Abstract
Single nucleotide polymorphisms (SNPs) are the largest source of sequence variation in the human genome. However, their functional significance is not well understood. We show that SNPs in the Wilson disease gene, ATP7B, that produce amino-acid substitutions K832R and R952K, modulate ATP7B properties in vitro and influence serum copper (Cu) status in vivo. Presence of R832 is associated with a lower ATP7B abundance and a diminished trafficking in response to elevated Cu. The K832R substitution alters surface exposure of amino acid residues in the actuator domain and increases its conformational flexibility. All SNP-related ATP7B variants (R832/R952, R832/K952, K832/K952, and K832/R952) have Cu-transport activity. However, the activity of ATP7B-K832/K952 is lower compared to other variants. In humans, the presence of K952 is associated with a higher fraction of exchangeable Cu in a serum. Thus, SNPs may modulate properties of ATP7B and the organism Cu status.
Keywords: copper, ATP7B, small nucleotide polymorphisms (SNPs), MD simulations
INTRODUCTION
Copper (Cu) is an essential metal that is required in numerous cellular processes. Cu is a cofactor of enzymes involved in respiration, detoxification of oxygen radicals, iron transport, and neurotransmitter synthesis.1, 2 Both insufficient and excessive Cu are detrimental; therefore, Cu homeostasis is tightly regulated through a network of Cu-transporting and Cu-utilizing proteins. ATP7B is a mammalian Cu-transporting P1B-type ATPase that plays a major role in balancing Cu levels in tissues. ATP7B is most highly expressed in the liver, and is also found in the brain, intestine, heart, kidneys, lungs, mammary glands, and placenta.3 In hepatocytes, ATP7B is localized to the trans-Golgi network (TGN), where it facilitates the incorporation of Cu into the secreted ferroxidase ceruloplasmin (Cp), the major Cu-binding protein in the serum.4 When the intracellular Cu levels in hepatocytes exceed cellular needs, ATP7B traffics from the TGN to vesicles located in the vicinity of the canalicular membrane. There, ATP7B sequesters excess Cu into vesicle for subsequent export into the bile.5, 6
Inactivation of ATP7B causes Wilson disease (WD), a potentially fatal disorder of Cu misbalance.7 Loss of ATP7B activity is associated with intracellular Cu accumulation, impaired incorporation of Cu into Cp, and an increased proportion of labile (i.e. non-Cp bound) Cu in the serum, which serves as one of the clinical markers of WD.8 WD manifestations range from mild hepatic inflammation and tremor to cirrhosis, fulminant liver failure, depression, and psychotic episodes. The exact cause of this phenotypic variability remains poorly understood. Over 600 WD-causing mutations have been identified in ATP7B, and increasing number of those mutations has been characterized. These studies revealed a spectrum of biochemical and cellular effects, ranging from a complete loss of ATP7B expression and function to potentially milder effects on ATP7B trafficking or stability. However, strong correlations between mutations and associated phenotypes have not been observed, suggesting existence of additional, modifying factors. In addition, increasing evidence suggests that Cu homeostasis is altered in several other disorders (e.g. Alzheimer’s disease, Parkinson’s disease, and non-alcoholic fatty liver disease), and the cause of Cu mis-balance in these disorders remains unclear.
Single nucleotide polymorphisms (SNPs) represent a rich source of variation in the human genome. Approximately 800 SNPs have been identified in ATP7B, the vast majority of which have not been studied in detail and have been thought completely benign.9 However, two common SNP-related amino acid substitutions (SNP-RAAS) in ATP7B—R832 (c.2495 A>G, rs1061472) and K952 (c.2855 G>A, rs732774) appear to be enriched in patients in patients with Alzheimer’s disease (AD). While 31% of the healthy population is homozygous for R832 and 30% for K952, 43% of AD patients are homozygous for R832 and 45% for K952.10 The higher frequency of these variants could be significant, as the ATP7B-R832 variant has a decreased Cu transport activity in vitro11 and AD patients with these variants have a higher fraction of labile Cu in the serum compared to healthy controls.10, 12, 13 Furthermore, the presence of Arg in the Drosophila melanogaster Cu-ATPase at a position equivalent to that of 832 of human ATP7B results in the loss of Cu-ATPase function.14 Here, we tested how the SNP-RAAS R832 and K952 affect human ATP7B in cells and found that they influence the biochemical and cellular behavior of ATP7B and, therefore, may impact Cu homeostasis in cells and tissues.
MATERIALS AND METHODS
Human subjects and serum sample collection
All procedures involving human subjects were compliant with the ethical standards of the Committee on Human Experimentation of IRCCS Istituto Centro San Giovanni di Dio-Fatebenefratelli and the Helsinki Declaration of 1975. Blind procedures for data collection and analysis were applied. Eight-one elderly volunteers were recruited and screened for conditions known to affect copper metabolism and biological indicators of neurological, psychiatric, and cardio-cerebro-vascular diseases. A mini-mental state examination (MMSE) test was conducted.15 Blood was drawn in the morning after an overnight fast, collected in a test tube (Vacutainer® BD Thrombine) containing an activator of coagulation, quickly frozen, and stored at −80°C.
Genotyping and serum analysis
Genomic DNA was purified from peripheral blood using the conventional method for DNA isolation (QIAamp DNA Blood Midi kit). SNP IDs are ID_C_1919004_30 (rs1061472) and ID_C_938208_30 (rs732774). Genotyping was performed using the TaqMan allelic discrimination assay (Applied Biosystems, Inc) as previously described12. Direct DNA bidirectional sequencing was performed for 15% of the PCR products, which were randomly selected and analyzed to confirm the genotypes.
Total serum Cu was measured using an Analyst 600 Perkin Elmer atomic absorption spectrophotometer following previously described methods.16 Cp concentration was measured with an immunoturbidimetry assay (Horiba ABX, Montpellier, France) automated on an ABX Pentra 400 (Horiba ABX, Montpellier, France), performed in duplicate. For each serum Cu and Cp pair, the amount of Cu bound to Cp, the amount of non-Cp Cu, and the Cu:Cp ratio were calculated, as previously described.17, 18 Sample subjects were stratified based on whether or not they were carriers of at least one of the alleles of interest. First, the Cu values of carriers of at least one K952 allele were compared to those from non-carriers. Second, Cu values of carriers of at least one R832 allele were compared to those from non-carriers. Lastly, Cu values of carriers of at least one R832 and one K952 allele were compared to those from non-carriers. Student’s t-test was used to determine the significance of difference between the groups described above. * p<0.05, ** p<0.01.
Conservation analysis of the ATP7B SNPs
NCBI BLASTP was used to identify animal orthologs of human ATP7B (ID: XP_005266487.1). A library of 165 unique sequences from vertebrate species was assembled and 11-residue amino acid (AA) sequences centered on the SNP-RAAS of interest were extracted. These sequences for each SNP-RAAS were compiled into a WebLogo.19
Generation of GFP-tagged ATP7B variants
The plasmid pYG7, encoding ATP7B with an N-terminal GFP tag, was used as a template.20 The four ATP7B variants—R832/R952, R832/K952, K832/K952, and K832/R852—were generated using a QuikChange XL Site-directed Mutagenesis kit (Stratagene), following the manufacturer’s protocol. The primers used were as follows: K832R 5’-GGCGATATCGTCA-GGGTGGTCCC-3’ and 5’-CCCAGGGACCACC-CTGACGATAT-3’; R952K 5’-GGTGTTGT-TCAGAAATACTTTCC-3’ and 5’-GTTAGGAAAGTATTTCTGAACAA-3’. Presence of the correct subsitutions for each variant was confirmed by sequencing the entire ATPB cDNA region. The identical purity and quantity of the plasmids was verified prior to transfection into cells by examining the 210–300 nm spectra.
Cell culture and transfection
HEK293A and skin fibroblast (YST) cells were cultured in complete medium consisting of Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cell cultures were maintained at 37°C in a humidified chamber (5% CO2). Cells were transfected in 6-well plates for 20 h using Lipofectamine LTX with PLUS reagent (Invitrogen), with 1 or 2 μg plasmid DNA as indicated and 6 μL Lipofectamine per well, according to the manufacturer’s protocol. HEK293A cells were used in experiments protein characterizing ATP7B abundance, stability, and trafficking; YST cells were used for determining ATP7B Cu transport activity.
Protein abundance of ATP7B variants
Transfected HEK293 cells were lysed in RIPA buffer (0.05 M tris-HCl, pH 7.0; 0.15 M NaCl; 0.25 % deoxycholic acid; 1% NP-40; 1 mM EDTA; Millipore) for 30 min on ice. Centrifugation at 3,000 g for 15 min removed cell debris. The supernatant was collected and used for further studies. Protein concentration was determined using BCA assay (Pierce). Thirty μg of protein were separated on 10% Laemmli gels and then transferred to PVDF (Millipore) membranes using CAPS, pH 11 transfer buffer. Membranes were blocked for 1 h at room temperature (RT) in 5% milk in phosphate-buffered saline (PBS), then incubated for overnight (16 h) in either 1:6000 anti-ATP7B (Abcam, ab124973) or 1:2000 anti-β-actin (Abcam, ab6276) diluted in PBS containing 0.2% Tween-20 (PBST) with 0.05% sodium azide. Membranes were then incubated for 1 h at room temperature (RT) in either 1:10,000 anti-mouse or anti-rabbit secondary antibodies (Santa Cruz) before imaging on an Alpha-Imager (ProteinSimple). The fluorescent intensities of protein bands were determined with Image-J (NIH) and ATP7B levels were normalized to a β-actin loading control.
Analysis of ATP7B degradation
HEK293A cells transfected with each of the ATP7B variants and 16 h post-transfection protein synthesis was blocked by addition of 50 μM cycloheximide (CHX). Cells were then incubated in the presence of CHX for 0, 6, 12, or 24 h to allow for protein degradation. Cells were lysed using RIPA buffer, as described above, 5 μg of total cell lysate was separated on an SDS-PAGE gel and ATP7B abundance was analyzed by Western blot. The blots were imaged on an AlphaImager and the fluorescent intensities of the protein bands were determined using Image-J. The 6, 12, and 24 hour intensities for each variant were normalized to the corresponding intensity at 0 h.
Modeling of the ATP7B SNP-RAAS and adaptive Poisson-Boltzmann surface calculations
The homology model of ATP7B21 was employed to model R832 and K952 variants using the PyMOL (Schrödinger, LLC) mutagenesis feature. Structural changes were visualized by using a space-filling model. The PDB coordinates for the solution NMR structure of the ATP7B A-domain were kindly provided by Dr. Banci (University of Florence). SWISS-MODEL22 was used to generate structural model for the R832 A-domain variant. The structures were used for Adaptive Poisson-Boltzmann Surface (APBS) calculations.23 The PDB2PQR24–26 web server was used to prepare PQR files using the AMBER-9927 and PARSE28, 29 force-fields. APBS calculations were done using the APBS Tools2 plug-in30 within PyMOL. The dielectric constants for the APBS calculations were set to 2.0 (protein) and 78.0 (solvent) with ion concentrations of 150 mM (radius of +1 = 2.0 and −1 = 1.8). The probe radius for surface tracing was 1.4 Å with a system temperature of 310 K.
All-Atom molecular dynamics simulations setup
The A-domain structural models (generated as detailed above) were submitted to CHARMM-GUI web-based server to generate the respective NAMD 2.9-based molecular dynamics system and input files.31–33 A rectangular water box was constructed around each A-domain with an edge distance of 10 Å. The system was supplemented with 150 mM NaCl (48 sodium and 42 chloride ions placed in each box using the Monte Carlo method).31 The initial equilibration was performed for 35 frames using the NVT (constant volume, constant temperature) ensemble followed by production runs for up to 200 ns (10,000 frames) using the NPT (constant pressure, constant temperature) ensemble at a constant temperature of 303.15 K. All simulations were performed on the Maryland Advanced Research Computing Core (MARCC) facility high performance blue crab cluster (http://www.marcc.jhu.edu). The resulting individual trajectory was further extracted into 200 frames using the catdcd feature (stride 500 frames) within the Visual Molecular Dynamics (VMD) program34 for further processing using a local Linux machine.
MD simulation trajectory analysis
The root-mean-square deviation (RMSD) calculations for the individual trajectories were calculated using the VMD program.34 The solvent accessible surface area (SASA) measurements were performed using the Correl function and the SURF probe feature (RPROBE radius 1.4 Å) within the CHARMM program version 42b. The Gaussian network model (GNM) frequencies were generated using the GNM feature within the molecular dynamics analysis suite MDAnalysis.35 The secondary structure calculation was performed by analyzing secondary structure elements in the definition of secondary structure of proteins (DSSP) program using the GROMACs package.36–38 Cross-correlation of motion heat maps were also calculated with MDAnalysis using scripts kindly provided by Anu Nagarajan (NINDS, NIH). The GNM frequencies were plotted using XMGRACE (http://plasma-gate.weizmann.ac.il/Grace/). The ribbon diagrams of the A-domain were displayed using UCSF chimera.39
Tyrosinase assay for determining the Cu transport activity of ATP7B variants
YST cells were seeded on glass cover slips at a density of 1.5 × 106 cells per well. Cells were co-transfected with the pTyr plasmid and with one of the ATP7B-SNP-RAAS plasmid, as described above. For activity measurements, cells were washed in PBS and then fixed for 30 seconds in an acetone:methanol mixture (1:1 v/v) that was pre-chilled at −20°C. This was followed by incubation in 0.1% sodium phosphate, pH 6.8 containing 0.15% (wt/vol) levo-3,4-dihydroxyl-L-phenylalanine (L-DOPA) for 4 h at RT. Coverslips were then mounted onto slides using Fluoromount-G (Electron Microscopy Science). The formation of dark colored DOPA-chrome was examined using a phase contrast microscopy. Forty pigment-containing areas per replicate were manually selected using the tracing tool within Image-J. The areas and intensities of pigmentation were quantified, and the intensities were normalized to the area. Relative ATP7B abundance was estimated by confocal microscopy using GFP-fluorescence as a proxy of ATP7B-GFP protein levels. Fluorescent intensity was quantified using Image-J and normalized to the area. Fluorescent intensity of control cells expressing the pTyr plasmid was used as a background control. Fluorescent intensity of the previously characterized TST-ATP7B was taken as 100%.40
Trafficking studies
Transfected HEK293 cells grown on glass coverslips were treated with either 25 μM tetra-thiomolybdate (TTM) or 100 μM CuSO4 for 4 h at 37°C. The cells were then fixed in 3% paraformaldehyde (PFA) in PBS for 12 min and permeabilized and blocked in 0.5% Triton X-100 (Sigma) and 0.5% BSA in PBS for 15 min. Cells were incubated with anti-TGN46 primary antibody (1:500 dilution, GeneTex) for 1 h at RT, and then incubated with donkey anti-sheep secondary antibody conjugated with AlexaFluor-555 (Invitrogen) for 1 h at RT. Coverslips were mounted using 3:1 Fluoromount: DAPI (Electron Microscopy Science). Stained cells were visualized using confocal microscopy (Zeiss) and images were processed using Image-J. For quantization of protein co-localization, at least 20 cells were used per each condition, and the loss co-localization between ATP7B-GFP and TGN46 was used as a measure of ATP7B trafficking to vesicles.
Statistical analyses
All values are reported as means ± standard error of the mean (SEM) and plotted in GraphPad Prism (GraphPad Software, Inc). Statistical analyses were performed in GraphPad Prism using either unpaired Student’s t-test or one-way ANOVA with Fisher’s least significant differences (LDS) test, as indicated. * p<0.05, ** p<0.001, and **** p<0.0001.
RESULTS
The presence of R832 and K952 correlates with changes in serum Cu indicators
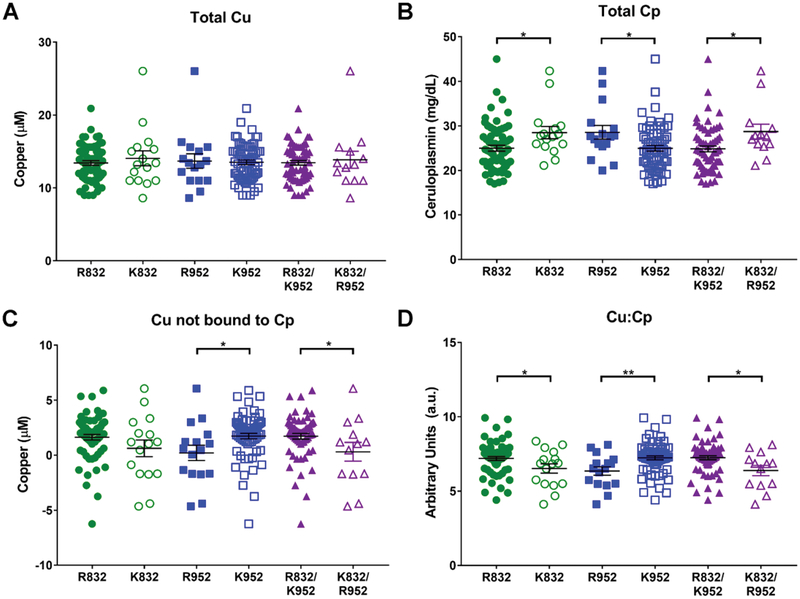
The A>G SNP at c.2495 in ATP7B results in an Arg at position 832 (R832), and the G>A SNP at c.2855 results in a Lys at position 952 (K952). To determine whether these substitutions in ATP7B impact Cu status in vivo, we characterized blood serum samples from individuals with different SNP combinations. Cp is an abundant serum protein containing a tightly bound, non-exchangeable Cu. Cp is secreted predominantly by the liver. Decreases in ATP7B activity lower Cu incorporation into CP and, subsequently, increase the fraction of non-Cp bound Cu (i.e. exchangeable Cu) in the serum.8 Consequently, to define the impact of ATP7b SNPs on Cu status, we measured four parameters: total serum Cu, total serum Cp, non-Cp Cu, and the Cu:Cp ratio. In this analysis, 81 healthy, elderly patients were recruited, and their demographic and biological variables pertaining to these subjects are reported in Table 1.
The indicators of Cu status were first compared for the carriers of the R832 allele and non-carriers (Fig. 1). The levels of total serum Cu (13.43±0.31 μM vs. 14.06±1.00 μM) and non-Cp Cu (1.63±0.25 μM vs. 0.62±0.73 μM) did not significantly differ between the carriers and non-carriers, respectively. However, total Cp levels (24.98±0.65 mg/dL vs. 28.48±1.34 mg/dL, p=0.022) were significantly lower in carriers of the R832 allele. Consequently, the Cu:Cp ratio (7.20±0.13 vs. 6.52±0.30, p=0.029) was increased in these individuals. Carriers of the K952 allele had Cu levels comparable to those of the non-carriers (13.52±0.32 μM vs. 13.69±0.96 μM). However, K952 carriers had significantly lower Cp levels (24.97±0.63 mg/dL vs. 28.55±1.49 mg/dL, p=0.019), significantly higher non-Cp Cu levels (1.74±0.25 μM vs. 0.21±0.67 μM, p=0.015), and a higher Cu:Cp ratio in the serum (7.24±0.13 vs. 6.35±0.27, p=0.003) than did the non-carriers.
Fig. 1.
Presence of R832 and K952 correlates with changes in serum Cu markers. (A) Total serum Cu concentration (mM) in healthy individuals with indicated genotypes. (B) Serum Cp (mg dL1) from healthy individuals with indicated genotypes; (C) concentration of non-Cp bound Cu in the serum of healthy individuals with indicated genotypes. (D) Ratio of total Cu to total Cp in the serum of healthy individuals with indicated genotypes. Normal, healthy levels range from 16–24 mM for Cu [3], 20–40 mg dL1 for Cp [3], 0.07–2.19 mM for non-Cp Cu [4], and 6.8 for Cu : Cp [4]. n = 16 for R832, n = 65 for K832, n = 16 for R952, n = 65 for K952, n = 13 for K832/R952, and n = 62 for R832/K952. Values are reported as means SEM. Significance was determined by Student’s t-test; *p o 0.05 and **p o 0.01.
To determine whether the presence of R832 and K952 had a combined effect, the Cu status was compared for R832/K952 carriers and non-carriers (K832/R952). Carriers of R832/K952 had serum readouts similar to those of K952 carriers. Although their total Cu levels were similar to non-carriers (13.45±0.32 μM vs. 13.86±1.13 μM), the R832/K952 carriers had significantly lower Cp levels (24.84±0.66 mg/dL vs. 28.70±1.61 mg/dL, p=0.021), higher non-Cp Cu levels (1.72±0.25 μM vs. 0.31±0.81 μM, p=0.041), and a higher Cu:Cp ratio (7.25±0.13 vs. 6.39±0.33, p=0.012). Thus, ATP7B SNPs appear to have functional consequences, as measured by Cu status in the serum. To directly test this prediction, we examined the effects of R832 and K952 on various properties of ATP7B protein In vitro.
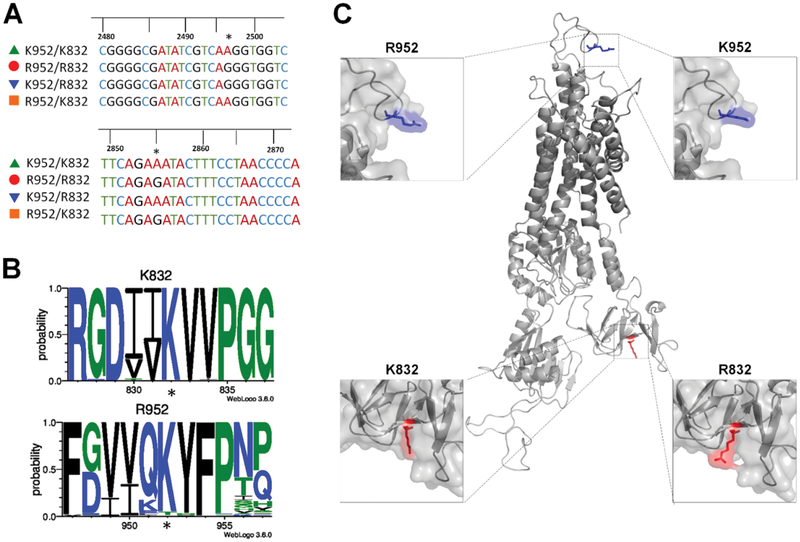
Substitution of a highly conserved K832 with R832 may impact ATP7B structure
Two SNPs produce four possible amino-acid (AA) combinations (Fig. 2A). To better understand their functional significance, we first examined conservation of the respective AA residues among ATP7B orthologs. Protein sequences from 165 vertebrate species were aligned with human ATP7B, and the results were compiled into an 11-AA long WebLogo (Fig. 2B). In a WebLogo, the size of the residue is indicative of the percentage of sequences in which that residue occurs at each given position.19 Lys at position 832 (K832) was present in all 165 sequences, suggesting that Lys in this position is a strongly preferred residue and substitutions of this residue may have structural and/or functional consequences. In contrast, Arg at position 952 (R952) was only observed in human ATP7B. The other ATP7B orthologs have mostly Lys at this position (K952).
Fig. 2.
Location, conservation, and potential impact of SNP-related substitutions on ATP7B structure (A) sequence of four generated ATP7B plasmids shows either an adenine (encoding Lys) or a guanine (encoding Arg) at positions 2495 and 2855, marked by asterisks. (B) Conservation logos of 11-AA sequences centered on K832 or R952 were generated using 165 animal ATP7B orthologues. The SNP related AA are marked with asterisks. K832 is well-conserved among ATP7B homologs, whereas R952 is specific for human ATP7B. (C) Homology model of ATP7B based on the crystal structure of L. pneumophila CopA [1]. Top panels illustrate location of the R/K variants at position 952. Bottompanels show location of the R/K variants at position 832 in the A-domain of ATP7B.
To predict the potential effects of R832 and K952 on ATP7B structure, we used an existing homology model of ATP7B23 (Fig. 2C). ATP7B is a large membrane protein with multiple domains that have distinct functions. The AA position 952 is located in the loop connecting transmembrane (TM) segments TM3 and TM4. This region faces the lumen of the TGN, and its specific function is unknown.41, 42 In the model, this region is predicted to be unstructured, and therefore the impact of R952K substitution on ATP7B structure is difficult to evaluate (Fig. 2C, top panels). In contrast, the AA residue 832 is located in the actuator (A)-domain, a domain that is critically involved in conformational transitions of the protein.23 The side-chain of Arg at the position 832 occludes a small “pocket” that is present in the A-domain containing K832, and this structural change could be consequential (Fig. 2C, bottom panels).
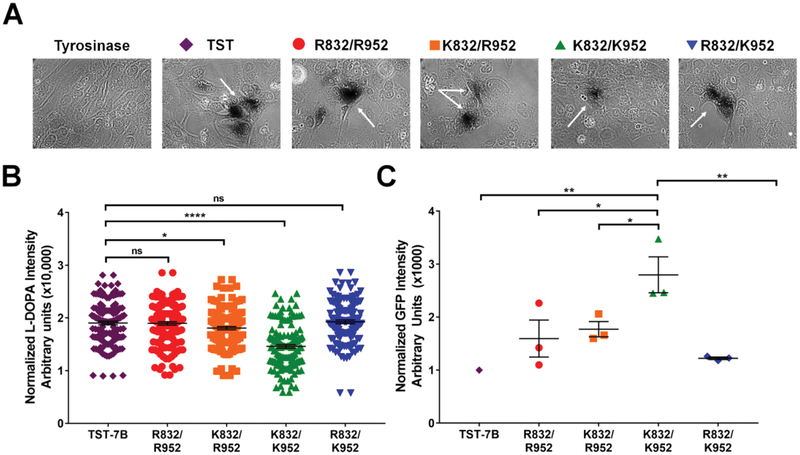
ATP7B-K952/K832 has a lower Cu-transport activity compared to other SNP-related variants
To evaluate functional significance of the SNP-related amino acid substitutions we compared Cu-transport activity of all four variants by measuring ATP7B-dependent activation of tyrosinase. Tyrosinase activity requires presence of copper cofactor, and cells expressing functional tyrosinase produce a dark brown pigment.40 YST cells, a fibroblast cell line lacking endogenous Cu-transporting ATPases, were co-transfected with the ATP7B-GFP variants and pTyr, a plasmid expressing apo-tyrosinase. The twin-strep tag (TST)-ATP7B construct, which contains R832/R952 and has been previously used in tyrosinase assays40, served as a positive control.
All four variants showed Cu-transport activity, as evidenced by the presence of the pigment in cells (Fig. 3A). The pigment intensity and pigment area were measured and normalized to the area to compare Cu-transport activity between variants quantitatively (Fig. 3B). The sequences of the ATP7B region for TST-ATP7B and the R832/R952 GFP-ATP7B are identical, and very similar pigment intensities (1.90±0.03 vs 1.89±0.03, p=0.911) confirmed their similar activity as well as the accuracy of the assay. The normalized pigment intensity of R832/K952 is also comparable to the TST-ATP7B control (1.93 ± 0.034 v 1.90 ± 0.03, p=0.649), while the pigment intensity of K832/R952 showed a small, but significant decrease compared to the control (1.81 ± 0.03 v 1.90 ± 0.03, p=0.042). The K832/K952 variant showed the most significant change—an approximately 20% decrease in normalized pigment intensity compared to TST-ATP7B (1.459±0.0347, p<0.0001).
Fig. 3.
ATP7B-K952/K832 has a lower Cu-transport activity compared to other variants. (A) YST cells were transfected with a plasmid expressing pTyrosinase (pTyr) alone (Tyrosinase) or co-transfected with pTyr and twin strep-tagged ATP7B (TST-ATP7B) or pTyr and one of the GFP-tagged ATP7B variants. The Cu transport activity of all ATP7B variants was evaluated by activation of tyrosinase (dark brown reaction product, marked by arrows, indicates tyrosinase activity). (B) Quantitation of tyrosinase activity. Pigment intensity is normalized to pigment area. (~) TST-ATP7B in purple, (K) R832/ R952 in red, (‘) K832/R952 in orange, (m) K832/K952 in green, and (.) R832/K952 in blue. n = 4, over 40 areas counted per n. (C) Quantitation of ATP7BGFP levels in cells used in the Cu-transport assay. GFP fluorescence intensity was normalized to the area, the fluorescence intensity of control cells expressing pTyr only, and finally to the fluorescent intensity of cells expressing TST-ATP7B, which was taken as 1. n = 3. All values are reported as means SEM. Significance was determined by one-way ANOVA with Fisher’s LSD test; *p o 0.05, **p o 0.01, and ****p o 0.0001.
To ensure that this decrease in Cu-transport activity was not due to low protein expression levels of the K832/K952 variant, we measured the GFP-ATP7B fluorescence in transfected cells. The K832/K952 and K832/R952 variants were more abundant (2.797±0.339 and 1.771±0.145, respectively) than the R832/R952 and R832/K952 variants (1.595±0.347 and 1.223±0.024, respectively; Fig. 3C). The lower transport activity of the K832/K952 variant compared to the other variants is therefore not due to lower levels of expression.
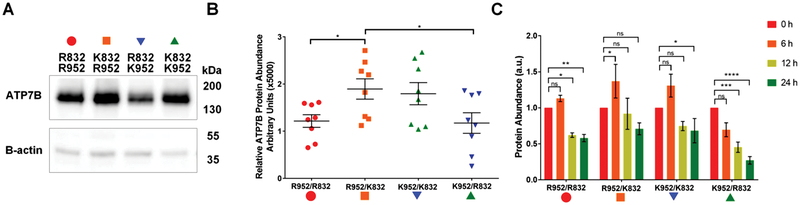
Presence of R832 enhances ATP7B degradation and decreases protein abundance
To further explore whether protein abundance may be influenced by SNP-related substitutions, we expressed in HEK293A cells the GFP-tagged ATP7B with each of possible AA combinations: R832/R952, R832/K952, K832/K952, and K832/R952. Quantitative real-time PCR (qRT-PCR) confirmed that the mRNA levels were comparable among cells expressing the different ATP7B variants (data not shown). The protein abundance of the ATP7B variants was evaluated by Western blotting. Analysis of bands intensity demonstrated that (similarly to findings in YST cells) presence of R832 was associated with lower ATP7B abundance independently of the residue present at position 952 (Fig. 4A, B). The R832/R952 variant was less abundant than the K832/R952 variant (1.22±0.13 and 1.9±0.21, respectively; p=0.0258), and the R832/K952 variant was less abundant than K832/K952 variant (1.172±0.218 and 1.795±0.236; p=0.0397). The ATP7B variant with both of the AD-associated SNP-RAAS, R832/K952, was the least abundant (1.172±0.218).
Fig. 4.
Presence of R832 is associated with lower ATP7B abundance. (A) HEK293A cells were transfected with ATP7B-GFP variants for 20 h, whole cell lysates were prepared and run on SDS-PAGE (20 mg of protein per lane).Western blot shows the relative abundance of each ATP7B variant. b-Actin is used as a loading control. (B) Densitometry of Western blots, n = 8. (K) R832/R952 in red, (‘) K832/R952 in orange, (.) R832/K952 in blue, and (m) K832/K952 in green. Intensities of ATP7B bands were normalized to b-actin and plotted. Means SEM are indicated by horizontal lines. Significance was determined by one-way ANOVA with Fisher’s LSD test; *po0.05. (C) The R832-containing ATP7B variants aremore susceptible to degradation than K832 variants. HEK293A cells were transfected, as described above, and then treated with 50 mM cycloheximide (CHX) for 0, 6, 12, or 24 h. For each time point,whole cell lysateswere prepared and equal amounts of total protein (5 mg per lane) were run on SDS-PAGE; ATP7B was visualized by Western blotting. The bars indicate the intensity of ATP7B bands at 6 h (orange), 12 h (yellow), and 24 h (green) relative to intensity at 0 time point (red) which is taken as 1. n = 4. Values are reported as means SEM. Significance was determined by one-way ANOVA with Fisher’s LSD test; *p o 0.05, **p o 0.01, ***p o 0.001, and ****p o 0.0001.
To determine whether difference in protein abundance was due to differences in ATP7B degradation, HEK293A cells expressing the ATP7B variants were treated with cycloheximide to block protein synthesis and then chased for 6, 12, or 24 h. Western blot analysis of cell lysates showed no significant changes in protein abundance after 6 h – for all variants (Fig. 4C). However, after 12 and 24 h the variants containing R832 showed lower protein abundances compared to the K832-containing variants (Fig. 4 C). The R832/R952 variant had significant decreases in abundance after 12 and 24 h (0.621±0.032, p=0.017 and 0.580±0.051, p=0.0088, respectively), whereas the K832/R952 variant showed significant changes only at 24 h (0.917±0.216, p=0.5923 and 0.708±0.083, p=0.0636, respectively). Similarly, the abundance of R832/K952 variant was decreased after 12 and 24 h of treatment (0.453±0.072, p=0.0008 and 0.271±0.052, p<0.0001), whereas K832/K952 showed a significant decrease only after 24 h (0.683±0.169, p=0.445). These results indicate that the ATP7B variants containing R832 are more susceptible to protein degradation than the variants containing K832, which may explain the observed differences in their abundance.
Residues at position 832 modulate ATP7B trafficking response to high Cu
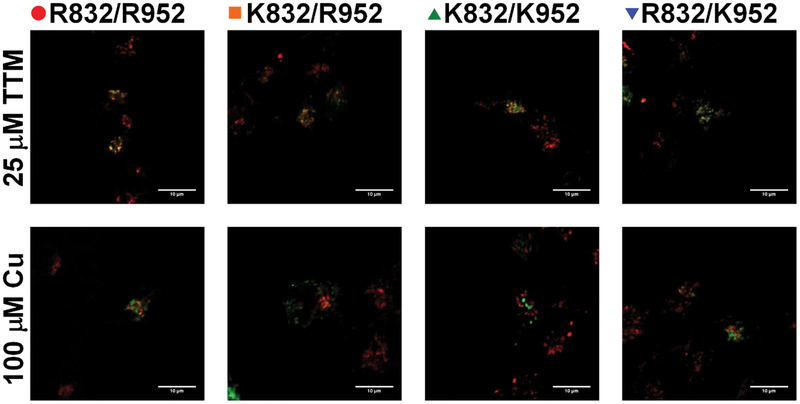
Hepatic ATP7B is targeted primarily to the TGN under basal or low Cu conditions and traffics to vesicles when Cu is elevated.43 Therefore, the loss of ATP7B retention in the TGN can be used as a measure of protein trafficking. To compare trafficking response of ATP7B variants, we measured their retention in the TGN under different Cu conditions. Cu depletion was achieved by treatment of HEK293A cells with the Cu chelator tetrathiomolybdate (TTM), and Cu elevation was produced by treating cells with CuSO4 (Fig. 5). Under Cu-limiting conditions, all variants were predominantly localized to the TGN, except for R832/K952-ATP7B, which was found in both TGN and vesicles. Upon Cu elevation, the variants with K832 behaved as expected, i.e. they trafficked normally out of the TGN as evidenced by the loss of co-localization between ATP7B and the TGN marker, TGN46 (Fig.5). In contrast, the variants with R832 trafficked to vesicles but remained close to the TGN and some retained an overlap with the TGN marker. Thus, the SNP-related substitutions do not disrupt ATP7B trafficking but may modulate the rate of ATP7B exit from the TGN.
Fig. 5.
Localization of ATP7B variants in HEK293 cells in low and high copper HEK293 cells were transfected with one of the indicated ATP7B-GFP variant (green), cells were treated for 4 h with either 25 mM TTM (top panels) or 100 mM CuSO4 (bottom panels) and immunostained for trans Golgi network marker TGN46 (red). Colocolization is indicated by yellow; the loss of colocalization is indicative of ATP7B trafficking (n = 4).
R832 affects the conformational dynamics of the A-domain
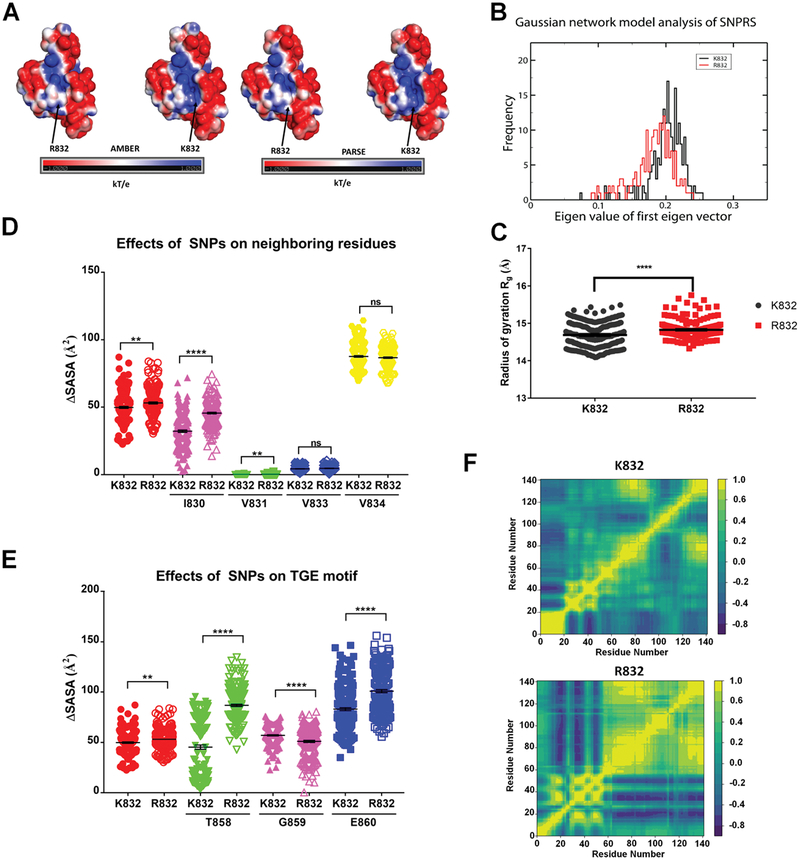
The decrease in abundance of the R832 variant proteins, as well as the putative effect of this substitution on ATP7B structure, suggested that Arg at position 832 may impact the structure of the A-domain, which has previously been shown to affect protein abundance.44 To test this hypothesis, we generated a model of the A-domain with R832 using the available NMR structure of the ATP7B A-domain with K83223 and compared the properties of each (Fig. 6A) Two different force-field calculations, AMBER-9927 and PARSE28, 29, both revealed a difference in the surface electrostatics of the domain depending on the residue present at position 832. The A-domain with K832 has a more positive net charge, whereas the A-domain with R832 has a more neutral net charge, with the largest difference between the domains occurring in the area immediately surrounding the 832 position.
Fig. 6.
R832 increases A-domain flexibility (A) surface electrostatics of the ATP7B A-domain [2] containing R832/K832 variants quantified using AMBER (left pair) or PARSE (right pair). (B) R832 increases the conformational flexibility of the A-domain. Gaussian network mode-based normal mode frequencies of (‘) K832 in black and (K) R832 in red (C) the radius of gyration of the A-domain containing either K832 (K, red) or R832 (‘, gray). (D) The variation of SASA values for AA residues in b sheet 3 during 200 ns simulation: R832 (red, filled circles), K832 (red, open circles): V833 (K, blue), V834 (yellow), I830 (green) and V831 (magenta). (E) Variation in SASA values for residues from the TGE motif in the R832 and K832 A domains: T858 (green), E860 (magenta), and G859 (blue). (F) Cross-correlation maps of AA motions in K832 (top) and R832 (bottom) A-domains during 200 ns simulation. Positive values (yellow) indicate motion in the same direction, whereas negative values (blue) indicate motion in the opposite direction. All values are reported as means SEM. Significance was determined by unpaired Student’s t-test; **p o 0.01, ****p o 0.0001.
To better understand the effect of the SNPRS on the secondary structure and biophysical properties of the A-domain, we performed all-atom MD simulations in explicit solvent (water/sodium chloride) that mimics the intracellular milieu. The backbone root mean square deviation (RMSD) trajectories show that both the R832 and K832 variants equilibrate past 90 nanoseconds (Fig. S1A). To analyze the conformational motions of the domain and the existence of stable state(s), we calculated the frequencies of the eigenvalues using the Gaussian network model (GNM).45, 46 Explicit solvent calculations for up to 200 nanoseconds were used to determine the impact of R832 on the GNM frequencies, which represent the rotational and translational motions of the domain.45 This analysis showed that both the K832 and R832 variants were present largely in one conformational state. However, the R832 variant has a broader distribution of eigenvalues, indicating a higher conformational flexibility (Fig. 6B). The increase in the intra-molecular motion is also supported by the larger radius of gyration seen in the R832 variant compared to the K832 variant (53.15±0.733 Å vs. 49.81±0.751 Å, p=0.0016; Fig. 6C).
Effects of the SNPRS on the secondary structural elements of the A-domain were analyzed using a definition of secondary structure of proteins (DSSP) plot. The third β-strand—the strand in which residue 832 is located—was relatively stable throughout the simulation in both variants (Fig. S1C). At the same time, the R832 SNPRS significantly altered the solvent accessibility surface area (SASA) for some of the residues in the vicinity of position 832. While V833 (4.29±0.153 Å2 vs. 4.63±0.140 Å2, p=0.099) and V834 (87.65±0.633 Å2 vs. 86.71±0.528 Å2, p=0.255) were not significantly changed, both I830 (32.30±0.918 Å2 vs. 45.62±0.655 Å2, p<0.0001) and V831 (0.039±0.008 Å2 vs. 0.099±0.020 Å2, p=0.005) had an increase in SASA in the R832 variant (Fig. 6D, S1B).
The A-domain contains the highly conserved TGE motif. This motif facilitates the dephosphorylation of the catalytic Asp and the conformational transitions of the full-length ATP7B protein.47 Increased structural fluctuations associated with the presence of Arg at position 832 could have long-range, allosteric effects on this motif. To examine this possibility, we analyzed the SASA changes for the residues that constitute the TGE motif (T858, G859, E860) and found that the surface exposure of G859 (57.01±0.638 Å2 vs. 51.03±1.078 Å2, p<0.0001) and E860 (83.05±1.431 Å2 vs. 100.90±1.582 Å2, p<0.0001) was higher in the R832-containing variant. The surface exposure of T858 was most significantly increased (45.31±2.102 Å2 vs 86.81±1.156 Å2, p<0.0001) in the R832 variant compared to the K832 variant (Fig. 6E, S1B)
To visualize the effect of the SNPRS on residue motions within the A-domain, we calculated the correlation matrix for all 140 residues in the domain and displayed them as a two-dimensional heat map (Fig. 6F). Positive cross-correlation values (yellow) indicate a strongly correlated motion of the residues in the same direction during the simulation. Negative values (blue) imply an anti-correlated motion of the residues, i.e. no communication between the residues. Position 832 (residue 40 on the map) lies within the segment of strong correlation for both variants. However, the long-range effects vary between the R832 and K832 variants, especially for residues 60–100 and 120–140. In the R832 variant, these regions show strong correlation; in the K832 variant, these regions have a less strong correlation. Taken together, the computational studies illustrate different MD of the A-domain with between the two variants, which may contribute to the distinct properties of the corresponding ATP7B variants in cells.
DISCUSSION
SNPs are typically considered benign, largely because they are frequently detected in the healthy population. In addition, SNPs often produce substitutions that preserve the main chemical properties of the corresponding amino acid residues, such as charge, bulk, and hydrophobicity. Our studies of two common SNP-dependent amino-acid substitutions in ATP7B have confirmed that ATP7B is active with any of the four amino-acid combinations at positions 832/952. At the same time, we found that these substitutions have measurable effects on both the structure and other properties of ATP7B. The presence of R832 and K952 modulates ATP7B abundance, trafficking, and Cu-transport activity to varying degrees. Analysis of markers of Cu status in human serum appears to correlate with these in vitro effects, although direct comparison of the Cu:Cp values with ATP7B abundance and activity is needed to draw firm conclusions.
These findings add to accumulating evidence that ATP7B SNPs influence cellular Cu homeostasis and, in combination with other factors, may contribute to human disease. Presence of R832 and K952 in ATP7B variants was previously linked to higher levels of labile Cu in the serum in AD patients.10, 13, 48 Previous in vitro studies measuring the ATP7B-driven Cu uptake in vesicles revealed that the R832 variant had lower Cu transport activity.11 Moreover, R832 was identified as a loss-of-function SNP in Drosophila melanogaster ATP7, an ATP7B homolog.11, 14 The Drodophila ATP7 protein containing Arg at the position equivalent to human R832 had lower expression levels than the control, and it was suggested that the presence of Arg in this position may promote protein degradation.14 Our studies in human cells show that R832 alters ATP7B protein dynamics, abundance, and trafficking, and that the K832/K952 variant has lower Cu-transport activity. We did not find that the inhibitory effect of R832 on ATP7B Cu transport activity in cells using the tyrosinase assay. This discrepancy could be due to the lower sensitivity of this assay in comparison to direct measurements of Cu uptake.
R832 is located in the A-domain, which is involved in conformational transitions and catalysis during the ATP7B enzymatic cycle.23 In addition ATP7B abundance and trafficking is regulated by a kinase-mediated phosphorylation of key Ser resides.4, 49, 50 Currently, it is not known whether R832K affects the phosphorylation state of ATP7B, and further study is needed. Our computational studies demonstrate that R832 alters the local structure, surface charge distribution, and dynamics of the A-domain. A higher conformational flexibility of the domain containing R832 may explain propensity to stronger degradation and decreased protein abundance of R832 variants compared to K832 containing proteins. Although differences between the ATP7B variants are rather small, in combination with other factors (or over long period of time) they may impact overall cellular copper balance.
K952 is located in the lumenal loop between TM segments 5 and 6. An equivalent loop in the homologous ATP7A has been shown to function in Cu release from the ATPase41, and the loop between TM3 and TM4 of ATP7B has been identified as a site of kinase-mediated phosphorylation.42 We observed that in cells the K832/K952 variant had a lower Cu-transport activity when compared to other ATP7B variants, which is consistent with potential negative effect on Cu release. The decrease in activity is not very large (about 20%); nevertheless, in Wilson disease patients with this genetic background presence of K952 may exacerbate the effects of otherwise mild ATP7B mutations.
Both R832 and K952 are common in the general population.10, 51 If these SNPs alter ATP7B structure or diminish protein function, how could they be present in high frequencies in the healthy population? Squitti, et al. have shown that healthy individuals are more likely to be heterozygous for R832 and K952, while AD patients are more likely to have both variants.10 Analysis of Cu status markers in the serum—total Cu, total Cp, and non-Cp Cu— revealed that both R832 and K952 may affect Cu homeostasis, even in healthy individuals. Although total Cu levels are comparable between the R832 and K952 carriers and non-carriers, presence of R832 appears to correlate with lower Cp levels, and carriers of K952 have both lower levels of Cp and higher levels of exchangeable Cu. In other words, presence of certain SNPs results in measurable changes to human Cu parameters.
Both R832 and K952 have been reported in WD patients alongside known WD mutations (Table 2). In Asian populations where WD prevalence is greater than that worldwide, R832 and K952 both appear with higher frequency. For example, the Han Chinese in Beijing (CHB) population has a WD prevalence of approximately 1 in 5400, compared to the worldwide prevalence of 1 in 30,000. R832 and K952 both appear in 65% of the CHB population, compared to 50% in the Utah Residents with Northern and Western European Ancestry (CEU) and the Indian (IND) populations.51, 52 We speculate that SNPs are generally benign but, in combination with a disease causing mutation, may exacerbate the manifestation of the disease.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by National Research Council [Aging Program 2012–2014, ‘A low-copper diet as a preventive strategy for cognitive disability in Aging’]; Italian Ministry of Health [5XMille project ‘Un metodo sensibile, diretto e preciso per misurare il rame Non-legato alla Ceruloplasmina nel siero per applicazione in ambiente clinico’ 09/02/2013 to 08/31/2015]; Italian Ministry of Health [‘Tolerability and efficacy of Zinc therapy in Mild Cognitive Impairment for treatment and prevention of Alzheimer’s disease: a prospective, randomized, double blind, parallel, placebo controlled Phase II clinical trial’ (Project Code: CO-2013–02358488)]; Canox4drug SpA [2013–2016 ‘Non-Ceruloplasmin copper in Alzheimer’s disease’ (Prot. 30/2013)], Italian Ministry of Health, Ricerca Corrente. This work was also funded in part by the National Institutes of Health grant R01 DK071865 (2SL).
Footnotes
CONFLICTS OF INTEREST
Dr. R. Squitti is a consultant Canox4drug SPA (Italy) and Igea Research Inc. (Miami, FL). Other authors have no conflicts of interest to declare.
REFERENCES
- 1.Lutsenko S, Copper trafficking to the secretory pathway, Metallomics, 2016, 8, 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Berghe PV and Klomp LW, New developments in the regulation of intestinal copper absorption, Nutr Rev, 2009, 67, 658–672. [DOI] [PubMed] [Google Scholar]
- 3.Michalczyk AA, Rieger J, Allen KJ, Mercer JF and Ackland ML, Defective localization of the Wilson disease protein (ATP7B) in the mammary gland of the toxic milk mouse and the effects of copper supplementation, Biochem J, 2000, 352 Pt 2, 565–571. [PMC free article] [PubMed] [Google Scholar]
- 4.Braiterman LT, Gupta A, Chaerkady R, Cole RN and Hubbard AL, Communication between the N and C termini is required for copper-stimulated Ser/Thr phosphorylation of Cu(I)-ATPase (ATP7B), J Biol Chem, 2015, 290, 8803–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutsenko S, Barnes NL, Bartee MY and Dmitriev OY, Function and regulation of human copper-transporting ATPases, Physiol Rev, 2007, 87, 1011–1046. [DOI] [PubMed] [Google Scholar]
- 6.Lalioti V, Peiro R, Perez-Berlanga M, Tsuchiya Y, Munoz A, Villalba T, Sanchez C and Sandoval IV, Basolateral sorting and transcytosis define the Cu+-regulated translocation of ATP7B to the bile canaliculus, J Cell Sci, 2016, 129, 2190–2201. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton JP, Koganti L, Muchenditsi A, Pendyala VS, Huso D, Hankin J, Murphy RC, Huster D, Merle U, Mangels C, Yang N, Potter JJ, Mezey E and Lutsenko S, Activation of liver X receptor/retinoid X receptor pathway ameliorates liver disease in Atp7B(−/−) (Wilson disease) mice, Hepatology, 2016, 63, 1828–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James SA, Volitakis I, Adlard PA, Duce JA, Masters CL, Cherny RA and Bush AI, Elevated labile Cu is associated with oxidative pathology in Alzheimer disease, Free Radic Biol Med, 2012, 52, 298–302. [DOI] [PubMed] [Google Scholar]
- 9.Squitti R, Siotto M, Arciello M and Rossi L, Non-ceruloplasmin bound copper and ATP7B gene variants in Alzheimer’s disease, Metallomics, 2016, 8, 863–873. [DOI] [PubMed] [Google Scholar]
- 10.Bucossi S, Polimanti R, Mariani S, Ventriglia M, Bonvicini C, Migliore S, Manfellotto D, Salustri C, Vernieri F, Rossini PM and Squitti R, Association of K832R and R952K SNPs of Wilson’s disease gene with Alzheimer’s disease, J Alzheimers Dis, 2012, 29, 913–919. [DOI] [PubMed] [Google Scholar]
- 11.Huster D, Kuhne A, Bhattacharjee A, Raines L, Jantsch V, Noe J, Schirrmeister W, Sommerer I, Sabri O, Berr F, Mossner J, Stieger B, Caca K and Lutsenko S, Diverse functional properties of Wilson disease ATP7B variants, Gastroenterology, 2012, 142, 947–956 e945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squitti R, Polimanti R, Bucossi S, Ventriglia M, Mariani S, Manfellotto D, Vernieri F, Cassetta E, Ursini F and Rossini PM, Linkage disequilibrium and haplotype analysis of the ATP7B gene in Alzheimer’s disease, Rejuvenation Res, 2013, 16, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucossi S, Mariani S, Ventriglia M, Polimanti R, Gennarelli M, Bonvicini C, Pasqualetti P, Scrascia F, Migliore S, Vernieri F, Rossini PM and Squitti R, Association between the c. 2495 A>G ATP7B Polymorphism and Sporadic Alzheimer’s Disease, Int J Alzheimers Dis, 2011, 2011, 973692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer SW, Wang J and Burke R, In Vivo Modeling of the Pathogenic Effect of Copper Transporter Mutations That Cause Menkes and Wilson Diseases, Motor Neuropathy, and Susceptibility to Alzheimer’s Disease, J Biol Chem, 2017, 292, 4113–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folstein MF, Folstein SE and McHugh PR, “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician, J Psychiatr Res, 1975, 12, 189–198. [DOI] [PubMed] [Google Scholar]
- 16.Siotto M, Simonelli I, Pasqualetti P, Mariani S, Caprara D, Bucossi S, Ventriglia M, Molinario R, Antenucci M, Rongioletti M, Rossini PM and Squitti R, Association Between Serum Ceruloplasmin Specific Activity and Risk of Alzheimer’s Disease, J Alzheimers Dis, 2016, 50, 1181–1189. [DOI] [PubMed] [Google Scholar]
- 17.Walshe JM and Clinical B Investigations Standing Committee of the Association of Clinical, Wilson’s disease: the importance of measuring serum caeruloplasmin non-immunologically, Ann Clin Biochem, 2003, 40, 115–121. [DOI] [PubMed] [Google Scholar]
- 18.Twomey PJ, Viljoen A, House IM, Reynolds TM and Wierzbicki AS, Copper:caeruloplasmin ratio, J Clin Pathol, 2007, 60, 441–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crooks GE, Hon G, Chandonia JM and Brenner SE, WebLogo: a sequence logo generator, Genome Res, 2004, 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Y, Nyasae L, Braiterman LT and Hubbard AL, NH2-terminal signals in ATP7B Cu-ATPase mediate its Cu-dependent anterograde traffic in polarized hepatic cells, Am J Physiol Gastrointest Liver Physiol, 2005, 289, G904–916. [DOI] [PubMed] [Google Scholar]
- 21.Schushan M, Bhattacharjee A, Ben-Tal N and Lutsenko S, A structural model of the copper ATPase ATP7B to facilitate analysis of Wilson disease-causing mutations and studies of the transport mechanism, Metallomics, 2012, 4, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex N and Peitsch MC, SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling, Electrophoresis, 1997, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 23.Banci L, Bertini I, Cantini F, Migliardi M, Natile G, Nushi F and Rosato A, Solution structures of the actuator domain of ATP7A and ATP7B, the Menkes and Wilson disease proteins, Biochemistry, 2009, 48, 7849–7855. [DOI] [PubMed] [Google Scholar]
- 24.Dolinsky TJ, Nielsen JE, McCammon JA and Baker NA, PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations, Nucleic Acids Res, 2004, 32, W665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G and Baker NA, PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations, Nucleic Acids Res, 2007, 35, W522–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unni S, Huang Y, Hanson RM, Tobias M, Krishnan S, Li WW, Nielsen JE and Baker NA, Web servers and services for electrostatics calculations with APBS and PDB2PQR, J Comput Chem, 2011, 32, 1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen AA and Pappu RV, Parameters of monovalent ions in the AMBER-99 forcefield: assessment of inaccuracies and proposed improvements, J Phys Chem B, 2007, 111, 11884–11887. [DOI] [PubMed] [Google Scholar]
- 28.Baker CM, Lopes PE, Zhu X, Roux B and Mackerell AD Jr., Accurate Calculation of Hydration Free Energies using Pair-Specific Lennard-Jones Parameters in the CHARMM Drude Polarizable Force Field, J Chem Theory Comput, 2010, 6, 1181–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang CL, Alexov E, Pyle AM and Honig B, Calculation of pKas in RNA: on the structural origins and functional roles of protonated nucleotides, J Mol Biol, 2007, 366, 1475–1496. [DOI] [PubMed] [Google Scholar]
- 30.Baker NA, Sept D, Joseph S, Holst MJ and McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome, Proc Natl Acad Sci U S A, 2001, 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL 3rd, MacKerell AD Jr., Klauda JB and Im W, CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field, J Chem Theory Comput, 2016, 12, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM and Karplus M, CHARMM: the biomolecular simulation program, J Comput Chem, 2009, 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jo S, Kim T, Iyer VG and Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM, J Comput Chem, 2008, 29, 1859–1865. [DOI] [PubMed] [Google Scholar]
- 34.Humphrey W, Dalke A and Schulten K, VMD: visual molecular dynamics, J Mol Graph, 1996, 14, 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- 35.Michaud-Agrawal N, Denning EJ, Woolf TB and Beckstein O, MDAnalysis: a toolkit for the analysis of molecular dynamics simulations, J Comput Chem, 2011, 32, 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joosten RP, te Beek TA, Krieger E, Hekkelman ML, Hooft RW, Schneider R, Sander C and Vriend G, A series of PDB related databases for everyday needs, Nucleic Acids Res, 2011, 39, D411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W and Sander C, Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features, Biopolymers, 1983, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 38.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE and Berendsen HJ, GROMACS: fast, flexible, and free, J Comput Chem, 2005, 26, 1701–1718. [DOI] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem, 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 40.Petris MJ, Strausak D and Mercer JF, The Menkes copper transporter is required for the activation of tyrosinase, Hum Mol Genet, 2000, 9, 2845–2851. [DOI] [PubMed] [Google Scholar]
- 41.Barry AN, Otoikhian A, Bhatt S, Shinde U, Tsivkovskii R, Blackburn NJ and Lutsenko S, The lumenal loop Met672-Pro707 of copper-transporting ATPase ATP7A binds metals and facilitates copper release from the intramembrane sites, J Biol Chem, 2011, 286, 26585–26594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartee MY, Ralle M and Lutsenko S, The loop connecting metal-binding domains 3 and 4 of ATP7B is a target of a kinase-mediated phosphorylation, Biochemistry, 2009, 48, 5573–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roelofsen H, Wolters H, Van Luyn MJA, Miura N, Kuipers F and Vonk RJ, Copper-induced apical trafficking of ATP7B in polarized hepatoma cells provides a mechanism for biliary copper excretion, Gastroenterology, 2000, 119, 782–793. [DOI] [PubMed] [Google Scholar]
- 44.Gupta A, Bhattacharjee A, Dmitriev OY, Nokhrin S, Braiterman L, Hubbard AL and Lutsenko S, Cellular copper levels determine the phenotype of the Arg875 variant of ATP7B/Wilson disease protein, Proc Natl Acad Sci U S A, 2011, 108, 5390–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall BA, Kaye SL, Pang A, Perera R and Biggin PC, Characterization of protein conformational states by normal-mode frequencies, J Am Chem Soc, 2007, 129, 11394–11401. [DOI] [PubMed] [Google Scholar]
- 46.Bahar I, Atilgan AR and Erman B, Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential, Fold Des, 1997, 2, 173–181. [DOI] [PubMed] [Google Scholar]
- 47.Tsivkovskii R, Eisses JF, Kaplan JH and Lutsenko S, Functional properties of the copper-transporting ATPase ATP7B (the Wilson’s disease protein) expressed in insect cells, J Biol Chem, 2002, 277, 976–983. [DOI] [PubMed] [Google Scholar]
- 48.Squitti R, Polimanti R, Siotto M, Bucossi S, Ventriglia M, Mariani S, Vernieri F, Scrascia F, Trotta L and Rossini PM, ATP7B variants as modulators of copper dyshomeostasis in Alzheimer’s disease, Neuromolecular Med, 2013, 15, 515–522. [DOI] [PubMed] [Google Scholar]
- 49.Pilankatta R, Lewis D and Inesi G, Involvement of protein kinase D in expression and trafficking of ATP7B (copper ATPase), J Biol Chem, 2011, 286, 7389–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanderwerf SM, Cooper MJ, Stetsenko IV and Lutsenko S, Copper specifically regulates intracellular phosphorylation of the Wilson’s disease protein, a human copper-transporting ATPase, J Biol Chem, 2001, 276, 36289–36294. [DOI] [PubMed] [Google Scholar]
- 51.Gupta A, Maulik M, Nasipuri P, Chattopadhyay I, Das SK, Gangopadhyay PK, Indian Genome Variation C and Ray K, Molecular diagnosis of Wilson disease using prevalent mutations and informative single-nucleotide polymorphism markers, Clin Chem, 2007, 53, 1601–1608. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y and Wu ZY, Wilson’s disease in Asia, Neurol Asia, 2011, 16, 103–109. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.