Abstract
Background:
Aortic valve stenosis (AVS) and coronary artery disease (CAD) have a significant genetic contribution and commonly co-exist. To compare and contrast genetic determinants of the two diseases, we investigated associations of the LPA and 9p21 loci, i.e. the two strongest CAD risk loci, with risk of AVS.
Methods:
We genotyped the CAD-associated variants at the LPA (rs10455872) and 9p21 loci (rs1333049) in the GeneCAST (Genetics of Calcific Aortic STenosis) Consortium and conducted a meta-analysis for their association with AVS. Cases and controls were stratified by CAD status. External validation of findings was undertaken in five cohorts including 7,880 cases and 851,152 controls.
Results:
In the meta-analysis including 4,651 cases and 8,231 controls the CAD-associated allele at the LPA locus was associated with increased risk of AVS (OR 1.37; 95%CI 1.24–1.52, p=6.9×10−10) with a larger effect size in those without CAD (OR 1.53; 95%CI 1.31–1.79) compared to those with CAD (OR 1.27; 95%CI 1.12–1.45). The CAD-associated allele at 9p21 was associated with a trend towards lower risk of AVS (OR 0.93; 95%CI 0.88–0.99, p=0.014). External validation confirmed the association of the LPA risk allele with risk of AVS (OR 1.37; 95%CI 1.27–1.47), again with a higher effect size in those without CAD. The small protective effect of the 9p21 CAD risk allele could not be replicated (OR 0.98; 95%CI 0.95–1.02).
Conclusions:
Our study confirms the association of the LPA locus with risk of AVS, with a higher effect in those without concomitant CAD. Overall, 9p21 was not associated with AVS.
Keywords: Aortic valve stenosis, risk factors, valvular heart disease, Lipoprotein (a), 9p21
Introduction
Progressive calcification of the aortic valve leaflets leads to a high prevalence of aortic valve stenosis (AVS) in the elderly population (>75 years) resulting in the most common form of degenerative valvular heart disease [1]. The long-time course before it becomes severe enough to cause symptoms and affect prognosis provides a significant opportunity to prevent its progression. As soon as patients become symptomatic, mortality increases up to 50% within two years [2] and surgical or percutaneous aortic valve replacement remain the only current therapeutic options [3]. With an increasingly ageing population in many countries, the need for other treatment options for AVS is gaining an ever-greater imperative. Development and progression of AVS is a multifactorial process involving several risk factors, e.g., smoking, high blood pressure, diabetes, and cholesterol. In addition, elevated levels of lipoprotein (a) (Lp(a)) [4] and predisposition for high LDL-cholesterol are associated with AVS [5]. Thus, development of AVS represents an active process involving lipid metabolism and inflammation [6] – largely replicating the risk factors underlying coronary artery disease (CAD). However, despite identification of risk factors and understanding of some of the biology of AVS [7] there has been little progress in development of preventative treatments.
The first genome-wide association study (GWAS) of aortic valve disease focused on calcification of the aortic valve using computed tomography (CT) scan analysis and identified one SNP at the LPA locus (rs10455872) on chromosome 6 reaching genome-wide significance [8]. Aortic valve calcification correlates with the progression of AVS [9], and subsequent studies have confirmed the association of the locus with AVS [4,10–12]. Recently, a GWAS of AVS identified two new loci on chromosome 1p21 and 2q22, the first of these (PALMD) also associates with bicuspid aortic valve disease and aortic root size, whereas the second variant (TEX41) also associates with coronary artery disease [13]. AVS and CAD are both age-related conditions, share several traditional risk factors and commonly co-exist [14,15]. Therefore, any analysis of genetic determinants of AVS needs to take into account the presence of CAD, and exclude the possibility that the association is a confounded association due to the presence of CAD. The aim of our study was to investigate the association of the LPA locus with hemodynamically significant AVS taking CAD into account and also to investigate whether the strongest non-lipid locus for CAD (9p21) shows an association with AVS.
Methods
Discovery study population
The discovery study consisted of three cohorts (Dundee, Leicester and Munich) from the Genetics of Calcific Aortic STenosis (GeneCAST) Consortium and the study flow chart is illustrated in the Supplementary Figure 1. All studies were approved by the respective institutional Ethics Committees and all patients consented to participate in the studies. All recruitments and analyses were performed in accordance with the Declaration of Helsinki.
In all three cohorts from the GeneCAST consortium AVS was defined by standard 2D echocardiography as thickening of the aortic valve leaflets and evidence of at least mild aortic stenosis with an increased peak velocity >2.5 m/s. In addition, patients with aortic valve replacement for severe AVS were included.
In GeneCAST Dundee patients were identified from the Genetics of Diabetes and Audit Research Tayside Study (GoDARTS) which has been described previously [16,17]. Briefly, a unique patient identifier to link demographic, echocardiographic, outcome and genotype databases was used. Patients with bicuspid aortic valves were excluded. Patients were categorized into CAD positive (+) or CAD negative (−). CAD+ was classified if they had a previous hospital admission due to CAD (primary or secondary diagnosis) as recorded by the Scottish Morbidity Record or their primary cause of death was due to CAD as recorded in the Scottish General Register Office of Births and Deaths based on the ICD-10 code.
In GeneCAST Leicester cases and controls were classified into CAD+ and CAD- groups depending on their reported medical history. Patients with congenital aortic valve abnormalities including bicuspid aortic valves, evidence of rheumatic heart disease including moderate or severe mitral stenosis or mitral valve repair/replacement, aortic valve endocarditis, or aortic valve replacement before the age of 50 years were excluded from the analysis.
In GeneCAST Munich all patients (cases and controls) had undergone coronary angiography. If narrowing of the coronary arteries was <25% patients were classified as CAD-. Exclusion criteria were identical to the criteria from the GeneCAST Leicester cohort as stated above.
External replication study populations
External validation was performed using data from five additional cohorts, deCODE, ASAP/DAVAACA, HUNT, Malmo Diet and Cancer Study and the UK Biobank. Details on the study cohorts have been described in detail previously [13]. In brief, the definition of AVS in deCODE was based on ICD-codes (I35.0, I35.2 for discharge diagnosis or the NOMESCO classification of surgical procedures codes (FMA, FMSA, FMD/FMSD and subcodes). The definition of CAD was taken from the deCODE genotype phenotype database [18]. In the data from Karolinska Institute ASAP Biobank and its subsequently collected DAVAACA Biobank CAD was defined as significant stenosis on coronary angiogram. For the ASAP part of the Biobanks, individuals with CAD were excluded from the Biobank, explaining the general low level of CAD+ subjects in the Karolinska Institute analysis. The AVS status was assessed from transthoracic echocardiography. In UK Biobank AVS cases were defined as subjects with a self-reported history of AVS, or hospitalization or death due to ICD9 424.1 or ICD10 I35.0. CAD cases in UK Biobank were defined as a subject with self-reported history of heart attack/myocardial infarction, percutaneous transluminal coronary angioplasty, stent implantation, coronary artery bypass graft; or hospitalization for ICD9 410–412, ICD10 I21-I24, I25.2, OPCS-4 K40-K46, K49, K50.1, K75, or the following cause of death (ICD10 I21-I24, I25.2). In the HUNT study AVS was defined as ICD-10 code I35.0 or I35.2 and CAD was defined as individuals with self-reported coronary artery bypass graft, coronary angioplasty, or stent placement, or those with ICD-10 codes I21 or I25.2 or ICD-9 codes 410 or 412. In the Malmo Diet and Cancer Study cases of AVS were retrieved from nationwide hospital registers [5] defined as ICD-9 424.1, ICD-10 I35.0 or I35.2. The definition of CAD was defined as a previous hospital diagnosis of myocardial infarction (ICD-9 410, ICD-10 I21), or if participants had undergone percutaneous coronary intervention or coronary artery bypass surgery.
Genotyping
In the GeneCAST Dundee cohort, samples were genotyped at the Affymetrix service laboratory on the Genome-Wide Human SNP Array 6.0. Genotype QC was via the standard protocol established for the Wellcome Trust Case Control Consortium 2 Study, as previously described [16]. In the GeneCAST Leicester cohort SNP genotyping was performed using the Fluidigm EP1™ system at Source BioScience genomics services (Nottingham, UK). SNP genotyping in the GeneCAST Munich cohort was performed with TaqMan® Gene Expression Master Mix and TaqMan® MGB Probes VIC and FAM (ThermoFisher, Life Technologies, Carlsbad, USA) for allele labelling on a ViiA7 qPCR instrument (ThermoFisher, Life Technologies, Carlsbad, USA). The primers used for LPA (rs10455872) and 9p21 (rs1333049) genotyping are provided in the Supplementary Appendix. In UKB, participants were genotyped in two phases with the first 50,000 using the BiLEVE array and the remainder using the main UK Biobank array [19]. In the Malmo Diet and Cancer study genotyping was performed using the Illumina Human Omni Express Exome BeadChip kit. In the Karolinska Institute analysis, the Illumina Human610-Quad BeadChip and Infinium Global Screening Arrays were used (416 and 922 individuals respectively). In the deCODE study genotyping was performed either using Illumina SNP chips or data was taken from whole genome sequencing. The HUNT study performed genotyping using the IlluminaCoreExome array.
Lp(a) Measurements
In a subgroup of AVS cases without CAD from the GeneCAST Munich cohort, plasma levels of Lp(a) were measured. As previously described [20], Lp(a) mass was analysed using an immunoturbimetry method and measurements were performed on an automated analyser (Cobas MIRA, Roche, Basel Switzerland). In addition, we retrospectively analysed the routinely assessed Lp(a) levels of patients undergoing transcatheter aortic valve replacement (TAVR) for severe aortic stenosis at the German Heart Center Munich, Munich, Germany between September 2016 and September 2018. Definition of CAD in these patients was based on coronary angiography prior to TAVR procedure.
Statistical analysis
Analyses of association of the two variants with AVS were performed using logistic regression in unadjusted models within each GeneCAST cohort stratified by CAD status. Results were combined using a fixed-effect inverse-variance weighted meta-analysis on the log-odds ratios, with consideration of study heterogeneity using the I2 statistic. In the external validation in UKB, due to the level of relatedness within UK Biobank, a family identifier was derived based on kinship scores, where any individuals found to be related (k>0.044) were joined into a family. All validation cohorts performed logistic regression analyses adjusting for age, sex, , the first five principal components statin use, array type in UKB (BiLEVE array versus main UK Biobank array) with family fitted as a random effect where required using mixed models. Lp(a) plasma levels were compared using unpaired Student t test. Analysis of Lp(a) quartiles was performed using one-way ANOVA and post trend analysis. Comparisons of allele frequencies were performed using two sample test of proportions. All analyses were conducted using Stata version 13.1.
Results
The baseline characteristics of the discovery study cohorts are summarised in the Supplementary Table 1. The cases in all cohorts are typical of clinical patients with degenerative AVS.
Association of the LPA locus with AVS
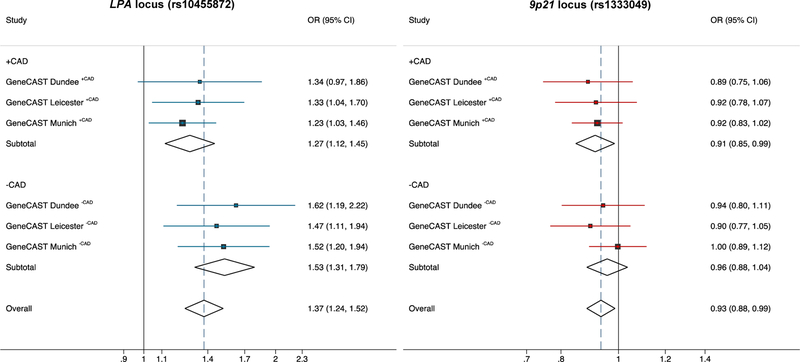
CAD risk allele (G) frequencies for rs10455872 for each cohort and association analysis of the LPA CAD risk allele with risk of AVS in each cohort are shown in Figure 1 and Table 1. Results were consistent across all three cohorts and the meta-analysis confirmed a strongly significant association of the LPA locus with risk of AVS (Odds ratio (OR) 1.37; 95% confidence interval (CI) 1.24–1.52, p=6.9×10−10) (Figure 1). Interestingly, the effect size for the LPA risk allele was lower in the cohorts with CAD (meta OR 1.27; 95% CI 1.12–1.45, p=2.8×10−4) compared to the cohorts without CAD (meta OR 1.53; 95% CI 1.31–1.79, p=1.2×10−7), although a test for interaction was not statistically significant.
Figure 1 – Analysis of the LPA and 9p21 loci with risk of AVS in the discovery cohorts.
The (CAD) risk alleles at the LPA locus (rs10455872) and at the 9p21 locus (rs1333049) were analysed for their association with risk of aortic valve stenosis (AVS) in three cohorts of the GeneCAST (Genetics of Calcific Aortic STenosis) Consortium. Both loci in the three cohorts were stratified for CAD status. In addition, a meta-analysis in the stratified groups as well as an overall meta-analysis was performed. The CAD risk allele at the LPA locus was concordantly associated with increased risk of AVS, whereas the 9p21 CAD risk allele was significantly associated with a lower risk of AVS. Odds Ratio; CI, Confidence interval.
Table 1 –
Effect allele frequencies and association of the LPA and 9p21 locus with risk of AVS
| rs10455872 (G) | rs1333049 (C) | ||||||
|---|---|---|---|---|---|---|---|
| Study | Cases/Controls | EAF (Cases/Controls) | OR 95% CI | p-value | EAF (Cases/Controls) | OR 95% CI | p-value |
| GeneCAST Dundee CAD+ | 320/1565 | 0.103/0.079 | 1.34 (0.97–1.86) | 0.076 | 0.483/0.511 | 0.89 (0.75.1.06) | 0.181 |
| GeneCAST Leicester CAD+ | 439/993 | 0.137/0.106 | 1.33 (1.04–1.70) | 0.021 | 0.489/0.511 | 0.92 (0.78–1.07) | 0.280 |
| GeneCAST Munich CAD+ | 2054/1340 | 0.095/0.079 | 1.23 (1.03–1.46) | 0.024 | 0.507/0.527 | 0.92 (0.83–1.02) | 0.102 |
| Subtotal CAD+ | 2813/3898 | 1.27 (1.12–1.45) | 2.8×10−4 | 0.91 (0.85–0.99) | 0.019 | ||
| GeneCAST Dundee CAD- | 340/2056 | 0.110/0.071 | 1.62 (1.19–2.22) | 0.002 | 0.468/0.483 | 0.94 (0.80–1.11) | 0.964 |
| GeneCAST Leicester CAD- | 520/783 | 0.102/0.072 | 1.47 (1.11–1.94) | 0.007 | 0.444/0.472 | 0.90 (0.77–1.05) | 0.470 |
| GeneCAST Munich CAD- | 978/1494 | 0.07/0.047 | 1.52 (1.20–1.94) | 6.5×10−4 | 0.472/0.473 | 1.00 (0.89–1.12) | 0.170 |
| Subtotal CAD- | 1838/4333 | 1.53 (1.31–1.79) | 1.2×10−7 | 0.96 (0.88–1.04) | 0.278 | ||
| Overall | 4651/8231 | 1.37 (1.24–1.52) | 6.9×10−10 | 0.93 (0.88–0.99) | 0.014 | ||
The effect allele of rs10455872 and rs1333049 is shown in brackets (effect allele). EAF = effect allele frequency. OR 95% CI = Odds ratio 95% confidence interval.
Association of the 9p21 locus with AVS
CAD risk allele (C) frequencies for rs1333049 for each cohort are shown in Figure 1 and Table 1. At the individual cohort level, there was no association of the CAD risk allele with risk of AVS. In the overall meta-analysis, the 9p21 CAD risk allele showed a trend towards a lower risk of AVS (OR 0.93; 95% CI 0.88–0.99, p=0.014). When stratified by CAD status, there was a small protective effect of the 9p21 CAD risk allele with lower risk of AVS in the cohorts with CAD (meta OR 0.91; 95% CI 0.85–0.99, p= 0.019), but not in the cohorts without CAD (meta OR 0.96; 95% CI 0.88–1.04, p=0.278) (Figure 1).
External validation
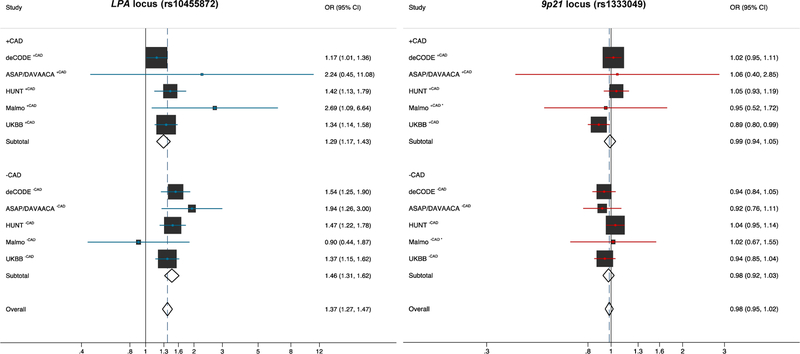
External validation in 7,880 cases and 851,192 controls for the association analysis of the LPA risk allele in five additional cohorts showed similar effect estimates with an overall OR of 1.37 (95% CI 1.27–1.47) (Figure 2 and Supplementary Table 2). Likewise, the ORs showed a higher effect size for the LPA risk allele in subjects without CAD compared to those with CAD OR 1.46 (95% CI 1.31–1.62) and OR 1.29 (95% CI 1.17–1.43), respectively. Analysis of the 9p21 CAD risk allele in the replication studies did not confirm an association with lower risk of AVS with an overall OR of 0.98 (95% CI 0.95–1.02) (Figure 2 and Supplementary Table 2).
Figure 2 – Replication analysis of the LPA and 9p21 loci with risk of AVS.
External validation for the association analysis of the LPA (rs10455872) and the 9p21 (rs1333049) risk locus in five additional cohorts comprising 7,880 cases and 851,192 controls. Again, both loci are stratified for CAD status. Analysis of the LPA risk allele showed similar effect estimates, whereas in the replication studies the 9p21 CAD risk allele did not show an association with risk of AVS.
Exploration for survival bias
Given the well-validated relationship between the 9p21 locus and CAD risk, one possibility for the finding of a potential inverse association of the CAD risk allele with AVS risk could be that a greater proportion of subjects carrying this allele develop CAD and die before they can develop AVS and thus impact on the association of the genotype with AVS. Similarly, the slightly reduced association of the LPA locus with AVS in the CAD+ group compared with the CAD- group could similarly represent a possible relative loss of the LPA risk allele in the CAD+ due to premature CAD death in carriers. To investigate these possibilities, we analysed risk allele frequencies by CAD status within AVS groups in GeneCAST, combined across all the cohorts. As shown in the Supplementary Figure 2, although the risk allele frequencies in those with AVS were higher for LPA and lower for 9p21 as might be expected from the observed associations with AVS, the association with CAD was observed in all four groups irrespective of AVS status without significant difference in the proportionate relationship (Supplementary Figure 2). This would argue against a substantial survival effect impacting on the findings.
Association of the LPA locus with circulating Lipoprotein (a) levels
To investigate whether LPA genotype was also associated with circulating lipoprotein (a) (Lp(a)) levels, we analysed a subgroup of 278 individuals with AVS from the GeneCAST Munich CAD- cohort with both available LPA genotype and Lp(a) plasma levels. Lp(a) levels were only available from heterozygous individuals (AG, n=41) and homozygous non-risk allele carriers (AA, n=237). The risk allele (G) was associated with significantly higher circulating Lp(a) levels (AG vs. AA: 88.22 ± 7.0 mg/dl vs. 23.96 ± 7.1 mg/dl; p<0.001).
Lipoprotein (a) levels and age at TAVR symptomatic severe AVS
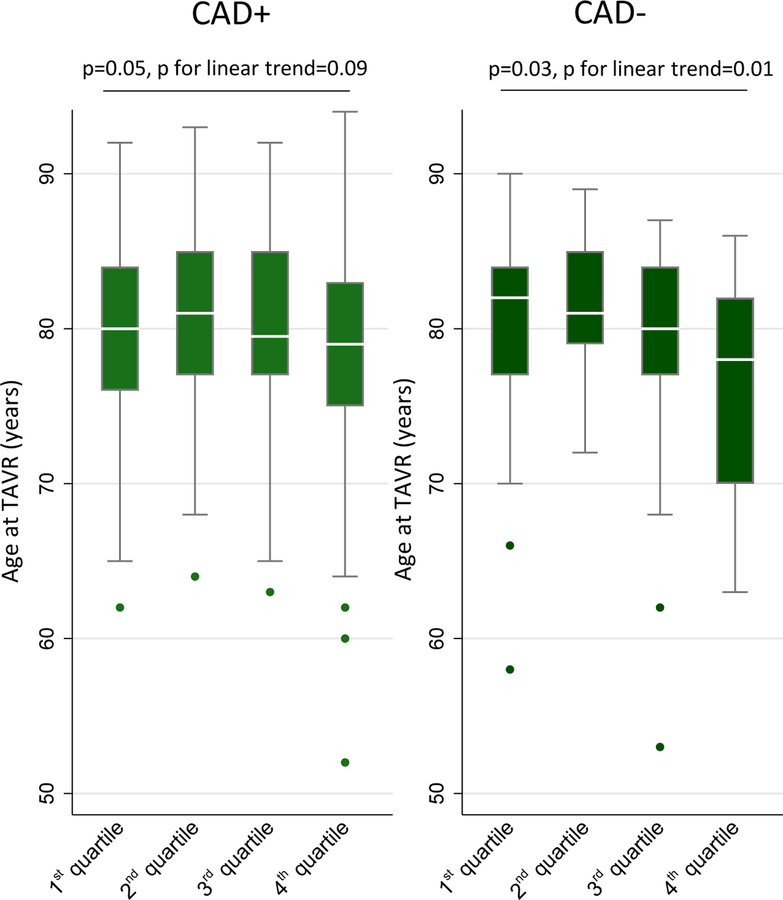
To analyse whether the observed difference in risk of AVS between patients with CAD and patients without CAD depends on Lp(a) plasma levels we analysed Lp(a) levels in patients with symptomatic severe AVS undergoing TAVR. After classification of 451 patients into quartiles by their Lp(a) level there was a linear decrease in the mean age at TAVR (from 80.1 years to 75.8 years, p for linear trend 0.01) but only in those without CAD (Figure 3). In patients with CAD, the mean age at TAVR only slightly differed between those with high and those with low Lp(a) level quartiles (from 79.6 years to 78.2 years, p for the trend 0.09).
Figure 3 – LP(a) levels in patients with symptomatic AVS undergoing TAVR.
Age at TAVR due to severe symptomatic AVS stratified by CAD status. Lp(a) levels were classified into quartiles from low (1st quartile) to high (4th quartile) Lp(a) serum level.
Discussion
In our analysis we confirmed a robust association of AVS with the LPA locus, independent of concomitant CAD, which was replicated in five external cohorts. Interestingly, the effect size of the LPA risk allele was higher in patients without concomitant CAD. Similar, patients without CAD and high Lp(a) levels developed symptomatic severe AVS earlier. In addition, we identified a potential protective effect of the CAD risk allele at 9p21. However, this finding did not replicate in the validation cohorts.
In 2013 Thanassoulis and colleagues [8] reported an association of aortic valve calcification with the LPA locus, specifically with rs10455872 [8]. Since then several subsequent cross-sectional and prospective studies have confirmed the association of this variant with AVS [4,10–12]. The risk (G) allele at rs10455872 is associated with increased plasma Lp(a) levels [4,10], as we also show here. Higher Lp(a) levels are also associated with risk of AVS[4,10] providing a mechanism by which genetic variation at the LPA locus affects AVS risk and Mendelian randomization analyses have supported this likely causality [4,10].
A particular challenge in identifying genetic determinants of AVS is that half of patients with AVS also have significant CAD [7,21] and therefore care is required in genetic studies of AVS to minimize the possibility that any association observed is not simply a proxy for an association with CAD. Based on the common co-occurrence and shared key risk factors of the two diseases Helgadottir and colleagues recently investigated common CAD associated variants in AVS cases[13]. A combined genetic risk score of 71 CAD variants was associated with risk of AVS. However, the significant result completely disappeared after adjusting for CAD [13]. Therefore, in the current study we took specific measures not previously utilized and avoiding any spurious association due to the co-occurrence of CAD. Indeed, the LPA locus and rs10455872 specifically have also been found to be strongly associated with CAD [22]. However, the consistent findings across the six comparisons, as well as the external validation in five additional cohorts, strongly suggest that there is a specific association of the LPA locus with AVS and that our finding as well as those of previous studies where CAD was not always accounted for is not due to confounding.
Our findings in this large study of patients with hemodynamically significant AVS add to the growing body of evidence, based on a strong genetic foundation, that Lp(a) is a viable therapeutic target for AVS [4,8,10,11]. The finding comes at a particularly relevant time as recently several lipid-lowering therapies have been developed, including PCSK9 (proprotein convertase subtilisin/kexin type 9) inhibitors [23] and antisense oligonucleotides [24] that lower plasma Lp(a) levels. Indeed, a loss of function mutation (R46L) in PCSK9 mimicking the effect of PCSK9 inhibitors have been found to be associated with lower Lp(a) level and reduced risk of AVS [25]. Modelling of the population impact of Lp(a) lowering has found that Lp(a) lowering among the top 20% of the population distribution of Lp(a) could prevent 1 in 7 cases of AVS [26].
Although, the weaker association of the LPA locus with risk of AVS in patients with CAD was not statistically different from that in patients without CAD, there was a consistent pattern across all cohorts (Figure 1 and 2) and it is therefore interesting to briefly speculate why this may be the case. As discussed earlier, we did not find evidence of a survival effect. It is however possible that earlier risk factor management and pharmacological intervention in patients with CAD, might have attenuated the subsequent LPA genotype-related risk of developing AVS. Although lipid-lowering trials of statins in patients with AVS have not shown a substantial reduction in disease progression [27–29], this might be explained by a too late medical intervention or a too short follow-up time. Furthermore, beneficial effects of RAS inhibition in patients with AVS have been shown [30] and although the data is controversial, RAS blockade may retard progression of aortic valve calcification and stenosis [31–33]. Our analysis in the TAVR cohort suggests that especially, in patients without CAD and thus less stringent management of risk factors, high Lp(a) levels seem to be a factor in AVS progression resulting in symptomatic AVS four years earlier than in patients with low Lp(a) levels. This again underlines the therapeutic potential of Lp(a) lowering therapies.
In contrast to the finding with rs10455872, we observed no association of the strongest CAD risk allele at 9p21 [34] and even a small protective effect in the meta-analysis of the GeneCAST cohorts. However, we could not replicate this small effect in five external cohorts, making it unlikely that it is of clinical relevance.
Our study has some limitations which need to be considered. First, we used a candidate approach only investigating the association of two risk variants with AVS. The findings, however, highlight the potential of genetic studies in this disease, especially when potential confounding by CAD is taken into consideration. Second, our study is a cross-sectional analysis not providing longitudinal data. Therefore, future studies are warranted testing further cardiovascular risk variants and their long-term impact on both AVS development and progression. In particular, variants which are associated with blood pressure, cholesterol traits and inflammation may help to further clarify the role of these processes in the pathogenesis of AVS. Our analysis here demonstrates that the GeneCAST cohorts have the strong potential to contribute to such analyses. Finally, our study cohorts were predominantly of Caucasian origin and whether the observed associations hold for other ethnic groups remains to be determined.
In conclusion, our study provides further evidence for a role of variation at the LPA locus in the aetiology of AVS which is distinct from the association of this locus with CAD. Especially, in patients without CAD the genetic contribution of the LPA risk allele and subsequent high Lp(a) levels result in faster AVS progression.
Supplementary Material
Acknowledgments
We acknowledge the support of the Health Informatics Centre, University of Dundee and NHS Tayside Dundee for providing the datasets for GeneCAST Dundee cohorts. We are grateful to nursing and clinical informatics staff in the Leicester NIHR Biomedical Research Centre for recruitment of the GeneCAST Leicester cohort. We thank Dr. Werner Koch for support in genotyping of the GeneCAST Munich cohort. We also thank Anna Helgadoóttir and Hilma Holm for providing us the data from deCODE genetics.
Funding Sources: Collection and genotyping of the GeneCAST Leicester cohorts were supported by the Leicester NIHR Biomedical Centre. NJS and CPN are funded by the British Heart Foundation and NJS is a NIHR Senior Investigator. IRM is supported by a NHS Education for Scotland/Chief Scientist Office Postdoctoral Clinical Lectureship [grant number: PCL17/07]. CCL acknowledges support from the British Heart Foundation [grant numbers: PG/16/32/32132 and PG/14/4/30539]. JGS was supported by the European Research Council, Swedish Heart-Lung Foundation, the Wallenberg Center for Molecular Medicine at Lund University, the Swedish Research Council, the Crafoord Foundation, governmental funding of clinical research within the Swedish National Health Service, Skåne University Hospital in Lund, and the Scania county.
Disclosures: Dr. Koenig reports personal fees from AstraZeneca, Novartis, Pfizer, The Medicines Company, GSK DalCor, Sanofi, Berlin-Chemie Kowa, Amgen, grants and non-financial support from Roche Diagnostics, Beckmann, Singulex and Abbott, all outside the submitted work. All other authors declare no conflict of interest.
References
- 1.Osnabrugge RL, Mylotte D, Head SJ et al. Aortic stenosis in the elderly: disease prevalence and number of candidates for transcatheter aortic valve replacement: a meta-analysis and modeling study. J Am Coll Cardiol 2013;62(11):1002–12. [DOI] [PubMed] [Google Scholar]
- 2.Otto CM, Prendergast B. Aortic-valve stenosis--from patients at risk to severe valve obstruction. N Engl J Med 2014;371(8):744–56. [DOI] [PubMed] [Google Scholar]
- 3.Baumgartner H, Falk V, Bax JJ et al. 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur Heart J 2017;38(36):2739–2791. [DOI] [PubMed] [Google Scholar]
- 4.Kamstrup PR, Tybjaerg-Hansen A, Nordestgaard BG. Elevated lipoprotein(a) and risk of aortic valve stenosis in the general population. J Am Coll Cardiol 2014;63(5):470–7. [DOI] [PubMed] [Google Scholar]
- 5.Smith JG, Luk K, Schulz CA et al. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA 2014;312(17):1764–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchareb R, Mahmut A, Nsaibia MJ et al. Autotaxin Derived From Lipoprotein(a) and Valve Interstitial Cells Promotes Inflammation and Mineralization of the Aortic Valve. Circulation 2015;132(8):677–90. [DOI] [PubMed] [Google Scholar]
- 7.Iung B, Vahanian A. Epidemiology of valvular heart disease in the adult. Nat Rev Cardiol 2011;8(3):162–72. [DOI] [PubMed] [Google Scholar]
- 8.Thanassoulis G, Campbell CY, Owens DS et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med 2013;368(6):503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen V, Cimadevilla C, Estellat C et al. Haemodynamic and anatomic progression of aortic stenosis. Heart 2015;101(12):943–7. [DOI] [PubMed] [Google Scholar]
- 10.Arsenault BJ, Boekholdt SM, Dube MP et al. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective Mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet 2014;7(3):304–10. [DOI] [PubMed] [Google Scholar]
- 11.Cairns BJ, Coffey S, Travis RC et al. A Replicated, Genome-Wide Significant Association of Aortic Stenosis With a Genetic Variant for Lipoprotein(a): Meta-Analysis of Published and Novel Data. Circulation 2017;135(12):1181–1183. [DOI] [PubMed] [Google Scholar]
- 12.Chen HY, Dufresne L, Burr H et al. Association of LPA Variants With Aortic Stenosis: A Large-Scale Study Using Diagnostic and Procedural Codes From Electronic Health Records. JAMA Cardiol 2017. [DOI] [PMC free article] [PubMed]
- 13.Helgadottir A, Thorleifsson G, Gretarsdottir S et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 2018;9(1):987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart BF, Siscovick D, Lind BK et al. Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol 1997;29(3):630–4. [DOI] [PubMed] [Google Scholar]
- 15.Yan AT, Koh M, Chan KK et al. Association Between Cardiovascular Risk Factors and Aortic Stenosis: The CANHEART Aortic Stenosis Study. J Am Coll Cardiol 2017;69(12):1523–1532. [DOI] [PubMed] [Google Scholar]
- 16.Parry HM, Donnelly LA, Van Zuydam N et al. Genetic variants predicting left ventricular hypertrophy in a diabetic population: a Go-DARTS study including meta-analysis. Cardiovasc Diabetol 2013;12:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris AD, Boyle DI, MacAlpine R et al. The diabetes audit and research in Tayside Scotland (DARTS) study: electronic record linkage to create a diabetes register. DARTS/MEMO Collaboration. BMJ 1997;315(7107):524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgadottir A, Gretarsdottir S, Thorleifsson G et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat Genet 2016;48(6):634–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudlow C, Gallacher J, Allen N et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine DM, Sloan BJ, Donner JE, Lorenz JD, Heinzerling RH. Automated measurement of lipoprotein(a) by immunoturbidimetric analysis. Int J Clin Lab Res 1992;22(3):173–8. [DOI] [PubMed] [Google Scholar]
- 21.Rajamannan NM, Evans FJ, Aikawa E et al. Calcific aortic valve disease: not simply a degenerative process: A review and agenda for research from the National Heart and Lung and Blood Institute Aortic Stenosis Working Group. Executive summary: Calcific aortic valve disease-2011 update. Circulation 2011;124(16):1783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke R, Peden JF, Hopewell JC et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med 2009;361(26):2518–28. [DOI] [PubMed] [Google Scholar]
- 23.Sabatine MS, Wasserman SM, Stein EA. PCSK9 Inhibitors and Cardiovascular Events. N Engl J Med 2015;373(8):774–5. [DOI] [PubMed] [Google Scholar]
- 24.Viney NJ, van Capelleveen JC, Geary RS et al. Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet 2016;388(10057):2239–2253. [DOI] [PubMed] [Google Scholar]
- 25.Langsted A, Nordestgaard BG, Benn M, Tybjaerg-Hansen A, Kamstrup PR. PCSK9 R46L Loss-of-Function Mutation Reduces Lipoprotein(a), LDL Cholesterol, and Risk of Aortic Valve Stenosis. J Clin Endocrinol Metab 2016;101(9):3281–7. [DOI] [PubMed] [Google Scholar]
- 26.Afshar M, Kamstrup PR, Williams K, Sniderman AD, Nordestgaard BG, Thanassoulis G. Estimating the Population Impact of Lp(a) Lowering on the Incidence of Myocardial Infarction and Aortic Stenosis-Brief Report. Arterioscler Thromb Vasc Biol 2016;36(12):2421–2423. [DOI] [PubMed] [Google Scholar]
- 27.Cowell SJ, Newby DE, Prescott RJ et al. A randomized trial of intensive lipid-lowering therapy in calcific aortic stenosis. N Engl J Med 2005;352(23):2389–97. [DOI] [PubMed] [Google Scholar]
- 28.Rossebo AB, Pedersen TR, Boman K et al. Intensive lipid lowering with simvastatin and ezetimibe in aortic stenosis. N Engl J Med 2008;359(13):1343–56. [DOI] [PubMed] [Google Scholar]
- 29.Chan KL, Teo K, Dumesnil JG, Ni A, Tam J, Investigators A. Effect of Lipid lowering with rosuvastatin on progression of aortic stenosis: results of the aortic stenosis progression observation: measuring effects of rosuvastatin (ASTRONOMER) trial. Circulation 2010;121(2):306–14. [DOI] [PubMed] [Google Scholar]
- 30.Nadir MA, Wei L, Elder DH et al. Impact of renin-angiotensin system blockade therapy on outcome in aortic stenosis. J Am Coll Cardiol 2011;58(6):570–6. [DOI] [PubMed] [Google Scholar]
- 31.Bull S, Loudon M, Francis JM et al. A prospective, double-blind, randomized controlled trial of the angiotensin-converting enzyme inhibitor Ramipril In Aortic Stenosis (RIAS trial). Eur Heart J Cardiovasc Imaging 2015;16(8):834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Brien KD, Shavelle DM, Caulfield MT et al. Association of angiotensin-converting enzyme with low-density lipoprotein in aortic valvular lesions and in human plasma. Circulation 2002;106(17):2224–30. [DOI] [PubMed] [Google Scholar]
- 33.Wakabayashi K, Tsujino T, Naito Y et al. Administration of angiotensin-converting enzyme inhibitors is associated with slow progression of mild aortic stenosis in Japanese patients. Heart Vessels 2011;26(3):252–7. [DOI] [PubMed] [Google Scholar]
- 34.Schunkert H, Gotz A, Braund P et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation 2008;117(13):1675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.