic trials [10].
Conclusion
We conducted a systematic review that includes automated segmentation using CT and MRI of the entire body. Our findings have implications for both research and clinical practice. Automated CT and MR image segmentation allows for an objective evaluation of diseases by identifying quantitative imaging markers that are widely used in research. Major challenges to implementation of automated segmentation tools to clinical practice remain. For all the anatomic regions evaluated in our study, there is continuing need for further validation studies across different centers, scanner platforms, and acquisition parameters. Increasing use of machine learning approaches has accelerated the development of robust automated segmentation techniques. For this field to advance, it is imperative that radiologists who are the custodians of medical imaging data help create larger databases. More studies on larger populations are needed to validate automated segmentation techniques. Publicly available databases can provide access to large number of images. A list of these databases can be found at (https://wiki.cancerimagingarchive.net/display/Public/RIDER+NEURO+MRI; https://en.wikipedia.org/wiki/List_of_neuroscience_databases; http://www.aylward.org/notes/open-access-medical-image-repositories). Eventually, automated segmentation techniques will be used in routine clinical practice to help improve patient care. Radiologists, biomedical engineers, and medical physicists will play a key role in transforming clinical radiology practice from being focused on qualitative image interpretation to a more objective quantitative identification of disease markers.
Acknowledgments
FUNDING
National Institute of Health: P30 AG021332 (LL).
Abbreviations
- ML
Machine Learning
- DSC
Dice Similarity Coefficient
- MICCAI
Medical Image Computing and Computer Assisted Intervention
- CNN
Convolutional Neural Network
- FGT
Fibroglandular Tissue
- BPE
Background Parenchymal Enhancement
Appendix A1: Initial Search Strategy
(“Automatic”[tw] OR “Automated”[tw]) AND (((“Image Analysis”[tw] OR “Image Analytics”[tw]) OR (“Segmentation”[tw] OR “Segmentations”[tw]) OR (“Volumetric”[tw] OR “Volumetrics”[tw])) OR ((“Neural Networks”[tw] OR “Neural Network”[tw] OR “Neural Network Models”[tw] OR “Neural Network Model”[tw] OR “Connectionist Models “[tw] OR “Connectionist Model”[tw] OR “Perceptrons”[tw] OR “Perceptron”[tw] OR “Convolutional Neural Network”[tw] OR “Convolutional Neural Networks”[tw] OR “CNN”[tw] OR “CNNs”[tw]) OR (“Machine Learning”[tw]) OR (“Automated Pattern Recognition”[tw] OR “Pattern Recognition System”[tw] OR “Pattern Recognition Systems”[tw]) OR (“Computer-Assisted Image Processing”[tw] OR “Computer-Aided Image Processing”[tw] OR “Computer-Assisted Image Analysis”[tw] OR “Computer-Assisted Image Analyses”[tw] OR “Computer-Aided Image Analysis”[tw] OR “Computer-Generated Image Analysis”[tw] OR “Image Reconstruction”[tw] OR “Image Reconstructions”[tw]) OR (“Finite Element Analysis”[tw] OR “Finite Element Analyses”[tw] OR “Finite Element Method”[tw] OR “Finite Element Methods”[tw] OR “FEM”[tw]) OR (“Cluster Analysis”[tw] OR “Cluster Analyses”[tw] OR “Clustering”[tw] OR “Clusterings”[tw] OR “Disease Clustering”[tw]) OR (“Principal Component Analyses”[tw])) OR (“Neural Networks (Computer)”[majr:noexp] OR “Machine Learning”[majr:noexp] OR “Pattern Recognition, Automated”[majr:noexp] OR “Image Processing, Computer-Assisted”[majr:noexp] OR “Finite Element Analysis”[majr:noexp] OR “Cluster Analysis”[majr:noexp] OR “Principal Component Analysis “[majr:noexp])) AND (“Cardiac-Gated Imaging Techniques”[mesh:noexp] OR “Cardiac-Gated Imaging Techniques”[tw] OR “Radiographic Image Enhancement”[mesh:noexp] OR “Radiographic Image Enhancement”[tw] OR “Radiography, Dual-Energy Scanned Projection”[mesh:noexp] OR “Dual-Energy Scanned Projection Radiography”[tw] OR “Tomography, X-Ray Computed”[mesh:noexp] OR “X-Ray Computed Tomography”[tw] OR “Xray Computed Tomography”[tw] OR “Computed X-Ray Tomography”[tw] OR “X-Ray Computerized Tomography”[tw] OR “Computerized X-Ray Tomography”[tw] OR “CT X-Ray”[tw] OR “CT Xray”[tw] OR “CT X-Rays”[tw] OR “X-Ray CT Scan”[tw] OR “X-Ray CT Scans”[tw] OR “X-Ray Computer Assisted Tomography”[tw] OR “X-Ray Computer Aided Tomography”[tw] OR “X-Ray CAT Scan”[tw] OR “Transmission Computed Tomography”[tw] OR “Transmission CT”[tw] OR “Cine Computed Tomography”[tw] OR “Cine Computerized Tomography”[tw] OR “Cine CT”[tw] OR “Cine CTs”[tw] OR “Electron Beam Computed Tomography”[tw] OR “Electron Beam CT”[tw] OR “Electron Beam Tomography”[tw] OR “Colonography, Computed Tomographic”[mesh:noexp] OR “Computed Tomographic Colonography”[tw] OR “CT Colonography”[tw] OR “Colonography, CT”[tw] OR “Virtual Colonoscopy”[tw] OR “Virtual Colonoscopies”[tw] OR “Four-Dimensional Computed Tomography”[mesh:noexp] OR “Four-Dimensional Computed Tomography”[tw] OR “Four-Dimensional Computerized Tomography”[tw] OR “Four-Dimensional CT”[tw] OR “4-D CT”[tw] OR “4D Computed Tomography”[tw] OR “4D CT”[tw] OR “Four-Dimensional Computed Tomography Scan”[tw] OR “Four-Dimensional Computed Tomography Scans”[tw] OR “Four-Dimensional CT Scan”[tw] OR “Four-Dimensional CT Scans”[tw] OR “4-D CT Scan”[tw] OR “4D CT Scan”[tw] OR “4D CT Scans”[tw] OR “Tomography, Spiral Computed”[mesh:noexp] OR “Spiral Computed Tomography”[tw] OR “Spiral Computerized Tomography”[tw] OR “Spiral Computer-Assisted Tomography”[tw] OR “Spiral CT”[tw] OR “Spiral CTs”[tw] OR “Spiral CT Scan”[tw] OR “Spiral CT Scans”[tw] OR “Spiral CAT Scan”[tw] OR “Spiral CAT Scans”[tw] OR “Helical Computed Tomography”[tw] OR “Helical Computerized Tomography”[tw] OR “Helical CT”[tw] OR “Helical CTs”[tw] OR “Helical CT Scan”[tw] OR “Helical CT Scans”[tw] OR “Multidetector Computed Tomography”[mesh:noexp] OR “Multidetector Computed Tomography”[tw] OR “Multidetector Computerized Tomography”[tw] OR “Multisection Computed Tomography”[tw] OR “Multislice Computed Tomography”[tw] OR “Multislice Computerized Tomography”[tw] OR “Multi-detectorRow Computed Tomography”[tw] OR “Multidetector-Row Computerized Tomography”[tw] OR “Multidetector CT”[tw] OR “Multisection CT”[tw] OR “Multislice CT”[tw] OR “Multislice CTs”[tw] OR “Multidetector-Row CT”[tw] OR “Radiographic Image Interpretation, Computer-Assisted”[mesh:noexp] OR “Radiographic Image Interpretation, Computer-Assisted”[tw] OR “Computer-Assisted Radiographic Image Interpretation”[tw] OR “Computer-Aided Radiographic Image Interpretation”[tw] OR “Magnetic Resonance Imaging”[mesh:noexp] OR “Magnetic Resonance Imaging”[tw] OR “Magnetic Resonance Imagings”[tw] OR “MR Imaging”[tw] OR “MR Imagings”[tw] OR “MRI”[tw] OR “MRIs”[tw] OR “Spin Echo Imaging”[tw] OR “Magnetization Transfer Contrast Imaging”[tw] OR “Steady-State Free Precession MR Imaging”[tw] OR “Steady-State Free Precession MRI”[tw] OR “Magnetic Resonance Tomography”[tw] OR “Magnetic Resonance Tomographies”[tw] OR “MR Tomography”[tw] OR “MR Tomographies”[tw] OR “Nuclear Magnetic Resonance Tomography”[tw] OR “NMR Tomography”[tw] OR “Proton Spin Tomography”[tw] OR “Zeugmatography”[tw] OR “Magnetic Resonance Imaging Scan”[tw] OR “Magnetic Resonance Imaging Scans”[tw] OR “MR Imaging Scan”[tw] OR “MR Imaging Scans”[tw] OR “MRI Scan”[tw] OR “MRI Scans”[tw] OR “Cholangiopancreatography, Magnetic Resonance”[mesh:noexp] OR “Magnetic Resonance Cholangiopancreatography”[tw] OR “Magnetic Resonance Cholangiopancreatographies”[tw] OR “MR Cholangiopancreatography”[tw] OR “Echo-Planar Imaging”[mesh:noexp] OR “Echo-Planar Imaging”[tw] OR “Echoplanar Imaging”[tw] OR “Echo-Planar Magnetic Resonance Imaging”[tw] OR “Echoplanar Magnetic Resonance Imaging”[tw] OR “Echo-Planar MR Imaging”[tw] OR “Echo-Planar MRI”[tw] OR “Echoplanar MRI”[tw] OR “Echoplanar MR Tomography”[tw] OR “Magnetic Resonance Angiography”[mesh:noexp] OR “Magnetic Resonance Angiography”[tw] OR “Magnetic Resonance Imaging Angiography”[tw] OR “MR Angiography”[tw] OR “MR Angiographies”[tw] OR “MRI Angiography”[tw] OR “MRI Angiographies”[tw] OR “Magnetic Resonance Imaging, Cine”[mesh:noexp] OR “Cine Magnetic Resonance Imaging”[tw] OR “Cine MR Imaging”[tw] OR “Cine MRI”[tw] OR “Cine MRIs”[tw] OR “Whole Body Imaging”[mesh:noexp] OR “Whole Body Imaging”[tw] OR “Whole Body Image”[tw] OR “Whole Body Images”[tw] OR “Whole Body Scanning”[tw] OR “Whole Body Scan”[tw] OR “Whole Body Scans”[tw] OR “Whole Body Screening”[tw] OR “Whole Body Screenings”[tw] OR “Whole Body Screen”[tw] OR “Computed Tomography”[tw] OR “Computed Tomographies”[tw] OR “Computerized Tomography”[tw] OR “Computerized Tomographies”[tw] OR “CT”[tw] OR “CTs”[tw] OR “Computed Tomography Imaging”[tw] OR “Computed Tomography Imagings”[tw] OR “Computerized Tomography Imaging”[tw] OR “CT Imaging”[tw] OR “CT Imagings”[tw] OR “Computed Tomography Image”[tw] OR “Computed Tomography Images”[tw] OR “Computerized Tomography Image”[tw] OR “Computerized Tomography Images”[tw] OR “CT Image”[tw] OR “CT Images”[tw] OR “Three-Dimensional Computed Tomography”[tw] OR “Three-Dimensional Computerized Tomography”[tw] OR “Three-Dimensional CT”[tw] OR “3-D Computed Tomography”[tw] OR “3-D CT”[tw] OR “3D Computed Tomography”[tw] OR “3D CT”[tw] OR “Three-Dimensional Computed Tomography Scan”[tw] OR “Three-Dimensional Computed Tomography Scans”[tw] OR “3D Computed Tomography Scan”[tw] OR “Three-Dimensional CT Scan”[tw] OR “Three-Dimensional CT Scans”[tw] OR “3-D CT Scan”[tw] OR “3-D CT Scans”[tw] OR “3D CT Scan”[tw] OR “3D CT Scans”[tw] OR “Three-Dimensional CAT Scan”[tw] OR “Cross-Sectional Imaging”[tw] OR “Cross-Sectional Diagnostic Imaging”[tw] OR “Cross-Sectional Computed Tomography”[tw] OR “Cross-Sectional Computerized Tomography”[tw] OR “Cross-Sectional CT”[tw] OR “Cross-Sectional Magnetic Resonance Imaging”[tw] OR “Cross-Sectional MRI”[tw]) AND (“Cardiac-Gated Imaging Techniques”[majr:noexp] OR “Cardiac-Gated Imaging Techniques”[tw] OR “Radiographic Image Enhancement”[majr:noexp] OR “Radiographic Image Enhancement”[tw] OR “Radiography, Dual-Energy Scanned Projection”[majr:noexp] OR “Dual-Energy Scanned Projection Radiography”[tw] OR “Tomography, X-Ray Computed”[majr:noexp] OR “X-Ray Computed Tomography”[tw] OR “Xray Computed Tomography”[tw] OR “Computed X-Ray Tomography”[tw] OR “X-Ray Computerized Tomography”[tw] OR “Computerized X-Ray Tomography”[tw] OR “CT X-Ray”[tw] OR “CT Xray”[tw] OR “CT X-Rays”[tw] OR “X-Ray CT Scan”[tw] OR “X-Ray CT Scans”[tw] OR “X-Ray Computer Assisted Tomography”[tw] OR “X-Ray Computer Aided Tomography”[tw] OR “X-Ray CAT Scan”[tw] OR “Transmission Computed Tomography”[tw] OR “Transmission CT”[tw] OR “Cine Computed Tomography”[tw] OR “Cine Computerized Tomography”[tw] OR “Cine CT”[tw] OR “Cine CTs”[tw] OR “Electron Beam Computed Tomography”[tw] OR “Electron Beam CT”[tw] OR “Electron Beam Tomography”[tw] OR “Colonography, Computed Tomographic”[majr:noexp] OR “Computed Tomographic Colonography”[tw] OR “CT Colonography”[tw] OR “Colonography, CT”[tw] OR “Virtual Colonoscopy”[tw] OR “Virtual Colonoscopies”[tw] OR “Four-Dimensional Computed Tomography”[majr:noexp] OR “Four-Dimensional Computed Tomography”[tw] OR “Four-Dimensional Computerized Tomography”[tw] OR “Four-Dimensional CT”[tw] OR “4-D CT”[tw] OR “4D Computed Tomography”[tw] OR “4D CT”[tw] OR “Four-Dimensional Computed Tomography Scan”[tw] OR “Four-Dimensional Computed Tomography Scans”[tw] OR “Four-Dimensional CT Scan”[tw] OR “Four-Dimensional CT Scans”[tw] OR “4-D CT Scan”[tw] OR “4D CT Scan”[tw] OR “4D CT Scans”[tw] OR “Tomography, Spiral Computed”[majr:noexp] OR “Spiral Computed Tomography”[tw] OR “Spiral Computerized Tomography”[tw] OR “Spiral Computer-Assisted Tomography”[tw] OR “Spiral CT”[tw] OR “Spiral CTs”[tw] OR “Spiral CT Scan”[tw] OR “Spiral CT Scans”[tw] OR “Spiral CAT Scan”[tw] OR “Spiral CAT Scans”[tw] OR “Helical Computed Tomography”[tw] OR “Helical Computerized Tomography”[tw] OR “Helical CT”[tw] OR “Helical CTs”[tw] OR “Helical CT Scan”[tw] OR “Helical CT Scans”[tw] OR “Multidetector Computed Tomography”[majr:noexp] OR “Multidetector Computed Tomography”[tw] OR “Multidetector Computerized Tomography”[tw] OR “Multisection Computed Tomography”[tw] OR “Multislice Computed Tomography”[tw] OR “Multislice Computerized Tomography”[tw] OR “Multidetector-Row Computed Tomography”[tw] OR “Multidetector-Row Computerized Tomography”[tw] OR “Multidetector CT”[tw] OR “Multisection CT”[tw] OR “Multislice CT”[tw] OR “Multislice CTs”[tw] OR “Multidetector-Row CT”[tw] OR “Radiographic Image Interpretation, Computer-Assisted”[majr:noexp] OR “Radiographic Image Interpretation, Computer-Assisted”[tw] OR “Computer-Assisted Radiographic Image Interpretation”[tw] OR “Computer-Aided Radiographic Image Interpretation”[tw] OR “Magnetic Resonance Imaging”[majr:noexp] OR “Magnetic Resonance Imaging”[tw] OR “Magnetic Resonance Imagings”[tw] OR “MR Imaging”[tw] OR “MR Imagings”[tw] OR “MRI”[tw] OR “MRIs”[tw] OR “Spin Echo Imaging”[tw] OR “Magnetization Transfer Contrast Imaging”[tw] OR “Steady-State Free Precession MR Imaging”[tw] OR “Steady-State Free Precession MRI”[tw] OR “Magnetic Resonance Tomography”[tw] OR “Magnetic Resonance Tomographies”[tw] OR “MR Tomography”[tw] OR “MR Tomographies”[tw] OR “Nuclear Magnetic Resonance Tomography”[tw] OR “NMR Tomography”[tw] OR “Proton Spin Tomography”[tw] OR “Zeugmatography”[tw] OR “Magnetic Resonance Imaging Scan”[tw] OR “Magnetic Resonance Imaging Scans”[tw] OR “MR Imaging Scan”[tw] OR “MR Imaging Scans”[tw] OR “MRI Scan”[tw] OR “MRI Scans”[tw] OR “Cholangiopancreatography, Magnetic Resonance”[majr:noexp] OR “Magnetic Resonance Cholangiopancreatography”[tw] OR “Magnetic Resonance Cholangiopancreatographies”[tw] OR “MR Cholangiopancreatography”[tw] OR “Echo-Planar Imaging”[majr:noexp] OR “Echo-Planar Imaging”[tw] OR “Echoplanar Imaging”[tw] OR “Echo-Planar Magnetic Resonance Imaging”[tw] OR “Echoplanar Magnetic Resonance Imaging”[tw] OR “Echo-Planar MR Imaging”[tw] OR “Echo-Planar MRI”[tw] OR “Echoplanar MRI”[tw] OR “Echoplanar MR Tomography”[tw] OR “Magnetic Resonance Angiography”[majr:noexp] OR “Magnetic Resonance Angiography”[tw] OR “Magnetic Resonance Imaging Angiography”[tw] OR “MR Angiography”[tw] OR “MR Angiographies”[tw] OR “MRI Angiography”[tw] OR “MRI Angiographies”[tw] OR “Magnetic Resonance Imaging, Cine”[majr:noexp] OR “Cine Magnetic Resonance Imaging”[tw] OR “Cine MR Imaging”[tw] OR “Cine MRI”[tw] OR “Cine MRIs”[tw] OR “Whole Body Imaging”[majr:noexp] OR “Whole Body Imaging”[tw] OR “Whole Body Image”[tw] OR “Whole Body Images”[tw] OR “Whole Body Scanning”[tw] OR “Whole Body Scan”[tw] OR “Whole Body Scans”[tw] OR “Whole Body Screening”[tw] OR “Whole Body Screenings”[tw] OR “Whole Body Screen”[tw] OR “Computed Tomography”[tw] OR “Computed Tomographies”[tw] OR “Computerized Tomography”[tw] OR “Computerized Tomographies”[tw] OR “CT”[tw] OR “CTs”[tw] OR “Computed Tomography Imaging”[tw] OR “Computed Tomography Imagings”[tw] OR “Computerized Tomography Imaging”[tw] OR “CT Imaging”[tw] OR “CT Imagings”[tw] OR “Computed Tomography Image”[tw] OR “Computed Tomography Images”[tw] OR “Computerized Tomography Image”[tw] OR “Computerized Tomography Images”[tw] OR “CT Image”[tw] OR “CT Images”[tw] OR “Three-Dimensional Computed Tomography”[tw] OR “Three-Dimensional Computerized Tomography”[tw] OR “Three-Dimensional CT”[tw] OR “3-D Computed Tomography”[tw] OR “3-D CT”[tw] OR “3D Computed Tomography”[tw] OR “3D CT”[tw] OR “Three-Dimensional Computed Tomography Scan”[tw] OR “Three-Dimensional Computed Tomography Scans”[tw] OR “3D Computed Tomography Scan”[tw] OR “Three-Dimensional CT Scan”[tw] OR “Three-Dimensional CT Scans”[tw] OR “3-D CT Scan”[tw] OR “3-D CT Scans”[tw] OR “3D CT Scan”[tw] OR “3D CT Scans”[tw] OR “Three-Dimensional CAT Scan”[tw] OR “Cross-Sectional Imaging”[tw] OR “Cross-Sectional Diagnostic Imaging”[tw] OR “Cross-Sectional Computed Tomography”[tw] OR “Cross-Sectional Computerized Tomography”[tw] OR “Cross-Sectional CT”[tw] OR “Cross-Sectional Magnetic Resonance Imaging”[tw] OR “Cross-Sectional MRI”[tw]) NOT (“Animals”[mh] NOT “Humans”[mh]) AND (“2013/01/23”[PDat] : “2018/01/23”)
Appendix A2: Secondary Search Strategy
(“tomography, x-ray computed”[MeSH Terms] OR (“tomography”[All Fields] AND “x-ray”[All Fields] AND “computed”[All Fields]) OR “x-ray computed tomography”[All Fields] OR (“computed”[All Fields] AND “tomography”[All Fields]) OR “computed tomography”[All Fields])) OR ((“magnetic resonance imaging”[MeSH Terms] OR (“magnetic”[All Fields] AND “resonance”[All Fields] AND “imaging”[All Fields]) OR “magnetic resonance imaging”[All Fields] OR “mri”[All Fields])) AND ((“tissue segmentation”[All fields] OR “tissue classification”[All fields] OR “machine learning”[All fields] OR “volumetry”[All fields] OR “neural networks”[All fields] OR “segmentation”[All fields]) AND (“automatic”[All Fields] OR “automated”[All Fields] OR “automatically”[All Fields]))
Appendix B: Manuscripts Included in Systematic Review
B1. Neurosegmentation
- 1.Abbasi S, Tajeripour F. Detection of brain tumor in 3D MRI images using local binary patterns and histogram orientation gradient. Neurocomputing. 2017; 219:526–535. [Google Scholar]
- 2.Adamson C, Da Costa AC, Beare R, Wood AG. Automatic intracranial space segmentation for computed tomography brain images. J. Digit. Imaging. 2013; 26(3):563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamson C, Beare R, Walterfang M, Seal M. Software Pipeline for Midsagittal Corpus Callosum Thickness Profile Processing: Automated Segmentation, Manual Editor, Thickness Profile Generator, Group-Wise Statistical Comparison and Results Display. Neuroinformatics. 2014; 12(4):595–614. [DOI] [PubMed] [Google Scholar]
- 4.Adler S, Wagstyl K, Gunny R, et al. Novel surface features for automated detection of focal cortical dysplasias in paediatric epilepsy. NeuroImage Clin. 2017; 14:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aghdasi N, Li Y, Berens A, et al. Efficient orbital structures segmentation with prior anatomical knowledge. J. Med. Imaging 2017; 4(3): 034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmed B, Brodley CE, Blackmon KE, et al. Cortical feature analysis and machine learning improves detection of MRI-negative focal cortical dysplasia. Epilepsy Behav. 2015; 48:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: Impact of cross-institutional training and testing: Impact. Med. Phys. 2018; 45(3):1150–1158. [DOI] [PubMed] [Google Scholar]
- 8.Amoroso N, Errico R, Bruno S, et al. Hippocampal unified multi-atlas network (HUMAN): protocol and scale validation of a novel segmentation tool. Phys. Med. Biol. 2015; 60(22):8851–8897. [DOI] [PubMed] [Google Scholar]
- 9.Anandh KR, Sujatha CM, Ramakrishnan S. A method to differentiate mild cognitive impairment and Alzheimer in MR images using eigen value descriptors. J. Med. Syst 2016; 40(1):25. [DOI] [PubMed] [Google Scholar]
- 10.Asman AJ, Huo Y, Plassard AJ, Landman BA. Multi-atlas learner fusion: An efficient segmentation approach for large-scale data. Med. Image Anal 2015; 26(1):82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asman AJ, Bryan FW, Smith SA, Reich DS, Landman BA. Groupwise multi-atlas segmentation of the spinal cord’s internal structure. Med. Image Anal 2014; 18(3):460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beheshti I, Demirel H, Initiative ADN, others. Probability distribution function-based classification of structural MRI for the detection of Alzheimer’s disease. Comput. Biol. Med 2015; 64:208–216. [DOI] [PubMed] [Google Scholar]
- 13.Benkarim OM, Piella G, González Ballester MA, Sanroma G. Discriminative confidence estimation for probabilistic multi-atlas label fusion. Med. Image Anal 2017; 42:274–287. [DOI] [PubMed] [Google Scholar]
- 14.Bhagwat N, Pipitone J, Winterburn JL, et al. Manual-protocol inspired technique for improving automated MR image segmentation during label fusion. Front. Neurosci 2016; 10:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bijar A, Khayati R, Peñalver Benavent A, Benavent AP. Increasing the contrast of the brain mr flair images using fuzzy membership functions and structural similarity indices in order to segment ms lesions. PLoS One. 2013; 8(6):e65469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brosch T, Tang LYW, Yoo Y, et al. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging. 2016; 35(5):1229–1239. [DOI] [PubMed] [Google Scholar]
- 17.Cabezas M, Oliver A, Roura E, et al. Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding. Comput. Methods Programs Biomed 2014; 115(3):147–161. [DOI] [PubMed] [Google Scholar]
- 18.Cabezas M, Oliver A, Valverde S, et al. BOOST: A supervised approach for multiple sclerosis lesion segmentation. J. Neurosci. Methods. 2014; 237:108–117. [DOI] [PubMed] [Google Scholar]
- 19.Chaddad A, Tanougast C. Quantitative evaluation of robust skull stripping and tumor detection applied to axial MR images. Brain Informatics. 2016; 3(1):53–61.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charron O, Lallement A, Jarnet D, et al. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput. Biol. Med 2018; 95:43–54. [DOI] [PubMed] [Google Scholar]
- 21.Chen M, Carass A, Oh J, et al. Automatic magnetic resonance spinal cord segmentation with topology constraints for variable fields of view. Neuroimage. 2013; 83:1051–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Dhar R, Heitsch L, et al. Automated quantification of cerebral edema following hemispheric infarction: application of a machine-learning algorithm to evaluate CSF shifts on serial head CTs. NeuroImage Clin. 2016; 12:673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Zhao B, Zhang J, Zheng Y. Automatic segmentation for brain MR images via a convex optimized segmentation and bias field correction coupled model. Magn. Reson. Imaging 2014; 32(7):941–955. [DOI] [PubMed] [Google Scholar]
- 24.Cordier N, Delingette H, Ayache N. A Patch-Based Approach for the Segmentation of Pathologies: Application to Glioma Labelling. IEEE Trans. Med. Imaging 2016; 35(4):1066–1076. [DOI] [PubMed] [Google Scholar]
- 25.Cui S, Mao L, Xiong S. Brain Tumor Automatic Segmentation Using Fully Convolutional Networks. J. Med. Imaging Heal. Informatics 2017; 7(7):1641–1647. [Google Scholar]
- 26.da Silva Senra Filho AC. A hybrid approach based on logistic classification and iterative contrast enhancement algorithm for hyperintense multiple sclerosis lesion segmentation. Med. Biol. Eng. Comput 2018; 56(6):1063–1076. [DOI] [PubMed] [Google Scholar]
- 27.Dadar M, Pascoal TA, Manitsirikul S, et al. Validation of a Regression Technique for Segmentation of White Matter Hyperintensities in Alzheimer’s Disease. IEEE Trans. Med. Imaging 2017; 36(8):1758–1768. [DOI] [PubMed] [Google Scholar]
- 28.Datta S, Narayana PA. A comprehensive approach to the segmentation of multichannel three-dimensional MR brain images in multiple sclerosis. NeuroImage Clin. 2013; 2(1):184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C. Multi-atlas skull-stripping. Acad. Radiol 2013; 20(12):1566–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupont SM, De Leener B, Taso M, et al. Fully-integrated framework for the segmentation and registration of the spinal cord white and gray matter. Neuroimage. 2017; 150:358–372. [DOI] [PubMed] [Google Scholar]
- 31.Fartaria MJ, Bonnier G, Roche A, et al. Automated detection of white matter and cortical lesions in early stages of multiple sclerosis. J. Magn. Reson. Imaging 2016; 43(6):1445–1454. [DOI] [PubMed] [Google Scholar]
- 32.Feng X, Deistung A, Dwyer MG, et al. An improved FSL-FIRST pipeline for subcortical gray matter segmentation to study abnormal brain anatomy using quantitative susceptibility mapping (QSM). Magn. Reson. Imaging 2017; 39:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galimzianova A, Pernuš F, Likar B, Špiclin Ţ. Stratified mixture modeling for segmentation of white-matter lesions in brain MR images. Neuroimage. 2016; 124:1031–1043. [DOI] [PubMed] [Google Scholar]
- 34.Gao J, Li C, Feng C, et al. Non-locally regularized segmentation of multiple sclerosis lesion from multi-channel MRI data. Magn. Reson. Imaging 2014; 32(8):1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghafoorian M, Karssemeijer N, Heskes T, et al. Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. NeuroImage Clin. 2017; 14:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghafoorian M, Karssemeijer N, van Uden IWM, et al. Automated detection of white matter hyperintensities of all sizes in cerebral small vessel disease. Med. Phys 2016; 43(12):6246–6258. [DOI] [PubMed] [Google Scholar]
- 37.Ghribi O, Sellami L, Ben Slima M, et al. An Advanced MRI Multi-Modalities Segmentation Methodology Dedicated to Multiple Sclerosis Lesions Exploration and Differentiation. IEEE Trans. Nanobioscience 2017; 16(8):656–665. [DOI] [PubMed] [Google Scholar]
- 38.Ghribi O, Sellami L, Slima M Ben, et al. Multiple sclerosis exploration based on automatic MRI modalities segmentation approach with advanced volumetric evaluations for essential feature extraction. Biomed. Signal Process. Control 2018; 40:473–487. [Google Scholar]
- 39.Gillebert CRCR, Humphreys GW, Mantini D. Automated delineation of stroke lesions using brain CT images. NeuroImage Clin. 2014; 4:540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giraud R, Ta V-TT, Papadakis N, et al. An optimized patchmatch for multi-scale and multi-feature label fusion. Neuroimage. 2016; 124:770–782. [DOI] [PubMed] [Google Scholar]
- 41.Glatz A, Bastin ME, Kiker AJ, et al. Automated segmentation of multifocal basal ganglia T2*-weighted MRI hypointensities. Neuroimage. 2015; 105:332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffis JC, Allendorfer JB, Szaflarski JP. Voxel-based Gaussian naive Bayes classification of ischemic stroke lesions in individual T1-weighted MRI scans. J. Neurosci. Methods 2016; 257:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gros C, De Leener B, Dupont SM, et al. Automatic spinal cord localization, robust to MRI contrasts using global curve optimization. Med. Image Anal 2018; 44:215–227. [DOI] [PubMed] [Google Scholar]
- 44.Guerrero R, Qin C, Oktay O, et al. White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. NeuroImage Clin. 2018; 17:918–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guizard N, Coupé P, Fonov VS, et al. Rotation-invariant multi-contrast non-local means for MS lesion segmentation. NeuroImage Clin 2015; 8:376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo D, Fridriksson J, Fillmore P, et al. Automated lesion detection on MRI scans using combined unsupervised and supervised methods. BMC Med. Imaging 2015; 15(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanning U, Sporns PB, Schmidt R, et al. Quantitative Rapid Assessment of Leukoaraiosis in CT: Comparison to Gold Standard MRI. Clin. Neuroradiol 2017; 29(1):109–115. [DOI] [PubMed] [Google Scholar]
- 48.Hao Y, Wang T, Zhang X, et al. Local label learning (LLL) for subcortical structure segmentation: application to hippocampus segmentation. Hum. Brain Mapp 2014; 35(6):2674–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harmouche R, Subbanna NK, Collins DL, Arnold DL, Arbel T. Probabilistic multiple sclerosis lesion classification Based on Modeling Regional Intensity Variability and Local Neighborhood Information. IEEE Trans Biomed Eng. 2015; 62(5):1281–1292 2014; 62:1281–1292. [DOI] [PubMed] [Google Scholar]
- 50.Harrigan RL, Panda S, Asman AJ, et al. Robust optic nerve segmentation on clinically acquired computed tomography. J. Med. Imaging 2014; 1(3):034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heckemann RA, Ledig C, Gray KR, et al. Brain extraction using label propagation and group agreement: pincram. PLoS One. 2015; 10(7):e0129211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hentschke CM, Beuing O, Paukisch H, et al. A system to detect cerebral aneurysms in multimodality angiographic data sets. Med. Phys 2014; 41(9):91904. [DOI] [PubMed] [Google Scholar]
- 53.Huang M, Yang W, Wu Y, et al. Brain tumor segmentation based on local independent projection-based classification. IEEE Trans. Biomed. Eng 2014; 61(10):2633–2645. [DOI] [PubMed] [Google Scholar]
- 54.Ismail M, Soliman A, Ghazal M, et al. A fast stochastic framework for automatic MR brain images segmentation. PLoS One. 2017; 12(11):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain S, Sima DM, Ribbens A, et al. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 2015; 8:367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji S, Ye C, Li F, et al. Automatic segmentation of white matter hyperintensities by an extended FitzHugh & Nagumo reaction diffusion model. J. Magn. Reson. Imaging 2013; 37(2):343–350. [DOI] [PubMed] [Google Scholar]
- 57.Jorge Cardoso M, Leung K, Modat M, et al. STEPS: Similarity and Truth Estimation for Propagated Segmentations and its application to hippocampal segmentation and brain parcelation. Med. Image Anal. 2013; 17(6):671–684. [DOI] [PubMed] [Google Scholar]
- 58.Juan-Albarracín J, Fuster-Garcia E, Manjón JV., et al. Automated glioblastoma segmentation based on a multiparametric structured unsupervised classification. PLoS One. 2015; 10(5): e0125143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karimaghaloo Z, Rivaz H, Arnold DL, Collins DL, Arbel T. Temporal hierarchical adaptive texture CRF for automatic detection of gadolinium-enhancing multiple sclerosis lesions in brain MRI. IEEE Trans. Med. Imaging 2015; 34(6):1227–1241. [DOI] [PubMed] [Google Scholar]
- 60.Karimian A, Jafari S. A new method to segment the multiple sclerosis lesions on brain magnetic resonance images. J. Med. Signals Sens 2015; 5(4):238–244. [PMC free article] [PubMed] [Google Scholar]
- 61.Kazemifar S, Drozd JJ, Rajakumar N, et al. Automated algorithm to measure changes in medial temporal lobe volume in Alzheimer disease. J. Neurosci. Methods 2014; 227:35–46. [DOI] [PubMed] [Google Scholar]
- 62.Kim EY, Johnson HJ. Robust multi-site MR data processing: iterative optimization of bias correction, tissue classification, and registration. Front. Neuroinform 2013; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim H, Caldairou B, Hwang JW, et al. Accurate cortical tissue classification on MRI by modeling cortical folding patterns. Hum. Brain Mapp. 2015; 36(9):3563–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korfiatis P, Kline TL, Erickson BJ. Automated segmentation of hyperintense regions in FLAIR MRI using deep learning. Tomography. 2016; 2(4):334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ledig C, Heckemann RA, Hammers A, et al. Robust whole-brain segmentation: Application to traumatic brain injury. Med. Image Anal 2015; 21(1):40–58. [DOI] [PubMed] [Google Scholar]
- 66.Lee DK, Yoon U, Kwak K, Lee JM. Automated segmentation of cerebellum using brain mask and partial volume estimation map. Comput. Math. Methods Med. 2015; 2015:167489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li S, Zhang X, Dai J, et al. An Improvement Method of Brain Extraction Tools for Magnetic Resonance Images. J. Med. Imaging Heal. Informatics 2014; 4(6):895–900. [Google Scholar]
- 68.Li XW, Li QL, Li SY, Li DY. Local manifold learning for multiatlas segmentation: Application to hippocampal segmentation in healthy population and Alzheimer’s disease. CNS Neurosci. Ther 2015; 21(10):826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Jia F, Qin J. Brain tumor segmentation from multimodal magnetic resonance images via sparse representation. Artif. Intell. Med 2016; 73:1–13. [DOI] [PubMed] [Google Scholar]
- 70.Liao CC, Ting HW, Xiao F. Atlas-Free Cervical Spinal Cord Segmentation on Midsagittal T2-Weighted Magnetic Resonance Images. J. Healthc. Eng 2017; 2017: 8691505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liberman G, Louzoun Y, Aizenstein O, et al. Automatic multi-modal MR tissue classification for the assessment of response to bevacizumab in patients with glioblastoma. Eur. J. Radiol 2013; 82(2):e87–e94. [DOI] [PubMed] [Google Scholar]
- 72.Liu H-T, Sheu TWH, Chang H-H. Automatic segmentation of brain MR images using an adaptive balloon snake model with fuzzy classification. Med. Biol. Eng. Comput 2013; 51(10):1091–1104. [DOI] [PubMed] [Google Scholar]
- 73.Loh WY, Connelly A, Cheong JL et al. No TitleA New MRI-Based Pediatric Subcortical Segmentation Technique (PSST). Neuroinformatics. 2016; 14(1):69–81. [DOI] [PubMed] [Google Scholar]
- 74.Maglietta R, Amoroso N, Boccardi M, et al. Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern Anal. Appl 2016; 19(2):579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahmood Q, Chodorowski A, Persson M. Automated MRI brain tissue segmentation based on mean shift and fuzzy c -means using a priori tissue probability maps. IRBM. 2015; 36(3):185–196. [Google Scholar]
- 76.Maji P, Roy S. Rough-fuzzy clustering and unsupervised feature selection for wavelet based MR image segmentation. PLoS One. 2015; 10(6):e0132081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manniesing R, Oei MTH, Oostveen LJ, et al. White matter and gray matter segmentation in 4D computed tomography. Sci. Rep 2017; 7(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martinez-Murcia FJ, Górriz JM, Ram’irez J, Ortiz A, The Alzheimer’s Disease Neuroimaging Initiative. A spherical brain mapping of MR images for the detection of Alzheimer’s disease. Curr. Alzheimer Res 2016; 13(5):575–588. [DOI] [PubMed] [Google Scholar]
- 79.Mechrez R, Goldberger J, Greenspan H. Patch-based segmentation with spatial consistency: application to MS lesions in brain MRI. J. Biomed. Imaging 2016; 2016:7952541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mehta R, Majumdar A, Sivaswamy J. BrainSegNet: a convolutional neural network architecture for automated segmentation of human brain structures. J. Med. Imaging 2017; 4(2):24003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meier DS, Guttmann CRG, Tummala S, et al. Dual-Sensitivity Multiple Sclerosis Lesion and CSF Segmentation for Multichannel 3T Brain MRI. J. Neuroimaging 2018; 28(1):36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michoux N, Guillet A, Rommel D, et al. Texture analysis of T2-weighted MR images to assess acute inflammation in brain MS lesions. PLoS One. 2015; 10(12):e0145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitra J, Bourgeat P, Fripp J, et al. Lesion segmentation from multimodal MRI using random forest following ischemic stroke. Neuroimage. 2014; 98:324–335. [DOI] [PubMed] [Google Scholar]
- 84.Moeskops P, de Bresser J, Kuijf HJ, et al. Evaluation of a deep learning approach for the segmentation of brain tissues and white matter hyperintensities of presumed vascular origin in MRI. NeuroImage Clin. 2018; 17:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moeskops P, Viergever MA, Mendrik AM, et al. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans. Med. Imaging 2016; 35(5):1252–1261. [DOI] [PubMed] [Google Scholar]
- 86.Nigro S, Cerasa A, Zito G, et al. Fully automated segmentation of the pons and midbrain using human T1 MR brain images. PLoS One. 2014; 9(1):e85618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Njeh I, Sallemi L, Ayed I Ben, et al. 3D multimodal MRI brain glioma tumor and edema segmentation: a graph cut distribution matching approach. Comput. Med. Imaging Graph 2015; 40:108–119. [DOI] [PubMed] [Google Scholar]
- 88.Nowinski WL, Gupta V, Qian G, et al. Automatic detection, localization, and volume estimation of ischemic infarcts in noncontrast computed tomographic scans: method and preliminary results. Invest. Radiol 2013; 48(9):661–670. [DOI] [PubMed] [Google Scholar]
- 89.Panda S, Asman AJ, Khare SP, et al. Evaluation of multiatlas label fusion for in vivo magnetic resonance imaging orbital segmentation. J. Med. Imaging 2014; 1(2):024002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016; 35(5):1240–1251. [DOI] [PubMed] [Google Scholar]
- 91.Pereira S, Pinto A, Oliveira J, et al. Automatic brain tissue segmentation in MR images using random forests and conditional random fields. J. Neurosci. Methods 2016; 270:111–123. [DOI] [PubMed] [Google Scholar]
- 92.Pipitone J, Park MTM, Winterburn J, et al. Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014; 101:494–512. [DOI] [PubMed] [Google Scholar]
- 93.Plassard AJ, McHugo M, Heckers S, Landman BA. Multi-scale hippocampal parcellation improves atlas-based segmentation accuracy In: Medical Imaging 2017: Image Processing; Vol 10133; 2017:101332D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Plassard AJ, Yang Z, Rane S, et al. Improving cerebellar segmentation with statistical fusion. Med. Imaging 2016 Image Process. 2016; 9784:97842R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Platero C, Tobar MC. A fast approach for hippocampal segmentation from T1-MRI for predicting progression in Alzheimer’s disease from elderly controls. J. Neurosci. Methods 2016; 270:61–75. [DOI] [PubMed] [Google Scholar]
- 96.Prakash RM, Kumari RSS. Spatial fuzzy c means and expectation maximization algorithms with bias correction for segmentation of MR brain images. J. Med. Syst 2017; 41(1):15. [DOI] [PubMed] [Google Scholar]
- 97.Price M, Cardenas VA, Fein G. Automated MRI cerebellar size measurements using active appearance modeling. Neuroimage. 2014; 103:511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Puonti O, Iglesias JE, Van Leemput K. Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. Neuroimage. 2016; 143:235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Puonti O, Iglesias JE, Van Leemput K. Fast, sequence adaptive parcellation of brain MR using parametric models. In: International Conference on Medical Image Computing and Computer-Assisted Intervention 2013:727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pustina D, Coslett HB, Turkeltaub PE, et al. Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Hum. Brain Mapp 2016; 37(4):1405–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rajchl M, Baxter JSH, McLeod AJ, et al. Hierarchical max-flow segmentation framework for multi-atlas segmentation with Kohonen self-organizing map based Gaussian mixture modeling. Med. Image Anal 2016; 27:45–56. [DOI] [PubMed] [Google Scholar]
- 102.Ranta ME, Chen M, Crocetti D, et al. Automated MRI parcellation of the frontal lobe. Hum. Brain Mapp 2014; 35(5):2009–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Resmi A, Thomas T, Thomas B. A novel automatic method for extraction of glioma tumour, white matter and grey matter from brain magnetic resonance images. Biomed Imaging Interv J 2013; 9(2):e21. [Google Scholar]
- 104.Rincón M, Diaz-López E, Selnes P, et al. Improved automatic segmentation of white matter hyperintensities in MRI based on multilevel lesion features. Neuroinformatics. 2017; 15(3):231–245. [DOI] [PubMed] [Google Scholar]
- 105.Romero JE, Coupé P, Giraud R, et al. CERES: A new cerebellum lobule segmentation method. Neuroimage. 2017; 147:916–924. [DOI] [PubMed] [Google Scholar]
- 106.Romero JE, Manjón JV, Tohka J, Coupé P, Robles M. NABS: non-local automatic brain hemisphere segmentation. Magn. Reson. Imaging 2015; 33(4):474–484. [DOI] [PubMed] [Google Scholar]
- 107.Roura E, Sarbu N, Oliver A, Valverde S, González-Villà S, Cervera R, Bargalló N, Lladó X. Automated detection of lupus white matter lesions in MRI. Frontiers in neuroinformatics. 2016. August 12;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roy S, Butman JA, Pham DL. Robust skull stripping using multiple MR image contrasts insensitive to pathology. Neuroimage. 2017; 146:132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sanjuán A, Price CJ, Mancini L, et al. Automated identification of brain tumors from single MR images based on segmentation with refined patient-specific priors. Front. Neurosci. 2013; 7:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Serag A, Wilkinson AG, Telford EJ, et al. SEGMA: An Automatic SEGMentation Approach for Human Brain MRI Using Sliding Window and Random Forests. Front. Neuroinform 2017; 11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Simões R, Mönninghoff C, Dlugaj M, et al. Automatic segmentation of cerebral white matter hyperintensities using only 3D FLAIR images. Magn. Reson. Imaging 2013; 31(7):1182–1189. [DOI] [PubMed] [Google Scholar]
- 112.Smeets D, Ribbens A, Sima DM, et al. Reliable measurements of brain atrophy in individual patients with multiple sclerosis. Brain Behav. 2016; 6(9):e00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Soltaninejad M, Yang G, Lambrou T, et al. Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in FLAIR MRI. Int. J. Comput. Assist. Radiol. Surg 2017; 12(2):183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Somasundaram K, Ezhilarasan K. Automatic Brain Portion Segmentation From Magnetic Resonance Images of Head Scans Using Gray Scale Transformation and Morphological Operations. J. Comput. Assist. Tomogr. 2015; 39(4):552–558. [DOI] [PubMed] [Google Scholar]
- 115.Stone JR, Wilde EA, Taylor BA, et al. Supervised learning technique for the automated identification of white matter hyperintensities in traumatic brain injury. Brain Inj. 2016; 30(12):1458–1468. [DOI] [PubMed] [Google Scholar]
- 116.Sweeney EM, Shinohara RT, Shiee N, et al. OASIS is Automated Statistical Inference for Segmentation, with applications to multiple sclerosis lesion segmentation in MRI. NeuroImage Clin. 2013; 2:402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tangaro S, Amoroso N, Boccardi M, et al. Automated voxel-by-voxel tissue classification for hippocampal segmentation: methods and validation. Phys. Medica 2014; 30(8):878–887. [DOI] [PubMed] [Google Scholar]
- 118.Tangaro S, Amoroso N, Brescia M, et al. Feature selection based on machine learning in MRIs for hippocampal segmentation. Comput. Math. Methods Med. 2015; 2015: 814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tomas-Fernandez X, Warfield SK. A model of population and subject (MOPS) intensities with application to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging 2015; 34(6):1349–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tong T, Wolz R, Coupé P, et al. Segmentation of MR images via discriminative dictionary learning and sparse coding: application to hippocampus labeling. Neuroimage. 2013; 76:11–23. [DOI] [PubMed] [Google Scholar]
- 121.Tustison NJ, Shrinidhi KL, Wintermark M, et al. Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with ANTsR. Neuroinformatics. 2015; 13(2):209–225. [DOI] [PubMed] [Google Scholar]
- 122.van Opbroek A;Ikram MA;Vernooji MW;deBrijne M Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans Med Imaging. 2015; 34(5):1018–1030. [DOI] [PubMed] [Google Scholar]
- 123.Van Schependom J, Jain S, Cambron M, et al. Reliability of measuring regional callosal atrophy in neurodegenerative diseases. NeuroImage Clin. 2016; 12:825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Visser E, Keuken MC, Douaud G, et al. Automatic segmentation of the striatum and globus pallidus using MIST: multimodal image segmentation tool. Neuroimage. 2016; 125:479–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Visser E, Keuken MC, Forstmann BU, Jenkinson M. Automated segmentation of the substantia nigra, subthalamic nucleus and red nucleus in 7 T data at young and old age. Neuroimage. 2016; 139:324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wachinger C, Golland P, Kremen W, et al. BrainPrint: A discriminative characterization of brain morphology. Neuroimage. 2015; 109:232–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wachinger C, Golland P, Reuter M. Brainprint: Identifying subjects by their brain. In: International Conference on Medical Image Computing and Computer-Assisted Intervention; 2014:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang H, Yushkevich PA. Groupwise segmentation with multi-atlas joint label fusion. In: International Conference on Medical Image Computing and Computer-Assisted Intervention 2013:711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Q, Lu L, Wu D, et al. Automatic segmentation of spinal canals in CT images via iterative topology refinement. IEEE Trans. Med. Imaging 2015; 34(8):1694–1704. [DOI] [PubMed] [Google Scholar]
- 130.Wang R, Li C, Wang J, et al. Automatic segmentation of white matter lesions on magnetic resonance images of the brain by using an outlier detection strategy. Magn. Reson. Imaging 2014; 32(10):1321–1329. [DOI] [PubMed] [Google Scholar]
- 131.Wang R, Li C, Wang J, et al. Automatic segmentation and quantitative analysis of white matter hyperintensities on FLAIR images using trimmed-likelihood estimator. Acad. Radiol. 2014; 21(12):1512–1523. [DOI] [PubMed] [Google Scholar]
- 132.Weier K, Fonov V, Lavoie K, Doyon J, Collins DL. Rapid automatic segmentation of the human cerebellum and its lobules (RASCAL) - Implementation and application of the patch-based label-fusion technique with a template library to segment the human cerebellum. Hum. Brain Mapp 2014; 35(10):5026–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Winston GP, Cardoso MJ, Williams EJ, et al. Automated hippocampal segmentation in patients with epilepsy: available free online. Epilepsia. 2013; 54(12):2166–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu G, Kim M, Sanroma G, et al. Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. Neuroimage. 2015; 106:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Z, Ye C, Bogovic JA, et al. Automated cerebellar lobule segmentation with application to cerebellar structural analysis in cerebellar disease. Neuroimage. 2016; 127:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yushkevich PA, Pluta J, Wang H, et al. Fast Automatic Segmentation of Hippocampal Subfields and Medial Temporal Lobe Subregions in 3 Tesla and 7 Tesla T2-Weighted Mri. Alzheimer’s Dement. 2016; 12(7):P126–P127. [Google Scholar]
- 137.Zhan T, Chen Y, Hong X, Lu Z, Chen Y. Brain Tumor Segmentation Using Deep Belief Networks and Pathological Knowledge. CNS Neurol. Disord. Targets (Formerly Curr. Drug Targets-CNS Neurol. Disord 2017; 16(2):129–136. [DOI] [PubMed] [Google Scholar]
- 138.Zhang J, Barboriak DP, Hobbs H, Mazurowski MA. A fully automatic extraction of magnetic resonance image features in glioblastoma patients. Med. Phys 2014; 41(4):1–13. [DOI] [PubMed] [Google Scholar]
- 139.Zhang J, Jiang W, Wang R, Wang L. Brain mr image segmentation with spatial constrained k-mean algorithm and dual-tree complex wavelet transform. J. Med. Syst 2014; 38(9):93. [DOI] [PubMed] [Google Scholar]
- 140.Zhang L, Wang Q, Gao Y, Wu G, Shen D. Automatic labeling of MR brain images by hierarchical learning of atlas forests. Med. Phys 2016; 43(3):1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang M, Lu Z, Feng Q, Zhang Y. Automatic Thalamus Segmentation from Magnetic Resonance Images Using Multiple Atlases Level Set Framework (MALSF). Sci. Rep 2017; 7(1):4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang Y, Wei H, Cronin MJ, et al. Longitudinal atlas for normative human brain development and aging over the lifespan using quantitative susceptibility mapping. Neuroimage. 2018; 171:176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao L, Jia K. Multiscale cnns for brain tumor segmentation and diagnosis. Comput. Math. Methods Med 2016; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhu H, Cheng H, Yang X, et al. Metric learning for multi-atlas based segmentation of hippocampus. Neuroinformatics. 2017; 15(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhuge Y, Krauze AV, Ning H, et al. Brain tumor segmentation using holistically nested neural networks in MRI images. Med. Phys. 2017; 44(10):5234–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
B2. Thoracic Segmentation
- 1.Abbas Q Segmentation of differential structures on computed tomography images for diagnosis lung-related diseases. Biomed Signal Process Control. 2017;33:325–34. [Google Scholar]
- 2.Albà X, Figueras I Ventura RM, Lekadir K, Tobon-Gomez C, Hoogendoorn C, Frangi AF. Automatic cardiac LV Segmentation in MRI using modified graph cuts with smoothness and interslice constraints. Magn Reson Med. 2014;72(6):1775–84. [DOI] [PubMed] [Google Scholar]
- 3.Albà X, Lekadir K, Pereañez M, Medrano-Gracia P, Young AA, Frangi AF. Automatic initialization and quality control of large-scale cardiac MRI segmentations. Med Image Anal. 2018;43:129–41. [DOI] [PubMed] [Google Scholar]
- 4.Anders K, Achenbach S, Petit I, Daniel WG, Uder M, Pflederer T. Accuracy of automated software-guided detection of significant coronary artery stenosis by CT angiography: Comparison with invasive catheterisation. Eur Radiol. 2013;23(5):1218–25. [DOI] [PubMed] [Google Scholar]
- 5.Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans Med Imaging. 2016;35(5):1207–16. [DOI] [PubMed] [Google Scholar]
- 6.Bragman FJS, McClelland JR, Jacob J, Hurst JR, Hawkes DJ. Pulmonary Lobe Segmentation with Probabilistic Segmentation of the Fissures and a Groupwise Fissure Prior. IEEE Trans Med Imaging. 2017;36(8):1650–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustamante M, Petersson S, Eriksson J, Alehagen U, Dyverfeldt P, Carlhäll CJ, et al. Atlas-based analysis of 4D flow CMR: Automated vessel segmentation and flow quantification. J Cardiovasc Magn Reson. 2015;17:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Islam A, Bhaduri M, Chan I, Li S. Unsupervised Freeview Groupwise Cardiac Segmentation Using Synchronized Spectral Network. IEEE Trans Med Imaging. 2016;35(9):2174–88. [DOI] [PubMed] [Google Scholar]
- 9.Cao Q, Broersen A, de Graaf MA, Kitslaar PH, Yang G, Scholte AJ, et al. Automatic identification of coronary tree anatomy in coronary computed tomography angiography. Int J Cardiovasc Imaging. 2017;33(11):1809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cochet H, Denis A, Komatsu Y, Jadidi AS, Aït Ali T, Sacher F, et al. Automated Quantification of Right Ventricular Fat at Contrast-enhanced Cardiac Multidetector CT in Arrhythmogenic Right Ventricular Cardiomyopathy. Radiology. 2015;275(3):683–91. [DOI] [PubMed] [Google Scholar]
- 11.Cruz-Aceves I, Avina-Cervantes JG, Lopez-Hernandez JM, Garcia-Hernandez MG, Ibarra-Manzano MA. Unsupervised Cardiac Image Segmentation via Multiswarm Active Contours with a Shape Prior. Comput Math Methods Med. 2013;2013:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vos BD, Wolterink JM, De Jong PA, Leiner T, Viergever MA, Isgum I. ConvNet-Based Localization of Anatomical Structures in 3-D Medical Images. IEEE Trans Med Imaging. 2017;36(7):1470–81. [DOI] [PubMed] [Google Scholar]
- 13.Eilot D, Goldenberg R. Fully automatic model-based calcium segmentation and scoring in coronary CT angiography. Int J Comput Assist Radiol Surg. 2014;9(4):595–608. [DOI] [PubMed] [Google Scholar]
- 14.Elattar MA, Wiegerinck EM, Planken RN, Vanbavel E, Van Assen HC, Baan J, et al. Automatic segmentation of the aortic root in CT angiography of candidate patients for transcatheter aortic valve implantation. Med Biol Eng Comput. 2014;52(7):611–8. [DOI] [PubMed] [Google Scholar]
- 15.Eslami A, Karamalis A, Katouzian A, Navab N. Segmentation by retrieval with guided random walks: Application to left ventricle segmentation in MRI. Med Image Anal. 2013;17(2):236–53. [DOI] [PubMed] [Google Scholar]
- 16.Feng C, Zhang S, Zhao D, Li C. Simultaneous extraction of endocardial and epicardial contours of the left ventricle by distance regularized level sets. Med Phys. 2016;43(6):2741–55. [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Boccalini S, Kitslaar PH, Budde RPJ, Attrach M, Tu S, et al. Quantification of aortic annulus in computed tomography angiography: Validation of a fully automatic methodology. Eur J Radiol. 2017;93:1–8. [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Kitslaar PH, Budde RPJ, Tu S, de Graaf MA, Xu L, et al. Automatic detection of aortofemoral vessel trajectory from whole-body computed tomography angiography data sets. Int J Cardiovasc Imaging. 2016;32(8):1311–22. [DOI] [PubMed] [Google Scholar]
- 19.Gill G, Bauer C, Beichel RR. A method for avoiding overlap of left and right lungs in shape model guided segmentation of lungs in CT volumes. Med Phys. 2014;41(10):101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill G, Beichel RR. An approach for reducing the error rate in automated lung segmentation. Comput Biol Med. 2016;76:143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel A, McColl R, King KS, Whittemore A, Peshock RM. Fully automated tool to identify the aorta and compute flow using phase-contrast MRI: Validation and application in a large population based study. J Magn Reson Imaging. 2014;40(1):221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Zhou C, Chan HP, Chughtai A, Wei J, Hadjiiski LM, et al. Automated iterative neutrosophic lung segmentation for image analysis in thoracic computed tomography. Med Phys. 2013;40(8):081912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajiaghayi M, Groves EM, Jafarkhani H, Kheradvar A. A 3-D active contour method for automated segmentation of the left ventricle from magnetic resonance images. IEEE Trans Biomed Eng. 2017;64(1):134–44. [DOI] [PubMed] [Google Scholar]
- 24.Han D, Shim H, Jeon B, Jang Y, Hong Y, Jung S, et al. Automatic coronary artery segmentation using active search for branches and seemingly disconnected vessel segments from coronary CT angiography. PLoS One. 2016;11(8):e0156837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmouche R, San Jose Estepar R, Ross JC. Automatic inspiratory and expiratory left and right lung segmentation for disease characterization [Internet]. Vol. 189, American Journal of Respiratory and Critical Care Medicine; 2014. p. A4354 Available from: http://www.atsjournals.org/doi/pdf/10.1164/ajrccm-conference.2014.189.1_MeetingAbstracts. [Google Scholar]
- 26.Hsu LY, Jacobs M, Benovoy M, Ta AD, Conn HM, Winkler S, et al. Diagnostic Performance of Fully Automated Pixel-Wise Quantitative Myocardial Perfusion Imaging by Cardiovascular Magnetic Resonance. JACC Cardiovasc Imaging. 2018;11(5):697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu H, Gao Z, Liu L, Liu H, Gao J, Xu S, et al. Automatic segmentation of the left ventricle in cardiac MRI using local binary fitting model and dynamic programming techniques. PLoS One. 2014;9(12):e114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaderka R, Gillespie EF, Mundt RC, Bryant AK, Sanudo-Thomas CB, Harrison AL, et al. Geometric and dosimetric evaluation of atlas based auto-segmentation of cardiac structures in breast cancer patients. Radiotherapy and Oncology. 2019;215–20. [DOI] [PubMed] [Google Scholar]
- 29.Kang HC, Kim B, Lee J, Shin J, Shin YG. Automatic left and right heart segmentation using power watershed and active contour model without edge. Biomed Eng Lett. 2014;4(4):355–61. [Google Scholar]
- 30.Kohlmann P, Strehlow J, Jobst B, Krass S, Kuhnigk JM, Anjorin A, et al. Automatic lung segmentation method for MRI-based lung perfusion studies of patients with chronic obstructive pulmonary disease. Int J Comput Assist Radiol Surg. 2015;10(4):403–17. [DOI] [PubMed] [Google Scholar]
- 31.Kumamaru KK, George E, Aghayev A, Saboo SS, Khandelwal A, Rodríguez-López S, et al. Implementation and performance of automated software for computing right-to-left ventricular diameter ratio from computed tomography pulmonary angiography images. J Comput Assist Tomogr. 2016;40(3):387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurugol S, Come CE, Diaz AA, Ross JC, Kinney GL, Black-Shinn JL, et al. Automated quantitative 3D analysis of aorta size, morphology, and mural calcification distributions. Med Phys. 2015;42(9):5467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurzendorfer T, Forman C, Schmidt M, Tillmanns C, Maier A, Brost A. Fully automatic segmentation of left ventricular anatomy in 3-D LGE-MRI. Comput Med Imaging Graph. 2017;59:13–27. [DOI] [PubMed] [Google Scholar]
- 34.Lassen B, Van Rikxoort EM, Schmidt M, Kerkstra S, Van Ginneken B, Kuhnigk JM. Automatic segmentation of the pulmonary lobes from chest CT scans based on Fissures, Vessels, and Bronchi. IEEE Trans Med Imaging. 2013;32(2):210–22. [DOI] [PubMed] [Google Scholar]
- 35.Lin K, Collins JD, Lloyd-Jones DM, Jolly MP, Li D, Markl M, et al. Automated Assessment of Left Ventricular Function and Mass Using Heart Deformation Analysis: Initial Experience in 160 Older Adults. Acad Radiol. 2016;23(3):321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto AJ, Bartholmai BJ, Wylam ME. Comparison of Total Lung Capacity Determined by Plethysmography with Computed Tomographic Segmentation Using CALIPER. J Thorac Imaging. 2017;32(2):101–6. [DOI] [PubMed] [Google Scholar]
- 37.Medrano-Gracia P, Cowan BR, Ambale-Venkatesh B, Bluemke DA, Eng J, Finn JP, et al. Left ventricular shape variation in asymptomatic populations: The multi-ethnic study of atherosclerosis. J Cardiovasc Magn Reson. 2014;16:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng Q, Kitasaka T, Nimura Y, Oda M, Ueno J, Mori K. Automatic segmentation of airway tree based on local intensity filter and machine learning technique in 3D chest CT volume. Int J Comput Assist Radiol Surg. 2017;12(2):245–61. [DOI] [PubMed] [Google Scholar]
- 39.Molaei S, Shiri ME, Horan K, Kahrobaei D, Nallamothu B, Najarian K. Deep Convolutional Neural Networks for left ventricle segmentation. In: Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS; 2017. p. 668–71. [DOI] [PubMed] [Google Scholar]
- 40.Moolan-Feroze O, Mirmehdi M, Hamilton M, Bucciarelli-Ducci C. Segmentation of the right ventricle using diffusion maps and Markov random fields. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2014. p. 682–9. [DOI] [PubMed] [Google Scholar]
- 41.Morais P, Queirós S, Heyde B, Engvall J, Hooge JD, Vilaça JL. Fully automatic left ventricular myocardial strain estimation in 2D short-axis tagged magnetic resonance imaging. Phys Med Biol. 2017;62(17):6899–919. [DOI] [PubMed] [Google Scholar]
- 42.Moses D, Sammut C, Zrimec T. Automatic segmentation and analysis of the main pulmonary artery on standard post-contrast CT studies using iterative erosion and dilation. Int J Comput Assist Radiol Surg. 2016;11(3):381–95. [DOI] [PubMed] [Google Scholar]
- 43.Nageswararao AV, Srinivasan S, Babu Peter S. Automatic hybrid ventricular segmentation of short-axis cardiac MRI images. Biomed Res. 2017;28(13):5816–24. [Google Scholar]
- 44.Noor NM, Than JCM, Rijal OM, Kassim RM, Yunus A, Zeki AA, et al. Automatic Lung Segmentation Using Control Feedback System: Morphology and Texture Paradigm. J Med Syst. 2015;39(3):22. [DOI] [PubMed] [Google Scholar]
- 45.Queirós S, Barbosa D, Engvall J, Ebbers T, Nagel E, Sarvari SI, et al. Multi-centre validation of an automatic algorithm for fast 4D myocardial segmentation in cine CMR datasets. Eur Heart J Cardiovasc Imaging. 2016;17(10):1118–27. [DOI] [PubMed] [Google Scholar]
- 46.Rebouças Filho PP, Cortez PC, da Silva Barros AC, Victor VH, Tavares RSJM. Novel and powerful 3D adaptive crisp active contour method applied in the segmentation of CT lung images. Med Image Anal. 2017;35:503–16. [DOI] [PubMed] [Google Scholar]
- 47.Rengier F, Wörz S, Melzig C, Ley S, Fink C, Benjamin N, et al. Automated 3D volumetry of the pulmonary arteries based on magnetic resonance angiography has potential for predicting pulmonary hypertension. PLoS One. 2016;11(9):e0162516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ringenberg J, Deo M, Devabhaktuni V, Berenfeld O, Boyers P, Gold J. Fast, accurate, and fully automatic segmentation of the right ventricle in short-axis cardiac MRI. Comput Med Imaging Graph. 2014;38(3):190–201. [DOI] [PubMed] [Google Scholar]
- 49.Rodrigues O, Rodrigues LO, Oliveira LSN, Conci A, Liatsis P. Automated recognition of the pericardium contour on processed CT images using genetic algorithms. Comput Biol Med. 2017;87:38–45. [DOI] [PubMed] [Google Scholar]
- 50.Ross JC, Kindlmann GL, Okajima Y, Hatabu H, Díaz AA, Silverman EK, et al. Pulmonary lobe segmentation based on ridge surface sampling and shape model fitting. Med Phys. 2013;40(12):121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shahzad R, Bos D, Budde RPJ, Pellikaan K, Niessen WJ, Van Der Lugt A, et al. Automatic segmentation and quantification of the cardiac structures from non-contrast-enhanced cardiac CT scans. Phys Med Biol. 2017;62(9):3798–813. [DOI] [PubMed] [Google Scholar]
- 52.Shahzad R, Tao Q, Dzyubachyk O, Staring M, Lelieveldt BPF, van der Geest RJ. Fully-automatic left ventricular segmentation from long-axis cardiac cine MR scans. Med Image Anal. 2017;39:44–55. [DOI] [PubMed] [Google Scholar]
- 53.Soliman A, Khalifa F, Elnakib A, El-Ghar MA, Dunlap N, Wang B, et al. Accurate lungs segmentation on CT chest images by adaptive appearance-guided shape modeling. IEEE Trans Med Imaging. 2017;36(1):263–76. [DOI] [PubMed] [Google Scholar]
- 54.Sugiura T, Takeguchi T, Sakata Y, Nitta S, Okazaki T, Matsumoto N, et al. Automatic model-based contour detection of left ventricle myocardium from cardiac CT images. Int J Comput Assist Radiol Surg. 2013;8(1):145–55. [DOI] [PubMed] [Google Scholar]
- 55.Sun K, Udupa JK, Odhner D, Tong Y, Zhao L, Torigian DA. Automatic thoracic anatomy segmentation on CT images using hierarchical fuzzy models and registration. Med Phys. 2016;43(3):1487–500. [DOI] [PubMed] [Google Scholar]
- 56.Takx RAP, De Jong PA, Leiner T, Oudkerk M, De Koning HJ, Mol CP, et al. Automated coronary artery calcification scoring in non-gated chest CT: Agreement and reliability. PLoS One. 2014;9(3):e91239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan LK, McLaughlin RA, Lim E, Abdul Aziz YF, Liew YM. Fully automated segmentation of the left ventricle in cine cardiac MRI using neural network regression. J Magn Reson Imaging. 2018;48(1):140–52. [DOI] [PubMed] [Google Scholar]
- 58.Tsadok Y, Petrank Y, Sarvari S, Edvardsen T, Adam D. Automatic segmentation of cardiac MRI cines validated for long axis views. Comput Med Imaging Graph. 2013;37(7–8):500–11. [DOI] [PubMed] [Google Scholar]
- 59.Tufvesson J, Hedström E, Steding-Ehrenborg K, Carlsson M, Arheden H, Heiberg E. Validation and Development of a New Automatic Algorithm for Time-Resolved Segmentation of the Left Ventricle in Magnetic Resonance Imaging. Biomed Res Int. 2015;2015:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B, Gu X, Fan C, Xie H, Zhang S, Tian X, et al. Sparse group composition for robust left ventricular epicardium segmentation. Comput Med Imaging Graph. 2015;46:56–63. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Pei M, Codella NCF, Kochar M, Weinsaft JW, Li J, et al. Left Ventricle: Fully Automated Segmentation Based on Spatiotemporal Continuity and Myocardium Information in Cine Cardiac Magnetic Resonance Imaging (LV-FAST). Biomed Res Int. 2015;2015:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Bhatia KK, Glocker B, Marvao A, Dawes T, Misawa K, et al. Geodesic patch-based segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2014. p. 666–73. [DOI] [PubMed] [Google Scholar]
- 63.Wei D, Sun Y, Ong SH, Chai P, Teo LL, Low AF. A comprehensive 3-D framework for automatic quantification of late gadolinium enhanced cardiac magnetic resonance images. IEEE Trans Biomed Eng. 2013;60(6):1499–508. [DOI] [PubMed] [Google Scholar]
- 64.Wei Y, Shen G, Li JJ. A fully automatic method for lung parenchyma segmentation and repairing. J Digit Imaging. 2013;26(3):483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolterink JM, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Išgum I. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal. 2016;34:123–36. [DOI] [PubMed] [Google Scholar]
- 66.Wolterink JM, Leiner T, Takx RAP, Viergever MA, Išgum I. Automatic Coronary Calcium Scoring in Non-Contrast-Enhanced ECG-Triggered Cardiac CT with Ambiguity Detection. IEEE Trans Med Imaging. 2015;34(9):1867–78. [DOI] [PubMed] [Google Scholar]
- 67.Xie Y, Padgett J, Biancardi AM, Reeves AP. Automated aorta segmentation in low-dose chest CT images. Int J Comput Assist Radiol Surg. 2014;9(2):211–9. [DOI] [PubMed] [Google Scholar]
- 68.Xu Z, Bagci U, Jonsson C, Jain S, Mollura DJ. Accurate and efficient separation of left and right lungs from 3D CT scans: A generic hysteresis approach. In: 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBC; 2014. 2014. p. 6036–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang G, Chen Y, Ning X, Sun Q, Shu H, Coatrieux JL. Automatic coronary calcium scoring using noncontrast and contrast CT images. Med Phys. 2016;43(5):2174–86. [DOI] [PubMed] [Google Scholar]
- 70.Zhang W, Wang X, Zhang P, Chen J. Global optimal hybrid geometric active contour for automated lung segmentation on CT images. Comput Biol Med. 2017;91:168–80. [DOI] [PubMed] [Google Scholar]
- 71.Zhen X, Zhang H, Islam A, Bhaduri M, Chan I, Li S. Direct and simultaneous estimation of cardiac four chamber volumes by multioutput sparse regression. Med Image Anal. 2017;36:184–96. [DOI] [PubMed] [Google Scholar]
- 72.Zhou H, Sun P, Ha S, Lundine D, Xiong G. Watertight modeling and segmentation of bifurcated Coronary arteries for blood flow simulation using CT imaging. Comput Med Imaging Graph. 2016;53:43–53. [DOI] [PubMed] [Google Scholar]
- 73.Zhou J, Yan Z, Lasio G, Huang J, Zhang B, Sharma N, et al. Automated compromised right lung segmentation method using a robust atlas-based active volume model with sparse shape composition prior in CT. Comput Med Imaging Graph. 2015;46:47–55. [DOI] [PubMed] [Google Scholar]
- 74.Zhu L, Gao Y, Appia V, Yezzi A, Arepalli C, Faber T, et al. A complete system for automatic extraction of left ventricular myocardium from ct images using shape segmentation and contour evolution. IEEE Trans Image Process. 2014;23(3):1340–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu L, Gao Y, Appia V, Yezzi A, Arepalli C, Faber T, et al. Automatic delineation of the myocardial wall from CT images via shape segmentation and variational region growing. IEEE Trans Biomed Eng. 2013;60(10):2887–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu L, Gao Y, Yezzi A, Tannenbaum A. Automatic segmentation of the left atrium from MR images via variational region growing with a moments-based shape prior. IEEE Trans Image Process. 2013;22(12):5111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhuang X, Bai W, Song J, Zhan S, Qian X, Shi W, et al. Multiatlas whole heart segmentation of CT data using conditional entropy for atlas ranking and selection. Med Phys. 2015;42(7):3822–33. [DOI] [PubMed] [Google Scholar]
- 78.Zhuang X, Shen J. Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Med Image Anal. 2016;31:77–87. [DOI] [PubMed] [Google Scholar]
B3. Abdominal Segmentation
- 1.Acosta O, Mylona E, Le Dain M, Voisin C, Lizee T, Rigaud B, et al. Multi-atlas-based segmentation of prostatic urethra from planning CT imaging to quantify dose distribution in prostate cancer radiotherapy. Radiother Oncol. 2017;125(3):492–9. [DOI] [PubMed] [Google Scholar]
- 2.Akhondi-Asl A, Hoyte L, Lockhart ME, Warfield SK. A logarithmic opinion pool based STAPLE algorithm for the fusion of segmentations with associated reliability weights. IEEE Trans Med Imaging. 2014;33(10):1997–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anter AMH AE; ElSoud MA; Azar AT Automatic liver parenchyma segmentation system from abdominal CT scans using hybrid techniques. International Journal of Biomedical Engineering and Technology.01 January 2015;17(2):148–167. [Google Scholar]
- 4.Cai J, Lu L, Zhang Z, Xing F, Yang L, Yin Q. Pancreas Segmentation in MRI using Graph-Based Decision Fusion on Convolutional Neural Networks. Med Image Comput Comput Assist Interv. 2016;9901:442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, He B, Fan Y, Fang C, Jia F. Comparison of liver volumetry on contrast-enhanced CT images: one semiautomatic and two automatic approaches. J Appl Clin Med Phys. 2016;17(6):118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra SS, Dowling JA, Greer PB, Martin J, Wratten C, Pichler P, et al. Fast automated segmentation of multiple objects via spatially weighted shape learning. Phys Med Biol. 2016;61(22):8070–84. [DOI] [PubMed] [Google Scholar]
- 7.Cheng R, Roth HR, Lay N, Lu L, Turkbey B, Gandler W, et al. Automatic magnetic resonance prostate segmentation by deep learning with holistically nested networks. J Med Imaging (Bellingham). 2017;4(4):041302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng R, Turkbey B, Gandler W, Agarwal HK, Shah VP, Bokinsky A, et al. Atlas based AAM and SVM model for fully automatic MRI prostate segmentation. Conf Proc IEEE Eng Med Biol Soc 2014;2014:2881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilali O, Puech P, Lakroum S, Diaf M, Mordon S, Betrouni N. Gland and Zonal Segmentation of Prostate on T2W MR Images. J Digit Imaging. 2016;29(6):730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu C, Oda M, Kitasaka T, Misawa K, Fujiwara M, Hayashi Y, et al. Multi-organ segmentation based on spatially-divided probabilistic atlas from 3D abdominal CT images. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):165–72. [DOI] [PubMed] [Google Scholar]
- 11.Clark T, Zhang J, Baig S, Wong A, Haider MA, Khalvati F. Fully automated segmentation of prostate whole gland and transition zone in diffusion-weighted MRI using convolutional neural networks. J Med Imaging (Bellingham). 2017;4(4):041307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derraz F, Forzy G, Delebarre A, Taleb-Ahmed A, Oussalah M, Peyrodie L, et al. Prostate contours delineation using interactive directional active contours model and parametric shape prior model. Int J Numer Method Biomed Eng. 2015;31(11). [DOI] [PubMed] [Google Scholar]
- 13.Dong C, Chen YW, Foruzan AH, Lin L, Han XH, Tateyama T, et al. Segmentation of liver and spleen based on computational anatomy models. Comput Biol Med. 2015;67:146–60. [DOI] [PubMed] [Google Scholar]
- 14.Farag A, Le L, Roth HR, Liu J, Turkbey E, Summers RM. A Bottom-Up Approach for Pancreas Segmentation Using Cascaded Superpixels and (Deep) Image Patch Labeling. IEEE Trans Image Process. 2017;26(1):386–99. [DOI] [PubMed] [Google Scholar]
- 15.Fechter T, Adebahr S, Baltas D, Ben Ayed I, Desrosiers C, Dolz J. Esophagus segmentation in CT via 3D fully convolutional neural network and random walk. Med Phys. 2017;44(12):6341–52. [DOI] [PubMed] [Google Scholar]
- 16.Gauriau R, Cuingnet R, Lesage D, Bloch I. Multi-organ localization combining global-to-local regression and confidence maps. Med Image Comput Comput Assist Interv. 2014;17(Pt 3):337–44. [DOI] [PubMed] [Google Scholar]
- 17.Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, et al. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans Med Imaging. 2018;37(8):1822–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gloger O, Bulow R, Tonnies K, Volzke H. Automatic gallbladder segmentation using combined 2D and 3D shape features to perform volumetric analysis in native and secretin-enhanced MRCP sequences. MAGMA. 2018;31(3):383–97. [DOI] [PubMed] [Google Scholar]
- 19.Gloger O, Tonnies K, Bulow R, Volzke H. Automatized spleen segmentation in non-contrast-enhanced MR volume data using subject-specific shape priors. Phys Med Biol. 2017;62(14):5861–83. [DOI] [PubMed] [Google Scholar]
- 20.Gloger O, Tonnies K, Mensel B, Volzke H. Fully automatized renal parenchyma volumetry using a support vector machine based recognition system for subject-specific probability map generation in native MR volume data. Phys Med Biol. 2015;60(22):8675–93. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y, Gao Y, Shao Y, Price T, Oto A, Shen D. Deformable segmentation of 3D MR prostate images via distributed discriminative dictionary and ensemble learning. Med Phys. 2014;41(7):072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadjiiski L, Chan HP, Cohan RH, Caoili EM, Law Y, Cha K, et al. Urinary bladder segmentation in CT urography (CTU) using CLASS. Med Phys. 2013;40(11):111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammon M, Cavallaro A, Erdt M, Dankerl P, Kirschner M, Drechsler K, et al. Model-based pancreas segmentation in portal venous phase contrast-enhanced CT images. J Digit Imaging. 2013;26(6):1082–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He B, Huang C, Sharp G, Zhou S, Hu Q, Fang C, et al. Fast automatic 3D liver segmentation based on a three-level AdaBoost-guided active shape model. Med Phys. 2016;43(5):2421. [DOI] [PubMed] [Google Scholar]
- 25.Hu P, Wu F, Peng J, Bao Y, Chen F, Kong D. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J Comput Assist Radiol Surg. 2017;12(3):399–411. [DOI] [PubMed] [Google Scholar]
- 26.Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61(24):8676–98. [DOI] [PubMed] [Google Scholar]
- 27.Huo Y, Liu J, Xu Z, Harrigan RL, Assad A, Abramson RG, et al. Robust Multicontrast MRI Spleen Segmentation for Splenomegaly Using Multi-Atlas Segmentation. IEEE Trans Biomed Eng. 2018;65(2):336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh HT, Karademir I, Oto A, Suzuki K. Computerized liver volumetry on MRI by using 3D geodesic active contour segmentation. AJR Am J Roentgenol. 2014;202(1):152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huynh HT, Le-Trong N, Bao PT, Oto A, Suzuki K. Fully automated MR liver volumetry using watershed segmentation coupled with active contouring. Int J Comput Assist Radiol Surg. 2017;12(2):235–43. [DOI] [PubMed] [Google Scholar]
- 30.Ji H, He J, Yang X, Deklerck R, Cornelis J. ACM-based automatic liver segmentation from 3-D CT images by combining multiple atlases and improved mean-shift techniques. IEEE J Biomed Health Inform. 2013;17(3):690–8. [DOI] [PubMed] [Google Scholar]
- 31.Jia HX Y; Song Y; Cai W; Fulham M; Feng DD Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing. 2018;31 January 2018;275():1358–1369. [Google Scholar]
- 32.Jin C, Shi F, Xiang D, Jiang X, Zhang B, Wang X, et al. 3D Fast Automatic Segmentation of Kidney Based on Modified AAM and Random Forest. IEEE Trans Med Imaging. 2016;35(6):1395–407. [DOI] [PubMed] [Google Scholar]
- 33.Jin C, Shi F, Xiang D, Zhang L, Chen X. Fast segmentation of kidney components using random forests and ferns. Med Phys. 2017;44(12):6353–63. [DOI] [PubMed] [Google Scholar]
- 34.Karasawa K, Oda M, Kitasaka T, Misawa K, Fujiwara M, Chu C, et al. Multi-atlas pancreas segmentation: Atlas selection based on vessel structure. Med Image Anal. 2017;39:18–28. [DOI] [PubMed] [Google Scholar]
- 35.Ke Y, Changyang L, Xiuying W, Ang L, Yuchen Y, Dagan F, et al. Automatic prostate segmentation on MR images with deep network and graph model. Conf Proc IEEE Eng Med Biol Soc 2016;2016:635–8. [DOI] [PubMed] [Google Scholar]
- 36.Khalifa F, Soliman A, Elmaghraby A, Gimel’farb G, El-Baz A. 3D Kidney Segmentation from Abdominal Images Using Spatial-Appearance Models. Comput Math Methods Med. 2017;2017:9818506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y, Ge Y, Tao C, Zhu J, Chapman AB, Torres VE, et al. Automated Segmentation of Kidneys from MR Images in Patients with Autosomal Dominant Polycystic Kidney Disease. Clin J Am Soc Nephrol. 2016;11(4):576–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kline TL, Korfiatis P, Edwards ME, Blais JD, Czerwiec FS, Harris PC, et al. Performance of an Artificial Multi-observer Deep Neural Network for Fully Automated Segmentation of Polycystic Kidneys. J Digit Imaging. 2017;30(4):442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korsager AS, Fortunati V, van der Lijn F, Carl J, Niessen W, Ostergaard LR, et al. The use of atlas registration and graph cuts for prostate segmentation in magnetic resonance images. Med Phys. 2015;42(4):1614–24. [DOI] [PubMed] [Google Scholar]
- 40.Langerak TR, Berendsen FF, Van der Heide UA, Kotte AN, Pluim JP. Multiatlas-based segmentation with preregistration atlas selection. Med Phys. 2013;40(9):091701. [DOI] [PubMed] [Google Scholar]
- 41.Langerak TR, van der Heide UA, Kotte AN, Berendsen FF, van Vulpen M, Pluim JP. Expert-driven label fusion in multi-atlas-based segmentation of the prostate using weighted atlases. Int J Comput Assist Radiol Surg. 2013;8(6):929–36. [DOI] [PubMed] [Google Scholar]
- 42.Lavdas I, Glocker B, Kamnitsas K, Rueckert D, Mair H, Sandhu A, et al. Fully automatic, multiorgan segmentation in normal whole body magnetic resonance imaging (MRI), using classification forests (CFs), convolutional neural networks (CNNs), and a multi-atlas (MA) approach. Med Phys. 2017;44(10):5210–20. [DOI] [PubMed] [Google Scholar]
- 43.Li D, Liu L, Chen J, Li H, Yin Y, Ibragimov B, et al. Augmenting atlas-based liver segmentation for radiotherapy treatment planning by incorporating image features proximal to the atlas contours. Phys Med Biol. 2017;62(1):272–88. [DOI] [PubMed] [Google Scholar]
- 44.Li G, Chen X, Shi F, Zhu W, Tian J, Xiang D. Automatic Liver Segmentation Based on Shape Constraints and Deformable Graph Cut in CT Images. IEEE Trans Image Process. 2015;24(12):5315–29. [DOI] [PubMed] [Google Scholar]
- 45.Liao M, Zhao YQ, Liu XY, Zeng YZ, Zou BJ, Wang XF, et al. Automatic liver segmentation from abdominal CT volumes using graph cuts and border marching. Comput Methods Programs Biomed. 2017;143:1–12. [DOI] [PubMed] [Google Scholar]
- 46.Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate MR segmentation. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):254–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao S, Gao Y, Shi Y, Yousuf A, Karademir I, Oto A, et al. Automatic prostate MR image segmentation with sparse label propagation and domain-specific manifold regularization. Inf Process Med Imaging. 2013;23:511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Huo Y, Xu Z, Assad A, Abramson RG, Landman BA. Multi-Atlas Spleen Segmentation on CT Using Adaptive Context Learning. Proc SPIE Int Soc Opt Eng. 2017;10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu F, Wu F, Hu P, Peng Z, Kong D. Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg. 2017;12(2):171–82. [DOI] [PubMed] [Google Scholar]
- 50.Lu L, Zhao J. An improved method of automatic colon segmentation for virtual colon unfolding. Comput Methods Programs Biomed. 2013;109(1):1–12. [DOI] [PubMed] [Google Scholar]
- 51.Mahapatra D, Buhmann JM. Prostate MRI segmentation using learned semantic knowledge and graph cuts. IEEE Trans Biomed Eng. 2014;61(3):756–64. [DOI] [PubMed] [Google Scholar]
- 52.Maklad AS, Matsuhiro M, Suzuki H, Kawata Y, Niki N, Satake M, et al. Blood vessel-based liver segmentation using the portal phase of an abdominal CT dataset. Med Phys. 2013;40(11):113501. [DOI] [PubMed] [Google Scholar]
- 53.Nelson ASB J; Lu M; Javorek A; Pirozzi S; Piper JW Evaluation of an atlas-based segmentation method for prostate MRI. International Journal of Radiation Oncology Biology Physics 2014;01 September 2014;1)():S419–S420. [Google Scholar]
- 54.Oh J, Martin DR, Hu X. Partitioned edge-function-scaled region-based active contour (p-ESRAC): automated liver segmentation in multiphase contrast-enhanced MRI. Med Phys. 2014;41(4):041914. [DOI] [PubMed] [Google Scholar]
- 55.Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, Sato Y. Abdominal multi-organ segmentation from CT images using conditional shape-location and unsupervised intensity priors. Med Image Anal. 2015;26(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okada TL MG; Hori M; Summers RM; Tomiyama N; Sato Y, editor Abdominal multi-organ CT segmentation using organ correlation graph and prediction-based shape and location priors. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2013. [DOI] [PubMed] [Google Scholar]
- 57.Peng J, Hu P, Lu F, Peng Z, Kong D, Zhang H. 3D liver segmentation using multiple region appearances and graph cuts. Med Phys. 2015;42(12):6840–52. [DOI] [PubMed] [Google Scholar]
- 58.Polymeri ES M; Hasani N; Fejne F; Kahl F; Geronymakis C; Edenbrandt L, editor Automatic 3D-segmentation of the prostate gland. Eur J Nucl Med Mol Imaging 2016. [Google Scholar]
- 59.Polymeri ES M; Kaboteh R; Enqvist O; Ulen J; Tragardh E; Poulsen MH; Simonsen JA; Hoilund-Carlsen PF; Edenbrandt L; Johnsson A, editor Analytical validation of an automated method for segmentation of the prostate gland in CT images. Eur J Nucl Med Mol Imaging; 2017. [Google Scholar]
- 60.Qiu W, Yuan J, Ukwatta E, Sun Y, Rajchl M, Fenster A. Fast globally optimal segmentation of 3D prostate MRI with axial symmetry prior. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):198–205. [DOI] [PubMed] [Google Scholar]
- 61.Roth HR, Lu L, Lay N, Harrison AP, Farag A, Sohn A, et al. Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. Med Image Anal. 2018;45:94–107. [DOI] [PubMed] [Google Scholar]
- 62.Saito A, Nawano S, Shimizu A. Joint optimization of segmentation and shape prior from level-set-based statistical shape model, and its application to the automated segmentation of abdominal organs. Med Image Anal. 2016;28:46–65. [DOI] [PubMed] [Google Scholar]
- 63.Saito A, Nawano S, Shimizu A. Fast approximation for joint optimization of segmentation, shape, and location priors, and its application in gallbladder segmentation. Int J Comput Assist Radiol Surg. 2017;12(5):743–56. [DOI] [PubMed] [Google Scholar]
- 64.Shahedi M, Cool DW, Bauman GS, Bastian-Jordan M, Fenster A, Ward AD. Accuracy Validation of an Automated Method for Prostate Segmentation in Magnetic Resonance Imaging. J Digit Imaging. 2017;30(6):782–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen J, Baum T, Cordes C, Ott B, Skurk T, Kooijman H, et al. Automatic segmentation of abdominal organs and adipose tissue compartments in water-fat MRI: Application to weight-loss in obesity. Eur J Radiol. 2016;85(9):1613–21. [DOI] [PubMed] [Google Scholar]
- 66.Song X, Cheng M, Wang B, Huang S, Huang X, Yang J. Adaptive fast marching method for automatic liver segmentation from CT images. Med Phys. 2013;40(9):091917. [DOI] [PubMed] [Google Scholar]
- 67.Tian Z, Liu L, Zhang Z, Fei B. PSNet: prostate segmentation on MRI based on a convolutional neural network. J Med Imaging (Bellingham). 2018;5(2):021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomoshige S, Oost E, Shimizu A, Watanabe H, Nawano S. A conditional statistical shape model with integrated error estimation of the conditions; application to liver segmentation in non-contrast CT images. Med Image Anal. 2014;18(1):130–43. [DOI] [PubMed] [Google Scholar]
- 69.Tong T, Wolz R, Wang Z, Gao Q, Misawa K, Fujiwara M, et al. Discriminative dictionary learning for abdominal multi-organ segmentation. Med Image Anal. 2015;23(1):92–104. [DOI] [PubMed] [Google Scholar]
- 70.Turkbey B, Fotin SV, Huang RJ, Yin Y, Daar D, Aras O, et al. Fully automated prostate segmentation on MRI: comparison with manual segmentation methods and specimen volumes. AJR Am J Roentgenol. 2013;201(5):W720–9. [DOI] [PubMed] [Google Scholar]
- 71.Udupa JK, Odhner D, Zhao L, Tong Y, Matsumoto MM, Ciesielski KC, et al. Body-wide hierarchical fuzzy modeling, recognition, and delineation of anatomy in medical images. Med Image Anal. 2014;18(5):752–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Cheng Y, Guo C, Wang Y, Tamura S. Shape-intensity prior level set combining probabilistic atlas and probability map constrains for automatic liver segmentation from abdominal CT images. Int J Comput Assist Radiol Surg. 2016;11(5):817–26. [DOI] [PubMed] [Google Scholar]
- 73.Wang L, Li D, Huang S. An improved parallel fuzzy connected image segmentation method based on CUDA. Biomed Eng Online. 2016;15(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D. Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging. 2013;32(9):1723–30. [DOI] [PubMed] [Google Scholar]
- 75.Wu W, Zhou Z, Wu S, Zhang Y. Automatic Liver Segmentation on Volumetric CT Images Using Supervoxel-Based Graph Cuts. Comput Math Methods Med. 2016;2016:9093721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiang D, Bagci U, Jin C, Shi F, Zhu W, Yao J, et al. CorteXpert: A model-based method for automatic renal cortex segmentation. Med Image Anal. 2017;42:257–73. [DOI] [PubMed] [Google Scholar]
- 77.Xie Q, Ruan D. Low-complexity atlas-based prostate segmentation by combining global, regional, and local metrics. Med Phys. 2014;41(4):041909. [DOI] [PubMed] [Google Scholar]
- 78.Xu Z, Asman AJ, Shanahan PL, Abramson RG, Landman BA. SIMPLE is a good idea (and better with context learning). Med Image Comput Comput Assist Interv. 2014;17(Pt 1):364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Z, Burke RP, Lee CP, Baucom RB, Poulose BK, Abramson RG, et al. Efficient multi-atlas abdominal segmentation on clinically acquired CT with SIMPLE context learning. Med Image Anal. 2015;24(1):18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Z, Gertz AL, Burke RP, Bansal N, Kang H, Landman BA, et al. Improving Spleen Volume Estimation Via Computer-assisted Segmentation on Clinically Acquired CT Scans. Acad Radiol. 2016;23(10):1214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Haas B, Fang R, Beadle BM, Garden AS, Liao Z, et al. Atlas ranking and selection for automatic segmentation of the esophagus from CT scans. Phys Med Biol. 2017;62(23):9140–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X, Yang JD, Hwang HP, Yu HC, Ahn S, Kim BW, et al. Segmentation of liver and vessels from CT images and classification of liver segments for preoperative liver surgical planning in living donor liver transplantation. Comput Methods Programs Biomed. 2018;158:41–52. [DOI] [PubMed] [Google Scholar]
- 83.Yang X, Ye X, Slabaugh G. Multilabel region classification and semantic linking for colon segmentation in CT colonography. IEEE Trans Biomed Eng. 2015;62(3):948–59. [DOI] [PubMed] [Google Scholar]
- 84.Yoruk U, Hargreaves BA, Vasanawala SS. Automatic renal segmentation for MR urography using 3D-GrabCut and random forests. Magn Reson Med. 2018;79(3):1696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang P, Liang Y, Chang S, Fan H. Kidney segmentation in CT sequences using graph cuts based active contours model and contextual continuity. Med Phys. 2013;40(8):081905. [DOI] [PubMed] [Google Scholar]
- 86.Zheng Y, Ai D, Mu J, Cong W, Wang X, Zhao H, et al. Automatic liver segmentation based on appearance and context information. Biomed Eng Online. 2017;16(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys. 2017;44(10):5221–33. [DOI] [PubMed] [Google Scholar]
B4. Musculoskeletal Segmentation
- 1.Ahn C, Bui TD, Lee YW, Shin J, Park H. Fully automated, level set-based segmentation for knee MRIs using an adaptive force function and template: data from the osteoarthritis initiative. Biomed Eng Online. 2016. August 24;15(1):99. doi: 10.1186/s12938-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida DF, Ruben RB, Folgado J, Fernandes PR, Audenaert E, Verhegghe B, et al. Fully automatic segmentation of femurs with medullary canal definition in high and in low resolution CT scans. Med Eng Phys. 2016;38(12):1474–80. [DOI] [PubMed] [Google Scholar]
- 3.Anas EM, Rasoulian A, Seitel A, Darras K, Wilson D, John PS, et al. Automatic Segmentation of Wrist Bones in CT Using a Statistical Wrist Shape + Pose Model. IEEE Trans Med Imaging. 2016;35(8):1789–801. [DOI] [PubMed] [Google Scholar]
- 4.Athertya JS, Saravana Kumar G. Automatic segmentation of vertebral contours from CT images using fuzzy corners. Comput Biol Med. 2016;72:75–89. [DOI] [PubMed] [Google Scholar]
- 5.Bowes MA, Vincent GR, Wolstenholme CB, Conaghan PG. A novel method for bone area measurement provides new insights into osteoarthritis and its progression. Ann Rheum Dis. 2015. March;74(3):519–25. doi: 10.1136/annrheumdis-2013-204052. [DOI] [PubMed] [Google Scholar]
- 6.Carballido-Gamio J, Bonaretti S, Saeed I, Harnish R, Recker R, Burghardt AJ, et al. Automatic multi-parametric quantification of the proximal femur with quantitative computed tomography. Quant Imaging Med Surg. 2015;5(4):552–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro-Mateos I, Pozo JM, Eltes PE, Rio LD, Lazary A, Frangi AF. 3D segmentation of annulus fibrosus and nucleus pulposus from T2-weighted magnetic resonance images. Phys Med Biol. 2014;59(24):7847–64. [DOI] [PubMed] [Google Scholar]
- 8.Chandra SS, Xia Y, Engstrom C, Crozier S, Schwarz R, Fripp J. Focused shape models for hip joint segmentation in 3D magnetic resonance images. Med Image Anal. 2014;18(3):567–78. [DOI] [PubMed] [Google Scholar]
- 9.Chandra SS, Surowiec R, Ho C, Xia Y, Engstrom C, Crozier S, Fripp J. Automated analysis of hip joint cartilage combining MR T2 and three-dimensional fast-spin-echo images. Magn Reson Med. 2016. January;75(1):403–13. doi: 10.1002/mrm.25598. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Belavy D, Yu W, Chu C, Armbrecht G, Bansmann M, et al. Localization and Segmentation of 3D Intervertebral Discs in MR Images by Data Driven Estimation. IEEE Trans Med Imaging. 2015;34(8):1719–29. [DOI] [PubMed] [Google Scholar]
- 11.Chen F, Liu J, Zhao Z, Zhu M, Liao H. Three-Dimensional Feature-Enhanced Network for Automatic Femur Segmentation. IEEE J Biomed Health Inform. 2019;23(1):243–52. [DOI] [PubMed] [Google Scholar]
- 12.Chu C, Bai J, Wu X, Zheng G. MASCG: Multi-Atlas Segmentation Constrained Graph method for accurate segmentation of hip CT images. Med Image Anal. 2015;26(1):173–84. [DOI] [PubMed] [Google Scholar]
- 13.Chu C, Belavy DL, Armbrecht G, Bansmann M, Felsenberg D, Zheng G. Fully Automatic Localization and Segmentation of 3D Vertebral Bodies from CT/MR Images via a Learning-Based Method. PLoS One. 2015;10(11):e0143327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu C, Chen C, Liu L, Zheng G. FACTS: Fully Automatic CT Segmentation of a Hip Joint. Ann Biomed Eng. 2015;43(5):1247–59. [DOI] [PubMed] [Google Scholar]
- 15.Dam EB, Lillholm M, Marques J, Nielsen M. Automatic segmentation of high- and low-field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging (Bellingham). 2015. April;2(2):024001. doi: 10.1117/1.JMI.2.2.024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadermayr M, Disch C, Müller M, Merhof D, Gess B. A comprehensive study on automated muscle segmentation for assessing fat infiltration in neuromuscular diseases. Magn Reson Imaging. 2018. May;48:20–26. doi: 10.1016/j.mri.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 17.Gaonkar B, Xia Y, Villaroman DS, Ko A, Attiah M, Beckett JS, et al. Multi-Parameter Ensemble Learning for Automated Vertebral Body Segmentation in Heterogeneously Acquired Clinical MR Images. IEEE J Transl Eng Health Med. 2017;5:1800412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S, Chaudhary V. Supervised methods for detection and segmentation of tissues in clinical lumbar MRI. Comput Med Imaging Graph. 2014;38(7):639–49. [DOI] [PubMed] [Google Scholar]
- 19.Hanaoka S, Masutani Y, Nemoto M, Nomura Y, Miki S, Yoshikawa T, et al. Landmark-guided diffeomorphic demons algorithm and its application to automatic segmentation of the whole spine and pelvis in CT images. Int J Comput Assist Radiol Surg. 2017;12(3):413–30. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Jian F, Wu H, Li H. An improved level set method for vertebra CT image segmentation. Biomed Eng Online. 2013;12:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, Griffith JF, Wang D, Shi L. Graph-Cut-Based Segmentation of Proximal Femur from Computed Tomography Images with Shape Prior. Journal of Medical and Biological Engineering. 2015;35(5):594–607. [Google Scholar]
- 22.Karlsson A, Rosander J, Romu T, Tallberg J, Grönqvist A, Borga M, Dahlqvist Leinhard O. Automatic and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water-fat MRI. J Magn Reson Imaging. 2015. June;41(6):1558–69. doi: 10.1002/jmri.24726. [DOI] [PubMed] [Google Scholar]
- 23.Kashyap S, Oguz I, Zhang H, Sonka M. Automated Segmentation of Knee MRI Using Hierarchical Classifiers and Just Enough Interaction Based Learning: Data from Osteoarthritis Initiative. Med Image Comput Comput Assist Interv. 2016. October;9901:344–351. doi: 10.1007/978-3-319-46723-8_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh J, Chaudhary V, Jeon EK, Dhillon G. Automatic spinal canal detection in lumbar MR images in the sagittal view using dynamic programming. Comput Med Imaging Graph. 2014;38(7):569–79. [DOI] [PubMed] [Google Scholar]
- 25.Korez R, Ibragimov B, Likar B, Pernus F, Vrtovec T. A Framework for Automated Spine and Vertebrae Interpolation-Based Detection and Model-Based Segmentation. IEEE Trans Med Imaging. 2015;34(8):1649–62. [DOI] [PubMed] [Google Scholar]
- 26.Lareau-Trudel E, Le Troter A, Ghattas B, Pouget J, Attarian S, Bendahan D, Salort-Campana E. Muscle Quantitative MR Imaging and Clustering Analysis in Patients with Facioscapulohumeral Muscular Dystrophy Type 1. PLoS One. 2015. July 16;10(7):e0132717. doi: 10.1371/journal.pone.0132717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Troter A, Fouré A, Guye M, Confort-Gouny S, Mattei JP, Gondin J, Salort-Campana E, Bendahan D. Volume measurements of individual muscles in human quadriceps femoris using atlas-based segmentation approaches. MAGMA. 2016. April;29(2):245–57. doi: 10.1007/s10334-016-0535-6. [DOI] [PubMed] [Google Scholar]
- 28.Lee H, Troschel FM, Tajmir S, Fuchs G, Mario J, Fintelmann FJ, Do S. Pixel-Level Deep Segmentation: Artificial Intelligence Quantifies Muscle on Computed Tomography for Body Morphometric Analysis. J Digit Imaging. 2017. August;30(4):487–498. doi: 10.1007/s10278-017-9988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JG, Gumus S, Moon CH, Kwoh CK, Bae KT. Fully automated segmentation of cartilage from the MR images of knee using a multi-atlas and local structural analysis method. Med Phys. 2014. September;41(9):092303. doi: 10.1118/1.4893533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Dou Q, Chen H, Fu CW, Qi X, Belavy DL, et al. 3D multi-scale FCN with random modality voxel dropout learning for Intervertebral Disc Localization and Segmentation from Multi-modality MR Images. Med Image Anal. 2018;45:41–54. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. 2018. April;79(4):2379–2391. doi: 10.1002/mrm.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Zhao J, Dai N, Qian H, Tang Y. Improve accuracy for automatic acetabulum segmentation in CT images. Biomed Mater Eng. 2014;24(6):3159–77. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Xie Y, Reeves AP. Automated 3D closed surface segmentation: application to vertebral body segmentation in CT images. Int J Comput Assist Radiol Surg. 2016;11(5):789–801. [DOI] [PubMed] [Google Scholar]
- 34.Makrogiannis S, Boukari F, Ferrucci L. Automated skeletal tissue quantification in the lower leg using peripheral quantitative computed tomography. Physiol Meas. 2018. April 3;39(3):035011. doi: 10.1088/1361-6579/aaafb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makrogiannis S, Fishbein KW, Moore AZ, Spencer RG, Ferrucci L. Image-Based Tissue Distribution Modeling for Skeletal Muscle Quality Characterization. IEEE Trans Biomed Eng. 2016. April;63(4):805–13. doi: 10.1109/TBME.2015.2474305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendoza CS, Safdar N, Okada K, Myers E, Rogers GF, Linguraru MG. Personalized assessment of craniosynostosis via statistical shape modeling. Med Image Anal. 2014;18(4):635–46. [DOI] [PubMed] [Google Scholar]
- 37.Neubert A, Yang Z, Engstrom C, Xia Y, Strudwick MW, Chandra SS, Fripp J, Crozier S. Automatic segmentation of the glenohumeral cartilages from magnetic resonance images. Medical physics. 2016. October 1;43(10):5370–9. [DOI] [PubMed] [Google Scholar]
- 38.Onal S, Chen X, Lai-Yuen S, Hart S. Automatic vertebra segmentation on dynamic magnetic resonance imaging. J Med Imaging (Bellingham). 2017;4(1):014504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Öztürk CN, Albayrak S. Automatic segmentation of cartilage in high-field magnetic resonance images of the knee joint with an improved voxel-classification-driven region-growing algorithm using vicinity-correlated subsampling. Comput Biol Med. 2016. May 1;72:90–107. doi: 10.1016/j.compbiomed.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Paproki A, Engstrom C, Chandra SS, Neubert A, Fripp J, Crozier S. Automated segmentation and analysis of normal and osteoarthritic knee menisci from magnetic resonance images--data from the Osteoarthritis Initiative. Osteoarthritis Cartilage. 2014. September;22(9):1259–70. doi: 10.1016/j.joca.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 41.Paproki A, Engstrom C, Strudwick M, Wilson KJ, Surowiec RK, Ho C, Crozier S, Fripp J. Automated T2-mapping of the Menisci From Magnetic Resonance Images in Patients with Acute Knee Injury. Acad Radiol. 2017. October;24(10):1295–1304. doi: 10.1016/j.acra.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 42.Pedoia V, Li X, Su F, Calixto N, Majumdar S. Fully automatic analysis of the knee articular cartilage T1ρ relaxation time using voxel-based relaxometry. J Magn Reson Imaging. 2016. April;43(4):970–80. doi: 10.1002/jmri.25065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prasoon A, Igel C, Loog M, Lauze F, Dam EB, Nielsen M. Femoral cartilage segmentation in knee MRI scans using two stage voxel classification. Conf Proc IEEE Eng Med Biol Soc 2013;2013:5469–72. doi: 10.1109/EMBC.2013.6610787. [DOI] [PubMed] [Google Scholar]
- 44.Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):246–53. PubMed PMID: 24579147. [DOI] [PubMed] [Google Scholar]
- 45.Ramme AJ, Guss MS, Vira S, Vigdorchik JM, Newe A, Raithel E, Chang G. Evaluation of Automated Volumetric Cartilage Quantification for Hip Preservation Surgery. J Arthroplasty. 2016. January;31(1):64–9. doi: 10.1016/j.arth.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Ruiz-Espana S, Domingo J, Diaz-Parra A, Dura E, D’Ocon-Alcaniz V, Arana E, et al. Automatic segmentation of the spine by means of a probabilistic atlas with a special focus on ribs suppression. Med Phys. 2017;44(9):4695–707. [DOI] [PubMed] [Google Scholar]
- 47.Shan L, Zach C, Charles C, Niethammer M. Automatic atlas-based three-label cartilage segmentation from MR knee images. Med Image Anal. 2014. October;18(7):1233–46. doi: 10.1016/j.media.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas MS, Newman D, Leinhard OD, Kasmai B, Greenwood R, Malcolm PN, Karlsson A, Rosander J, Borga M, Toms AP. Test-retest reliability of automated whole body and compartmental muscle volume measurements on a wide bore 3T MR system. Eur Radiol. 2014. September;24(9):2279–91. doi: 10.1007/s00330-014-3226-6. [DOI] [PubMed] [Google Scholar]
- 49.Wlodarczyk J, Czaplicka K, Tabor Z, Wojciechowski W, Urbanik A. Segmentation of bones in magnetic resonance images of the wrist. Int J Comput Assist Radiol Surg. 2015;10(4):419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu D, Sofka M, Birkbeck N, Zhou SK. Segmentation of multiple knee bones from CT for orthopedic knee surgery planning. Med Image Comput Comput Assist Interv. 2014;17(Pt 1):372–80. [DOI] [PubMed] [Google Scholar]
- 51.Xia Y, Fripp J, Chandra SS, Schwarz R, Engstrom C, Crozier S. Automated bone segmentation from large field of view 3D MR images of the hip joint. Phys Med Biol. 2013;58(20):7375–90. [DOI] [PubMed] [Google Scholar]
- 52.Xia Y, Chandra SS, Engstrom C, Strudwick MW, Crozier S, Fripp J. Automatic hip cartilage segmentation from 3D MR images using arc-weighted graph searching. Phys Med Biol. 2014. December 7;59(23):7245–66. doi: 10.1088/0031-9155/59/23/7245. [DOI] [PubMed] [Google Scholar]
- 53.Xin C, Graham J, Hutchinson C, Muir L. Automatic generation of statistical pose and shape models for articulated joints. IEEE Trans Med Imaging. 2014;33(2):372–83. [DOI] [PubMed] [Google Scholar]
- 54.Yang YX, Chong MS, Lim WS, Tay L, Yew S, Yeo A, Tan CH. Validity of estimating muscle and fat volume from a single MRI section in older adults with sarcopenia and sarcopenic obesity. Clin Radiol. 2017. May;72(5):427.e9–427.e14. doi: 10.1016/j.crad.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 55.Yang Z, Fripp J, Chandra SS, Neubert A, Xia Y, Strudwick M, et al. Automatic bone segmentation and bone-cartilage interface extraction for the shoulder joint from magnetic resonance images. Phys Med Biol. 2015;60(4):1441–59. [DOI] [PubMed] [Google Scholar]
- 56.Yao J, Burns JE, Forsberg D, Seitel A, Rasoulian A, Abolmaesumi P, et al. A multi-center milestone study of clinical vertebral CT segmentation. Comput Med Imaging Graph. 2016;49:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Q, Lu Z, Feng Q, Ma J, Yang W, Chen C, et al. Adaptive segmentation of vertebral bodies from sagittal MR images based on local spatial information and Gaussian weighted chi-square distance. J Digit Imaging. 2013;26(3):578–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng G, Chu C, Belavy DL, Ibragimov B, Korez R, Vrtovec T, et al. Evaluation and comparison of 3D intervertebral disc localization and segmentation methods for 3D T2 MR data: A grand challenge. Med Image Anal. 2017;35:327–44. [DOI] [PubMed] [Google Scholar]
B5. Breast Segmentation
- 1.Agner SC, Xu J, Madabhushi A. Spectral embedding based active contour (SEAC) for lesion segmentation on breast dynamic contrast enhanced magnetic resonance imaging. Medical physics. 2013. March 1;40(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Faris AQ, Ngah UK, Isa NA, Shuaib IL. Computer-aided segmentation system for breast MRI tumour using modified automatic seeded region growing (BMRI-MASRG). Journal of digital imaging. 2014. February 1;27(1):133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DalmƖş MU, Litjens G, Holland K, Setio A, Mann R, Karssemeijer N, Gubern Mérida A. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Medical physics. 2017. February 1;44(2):533–46. [DOI] [PubMed] [Google Scholar]
- 4.DalmƖş MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern-Mérida A. Fully automated detection of breast cancer in screening MRI using convolutional neural networks. Journal of Medical Imaging. 2018. January;5(1):014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doran SJ, Hipwell JH, Denholm R, Eiben B, Busana M, Hawkes DJ, Leach MO, Silva ID. Breast MRI segmentation for density estimation: Do different methods give the same results and how much do differences matter?. Medical physics. 2017. September;44(9):4573–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ertas G, Doran SJ, Leach MO. A computerized volumetric segmentation method applicable to multicentre MRI data to support computer-aided breast tissue analysis, density assessment and lesion localization. Medical & biological engineering & computing. 2017. January 1;55(1):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fooladivanda A, Shokouhi SB, Ahmadinejad N. Localized-atlas-based segmentation of breast MRI in a decision-making framework. Australasian physical & engineering sciences in medicine. 2017. March 1;40(1):69–84. [DOI] [PubMed] [Google Scholar]
- 8.Gubern-Merida A, Kallenberg M, Mann RM, Marti R, Karssemeijer N. Breast segmentation and density estimation in breast MRI: a fully automatic framework. IEEE journal of biomedical and health informatics. 2015. January;19(1):349–57. [DOI] [PubMed] [Google Scholar]
- 9.Hu L, Cheng Z, Wang M, Song Z. Image manifold revealing for breast lesion segmentation in DCE-MRI. Bio-medical materials and engineering. 2015. January 1;26(s1):S1353–60. [DOI] [PubMed] [Google Scholar]
- 10.Ivanovska T, Laqua R, Wang L, Liebscher V, Völzke H, Hegenscheid K. A level set based framework for quantitative evaluation of breast tissue density from MRI data. PloS one. 2014. November 25;9(11):e112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janaki SD, Geetha K. Automatic segmentation of lesion from breast DCE-MR image using artificial fish swarm optimization algorithm. Polish Journal of Medical Physics and Engineering. 2017. June 27;23(2):29–36. [Google Scholar]
- 12.Jiang L, Hu X, Xiao Q, Gu Y, Li Q. Fully automated segmentation of whole breast using dynamic programming in dynamic contrast enhanced MR images. Medical physics. 2017. June;44(6):2400–14. [DOI] [PubMed] [Google Scholar]
- 13.Khalvati F, Gallego-Ortiz C, Balasingham S, Martel AL. Automated segmentation of breast in 3-D MR images using a robust atlas. IEEE transactions on medical imaging. 2015. January;34(1):116–25. [DOI] [PubMed] [Google Scholar]
- 14.Lin M, Chen JH, Wang X, Chan S, Chen S, Su MY. Template based automatic breast segmentation on MRI by excluding the chest region. Medical physics. 2013. December 1;40(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milenković J, Chambers O, Mušič MM, Tasič JF. Automated breast-region segmentation in the axial breast MR images. Computers in biology and medicine. 2015. July 1;62:55–64. [DOI] [PubMed] [Google Scholar]
- 16.Tagliafico A, Bignotti B, Tagliafico G, Tosto S, Signori A, Calabrese M. Quantitative evaluation of background parenchymal enhancement (BPE) on breast MRI. A feasibility study with a semi-automatic and automatic software compared to observer-based scores. The British journal of radiology. 2015. December;88(1056):20150417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakran S, Chatterjee S, Singhal M, Gupta RK, Singh A. Automatic outer and inner breast tissue segmentation using multi-parametric MRI images of breast tumor patients. PloS one. 2018. January 10;13(1):e0190348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wengert GJ, Helbich TH, Vogl WD, Baltzer P, Langs G, Weber M, Bogner W, Gruber S, Trattnig S, Pinker K. Introduction of an Automated User–Independent Quantitative Volumetric Magnetic Resonance Imaging Breast Density Measurement System Using the Dixon Sequence: Comparison With Mammographic Breast Density Assessment. Investigative radiology. 2015. February 1;50(2):73–80. [DOI] [PubMed] [Google Scholar]
- 19.Wengert GJ, Pinker Domenig K, Helbich TH, Vogl WD, Clauser P, Bickel H, Marino MA, Magometschnigg HF, Baltzer PA. Influence of fat–water separation and spatial resolution on automated volumetric MRI measurements of fibroglandular breast tissue. NMR in Biomedicine. 2016. June;29(6):702–8. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Weinstein SP, Conant EF, Kontos D. Automated fibroglandular tissue segmentation and volumetric density estimation in breast MRI using an atlas aided fuzzy C means method. Medical physics. 2013. December 1;40(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
B6. Adipose Tissue Segmentation
- 1.Addeman BT, Kutty S, Perkins TG, Soliman AS, Wiens CN, McCurdy CM, Beaton MD, Hegele RA, McKenzie CA. Validation of volumetric and single slice MRI adipose analysis using a novel fully automated segmentation method. Journal of Magnetic Resonance Imaging. 2015. January;41(1):233–41. [DOI] [PubMed] [Google Scholar]
- 2.Commandeur F, Goeller M, Betancur J, Cadet S, Doris M, Chen X, Berman DS, Slomka PJ, Tamarappoo BK, Dey D. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE transactions on medical imaging. 2018. August;37(8):1835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decazes P, Rouquette A, Chetrit A, Vera P, Gardin I. Automatic measurement of the total visceral adipose tissue from computed tomography images by using a multi-atlas segmentation method. Journal of computer assisted tomography. 2018. January 1;42(1):139–45. [DOI] [PubMed] [Google Scholar]
- 4.Ding X, Terzopoulos D, Diaz Zamudio M, Berman DS, Slomka PJ, Dey D. Automated pericardium delineation and epicardial fat volume quantification from noncontrast CT. Medical physics. 2015. September 1;42(9):5015–26. [DOI] [PubMed] [Google Scholar]
- 5.Fallah F, Machann J, Martirosian P, Bamberg F, Schick F, Yang B. Comparison of T1-weighted 2D TSE, 3D SPGR, and two-point 3D Dixon MRI for automated segmentation of visceral adipose tissue at 3 Tesla. Magnetic Resonance Materials in Physics, Biology and Medicine. 2017. April 1;30(2):139–51. [DOI] [PubMed] [Google Scholar]
- 6.Kim YJ, Park JW, Kim JW, Park CS, Gonzalez JP, Lee SH, Kim KG, Oh JH. Computerized automated quantification of subcutaneous and visceral adipose tissue from computed tomography scans: development and validation study. JMIR medical informatics. 2016. January;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullberg J, Hedström A, Brandberg J, Strand R, Johansson L, Bergström G, Ahlström H. Automated analysis of liver fat, muscle and adipose tissue distribution from CT suitable for large-scale studies. Scientific reports. 2017. September 5;7(1):10425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundström E, Strand R, Forslund A, Bergsten P, Weghuber D, Ahlström H, Kullberg J. Automated segmentation of human cervical-supraclavicular adipose tissue in magnetic resonance images. Scientific reports. 2017. June 8;7(1):3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mihl C, Loeffen D, Versteylen MO, Takx RA, Nelemans PJ, Nijssen EC, Vega-Higuera F, Wildberger JE, Das M. Automated quantification of epicardial adipose tissue (EAT) in coronary CT angiography; comparison with manual assessment and correlation with coronary artery disease. Journal of cardiovascular computed tomography. 2014. May 1;8(3):215–21. [DOI] [PubMed] [Google Scholar]
- 10.Nemoto M, Yeernuer T, Masutani Y, Nomura Y, Hanaoka S, Miki S, Yoshikawa T, Hayashi N, Ohtomo K. Development of automatic visceral fat volume calculation software for CT volume data. Journal of obesity. 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norlén A, Alvén J, Molnar D, Enqvist O, Norrlund RR, Brandberg J, Bergström G, Kahl F. Automatic pericardium segmentation and quantification of epicardial fat from computed tomography angiography. Journal of Medical Imaging. 2016. September;3(3):034003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popuri K, Cobzas D, Esfandiari N, Baracos V, Jägersand M. Body composition assessment in axial CT images using FEM-based automatic segmentation of skeletal muscle. IEEE transactions on medical imaging. 2016. February;35(2):512–20. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigues ÉO, Morais FF, Morais NA, Conci LS, Neto LV, Conci A. A novel approach for the automated segmentation and volume quantification of cardiac fats on computed tomography. Computer methods and programs in biomedicine. 2016. January 1;123:109–28. [DOI] [PubMed] [Google Scholar]
- 14.Sadananthan SA, Prakash B, Leow MK, Khoo CM, Chou H, Venkataraman K, Khoo EY, Lee YS, Gluckman PD, Tai ES, Velan SS. Automated segmentation of visceral and subcutaneous (deep and superficial) adipose tissues in normal and overweight men. Journal of Magnetic Resonance Imaging. 2015. April;41(4):924–34. [DOI] [PubMed] [Google Scholar]
- 15.Shahzad R, Bos D, Metz C, Rossi A, Kirişli H, van der Lugt A, Klein S, Witteman J, de Feyter P, Niessen W, van Vliet L. Automatic quantification of epicardial fat volume on non enhanced cardiac CT scans using a multi atlas segmentation approach. Medical physics. 2013. September;40(9):091910. [DOI] [PubMed] [Google Scholar]
- 16.Spearman JV, Meinel FG, Schoepf UJ, Apfaltrer P, Silverman JR, Krazinski AW, Canstein C, De Cecco CN, Costello P, Geyer LL. Automated quantification of epicardial adipose tissue using CT angiography: evaluation of a prototype software. European radiology. 2014. February 1;24(2):519–26. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, Xu B, Freeland Graves J. Automated quantification of abdominal adiposity by magnetic resonance imaging. American Journal of Human Biology. 2016. November;28(6):757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thörmer G, Bertram HH, Garnov N, Peter V, Schütz T, Shang E, Blüher M, Kahn T, Busse H. Software for automated MRI based quantification of abdominal fat and preliminary evaluation in morbidly obese patients. Journal of Magnetic Resonance Imaging. 2013. May;37(5):1144–50. [DOI] [PubMed] [Google Scholar]
- 19.Valentinitsch AC Karampinos D, Alizai H, Subburaj K, Kumar D, M. Link T, Majumdar S. Automated unsupervised multi parametric classification of adipose tissue depots in skeletal muscle. Journal of Magnetic Resonance Imaging. 2013. April;37(4):917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang YX, Chong MS, Tay L, Yew S, Yeo A, Tan CH. Automated assessment of thigh composition using machine learning for Dixon magnetic resonance images. Magnetic Resonance Materials in Physics, Biology and Medicine. 2016. October 1;29(5):723–31. [DOI] [PubMed] [Google Scholar]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McBee MP, Awan OA, Colucci AT, Ghobadi CW, Kadom N, Kansagra AP, Tridandapani S, Auffermann WF. Deep learning in radiology. Academic Radiology. 2018. November 1;25(11):1472–80. [DOI] [PubMed] [Google Scholar]
- 2.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Zeitschrift für Medizinische Physik. 2018. December 13. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Chen JH, Chang KT, Park VY, Kim MJ, Chan S, Chang P, Chow D, Luk A, Kwong T, Su MY. Automatic Breast and Fibroglandular Tissue Segmentation in Breast MRI Using Deep Learning by a Fully-Convolutional Residual Neural Network U-Net. Academic Radiology. 2019. January 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanzione A, Cuocolo R, Cocozza S, Romeo V, Persico F, Fusco F, Longo N, Brunetti A, Imbriaco M. Detection of Extraprostatic Extension of Cancer on Biparametric MRI Combining Texture Analysis and Machine Learning: Preliminary Results. Academic Radiology. 2019. January 1. [DOI] [PubMed] [Google Scholar]
- 5.England JR, Cheng PM. Artificial intelligence for medical image analysis: a guide for authors and reviewers. American Journal of Roentgenology. 2019. March;212(3):513–9. [DOI] [PubMed] [Google Scholar]
- 6.Tustison NJ, Avants BB, Lin Z, Feng X, Cullen N, Mata JF, Flors L, Gee JC, Altes TA, Mugler JP III, Qing K. Convolutional Neural Networks with Template-Based Data Augmentation for Functional Lung Image Quantification. Academic Radiology. 2019. March 1;26(3):412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezinque A, Moriarity A, Farrell C, Peabody H, Noyes SL, Lane BR. Determination of prostate volume: A comparison of contemporary methods. Academic Radiology. 2018. December 1;25(12):1582–7. [DOI] [PubMed] [Google Scholar]
- 8.Turco D, Valinoti M, Martin EM, Tagliaferri C, Scolari F, Corsi C. Fully Automated Segmentation of Polycystic Kidneys From Noncontrast Computed Tomography: A Feasibility Study and Preliminary Results. Academic Radiology. 2018. July 1;25(7):850–5. [DOI] [PubMed] [Google Scholar]
- 9.Brown M, Browning P, Wahi-Anwar MW, Murphy M, Delgado J, Greenspan H, Abtin F, Ghahremani S, Yaghmai N, da Costa I, Becker M. Integration of Chest CT CAD into the Clinical Workflow and Impact on Radiologist Efficiency. Academic Radiology. 2018. August 8. [DOI] [PubMed] [Google Scholar]
- 10.Lenchik L, Boutin RD. Sarcopenia: Beyond Muscle Atrophy and into the New Frontiers of Opportunistic Imaging, Precision Medicine, and Machine Learning. Semin Musculoskelet Radiol. 2018; 22(3):307–322. [DOI] [PubMed] [Google Scholar]
- 11.Amini B, Boyle SP, Boutin RD, Lenchik L. Approaches to Assessment of Muscle Mass and Myosteatosis on Computed Tomography (CT): A Systematic Review. J Gerontol A Biol Sci Med Sci. 2019. February 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers RM. Progress in fully automated abdominal CT interpretation. American Journal of Roentgenology. 2016. July;207(1):67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Medical image analysis. 2017. December 1;42:60–88. [DOI] [PubMed] [Google Scholar]
- 14.Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annual review of biomedical engineering. 2017. June 21;19:221–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aganj I, Harisinghani MG, Weissleder R, Fischl B. Unsupervised medical image segmentation based on the local center of mass. Scientific reports. 2018. August 29;8(1):13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma N, Aggarwal LM. Automated medical image segmentation techniques. J Med Phys. 2010;35(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Commowick O, Warfield SK. A continuous STAPLE for scalar, vector, and tensor images: an application to DTI analysis. IEEE Transactions on Medical Imaging. 2009. June;28(6):838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, Shekelle P, Stewart LA. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Systematic reviews. 2015. December;4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Heart, Lung and Blood Institute (NHLBI) Case Series Quality Assessment Tool https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools[nhlbi.nih.gov]. Accessed January 12, 2019.
- 20.Withey DJ, Koles ZJ. A review of medical image segmentation: methods and available software. International Journal of Bioelectromagnetism. 2008. January;10(3):125–48. [Google Scholar]
- 21.Pitas I Digital image processing algorithms. Prentice-Hall, Inc.; 1993. [Google Scholar]
- 22.Bezdek JC, Hall LO, Clarke LP. Review of MR image segmentation techniques using pattern recognition. Med Phys. 1993;20(4):1033–1048. [DOI] [PubMed] [Google Scholar]
- 23.Clarke LP, Velthuizen RP, Phuphanich S, Schellenberg JD, Arrington JA, Silbiger M. MRI: stability of three supervised segmentation techniques. Magnetic resonance imaging. 1993;11(1):95–106. [DOI] [PubMed] [Google Scholar]
- 24.Jain AK, Duin RPW, Jianchang M. Statistical pattern recognition: a review. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2000;22(1):4–37. [Google Scholar]
- 25.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Transactions on Medical Imaging. 2001;20(1):45–57. [DOI] [PubMed] [Google Scholar]
- 26.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annual review of biomedical engineering. 2000;2:315–337. [DOI] [PubMed] [Google Scholar]
- 27.Boykov YY, Jolly M. Interactive graph cuts for optimal boundary & region segmentation of objects in N-D images. Paper presented at: Proceedings Eighth IEEE International Conference on Computer Vision ICCV; 2001; 7–14 July 2001, 2001. [Google Scholar]
- 28.Niessen WJ, Vincken KL, Weickert J, Romeny BMTH, Viergever MA. Multiscale Segmentation of Three-Dimensional MR Brain Images. Int J Comput Vision. 1999;31(2–3):185–202. [Google Scholar]
- 29.Cabezas M, Oliver A, Lladó X, Freixenet J, Bach Cuadra M. A review of atlas-based segmentation for magnetic resonance brain images. Computer Methods and Programs in Biomedicine. 2011;104(3):e158–e177. [DOI] [PubMed] [Google Scholar]
- 30.Cootes TF, Taylor CJ, Cooper DH, Graham J. Active Shape Models-Their Training and Application. Computer Vision and Image Understanding. 1995;61(1):38–59. [Google Scholar]
- 31.Cootes TF, Edwards GJ, Taylor CJ. Active Appearance Models. IEEE Trans Pattern Anal Mach Intell. 2001;23(6):681–685. [Google Scholar]
- 32.García-Lorenzo D1, Francis S, Narayanan S, Arnold DLCD. Review of Automated segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal. 2013;17(1):1–18. [DOI] [PubMed] [Google Scholar]
- 33.Danelakis A, Theoharis T, Verganelakis DA. Survey of automated multiple sclerosis lesion segmentation techniques on magnetic resonance imaging. Comput Med Imaging Graph. 2018;70:83–100. [DOI] [PubMed] [Google Scholar]
- 34.Datta S, Narayana PA. A comprehensive approach to the segmentation of multichannel three-dimensional MR brain images in multiple sclerosis. NeuroImage Clin. 2013;2(1):184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González-Villà S, Oliver A, Valverde S, Wang L, Zwiggelaar R, Lladó X. A review on brain structures segmentation in magnetic resonance imaging. Artif Intell Med. 2016;73:45–69. [DOI] [PubMed] [Google Scholar]
- 36.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2015. [Google Scholar]
- 37.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. Journal of Digital Imaging. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickie DA, Shenkin SD, Anblagan D, Lee J, Blesa Cabez M, Rodriguez D, et al. Whole Brain Magnetic Resonance Image Atlases: A Systematic Review of Existing Atlases and Caveats for Use in Population Imaging. Front Neuroinform. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cover GS, Herrera WG, Bento MP, Appenzeller S, Rittner L. Computational methods for corpus callosum segmentation on MRI: A systematic literature review. Comput Methods Programs Biomed. 2018;154:25–35. [DOI] [PubMed] [Google Scholar]
- 40.Rak M, Tönnies KD, Rak MTK. On computerized methods for spine analysis in MRI: a systematic review. Int J Comput Assist Radiol Surg. 2016;11(8):1445–65. [DOI] [PubMed] [Google Scholar]
- 41.Dolz J, Desrosiers C, Ayed I Ben. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. Neuroimage. 2018;170:456–70. [DOI] [PubMed] [Google Scholar]
- 42.Gordillo N, Montseny ESP. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging. 2013;31(8):1426–38. [DOI] [PubMed] [Google Scholar]
- 43.Garccia-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL. Review of Automated segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal. 2013;17(1):1–18. [DOI] [PubMed] [Google Scholar]
- 44.Despotović I, Goossens BPW. MRI segmentation of the human brain: challenges, methods, and applications. Comput Math Methods Med. 2015;2015:450341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dill V, Franco AR PM. Automated methods for hippocampus segmentation: the evolution and a review of the state of the art. Neuroinformatics. 2015;13(2):133–50. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Parra LC. Fully automated whole-head segmentation with improved smoothness and continuity, with theory reviewed. PLoS One. 2015;10(5):1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunther Helms. Segmentation of human brain using structural MRI. MAGMA. 2016;29(2):111–24. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, Dou Q, Yu L, Qin J, Heng P-A. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage. 2018;170:446–55. [DOI] [PubMed] [Google Scholar]
- 49.De Leener B, Taso M, Cohen-Adad JCV. Segmentation of the human spinal cord. MAGMA. 2016;29(2):125–53. [DOI] [PubMed] [Google Scholar]
- 50.Prados F, Cardoso MJ, Yiannakas MC, Hoy LR, Tebaldi E, Kearney H, Liechti MD, Miller DH, Ciccarelli O, Wheeler-Kingshott CA2 OS. Fully automated grey and white matter spinal cord segmentation. Sci Rep. 2016;6:36151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dupont SM, De Leener B, Taso M, Le Troter A, Nadeau S, Stikov N, et al. Fully-integrated framework for the segmentation and registration of the spinal cord white and gray matter. Neuroimage. 2017;150:358–72. [DOI] [PubMed] [Google Scholar]
- 52.Berndt B, Landry G, Schwarz F, Tessonnier T, Kamp F, Dedes G, Thieke C, Würl M, Kurz C, Ganswindt U, Verhaegen F, Debus J, Belka C, Sommer W, Reiser M, Bauer JPK. Application of single- and dual-energy CT brain tissue segmentation to PET monitoring of proton therapy. Phys Med Biol. 2017;62(6):2427–48. [DOI] [PubMed] [Google Scholar]
- 53.Qian X, Lin Y, Zhao Y, Yue X, Lu BWJ. Objective Ventricle Segmentation in Brain CT with Ischemic Stroke Based on Anatomical Knowledge. Biomed Res Int. 2017;2017:8690892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel A, van Ginneken B, Meijer FJA, van Dijk EJ, Prokop MMR. Robust cranial cavity segmentation in CT and CT perfusion images of trauma and suspected stroke patients. Med Image Anal. 2017;36:216–28. [DOI] [PubMed] [Google Scholar]
- 55.Hanaoka S, Masutani Y, Nemoto M, Nomura Y, Miki S, Yoshikawa T, Hayashi N, Ohtomo KSA. Landmark-guided diffeomorphic demons algorithm and its application to Automated segmentation of the whole spine and pelvis in CT images. Int J Comput Assist Radiol Surg. 2017;12(3):413–30. [DOI] [PubMed] [Google Scholar]
- 56.Kim KLS. Vertebrae localization in CT using both local and global symmetry features. Comput Med Imaging Graph. 2017;58:45–55. [DOI] [PubMed] [Google Scholar]
- 57.Slomka PJ, Dey D, Sitek A, Motwani M, Berman DS, Germano G. Cardiac imaging: working towards fully-automated machine analysis & interpretation. Expert Rev Med Devices. 2017;14(3):197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moccia S, De Momi E, El Hadji S, Mattos LS. Blood vessel segmentation algorithms — Review of methods, datasets and evaluation metrics. Comput Methods Programs Biomed [Internet]. 2018;158:71–91. Available from: 10.1016/j.cmpb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Wang L, Chitiboi T, Meine H, Günther M, Hahn HK. Principles and methods for Automated and semi-Automated tissue segmentation in MRI data. Magn Reson Mater Physics, Biol Med. 2016;29(2):95–110. [DOI] [PubMed] [Google Scholar]
- 60.Peng P, Lekadir K, Gooya A, Shao L, Petersen SE, Frangi AF. A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. Magn Reson Mater Physics, Biol Med. 2016;29(2):155–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krittanawong C, Johnson KW, Rosenson RS, Wang Z, Aydar M, Baber U, et al. Deep learning for cardiovascular medicine: a practical primer. Eur Heart J. 2019;1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin Y, Zheng H, Huang X, Yang J, Zhu YM. Pulmonary nodule segmentation with CT sample synthesis using adversarial networks. Med Phys. 2019;46(3):1218–29. [DOI] [PubMed] [Google Scholar]
- 63.Huang X, Sun W, Tseng T-L (Bill), Li C, Qian W. Fast and Fully-Automated Detection and Segmentation of Pulmonary Nodules in Thoracic CT Scans Using Deep Convolutional Neural Networks. Comput Med Imaging Graph [Internet]. 2019; Available from: https://linkinghub.elsevier.com/retrieve/pii/S0895611118305366. [DOI] [PubMed] [Google Scholar]
- 64.Torres HR, Queiros S, Morais P, Oliveira B, Fonseca JC, Vilaca JL. Kidney segmentation in ultrasound, magnetic resonance and computed tomography images: A systematic review. Comput Methods Programs Biomed. 2018;157:49–67. [DOI] [PubMed] [Google Scholar]
- 65.Gotra A, Sivakumaran L, Chartrand G, Vu KN, Vandenbroucke-Menu F, Kauffmann C, et al. Liver segmentation: indications, techniques and future directions. Insights Imaging. 2017;8(4):377–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kline TL, Edwards ME, Garg I, Irazabal MV, Korfiatis P, Harris PC, et al. Quantitative MRI of kidneys in renal disease. Abdom Radiol (NY). 2018;43(3):629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huo Y, Liu J, Xu Z, Harrigan RL, Assad A, Abramson RG, et al. Robust Multicontrast MRI Spleen Segmentation for Splenomegaly Using Multi-Atlas Segmentation. IEEE Trans Biomed Eng. 2018;65(2):336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J, Huo Y, Xu Z, Assad A, Abramson RG, Landman BA. Multi-Atlas Spleen Segmentation on CT Using Adaptive Context Learning. Proc SPIE Int Soc Opt Eng. 2017;10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu Y, Liu S, Li H, Yang D. Automated and hierarchical segmentation of the human skeleton in CT images. Phys Med Biol. 2017;62(7):2812–33. [DOI] [PubMed] [Google Scholar]
- 70.Chandra SS, Xia Y, Engstrom C, Crozier S, Schwarz R, Fripp J. Focused shape models for hip joint segmentation in 3D magnetic resonance images. Med Image Anal. 2014;18(3):567–78. [DOI] [PubMed] [Google Scholar]
- 71.SpineWeb. SpineWeb: Collaborative Platform for Research on Spine Imaging and Image Analysis 2018. [Available from: http://spineweb.digitalimaginggroup.ca/spineweb/index.php?n=Main.Datasets.
- 72.Yao J, Burns JE, Forsberg D, Seitel A, Rasoulian A, Abolmaesumi P, et al. A multi-center milestone study of clinical vertebral CT segmentation. Comput Med Imaging Graph. 2016;49:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanaoka S, Masutani Y, Nemoto M, Nomura Y, Miki S, Yoshikawa T, et al. Landmark-guided diffeomorphic demons algorithm and its application to Automated segmentation of the whole spine and pelvis in CT images. Int J Comput Assist Radiol Surg. 2017;12(3):413–30. [DOI] [PubMed] [Google Scholar]
- 74.Zheng G, Chu C, Belavy DL, Ibragimov B, Korez R, Vrtovec T, et al. Evaluation and comparison of 3D intervertebral disc localization and segmentation methods for 3D T2 MR data: A grand challenge. Med Image Anal. 2017;35:327–44. [DOI] [PubMed] [Google Scholar]
- 75.Norman B, Pedoia V, Majumdar S. Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology. 2018. July;288(1):177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gadermayr M, Disch C, Müller M, Merhof D, Gess B. A comprehensive study on automated muscle segmentation for assessing fat infiltration in neuromuscular diseases. Magn Reson Imaging. 2018. May;48:20–26. [DOI] [PubMed] [Google Scholar]
- 77.Ambellan F, Tack A, Ehlke M, Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med Image Anal. 2019. February;52:109–118. [DOI] [PubMed] [Google Scholar]
- 78.Pedoia V, Li X, Su F, Calixto N, Majumdar S. Fully Automated analysis of the knee articular cartilage T1ρ relaxation time using voxel-based relaxometry. J Magn Reson Imaging. 2016. April;43(4):970–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. 2018. April;79(4):2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barnard R, Tan J, Roller B, Chiles C, Weaver AA, Boutin RD, Kritchevsky SB, Lenchik L. New Machine Learning for Automatic Paraspinous Muscle Area and Attenuation Measures on Low-Dose Chest CT Scans. Academic Radiology. 2019. [in press]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karlsson A, Rosander J, Romu T, et al. Automated and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water-fat MRI. J Magn Reson Imaging. 2015. June;41(6):1558–69. [DOI] [PubMed] [Google Scholar]
- 82.Yang YX, Chong MS, Lim WS, Tay L, Yew S, Yeo A, Tan CH. Validity of estimating muscle and fat volume from a single MRI section in older adults with sarcopenia and sarcopenic obesity. Clin Radiol. 2017. May;72(5):427.e9–427.e14. [DOI] [PubMed] [Google Scholar]
- 83.Lee H, Troschel FM, Tajmir S, Fuchs G, Mario J, Fintelmann FJ, Do S. Pixel-Level Deep Segmentation: Artificial Intelligence Quantifies Muscle on Computed Tomography for Body Morphometric Analysis. J Digit Imaging. 2017. August;30(4):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhl CK, Kooijman H, Gieseke J, Schild HH. Effect of B1 inhomogeneity on breast MR imaging at 3.0 T. Radiology. 2007. September;244(3):929–30. [DOI] [PubMed] [Google Scholar]
- 85.Wang L, Chitiboi T, Meine H, Günther M, Hahn HK. Principles and methods for Automated and semi-Automated tissue segmentation in MRI data. Magnetic Resonance Materials in Physics, Biology and Medicine. 2016. April 1;29(2):95–110. [DOI] [PubMed] [Google Scholar]
- 86.Codari M, Schiaffino S, Sardanelli F, Trimboli RM. Artificial Intelligence for Breast MRI in 2008–2018: A Systematic Mapping Review. American Journal of Roentgenology. 2019. February;212(2):280–92. [DOI] [PubMed] [Google Scholar]
- 87.Wu S, Weinstein SP, Conant EF, Kontos D. Automated fibroglandular tissue segmentation and volumetric density estimation in breast MRI using an atlas aided fuzzy C means method. Medical physics. 2013. December 1;40(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosado-Toro JA, Barr T, Galons JP, Marron MT, Stopeck A, Thomson C, Thompson P, Carroll D, Wolf E, Altbach MI, Rodríguez JJ. Automated breast segmentation of fat and water MR images using dynamic programming. Academic radiology. 2015. February 1;22(2):139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wengert GJ, Helbich TH, Vogl WD, Baltzer P, Langs G, Weber M, Bogner W, Gruber S, Trattnig S, Pinker K. Introduction of an Automated User–Independent Quantitative Volumetric Magnetic Resonance Imaging Breast Density Measurement System Using the Dixon Sequence: Comparison With Mammographic Breast Density Assessment. Investigative radiology. 2015. February 1;50(2):73–80. [DOI] [PubMed] [Google Scholar]
- 90.DalmƖş MU, Litjens G, Holland K, Setio A, Mann R, Karssemeijer N, Gubern Mérida A. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Medical physics. 2017. February 1;44(2):533–46. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, Morrell G, Heibrun ME, Payne A, Parker DL. 3D multi-parametric breast MRI segmentation using hierarchical support vector machine with coil sensitivity correction. Academic radiology. 2013. February 1;20(2):137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DalmƖş MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern-Mérida A. Fully automated detection of breast cancer in screening MRI using convolutional neural networks. Journal of Medical Imaging. 2018. January;5(1):014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging. 2017;30:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 95.Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, Yang GZ. Deep Learning for Health Informatics. IEEE J Biomed Health Inform. 2017;21:4–21. [DOI] [PubMed] [Google Scholar]
- 96.Ronneberger O, Fischer P, Brox T, editors. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2015: Springer. [Google Scholar]
- 97.Shi Y, Cheng K, Liu Z. Hippocampal subfields segmentation in brain MR images using generative adversarial networks. Biomedical engineering online. 2019. December;18(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xue Y, Xu T, Zhang H, Long LR, Huang X. Segan: Adversarial network with multi-scale l 1 loss for medical image segmentation. Neuroinformatics. 2018;16:383–392. [DOI] [PubMed] [Google Scholar]