Abstract
Background:
Atopic dermatitis (AD) is a common chronic relapsing skin disease. Genetic variants have been associated with skin barrier function and immune regulation. Thymic stromal lymphopoietin (TSLP), an immune regulator, has been previously associated with AD.
Objective:
The goal of this study was to fine map TSLP and evaluate associations with the onset and persistence of AD.
Methods:
TSLP variation was determined using targeted massively parallel sequencing in a longitudinal cohort of children with AD. Evaluations included linkage disequilibrium (LD) and the persistence of AD over as many as 10 years of follow-up. The association between the presence of AD and rs1898671 variation was evaluated in a second independent cohort.
Results:
The minor variant frequency for rs1898671 was 23.5% (95% CI: 21.4, 25.8). This variant was not in LD with other TSLP variants in the longitudinal cohort (N=741). White children with AD were less likely to have rs1898671 variant (1.41 (1.20, 1.66)) than a genomAD control. Children with AD and the rs1898671 variant during follow-up were more likely to have a remission than children who were wildtype for rs1898671 (OR: 1.56; 95% CI: 1.26, 1.91). In the second cohort (N=585), the rs1898671 variant was less prevalent in those with AD than those without. The protective effect was found to be greater in rs1898671 heterozygotes (OR 1.91 (1.34, 2.75)) than homozygotes (OR: 1.28 (0.61, 2.70)).
Conclusion:
TSLP and specifically rs1898671 is important in the pathogenesis of AD and could represent a potential clinical target for the development of therapies to treat individuals with AD.
Keywords: Atopic Dermatitis, eczema, gene association, genetics, filaggrin, thymic stromal lymphopoietin
Introduction
Atopic dermatitis (AD) is a common chronic relapsing disease that manifests as itchy, typically excoriated skin lesions that are often concentrated around the flexor surfaces of the extremities.1,2 It was once thought to be solely a childhood illness, but recent studies show that AD can be a life-long disease that often persists into adulthood and even the initial diagnosis of AD can occur in adulthood.3–7 In the US, the yearly prevalence of AD is about 10% in both children and adults with an annual cost of more than 4 billion dollars.8–10 Despite our increasing understanding of the epidemiology of AD, the genetic risk factors that predispose to its development remain incompletely defined.
A family history of AD and other atopic illnesses such as asthma and seasonal allergies are associated with an increased risk of developing AD as a child.11, 12 The heritability of AD is estimated to be as high as 84%.13 The most commonly reported AD associated genetic variants are loss of function variants (LoF) in a skin barrier protein called filaggrin (FLG).14 However, FLG LoF variants are only found in about 25–30% of those of European and Asian ancestry with AD and in only about 12% of individuals of American-African ancestry with AD exhibit FLG LoF variants.3, 14–16
Several studies have demonstrated that the heritability of AD is also linked to variations in genes that result in immune dysregulatrion.17 TSLP is the gene that encodes for thymic stromal lymphopoietin (TSLP) protein, which is thought to be a master initiator of allergic inflammation at multiple barrier surfaces.18–21 TSLP protein, predominantly expressed from stressed epithelial cells, promotes type 2 inflammation via a variety of cellular pathways.18, 19 TSLP expression occurs upon skin skin barrier (e.g., FLG abnormality) disruption and subsequently promotes the type 2 immune responses that results in inflammation leading to AD.14, 19, 21 However, which genetic variations in TSLP play a major role in the pathogenesis of AD remains poorly defined.
Increased expression of TSLP has been strongly associated with AD as well as other allergic diseases including asthma, allergic rhinitis, and food allergy.18–25 A 2013 publication by Noti et al demonstrated that TSLP variation was associated with the production of a specific lineage of circulating basophils in patients with eosinophilic esophagitis.26 TSLP also has been shown to be upregulated in keratinocytes in response to local pro-inflammatory triggers.27 In 2010, Gao et al reported an association between two TSLP single nucleotide polymorphisms (SNPs) (rs1898671, rs2416259) and eczema herpeticum, which is an infectious complication of AD most often seen in those with severe AD.22 Recently, siRNA-mediated knockdown of FLG expression was shown to induce TSLP expression in epidermal keratinocytes.28 Using chip-based technology and tagging SNPs, we previously demonstrated that TSLP variant rs1898671 is associated with less persistent AD.29 The goal of this study was to more carefully evaluate the association between TSLP variants using fine mapping of the region to identify other potentially causal SNPs and to investigate SNPs in linkage disequilibrium with the tagging SNP rs1898671. We evaluated haploblocks in the region of rs1898671 to further investigate the association with AD onset and persistence.
Methods:
Population
The Pediatric Eczema Elective Registry (PEER; www.thepeerprogram.com) is a United States nationwide cohort of more than 8,000 subjects with pediatric-onset AD. The current study represents the subcohort of PEER children who provided a genetic sample (PEER DNA cohort).30, 31 Both self-described race and ancestry informative markers were previously used to define race and were found in this cohort to be highly correlated.30 PEER DNA enrollment occurred between November of 2004 and January 2015. At the time of enrollment, children were two to seventeen years old, had a physician-confirmed diagnosis of AD, and had used pimecrolimus cream for at least 6 months.30 Subjects were followed for up to 10 years and during that time were not required to (and most did not) continue therapy with pimecrolimus.32 Full details of the PEER cohort have been previously reported.3, 30, 32
A second cohort, not related to the PEER cohort, called the Genetics of Atopic Dermatitis (GAD) cohort was also studied to evaluate the effect of TSLP variation on the likelihood of having AD. All subjects were examined by a dermatologist from the following Dermatology practice locations: University of Pennsylvania Perelman School of Medicine, Children’s Hospital of Philadelphia, Pennsylvania State University/Hershey Medical Center and Washington University at St Louis School of Medicine. All subjects had a history and an exam consistent with AD (cases) or no history of AD (controls). There was no age limitation, but enrollment was restricted to African-Americans or white patients. All patients provided written informed consent approved by their appropriate Institutional Review Board.
Genetic analysis
DNA was collected using Oragene DNA collection kits (DNA Genotek, Ottawa Canada) as previously reported.30 For the PEER Cohort TSLP and FLG gene was sequenced for all subjects using targeted massively parallel sequencing (MPS). The average coverage for TSLP by region varied from 115 to 181. The reliability of this technique for FLG sequencing, was previously reported.15 Raw sequencing data were aligned and mapped to the reference genome GRCh37 using the Burrows-Wheeler Aligner.33 Single nucleotide variant and insertion/deletion (indel) calling was accomplished using the Genome Analysis Toolkit (GATK) HaplotypeCaller, after following GATK Best Practices realignment and recalibration.34–37 For the GAD cohort, TSLP rs1898671 was assayed using TaqMan technique. Population based gene variation was confirmed using Genome Aggregation Database, gnomAD, (https://gnomad.broadinstitute.org/). Tissue specific gene expression in the skin was confirmed using Genotype-Tissue Expression (GTEx) portal (https://gtexportal.org/home/snp/rs1898671).
Outcome:
AD resolution without therapy (remission) was evaluated in the PEER Cohort based on the self-reported outcome of whether or not a child’s skin was AD symptom-free during the previous six-months while the child was not using medication to treat their AD.30 AD disease activity was based on the survey question: “During the last six months would you say that your child’s skin disease (AD) has shown: complete disease control, good disease control, limited disease control, or uncontrolled disease”. Symptom free was defined as an affirmative response to “complete disease control”. This response has been shown to correlate with other tools used to evaluate symptom control and is likely a marker of long-term disease severity.38 The absence of medication use was determined by series of questions and pictures asking about specific medications as well as the quantity of the medication prescribed. A single Patient Oriented Eczema Measure (POEM) score (0–7 (mild); 8–16 (moderate); and 17–28 (severe)), a tool used to measure eczema severity as reflected by the patient, was obtained on enrollment into the GAD cohort.39
Haplotype Blocks:
Linkage disequilibrium (LD) was assessed for the full PEER DNA Cohort and separately for both the African American and white sub cohorts. LD was first estimated using R2 values between rs1898671 and all other co-occurring SNPs. The software Haploview (https://www.broadinstitute.org/haploview/haploview) was used to visualize haplotype blocks and estimate haplotype frequencies assuming the Gabriel algorithm.40
Statistical Analyses:
Covariate frequencies were summarized using means or medians as appropriate. Initial comparisons were conducted using Chi-Square or logistic regression. The association between the “remission” outcome and the TSLP variant was evaluated using an additive model for the variant and generalized estimating equations (GEE) for binary outcomes assuming an independence working correlation structure to account for the correlation among repeated measures per participant (one survey every six months for up to 10 years). Initial enrollment occurred over a ten-year period and observation is still ongoing for some of the subjects. At any given survey, approximately 60% of subjects responded. Missingness was evaluated visually and felt to be completely at random and consistent with GEE modeling specifications. Weighted GEE models to account for survey response missingness were also conducted and the results were nearly identical (not presented). All analyses were conducted with Stata version 15.1 (StataCorp, College Station, Texas) or R (https://www.r-project.org/).
Results:
The PEER DNA cohort included an analysis of 741 children with an MPS analysis of the TSLP gene. From the full PEER DNA cohort of 741 children, 326 children were of African-American ancestry, 379 were white, 21 children of other ancestries, and 15 children multiple ancestries. 53.2% (394) of the children were female, the average age of AD onset was 1.99 (sd: 2.76) years, and 78.6% and 56.4% of the children had completed five and the full 10 years of PEER enrollment, respectively. Overall 44.4% of children had at least one six-month period of AD remission. MPS revealed 156 variants in TSLP. This study focused on rs1898671.
The minor variant frequency (MVF) for rs1898671 (C substituted by T) was 23.5% (95% CI: 21.4, 25.8) for the full cohort, 9.8% (7.6, 12.4) for children of African-American ancestry, and 34.9% (31.6, 38.3) for white children. For comparison, we obtained population based frequencies from the Genome Aggregation Database, gnomAD, (https://gnomad.broadinstitute.org/), and found that European (non-Finnish) controls have MVF 27.5% (26.3, 29.7) and African controls have 9.8% (8.7, 11.1). In other words, white PEER DNA children with AD are less likely to have rs1898671 variant (1.41 (1.20, 1.66)) than the more general population but African-American children are equally likely (1.00 (0.73, 1.13) as the genomAD African population. Per GTEx (https://gtexportal.org/home/snp/rs1898671), the measured TSLP mRNA in skin of the lower leg and suprapubic regions is increased by rs1898671 variation.
At enrollment children, with rs1898671 were more likely to have a food, animal and medication allergies than children in PEER who did not have the variant, but were not more likely to have asthma, seasonal allergies, concomitant FLG LoF variant, or an earlier AD disease onset (Table 1). Compared to white children, African-American children with the rs1898671 variant also had many more additional TSLP variants (Table 2). However, these variants do not appear to be in linkage with rs1898671 (Table 2). MVF for all of the other variants as well as the most common (≥ 10%) TSLP variants not co-occuring with rs1898671 are reported in Table 2 and Supplement Table 1–3). Overall very few common variants were noted. One haplotype block was identified for the white children (Figure 1) and no haplotype blocks were identified for African-American children.
Table 1:
Phenotype characteristics by rs1898671 (C>T) genotype from the PEER cohort (chi –square p-value)
| Frequency of those without rs1898671 and attribute |
Frequency of those with rs1898671 and attribute |
|
|---|---|---|
| Sex (female) | 51.2% | 54.5% |
| African-American | 59.6% | 20.3% p<0.0001 |
| Asthma | 52.9% | 55.2% |
| Seasonal allergies | 69.4% | 68.5% |
| Food allergy | 28.5% | 37.3% p=0.012 |
| Peanut allergy | 14.1% | 16.6% |
| Milk allergy | 6.7% | 10.8% p=0.047 |
| Animal allergy | 26.5% | 36.3% p=0.004 |
| Dog allergy | 17.3% | 26.1% p=0.004 |
| Cat allergy | 22.4% | 29.8% p=0.023 |
| Medication Allergy | 48.2% | 51.7% p=0.004 |
| Any FLG variant | 22.6% | 25.8% |
| Age AD onset | 2.02 years | 1.89 years |
Table 2:
Linkage disequilibrium as estimated by R2 for TSLP variants if they co-occurred with rs1898671. R2 is listed for the full PEER DNA cohort and then for children of African-American ancestry or Caucasian ancestry. Variants are listed by RSID and in order of location on the TSLP gene per GRCh37.
| RSID | Location | Full Peer R2 | African American R2 |
African American MVF (%) |
White R2 | White MVF (%) |
|---|---|---|---|---|---|---|
| 375939272 | 110406225 | 0.0002 | 0.0010 | 0.5 (0.1,1.3) | ||
| 189331165 | 110406489 | 0.0006 | 0.0002 | 0.6 (0.2,1.6) | ||
| 560721535 | 110406605 | 0.0023 | 0.0020 | 0.5 (0.1,1.3) | ||
| 185194032 | 110406650 | 0.0088 | 0.0109 | 0.2 (0.0,0.8) | 0.0071 | 33.1(29.9,34.4) |
| 186519906 | 110406923 | 0.0020 | 0.0079 | 0.2 (0.0,0.8) | 0.0005 | 0.1 (0.0,0.7) |
| 188567463 | 110407103 | 0.0002 | 0.0010 | 0.5 (0.1,1.3) | 0.0005 | 0.1 (0.0,0.7) |
| 532579260 | 110407131 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) | ||
| 755621192 | 110407189 | 0.0000 | 0.0031 | 0.3 (0.0,1.1) | ||
| 141800763 | 110407492 | 0.0000 | 0.0031 | 0.3 (0.0,1.1) | ||
| 1898671 | 110408002 | reference | reference | 9.8 (7.6, 12.4) | reference | 34.9 (31.6, 38.3) |
| 148296322 | 110408003 | 0.0006 | 0.0002 | 0.6 (0.2,1.6) | ||
| 1016936592 | 110408158 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) | ||
| 560518251 | 110408275 | 0.0023 | 0.0013 | 0.3 (0.0,1.1) | 0.0029 | 0.1 (0.0,0.7) |
| 148396476 | 110408710 | 0.0006 | 0.0002 | 0.6 (0.2,1.6) | ||
| 374175217 | 110408804 | 0.0031 | 0.0026 | 0.6 (0.2,1.6) | ||
| 368498616 | 110408815 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) | ||
| 571508866 | 110409065 | 0.0002 | 0.0006 | 0.2 (0.0,0.8) | 0.0023 | 0.4 (0.0,1.0) |
| 201709945 | 110409391 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) | ||
| 140872604 | 110409408 | 0.0076 | 0.0109 | 0.2 (0.0,0.8) | ||
| 1046714887 | 110409740 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) | ||
| 532541421 | 110409819 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) | ||
| 540379350 | 110409894 | 0.0000 | 0.0031 | 0.3 (0.0,1.1) | ||
| 532704183 | 110411182 | 0.0023 | 0.0018 | 0.5 (0.1,1.3) | ||
| 141535387 | 110411311 | 0.0031 | 0.0329 | 0.5 (0.1,1.3) | ||
| 374549851 | 110411336 | 0.0002 | 0.0010 | 0.5 (0.1,1.3) | ||
| 192309314 | 110412049 | 0.0006 | 0.0002 | 0.6 (0.2,1.6) | ||
| 146408762 | 110412321 | 0.0020 | 0.0219 | 0.3 (0.0,1.1) | ||
| 11466749 | 110412585 | 0.0308 | 0.0132 | 11.0(8.7,13.7) | 0.0910 | 15.1 (12.7,17.7) |
| 115625984 | 110413287 | 0.0006 | 0.0002 | 0.6 (0.2,1.6) | ||
| 966395563 | 110413332 | 0.0015 | 0.0013 | 0.3 (0.0,1.1) |
Figure 1:
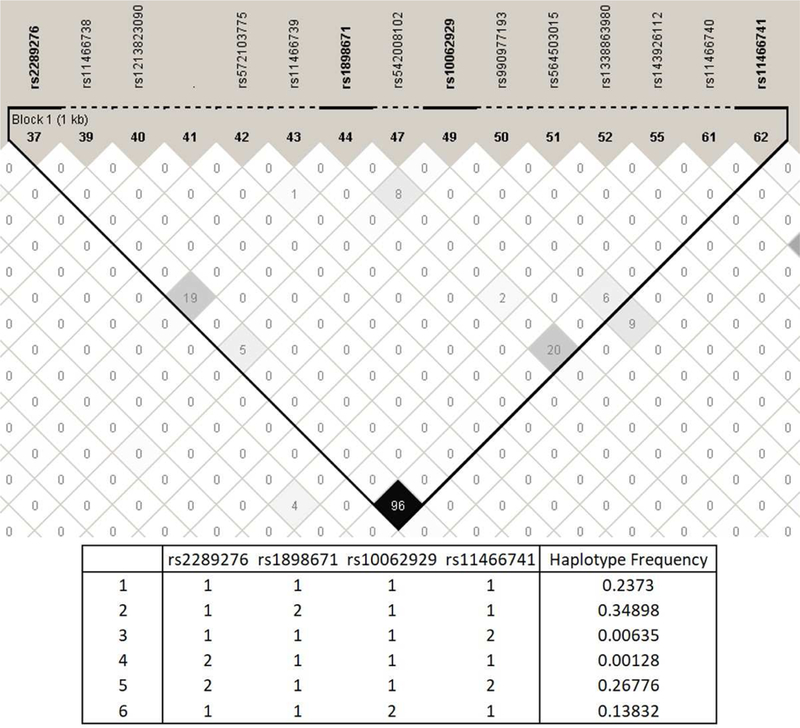
rs1898671 TSLP Haplotypes in white PEER cohort children including the only haploblock (1=not present, 2=variant present)
Children with the rs1898671 variant, were more likely to have a remission period (Table 3 and Figure 2). Children with the rs1898671 variant at any given survey were more likely to have a remission than children who were wildtype for rs1898671 (OR: 1.56; 95% CI: 1.26, 1.91) (Table 3)). This effect was greater for African-American than white children and appears to be greater in those with a FLG LoF variant (Table 3). In addition to rs1898671, the SNPs in the haplotype for the white children included rs10062929, rs2289276, and rs11466741 (Figure 2). Neither of the haplotypes nor the individual SNPs were associated with remission (Table 3).
Table 3:
The association of rs1898671 and remission in the full PEER cohort and among those who are African-American and white and based on the presence of FLG LoF. The odd ratios are adjusted for age onset, sex, race if appropriate and the presence of FLG LoF when appropriate.
| PEER: White or African- American |
FLG LoF | No FLG LoF | |
|---|---|---|---|
| Combined | 1.56 (1.26,1.91) p<0.0001 N=705 |
1.98 (1.33,2.98) p=0.001 N=170 |
1.43 (1.12,1.82) p=0.003 N=535 |
| White | 1.20 (0.94,1.53) N=379 |
1.81 (1.16,2.84) p=0.007 N=120 |
1.19 (0.94,1.53) N=259 |
| African-American | 1.83 (1.12,2.98) p=0.018 N=326 |
_______* N=50 |
1.94 (1.18,3.20) p=0.011 N=276 |
insufficient sample for estimate.
Figure 2:
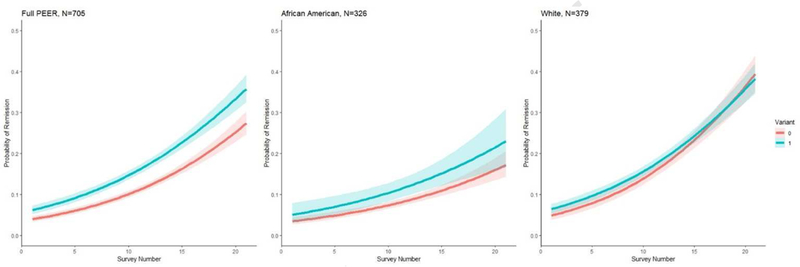
Likelihood of remission (overtime, each survey represents approximately a six-month interval) for children with and without the rs1898671 variant in the PEER cohort for the full cohort, white children, and children of African ancestry.
At this time, the GAD cohort includes 585 individuals including 337 individuals with AD and 238 individuals without AD, respectively. The average age of enrollment was 33.6 years (sd: 21.7), the average age of disease onset for those with AD was 6.8 years (sd: 13.8). 56.7% were female and 49.2% were African-American. The rs1898671 MVF was 21.6% (18.9, 23.7) for the full GAD, 12.0% (9.4, 14.9) for African-Americans and 31.9% (27.8, 36.1) for whites. Individuals with rs1898671 were less likely to have AD (1.48 (1.12, 1.96)). This effect is not significantly different between African-Americans and for whites (p=0.150). The rs1898671 variant AD protective effect was found to be greater in heterozygotes (1.91 (1.34, 2.75)) than homozygotes (1.28 (0.61, 2.70)). Individuals with AD and the rs1898671 variant tend to have a later age of onset of their AD (wildype-6.2 years (sd: 11.8), heterozygous-7.6 years (sd:16.6) years and homozygous-9.0 years (sd:19.6)). The median POEM score was of 12 (moderate eczema severity) (25%: 6 (mild), 75%:17 (severe)) for GAD group with AD and was not associated with rs1898671 variation.
Discussion:
TSLP is important for the activation of type 2 inflammation at barrier surfaces. It has previously been shown to be associated with multiple allergic disorders. Based on prior studies, mostly chip based studies, we focused on rs1898671 to examine its association with AD. We replicated prior findings using massively parallel sequencing and demonstrated that rs1898671 variant, which was originally selected as a tagging SNP, is not in linkage disequilibrium with other TSLP variants found in the PEER DNA cohort.22 It is more frequently found in white individuals than those of African-American ancestry. We can now confirm that children with AD who have rs1898671 have a less severe course of AD as manifested by more frequent remissions. The variant may also be associated with decreased incidence of AD, as noted in both the white PEER DNA cohort as compared to gnomAD and the first assessment of the GAD cohort, which is a new cohort of individuals with AD that is not related to the PEER cohort. Based on the clinical observations and GTEx, the rs1898671 variant may have a direct effect on the production of TSLP in the skin. Finally, the effect of FLG LoF, which has been shown to increase the prevalence and persistence of AD, appears to be modified by rs1898671.15, 30 These findings are unique and important because we can now confirm that the rs1898671 variant and not another TSLP variant in linkage disequilibrium (LD) with re1898671 is associated with both an decreased risk of AD and decreased AD persistence/severity.
The rs1898671 variant is located in an intron of the TSLP gene on chromosome 5(110408002 (GRCh37)). It represents a single nucleotide change from C to T and potentially falls on two TSLP transcripts. The TSLP gene is transcribed into a long form (exons one to four) and short form (exons three and four) protein.41, 42 Increased TSLP production is associated with rs1898671. The rs1898671 variant is within about 300 bp of exon three and the initiation sequence for the short form of the TSLP protein (sfTSLP). sfTSLP protein is constitutively expressed at the mRNA level in human keratinocytes and is likely the predominate form produced by keratinocytes.41 It appears to act as an antimicrobial peptide creating a skin barrier defense and is not secreted in response to inflammatory influences.41, 42 The exact functions of the two forms of TSLP are not fully understood. It is possible that the isoforms act in antagonistic ways; however, due to technical difficulties, in most studies, these two proteins are often not differentiated.41–43
Gao et al reported an association between two TSLP SNPs (rs1898671, rs2416259) and eczema herpeticum.22 In addition, we previously noted that rs1898671 was associated with less persistent AD and was not associated with the timing of onset of AD. 26, 44 These studies relied on genotyping techniques and could not properly differentiate rs189867 from other variants that could potentially be in LD. In both studies, the presence of the rs1898671 appeared to be associated with a milder clinical phenotype. A recent study in Korea of another set of TSLP SNPs showed an alteration in the onset of AD and co-morbidities like asthma and allergic rhinitis in those with TSLP variation.45 TSLP variation has been associated with eosinophilic esophagitis another illness that is associated with AD.26, 46 TSLP has also been shown in experimental human keratinocyte models and mouse models to be associated with skin inflammation like that seen in AD.23
Our study does have limitations. The PEER study is a cohort study and follow-up data are obtained by survey. It is possible that the survey data do not properly reflect the severity of AD. However, it is unlikely that misclassification of severity is different between genotypes (which are unknown to the participant) thereby minimizing this potential bias. Outcome data, like POEM, were obtained during an office visit usually scheduled for AD treatment. It is likely that the POEM results are biased towards more severe findings (i.e., differential information bias) and, since the GAD cohort is a one-time evaluation, could explain why no clinical differences in POEM were noted in our study. It is also important to note that neither PEER nor GAD are random samples of the US population so it is possible that our results are not generalizable. However, many of the rs1898671 findings first noted in PEER were reproduced in the GAD cohort. As discussed isoforms of TSLP exist that have different functions. Differentiating the isoform of TSLP is technically difficult, was not assessed in this report, and is necessary to properly differentiate TSLP function. In addition, allergen reactivity testing and measurements of serum IgE, which could be influenced by rs1898671 variation was not measured in this study.
In summary, rs1898671 genetic variation in those with AD appears to have a direct influence on the persistence of AD and, in whites, it appears to decrease the incidence of AD. Others have shown that the presence of rs1898671 in those with AD diminishes the likelihood of AD associated HSV infection and other atopic illnesses, such as asthma and eosinophilic esophogitis.22 We demonstrated that rs1898671 is not in LD with other co-existing TSLP variants; thus it is likely to have a direct effect on TSLP production or function. Based on the location of rs1898671 and GTEx database, it is likely that the variant has an effect on the function or production of the sfTSLP.
Current biologic therapies for AD due target the general immunologic pathway that includes lymphocyte activation by TSLP. Clinicians may begin to use FLG LoF genotyping, TSLP genotyping, or the AD polygenic risk score to personalize AD treatment as part of their treatment plan30, 31, 47. However, at this time it is best to proceed with caution. Very few studies have formally evaluated the influence of genetic variation on longitudinal AD severity and very few have evaluated the influence that genetic variation has on treatment30, 31. Studies have also shown that genetic testing for genes associated with AD is technically difficult16, 48, 49 Additional study is necessary before clinical personalized treatment is based AD genetic testing. Based on our current findings it is fair to conclude that TSLP, and specifically rs1898671 is an important genetic variant in the pathogenesis and long-term prognosis of AD and could represent a potential clinical target for the development of therapies to treat individuals with AD.
Supplementary Material
Acknowledgments
Funding Source:
This project has been funded in whole or in part by R01-AR060962 and R01-AR070873 from NIAMS (PI: David Margolis). The PEER studies if funded by Valeant Pharmaceuticals (Pi: David Margolis).
Andrea L. Zaenglein reports research grants from Pfizer, Incyte, Abbvie, and Dermavant. Zelma Chiesa Fuxench has served as a consultant for the Asthma and Allergy Foundation of American and the National Eczema Association; has received research grants from Regeneron, Sanofi, Tioga, Vanda, Realm therapeutics and Leo for work related to atopic dermatitis; and has received honoraria for continuing medical education work in atopic dermatitis sponsored by educational grants from Sanofi and Regeneron. David Margolis is a consultant for Pfizer, Leo, and Sunovion with respect to studies of atopic dermatitis and serves on an advisory board for the National Eczema Association.
Abbreviations:
- (AD)
Atopic dermatitis
- (CI)
Confidence interval
- (FLG)
Filaggrin
- (GEE)
Generalized estimating equations
- (GAD)
Genetics of Atopic Dermatitis
- (LD)
Linkage disequilibrium
- (LoF)
Loss of function
- (MPS)
Massively parallel sequencing
- (MVF)
Minor variant frequency
- (OR)
Odds Ratio
- (POEM)
Patient Oriented Eczema Measure
- (PEER)
Pediatric Eczema Elective Registry
- (SNP)
Single nucleotide polymorphisms
- (TSLP)
Thymic stromal lymphopoietin
Footnotes
Potential conflicts of interest: Nandita Mitra reports no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akdis CA, Akdis M, Bieber T, Bindslev-Jensen C, Boguniewicz M, Eigenmann P et al. Diagnosis and treatment of atopic dermatitis in children and adults: European Academy of Allergology and Clinical Immunology/American Academy of Allergy, Asthma and Immunology/PRACTALL Consensus Report. . Allergy 2006;61:969–87. [DOI] [PubMed] [Google Scholar]
- 2.Charman CR, Williams HC. Epidemiology In: Bieber Tand Leung DYM editors. Atopic Dermatitis. New York: Marcel Dekker, Inc.; 2002. p. 21–42. [Google Scholar]
- 3.Margolis JS, Abuabrar K, Bilker W, Hoffstad O, Margolis DJ Persistance of mild of mild to moderate atopic dermatitis. JAMA Dermatology 2014;150:593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abuabara K, Margolis DJ, Abuabara K, Margolis DJ. Do children really outgrow their eczema, or is there more than one eczema? Journal of Allergy & Clinical Immunology 2013;132:1139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abuabara K, Magyari A, McCulloch CE, Linos E, Margolis DJ, Langan SM. Prevalence of Atopic Eczema Among Patients Seen in Primary Care: Data From The Health Improvement Network. Ann Intern Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiesa Fuxench ZC, Block JK, Boguniewicz M, Boyle J, Fonacier L, Gelfand JM et al. Atopic Dermatitis in America Study: A Cross-Sectional Study Examining the Prevalence and Disease Burden of Atopic Dermatitis in the US Adult Population. J Invest Dermatol 2019;139:583–90. [DOI] [PubMed] [Google Scholar]
- 7.Silverberg JI, Hanifin JM. Adult eczema prevalence and associations with asthma and other variables: A US population-based study. Journal of Allergy & Clinical Immunology 2013;132:1132–8. [DOI] [PubMed] [Google Scholar]
- 8.Ellis CN, Drake LA, Prendergast MM, Abramovits W, Boguniewicz M, Daniel CR et al. Cost of atopic dermatitis and eczema in the United States. J Am Acad Dermatol 2002;46:361–70. [DOI] [PubMed] [Google Scholar]
- 9.Lim HW, Collins SAB, Resneck JS Jr., Bolognia JL, Hodge JA, Rohrer TA et al. The burden of skin disease in the United States. J Am Acad Dermatol 2017;76:958–72.e2. [DOI] [PubMed] [Google Scholar]
- 10.Lim HW, Collins SAB, Resneck JS Jr., Bolognia J, Hodge JA, Rohrer TA et al. A risk adjustment approach to estimating the burden of skin disease in the United States. J Am Acad Dermatol 2018;78:129–40. [DOI] [PubMed] [Google Scholar]
- 11.Eichenfield LF, Tom WL, Chamlin SL, Feldman SR, Hanifin JM, Simpson EL et al. Guidelines of care for the management of atopic dermatitis: section 1. Diagnosis and assessment of atopic dermatitis. [Review]. Journal of the American Academy of Dermatology 2014;70:338–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung DY, Bieber T. Atopic dermatitis. [Review] [100 refs]. Lancet 2003;361:151–60. [DOI] [PubMed] [Google Scholar]
- 13.Ober C, Yao TC. The genetics of asthma and allergic disesae: A 21st century perspective. Immunological Reviews 2011;242:10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown SJ, McLean WH. One remarkable molecule: filaggrin. J Invest Dermatol 2012;132:751–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margolis DJ, Mitra N, Gochnauer H, Wubbenhorst B, D’Andrea K, Kraya A et al. Uncommon Filaggrin Variants Are Associated with Persistent Atopic Dermatitis in African Americans. J Invest Dermatol 2018;138:1501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong X, Denil S, Foo JN, Chen H, Tay ASL, Haines RL et al. Array-based sequencing of filaggrin gene for comprehensive detection of disease-associated variants. J Allergy Clin Immunol 2018;141:814–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boguniewicz M, Leung DY. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. [Review]. Immunological Reviews 2011;242:233–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demehri S, Morimoto M, Holtzman MJ, Kopan R. Skin-derived TSLP triggers progression from epidermal-barrier defects to asthma. Plos Biology 2009;7:e1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esnault S, Rosenthal LA, Wang DS, Malter JS. Thymic Stromal Lymphopoietin (TSLP) as a Bridge between Infection and Atopy. Int J Clin Exp Pathol 2008;1:325–30. [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Hener P, Zhang Z, Ganti KP, Metzger D, Chambon P. Induction of thymic stromal lymphopoietin expression in keratinocytes is necessary for generating an atopic dermatitis upon application of the active vitamin D3 analogue MC903 on mouse skin. J Invest Dermatol 2009;129:498–502. [DOI] [PubMed] [Google Scholar]
- 21.Soumelis V, Reche PA, Kanzler H, Yuan W, Edward G, Homey B et al. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nature Immunology 2002;3:673–80. [DOI] [PubMed] [Google Scholar]
- 22.Gao PS, Rafaels NM, Mu D, Hand T, Murray T, Boguniewicz M et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. Journal of Allergy & Clinical Immunology 2010;125:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim BS, Siracusa MC, Saenz SA, Noti M, Monticelli LA, Sonnenberg GF et al. TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Science Translational Medicine 2013;5:170ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin AM, Mathias RA, Huang L, Roth LA, Daley D, Myers RA et al. A meta-analysis of genome-wide association studies for serum total IgE in diverse study populations. Journal of Allergy & Clinical Immunology 2013;131:1176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nature Genetics 2010;42:289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis Nature Medicine 2013;19:1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. Journal of Investigative Dermatology 2013;133:154–63. [DOI] [PubMed] [Google Scholar]
- 28.Lee KH, Cho KA, Kim JY, Kim JY, Baek JH, Woo SY et al. Filaggrin knockdown and toll-like receptor 3 (TLR3) stimulation enchanced the production of thymic stromal lymphopoietin (TSLP) from epidermal layers. Experimental Dermatology 2011;20:149–51. [DOI] [PubMed] [Google Scholar]
- 29.Margolis DJ, Kim B, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M et al. Thymic stromal lymphopoietin variation, filaggrin loss-of-function, and persistence of atopic dermatitis. JAMA Dermatology 2013;150:254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margolis DJ, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, Campbell LE et al. The persistence of atopic dermatitis and filaggrin (FLG) mutations in a US longitudinal cohort. J Allergy Clin Immunol 2012;130:912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang J, Mitra N, Hoffstad O, Margolis DJ. Association of Filaggrin Loss of Function and Thymic Stromal Lymphopoietin Variation With Treatment Use in Pediatric Atopic Dermatitis. JAMA Dermatol 2017;153:275–81. [DOI] [PubMed] [Google Scholar]
- 32.Margolis DJ, Abuabara K, Hoffstad OJ, Wan J, Raimondo D, Bilker WB. Association Between Malignancy and Topical Use of Pimecrolimus. JAMA Dermatol 2015;151:594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate long-read aligment with Burrows-Wheeler Transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013; 43:11.0.1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2018:201178. [Google Scholar]
- 38.Chang J, Bilker WB, Hoffstad O, Margolis DJ. Cross-sectional comparisons of patient-reported disease control, disease severity and symptom frequency in children with atopic dermatitis. Br J Dermatol 2017;177:e114–e5. [DOI] [PubMed] [Google Scholar]
- 39.Charman CR, Venn AJ, Williams HC. The patient-oriented eczema measure: development and initial validation of a new tool for measuring atopic eczema severity from the patients’ perspective. Arch Dermatol 2004;140:1513–9. [DOI] [PubMed] [Google Scholar]
- 40.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–9. [DOI] [PubMed] [Google Scholar]
- 41.Bjerkan L, Schreurs O, Engen SA, Jahnsen FL, Baekkevold ES, Blix IJ et al. The short form of TSLP is constitutively translated in human keratinocytes and has characteristics of an antimicrobial peptide. Mucosal Immunol 2015;8:49–56. [DOI] [PubMed] [Google Scholar]
- 42.Bjerkan L, Sonesson A, Schenck K. Multiple Functions of the New Cytokine-Based Antimicrobial Peptide Thymic Stromal Lymphopoietin (TSLP). Pharmaceuticals (Basel) 2016;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin SC, Cheng FY, Liu JJ, Ye YL. Expression and Regulation of Thymic Stromal Lymphopoietin and Thymic Stromal Lymphopoietin Receptor Heterocomplex in the Innate-Adaptive Immunity of Pediatric Asthma. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan J, Mitra N, Hoffstad OJ, Margolis DJ. Influence of FLG mutations and TSLP polymorphisms on atopic dermatitis onset age. Ann Allergy Asthma Immunol 2017;118:737–8.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ko EJ, Heo WI, Park KY, Lee MK, Seo SJ. Genetic polymorphism of thymic stromal lymphopoietin in Korean patients with atopic dermatitis and allergic march. J Eur Acad Dermatol Venereol 2018;32:E468–E70. [DOI] [PubMed] [Google Scholar]
- 46.Fahey LM, Chandramouleeswaran PM, Guan SB, Benitez AJ, Furuta GT, Aceves SS et al. Food allergen triggers are increased in children with the TSLP risk allele and eosinophilic esophagitis. Clin Transl Gastroenterol 2018;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abuabara K, You Y, Margolis DJ, Hoffmann TJ, Risch N, Jorgenson E. Genetic ancestry does not explain increased atopic dermatitis susceptibility or worse disease control among African Americans in two large US cohorts. J Allergy Clin Immunol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margolis DJ, Apter AJ, Mitra N, Gupta J, Hoffstad O, Papadopoulos M et al. Reliability and validity of genotyping filaggrin null mutations. Journal of Dermatologic Science 2013;70:67–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margolis DJ, Mitra N, Wubbenhorst B, Nathanson KL. Filaggrin sequencing and bioinformatics tools. Arch Dermatol Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


