Abstract
The serotonin 1A receptor (5-HT1A) system has been extensively implicated in modulating mood and behavior. Notably, 5-HT1A levels in humans display remarkable variation, and differences in receptor levels have been linked with a variety of psychiatric disorders. Further, reduction of receptor levels by 30–50% in mice suggests that changes in receptor levels that model existing human variation are sufficient to drive behavioral alterations. As a result, genetic mechanisms that modulate human 5-HT1A levels may be important for explaining individual differences in mood and behavior, representing a potential source of psychiatric disease risk. One common genetic variant implicated in differential 5-HT1A levels is the G/C single nucleotide polymorphism (SNP) rs6295, located upstream of the human 5-HT1A gene. This SNP differentially binds the transcription factor, NUDR/Deaf1, leading to cell-type specific effects on transcription in vitro. To investigate the direct effects of this SNP in the heterogeneous cellular context of the brain, we generated humanized transgenic mice using a design that maximized the local transcriptional landscape of the human HTR1A gene while also controlling for effects of genomic insertion location. We integrated a 180 kb human bacteria artificial chromosome (BAC) transgene containing G- and C-alleles of rs6295 flanked by FRT or loxP sites. Subsequent deletion of each allele by Cre- or Flp-recombinase resulted in rs6295G and C alleles in the same genomic location. These alleles were bred onto a 5-HT1A null mouse such that the human BAC was the sole source of 5-HT1A in these mice. We generated three separate lines, two of which had detectable human 5-HT1A levels in the brain, although none displayed expression in the raphe. Of these, one line exhibited rs6295-dependent differences in 5-HT1A levels and differences in behavior, even though the overall levels were considerably lower than native expression levels. The linedependent effect of rs6295 on protein levels and behavior may depend upon differences in background genetic factors or different insertion sites across each line. This work confirms that relatively subtle differences in 5-HT1A levels can contribute to differences in behavior and highlights the challenges of modeling human noncoding genetic variation in mice.
Keywords: rs6295; 5-HT1A; HTR1A; humanized; genetic variation; depression, anxiety; BAC transgenic
Graphical Abstract
INTRODUCTION
Serotonergic systems have been extensively implicated in mood and anxiety disorders, which represent the most common and costly psychiatric disorders. Convergent evidence from humans and animal models suggests that the serotonin 1a receptor plays a primary role in modulating the effect of serotonin on stress and anxiety-related behaviors. Clinically, 5-HT1A agonists, such as buspirone, are prescribed as anxiolytics and as augmentation for antidepressants.1,2 Likewise, pharamacological and genetic manipulations of the 5-HT1A system in rodents alters anxiety and depression-related behaviors.3,4 One particularly striking feature of the 5-HT1A system is that relatively small reductions in receptor levels (10–40%) in particular brain regions are sufficient to alter behavior.5–7 For instance, a selective 32% decrease in 5-HT1A autoreceptor levels in the raphe in adulthood, which mimics the natural variation in receptor levels observed in humans,8 increases both resilience to stress and likelihood of SSRI responsiveness.5,7 Likewise, a ∼40% decrease selectively during postnatal development increases later life anxiety-related behavior.6 This suggests that the 5-HT1A system represents a sensitive substrate in which variation in receptor levels can tune behavior. As such, factors that alter 5-HT1A levels may shape individual differences in behavior and psychiatric disease risk.
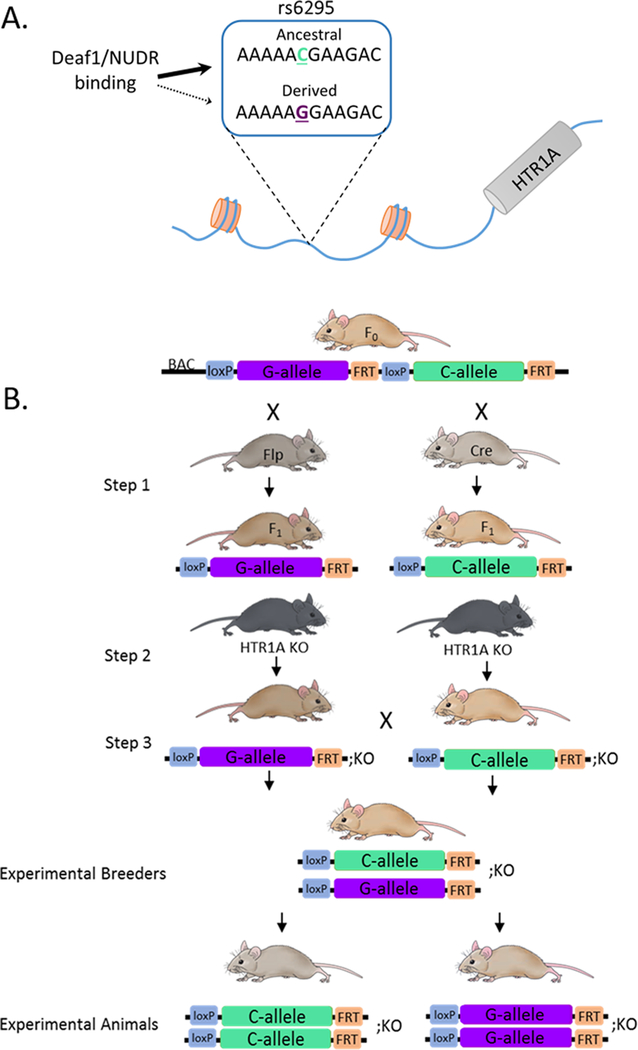
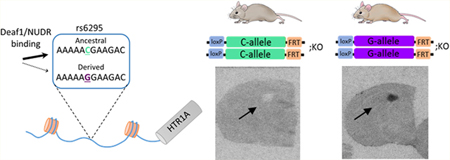
One potential genetic factor that has been implicated in modulating human 5-HT1A levels is a common G/C single nucleotide polymorphism (SNP; rs6925) located 1019 base pairs (bp) upstream of the serotonin 1a receptor (HTR1A) translation start site (Figure 1A).9 A number of gene association studies have found that the rs6295 G-allele is associated with elevated incidence of suicide attempts and depression and reduced antidepressant responsiveness, although replication of these effects have not been completely consistent.10–13 Beyond these epidemiological findings, there is evidence that rs6295 is associated with differences in 5-HT1A levels assessed via PET imaging and in post-mortem tissue samples,10,14 and in vitro work suggests that it may functionally alter 5-HT1A receptor levels in a cell-type specific fashion through altered binding of transcription factors.9,15,16 Specifically, the derived G-allele fails to bind the transcription factor NUDR/Deaf1.15,17 This leads to greater reporter expression from the G-allele in raphe-derived neurons where NUDR/Deaf1 acts as a repressor, but lower levels in forebrain-derived neurons where it acts as a transcriptional activator.15,17 This leads to a model in which the C-allele drives higher levels of 5-HT1A in forebrain neurons but lower levels in raphe neurons,9 a model that is supported by recent work in post-mortem human tissue.10
Figure 1.
Generation of rs6295 humanized mice. (A) The common G/C single nucleotide polymorphism rs6295 is located upstream of the human HTR1A gene. The transcription factor Deaf1/NUDR binds the C allele but does not bind the G allele. (B) Humanized mice were generated by inserting into the mouse genome a modified bacteria artificial chromosome (BAC) consisting of 180 kb of human DNA containing a duplicated human HTR1A gene containing each allele of rs6295. The rs6295G and rs6295C alleles were isolated by crossing the founder mouse to Cre- and FlpO-recombinase expressing mice (step 1). The offspring of this cross were then bred onto a mouse HTR1A−/− (KO) background (step 2) so that the only source of 5-HT1A in these mice was from human HTR1A locus. Three separate lines were generated.
Despite the strong convergent evidence that rs6295 alters 5-HT1A levels and is associated with psychiatric disease phenotypes, the direct effects of this SNP within the heterogeneous context of the brain remain unknown. To address this, we generated a humanized mouse model of rs6295 by inserting a 180 kb bacteria artificial chromosome (BAC) containing the human HTR1A gene into the mouse genome, an approach designed to include relatively distal regulatory elements (Figure 1B). These mice have three important characteristics: (1) the rs6295G and C alleles are present in the same genomic location, (2) we controlled for linked genetic variation in the humanized locus by exclusively varying rs6295, and (3) the only source of 5-HT1A in these animals comes from the human locus. We used this model to investigate the hypothesis that rs6295 directly modulates 5-HT1A levels in a brain region specific manner and that these differences in 5-HT1A levels contribute to differences in stress-coping and anxiety-related behaviors. We found that independent insertion sites led to distinct patterns of 5-HT1A expression in these mice and that the effects of rs6295 are dependent upon genomic insertion site or genetic background or both. This highlights the complexity of trying to humanize a mouse model to study noncoding variants but supports the hypothesis that rs6295 can directly modulate 5-HT1A levels and confirms that relatively subtle alterations in 5-HT1A levels contribute to behavior.
RESULTS AND DISCUSSION
Characterization of Human (h)5-HT1A Distribution and Function in rs6295GC Mice.
We used a BAC-transgenic strategy to examine the functional effects of rs6295 in the brain. BAC vectors accommodate dispersed regulatory sequences across relatively large regions of the genome (up to hundreds of kb).18 This strategy addressed a number of limitations of previous post-mortem human studies and provided the opportunity to examine the effects of rs6295 within the heterogeneous cellular context of the mammalian brain.
5-HT1A is widely expressed in the human and mouse brain, and many of the critical regulatory elements for the expression of HTR1A are well described and conserved between mice and humans.19 In addition, the general distribution of 5-HT1A receptors is similar between humans and mice, with enrichment in corticolimbic regions, the raphe, and parts of the hypothalamus.20–22 As such, we anticipated that expression patterns from our BAC would at least partially recapitulate human and mouse expression patterns, acknowledging that patterns of transcription factor expression might differ between species. Specifically, the BAC transgenic approach has previously been used to recapitulate region-specific gene expression patterns across hundreds of mouse genes in the GENSAT project, which reported that screening 3–4 founders produces an accurately expressing line in ∼85% of the vectors, although the cellular specificity of BAC-transgene expression was not rigorously investigated in most cases.23 In addition, multiple examples of faithful patterns of gene expression have previously been demonstrated for other humanized genetic loci, albeit largely in non-neural tissues.24 Thus, it was surprising that of the 3 founders generated, each line exhibited distinct patterns of 5-HT1A expression, ranging from no adult expression in one line and weak to moderate levels in the other two lines. All three lines generated appeared to contain complete or nearly complete BACs (Supplementary Figure 2), although it was substantially more difficult to amplify the BAC in line B.
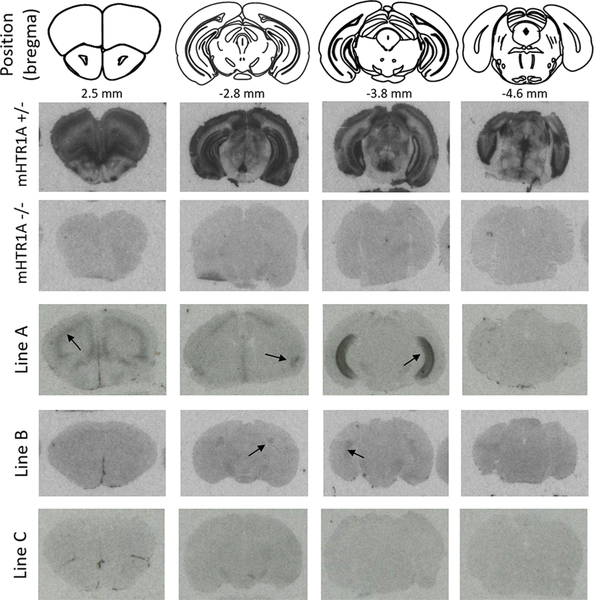
We employed hHTR1A, rs6295XX, and h5-HT1A to indicate the human HTR1A gene, rs6295 genotype, and protein, respectively, and mHTR1A/m5-HT1A to indicate the mouse gene/protein. rs6295GC is the most common genotype in human populations, so we used this genotype to compare our humanized lines (rs6295GC; mHTR1A−/−) to mHTR1A+/− and mHTR1A−/− mice. We mapped the location of 5-HT1A receptors in each line via I125-MPPI autoradiography. This revealed distinct patterns of h5-HT1A in each of our lines (Figure 2). Line B exhibited very low levels of h5-HT1A that were limited to regions of the hippocampus and claustrum, while line A had higher levels of h5-HT1A that were observable in the hippocampus, amygdala, and prefrontal cortex. Line C was indistinguishable from mHTR1A−/− animals. Due to this lack of h5-HT1A expression, we did not proceed with further analysis of this line.
Figure 2.
Human 5-HT1A expression varies across transgenic lines. Autoradiography revealed different expression patterns across the three lines created, most likely due to insertion effects of the BAC. Representative images from each line are shown, as well as from mHTR1A−/+ and mHTR1a−/− mice. Arrows indicate visually detectable h5-HT1A expression.
Epigenetic silencing of transgenes has previously been observed in mice and is influenced by insertion location, copy number, and transgene sequence.25 However, BAC transgenes are, in general, less prone to silencing, which is thought be attributable to the long flanking sequences.26 For short multicopy transgenes, tail-to-tail orientation and copy number are both predictive of epigenetic silencing.27 While integration site effects likely explain differences in expression and silencing across lines, we did not observe marked differences in transgene expression between F3 rs6295GC (Figure 1) and F4–F6 rs6295GG or CC mice (Figure 2), although it was not possible to compare these generations to the earliest ones because of our breeding strategy; the hHTR1A transgene was not on a mHTR1A −/− background until F3.
We also verified that there is a lack of functional h5-HT1A in the raphe of our transgenic lines. The 5-HT1A agonist 8-OH-DPAT induces hypothermia in mice. This physiological effect is mediated by autoreceptors located on serotonergic neurons in the raphe.28 We found that 8-OH-DPAT failed to elicit a hypothermic response in any of our humanized lines (Supplementary Figure 3). Because expression from the hHTR1A locus was considerably lower than from the mHTR1A locus, we compared our humanized animals to mHTR1A+/− animals, enabling us to use a higher dose of 8-OH-DPAT and maximizing the likelihood of identifying an effect. 8-OH-DPAT induced hypothermia in m5HT1A+/− mice, but mHTR1A−/− mice with one or two rs6295 alleles were indistinguishable from mHTR1A−/− mice for both line A (n = 6–13/grp) and line B (n = 6–18/grp). Full details from statistical analyses are available in Supplementary Table 4. As a result, we concluded that line A and B do not express functional h5-HT1A autoreceptors, a result that mirrors the lack of visually identifiable receptors in the raphe in Figure 2.
h5-HT1A Levels in rs6295CC and rs6295GG Mice.
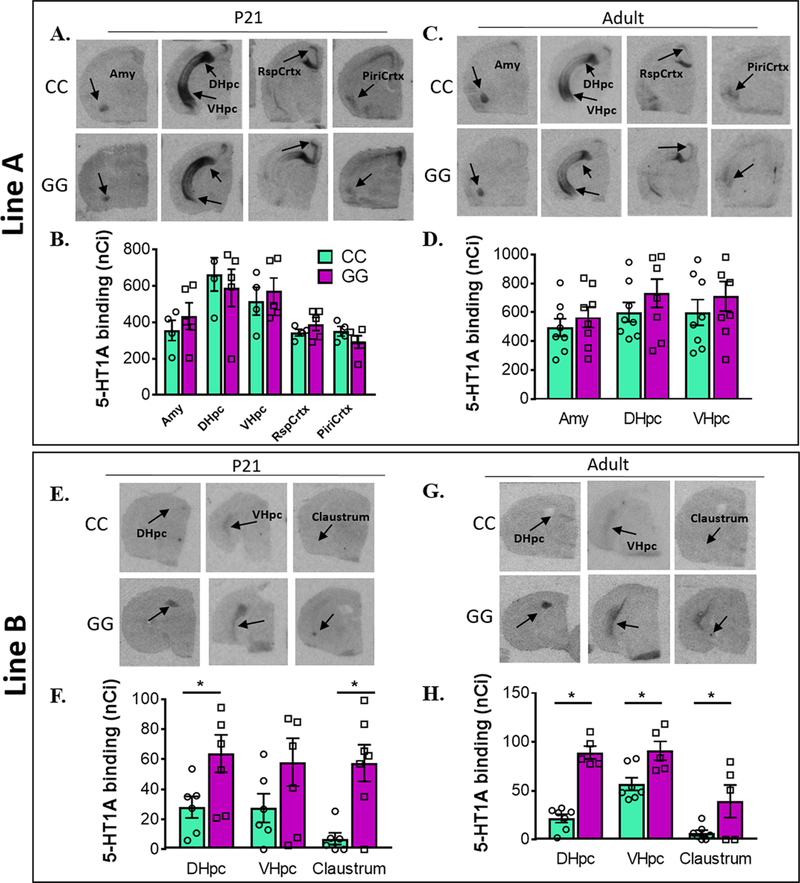
We used I125-MPPI autoradiography to investigate potential differences in h5-HT1A levels in rs6295CC and rs6295GG mice. Because small changes in m5-HT1A levels during development are known to have long lasting effects into adulthood, we examined levels at two developmental time points, postnatal day (P) 21 and P60–75.6 Lines A and B both exhibited h5-HT1A in the ventral hippocampus but differed in detectable receptor expression in the claustrum, cortex, and amygdala. In addition, relative levels of expression differed dramatically between the lines, with receptor levels in line B approximately 1/10th of those observed in line A (Figure 3). When we compared GG and CC animals within each line, we found that line B exhibited rs6295-dependent differences in h5-HT1A, while line A did not (Figure 3). Specifically, line B P21 rs6295GG animals had higher h5-HT1A protein levels compared to rs6295CC animals in the dorsal hippocampus/subiculum (CC = 7, GG = 6, p = 0.001) and claustrum (CC = 4, GG = 6, p = 0.001). In adulthood, line B rs6295GG animals had higher h5-HT1A receptor levels in the dorsal (CC = 7, GG = 5, p = 0.001) and ventral hippocampus (p = 0.035) and the claustrum (CC = 5, GG = 3, p = 0.007). Because these differences were evident during adolescence and adulthood, it suggests that they may be relatively stable across development.
Figure 3.
Differences in receptor density between genotypes is line specific. (A–D) Autoradiography images (A, C) and quantification (B, D) of h5-HT1A expression in rs6295 GG and CC animals from line A at P21 (A, B) and in adulthood (C, D). No significant differences in h5-HT1A levels were detected in any brain region at either time point. (E–H) Autoradiography images (E, G) and quantification (F, H) of h5-HT1A expression in rs6295 GG and CC animals from line B at P21 (E, F) and in adulthood (G, H). At P21, GG mice have higher levels of h5-HT1A in the dorsal hippocampus (DHpc) (p = 0.001) and the claustrum (p = 0.001), but no difference in the ventral hippocampus (VHpc) (p = 0.5290). In adulthood, GG animals have higher h5-HT1A expression in the DHpc (p = 0.0001), VHpc (p = 0.0350), and claustrum (p = 0.0070). Group sizes and statistical analyses available in Supplementary Table 4.
hHTR1A mRNA Expression.
To explore if the differences observed in 5-HT1A receptor protein are accompanied by differences in hHTR1A mRNA, we used quantitative-PCR (qPCR) to examine the expression of hHTR1A using mGAPDH as a control gene. We also examined expression of mDeaf1 as a potential confounding factor, reasoning that differences in mDeaf1 levels, as an important transcription factor involved in HTR1A expression, could contribute to differences in hHTR1A mRNA. We did not observe any main or interacting effects of genotype (details available in Supplementary Table 4; n = 5–9/grp). In line A, the only significant effect observed was a main effect of brain region on hHTR1A mRNA at p21 (p < 0.0001). In line B, there was a main effect of brain region at both time points (p < 0.05) and a main effect of sex at P21 (p = 0.009) on hHTR1A (Supplementary Figure 4). Deaf1 mRNA also differed as a function of brain region (p < 0.05, line A P21 and P60, line B P21 only), but we did not observe any effect of genotype, sex, or their interaction.
While it was surprising that differences in detectable receptor protein levels were not reflected at the level of the mRNA, it is worth noting that levels of HTR1A mRNA in these samples are quite low, often amplifying with a Ct > 30, which may result in less accurate quantitation. In addition, although hHTR1A mRNA is detectable in the PFC of line B via qPCR, the receptor protein is not detectable above background in this area when using autoradiography. This could be explained by trafficking of the receptor protein to terminals in another brain region or by a failure to insert into the membrane. Regardless, it suggests that there is not a direct correspondence between mRNA and protein levels at the tissue level, although we did not examine this at the individual cell level.
rs6295-Mediated Behavioral Differences.
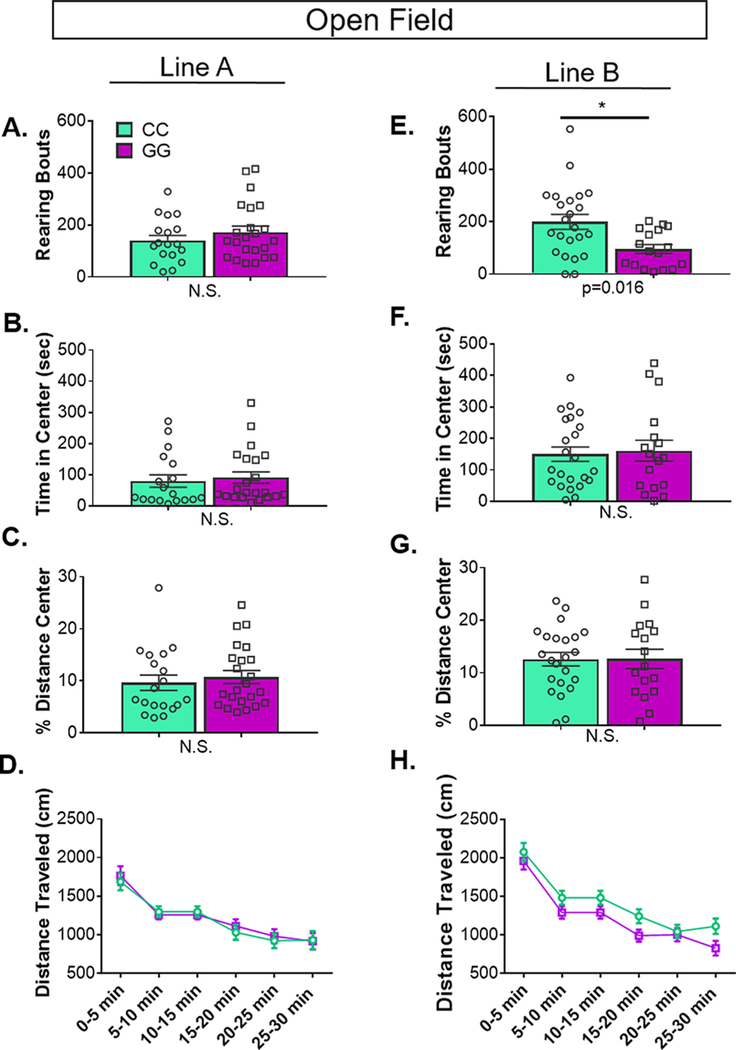
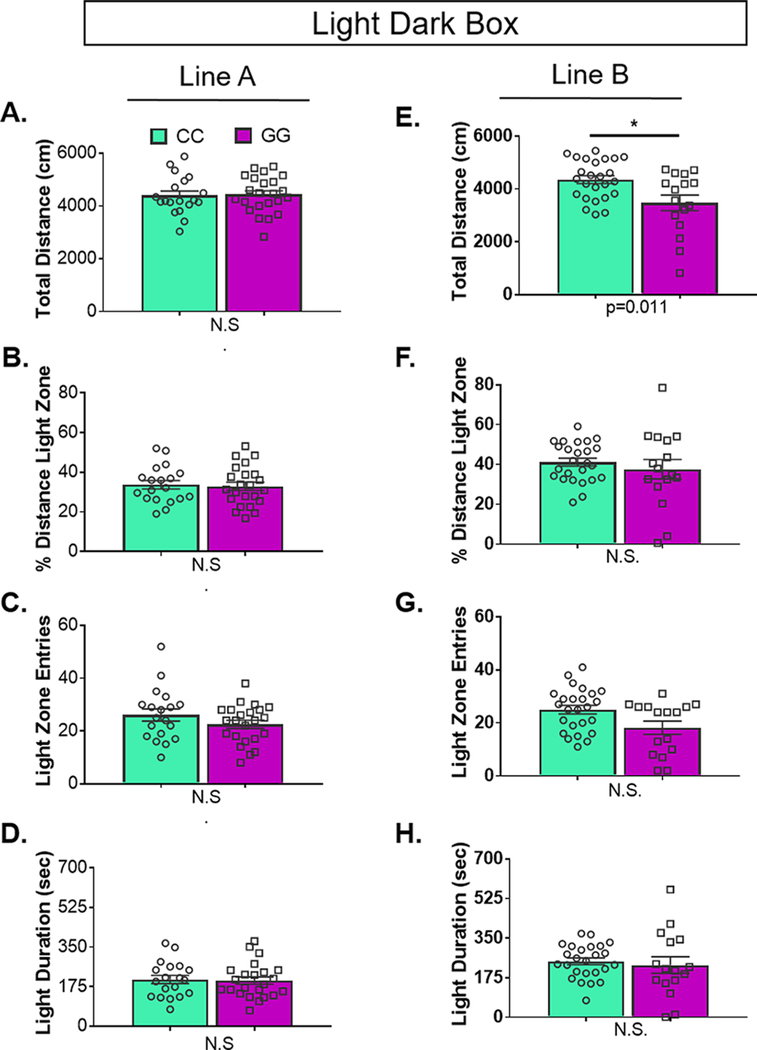
As the receptor protein is a better indicator of putative functional differences than the mRNA, we examined whether rs6295-mediated differences in h5-HT1A corresponded with differences in anxiety or stress-coping, as detailed below (n = 17–24/grp). We found that, mirroring receptor levels as detected by autoradiography, there were no genotype-dependent differences in line A, but line B exhibited subtle differences in anxiety-related and locomotor behaviors. In particular, line B GG animals reared ∼20% less in the open field and displayed ∼50% decreased locomotion in the light–dark box (Figure 3 and 4). Previous studies of transgenic lines with reduced or absent 5-HT1A receptors have observed a 50–75% decrease in locomotion in the open field and a ∼ 60% reduction in rearing, although there is significant variation in the reported literature, potentially reflecting differences in testing conditions or differences in genetic background across independently generated transgenic lines.3,4,6,7,29,30 No genotype-dependent differences were observed in either line for depression-related tests. Together, these data indicate continuity; despite extremely low levels of receptor expression, in instances in which we observed SNP-dependent differences in h5-HT1A, there is a corresponding, albeit subtle difference in behavior. This further confirms the sensitivity of 5-HT1A-mediated behaviors to small differences in receptor levels. Full results of statistical analyses are available in Supplementary Table 4.
Figure 4.
rs6295-dependent differences in rearing in open field are line specific. In open field, line A had no statistically significant difference in (A) rearing, (B) time spent in the center, or (C) percentage distance traveled in the center between genotypes. (D) Likewise, there was no main or interacting effect of genotype on distance traveled over time. (E) In line B, CC animals reared more than GG animals (p = 0.016). There were no differences in other measures of anxiety, specifically (F) time spent in the center or (G) percentage distance traveled in the center between genotypes. (H) There was no main or interacting effect of genotype on distance traveled over time (group sizes and statistics available in Supplementary Table 4). Values represent mean ± SEM.
Open Field.
rs6295CC animals reared more than rs6295GG animals only in line B (Figure 4; line B, p = 0.016). No differences were evident in the percent distance traveled in the center, the time in center, or the total distance traveled. As previously observed, locomotion decreased with time (line A, p < 0.00001; line B, p < 0.00001), independent of genotype.
Light–Dark Box.
Two animals in line B failed to enter the dark chamber and were omitted from analysis for failure to complete the test. Because data from line B violated the assumption of equal variances, we used a regression to test if genotype significantly predicted behavior. The results of the regression indicate that two predictors explained 22.5% of the variance (R2 = 0.22, p = 0.012). Genotype significantly predicted total distance traveled (CC > GG, β = −0.445, p = 0.011) (Figure 5). Genotype did not significantly predict percent distance traveled in light, light zone entries, or total time in the light chamber. We analyzed line A using an ANOVA. There were no genotype differences for total distance traveled, percent distance traveled in light, light zone entries, or total time in the light chamber.
Figure 5.
Genotype-dependent differences in light–dark box behaviors is line specific. (A–D) In line A, there were no differences between genotypes in any of the metrics examined. In contrast, in line B, rs6295CC animals traveled farther (p = 0.011). There were no differences in (G) light zone entries, (F) the percent distance traveled in the light, or (H) light zone duration between genotypes. Values represent mean ± SEM. Group sizes and statistics are available in Supplementary Table 4.
Zero Maze.
There were no genotype-dependent differences in performance on the zero maze (Supplementary Figure 5). Specifically, rs6295CC and GG animals did not differ in the percent time in the open area or the number of entries into the open areas.
Novelty Suppressed Feeding.
There was no difference in rs6295CC and rs6295GG mice in the latency to eat food in the novelty suppressed feeding test (Supplementary Figure 6). Likewise there was no main effect of genotype for latency to eat in the home cage, the amount eaten, or weight loss.
Forced Swim Test.
Forced swim test was performed on two consecutive days (Supplementary Figure 7). Previous studies suggest that the second test may be more sensitive for detecting antidepressant effects in both rats and mice.7,31,32 As has been previously observed, there was a significant main effect of time on float duration on day 1, with increased floating later in the test for both lines (line A, p < 0.0001; line B, p < 0.0001). However, there was no genotype-dependent differences in floating in forced swim test on either line.
Tail Suspension Test.
There was no difference in time spent immobile between rs6295CC and rs6295GG mice in tail suspension test (Supplementary Figure 8). In line A, there was no difference in immobile time between genotypes in either line.
The behavioral differences observed are quite subtle. It is possible, that this reflects a binary presence or absence of 5-HT1A. However, when considered within the context of other studies, this seems unlikely. For instance, there are a number of studies that have partially decreased 5-HT1A levels, and these studies observed behavioral changes within a similar magnitude.5–7 Together with the present study, this would suggest that the 5-HT1A system has the capacity to serve as a behavioral rheostat, modulating traits related to mood and anxiety rather than acting in an all-or-none capacity.
Although the direction of our behavioral effects are consistent with epidemiological evidence suggesting that the rs6295G allele is a risk allele, the relationship between our observed 5-HT1A differences and behavior are not immediately evident. It is not clear which cell types are expressing 5-HT1A in our transgenic mice, but the ventral hippocampus has been broadly implicated in modulating anxiety,33,34 and depending upon the cell type expressing 5-HT1A, differences in receptor levels could modulate anxiety by altering hippocampal activity. Alternatively, hippocampal 5-HT1A signaling during early postnatal development is required for normal synaptogenesis, and differences in anxiety could reflect long-term impacts of genotype-dependent differences in hippocampal development.35
One unanticipated result of this study is that rs6295 modulates h5-HT1A and behavior in only one of our lines. The rs6295G and C allele were in the same genomic location within each line. However, at least two factors differed across lines, which may explain the difference in penetrance of rs6295 between line A and B. First, each line had a different genomic insertion site. It is possible that differences in insertion site and accompanying genomic linkages could lead to a different subset of transcription factors being used to transcribe hHTR1A in each line. If transcription is mediated by factors that differ in their sensitivity to rs6295 (e.g., if transcription is Deaf1 dependent only in line B), then this could explain SNP effects occurring exclusively in line B. Alternatively or additionally, differences in penetrance could be explained by epistatic interactions due to different genetic backgrounds across each line. Each B6SJL F1 hybrid founder mouse was bred to Cre- and Flp-expressing mice and then bred to a mHTR1A−/− line, which also contained a mixed background. Each breeding step resulted in offspring carrying a random 50% of each parental genome. Thus, genetic assortment from different genetic backgrounds at each breeding step (Figure 1) resulted in different genetic backgrounds between our lines, potentially explaining the differential penetrance of rs6296. Background genetic effects have been shown to moderate the penetrance of other genetic variants. For example, a serotonin transporter (SERT) Ala56 genetic variant that is overtransmitted in autism, causes multiple biochemical, physiological, and behavioral changes in mice with a 129S6/S4 genetic background,36 but these phenotypes are not evident when the same allele is bred onto a C57Bl/6 background.37 Likewise, genetic background modifies Alzheimer’s disease-associated symptoms in mice carrying the 5XFAD transgene.38 While we cannot distinguish between background genetic effects and differences in transcription complexes across lines, we were able to rule out differences in Deaf1 expression as a potential explanation for line differences in expression of hHTR1A (Supplementary Figure 4).
Advances in transcriptomic, genomic, and genome architecture approaches have expanded the number of putative functional genetic variants implicated in the molecular pathophysiology of psychiatric illness, many of which appear to be within regulatory regions.38–41 However, a mechanistic understanding of these variants will require experimental approaches, such as those employed here to investigate rs6295. While in vitro gene expression and transcription factor binding assays are essential for determining whether a genetic variant, such as a SNP, can affect gene expression, these assays have limited utility for extrapolating how a variant may affect expression in vivo. rs6295 is an excellent example of this; the Gallele increases transcription in some cell lines but decreases it in others.10,15–17,19 Thus, there is a clear need to model functional variants, such as rs6295, in a way that reveals their effects on brain development, function, and behavior.
Our results highlight a number of considerations when pursuing functional investigation of putative genetic variants. Genetic background, integration site effects, and potential species differences in transcription factor expression and binding can all be mitigated by examining genetic variation in human induced pluripotent stem cells (iPSCs) or organoids. Although not widely available when we initiated these experiments, advances in CRISPR/Cas9-mediated genome engineering enable the modification of a single genetic variant, enabling comparison of allelic effects on the same genetic background and in a proper genomic location.42–44 While this approach is powerful and biologically informative, it falls short of providing a model in which genotype–phenotype relationships can be examined at a behavioral level. Further, human populations are not genetically identical, requiring assessment of multiple cell/organoid lines in order to estimate the effects of epistatic interactions on the resulting phenotype.
Thus, mouse models remain the most tractable option for examining behavioral effects of human noncoding variants. However, based on our results, we would recommend introducing the genetic variant into the correct genomic location. This can be achieved through homologous recombination of a BAC transgene into the mouse genome, either at the endogenous locus or at commonly used integration site, such as the ROSA or HPRT loci.40,45 While feasible, this approach can be difficult, costly, and time-consuming. In addition, recent advances have taken advantage of selective breeding designs to better assess the effects of genetic background on allelic penetrance, an approach that has promise for enhancing the translatability of humanized mouse models.38 However, this approach relies on F1 hybrids and requires a single, dominant disease-associated variant. As we sought to model existing biallelic human genotypes, this approach was not feasible with our study design. Alternatively, backcrossing onto a known genetic background may provide a way to control for genetic background; optimally this should be done on at least two different backgrounds in order to examine whether epistatic interaction can affect penetrance.
Finally, moving transgenes between species will always present challenges, as it can be difficult to predict whether the mouse transcription complexes will interact with the human allele the same way the human complexes do. Thus, in some instances, it may be advantageous to modify the endogenous mouse sequence rather than attempting to fully humanize a locus. With this in mind, we explored whether an analogous SNP could be introduced into the native mHTR1A promoter region. There are two putative Deaf1 binding sites in the mHTR1A locus, and previous work suggests one of them is functional.46 The sites do not exhibit direct homology with the human locus, and as a result, there is not a clear single nucleotide change that would model rs6295. Instead, we tested a few different putative mutants, comparing their effects on expression with the rs6295G and C allele.16 As detailed in Phillipe et al.,16 none of our mutations resulted in consistent replication of the effects of rs6295G allele across different cell types in vitro. Thus, mutating the endogenous mouse Deaf1 binding site(s) as a model for rs6295 would have significant limitations.
In sum, our approach had a number of advantages, and modified versions of our dual Cre–Flp strategy (Supplementary Figure 1) are an innovative way to ensure that two variants or haplotypes can be directly compared. As used here, this approach highlights the complexity of modeling human cis-regulatory differences in mice, an important consideration as the majority of GWAS “hits” fall in noncoding genomic regions. Importantly, our results suggest a strong need to investigate potential gene variant effects within an appropriate genomic context, preferably through homologous recombination at the locus-of-interest. Despite this, our data do support a role for rs6295 in directly modulating receptor levels and further support the finding that subtle differences in 5-HT1A levels can be behaviorally meaningful, making the 5-HT1A system an ideal system for investigating mechanisms that contribute to individual differences in receptor levels in order to further our understanding of psychiatric disease risk.
METHODS
Generation of rs6295 Humanized Mice.
We generated transgenic humanized mice carrying a genomically integrated BAC (bacterial artificial chromosome) of ∼180 kb containing a duplicated version of the human HTR1A gene flanked by FRT and loxP sites. The cloning and recombineering strategies employed to accomplish this are diagramed in Supplementary Figure 1. BAC no. RP11–158J3 (BACPAC Resources) was selected as it had no other protein-coding genes and HTR1A is located in the middle. It was electroporated into SW102 cells for subsequent recombineering (National Cancer Institute, Biological Resources Branch).47,48 There is a loxP site in the RP11 backbone, so we used p23loxZeo (gift from Dr. Kenji Tanaka) to replace the loxP site with a zeocin resistance cassette. Next, we replaced the HTR1A gene-containing-region (6054 bp) with a rpsL+−kana marker (Addgene no. 20871) containing a positive kanamycin resistance marker and a negative streptomycin-sensitive marker.49 The 5′ (ATG – 1944 bp) and 3′ (ATG +4110 bp) ends of this region were chosen because they were the closest regions with low homology outside of known regulatory regions and the 3′ UTR. In parallel, a plasmid containing a duplicated version of the same HTR1A-containing region was also generated. The initial rs6295G allele flanked by a SalI-loxP site (5′ end) and an FRT-Xho1 site (3′ end) with 100 bp flanking homology domains was generated via gene synthesis and cloned into pUCminusMCS (pUCrs6295G, Blue Heron Biotechnology). This plasmid was modified via targeted mutagenesis to alter the G-allele to rs6295C, resulting in identical sequences except for rs6295. pUCrs6295G was linearized via Sal1 and the rs6295C allele was removed from its backbone via Sal1 and Xho1 digestion. This was then ligated together to produce pUCrs6295C:rs6295G, which was then recombined into the BAC, replacing the rpsL+−kana marker. Integrity of HTR1A, loxP, and FRT sites were confirmed via Sanger sequencing.
Three BAC-transgenic founder mice on a B6SJL F1 hybrid background were generated at the Duke Neurotransgenic Facility and shipped to Columbia University. Upon receipt, founders and their offspring underwent successive breeding. Rates of transgene inheritance in F1 animals are shown in Supplementary Table 1. The founder mice were crossed sequentially to Cre (B6.Cg-Edil3Tg(Sox2‑cre)1Amc/J; JAX no. 008454) and FlpO (B6.Cg-TG(Pgk1-flpo)10Sykr/J; JAX no. 011065) recombinase-expressing mice, which resulted in F1 generation mice who had one copy of the rs6295G allele or one copy of the rs6295C allele, respectively. Excision was confirmed via a lack of overlapping peaks in Sanger sequencing reaction and Taqman SNP genotyping (assay no. 4351379; ThermoScientific Fisher). F1 mice were bred onto an mHTR1A KO (−/−) generated from 5HT1A flox mice.50 F2 mice were bred back to each other, resulting in F3 rs6295GC; mHTR1A−/− animals for breeding experimental animals. F3 rs6295GC; mHTR1A−/− animals were bred together to generate rs6295GG and CC experimental mice (Figure 1). Throughout the rest of this manuscript, rs6295GG or CC mice are on a mHTR1A−/− background unless otherwise specifically stated.
Husbandry.
Mice were housed three to five per cage on a 12-h dark–light cycle (06:00–18:00) at 20–25 °C. Ad libitum food and water were provided. All behavioral experiments took place during the light cycle. All animal procedures were approved by Columbia University’s and University of Colorado’s Institutional Animal Care and Use Committee.
Verification of BAC Integrity.
In order to determine whether the entire BAC integrated into the genome, we developed a series of 19 primer pairs designed to amplify small fragments tiled every ∼10 kb across the entire BAC (Supplementary Table 2). Each reaction consisted of 1 μL of primer pair (10 pmol), 0.5 μL of DNA, 0.2 μL of Taq DNA polymerase, 3.3 μL of ddH2O, and 5μL of premix B (Epicenter Biotechnologies) with reaction conditions 95 °C for 1 min; 35 × (95 °C for 15 s, 59 °C anneal for 30 s, 72 °C for 1 min). Bands were resolved on a 3% agarose gel and compared with a positive control PCR amplified from purified BAC-only DNA.
Genotyping.
Toes were taken at postnatal day 7 and digested using 97 μL of DirectPCR Lysis Reagent-Mouse Tail (Viagen Biotech) and 3 μL of proteinase K (20 mg/μL Gold Bio Technology Inc.). In order to identify mHTR1A−/− mice, genotyping was performed using primers 5′-CACTTGGTAGCTGAAGGTCACG, 5′-TAGCTGGAGCCT-CTGAGCGC, and 5′-GTACCGGGCGAAGCACTGC. Each reaction consisted of 1 μL of primer pair (10 pmol), 1 μL of DNA, 1.5 μL of ddH2O, and 5μL of Econotaq (Lucigen Corporation) with reaction conditions 95 °C for 1 min, 95 °C for 15 s, 64 °C for 15 s, 35 × (95 °C for 15 s, 59 °C anneal for 30 s, 72 °C for 1 min). Bands were resolved on a 3% agarose gel.
rs6295 genotyping was performed using taqman SNP genotyping (assay no. 4351379; ThermoScientific Fisher) on a Applied Biosystems QuantStudio 3 qPCR machine. Samples were genotyped in duplicate in MicroAmp Fast Optical 96 well Reaction Plate (Applied Bio Systems) in a reaction containing 5 μL of DNA, 0.3 μL of probe SNP Genotyping Assay, 3.4 μL of Nuclease free H2O, 4.3 μL of Taqman Fast master mix (Life Technologies), and 2 μL of ddH2O per well for a total volume of 15 μL. The cycling conditions were preholding stage 60 °C for 30 s, holding stage 95 °C for 10 min, 40 × (95 °C for 15 s, 60 °C for 1 min), and post-holding stage 60 °C for 30 s. Results were compared with positive controls for known rs6295CC, rs6295GG, and rs6295GC samples, as verified by Sanger sequencing.
Tissue Collection and Microdissection.
Experimental mice were sacrificed as adults (postnatal day 60–75; P60) via cervical dislocation followed by decapitation. The left hemisphere was excised, flash frozen, and stored −80 °C. The prefrontal cortex and the entire hippocampus of the right hemisphere were microdissected by hand and stored at −80 °C.
Autoradiography.
Complete brains from rs6295GC mice and left hemispheres from rs6295GG and rs6295CC mice were cryosectioned at a thickness of 20 μm and thaw mounted onto Superfrost Plus slides (Fisher Scientific). Sections were maintained at −80 °C until processing. Sections were processed for 4-(2′-methopxyphenyl)-1-[2′-(n-2″-pyridinyl)-p-[125I]iodobenzamido]ethylpiperazine (125I-MPPI) binding as previously described in other studies.7 All experimental and control brains for a given line were processed and exposed to film as a single batch. Receptor levels were quantified as follows. Films were scanned at 1600 dpi (Epson Perfection V600 Photo Scanner), and binding intensity was gauged using FIJI software by an experimenter blind to genotype.51 Levels of 5-HT1AR binding were determined by measuring intensity of the region of interest and subtracting a “background” level from a nearby region in the same section that lacked specific binding. Each region of interest was sampled at least 3 times, and the average intensity was used. Measurements were only used if the intensity was on the linear portion of the standard curve obtained from an ARC146-F 14C standard (ARC, St Louis, MO).
RNA Extraction and cDNA Generation.
Microdissected hippocampus and prefrontal cortex were processed for total purified RNA. The protocol outlined for the Norgen Total RNA/gDNA kit (Norgen Biotek no. 48700, Thorold, ON, Canada) was used to extract RNA from tissue samples with the following modifications: 600 μL rather than 300 μL of lysis buffer was used, and the final elution of RNA was performed twice with 25 μL of elution solution rather than a single 50 μL elution. Total RNA (2 μL) for each sample was then nanodropped to determine RNA concentration (Agilent Technologies, Santa Clara, CA). All RNA samples were then standardized to 25 ng/μL by dilution with RNase-free water (Norgen). High Capacity cDNA Reverse Transcription kit (Applied Bio Systems) was used to generate cDNA. cDNA for each sample was produced using a mixture of 2 μL of 10× TR buffer, 0.8 μL of dNTP, 2 μL of random primers, 1 μL of Transcriptase, 1 μL of RNA inhibitor, and 3.2 μL of ddH2O. RNA (250 ng) was then added, and cDNA was generated using reaction conditions 25 °C for 10 min, 2 × (37 °C for 120 min, 85 °C for 5 min). Samples were stored at −20 °C until processing.
Quantitative RT-PCR (qRT-PCR).
We examined gene expression in rs6295GG and CC mice. Human HRT1A (IDT; Hs.PT.58.50475698.g), mouse Deaf 1 (IDT;Mm.PT.58.29654925), and mouse GAPDH (Taqman Mm99999915-g1, cat no. 4331182) were quantified using qPCR. Efficiencies and primer sequences, where available, are provided in Supplementary Table 3. Samples and probes were processed in quadruplicate in MicroAmp Fast Optical 96 well Reaction Plate (Applied Bio Systems) with 0.25 μL of cDNA, 0.25 μL of probe, 2.5 μL of Taqman Fast master mix (Life Technologies), and 2 μL of ddH2O per well for a total volume of 5 μL. The plate was covered with Optical Adhesive Film (Applied Bio Systems), vortexed, and centrifuged prior to PCR amplification in an Applied Biosystems QuantStudio 3 qPCR machine (Applied Bio Systems). DNA was amplified in a qPCR reaction with and without addition of cDNA to ensure there was no contamination from genomic DNA. Cycling conditions were as follows: 50 °C for 2 min, 95 °C for 20 s, and 40 cycles of 95 °C for 1 s and 60 °C for 20 s.
The mean cycle threshold (Ct) value was taken for each sample in quadruplicate. Quadruplicates that had a standard deviation greater than 0.30 were checked for an obvious outlier, and if none could be visually identified, the sample was excluded from analysis. <di>2ΔΔCt </di>method was used for analysis, with the hippocampus of CC individuals used as the control group. A two-way RM-ANOVA was used to examine the effects of genotype and sex with brain region as a repeated measure.
8-OH-DPAT Induced Hypothermia.
To examine whether our humanized lines expressed functional 5-HT1A autoreceptors, we compared 8-OH-DPAT hypothermic responses in animals with one endogenous copy of the mouse allele, one copy of the human allele on an m5-HT1A−/− background, and m5HT1A−/− mice. 8-OH-DPAT induced hypothermia is mediated exclusively by 5-HT1A autoreceptors.28 Body temperature was measured rectally three times to establish a baseline temperature. 8-OH-DPAT (5 mg/kg; Sigma) was administered via intraperitoneal (ip) injection. Temperature was then measured every 10 min for 60 min. Change in temperature relative to baseline was compared using a RM-ANOVA with genotype and sex as the between subject factors and time as the within subject factor.
Behavioral Phenotyping.
Adult mice (postnatal day 60–75) underwent a battery of depression and anxiety-related behavior tests. Prior to testing, female mice were condensed into the same cage; male mice were housed with the same animals they were weaned with to avoid eliciting aggression. The tests were administered in the following order: open field under low light, light−dark box, elevated zero maze, novelty suppressed feeding, forced swim test, and tail suspension test. All testing apparatuses were cleaned with disinfectant and allowed to dry between animals.
Open Field.
Open field behavior was tested in square chambers (16 × 16 in.2) with metal floors, no bedding, and clear walls (Med Associates, Vermont, USA). Low light conditions consisted of ∼30 lx in the center of the open field chamber. On testing day, mice were given 30 min to acclimate in the behavior room and lighting conditions. Animals were placed in the center of the box and allowed to freely explore the test chamber for 30 min. Locomotion and rearing were recorded via beam-breaks using the software Activity Monitor (Med Associates).
Light–Dark Box.
Testing was conducted in the open field chambers described above with a dark plastic box insert opaque to visible light but transparent to infrared motion tracking beams in half of the chamber. This was joined via an open doorway to the light side of the chamber, which was brightly lit with a fluorescent bulb (∼400 lx). Mice were allowed to acclimate to the behavior room for 30 min prior to behavior testing. For testing, mice were placed in the light side of the chamber facing away from the door and were allowed to freely explore for 10 min. Locomotion, rearing, time, and entries into each chamber were recorded via beam-breaks using Activity Monitor. Mice were omitted from analysis if they failed to enter the dark chamber.
Elevated Zero Maze.
An elevated zero maze was designed with Tinkecard software and 3D printed with light gray Makerbot filament at Columbia University Engineering Library on a Makerbot printer with the following dimensions: 13.5 in. high, 21.5 in. outer diameter, 2 in. arm width, 6 in. tall closed arm walls, with 1 cm ridges lining the edges of the open arms. On test day, mice were given 30 min to acclimate in the behavior room before being placed on the open portion of the maze. Mice were allowed to explore the maze for 10 min. Behavior was recorded and scored using Noldus Ethovision XT software. Independent samples t tests were used to determine if there were statistically significant differences between genotypes in open arm entries and percent time spent in the open arms.
Novelty Suppressed Feeding (NSF).
NSF testing took place in a shallow white arena (17.875 in. long × 1.670 in. wide × 7.5 in. deep) with a lux of 1200–1300 in center of the box; 13.5 h prior to testing, mice were weighed, and food was removed from the home cage. On testing day, mice were weighed prior to testing. Animals were removed from their home cage and transferred to the arena with corn bedding lining the bottom. A single pellet of food was placed on a white paper platform positioned in the center of the crate and animals were able to explore for up to 6 min. The time at which the mouse took its first bite of food was recorded, and the food pellet was removed. After 6 min or after the mice took a bit of food, they were transferred back to their home cages and allowed to explore for up to 5 min. The time at which the mouse took its first bite of food in the home cage was recorded. After 5 min, the food was removed from the cage and weighed to determine the amount eaten. The mouse was weighed and change in weight as a percentage of body weight was calculated for each animal. Independent samples t tests were used to determine if there were statistically significant differences between genotypes in latency to eat in the home and anxiety cages, food eaten/body mass, and percentage weight change.
Forced Swim Test (FST).
The FST paradigm was comprised of a plastic container 7.5 in. wide and 9 in. deep filled with 23–25 °C water. Mice were placed in the bucket and filmed for 6 min. The height of water was selected so that animal was prevented from touching the bottom of the plastic tank and also to prevent its escape. This protocol was repeated 24 h later as previous research suggests that the second testing session may be more sensitive for detecting antidepressant effects.7,32,52 Behavior was recorded and scored using an automated Viewpoint Videotrack software package (Montreal, Canada). Floating was examined in 1 min bins and analyzed using a two way repeated measures ANOVA with time bin as a within-subject measure and genotype as a between-subject measure.
Tail Suspension Test (TST).
Mice were suspended by their tail using tape to secure them to a horizontal bar. The animals were suspended for 6 min, and immobility during this period was assessed using an automated Viewpoint Videotrack software package.
Statistical Analysis.
All statistical analyses were carried out using SPSS 24. Outliers were identified by finding the interquartile range (IQR). IQR×1.5 was then added to quartile 3 (Q3) to get the upper range, and IQR×1.5 subtracted from quartile 1 (Q1) gave the lower range; anything outside of these ranges was excluded as an outlier. Residual normalcy was checked, a Kolmogorov–Smirnov value >0.200 was considered normal. When performing ANOVAs, if the residuals were not normally distributed, the data was transformed until a normal distribution could be achieved. If transformation was not successful in generating a normal distribution, analysis was done using an ANOVA with bootstrapping simple sampling method (5000 repetitions, 95% confidence interval) with genotype and sex as the dependent variables. In all cases where two way ANOVA was appropriate, sphericity was assumed if sphericity was p > 0.05. If this requirement was not achieved, Greenhouse–Geisser F and p-values are reported. Likewise, if homogeneity of variance was violated, we instead used a regression. Details are provided in Supplementary Table 4.
Supplementary Material
ACKNOWLEDGMENTS
We thank the animal care staff at New York State Psychiatric Institute and at University of Colorado Boulder. We are also grateful to Meghin Sadsad, Eric Klein, Katelyn Gordon, and Kylia Ahuna for providing additional experimental and analytical support. We also thank Paul Albert for thoughtful discussion of experimental design and interpretation of results and Kenji Tanaka for advice on transgenic design.
Funding
This work was made possible through the support of American Foundation for Suicide Prevention, R00 MH102352 and R00 MH102352-S1 to Z.R.D. and R37 MH068542 to R.H.
Footnotes
ASSOCIATED CONTENT
S Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.8b00638.
Explanatory figures and negative results and supplementary tables including genetic transmission data and primer information (PDF)
Detailed results from all statistical tests (XLSX)
The authors declare no competing financial interest.
REFERENCES
- (1).Howland RH (2015) Buspirone: Back to the Future. J. Psychosoc Nurs Ment Health Serv 53 (11), 21–24. [DOI] [PubMed] [Google Scholar]
- (2).Trivedi MH, Fava M, Wisniewski SR, Thase ME, Quitkin F, Warden D, Ritz L, Nierenberg AA, Lebowitz BD, Biggs MM, et al. (2006) Medication Augmentation after the Failure of SSRIs for Depression. N. Engl. J. Med 354 (12), 1243–1252. [DOI] [PubMed] [Google Scholar]
- (3).Heisler LK, Chu HM, Brennan TJ, Danao JA, Bajwa P, Parsons LH, and Tecott LH (1998) Elevated Anxiety and Antidepressant-like Responses in Serotonin 5-HT1A Receptor Mutant Mice. Proc. Natl. Acad. Sci. U. S. A 95 (25), 15049–15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhuang X, Gross C, Santarelli L, Compan V, Trillat AC, and Hen R. (1999) Altered Emotional States in Knockout Mice Lacking 5-HT1A or 5-HT1B Receptors. Neuropsychopharmacology 21, 52S–60S. [DOI] [PubMed] [Google Scholar]
- (5).Bortolozzi A, Castane A, Semakova J, Santana N, Alvarado G, Cortes R, Ferres-Coy A, Fernandez G, Carmona MC, Toth M, et al. (2012) Selective siRNA-Mediated Suppression of 5-HT1A Autoreceptors Evokes Strong Anti-Depressant-like Effects. Mol. Psychiatry 17 (6), 612–623. [DOI] [PubMed] [Google Scholar]
- (6).Donaldson ZR, Piel DA, Santos TL, Richardson-Jones J, Leonardo ED, Beck SG, Champagne FA, and Hen R. (2014) Developmental Effects of Serotonin 1A Autoreceptors on Anxiety and Social Behavior. Neuropsychopharmacology 39 (2), 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Richardson-Jones JW, Craige CP, Guiard BP, Stephen A, Metzger KL, Kung HF, Gardier AM, Dranovsky A, David DJ, Beck SG, et al. (2010) 5-HT1A Autoreceptor Levels Determine Vulnerability to Stress and Response to Antidepressants. Neuron 65 (1), 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Drevets WC, Thase ME, Moses-Kolko EL, Price J, Frank E, Kupfer DJ, and Mathis C. (2007) Serotonin-1A Receptor Imaging in Recurrent Depression: Replication and Literature Review. Nucl. Med. Biol 34 (7), 865–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Albert PR (2012) Transcriptional Regulation of the 5-HT1A Receptor: Implications for Mental Illness. Philos. Trans. R. Soc., B 367 (1601), 2402–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Donaldson ZR, le Francois B, Santos TL, Almli LM, Boldrini M, Champagne FA, Arango V, Mann JJ, Stockmeier CA, Galfalvy H, et al. (2016) The Functional Serotonin 1a Receptor Promoter Polymorphism, rs6295, Is Associated with Psychiatric Illness and Differences in Transcription. Transl. Psychiatry 6, e746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Drago A, Ronchi DD, and Serretti A. (2008) 5-HT1A Gene Variants and Psychiatric Disorders: A Review of Current Literature and Selection of SNPs for Future Studies. Int. J. Neuropsychopharmacol 11 (5), 701–721. [DOI] [PubMed] [Google Scholar]
- (12).Lemonde S, Turecki G, Bakish D, Du L, Hrdina PD, Bown CD, Sequeira A, Kushwaha N, Morris SJ, Basak A, et al. (2003) Impaired Repression at a 5-Hydroxytryptamine 1A Receptor Gene Polymorphism Associated with Major Depression and Suicide. J. Neurosci 23 (25), 8788–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Strobel A, Gutknecht L, Rothe C, Reif A, Mössner R, Zeng Y, Brocke B, and Lesch K-P (2003) Allelic Variation in 5-HT1A Receptor Expression Is Associated with Anxiety- and Depression-Related Personality Traits. J. Neural Transm (Vienna) 110 (12), 1445–1453. [DOI] [PubMed] [Google Scholar]
- (14).Kautzky A, James GM, Philippe C, Baldinger-Melich P, Kraus C, Kranz GS, Vanicek T, Gryglewski G, Wadsak W, Mitterhauser M, et al. (2017) The Influence of the rs6295 Gene Polymorphism on Serotonin-1A Receptor Distribution Investigated with PET in Patients with Major Depression Applying Machine Learning. Transl. Psychiatry 7 (6), e1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Czesak M, Lemonde S, Peterson EA, Rogaeva A, and Albert PR (2006) Cell-Specific Repressor or Enhancer Activities of Deaf-1 at a Serotonin 1A Receptor Gene Polymorphism. J. Neurosci 26 (6), 1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Philippe TJ, Vahid-Ansari F, Donaldson ZR, Le François B, Zahrai A, Turcotte-Cardin V, Daigle M, James J, Hen R, Merali Z, et al. (2018) Loss of MeCP2 in Adult 5-HT Neurons Induces 5-HT1A Autoreceptors, with Opposite Sex-Dependent Anxiety and Depression Phenotypes. Sci. Rep 8 (1), 5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Le François B, Czesak M, Steubl D, and Albert PR (2008) Transcriptional Regulation at a HTR1A Polymorphism Associated with Mental Illness. Neuropharmacology 55 (6), 977–985. [DOI] [PubMed] [Google Scholar]
- (18).Schmidt EF, Kus L, Gong S, and Heintz N. (2013) BAC Transgenic Mice and the GENSAT Database of Engineered Mouse Strains. Cold Spring Harb Protoc 2013 (3), pdb.top073692. [DOI] [PubMed] [Google Scholar]
- (19).Albert PR, and Fiori LM (2014) Transcriptional Dys-Regulation in Anxiety and Major Depression: 5-HT1A Gene Promoter Architecture as a Therapeutic Opportunity. Curr. Pharm. Des 20 (23), 3738–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pazos A, Cortes R, and Palacios JM (1985) Quantitative Autoradiographic Mapping of Serotonin Receptors in the Rat Brain. II. Serotonin-2 Receptors. Brain Res 346 (2), 231–249. [DOI] [PubMed] [Google Scholar]
- (21).Pazos A, Probst A, and Palacios JM (1987) Serotonin Receptors in the Human Brain–III. Autoradiographic Mapping of Serotonin-1 Receptors. Neuroscience 21 (1), 97–122. [DOI] [PubMed] [Google Scholar]
- (22).Pazos A, and Palacios JM (1985) Quantitative Autoradiographic Mapping of Serotonin Receptors in the Rat Brain. I. Serotonin-1 Receptors. Brain Res 346 (2), 205–230. [DOI] [PubMed] [Google Scholar]
- (23).Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, and Gerfen CR (2007) Targeting Cre Recombinase to Specific Neuron Populations with Bacterial Artificial Chromosome Constructs. J. Neurosci 27 (37), 9817–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lee SM, Bishop KA, Goellner JJ, O’Brien CA, and Pike JW (2014) Mouse and Human BAC Transgenes Recapitulate Tissue-Specific Expression of the Vitamin D Receptor in Mice and Rescue the VDR-Null Phenotype. Endocrinology 155 (6), 2064–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kaundal R, Yang Y, Nottoli T, and Chi T. (2014) Towards the Molecular Mechanisms of Transgenerational Epigenetic Inheritance, Transgenerational Epigenetics, pp 75–85, Elsevier. [Google Scholar]
- (26).Calero-Nieto FJ, Bert AG, and Cockerill PN (2010) Transcription-Dependent Silencing of Inducible Convergent Transgenes in Transgenic Mice. Epigenet. Chromatin 3 (1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Garrick D, Fiering S, Martin DI, and Whitelaw E. (1998) Repeat-Induced Gene Silencing in Mammals. Nat. Genet 18 (1), 56–59. [DOI] [PubMed] [Google Scholar]
- (28).Martin KF, Phillips I, Hearson M, Prow MR, and Heal DJ (1992) Characterization of 8-OH-DPAT-Induced Hypothermia in Mice as a 5-HT1A Autoreceptor Response and Its Evaluation as a Model to Selectively Identify Antidepressants. Br. J. Pharmacol 107 (1), 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Garcia-Garcia AL, Meng Q, Richardson-Jones J, Dranovsky A, and Leonardo ED (2016) Disruption of 5-HT Function in Adolescence but Not Early Adulthood Leads to Sustained Increases of Anxiety. Neuroscience 321, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ramboz S, Oosting R, Amara DA, Kung HF, Blier P, Mendelsohn M, Mann JJ, Brunner D, and Hen R. (1998) Serotonin Receptor 1A Knockout: An Animal Model of Anxiety-Related Disorder. Proc. Natl. Acad. Sci. U. S. A 95 (24), 14476–14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Borsini F, Podhorna J, and Marazziti D. (2002) Do Animal Models of Anxiety Predict Anxiolytic-like Effects of Antidepressants? Psychopharmacology (Berl.) 163 (2), 121–141. [DOI] [PubMed] [Google Scholar]
- (32).Dulawa SC, Holick KA, Gundersen B, and Hen R. (2004) Effects of Chronic Fluoxetine in Animal Models of Anxiety and Depression. Neuropsychopharmacology 29 (7), 1321–1330. [DOI] [PubMed] [Google Scholar]
- (33).Jimenez JC, Su K, Goldberg AR, Luna VM, Biane JS, Ordek G, Zhou P, Ong SK, Wright MA, Zweifel L, et al. (2018) Anxiety Cells in a Hippocampal-Hypothalamic Circuit. Neuron 97 (3), 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kheirbek MA, Drew LJ, Burghardt NS, Costantini DO, Tannenholz L, Ahmari SE, Zeng H, Fenton AA, and Hen R. (2013) Differential Control of Learning and Anxiety along the Dorsoventral Axis of the Dentate Gyrus. Neuron 77 (5), 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mogha A, Guariglia SR, Debata PR, Wen GY, and Banerjee P. (2012) Serotonin 1A Receptor-Mediated Signaling through ERK and PKCα Is Essential for Normal Synaptogenesis in Neonatal Mouse Hippocampus. Transl. Psychiatry 2 (1), e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Veenstra-VanderWeele J, Muller CL, Iwamoto H, Sauer JE, Owens WA, Shah CR, Cohen J, Mannangatti P, Jessen T, Thompson BJ, et al. (2012) Autism Gene Variant Causes Hyperserotonemia, Serotonin Receptor Hypersensitivity, Social Impairment and Repetitive Behavior. Proc. Natl. Acad. Sci. U. S. A 109 (14), 5469–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kerr TM, Muller CL, Miah M, Jetter CS, Pfeiffer R, Shah C, Baganz N, Anderson GM, Crawley JN, Sutcliffe JS, et al. (2013) Genetic Background Modulates Phenotypes of Serotonin Transporter Ala56 Knock-in Mice. Mol. Autism 4 (1), 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Neuner SM, Heuer SE, Huentelman MJ, O’Connell KMS, and Kaczorowski CC (2018) Harnessing Genetic Complexity to Enhance Translatability of Alzheimer’s Disease Mouse Models: A Path toward Precision Medicine. Neuron, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gallagher MD, and Chen-Plotkin AS (2018) The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet 102 (5), 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Schmouth J-F, Bonaguro RJ, Corso-Diaz X, and Simpson EM (2012) Modelling Human Regulatory Variation in Mouse: Finding the Function in Genome-Wide Association Studies and Whole-Genome Sequencing. PLoS Genet 8 (3), e1002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, and Yang J. (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet 101 (1), 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bassett AR (2017) Editing the Genome of hiPSC with CRISPR/Cas9: Disease Models. Mamm. Genome 28 (7–8), 348–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hockemeyer D, and Jaenisch R. (2016) Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 18 (5), 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Liang X, Potter J, Kumar S, Ravinder N, and Chesnut JD (2017) Enhanced CRISPR/Cas9-Mediated Precise Genome Editing by Improved Design and Delivery of gRNA, Cas9 Nuclease, and Donor DNA. J. Biotechnol 241, 136–146. [DOI] [PubMed] [Google Scholar]
- (45).Devoy A, Bunton-Stasyshyn RK, Tybulewicz VL, Smith AJ, and Fisher EM (2012) Genomically Humanized Mice: Technologies and Promises. Nat. Rev. Genet 13 (1), 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Czesak M, Le Francois B, Millar AM, Deria M, Daigle M, Visvader JE, Anisman H, and Albert PR (2012) Increased Serotonin-1A (5-HT1A) Autoreceptor Expression and Reduced Raphe Serotonin Levels in Deformed Epidermal Autoregulatory Factor-1 (Deaf-1) Gene Knock-out Mice. J. Biol. Chem 287 (9), 6615–6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, and Copeland NG (2001) A Highly Efficient Escherichia Coli-Based Chromosome Engineering System Adapted for Recombinogenic Targeting and Subcloning of BAC DNA. Genomics 73 (1), 56–65. [DOI] [PubMed] [Google Scholar]
- (48).Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, and Court DL (2000) An Efficient Recombination System for Chromosome Engineering in Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A 97 (11), 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wang S, Zhao Y, Leiby M, and Zhu J. (2009) A New Positive/negative Selection Scheme for Precise BAC Recombineering. Mol. Biotechnol 42 (1), 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Samuels BA, Anacker C, Hu A, Levinstein MR, Pickenhagen A, Tsetsenis T, Madronal N, Donaldson ZR, Drew LJ, Dranovsky A, et al. (2015) 5-HT1A Receptors on Mature Dentate Gyrus Granule Cells Are Critical for the Antidepressant Response. Nat. Neurosci 18 (11), 1606–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 9 (7), 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Borsini F, Lecci A, Sessarego A, Frassine R, and Meli A. (1989) Discovery of Antidepressant Activity by Forced Swimming Test May Depend on Pre-Exposure of Rats to a Stressful Situation. Psychopharmacology (Berl.) 97 (2), 183–188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.