Abstract
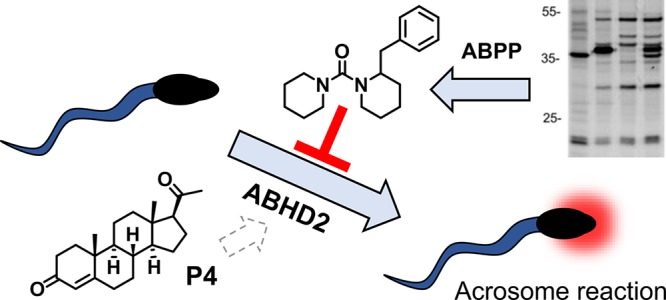
ABHD2 is a serine hydrolase that belongs to the subgroup of the α,β-hydrolase fold-containing proteins, which is involved in virus propagation, immune response, and fertilization. Chemical tools to selectively modulate the activity of ABHD2 in an acute setting are highly desired to investigate its biological role, but are currently lacking. Here, we report a library-versus-library screening using activity-based protein profiling (ABPP) to evaluate in parallel the selectivity and activity of a focused lipase inhibitor library against ABHD2 and a panel of closely related ABHD proteins. This screen resulted in the rapid identification of novel inhibitors for ABHD2. The selectivity of the inhibitor was further investigated in native mouse testis proteome by competitive ABPP, revealing a highly restricted off-target profile. The progesterone-induced acrosome reaction was reduced in a dose-dependent manner by the newly identified inhibitor, which provides further support for the key-role of ABHD2 in the P4-stimulated acrosome reaction. On this basis, the ABHD2 inhibitor is an excellent starting point for further optimization of ABHD2 inhibitors that can modulate sperm fertility and may lead to novel contraceptives.
The protein family containing the α,β-hydrolase fold is one of the largest and most diverse protein families, covering most, albeit not all, serine hydrolases. The α,β-hydrolase fold family includes lipases, esterases, epoxidases, peroxide hydrolases, and dehalogenases.1,2 A subgroup of the the α,β-hydrolase family consists of 22 members sharing the ABHD (α/β hydrolase fold domain) nomenclature. They generally have a GXSXG motif, and conserved structural features shared by this subfamily predict common roles in lipid biosynthesis and metabolism.3
Deregulated lipid metabolism is the underlying cause of many human disorders.4,5 In line with this, ABHD11 is deleted in the Williams-Beuren syndrome,6 and mutations in ABHD12 have been shown to be causative in polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract (PHARC).7 The α,β-hydrolase domain-containing protein 2 (ABHD2) is essential in hepatitis B virus propagation8 and is associated with chronic diseases that involve monocyte/macrophage recruitment, such as emphysema and atherosclerosis.9−11 ABHD2 has been identified as a progesterone (P4)-dependent 2-arachidonoylglycerol (2-AG) hydrolase.12 2-AG is an endogenous inhibitor of the CatSper channel in spermatozoa. Upon progesterone stimulation, ABHD2 hydrolyzes 2-AG, leading to a CatSper opening that triggers sperm hyperactivation and makes spermatozoa fertile. Despite the potentially important role of ABHD2 during various (patho)physiological processes, such as virus propagation, atherosclerosis, and fertilization, selective small molecule inhibitors are currently not available to study the biological role of this enzyme in an acute setting.
Activity-based protein profiling is a valuable technology to rapidly identify novel inhibitors. It makes use of activity-based probes (ABPs) that assess the functional state of enzymes in complex proteomes.13−15 We have previously shown that the β-lactone-based ABPs MB064 and MB108 target a broad range of serine hydrolases and can be used for inhibitor discovery and profiling.16−18 Here, we describe the target scope of MB064 and MB108 and map the activity of ABHD2 in eight different mouse tissues. Next, we tested 200+ lipase-directed inhibitors on ABHD2 and related ABHD-proteins in a library-versus-library screen. Such a screen facilitated the rapid identification of selective inhibitors for ABHD2, which reduced the acrosome reaction in mouse spermatozoa.
Results
Identification of α,β-Hydrolase Fold-Containing Proteins Targeted by MB064 and MB108
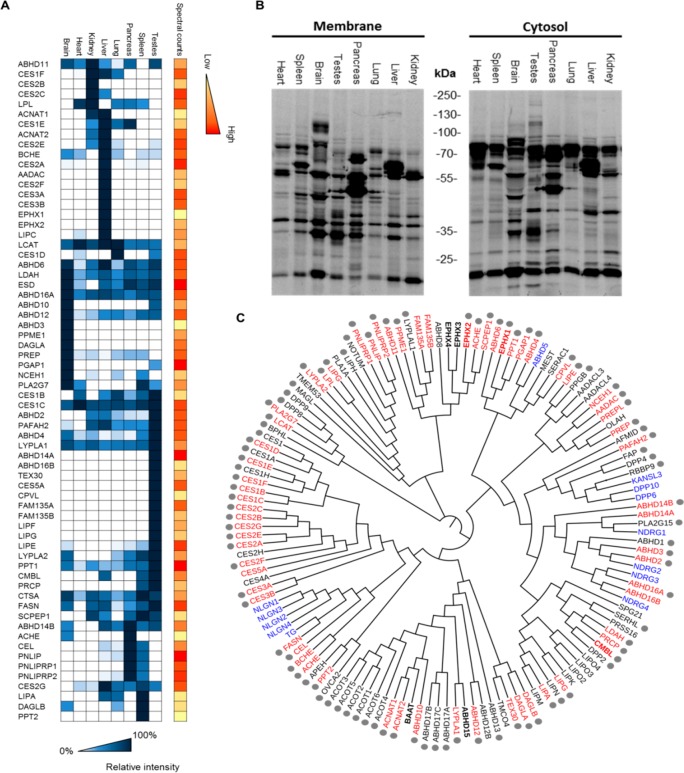
Previously, a large variety in expression for α,β-hydrolase fold family members across different tissue types has been observed by in situ hybridization and in global proteomics studies.3,13,19,20 Therefore, it was investigated whether this tissue-specific expression was mirrored by enzyme activity as measured by MB064. The labeling profile of MB064 in the mouse cytosol and membrane fractions of the brain, heart, kidney, liver, lung, pancreas, spleen, and testis revealed a highly diverse labeling pattern across different tissue types (Figure 1B). To determine the identity of the proteins, a tissue-wide chemoproteomic screen was performed by using MB108, a biotinylated analog of MB064 (Figure 1A).16 As a reference for probe targets, a list of α,β-hydrolase fold enzymes was retrieved from the ESTHER database (http://bioweb.ensam.inra.fr/esther).21 A phylogenetic tree of 136 α,β-hydrolase fold containing enzymes was generated using multiple sequence alignment (Muscle) and ClustalW2 omega phylogeny (Figure 1C).22 The α,β-hydrolase fold-containing proteins detected in this chemoproteomic screen are highlighted in red. The screen revealed that 66 out of the 136 known mouse α,β-hydrolase fold proteins, including ABHD2, were detected by MB108 in one or more tissues. In addition, a number of hydrolases and transferases that did not contain the α,β-hydrolase fold motif, but had a catalytic nucleophile that can attack the β-lactone, were also targeted (SI Figure 1). Interestingly, ABHD2 activity was most abundant in testis followed by kidney and liver.
Figure 1.
Detection of α,β-hydrolase fold enzyme activity in the mouse brain proteome, by using β-lactone-based ABPs MB064 and MB108. (A) Chemoproteomic screen of α,β-hydrolase fold enzyme activity in the mouse proteome, by using β-lactone-based activity-based probe MB108 (10 μM). Mean of the average spectral counts over three replicate experiments. Heatmap: proteins are ordered by hierarchical clustering. Intensities in blue represent the relative activity per protein in each tissue. Intensities in orange are the spectral counts in the tissue where the protein shows highest activity. (B) In-gel fluorescence readout of mouse tissues labeled by ABP MB064 (2 μM) in the membrane and cytosolic mouse proteomes. (C) Phylogenetic tree of α,β-hydrolase fold proteins generated by multiple sequence alignment (Muscle) and ClustalW2 omega phylogeny. Proteins highlighted in red are targeted by MB108. All proteins are serine hydrolases except those in bold or highlighted in blue. Blue proteins lack a catalytic triad. Bold proteins have a cysteine or asparagine as a catalytic residue. Gray dots indicate organophosphate-targeted proteins.
It is well-known that α,β-hydrolase fold proteins react well with organophosphates.23 Therefore, organophosphate-based ABPs are widely applied tools to study α,β-hydrolase fold protein enzymatic activity.24 For example, Bachovchin and colleagues have previously targeted >80% of mammalian metabolic serine hydrolases by using a single fluorophosphonate (FP)-based ABP.13 To study whether β-lactone-probe targets are complementary to proteins that are known to interact with organophospates, the FP-probe targets are marked with a gray dot in Figure 1C. A large overlap between the targets of organophosphates and MB108 was observed. Of note, MB108 targeted DAGLα, FAM135, FAM135B, CES5A, and ABHD14A, which were not detected in studies with organophosphate-based probes.13,23 In conclusion, the β-lactone-based ABPs, MB064, and MB108 are powerful broad-spectrum tools to study the ABHD2 activity and detect many members of the α,β-hydrolase fold family (∼50%), including several members that were not detected by organophosphate-based probes.13,23
Gel-Based Screen to Identify Novel Inhibitors for ABHD Family Members
To identify selective ABHD2 inhibitors, a competitive screen between ABHD proteins and a targeted lipase inhibitor library with 207 members (Supporting Table 2) was made. Chemoproteomic ABPP is highly sensitive and allows one to study native enzymes with relatively low abundance and activity in tissues or cells. However, chemoproteomics is expensive and time-consuming, and thus it is not suitable for high-throughput screening of inhibitor libraries. Therefore, a gel-based assay using MB064 was employed to identify novel inhibitors for ABHD2 and its family members. It is still sometimes challenging to correlate enzymes to specific fluorescent bands on SDS-PAGE in native proteomes, and native enzymes may show large variations in labeling intensities, due to unequal tissue distribution. Thus, to avoid these limitations we expressed recombinant proteins to generate protein libraries for inhibitor screening.
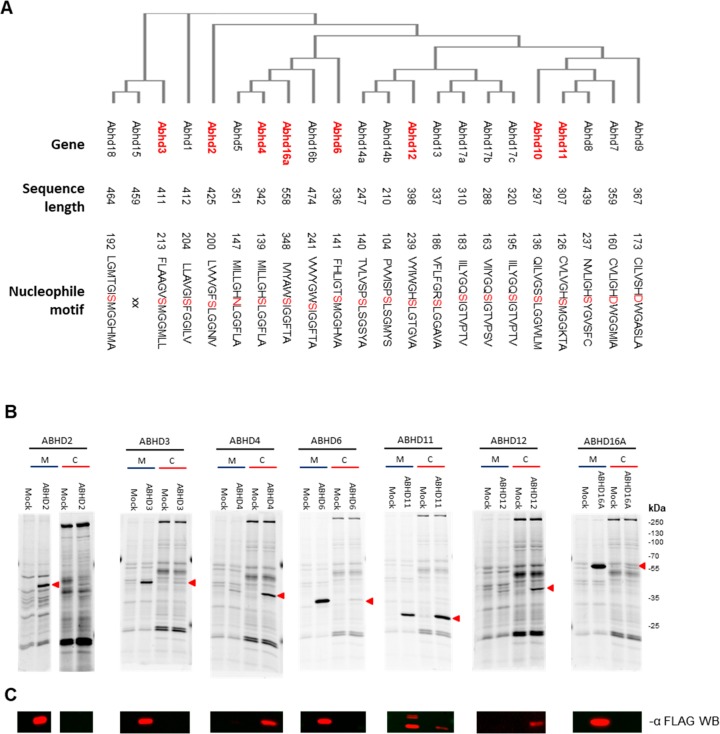
The human enzymes sharing the ABHD nomenclature corresponding to the enzymes that were detected in the chemoproteomic mouse tissue screen (ABHDs 3, 4, 6, 11, 12, and 16A) were recombinantly expressed in HEK293T (Figure 2A). The constructs were equipped with a FLAG-tag to monitor protein expression and gel migration. Instead, ABHD2 was expressed in U2OS cells because the efficiency of expression of its construct in HEK293T cells was too low. No expression or labeling of ABHD10, 14A, 14B, and 16B in HEK293T nor U2OS could be achieved for as yet unclear reasons (SI Figures 1, 2). Labeling with MB064 (2 μM) of lysates from ABHD transfected cells compared to mock transfected cells was used to monitor protein activity. To establish a multiple protein assay, recombinantly expressed proteins were pooled in protein libraries. ABHD3, 4, and 12 clustered around a molecular weight of ∼40 kDa. Therefore, two protein libraries were required to resolve all expressed ABHDs using 1D SDS-PAGE. Protein library 1 contained ABHD2, 3, and 12, and protein library 2 consisted of ABHD4, 6, 11, and 16A (Figure 3A,B).
Figure 2.
(A) Phylogenetic tree of the α,β-hydrolase fold family members sharing the ABHD nomenclature. The tree is generated by multiple sequence alignment (Muscle) and ClustalW2 omega phylogeny. Proteins used for the inhibitor screen are highlighted in red. (B) Labeling of ABHD proteins overexpressed in HEK293T or U2OS cells. Protein labeling of ABHD transfected cells was compared to mock transfected cells in the membrane (M) and cytosolic (C) fractions. Proteins (1 mg mL–1) were labeled by ABP MB064 (2 μM). Overexpressed proteins are marked by a red triangle. (C) Anti-FLAG Western blot for confirmation of protein expression and localization of the transfected ABHD. Cutout at the molecular weight indicated by the red triangle is represented. Full gels and blot are given in SI Figures 2 and 3.
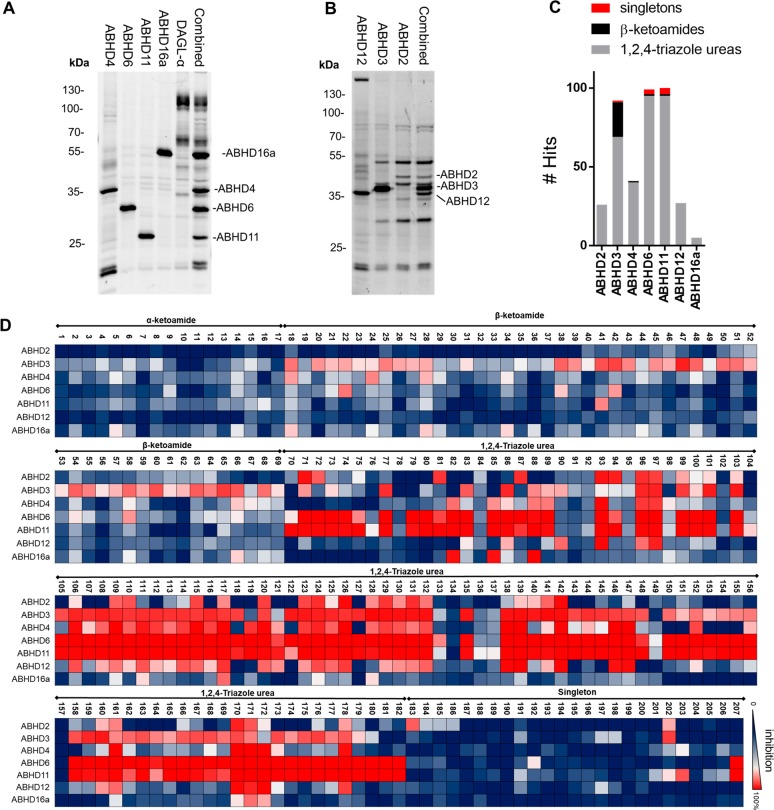
Figure 3.
(A) Fluorescent labeling (MB064, 2 μM) of HEK293T/and or U2OS lysates that overexpress ABHD4, 6, 11, 16A and DAGL-α, and a combination thereof to form protein library 1. (B) Fluorescent labeling (MB064, 2 μM) of HEK293T lysates that overexpress ABHD12, 3, 2 and a combination thereof to form protein library 2. (C) Graphical representation of the number of hits per chemotype per enzyme. (D) Heatmap overview of the inhibitor screen.
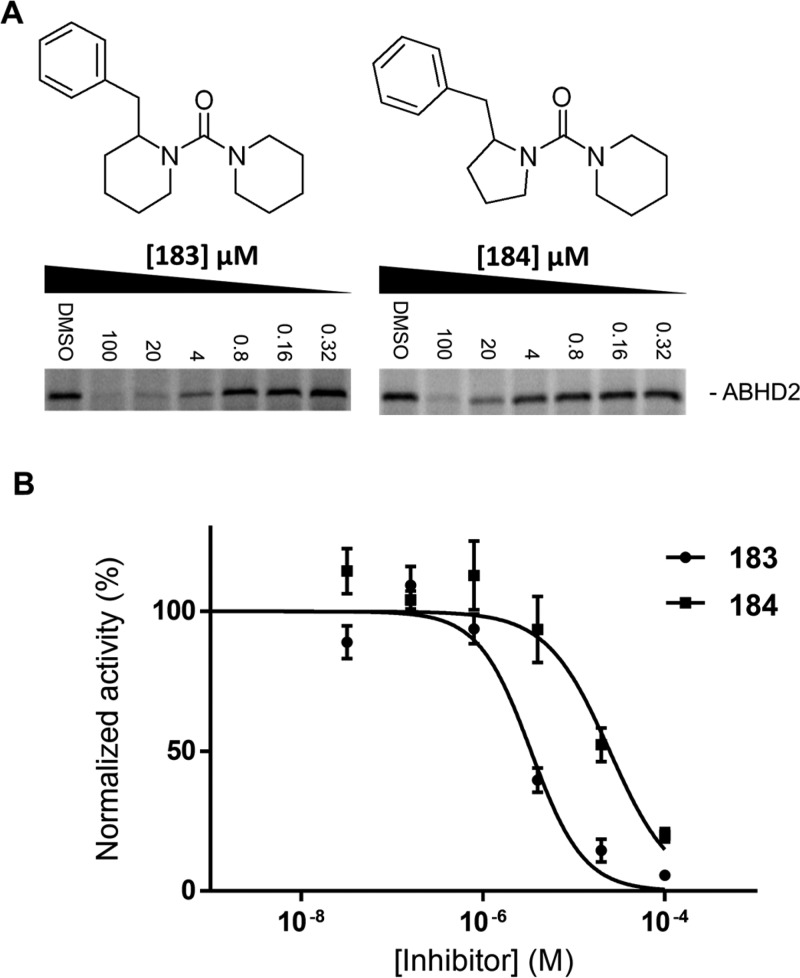
The lipase-focused inhibitor library contained 207 compounds that can be divided in 3 major classes (α-ketoamides, β-ketoamides, and 1,2,4-triazole ureas), and a set of unique chemotypes (singletons). Screening of the compound library at a concentration of 10 μM against the 2 protein libraries generated ∼1500 protein–inhibitor interaction data points and revealed that all proteins were hit by one or more compounds (Figure 3C,D). Inhibitors were qualified as a hit when protein labeling was reduced >70% compared to the vehicle-treated control. A clear distinction between target profiles of different inhibitor classes was detected (Figure 3C). The α-ketoamides were inactive against all tested ABHD family members. The β-ketoamides provided an excellent chemotype to inhibit ABHD3: 21 hits were identified, 13 of which were considered selective (<50% inhibition of the other ABHDs). Compounds 47 and 60 were the most potent ABHD3 inhibitors that remained selective over the other measured ABHDs. Inhibitors from the 1,2,4-triazole-urea class were more reactive. This class effectively targeted ABHD6 and ABHD11 (>70% of the compounds were classified as hits). No selective inhibitors were found for ABHD4, 6, 11, 12, or 16A, yet the nonselective compounds may still represent interesting starting points for hit optimization programs. Compound 183 and 184 from the singletons reduced the labeling of ABHD2 by >70% without reducing the labeling of other ABHD’s by >50%. In a concentration–response experiment, a modest pIC50 of 5.50 ± 0.06 was determined for 183 (Figure 4A,B). The structurally related compound 184, with a pyrrolidine moiety, had a 10-fold lower activity (pIC50 of 4.60 ± 0.10).
Figure 4.
Structure–activity relationships of lead inhibitors for ABHD. (A) Dose-dependent effect of 183 and 184 on ABHD2 labeling by MB064 (2 μM) on gel. (B) Dose–response curve of inhibitor 183 (pIC50 = 5.50 ± 0.06) and 184 (pIC50 = 4.60 ± 0.10).
Inhibitor Selectivity
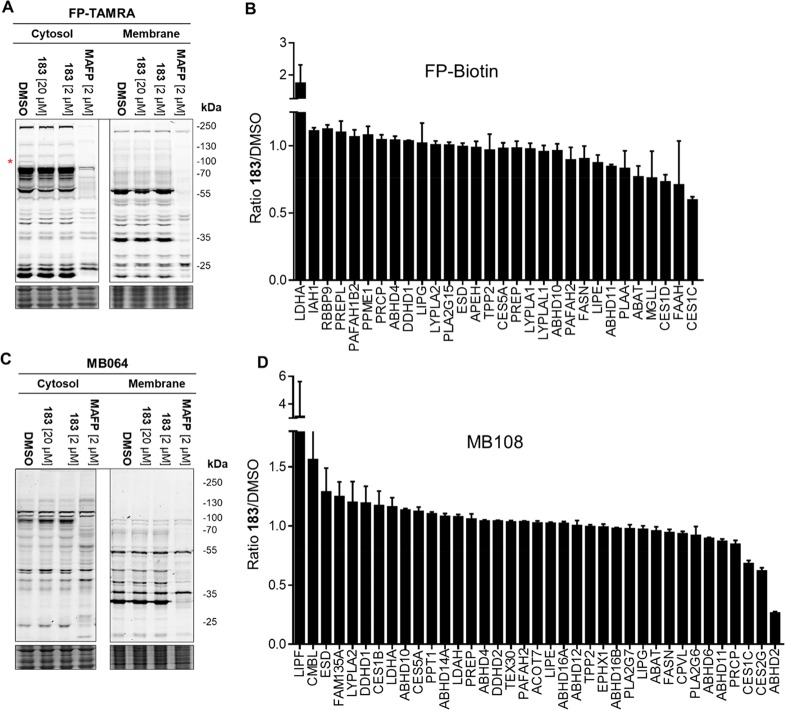
To investigate the selectivity of inhibitor 183 in more detail in a native mouse proteome, competitive ABPP with broad-spectrum FP-based serine hydrolase probes (FP-TAMRA and FP-biotin) and the ABPs MB064 and MB108 was employed.17 The fluorophosphonate was added to broaden the scope of the selectivity profile.13 Because of the important role in sperm fertility and the high expression of ABHD2 in the testis, the selectivity of ABHD2 inhibitor 183 (20 μM, 30 min) was investigated in the mouse testis proteome using MB064 and TAMRA-FP (Figure 5A,C). Inhibitor 183 reduced labeling of one fluorescent band in the cytosolic proteome, as determined by FP-TAMRA. The competitive chemoproteomic assay confirmed ABHD2 inhibition, with no other targets being detected (Figure 5B,D). Together, these studies indicate that inhibitor 183 shows a highly restrictive off-target profile and can be used to inhibit ABHD2 in the testis proteome in an acute fashion.
Figure 5.
Chemoproteomic assay for ABHD2 selectivity in mouse testis. (A) Gel-based competitive ABPP with inhibitor 183 (2 and 20 μM) and MAFP (2 μM) against TAMRA-FP (2 μM) in the mouse testis cytosolic and membrane fractions. (* indicates a band reduced in 183 treated vs control.) (B) Competitive chemoproteomic selectivity assay of inhibitor 183 (20 μM) in the testis proteome using ABP FP-biotin (10 μM). (C) Gel-based competitive ABPP with inhibitor 183 (2 and 20 μM) and MAFP (2 μM) against MB064 (2 μM) in the mouse testis cytosolic and membrane fractions. (D) Competitive chemoproteomic selectivity assay of inhibitor 183 (20 μM) in the mouse testis proteome using ABP MB108 (10 μM).
Compound 183 Reduces Acrosome Reaction and Inhibits ABHD2 Activity in Vitro
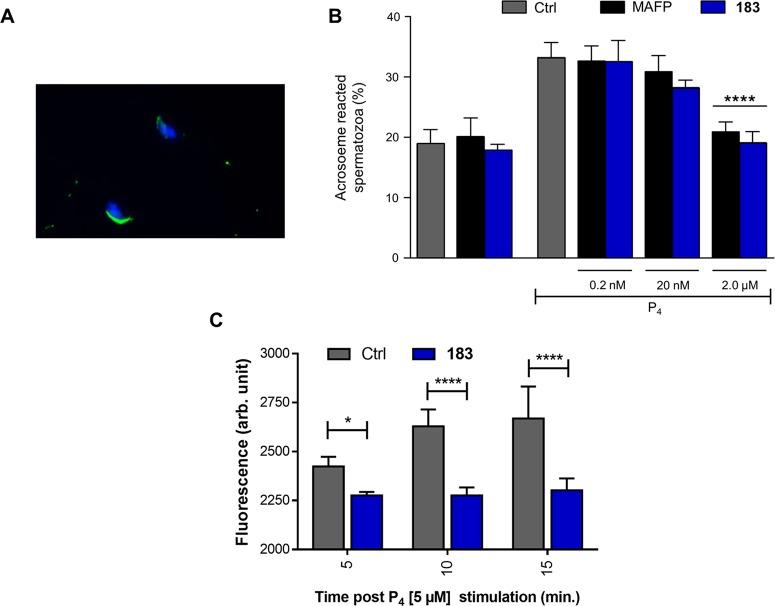
The acrosome reaction (AR) is a crucial process that enables capacitated spermatozoa to fertilize mammalian eggs.25,26 AR precedes sperm penetration through the zona pellucida and fusion with the oocyte membrane. This process is calcium-dependent, and it is induced in mammals in capacitated spermatozoa through the stimulation by several molecules, including the steroid hormone progesterone (P4).12,27 Intracellular calcium elevation triggers the fusion between the sperm plasma membrane and the outer acrosomal membrane. In order to monitor the dynamic of such a key event, Peanut Agglutinin-Fluorescein isothiocyanate (PNA-FITC) that selectively binds to the sugar residues present in the acrosome of spermatozoa has been used. Therefore, PNA-FITC staining allowed one to distinguish sperm with intact acrosome (Figure 6A) from AR ones.28
Figure 6.
(A) Representative image of one acrosome intact and one AR spermatozoa with the acrosomal enzymatic content and the nuclei stained with the PNA-FITC (green fluorescence) and DAPI (blue fluorescence), respectively. (B) Incidence of AR recorded on in vitro capacitated spermatozoa exposed to progesterone (P4:3 μM) in the absence (Ctrl) or presence of increasing concentrations of MAFP and 183 (from 0.2 nM to 2 μM). The induction of AR promoted by P4 was prevented in spermatozoa treated with 2 μM of MAFP or 183. In order to exclude any toxic effect, in vitro capacitated spermatozoa were also incubated with 2 μM of both the inhibitors (MAFP and 183 groups) without any treatment of P4. Data are expressed as mean with SD of n = 3 independent experiments. One-way ANOVA followed by post hoc Tukey’s multiple comparisons test. (C) Intracellular calcium analysis. Stimulation with P4 increases intracellular calcium levels in mouse spermatozoa as measured using Fluo-3 AM. 183 (2 μM) blocks the rise in intracellular calcium levels 5, 10, and 15 min after P4 stimulation. The data are expressed as florescence arbitrary units (Arb. Units) and are the mean ± standard deviation of three independent samples (full time course and positive control with ionomycin is given in SI Figure S9). *P < 0.05, **P < 0.01, ****P < 0.0001
In vitro capacitated spermatozoa exposed to P4 (3 μM) significantly increased the incidence of AR (from approximately 20% to 35%). ABHD2 inhibitor 183 reduced P4-induced AR in a concentration-dependent manner (Figure 6B). The broad spectrum serine hydrolase inhibitor MAFP that was used as a positive control exerted similar inhibitory action.12 Next, intracellular calcium measurements in mouse sperm were performed. Treatment with 183 (2 μM) blocks the calcium increase induced by P4, indicating a role of ABHD2 in the regulation of intracellular calcium levels in mouse sperm (Figure 6C). To further corroborate these functional data, ABHD2 was overexpressed in U2OS cells, and its activity was measured in vitro by a monoacylglycerol-lipase ABHD2 activity assay. Of note, ABHD2 activity was found to be enhanced by P4, and to be reduced by MAFP (used as a positive control)12 and dose-dependently by 183 (Figure 7).
Figure 7.
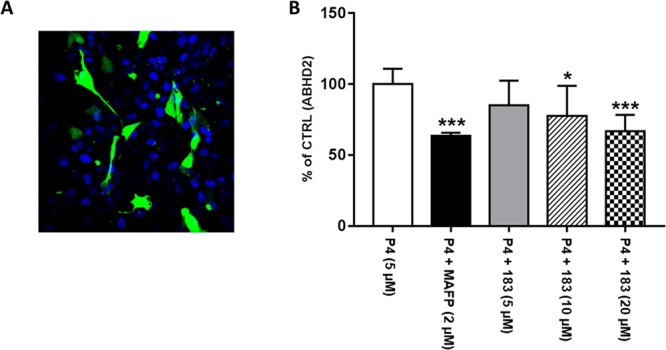
(A) Representative image of U2OS cells transfected with ABHD2-GFP (green) plasmid. (B) Effect of progesterone (P4) on ABHD2 in vitro activity, when added directly to the culture medium alone or in the presence of MAFP and 183. Data are expressed as mean with SD of n = 3 independent experiments. *p < 0.05 and ***p < 0.001.
Discussion
The target profile of the β-lactone containing ABPs (MB108 and MB064) across the mouse proteome (spleen, pancreas, liver, testis, brain, kidney, heart, and lung) was mapped. The ABPs interacted with ABHD2 and almost 50% of the α,β-hydrolase fold superfamily, including FAM135A, FAM135B, ABHD14B, and CES5A, which were not previously detected by organophosphates.13,23 The broad interaction landscape of the β-lactone-containing ABPs enabled the parallel screening of a targeted serine hydrolase inhibitor library for ABHD2 and multiple other ABHD enzymes. This led to the rapid identification of compound 183 as a novel inhibitor for ABHD2.
Recently, ABHD2 was shown to be highly expressed in spermatozoa, to bind P4, and to act as a P4-dependent 2-AG hydrolase.12 It finely tunes endogenous inhibition of the sperm calcium channel (Catsper) by 2-AG. Indeed, depletion of 2-AG by the hydrolytic activity of ABHD2 enables calcium influx via Catsper, leading to sperm activation in humans. A previous report indicated that murine Catsper is insensitive to P4, and may therefore lack regulation by ABHD2.29 Differential localization of ABHD2 in murine (restricted to the acrosomal region) compared to human (acrosomal region and flagellum) spermatozoa may explain the lack of murine Catsper regulation by P4, while still exhibiting a P4-induced acrosome reaction.12 The percentage of murine P4-stimulated acrosome reacted spermatozoa was reduced by the newly identified ABHD2 inhibitor 183. In addition, we have shown that the P4 induced calcium increase in mouse spermatozoa is blocked by the inhibition of ABHD2 using 183. A different calcium channel from Catsper could be involved in a ABHD2 regulated calcium flux in mouse sperm. Further research is required to identify the molecular components that constitute the pathway downstream of ABHD2 that regulates the acrosome reaction in mouse spermatozoa. Of note, the previously used inhibitor MAFP12 is known to target numerous endocannabinoid hydrolases, including fatty acid amide hydrolase (FAAH), monoacylglycerol lipase (MAGL), and ABHD6,30 whereas the ABHD2 inhibitor 183 displayed a highly restricted off-target profile in the mouse testis. Therefore, here we have provided further support to the key-role of ABHD2 in the P4-stimulated acrosome reaction. On this basis, 183 seems to represent an excellent starting point for further optimization of ABHD2 inhibitors that can modulate sperm fertility by preventing a sperm–egg recognition mechanism and may lead to a novel formulation of contraceptives. On a final note, it should be noted that the cross-talk between P4 and the ABHD2-dependent endocannabinoid metabolism appears to be of broad interest, and clearly extends previous data on the ability of P4 and strictly related steroid hormones to control other elements of endocannabinoid signaling, like the anandamide-hydrolase FAAH,31,32 and the endocannabinoid-binding CB1 receptors.33 Overall, this cross-talk brings about an unprecedented type of regulation, whereby 2-AG signaling is continuously “on” (to block Catsper), unless P4 terminates it by stimulating 2-AG hydrolysis by ABHD2.12 Therefore, the widely accepted dogma of the “on demand” synthesis of 2-AG, that suggests that this compound acts only upon stimulus-dependent release from phospholipid precursors, might not hold true in all tissues, extending previous observations on anandamide (AEA).34
Experimental Methods
Cell Culture
Cell culture was performed as described previously.17 In brief, HEK293T or U2OS cells were grown in DMEM with stable glutamine and phenol red (PAA), 10% newborn calf serum, penicillin, and streptomycin. Cell passage was performed every 2–3 days by resuspension in medium and seeding to appropriate confluence.
Transfection and Cell Lysis
24 h prior to transfection, 107 cells were seeded in a 15 cm Petri dish. A 3:1 mixture of polyethylenimine (60 μg) and plasmid DNA (20 μg) in 2 mL serum free medium was added. The medium was refreshed after 24 h, and after 72 h, HEK293T (48 h for U2OS) cells were harvested in 20 mL medium. Cells were isolated by centrifugation for 10 min at 1000 rpm and subsequent aspiration of the medium. The cell pellet was flash-frozen in liquid nitrogen and stored at −80 °C until use.
Cell pellets were slowly thawed on ice and suspended in lysis buffer (20 mM HEPES pH 7.2, 2 mM DTT, 0.25 M sucrose, 1 mM MgCl2, 25 U/mL Benzonase). Three pulses with a polytrone (3 × 7 s) were used to homogenize the suspension. After homogenization, the suspension was allowed to incubate for 10 min on ice. Ultracentrifugation (100,000 × g, 45 min, 4 °C, Beckman Coulter, Type Ti70 rotor) was used to separate the cytosolic and membrane fractions. The pellet (membrane fraction) was suspended in storage buffer (20 mM HEPES pH 7.2, 2 mM DTT). The total protein concentration was determined with Quick Start Bradford assay (Biorad) or Qubit protein assay (Invitrogen). The lysates were flash-frozen in liquid nitrogen and stored at −80 °C until use.
Preparation of Mouse Tissue Proteome
Mouse tissue was isolated according to guidelines approved by the ethical committee of Leiden University (DEC#13191). Mouse tissues were dounce homogenized in pH 7.2 lysis buffer A (20 mM HEPES pH 7.2, 2 mM DTT, 1 mM MgCl2, 25 U/mL Benzonase) and incubated for 5 min on ice. The suspension was centrifuged (2500 × g, 3 min, 4 °C) to remove debris. The supernatant was collected and subjected to ultracentrifugation (100,000 × g, 45 min, 4 °C, Beckman Coulter, Type Ti70 rotor). This yielded the membrane fraction as a pellet and the cytosolic fraction in the supernatant. The membrane fraction was suspended in a storage buffer (20 mM HEPES pH 7.2, 2 mM DTT). The total protein concentration was determined with a Quick Start Bradford assay (Biorad) or Qubit protein assay (Invitrogen). Membranes and supernatant were flash-frozen in liquid nitrogen and stored in aliquots at −80 °C until use.
Gel-Based Tissue Screen (MB064)
The membrane or lysate fraction of mouse brain, heart, kidney, liver, lung, pancreas, spleen, or testis was diluted to 1 mg mL–1. Proteome (1 mg mL–1, 20 μL) was incubated at 37 °C for 15 min with MB064 (1 μM, final concentration). After 15 min the reactions were quenched with 10 μL standard 3 × SDS-PAGE sample buffer. The samples were directly loaded and resolved on SDS PAGE gel (10% acrylamide). The gels were scanned using a ChemiDoc MP system (Cy3 settings, 605/50 filter) and analyzed using Image lab 4.1.
Chemoproteomic Tissue Screen
The membrane or lysate fraction of mouse brain, heart, kidney, liver, lung, pancreas, spleen, or testis was diluted to 2 mg mL–1. The proteomes (490 μL) were incubated with 10 μL of 500 μM ABP MB108 (final concentration 10 μM) for 1 h at rt. The labeling reaction was quenched, and excess probe was removed by chloroform methanol precipitation. Precipitated proteome was suspended in 500 μL 6 M urea/25 mM ammonium bicarbonate and allowed to incubate for 15 min. 5 μL of 1 M DTT was added, and the mixture was heated to 65 °C for 15 min. The sample was allowed to cool to rt before 40 μL (0.5 M) iodoacetamide was added, and the sample was alkylated for 30 min in the dark. 140 μL 10% (wt/vol) SDS was added and the proteome was heated for 5 min at 65 °C. The sample was diluted with 6 mL PBS. 100 μL of 50% slurry of Avidin–Agarose from egg white (Sigma-Aldrich) was washed with PBS and added to the proteome sample. The beads were incubated with the proteome for 3 h.
Beads were isolated by centrifugation and washed with 0.5% (wt/vol) SDS and PBS (3×). The proteins were digested overnight with sequencing grade trypsin (Promega) in 100 μL Pd buffer (100 mM Tris Ph 7.0, 100 mM NaCl, 1 mM CaCl2, 2% ACN, and 500 ng trypsin) at 37 °C with vigorous shaking. The pH was adjusted with formic acid to pH 3 and the beads were removed. The samples were further purified and measured as described previously.16
Enzyme Mixes for Inhibitor Screen
Protein library 1 is made in standard assay buffer (20 mM HEPES pH 7.2, 2 mM DTT) and contains the following transiently transfected proteins (concentrations are final concentrations): 4 mg mL–1 cytosolic fraction of ABHD12 transfected HEK293T cell lysate, 0.3 mg mL–1 membrane fraction ABHD3 transfected HEK293T cell lysate, and 1 mg mL–1 membrane fraction of ABHD2 transfected U2OS cell lysate.
Protein library 2 is made in standard assay buffer (20 mM HEPES pH 7.2, 2 mM DTT) and contains the following transiently transfected proteins (concentrations are final concentrations): 1 mg mL–1 cytosolic fraction of ABHD4 transfected HEK293T cell lysate, 0.25 mg mL–1 membrane fraction ABHD6 and 11 transfected HEK293T cell lysate, 0.2 mg mL–1 membrane fraction ABHD16A transfected HEK293T cell lysate, and finally 0.5 mg mL–1 membrane fraction DAGL-α transfected HEK293T cell lysate.
Inhibitor Screen
Enzyme mix 1 or 2 (19.5 μL) was incubated with DMSO (0.5 μL) or inhibitor (0.5 μL, 400 μM; 10 μM final concentration) for 30 min at 37 °C. Next, MB064 (0.5 μL, 80 μM; 2 μM final concentration) was incubated for 15 min at 37 °C. The reactions were quenched with 10 μL standard 3×SDS PAGE sample buffer. The samples were directly loaded and resolved on SDS PAGE gel (10% acrylamide). The gels were scanned using a ChemiDoc MP system (Cy3 settings, 605/50 filter), and the percentage inhibition was determined by comparison with DMSO (100%) using Image Lab 4.1. Control for protein loading was performed by Coomassie staining and adjustment for total lane intensity.
Gel-Based Dose–Response Analysis
Dose response analysis was performed as described previously.16 In brief, transfected proteome (19.5 μL) was incubated with DMSO (0.5 μL) or inhibitor (0.5 μL) at the indicated final concentrations for 30 min at 37 °C. Next, MB064 (0.5 μL, 80 μM; 2 μM final concentration) was incubated for 15 min at 37 °C. The reactions were quenched with 10 μL standard 3×SDS PAGE sample buffer. The samples were directly loaded and resolved on SDS PAGE gel (10% acrylamide). The gels were scanned using a ChemiDoc MP system (Cy3 settings, 605/50 filter); the percentage of activity remaining was determined by measuring the integrated optical intensity of the fluorescent protein bands using Image Lab 4.1. The relative intensity was compared to the vehicle treated proteins, which were set to 100%. Correction for protein loading was performed by Coomassie staining. IC50 values were determined by plotting log(inhibitor) vs normalized response (variable slope); dose–response curve was generated using Prism software (GraphPad).
Selectivity in Native Mouse Proteome
Mouse testis or brain cytosolic or membrane fractions were diluted to a total protein concentration of 2 mg mL–1. Proteome (19.5 μL) was incubated with DMSO (0.5 μL) or inhibitor (0.5 μL, 20 μM final concentration) for 30 min at 37 °C. Next, MB064 or TAMRA-FP (0.5 μL, 80 μM; 2 μM final concentration) was incubated for 15 min at 37 °C. The reactions were quenched with 10 μL standard 3×SDS PAGE sample buffer and analyzed as described above.
Chemoproteomic Selectivity Assay in Native Mouse Proteome
Mouse testis or brain cytosolic or membrane fraction was diluted to a total protein concentration of 2 mg mL–1. 5 μL of 1 mM inhibitor (20 μM final concentration) or DMSO was added to 240 μL proteome and allowed to incubate for 30 min at 37 °C. FP-Biotin or MB108 (5 μL, 500 μM) was added and incubated for 30 min at 37 °C. The samples were further processed and analyzed as described previously.16
Monoacylglycerol-Lipase ABHD2 Activity
Monoacylglycerol-lipase ABHD2 activity was assayed in transfected U2OS cell membrane (100,000g fraction; 100 μg for each point). Membranes were preincubated with serine protease/hydrolase inhibitor, MAFP (2.0 μM) (Cayman Chemicals, Ann Arbor, MI, U.S.A.) or 183 (5, 10, 20 μM), for 15 min at 37 °C. 2-Oleoyl-[3H]glycerol (10 μM, 10,000 cpm for each points (ARC, St. Louis, MO, U.S.A.)) was added and incubated for 30 min at 37 °C. The reaction was terminated by addition of a 2:1 (v/v) mixture of chloroform/methanol and centrifuged at 1600 rpm for 5 min, to induce phase separation. [3H]Glycerol release in the aqueous phase was measured by scintillation counting.
Acrosome Reaction
In each experimental group, two C57BL/6NBD male mice were used. Spermatozoa were retrieved from epididymis and vas deferens, and were incubated in TYH solution under the reported experimental conditions.35 Each assay was performed in triplicate.
Intracellular Calcium Levels
The spermatozoa were recovered from 6 mice epididymis and incubated at 37 °C, 5% CO2; stained with Fluo-3 AM; following a protocol by Bernabò et al.36 under control (Ctrl) conditions or in the presence of the inhibitor (2 μM). After 30 min, P4 was added at the final concentration of 5 μM. As a positive control, both samples were added to Ionomycin 10 μM. The fluorescence was assessed every 5 min with a fluorimeter (Fluoroskan 96 well Reader, PerkinElmer). The data are expressed as Florescence Arbitrary Units and are the mean ± standard deviation of three independent samples.
Acknowledgments
The authors express their gratitude to F. Fezza (Tor Vergata University of Rome, Italy) and X. Ruf for their kind support with the in vitro activity assays, and to the Italian Ministry of Education, University and Research (MIUR) for financial support under the competitive PRIN 2015 grant to M. Maccarrone.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00640.
Author Present Address
□ Department of Chemistry, Imperial College London, Molecular Sciences Research Hub, White City Campus, London W12 OBZ, UK
Author Contributions
¶ M.M. and M.v.d.S. are equally senior authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Nardini M.; Dijkstra B. W. (1999) Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9, 732–737. 10.1016/S0959-440X(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Ollis D. L.; Cheah E.; Cygler M.; Dijkstra B.; Frolow F.; Franken S. M.; Harel M.; Remington S. J.; Silman I.; Schrag J.; Sussman J. L.; Verschueren K. H. G.; Goldman A. (1992) The alpha/beta hydrolase fold. Protein Eng., Des. Sel. 5, 197–211. 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Lord C. C.; Thomas G.; Brown J. M. (2013) Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1831, 792–802. 10.1016/j.bbalip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G. S. (2006) Inflammation and metabolic disorders. Nature 444, 860–867. 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Nomura D. K.; Morrison B. E.; Blankman J. L.; Long J. Z.; Kinsey S. G.; Marcondes M. C.; Ward A. M.; Hahn Y. K.; Lichtman A. H.; Conti B.; Cravatt B. F. (2011) Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science 334, 809–813. 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G.; Ucla C.; Guipponi M.; Reymond A. (2002) Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum. Genet. 110, 429–438. 10.1007/s00439-002-0710-x. [DOI] [PubMed] [Google Scholar]
- Fiskerstrand T.; H’Mida-Ben Brahim D.; Johansson S.; M’Zahem A.; Haukanes B. I.; Drouot N.; Zimmermann J.; Cole A. J.; Vedeler C.; Bredrup C.; Assoum M.; Tazir M.; Klockgether T.; Hamri A.; Steen V. M.; Boman H.; Bindoff L. A.; Koenig M.; Knappskog P. M. (2010) Mutations in ABHD12 cause the neurodegenerative disease PHARC: An inborn error of endocannabinoid metabolism. Am. J. Hum. Genet. 87, 410–417. 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.; Yang J.; Wang S. (2011) Antisense oligonucleotides targeting abhydrolase domain containing 2 block human hepatitis B virus propagation. Oligonucleotides 21, 77–84. 10.1089/oli.2011.0280. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Oike Y.; Hoshii T.; Maekawa H.; Ogawa H.; Suda T.; Araki K.; Yamamura K. (2005) Increase of smooth muscle cell migration and of intimal hyperplasia in mice lacking the alpha/beta hydrolase domain containing 2 gene. Biochem. Biophys. Res. Commun. 329, 296–304. 10.1016/j.bbrc.2005.01.127. [DOI] [PubMed] [Google Scholar]
- Miyata K.; Nakayama M.; Mizuta S.; Hokimoto S.; Sugamura K.; Oshima S.; Oike Y.; Sugiyama S.; Ogawa H.; Yamamura K. (2008) Elevated mature macrophage expression of human ABHD2 gene in vulnerable plaque. Biochem. Biophys. Res. Commun. 365, 207–213. 10.1016/j.bbrc.2007.10.127. [DOI] [PubMed] [Google Scholar]
- Jin S.; Zhao G.; Li Z.; Nishimoto Y.; Isohama Y.; Shen J.; Ito T.; Takeya M.; Araki K.; He P.; Yamamura K. (2009) Age-related pulmonary emphysema in mice lacking alpha/beta hydrolase domain containing 2 gene. Biochem. Biophys. Res. Commun. 380, 419–424. 10.1016/j.bbrc.2009.01.098. [DOI] [PubMed] [Google Scholar]
- Miller M. R.; Mannowetz N.; Iavarone A. T.; Safavi R.; Gracheva E. O.; Smith J. F.; Hill R. Z.; Bautista D. M.; Kirichok Y.; Lishko P. V. (2016) Unconventional endocannabinoid signaling governs sperm activation via the sex hormone progesterone. Science 352, 555–559. 10.1126/science.aad6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin D. A.; Ji T.; Li W.; Simon G. M.; Blankman J. L.; Adibekian A.; Hoover H.; Niessen S.; Cravatt B. F. (2010) Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl. Acad. Sci. U. S. A. 107, 20941–20946. 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esbroeck A. C. M.; Janssen A. P. A.; Cognetta A. B. 3rd; Ogasawara D.; Shpak G.; van der Kroeg M.; Kantae V.; Baggelaar M. P.; de Vrij F. M. S.; Deng H.; Allara M.; Fezza F.; Lin Z.; van der Wel T.; Soethoudt M.; Mock E. D.; den Dulk H.; Baak I. L.; Florea B. I.; Hendriks G.; De Petrocellis L.; Overkleeft H. S.; Hankemeier T.; De Zeeuw C. I.; Di Marzo V.; Maccarrone M.; Cravatt B. F.; Kushner S. A.; van der Stelt M. (2017) Activity-based protein profiling reveals off-target proteins of the FAAH inhibitor BIA 10–2474. Science 356, 1084–1087. 10.1126/science.aaf7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K. L.; Tsuboi K.; Adibekian A.; Pugh H.; Masuda K.; Cravatt B. F. (2012) DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat. Chem. Biol. 8, 999–1007. 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; Chameau P. J.; Kantae V.; Hummel J.; Hsu K. L.; Janssen F.; van der Wel T.; Soethoudt M.; Deng H.; den Dulk H.; Allara M.; Florea B. I.; Di Marzo V.; Wadman W. J.; Kruse C. G.; Overkleeft H. S.; Hankemeier T.; Werkman T. R.; Cravatt B. F.; van der Stelt M. (2015) Highly Selective, Reversible Inhibitor Identified by Comparative Chemoproteomics Modulates Diacylglycerol Lipase Activity in Neurons. J. Am. Chem. Soc. 137, 8851–8857. 10.1021/jacs.5b04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; Janssen F. J.; van Esbroeck A. C.; den Dulk H.; Allara M.; Hoogendoorn S.; McGuire R.; Florea B. I.; Meeuwenoord N.; van den Elst H.; van der Marel G. A.; Brouwer J.; Di Marzo V.; Overkleeft H. S.; van der Stelt M. (2013) Development of an activity-based probe and in silico design reveal highly selective inhibitors for diacylglycerol lipase-alpha in brain. Angew. Chem., Int. Ed. 52, 12081–12085. 10.1002/anie.201306295. [DOI] [PubMed] [Google Scholar]
- van Rooden E. J.; Florea B. I.; Deng H.; Baggelaar M. P.; van Esbroeck A. C. M.; Zhou J.; Overkleeft H. S.; van der Stelt M. (2018) Mapping in vivo target interaction profiles of covalent inhibitors using chemical proteomics with label-free quantification. Nat. Protoc. 13, 752–767. 10.1038/nprot.2017.159. [DOI] [PubMed] [Google Scholar]
- Sharma K.; Schmitt S.; Bergner C. G.; Tyanova S.; Kannaiyan N.; Manrique-Hoyos N.; Kongi K.; Cantuti L.; Hanisch U. K.; Philips M. A.; Rossner M. J.; Mann M.; Simons M. (2015) Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831. 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M.; Fagerberg L.; Hallstrom B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson A.; Kampf C.; Sjostedt E.; Asplund A.; Olsson I.; Edlund K.; Lundberg E.; Navani S.; Szigyarto C. A.; Odeberg J.; Djureinovic D.; Takanen J. O.; Hober S.; Alm T.; Edqvist P. H.; Berling H.; Tegel H.; Mulder J.; Rockberg J.; Nilsson P.; Schwenk J. M.; Hamsten M.; von Feilitzen K.; Forsberg M.; Persson L.; Johansson F.; Zwahlen M.; von Heijne G.; Nielsen J.; Ponten F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Lenfant N.; Hotelier T.; Velluet E.; Bourne Y.; Marchot P.; Chatonnet A. (2012) ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. 41, D423–D429. 10.1093/nar/gks1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenfant N.; Bourne Y.; Marchot P.; Chatonnet A. (2016) Relationships of human alpha/beta hydrolase fold proteins and other organophosphate-interacting proteins. Chem.-Biol. Interact. 259, 343–351. 10.1016/j.cbi.2016.04.027. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Patricelli M. P.; Cravatt B. F. (1999) Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U. S. A. 96, 14694–14699. 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrat C.; Serres C.; Jouannet P. (2000) The acrosome reaction in human spermatozoa. Biol. Cell 92, 255–266. 10.1016/S0248-4900(00)01072-8. [DOI] [PubMed] [Google Scholar]
- Osman R. A.; Andria M. L.; Jones A. D.; Meizel S. (1989) Steroid induced exocytosis: the human sperm acrosome reaction. Biochem. Biophys. Res. Commun. 160, 828–833. 10.1016/0006-291X(89)92508-4. [DOI] [PubMed] [Google Scholar]
- Mannowetz N.; Miller M. R.; Lishko P. V. (2017) Regulation of the sperm calcium channel CatSper by endogenous steroids and plant triterpenoids. Proc. Natl. Acad. Sci. U. S. A. 114, 5743–5748. 10.1073/pnas.1700367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. P.; Fazeli A.; Voorhout W. F.; Marks A.; Bevers M. M.; Colenbrander B. (1996) Use of peanut agglutinin to assess the acrosomal status and the zona pellucida-induced acrosome reaction in stallion spermatozoa. J. Androl. 17, 674–682. [PubMed] [Google Scholar]
- Lishko P. V.; Botchkina I. L.; Kirichok Y. (2011) Progesterone activates the principal Ca2+ channel of human sperm. Nature 471, 387–391. 10.1038/nature09767. [DOI] [PubMed] [Google Scholar]
- Hoover H. S.; Blankman J. L.; Niessen S.; Cravatt B. F. (2008) Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg. Med. Chem. Lett. 18, 5838–5841. 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarrone M.; Bari M.; Di Rienzo M.; Finazzi-Agro A.; Rossi A. (2003) Progesterone activates fatty acid amide hydrolase (FAAH) promoter in human T lymphocytes through the transcription factor Ikaros. Evidence for a synergistic effect of leptin. J. Biol. Chem. 278, 32726–32732. 10.1074/jbc.M302123200. [DOI] [PubMed] [Google Scholar]
- Grimaldi P.; Pucci M.; Di Siena S.; Di Giacomo D.; Pirazzi V.; Geremia R.; Maccarrone M. (2012) The faah gene is the first direct target of estrogen in the testis: role of histone demethylase LSD1. Cell. Mol. Life Sci. 69, 4177–4190. 10.1007/s00018-012-1074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee M.; Vitiello S.; Bellocchio L.; Hebert-Chatelain E.; Monlezun S.; Martin-Garcia E.; Kasanetz F.; Baillie G. L.; Panin F.; Cathala A.; Roullot-Lacarriere V.; Fabre S.; Hurst D. P.; Lynch D. L.; Shore D. M.; Deroche-Gamonet V.; Spampinato U.; Revest J. M.; Maldonado R.; Reggio P. H.; Ross R. A.; Marsicano G.; Piazza P. V. (2014) Pregnenolone can protect the brain from cannabis intoxication. Science 343, 94–98. 10.1126/science.1243985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggelaar M. P.; Maccarrone M.; van der Stelt M. (2018) 2-Arachidonoylglycerol: A signaling lipid with manifold actions in the brain. Prog. Lipid Res. 71, 1–17. 10.1016/j.plipres.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Lybaert P.; Danguy A.; Leleux F.; Meuris S.; Lebrun P. (2009) Improved methodology for the detection and quantification of the acrosome reaction in mouse spermatozoa. Histol. Histopathol. 24, 999–1007. [DOI] [PubMed] [Google Scholar]
- Bernabò N.; Berardinelli P.; Mauro A.; Russo V.; Lucidi P.; Mattioli M.; Barboni B. (2011) The role of actin in capacitation-related signaling: an in silico and in vitro study. BMC Syst. Biol. 5, 47. 10.1186/1752-0509-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.