Abstract
Mitogen-activated protein kinase kinase (MEK) inhibitors have failed to show clinical benefit in Kirsten rat sarcoma (KRAS)-mutant lung cancer due to various resistance mechanisms. To identify differential therapeutic sensitivities between epithelial and mesenchymal lung tumors, we performed in vivo small hairpin RNA (shRNA) screens, proteomic profiling, and analysis of patient tumor datasets, which revealed an inverse correlation between mitogen-activated protein kinase (MAPK) signaling dependency and a zinc finger E-box-binding homeobox 1 (ZEB1)-regulated epithelial-to-mesenchymal transition (EMT). Mechanistic studies determined that MAPK signaling dependency in epithelial lung cancer cells is due to the scaffold protein interleukin 17 receptor D (IL17RD), which is directly repressed by ZEB1. Lung tumors in multiple Kras mutant murine models with increased ZEB1 displayed low IL17RD expression, accompanied by MAPK-independent tumor growth and therapeutic resistance to MEK inhibition. Suppression of ZEB1 function with miR-200 expression or the histone deacetylase (HDAC) inhibitor mocetinostat sensitized resistant cancer cells to MEK inhibition and markedly reduced in vivo tumor growth, showing a promising combinatorial treatment strategy for KRAS-mutant cancers. In human lung tumor samples, high ZEB1 and low IL17RD expression correlated with low MAPK signaling, presenting potential markers that predict patient response to MEK inhibitors.
One Sentence Summary:
Suppression of ZEB1 through miR-200 expression or HDAC inhibition with mocetinostat sensitizes resistant KRAS-mutant lung cancers to MEK inhibition.
Introduction
Approximately 30% of all cancers, including lung adenocarcinoma, have an activating KRAS mutation, resulting in aberrant signaling through the mitogen-activated protein kinase (MAPK) pathway to promote tumor initiation and progression (1–4). KRAS-mutant cancers respond poorly to conventional treatments, and no existing drug can effectively inhibit the range of oncogenic changes observed in the RAS protein (5–7). Thus, alternative therapeutic strategies have been developed targeting downstream effectors of mutant KRAS through the use of MEK inhibitors to block MAPK signaling (8, 9). However, recent pre-clinical and clinical trials have demonstrated poor responses to MEK inhibitors due to the development of drug resistance (10–13).
Previous studies have demonstrated that resistance to MAPK disruption in lung cancer can occur due to an epithelial-to-mesenchymal transition (EMT)-associated fibroblast growth factor receptor 1 (FGFR1) feedback re-activation of MAPK signaling (14–16). Earlier work using the KrasLA1-G12D (Kras) and KrasLA1-G12D;p53R172H (KP) murine models demonstrated that EMT is epigenetically regulated by a double-negative feedback loop between the ZEB1 transcription factor and the miR-200 family of microRNAs (17–21), whereby increased ZEB1 expression induces EMT and miR-200 expression reverts cells to an epithelial phenotype. In addition to therapy resistance and enhanced tumor progression, high ZEB1 expression in cancer cells results in metastatic disease, contributing to poor overall patient outcome (17, 22–27). Despite the importance of ZEB1 as a transcriptional repressor, pharmacologically targeting ZEB1 presents numerous challenges owing to its nuclear localization and pleiotropic effects. Thus, uncovering the contrasting sensitivities to specific targeted therapies between epithelial and mesenchymal tumor cells will facilitate the design of combinatorial treatment strategies in conjunction with MEK inhibitors.
Our study identifies distinct subpopulations of lung cancer cells with differential MEK inhibitor sensitivities as defined by ZEB1 and IL17RD expression, presenting potential markers associated with sensitivity to treatment. Therapeutically, suppression of ZEB1 through expression of miR-200 or HDAC inhibition with mocetinostat sensitized resistant KRAS-mutant lung cancers to MEK inhibitors, demonstrating a prospective combinatorial therapeutic strategy to overcome epigenetically-mediated resistance across a broad range of KRAS-mutant cancers.
Results
Epithelial lung cancer cells exhibit increased MAPK signaling and dependency for tumor growth
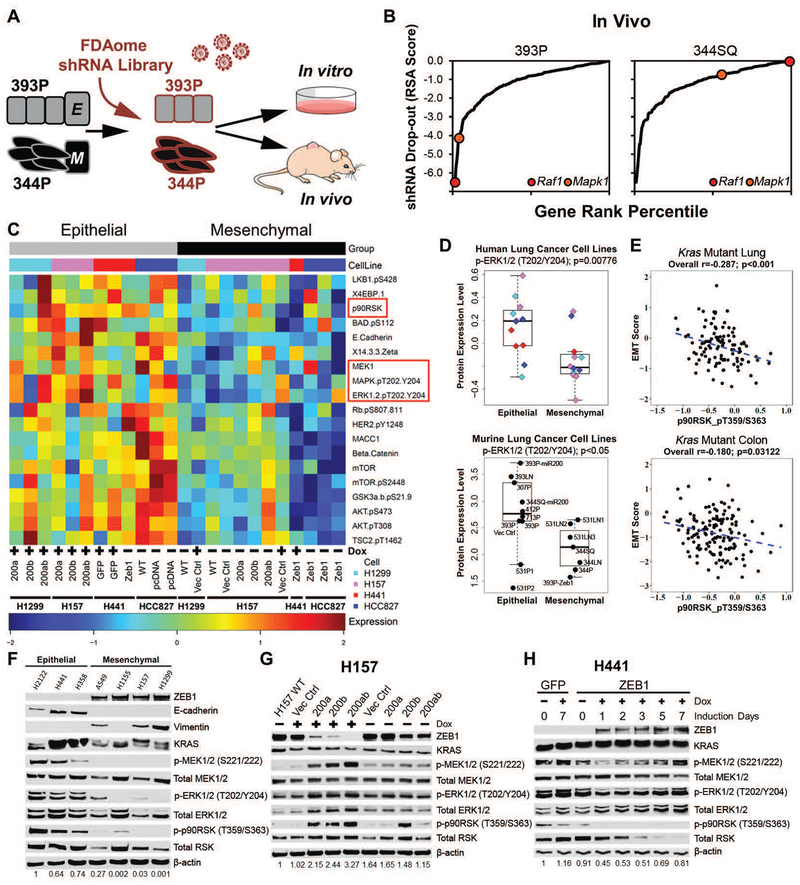
To identify therapeutically targetable genes essential for tumor formation and growth of epithelial or mesenchymal KRAS-mutant tumors, we used a previously developed in vivo loss-of-function screen approach with a barcoded, pooled shRNA library (28) targeting approximately 200 genes that have FDA-approved pharmacological inhibitors (FDAome). The shRNA library was transduced into representative epithelial (393P) and mesenchymal (344P) murine lung cancer cell lines derived from the KP genetically engineered mouse model (GEMM) (17) and implanted subcutaneously in nude mice or cultured in vitro (Fig. 1A). Tumors were harvested, shRNA barcodes were quantified by deep sequencing and referenced with the respective in vitro cell population, and the phenotypic impact of gene knockdown was inferred by the redundant shRNA activity (RSA) algorithm (29, 30), where a lower rank of the shRNA barcodes signified dropout from the population and greater dependency on the gene for tumorigenesis (tables S1 and S2 in data file S1). Although both cell line models have activating KrasG12D and p53 mutations, comparison of the results revealed that epithelial 393P tumors were more reliant on MAPK genes for both in vitro and in vivo growth, whereas mesenchymal 344P tumor growth was independent of MAPK signaling (Fig. 1B, fig. S1A; table S2 in data file S1).
Fig. 1. Epithelial tumors have greater MAPK signaling dependency for growth.
(A) Experimental design for FDAome shRNA drop-out screens in epithelial (393P) and mesenchymal (344P) murine lung cancer cell lines implanted subcutaneously in vivo (nude mice) and grown in parallel in vitro (20 doublings).
(B) Gene rank analysis highlighting the behavior of RAF1 and MAPK1 genes in the FDAome in vivo screens executed in epithelial (393P) and mesenchymal (344P) murine lung cancer cell lines. ShRNA dropout score was calculated as the log of the redundant shRNA activity (RSA) value shown in table S1 in data file S1.
(C) Heatmap of reverse phase protein array (RPPA) profile showing statistically significant (p<0.05) differentially regulated proteins in isogenically-matched epithelial and mesenchymal human lung cancer cell lines. Cell lines expressed doxycycline (Dox) inducible miR-200 or ZEB1, as indicated. ZEB1 was constitutively expressed in HCC827 cells, rather than Dox-inducible.
(D) Dot plot of ERK phosphorylation (T202/Y204) from RPPA dataset in a panel of mesenchymal and epithelial murine (epithelial: 393P, 393P Vector Control, 393P-miR200, 393LN, 307P, 412P, 713P, 531P1, 531P2, 344SQ-miR200; mesenchymal: 531LN1, 531LN2, 531LN3, 344P, 344LN, 344SQ, 393P-Zeb1) and human lung cancer cells.
(E) Cluster plot analysis of Spearman’s rank correlation between EMT score and the activated downstream MAPK signaling molecule p-p90RSK (T359/S363) in KRAS mutant lung and colorectal cancer patient samples from TCGA RPPA datasets.
(F) Western blot of the EMT markers ZEB1, E-cadherin, and vimentin and MAPK signaling molecules in a panel of human KRAS mutant lung cancer cell lines. Numbers at the bottom indicate relative quantification of western blot signal for p-ERK, relative to β-actin.
(G and H) Western blot of indicated proteins in (G) H157 cells after Dox-induced miR-200 expression for 7 days and (H) H441 cells after Dox-induced ZEB1 expression at the indicated time points. Numbers at the bottom indicate relative quantification of western blot signal for p-ERK, relative to β-actin.
Complementing the in vivo shRNA screen, we sought to elucidate the specific differences in protein signaling pathway rewiring between epithelial and mesenchymal lung cancer cells. Using reverse phase protein arrays (RPPA) to analyze changes in cell signaling proteins in a high throughput manner (31, 32), we screened a panel of previously characterized (17, 22) isogenic murine and human epithelial lung cancer cell lines expressing ZEB1 or mesenchymal lung cancer cell lines expressing miR-200 and observed an increase in MAPK pathway signaling in cells with an epithelial phenotype (Fig. 1C,D; tables S3–8 in data file S1).
Next we translated the shRNA and RPPA findings by analysis of tumor samples from multiple cancer types in which RAS or rapidly accelerated fibrosarcoma (RAF) are commonly mutated to activate MAPK signaling, including lung adenocarcinoma (30%), pancreatic (90%), colorectal (45%), and melanoma (70%) (4, 33). Cross-platform correlation analyses of a previously-derived EMT gene signature score (34, 35) or ZEB1 mRNA expression with MAPK signaling proteins as measured by RPPA from the TCGA datasets showed a significant (p<0.001) inverse correlation between EMT score and phosphorylated p90 ribosomal s6 kinase (p90RSK) in total lung and colorectal cancer patient samples (fig. S1B). Total melanoma samples did not show a statistically significant correlation between EMT score or ZEB1 and p90RSK, and no correlation between EMT and MAPK signaling was observed in total pancreatic cancer samples (fig. S1B). However, when patient samples across the four different tumor types were stratified for KRAS, NRAS, and BRAF mutations, there was an inverse correlation between EMT score and MAPK signaling (Fig. 1E and fig. S1B), though there was no significant correlation for KRAS-mutant pancreatic cancer and NRAS/BRAF-mutant melanoma (fig. S1B).
Validation of the bioinformatic datasets by western blotting in a representative panel of RAS mutant human NSCLC cell lines showed higher activity of endogenous phosphorylated MAPK molecules MEK, extracellular signal-regulated kinases (ERK), p90RSK in epithelial cell lines when stratified by the EMT markers ZEB1, E-cadherin, and vimentin and normalized to β-actin expression (Fig. 1F). Additionally, suppression of ZEB1 through inducible miR-200 expression in mesenchymal cells increased MAPK signaling in a time-dependent manner, whereas induced ZEB1 expression in epithelial cells reduced MAPK signaling in a dynamic manner, at both the upstream and downstream components of the MAPK pathway (Fig. 1G,H and fig. S1C,D). To determine if miR-200 activation of MAPK signaling was due to miR-200 repression of ZEB1, ZEB1 was knocked down in mesenchymal lung cancer cells, which similarly upregulated MAPK pathway signaling (fig. S1E), confirming the importance of ZEB1 in regulating MAPK signaling. Overexpression of each individual miR-200 family member increased activated MAPK proteins, with miR-200c having the greatest effect, consistent with our previous work demonstrating that miR-200c binds most strongly to the ZEB1 3’-UTR (fig. S1F) (35).
Activation of MEK/ERK signaling in epithelial cells is driven by the RAS-RAF-MEK-ERK signaling cascade
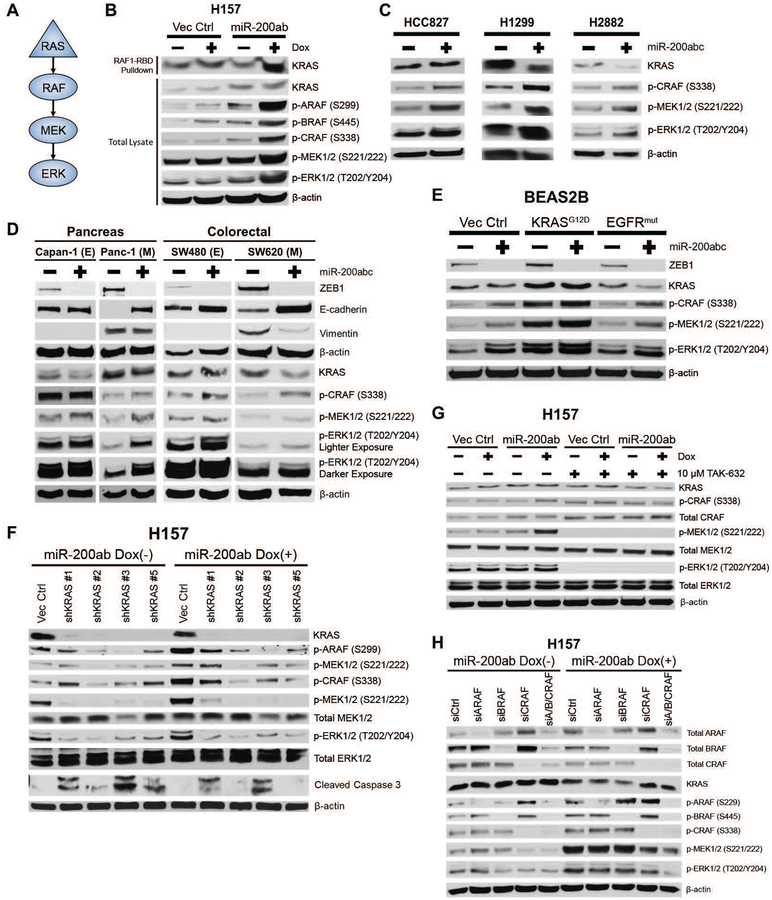
Because MEK and ERK are activated by multiple signaling molecules, we next sought to establish the pathway responsible for differential MAPK activation. Analysis of the canonical RAS-RAF-MEK-ERK signaling cascade (Fig. 2A) after reversion of the mesenchymal H157 and H1155 KRAS mutant cells to an epithelial state through induced, stable, or transient expression of miR-200, showed an increase in KRAS binding to the RAS binding domain (RBD) of RAF1 (CRAF), followed by activation of MAPK signaling (Fig. 2B and fig. S2A,B). We observed that MAPK signaling is activated by miR-200 regardless of the driver mutation present, as miR-200 expression activated MAPK signaling in EGFR mutant (HCC827), NRAS mutant (H1299), EGFR/RAS wild-type (WT) (H2882), and murine 344SQ KP lung cancer cells (Fig. 2C and fig. S2C,D). Consistent with our TCGA dataset correlation analyses, MAPK activation in epithelial cancer cells was not restricted to lung cancer cells, as pancreatic and colorectal cancer cell lines showed higher basal MAPK signaling activity in epithelial cells compared to mesenchymal cells, which was increased upon miR-200 expression (Fig. 2D). Moreover, miR-200 expression upregulated MAPK signaling in epithelial, non-transformed BEAS2B cells at baseline or with constitutive expression of mutant KRASG12D or EGFRΔ722–726 (36) (Fig. 2E), demonstrating that this process is independent of oncogenic and co-mutational events.
Fig. 2. RAS-RAF-MEK-ERK regulate MAPK signaling in epithelial cancer cells.
(A) Schematic of RAS-RAF-MEK-ERK MAPK pathway signaling.
(B) Top: Western blot of KRAS from RAS-GTP pulldown with the RAS binding domain (RBD) of RAF1 after miR-200ab induction in mesenchymal H157 lung cancer cell line. Bottom: Western blots of whole cell lysates of MAPK signaling molecules in H157 cells with induced miR-200 expression.
(C) Western blots of MAPK signaling proteins in HCC827 (EGFR mutant), H1299 (NRAS mutant), and H2882 (EGFR and NRAS WT) human lung cancer cell lines after 96-hour transient expression of miR-200abc.
(D) Western blot of ZEB1, E-cadherin, and vimentin and MAPK signaling proteins in epithelial (E) and mesenchymal (M) KRAS mutant pancreatic and colorectal cancer cell lines after 6-day transient expression of miR-200a/b/c.
(E) Western blots of MAPK signaling proteins in non-transformed BEAS2B epithelial human lung cell lines expressing mutant KRAS or mutant EGFR in conjunction with transient expression of miR-200a/b/c.
(F – H) Western blots of MAPK signaling proteins in H157 cells with induced miR-200ab expression after (F) KRAS knockdown by shRNA, (G) inhibition of pan-RAF enzymatic activity for 24 hours with TAK-632, and (H) 48-hour siRNA-mediated knockdown of RAF isoforms individually or in combination.
To verify that the RAS-RAF-MEK pathway is necessary to activate p-ERK, we transiently knocked down KRAS by shRNA after miR-200 induction in the mesenchymal H157 model and observed a reduction in the miR-200-dependent MAPK signaling (Fig. 2F). Additionally, KRAS knockdown caused activation of cleaved caspase-3, indicating that cell survival is still dependent on KRAS despite the suppression of downstream MAPK signaling by ZEB1. Similarly, chemical inhibition of pan-RAF enzymatic activity and siRNA knockdown of RAF isoforms individually or in combination demonstrated that MAPK activation was predominantly dependent on the enzymatic function of the CRAF isoform (Fig. 2G,H). Knockdown of all three RAF isoforms together produced the best suppression of p-MEK/p-ERK in miR-200-expressing cells, suggesting that miR-200-dependent MAPK signaling acts through RAF heterodimerization (Fig. 2H).
In addition to the RAS-RAF pathway, we also examined the alternative focal adhesion kinase (FAK)/Ras-related C3 botulinum toxin substrate (RAC)/p21-activated kinases (PAK) signaling pathway for differential regulation of p-MEK/p-ERK activation (fig. S2E). Stable or induced miR-200 expression in H157 cells decreased p-FAK, consistent with our previous work (25), but increased p-PAK2 (fig. S2F–I). The MEK (S298) phosphorylation site downstream of PAK1/2 also increased in a time-dependent manner but at a slower rate than MEK (S221/222) (fig. S2G). Small molecule inhibition of FAK had no effect on MAPK signaling, whereas siRNA-mediated knockdown of PAK1 and/or PAK2 showed only partial contribution of PAK2 in activating MAPK signaling, but to a much lower degree than RAS-RAF (fig. S2H,I). Taken together, these data suggest that the miR-200-dependent increase in MAPK signaling is dependent on the RAS-RAF-MEK-ERK pathway, rather than activation of a bypass signaling mechanism.
Regulation of MAPK pathway signaling is linked to ZEB1 by differential expression of the scaffold protein IL17RD
We next sought to ascertain the mechanism of MAPK activation in epithelial cells with high miR-200 expression vs suppression of signaling in mesenchymal cells with low miR-200 and high ZEB1 expression. To determine if signaling was due to upstream growth factor stimulation of receptor tyrosine kinases or other mitogen-activated receptors, the KRAS mutant H157 cells with or without miR-200 expression were cultured in serum-free medium for 24 hours and then stimulated with either fetal bovine serum (FBS)-containing medium or epithelial growth factor (EGF) for 4 hours. In the mesenchymal control cells, there was a marked reduction in MAPK signaling under serum-free conditions compared to FBS stimulation, and EGF only marginally activated signaling (Fig. 3A). However, cells with miR-200 induction continued to have increased MAPK signaling regardless of culture conditions (Fig. 3A), suggesting that miR-200-dependent activation of the MAPK pathway in KRAS mutant cells occurs through an intracellular mechanism, independent of extrinsic factors.
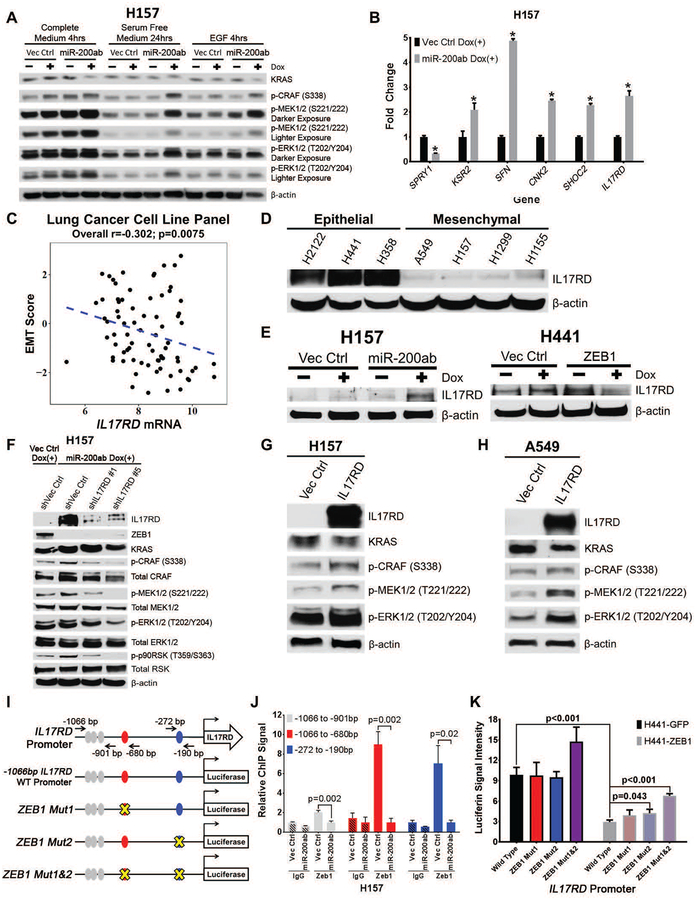
Fig. 3. ZEB1 regulates MAPK signaling by suppressing the scaffold protein IL17RD.
(A) Western blot of MAPK signaling molecules in H157 cells +/− miR-200 expression after 24 hours of serum-free starvation, followed by stimulation with complete serum medium, serum-free medium, or EGF for 4 hours.
(B) QPCR analysis for relative expression of SPRY1, KSR2, 14-3-3σ/SFN, CNK2, SHOC2, and IL17RD in H157 cells with inducible miR-200 expression. *p<0.05.
(C) Cluster plot analysis of Spearman’s rank correlation between IL17RD gene expression and EMT scores of 77 human lung cancer cell lines.
(D and E) Western blots of IL17RD in (D) a panel of human epithelial and mesenchymal KRAS mutant lung cancer cell lines and (E) H157 or H441 cells with induced miR-200 or ZEB1 expression, respectively.
(F) Western blot of IL17RD and MAPK signaling molecules after transient shRNA knockdown of IL17RD in H157 cells with induced miR-200 expression for 72 hours.
(G and H) Western blots of IL17RD and MAPK signaling molecules after 48-hour constitutive expression of IL17RD in (G) H157 and (H) A549 cells.
(I) Schematic of human IL17RD promoter region containing predicted ZEB1 binding sites represented by color-coded ellipses. Black arrows indicate location of genomic region used for qPCR amplification, containing potential ZEB1 binding sites in the IL17RD promoter after ZEB1 chromatin immunoprecipitation (ChIP). The IL17RD promoter was cloned 1,066 base pairs upstream of the transcriptional start site and inserted into a luciferase reporter vector. Mutations of potential ZEB1 binding sites indicated with yellow X.
(J) Fold enrichment by qPCR analysis of IL17RD promoter segments containing potential ZEB1 binding sites after endogenous ZEB1 ChIP in H157 cells with inducible vector control or miR-200 expression, using ZEB1 antibody or IgG control antibody. Promoter regions analyzed by qPCR labeled with black arrows in (I).
(K) Relative luciferase activity of IL17RD promoter reporter constructs in (I) transfected into epithelial H441 cells with induced GFP control or ZEB1 expression. Relative luciferin signal was normalized to promoter-less vector control signal.
Because miR-200-mediated activation of MEK/ERK signaling occurs predominantly through the RAS-RAF signaling junction, is dependent on multiple RAF isoforms, and is intracellularly regulated, our data implied that regulation of MAPK by ZEB1 depends on changes in inhibitory or scaffolding proteins. We performed qPCR analysis for expression of MAPK inhibitory and scaffold genes reported to interact with RAF (37), with and without miR-200 induction (Fig. 3B and fig. S3A–C). Inhibitory genes with at least 50% decrease or scaffold genes with at least 2-fold increase in expression after miR-200 induction were selected for further analysis (Fig. 3B). Increased expression of Sprouty homolog 4 (SPRY4) and various dual-specificity phosphatase (DUSP) genes (fig. S3A,B) after miR-200 induction is consistent with feedback inhibition responses induced by MAPK pathway activation (38–42). Because SPRY1 was the only inhibitory gene significantly (p<0.01) downregulated by over 50% with miR-200 expression, we tested the role of SPRY1 by siRNA-mediated knockdown in H157 cells or transiently expressed SPRY1 in miR-200-expressing H157 cells, but observed no relevant effect on MAPK signaling in either case (fig. S3D,E). QPCR analysis of various RAF-associated scaffolding genes (Fig. 3B and fig. S3C) identified Kinase suppressor of ras 2 (KSR2), Stratifin (SFN/14-3-3σ), Connector enhancer of kinase suppressor of ras 2 (CNK2), soc-2 suppressor of clear homolog (SHOC2), and IL17RD as potential targets that were significantly (p<0.01) upregulated at least 2-fold upon miR-200 expression (Fig. 3B). Western blot validation of the qPCR data showed no observable differences in SHOC2 or CNK2 protein expression (fig. S3F). Although miR-200 expression increased 14-3-3σ protein and KSR2 mRNA, knockdown of either gene after miR-200 induction had no effect on MAPK signaling (fig. S3G,H).
We next focused our analysis on the regulatory role of IL17RD on MAPK signaling. Correlation analysis of IL17RD mRNA expression to our previously reported 76-gene EMT signature score (34) in a panel of 77 human lung cancer cell lines showed a statistically significant (p=0.0075) inverse correlation between IL17RD and EMT signature (Fig. 3C). Validation of the correlation analysis and IL17RD qPCR data by western blotting in a panel of epithelial and mesenchymal RAS mutant human lung, pancreatic, and colorectal cancer cell lines confirmed higher basal IL17RD expression in epithelial cells compared to mesenchymal cells (Fig. 3D and fig. S3I), with induction of IL17RD upon miR-200 expression in multiple models and suppression of IL17RD after ZEB1 expression in epithelial H441 cells (Fig. 3E and fig. S3I,J). Knockdown of IL17RD by shRNA in miR-200 expressing H157 cells reduced MAPK signaling (Fig. 3F), whereas overexpression of IL17RD in mesenchymal H157 and A549 cells increased MAPK signaling (Fig. 3G,H). Although IL17RD was initially implicated in inhibiting fibroblast growth factor (FGF)-induced MAPK signaling (43), in FGF-independent contexts IL17RD has been shown to promote MAPK signaling (44, 45). Consistent with these reports, analysis of FGFR1 protein revealed a reciprocal expression to IL17RD, where mesenchymal cells had greater expression of FGFR1 compared to epithelial cells (fig. S3I,K) and transient IL17RD overexpression caused a decrease in FGFR1 protein (fig. S3L).
Considering the negative correlation between IL17RD and EMT, we next sought to define the regulatory mechanism of IL17RD gene expression by ZEB1 or miR-200. Analysis of the human IL17RD promoter region using the JASPAR transcription factor database (http://jaspar.genereg.net/) predicted multiple potential ZEB1 E-box recognition sites (Fig. 3I) responsible for transcriptional repression (21, 22, 46, 47). ZEB1 chromatin immunoprecipitation (ChIP) qPCR assays of the endogenous IL17RD promoter in mesenchymal H157 cells with or without miR-200 expression confirmed direct binding of ZEB1 to the IL17RD promoter, with the highest signal occurring 190 and 680 base pairs upstream of the IL17RD transcriptional start site (Fig. 3J). Luciferase reporter assays with the wild-type IL17RD promoter region confirmed transcriptional repression by ZEB1, which was abrogated by introduction of mutations into the two main ZEB1 binding sites (as assessed by ChIP) individually and in combination (Fig. 3K).
Mesenchymal lung cancer cells and tumors are resistant to MEK inhibition
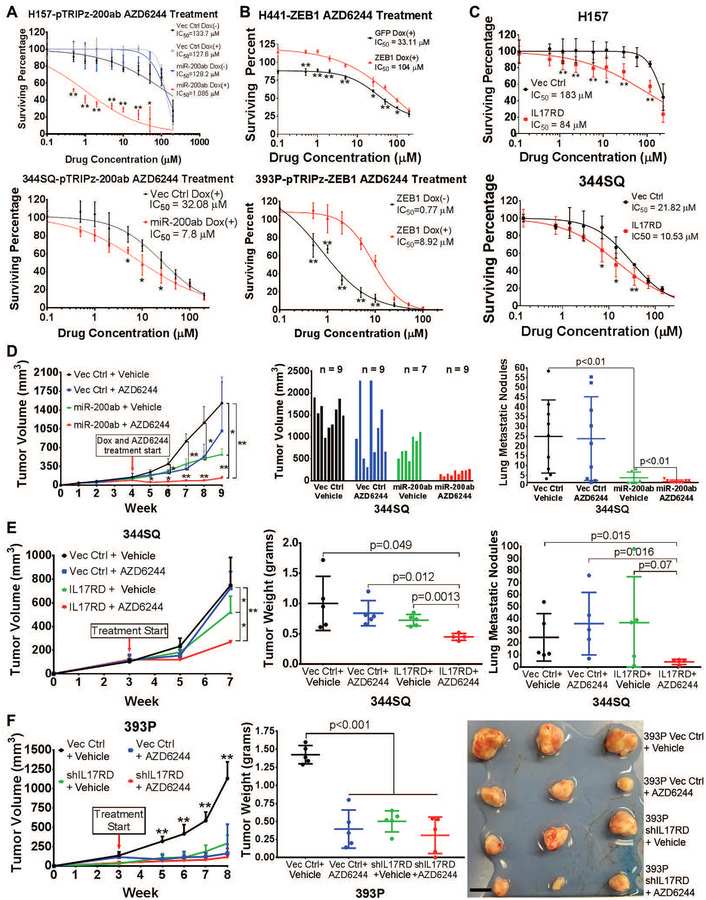
To functionally validate our shRNA screen and mechanistic findings, we treated a panel of human and murine KRAS mutant lung cancer cell lines, stratified as either epithelial or mesenchymal based on previous profiling (17, 22, 24), with MEK inhibitors. Although we did not observe a significant difference in baseline proliferation rate between epithelial and mesenchymal murine lung cancer cells (fig. S4A), mesenchymal cells were more resistant to both of the MEK inhibitors selumetinib (AZD6244) and trametinib (Fig. 4A,B and fig. S4B–D).
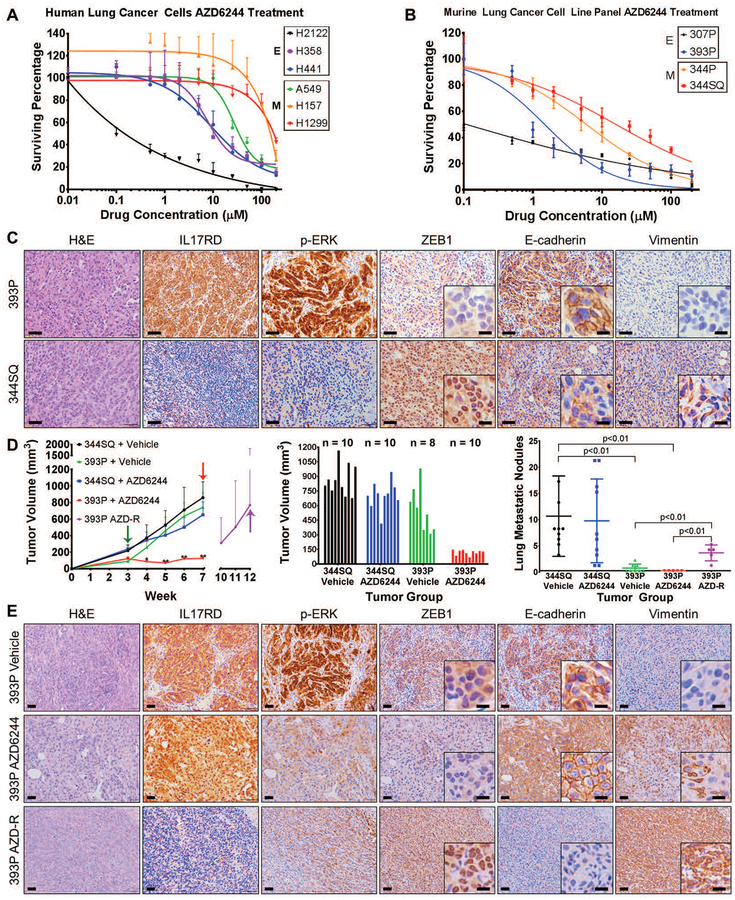
Fig. 4. Lung cancer cells and tumors with high ZEB1 are resistant to MEK inhibition.
(A and B) In vitro cell survival response after 72-hour selumetinib (AZD6244) treatment in a panel of epithelial and mesenchymal (A) human and (B) murine RAS mutant lung cancer cell lines.
(C) H&E, IL17RD, p-ERK, ZEB1, E-cadherin, and vimentin IHC stains of primary tumor tissues generated by subcutaneous injection of epithelial 393P and mesenchymal 344SQ murine lung cancer cell lines in syngeneic wild-type mice (n=8 tumors per group). Scale bars, 100 μm. Inset scale bars, 20 μm.
(D) Left: In vivo volume measurements at the indicated time points for 393P and 344SQ subcutaneous tumors in syngeneic wild-type mice after daily treatment with 25 mg/kg AZD6244 MEK inhibitor or vehicle control. Treatment start time denoted by green arrow, endpoint denoted by red arrows, and resistant tumors denoted by purple arrow. Treatment performed in experimental duplicate with 4 to 5 mice per replicate in each treatment group. Sample size is as indicated in the middle graph. Tumor volume data plotted as the mean and standard deviation. Purple data points represent 393P tumors that were initially responsive to MEK inhibition and developed resistance over time. Middle: Tumor volume measurements at Week 7 (at the endpoint, after 4 weeks of AZD6244 treatment). Right: Quantification of lung metastatic surface nodules in the indicated experimental groups at Week 7 of the experiment or at Week 12 in 393P tumors that developed resistance to AZD6244 (393P AZD-R). *: p<0.05, **: p<0.01.
(E) Stains for the indicated markers in 393P tumors in mice that received vehicle or AZD6244 until Week 7 of the experiment in (D) (top and middle panels), or in tumors that developed resistance to AZD6244 (393P AZD-R) at Week 12 of the experiment in (D) (bottom panel). Scale bars, 50 μm. Inset scale bars, 20 μm. Images are representative of n=5 tissues per group.
To validate our in vitro findings in an in vivo context, we first analyzed primary syngeneic tumors generated by implantation of epithelial 393P and mesenchymal 344SQ murine lung cancer cells in syngeneic WT mice (17). Immunohistochemical (IHC) staining of tumor tissues for IL17RD, p-ERK, and the EMT markers (nuclear ZEB1, membranous E-cadherin, and vimentin) showed that low ZEB1 and vimentin staining were associated with high IL17RD, p-ERK, and membrane-localized E-cadherin signals, whereas high ZEB1 and vimentin expression were associated with an opposing pattern (Fig. 4C). Consistent with these findings, in vivo administration of the MEK inhibitor selumetinib (AZD6244) significantly (p<0.01) reduced tumor growth only in epithelial 393P tumors, whereas mesenchymal 344SQ tumors were highly resistant to treatment (Fig. 4D; table S9 in data file S1). However, despite the pronounced efficacy, 80% of 393P tumors eventually developed resistance to MEK inhibition, reaching comparable sizes to control tumors by 12 weeks, and developed a higher number of metastatic lung lesions compared to untreated control or drug-sensitive tumors (Fig. 4D). IHC analysis of the vehicle control (393P Vehicle) and AZD6244-sensitive (393P AZD6244) tumors after 7 weeks, and the AZD6244-resistant (393P AZD-R) primary tumors after 12 weeks confirmed the efficacy of AZD6244 in suppressing p-ERK and revealed that the resistant tumors had undergone EMT (displaying high nuclear ZEB1, low E-cadherin, and high vimentin expression compared to control tissues) and lacked expression of IL17RD (Fig. 4E). We also observed increased vimentin even in AZD6244-sensitive 393P tumors after MEK inhibition. In addition to IHC stains of the tumor tissue, cell lines were generated from 393P vehicle control and AZD6244-resistant tumors, and western blot analyses confirmed the tumor IHC stains (fig. S4E).
IL17RD is sufficient to sensitize mesenchymal lung tumors to MEK inhibition
To determine if sensitivity to MEK inhibition in epithelial lung cancer cells was a result of high miR-200 as opposed to secondary factors in the basal state of epithelial cell lines, we induced miR-200 expression in drug-resistant, mesenchymal human and murine lung cancer cells and observed increased sensitivity to MEK inhibition (Fig. 5A and fig. S4F). Conversely, induced ZEB1 expression in epithelial cells increased resistance to MEK inhibitor treatments (Fig. 5B and fig. S4G). Because increased MAPK signaling in epithelial cells is mechanistically dependent upon IL17RD, we constitutively expressed IL17RD in human and murine mesenchymal cells and found it was sufficient to sensitize the resistant lines to MEK inhibitor (Fig. 5C).
Fig. 5. MiR-200 or IL17RD sensitizes mesenchymal lung tumors to MEK inhibition.
(A-C) In vitro cell survival response after 72 hours of AZD6244 treatment in:
(A) Mesenchymal human H157 (top) and murine 344SQ (bottom) cells with or without inducible miR-200 expression. *: p<0.05, **: p<0.01.
(B) Epithelial human H441 (top) and murine 393P (bottom) cells with or without inducible ZEB1 expression. *: p<0.05, **: p<0.01.
(C) Mesenchymal human H157 (top) and murine 344SQ (bottom) cells with stable vector control or IL17RD expression. *: p<0.05, **: p<0.01.
(D) Left: In vivo subcutaneous tumor volume measurements at the indicated time points for 344SQ tumors +/− doxycycline-inducible miR-200 expression, treated daily with AZD6244 or vehicle control. Starting time of treatment denoted by red arrow. Treatment performed in experimental duplicate with 3 to 5 mice per replicate and total sample size denoted in the middle graph. Tumor volume data plotted as the mean and standard deviation. Middle: Final tumor volume measurements at Week 9 of experiment (5 weeks of treatment). Right: Quantification of lung metastatic surface nodules in the indicated experimental groups at Week 9 of experiment. *: p<0.05, **: p<0.01.
(E) Left: In vivo subcutaneous tumor volume measurements at the indicated time points for 344SQ tumors +/− IL17RD expression, treated daily with AZD6244 or vehicle control. Starting time of treatment denoted by red arrow; n=4 to 5 mice per treatment group. Tumor volume data plotted as the mean and standard deviation. *: p<0.05, **: p<0.01. Middle: Final tumor weight measurements at Week 7 of experiment (4 weeks of treatment). Right: Quantification of lung metastatic surface nodules in the indicated experimental groups at Week 7 of experiment.
(F) Left: In vivo subcutaneous tumor volume measurements at the indicated time points for 393P tumors +/− IL17RD knockdown, treated daily with AZD6244 or vehicle control starting at the time indicated by the red arrow; n = 5 mice per treatment group. Tumor volume data plotted as the mean and standard deviation. Middle: Final tumor weight measurements at Week 8 of experiment (5 weeks of treatment). Right: Images of 3 representative primary subcutaneous tumors for each of the indicated treatment groups. Scale bar, 1 cm. **: p<0.01.
To validate our findings in an in vivo context, we first induced miR-200 expression in 344SQ syngeneic tumors and treated them with AZD6244. Although miR-200 increased p-ERK in the tumors (fig. S5A), miR-200 expression alone was sufficient to decrease tumor growth, and combination with AZD6244 treatment had an additive effect that reduced average tumor size by over 90% (Fig. 5D; table S10 in data file S1), similar to the treatment response of epithelial 393P tumors that have high basal expression of miR-200 (Fig. 4D). Constitutive expression of IL17RD in 344SQ cells and tumors increased MAPK signaling with no effect on ZEB1 (fig. S5B,C), moderately decreased tumor volume, and re-sensitized the tumors to AZD6244 treatment, though not to the same degree as induced miR-200 expression (Fig. 5E; table S11 in data file S1). Although IL17RD expression alone did not significantly suppress metastases, combination of IL17RD expression with MEK inhibition reduced lung metastatic lesions (Fig. 5E). Knockdown of IL17RD in epithelial 393P cells reduced MAPK signaling with no change in ZEB1 expression (fig. S5D,E), reduced in vitro cell growth rate as measured by colorimetric absorption over time (fig. S5F), and further sensitized 393P cells to in vitro MEK inhibition (fig. S5G). Knockdown of IL17RD in 393P tumors significantly (p<0.01) reduced in vivo tumor growth to a similar degree as MEK inhibition alone (Fig. 5F; table S12 in data file S1), emphasizing the dependency on the scaffold protein for MAPK signaling and tumor growth. Combination of IL17RD knockdown and MEK inhibitor had a slight additive effect on reducing tumor growth overall, and two of the tumor samples regressed completely in the combination treatment group (Fig. 5F).
ZEB1 expression in lung tumors of KRAS mutant mouse models correlates with decreased IL17RD and MAPK signaling and MEK inhibitor resistance
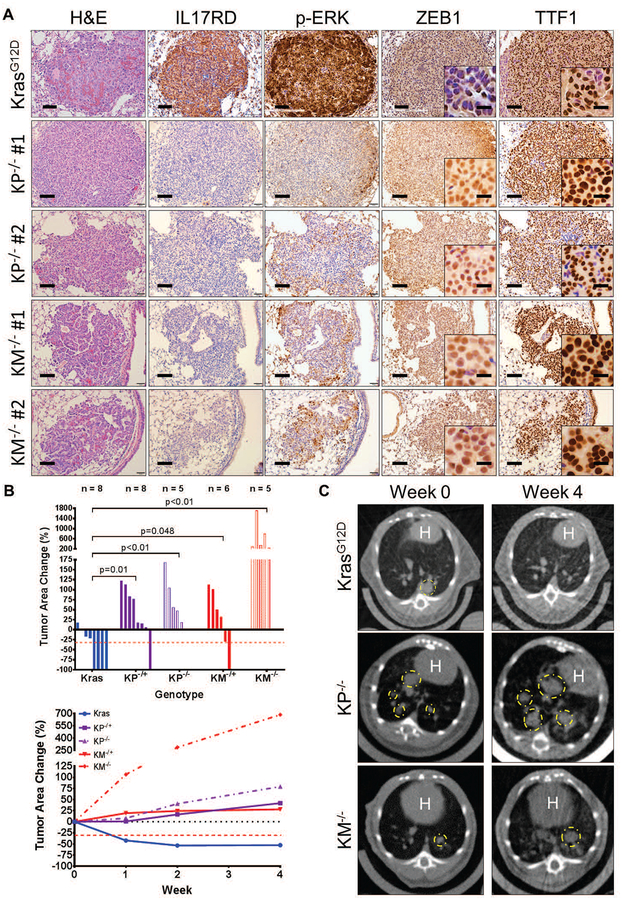
Murine co-clinical trials with selumetinib in combination with docetaxel have shown that the co–mutational status of tumors, such as loss of p53, contributed to therapy resistance (10). Previous studies have reported that Kras;p53 mutant lung tumors compared to mutant KrasG12D tumors display heterogeneous regions of mesenchymal tumor cells, as assessed by changes in expression of EMT markers, most notably ZEB1 (24). These findings suggest that Kras;p53 mutant lung tumor regions with high expression of ZEB1 contain tumor cell subpopulations that are resistant to MEK inhibition. Thus, we next analyzed lung tissues with autochthonous tumors from conditional KrasLSL-G12D (KrasG12D), heterozygous KrasLSL-G12D;p53fll+ (KP−/+), and homozygous KrasLSL-G12D;p53fl/fl (KP−/−) mice, generated through intratracheal administration of adenovirus expressing Cre recombinase (48). Histological analyses demonstrated that KP−/+ and KP−/− mice (4 – 6 months post-induction) displayed multiple tumor regions that stained positively for TTF1 and displayed high ZEB1 expression compared to KrasG12D mice (Fig. 6A and fig. S6). Similar to our observations in syngeneic primary tumors (Fig. 4C), tumor regions with high ZEB1 expression had low amounts of IL17RD and p-ERK, whereas tumor regions with low ZEB1 expression had high IL17RD and p-ERK (Fig. 6A and fig. S6).
Fig. 6. ZEB1 expression correlates with decreased IL17RD, decreased MAPK signaling, and MEK inhibitor resistance in KRAS mutant mouse lung tumors.
(A) H&E, IL17RD, p-ERK, ZEB1, and TTF1 IHC stains of lung tumor tissue from KrasG12D, KP−/−, and KM−/− mice (n=6 tissues per group). KP and KM lung tumor images show regions from two different mice. Scale bars, 100 μm. Inset scale bars, 20 μm.
(B) Top: Percent change in overall lung tumor area of age-matched KrasG12D, KP−/+, KP−/−, KM−/+, and KM−/− mice after 4 weeks of daily treatment with 25 mg/kg AZD6244, as assessed by micro-CT imaging of mouse lungs. Bottom: Trendlines of percent change in overall tumor area at indicated time points for each cohort over 4 weeks of daily treatment with AZD6244. Mouse sample sizes are indicated in the top graph. Percent change in tumor volume data plotted as the mean.
(C) Representative cross-sectional micro-CT images of KrasG12D, KP−/−, and KM−/− mouse lungs before AZD6244 administration (Week 0) and at the treatment endpoint (Week 4). Yellow circles outline representative target lesions. White “H” indicates mouse heart.
To assess whether our findings in the KP mice were due to ZEB1 expression driven by diminished miR-200 expression rather than a secondary effect of p53 loss, we generated a mouse model with conditional miR-141/200c expression by crossing mice with loxP sites flanking the miR-141/200c stem-loop in the miR-141/200c locus (49, 50) with the KrasLSL-G12D mice. Intratracheal delivery of adenoviral-Cre produced lung adenocarcinoma with an activated KrasG12D and heterozygous or homozygous loss of miR-141/200c (KM−/+ or KM−/−). Mice expressing the targeted alleles were validated by PCR amplification of the stem-loop loxP site (fig. S7A). Additionally, the targeting vector inserts a promoter-less lacZ gene driven by the endogenous miR-141/200c promoter activity after Cre-mediated removal, further validating successful locus targeting in the airway epithelium of miR-141/200c (M) and KM tumor tissues (fig. S7B). Verification of miR-141/200c knockout by qPCR analysis of KM lung tumor tissues showed reduction in miR-141/200c compared to KrasG12D tumors, with no compensatory effect by other miR-200 family members from the miR-200b/a/429 locus (fig. S7C). Overall tumor area in KM mice were smaller than KP tumors (fig. S6 and S7D,E). Analysis of murine lung tissues at 1 to 6 months post-induction revealed greater lung tumor numbers but similar overall tumor area in KM mice compared to KrasG12D mice when measured by micro-CT scans and H&E-stained lung tissues (fig. S7E). No tumor growth was observed in mice harboring only the loss of miR-141/200c without an activated Kras allele within the 6 month time course (fig. S6 and S7D,E). IHC analyses of KM tumor tissues showed higher numbers of ZEB1-expressing tumor regions with low amounts of IL17RD and p-ERK relative to KrasG12D tumors (Fig. 6A and fig. S6). We also observed tumor heterogeneity in heterozygous KM−/+ mice similar to that of the KP−/+ mice, with certain tumor regions displaying low ZEB1 expression with high IL17RD and p-ERK (fig. S6). However, homozygous KM−/− lung tumors displayed fewer regions of heterogeneity than heterozygous mice. IHC stains for TTF1 in tumor tissues from the various Kras mutant models demonstrated that, at 4 to 6 months post-induction, all lung tumor tissues were pathologically well-differentiated (Fig. 6A) and confirmed that tumor regions in KM mice were of lung origin.
We next assessed the sensitivity of our murine lung cancer models to therapeutic MEK inhibition. Three months post-induction of lung tumors in KrasG12D, KP, and KM animals by adeno-Cre, tumor formation was confirmed by micro-CT scans and daily doses of single-agent AZD6244 were administered. Micro-CT image analysis of changes in overall lung tumor area as well as H&E lung tissue sections demonstrated that mice with the single KrasG12D mutation, which presented with low ZEB1 and high p-ERK, were sensitive to MEK inhibition, whereas KP and KM lung tumors were more resistant over 4 weeks of treatment (Fig. 6B,C and fig. S8A,B; table S13 in data file S1). We also observed that MEK inhibitor response in the KP and KM mice was dependent on zygosity, as none of the homozygous KP and KM mice showed reduction in tumor size after treatment. Due to the heterogeneous nature of the KP−/+ and KM−/+ mice, we analyzed the lung tumor tissues from the heterozygous mice after MEK inhibitor treatment and observed that the residual TTF1 positive tumor tissues expressed high amounts of ZEB1 and vimentin, with low expression of IL17RD, p-ERK, and E-cadherin (fig. S8C), showing a similar pattern to the syngeneic 393P AZD6244-resistant tumors (Fig. 4E). Taken overall, these data demonstrate that ZEB1 expression inversely correlates with IL17RD expression and MAPK signaling in autochthonous Kras mutant lung cancers, resulting in in vivo resistance to MEK inhibitors.
Mocetinostat suppresses ZEB1 function, derepresses IL17RD expression, and sensitizes resistant tumor cells to MEK inhibition
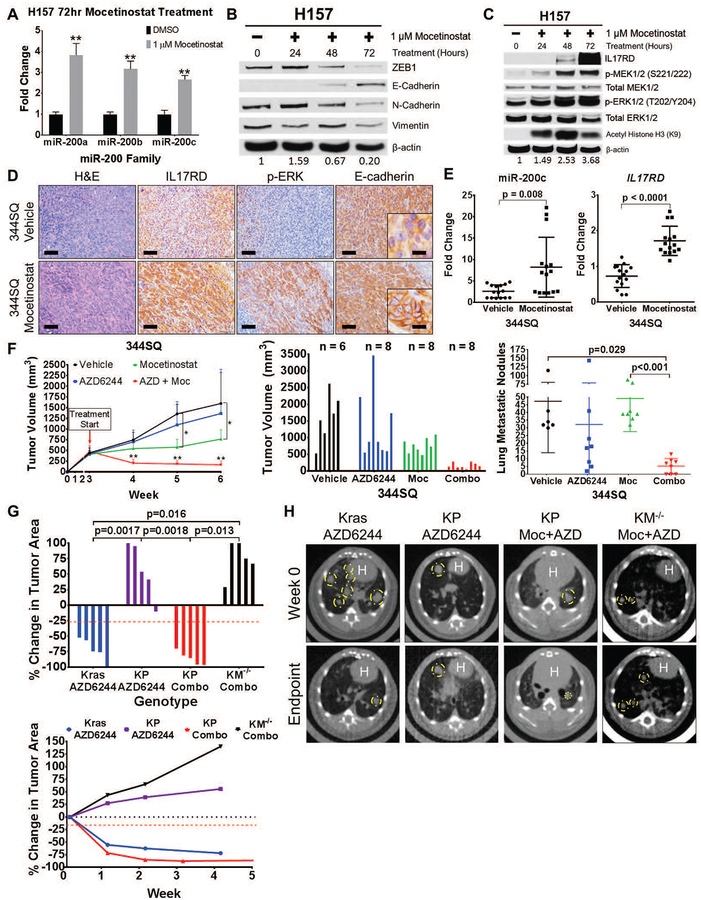
Identifying combinatorial treatment strategies with MEK inhibitors to reduce tumor growth and progression is vital for improving patient survival in KRAS mutant lung cancer. Previous studies have shown that the class I histone deacetylase (HDAC) inhibitor mocetinostat reverts the EMT phenotype through restoring the expression of epithelial microRNAs, including the miR-200 family (51). Treatment of mesenchymal H157 cells with mocetinostat induced miR-200 expression and caused the cells to adopt a more epithelial phenotype at the molecular level, resulting in increased IL17RD expression, increased MAPK signaling, and decreased FGFR1 (Fig. 7A–C and fig. S9A). To verify that the observed changes were independent of secondary effects due to histone acetylation, H157 cells were treated with the pan-HDAC inhibitor trichostatin A (TSA) and showed no observable changes in either EMT markers or MAPK signaling molecules (fig. S9B). Mocetinostat treatment of syngeneic 344SQ tumors produced an increase in IL17RD and p-ERK by IHC analysis, concomitant with an increase in E-cadherin localization to the cell membrane (Fig. 7D). Although we did not observe a changes in ZEB1 or vimentin expression by IHC after mocetinostat treatment (fig. S9C), qPCR analysis of the mocetinostat-treated 344SQ tumors showed an upregulation of both miR-200c and Il17rd, demonstrating functional suppression of ZEB1 activity (Fig. 7E). As previously reported, these findings suggest that in addition to direct promoter binding, ZEB1 can functionally repress expression of epithelial-associated genes through recruitment of histone deacetylases (51–55).
Fig. 7. Mocetinostat increases miR-200 and IL17RD expression and sensitizes resistant tumor cells to MEK inhibition.
(A) QPCR analysis for relative expression of miR-200a, miR-200b, and miR-200c in H157 cells after 72 hours of treatment with mocetinostat. **: p<0.01.
(B) Western blots of EMT markers in H157 cells after treatment with mocetinostat for the indicated time course.
(C) Western blots of IL17RD and MAPK signaling proteins in H157 cells after treatment with mocetinostat for the indicated time course. Acetylated histone (H3K9) is included to confirm HDAC inhibition by mocetinostat.
(D) H&E, IL17RD, p-ERK, and E-cadherin IHC stains of primary 344SQ tumor tissues in syngeneic wild-type mice after 3-week treatment with 80 mg/kg mocetinostat or vehicle control daily (n=5 tumors per group). Scale bars, 100 μm, inset scale bars, 20 μm.
(E) QPCR analysis of miR-200c and IL17RD expression in 344SQ tumor tissues treated with mocetinostat or vehicle control from (D). Technical triplicates of 5 tumor tissue samples from each treatment group were analyzed for each qPCR.
(F) Left: Tumor volume measurements at the indicated time points for 344SQ subcutaneous syngeneic tumors treated daily with single agent 25 mg/kg AZD6244 (AZD), single agent 80 mg/kg mocetinostat (Moc), or both drugs in combination (Combo). Starting time of treatment denoted by red arrow; n=5 mice per treatment group. Tumor volume data plotted as the mean and standard deviation. Middle: Final tumor volume measurements at Week 6 of experiment (3 weeks of treatment). Right: Quantification of lung metastatic surface nodules in the indicated treatment groups at Week 6 of experiment.
(G) Top: Percent change in overall tumor area of KrasLA1-G12D (Kras), KrasLA1-G12D;p53R172HΔG/+ (KP), and KM−/− mice after 4 to 5 weeks of daily treatment with single agent 25 mg/kg AZD6244 or daily combinatorial treatment with AZD6244 and 80 mg/kg mocetinostat (Combo). Bottom: Trendline of percent change in overall tumor area at the indicated time points for Kras, KP, and KM mice with single agent AZD6244 treatment or combinatorial treatment with AZD6244 and mocetinostat; n=5 mice per treatment group. Percent change in tumor volume data plotted as the mean.
(H) Representative micro-CT images of Kras, KP, and KM−/− mice before treatment (Week 0) and at treatment endpoint (Week 4 or Week 5). White “H” indicates location of mouse heart.
Because epithelial cancer cells with low ZEB1 are sensitive to MEK inhibition, suppression of ZEB1 in mesenchymal human and murine lung cancer cells in vitro with mocetinostat increased sensitivity to selumetinib treatment (fig. S9D). In vivo combinatorial administration of mocetinostat and selumetinib in mice bearing syngeneic 344SQ mesenchymal tumors had an additive effect on reducing average tumor size compared to single agent AZD6244 treatment (Fig. 7F; table S14 in data file S1). Mocetinostat alone reduced tumor growth rate, with no observed decrease in the number and size of lung metastatic nodules. However, lung metastatic nodules were reduced in both number and size with dual HDAC and MEK inhibition (Fig. 7F). To test the therapeutic effect on heterogeneous autochthonous tumors that more accurately mimic the p53 mutational status observed in lung cancer patients, we treated the spontaneous KrasLA1-G12D;p53R172HΔG/+ (KP) mice between 7 to 8 months of age with AZD6244 alone or in combination with mocetinostat. Comparable with the adeno-Cre conditional lung tumor mouse models, lung tumors in KrasLA1-G12D (Kras) animals had low expression of ZEB1, correlating with high p-ERK (fig. S9E). Similarly, KP mice exhibited heterogeneity, with the majority of tumor regions expressing high expression of ZEB1, correlating with low phosphorylation of ERK, but a few regions had low ZEB1 and high p-ERK (fig. S9E). Consistent with the adeno-Cre inducible Kras mice, single mutant Kras mice were responsive to MEK inhibition, whereas KP tumors were resistant (Fig. 7G; table S15 in data file S1). However, micro-CT image analysis demonstrated that combinatorial treatment with mocetinostat and AZD6244 reduced overall tumor area in KP, but was unable to reverse drug resistance in the KM mice (Fig. 7G,H; table S15 in data file S1), suggesting that mocetinostat induction of miR-200 is necessary to promote MEK inhibitor sensitivity. Our results present a promising combinatorial therapy with MEK inhibitors to treat KRAS mutant lung cancers.
Patients with poorly differentiated lung adenocarcinomas display high ZEB1 expression and lower phosphorylation of ERK
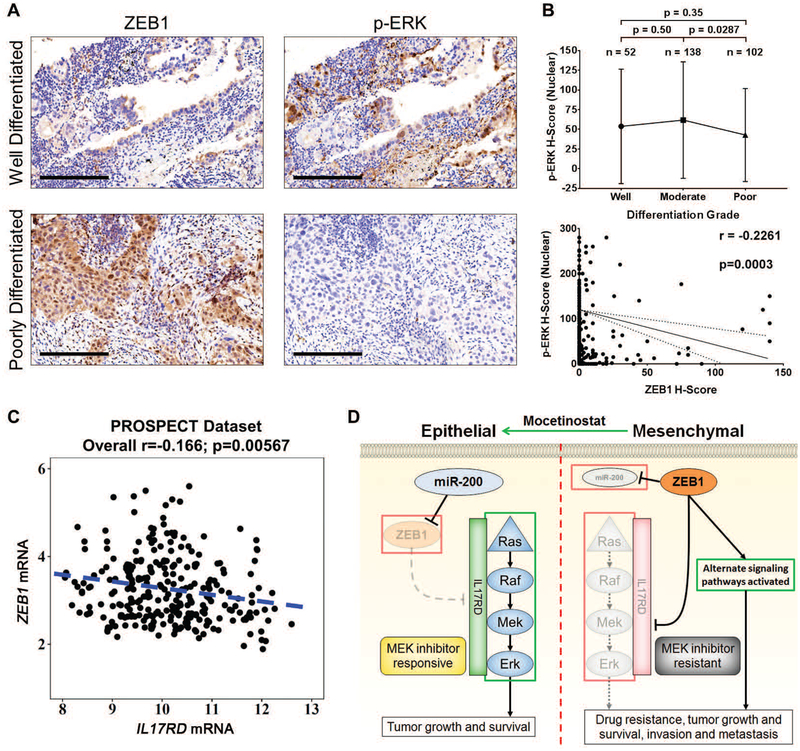
Finally, to further evaluate the clinical context of our findings beyond TCGA analyses, we determined the correlation between p-ERK, IL17RD, and ZEB1 expression in human lung cancer patient samples. Pathological assessment of cytoplasmic and nuclear p-ERK H-scores by IHC staining (Fig. 8A) in a cohort of patient lung tumor tissues (n = 451) showed higher expression of p-ERK in patients with lung adenocarcinoma (ADC) versus patients with squamous cell carcinoma (SCC) (fig. S10A,B), consistent with the frequent activation of this pathway in adenocarcinoma histology. However, pathological analyses revealed that poorly differentiated ADC tumors have lower amounts of nuclear p-ERK when compared to moderately differentiated ADC tumors (Fig. 8B). Cytoplasmic p-ERK showed no statistically significant changes between differentiation grades (fig. S10C).
Fig. 8. Lung adenocarcinoma patients with high ZEB1 have lower phosphorylation of ERK.
(A) Examples of well- and poorly-differentiated human lung ADC tissue sections stained by IHC for ZEB1 and p-ERK. Scale bars, 200 μm.
(B) Top: Nuclear H-scores of p-ERK in ADC specimens of different differentiation grades. Bottom: Cluster plot analysis of Spearman’s rank correlation between ZEB1 and nuclear p-ERK H-scores in ADC specimens. Solid line indicates the linear correlation and dotted lines represent 90% confidence bands.
(C) Cluster plot analysis of Spearman’s rank correlation between ZEB1 and IL17RD mRNA expression in human lung cancer patient tumor samples from PROSPECT dataset.
(D) Proposed model demonstrating differential MAPK signaling pathway activation and sensitivity to MEK inhibitor treatment between epithelial and mesenchymal lung cancer cells due to ZEB1 regulation of IL17RD expression.
Because ZEB1 is important in regulating MAPK signaling, we next wanted to determine the correlation between ZEB1 and p-ERK expression in patient lung tumors. Using data from ZEB1 IHC stains performed in a previous study (24), we compared H-score values of ZEB1 and p-ERK (Fig. 8A) in each patient tumor sample and observed a significant (p=0.0003) negative correlation between ZEB1 and nuclear/cytoplasmic p-ERK in ADC histologic subtypes but not in SCC samples (Fig. 8B and fig. S10D–F). Analysis of ZEB1 H-score and p-MEK did not show any significant correlation in patient tumor samples (fig. S10G–I). Due to the lack of optimized IL17RD antibodies that produced IHC stains of clinically acceptable quality for scoring, we analyzed gene expression profiles in patient lung tumor samples from the PROSPECT dataset and observed a significant (p=0.00567) inverse correlation between IL17RD and ZEB1 mRNA levels (Fig. 8C). However, IL17RD expression was not independently prognostic of overall patient survival in this dataset (fig. S10J). Collectively, our data support a model in which suppression of ZEB1 function in lung cancers promotes MAPK signaling/dependency through upregulation of IL17RD, thereby sensitizing KRAS mutant lung cancers to MEK inhibition (Fig. 8D).
Discussion
The poor response to conventional chemotherapy or MEK inhibitor therapy in patients with KRAS mutant cancers emphasizes the need to identify the underlying resistance mechanisms, build appropriate biomarkers for patient selection, and develop rational combinatorial treatment strategies (12, 13). Tumor heterogeneity and the emergence of resistant subpopulations are likely explanations accounting for some of the observed resistance. By carrying out the in vivo FDAome shRNA dropout screen in conjunction with proteomic profiling, we not only identified differential MAPK signaling activation/dependency during a ZEB1-regulated EMT, but uncovered alternative therapeutic sensitivities in resistant lung tumors which merit potential future investigations. The finding that MAPK signaling inversely correlates with EMT signatures, most notably ZEB1, in human patient samples and tumor models with various genetic backgrounds, including wild-type and mutant KRAS, suggests that this process may have a larger role in development and presents a potential marker to predict MEK inhibitor sensitivity in a broader patient population.
Mechanistically, we identified IL17RD as the factor involved in miR-200-dependent activation of the MAPK pathway in epithelial cells and revealed direct ZEB1 repression of the IL17RD promoter, linking ZEB1 to MAPK signaling regulation and dependency. Re-expression of IL17RD alone activated MAPK signaling, sensitized ZEB1-high tumors to MEK inhibition, and reduced lung metastasis, whereas IL17RD knockdown in ZEB1-low tumors reduced tumor growth to the same degree as MEK inhibitor monotherapy. Considering that stable manipulation of IL17RD did not alter ZEB1 expression or produce feedback regulation of MAPK signaling, our data suggest that increased ZEB1 expression is still necessary to promote MAPK-independent tumor growth after IL17RD suppression. Our finding that IL17RD drives MAPK signaling in epithelial tumors presents a unifying mechanism along with the FGFR1 feedback model of resistance (15, 16). IL17RD not only has a reciprocal expression to FGFR1 and ZEB1, but the scaffold protein negatively regulates FGFR1-asociated feedback activation of MAPK signaling (43, 45, 56). Due to its correlation with MAPK signaling and inverse correlation with ZEB1 in murine models and human patient datasets, IL17RD may serve as an additional marker to predict MEK inhibitor sensitivity. However, the optimization of IL17RD detection antibodies for pathological purposes will be critical to further assess its role in predicting patient response to clinical MEK inhibitors. Because IL17RD is involved during development (43, 56), is a negative regulator of EMT (57, 58), and causes metastasis to become predominantly dependent upon MAPK signaling, the broader role of IL17RD in counter regulation and restructuring of global cell signaling patterns warrants future studies. Lastly, in addition to ZEB1 regulating IL17RD-mediated MAPK signaling, validation of our RPPA data revealed a continuous decrease in p90RSK with ZEB1 expression, which did not follow the same pattern as upstream MAPK signaling molecules. This correlation suggests another potential mode of ZEB1 regulation of MAPK signaling at the most downstream component of the MAPK pathway, consistent with the observed correlation between p90RSK and EMT in the TCGA datasets.
Because our results demonstrate an epigenetic basis for the rewiring of signal transduction pathways in the tumor cell subpopulations with high ZEB1 expression, consistent with findings from earlier studies (24, 25, 34, 59), the application of combinatorial treatments is necessary to effectively prevent the outgrowth of resistant subpopulations. Separate studies have proposed a combination of MEK inhibitors with RTK- or FGFR1-specific inhibitors to reduce primary tumor growth (15, 16). However, our in vivo shRNA screen suggests that the emergent MEK inhibitor-resistant population is in a ZEB1-high, MAPK-independent state, which drives tumor cell invasion and metastasis (17, 22–26, 35), major causes of lung cancer patient deaths (27, 60). In light of these data, we advocate an alternative combination based on the effects of the class I HDAC inhibitor mocetinostat to suppress the epigenetic function of ZEB1 and recapitulate the functional and molecular changes observed with forced miR-200 expression. We demonstrate that the combination of selumetinib with mocetinostat, currently undergoing testing in clinical trials (https://clinicaltrials.gov/ct2/results?term=mocetinostat), not only decreases primary tumor growth in the syngeneic and various autochthonous models, but reduces metastatic disease as well, providing pre-clinical rationale for a combinatorial treatment strategy in patients.
Extending our findings to pre-clinical murine models to more accurately model tumor heterogeneity, we used our syngeneic KP tumor models, multiple previously characterized Kras mutant mouse models of lung cancer, and our conditional KrasLSL-G12D;miR-141/200cflfl (KM) mice. These models confirmed that: 1) loss of miR-200 de-represses ZEB1 expression, which is sufficient to suppress IL17RD and MAPK signaling, 2) miR-200c is the predominant miR-200 family member regulating ZEB1 expression, consistent with in vitro overexpression of individual miR-200 family members, and 3) cancer cells with low ZEB1 expression within the heterogeneous tumor tissue are responsive to single agent MEK inhibitor treatment, whereas the cell subpopulations with high ZEB1 expression are resistant and form the basis for outgrowth over time. Furthermore, our in vivo tumor growth assays demonstrate that MEK inhibitor resistance is both innate and acquired, correlating with expression of ZEB1. An independent study using murine models of pancreatic cancer demonstrated that ZEB1 knockout in KP mice enriched gene expression patterns associated with Kras addiction (61), indirectly confirming our in vivo findings. Because miR-141/200c knockout did not affect other miR-200 family members, further studies with conditional miR-141/200c and KM mutant mice should not only elucidate the non-redundant roles of other miR-200 members on tumor progression, but also provide the field a means to understand various biological and developmental processes.
Additionally important for translational therapeutic development, the combined RPPA and shRNA library screen data provide numerous additional drug targets that warrant further investigation to optimize combination strategies. A major advantage of applying the shRNA library screen to immunocompetent syngeneic models is the capability to detect target genes that may synergize with or antagonize immunotherapies. These combined high-throughput analyses will need to be further complemented with in vivo studies delineating changes in the tumor immune microenvironment as part of the systematic characterization of tumor progression and drug resistance.
Finally, the ability to predict sensitivity to MEK inhibition based upon ZEB1 status will allow for the establishment of other drug combinations – in addition to our proposed strategy – that target the resistant cancer cell subpopulations. Extending our pre-clinical therapeutic strategies to different RAS mutant cancer types, such as pancreas and colon, may further improve the treatment options for cancer patients. These results emphasize the importance of epigenetic changes and the need for biomarkers beyond the standard DNA mutation profiling to select patients who will likely benefit from single agent therapy versus combinations.
Materials and Methods
Study Design
The objective of the study was to understand the mechanisms of MEK inhibitor resistance in KRAS mutant lung cancers and identify therapeutic targets in the resistant cell population. We used an in vivo shRNA screen in epithelial or mesenchymal murine lung cancer cell lines, targeting genes with FDA-approved compounds. Concurrently, we performed an RPPA proteomic profile on murine and human epithelial and mesenchymal lung cancer cell lines with manipulated miR-200 and ZEB1 expression. These initial analyses revealed enhanced MAPK signaling activity and dependency in epithelial lung cancer cells and tumors. Western blots and qPCR analyses verified our in vivo screen and RPPA profile and mechanistically identified IL17RD as the scaffold protein responsible for enhanced MAPK signaling in epithelial lung cancer cells. Luciferase reporter assays and chromatin immunoprecipitation were performed in technical and biological triplicates to verify direct ZEB1 suppression of IL17RD transcriptional activity.
Translating our findings therapeutically, human and murine epithelial and mesenchymal lung cancer cells were treated in vitro with the MEK inhibitors selumetinib and trametinib in experimental triplicates. Findings from our mechanistic studies were validated in vivo by performing IHC analysis on multiple Kras mutant mouse models of lung tumors (n=6 to 10). To therapeutically validate our findings in vivo, we treated syngeneic epithelial and mesenchymal murine lung tumors as well as multiple Kras GEMMs with selumetinib (n=5 to 10 per group) unblinded in age-matched mice. We demonstrated that IL17RD was necessary to sensitize resistant tumors to MEK inhibition by overexpressing and knocking down IL17RD in mesenchymal and epithelial mouse lung tumors, respectively, and treated age-matched mice unblinded with selumetinib (n=4 to 5 mice per group). To determine if reversal of a ZEB1-driven EMT sensitized resistant lung cancers to MEK inhibition, syngeneic tumors and multiple KRAS GEMMs were treated unblinded with the class I HDAC inhibitor mocetinostat in combination with selumetinib (n=5 to 8 mice per group). The numbers of mice were selected based on resource availability and based on previous analyses to obtain statistical power with a minimum number of mice. Endpoints of in vivo experiments were determined when veterinary technicians flagged mice for euthanasia based on primary tumor volume >1.5 cm3. Data points from in vivo experiments were excluded if the animal died before the completion of the experiment or if the tumor regressed inexplicably.
Human lung tumor specimens were analyzed for markers by IHC, and H-scores were assessed by two pathologists unblinded. For nuclear and cytoplasmic IHC, the percentage of staining intensity was scored as 0 (no staining), 1+ (weak staining), 2+ (moderate staining), and 3+ (strong staining), and the extension of the expression for each sample was reported as percentage of positive cells. The final score (H-score) was obtained by multiplying the intensity and reactivity extension values (range, 0–300). Human lung tumor tissue numbers were based on availability of samples, and all patient identities remained confidential.
Statistics
For analysis of RPPA data, a linear mixed model was applied to compare protein expression on a protein-by-protein basis between epithelial and mesenchymal groups; the model includes cell line effects as a random effect factor. The resulting p-values were modeled by a Beta-Uniform Model (BUM). To identify protein markers differentially regulated in epithelial and mesenchymal phenotypes, we used a False Discovery Rate (FDR) of 0.05 as the cutoff. The Pearson correlation was used for distance matrix calculation, and Ward method was applied as linkage rule for the hierarchical clustering. Spearman’s Rank Correlation was used to correlate gene expression and IHC H-score values in human patient samples. Kaplan-Meier analysis was assessed by log-rank test. Statistical significance for all other experiments were assessed using two-tailed student’s t-test, and statistical significance was accepted as having a p-value less than 0.05.
Supplementary Material
Materials and methods.
Fig. S1. Epithelial tumor cells exhibit greater MAPK signaling
Fig. S2. MAPK signaling in epithelial cells is only partially dependent on FAK-PAK signaling
Fig. S3. IL17RD expression in epithelial cancer cells correlates with MAPK signaling and inversely correlates with FGFR1
Fig. S4. Zeb1-expressing lung cancer cells are resistant to MEK inhibition
Fig. S5. MiR-200 sensitizes mesenchymal lung tumors to MEK inhibition through IL17RD upregulation
Fig. S6. ZEB1 expression correlates with decreased IL17RD and decreased MAPK signaling in Kras mutant mice lung tumors
Fig. S7. KrasG12D;miR-141/200c−/+ (KM) mouse lung tumor characterization display high ZEB1, low MAPK signaling, and tumor heterogeneity
Fig. S8. ZEB1 expression in MEK inhibitor-treated lung tumors of Kras mutant mice correlates with resistance
Fig. S9. Mocetinostat recapitulates miR-200 expression sensitizes resistant tumor cells to MEK inhibition
Fig. S10. ZEB1 inversely correlates with ERK phosphorylation in lung adenocarcinoma patients
Data file S1 contains the following supplementary tables:
Table S1. Quality control metrics for the shRNA screens
Table S2. Gene level dropout scores for each of the four screens described in Fig. 1A
Table S3. RPPA dataset of protein expression fold change between epithelial and mesenchymal human lung cancer cell lines
Table S4. RPPA dataset of protein expression fold change between epithelial and mesenchymal murine lung cancer cell lines
Table S5. RPPA dataset of protein expression fold change in H157 cells (mesenchymal) following induced miR-200 expression (epithelial)
Table S6. RPPA dataset of protein expression fold change in H1299 cells (mesenchymal) following induced miR-200 expression (epithelial)
Table S7. RPPA dataset of protein expression fold change in HCC827 cells (epithelial) with constitutive ZEB1 expression (mesenchymal)
Table S8. RPPA dataset of protein expression fold change in H441 cells (epithelial) following induced ZEB1 expression (mesenchymal)
Table S9. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 4D
Table S10. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 5D
Table S11. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 5E
Table S12. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 5F
Table S13. Individual mouse lung tumor area measurements and percent change in lung tumor area from experiment in Fig. 6B
Table S14. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 7F
Table S15. Individual mouse lung tumor area measurements and percent change in lung tumor area from experiment in Fig. 7G
Table S16. Supplementary material table listing all commercially used antibodies, primers, shRNA, and cDNA ORFs including catalog numbers and sequences
Acknowledgements:
We thank Dr. G. Goodall (University of Adelaide, Australia) for the kind gift of the pTRIPz-miR-200 expression constructs. We thank Dr. Young-Ho Ahn (Ewha Womans University School of Medicine, South Korea) for the H441 inducible ZEB1 cells. We thank Charles Kingsley for providing the micro-CT images of the mouse lungs. We thank Dr. Scott Woodman for thoughtful comments on the manuscript.
Funding: This work was supported by the NIH R37 CA214609-01A1, CPRIT RP150405, CPRIT-MIRA RP160652-P3, Uniting against Lung Cancer/Lung Cancer Research Foundation award, Rexanna’s Foundation for Fighting Lung Cancer to D.L.G., and LUNGevity Foundation award (L.A.B. and D.L.G.). DP was supported by a CPRIT Graduate Scholar Training Grant (RP140106). JW and PT are supported by Lung SPORE (P50 CA070907), Cancer Center Support Grant (CCSG CA016672) and Mary K Chapman Foundation. LAB and DLG are R Lee Clark Fellows of the University of Texas MD Anderson Cancer Center, supported by the Jeane F Shelby Scholarship Fund. Y.Y. is supported by a Mayo Clinic CBD platform award (93059043). The work was also supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Cancer Moon Shots Program. This work is dedicated to the memory of Dr. Waun Ki Hong.
Footnotes
Competing Interests: D.L.G. declares advisory board work for Janssen, AstraZeneca, GlaxoSmithKline and Sanofi. D.L.G. receives research grant funding from AstraZeneca, Janssen, and Takeda. L.A.B. declares consulting work for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar, SA. L.A.B. receives research grant funding from AbbVie, AstraZeneca, GenMab, Tolero Pharmaceuticals. All other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or supplementary materials.
References and Notes:
- 1.Sharma SV, Haber DA, Settleman J, Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer 10, 241–253 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Heymach JV, Lippman SM, Lung cancer. N Engl J Med 359, 1367–1380 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodenhuis S, Slebos RJ, Clinical significance of ras oncogene activation in human lung cancer. Cancer Res 52, 2665s–2669s (1992). [PubMed] [Google Scholar]
- 4.Wang Y, Kaiser CE, Frett B, Li H.-y., Targeting Mutant KRAS for Anticancer Therapeutics: A Review of Novel Small Molecule Modulators. Journal of Medicinal Chemistry 56, 5219–5230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S, Lafitte JJ, Sculier JP, The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. British journal of cancer 92, 131–139 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, Liao ML, Bischoff H, Reck M, Sellers MV, Watkins CL, Speake G, Armour AA, Kim ES, Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 28, 744–752 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ, Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji H, Wang Z, Perera SA, Li D, Liang MC, Zaghlul S, McNamara K, Chen L, Albert M, Sun Y, Al-Hashem R, Chirieac LR, Padera R, Bronson RT, Thomas RK, Garraway LA, Janne PA, Johnson BE, Chin L, Wong KK, Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res 67, 4933–4939 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, Saborowski M, Kastenhuber E, Fellmann C, Ohara K, Morikami K, Miura T, Lukacs C, Ishii N, Lowe S, Rosen N, Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, Gao P, Woo MS, Xu C, Yanagita M, Altabef A, Wang S, Lee C, Nakada Y, Pena CG, Sun Y, Franchetti Y, Yao C, Saur A, Cameron MD, Nishino M, Hayes DN, Wilkerson MD, Roberts PJ, Lee CB, Bardeesy N, Butaney M, Chirieac LR, Costa DB, Jackman D, Sharpless NE, Castrillon DH, Demetri GD, Janne PA, Pandolfi PP, Cantley LC, Kung AL, Engelman JA, Wong KK, A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 483, 613–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, Schlicker A, Wessels LF, Smit EF, Thunnissen E, Halonen P, Lieftink C, Beijersbergen RL, Di Nicolantonio F, Bardelli A, Trusolino L, Bernards R, Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports 7, 86–93 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Blumenschein GR Jr., Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T, Dunphy F, Udud K, Ahn MJ, Hanna NH, Kim JH, Mazieres J, Kim SW, Baas P, Rappold E, Redhu S, Puski A, Wu FS, Janne PA, A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC). Ann Oncol 26, 894–901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jänne PA, van den Heuvel MM, Barlesi F, Cobo M, Mazieres J, Crinò L, Orlov S, Blackhall F, Wolf J, Garrido P, Poltoratskiy A, Mariani G, Ghiorghiu D, Kilgour E, Smith P, Kohlmann A, Carlile DJ, Lawrence D, Bowen K, Vansteenkiste J, Selumetinib Plus Docetaxel Compared With Docetaxel Alone and Progression-Free Survival in Patients With KRAS-Mutant Advanced Non–Small Cell Lung Cancer. Jama 317, 1844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J, A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, Oizumi S, Nishimura M, Faber AC, Yano S, Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov 6, 754–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manchado E, Weissmueller S, Morris J. P. t., Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, Engelman JA, Rudin CM, Rosen N, Lowe SW, A combinatorial strategy for treating KRAS-mutant lung cancer. Nature 534, 647–651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM, Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 23, 2140–2151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ, The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10, 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Gregory PA, Bracken CP, Smith E, Bert AG, Wright JA, Roslan S, Morris M, Wyatt L, Farshid G, Lim YY, Lindeman GJ, Shannon MF, Drew PA, Khew-Goodall Y, Goodall GJ, An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell 22, 1686–1698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, Girard L, Behrens C, Wistuba II, Gazdar AF, Hayward NK, Minna JD, ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest 126, 3219–3235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T, The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11, 1487–1495 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ, Flores ER, Kurie JM, ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest 122, 3170–3183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL, The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene 35, 173–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng DH, Ungewiss C, Tong P, Byers LA, Wang J, Canales JR, Villalobos PA, Uraoka N, Mino B, Behrens C, Wistuba II, Han RI, Wanna CA, Fahrenholtz M, Grande-Allen KJ, Creighton CJ, Gibbons DL, ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene 36, 1925–1938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungewiss C, Rizvi ZH, Roybal JD, Peng DH, Gold KA, Shin DH, Creighton CJ, Gibbons DL, The microRNA-200/Zeb1 axis regulates ECM-dependent beta1-integrin/FAK signaling, cancer cell invasion and metastasis through CRKL. Sci Rep 6, 18652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y, Ahn YH, Chen Y, Tan X, Guo L, Gibbons DL, Ungewiss C, Peng DH, Liu X, Lin SH, Thilaganathan N, Wistuba II, Rodriguez-Canales J, McLendon G, Creighton CJ, Kurie JM, ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J Clin Invest 124, 2696–2708 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel RL, Miller KD, Jemal A, Cancer statistics, 2016. CA Cancer J Clin 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Carugo A, Genovese G, Seth S, Nezi L, Rose JL, Bossi D, Cicalese A, Shah PK, Viale A, Pettazzoni PF, Akdemir KC, Bristow CA, Robinson FS, Tepper J, Sanchez N, Gupta S, Estecio MR, Giuliani V, Dellino GI, Riva L, Yao W, Di Francesco ME, Green T, D’Alesio C, Corti D, Kang Y, Jones P, Wang H, Fleming JB, Maitra A, Pelicci PG, Chin L, DePinho RA, Lanfrancone L, Heffernan TP, Draetta GF, In Vivo Functional Platform Targeting Patient-Derived Xenografts Identifies WDR5-Myc Association as a Critical Determinant of Pancreatic Cancer. Cell reports 16, 133–147 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Konig R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, Chanda SK, A probability-based approach for the analysis of large-scale RNAi screens. Nat Methods 4, 847–849 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D, Smith Q, Beijersbergen RL, Ghazal P, Shamu CE, Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods 6, 569–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardnell RJ, Behrens C, Diao L, Fan Y, Tang X, Tong P, Minna JD, Mills GB, Heymach JV, Wistuba II, Wang J, Byers LA, An Integrated Molecular Analysis of Lung Adenocarcinomas Identifies Potential Therapeutic Targets among TTF1-Negative Tumors, Including DNA Repair Proteins and Nrf2. Clin Cancer Res 21, 3480–3491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Davies MA, Liu W, Coombes K, Meric-Bernstam F, Bedrosian I, McGahren M, Agarwal R, Zhang F, Overgaard J, Alsner J, Neve RM, Kuo WL, Gray JW, Borresen-Dale AL, Mills GB, A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics 6, 129–151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling 6, pl1–pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, Nilsson MB, Gudikote J, Tran H, Cardnell RJ, Bearss DJ, Warner SL, Foulks JM, Kanner SB, Gandhi V, Krett N, Rosen ST, Kim ES, Herbst RS, Blumenschein GR, Lee JJ, Lippman SM, Ang KK, Mills GB, Hong WK, Weinstein JN, Wistuba II, Coombes KR, Minna JD, Heymach JV, An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 19, 279–290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal JD, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Jones S, Suraokar M, Welsh JW, Erez B, Wistuba II, Chen L, Peng D, Wang S, Ullrich SE, Heymach JV, Kurie JM, Qin FX, Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 5, 5241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Guo L, Creighton CJ, Lu Q, Gibbons DL, Yi ES, Deng B, Molina JR, Sun Z, Yang P, Yang Y, A genetic cell context-dependent role for ZEB1 in lung cancer. Nat Commun 7, 12231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An S, Yang Y, Ward R, Liu Y, Guo XX, Xu TR, Raf-interactome in tuning the complexity and diversity of Raf function. FEBS J 282, 32–53 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Bluthgen N, Legewie S, Kielbasa SM, Schramme A, Tchernitsa O, Keil J, Solf A, Vingron M, Schafer R, Herzel H, Sers C, A systems biological approach suggests that transcriptional feedback regulation by dual-specificity phosphatase 6 shapes extracellular signal-related kinase activity in RAS-transformed fibroblasts. Febs Journal 276, 1024–1035 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Owens DM, Keyse SM, Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Zhao Z, Chen CC, Rillahan CD, Shen RL, Kitzing T, McNerney ME, Diaz-Flores E, Zuber J, Shannon K, Le Beau MM, Spector MS, Kogan SC, Lowe SW, Cooperative loss of RAS feedback regulation drives myeloid leukemogenesis. Nat Genet 47, 539–U150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karreth FA, Tuveson DA, Modelling oncogenic Ras/Raf signalling in the mouse. Curr Opin Genet Dev 19, 4–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K, A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell 10, 459–472 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furthauer M, Lin W, Ang SL, Thisse B, Thisse C, Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol 4, 170–174 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Ren Y, Cheng L, Rong Z, Li Z, Li Y, Zhang X, Xiong S, Hu J, Fu XY, Chang Z, hSef potentiates EGF-mediated MAPK signaling through affecting EGFR trafficking and degradation. Cell Signal 20, 518–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovalenko D, Yang X, Nadeau RJ, Harkins LK, Friesel R, Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J Biol Chem 278, 14087–14091 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, Kirchner T, Behrens J, Brabletz T, The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 68, 537–544 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Thiery JP, Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2, 442–454 (2002). [DOI] [PubMed] [Google Scholar]
- 48.DuPage M, Dooley AL, Jacks T, Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, Martinez M, Nefadov M, Abbas AK, Weiss A, Lanier LL, de Jong PJ, Bluestone JA, Srivastava D, McManus MT, A resource for the conditional ablation of microRNAs in the mouse. Cell reports 1, 385–391 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao H, Jheon A, Li X, Sun Z, Wang J, Florez S, Zhang Z, McManus MT, Klein OD, Amendt BA, The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development 140, 3348–3359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meidhof S, Brabletz S, Lehmann W, Preca BT, Mock K, Ruh M, Schuler J, Berthold M, Weber A, Burk U, Lubbert M, Puhr M, Culig Z, Wellner U, Keck T, Bronsert P, Kusters S, Hopt UT, Stemmler MP, Brabletz T, ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med 7, 831–847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kakihana M, Ohira T, Chan D, Webster RB, Kato H, Drabkin HA, Gemmill RM, Induction of E-cadherin in lung cancer and interaction with growth suppression by histone deacetylase inhibition. J Thorac Oncol 4, 1455–1465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, Helfrich B, Dziadziuszko R, Chan DC, Sugita M, Chan Z, Baron A, Franklin W, Drabkin HA, Girard L, Gazdar AF, Minna JD, Bunn PA Jr., Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res 66, 944–950 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Lin X, Sun B, Liang M, Liang Y-Y, Gast A, Hildebrand J, Brunicardi FC, Melchior F, Feng X-H, Opposed Regulation of Corepressor CtBP by SUMOylation and PDZ Binding. Molecular Cell 11, 1389–1396 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn CO, Heidecke CD, Friess H, Buchler M, Evert M, Lerch MM, Weiss FU, Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 61, 439–448 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Tsang M, Friesel R, Kudoh T, Dawid IB, Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol 4, 165–169 (2002). [DOI] [PubMed] [Google Scholar]
- 57.He Q, Gong Y, Gower L, Yang X, Friesel RE, Sef Regulates Epithelial-Mesenchymal Transition in Breast Cancer Cells. J Cell Biochem 117, 2346–2356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hori S, Wadhwa K, Pisupati V, Zecchini V, Ramos-Montoya A, Warren AY, Neal DE, Gnanapragasam VJ, Loss of hSef promotes metastasis through upregulation of EMT in prostate cancer. Int J Cancer 140, 1881–1887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J, Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26, 207–221 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Cataldo VD, Gibbons DL, Pérez-Soler R, Quintás-Cardama A, Treatment of Non–Small-Cell Lung Cancer with Erlotinib or Gefitinib. New England Journal of Medicine 364, 947–955 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, Brunton VG, Pilarsky C, Winkler TH, Brabletz S, Stemmler MP, Brabletz T, The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 19, 518–529 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods.
Fig. S1. Epithelial tumor cells exhibit greater MAPK signaling
Fig. S2. MAPK signaling in epithelial cells is only partially dependent on FAK-PAK signaling
Fig. S3. IL17RD expression in epithelial cancer cells correlates with MAPK signaling and inversely correlates with FGFR1
Fig. S4. Zeb1-expressing lung cancer cells are resistant to MEK inhibition
Fig. S5. MiR-200 sensitizes mesenchymal lung tumors to MEK inhibition through IL17RD upregulation
Fig. S6. ZEB1 expression correlates with decreased IL17RD and decreased MAPK signaling in Kras mutant mice lung tumors
Fig. S7. KrasG12D;miR-141/200c−/+ (KM) mouse lung tumor characterization display high ZEB1, low MAPK signaling, and tumor heterogeneity
Fig. S8. ZEB1 expression in MEK inhibitor-treated lung tumors of Kras mutant mice correlates with resistance
Fig. S9. Mocetinostat recapitulates miR-200 expression sensitizes resistant tumor cells to MEK inhibition
Fig. S10. ZEB1 inversely correlates with ERK phosphorylation in lung adenocarcinoma patients
Data file S1 contains the following supplementary tables:
Table S1. Quality control metrics for the shRNA screens
Table S2. Gene level dropout scores for each of the four screens described in Fig. 1A
Table S3. RPPA dataset of protein expression fold change between epithelial and mesenchymal human lung cancer cell lines
Table S4. RPPA dataset of protein expression fold change between epithelial and mesenchymal murine lung cancer cell lines
Table S5. RPPA dataset of protein expression fold change in H157 cells (mesenchymal) following induced miR-200 expression (epithelial)
Table S6. RPPA dataset of protein expression fold change in H1299 cells (mesenchymal) following induced miR-200 expression (epithelial)
Table S7. RPPA dataset of protein expression fold change in HCC827 cells (epithelial) with constitutive ZEB1 expression (mesenchymal)
Table S8. RPPA dataset of protein expression fold change in H441 cells (epithelial) following induced ZEB1 expression (mesenchymal)
Table S9. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 4D
Table S10. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 5D
Table S11. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 5E
Table S12. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 5F
Table S13. Individual mouse lung tumor area measurements and percent change in lung tumor area from experiment in Fig. 6B
Table S14. Individual mouse subcutaneous tumor volume measurements from experiment in Fig. 7F
Table S15. Individual mouse lung tumor area measurements and percent change in lung tumor area from experiment in Fig. 7G
Table S16. Supplementary material table listing all commercially used antibodies, primers, shRNA, and cDNA ORFs including catalog numbers and sequences