SUMMARY
The RNA binding protein DAZL is essential for gametogenesis, but its direct in vivo functions, RNA targets, and the molecular basis for germ cell loss in Dazl-null mice are unknown. Here, we mapped transcriptome-wide DAZL-RNA interactions in vivo, revealing DAZL binding to thousands of mRNAs via polyA-proximal 3′ UTR interactions. In parallel, fluorescence-activated cell sorting and RNA-seq identified mRNAs sensitive to DAZL deletion in male germ cells. Despite binding a broad set of mRNAs, integrative analyses indicate that DAZL post-transcriptionally controls only a subset of its mRNA targets, namely those corresponding to a network of genes that are critical for germ cell proliferation and survival. In addition, we provide evidence that polyA sequences have key roles in specifying DAZL-RNA interactions across the transcriptome. Our results reveal a mechanism for DAZL-RNA binding and illustrate that DAZL functions as a master regulator of a post-transcriptional mRNA program essential for germ cell survival.
In Brief
Combining transgenic mice, FACS, and multiple RNA-profiling methods, Zagore et al. show that DAZL binds thousands of mRNAs via GUU sites upstream of polyA tails. Loss of DAZL results in decreased mRNA levels for a network of genes that are essential for germ cell proliferation and differentiation.
Graphical Abstract
INTRODUCTION
RNA binding proteins (RBPs) are potent post-transcriptional regulators of gene expression. In the nucleus, RBPs can alter pre-mRNA processing to generate mRNAs with different coding and non-coding sequences. In the cytoplasm, RBPs influence mRNA localization, translation, and stability, typically through interactions with 3′ untranslated regions (3′ UTRs). The combined effects of RBPs can greatly alter the cellular proteome. Despite widespread recognition of the importance of RBPs in development, immunity, and disease (Scotti and Swanson, 2016), our understanding of how specific RBPs modulate transcriptional programs to control cell fate is extremely limited.
Regulation of mRNA processing and translation is essential during spermatogenesis, the highly ordered process of postnatal male germ cell development that yields haploid spermatozoa (Licatalosi, 2016). The first few postnatal days are critical for the establishment of spermatogonial stem cells required for continued sperm production throughout life (Manku and Culty, 2015). Many genes critical for spermatogonial proliferation and differentiation have been identified, including genes encoding cell-cycle checkpoint factors, regulators of the DNA damage response, transcription factors, and RBPs that affect mRNA export and stability (Pan et al., 2009; Saga, 2010; Song and Wilkinson, 2014). However, how these genes are coordinately regulated to ensure proper germ cell development is not well understood.
The importance of RBPs in gametogenesis is well illustrated by the DAZ (deleted in azoospermia) family of RBPs. These germ cell-restricted RBPs (DAZ, DAZL, and BOULE) are necessary for gametogenesis in worms, flies, mice, and humans (Vangompel and Xu, 2011). Their significance was first demonstrated in the 1990s, when Daz was discovered in a region of the Y chromosome deleted in 10%–15% of men with azoospermia (Reijo et al., 1995, 1996). Deletion of DAZL in mice leads to dramatic germ cell loss (Ruggiu et al., 1997; Lin and Page, 2005). Transgenic expression of human DAZL or DAZ partially rescues the phenotype of Dazl knockout (KO) mice (Vogel et al., 2002), indicating functional conservation of DAZ family RBPs across species. Despite the clear biological importance of these RBPs, many critical questions remain, including the identities of their direct RNA targets, how these RNAs are regulated, and why loss of this regulation results in germ cell defects. In this study, we provide answers to these long-standing questions.
The cytoplasmic localization, co-sedimentation with polyribosomes, and association with polyadenylated (polyA+) RNA of DAZL suggest potential roles in regulating germ cell mRNA stability or translation (Ruggiu et al., 1997; Tsui et al., 2000). In addition, yeast two-hybrid analysis of DAZL interactors identified RBPs with cytoplasmic roles in mRNA regulation, including PUM2, QK3, and the polyA-binding protein PABPC1 (Moore et al., 2003; Fox et al., 2005; Collier et al., 2005). However, the scarcity and variable number of germ cells present in Dazl KO mice (Ruggiu et al., 1997; Schrans-Stassen et al., 2001; Saunders et al., 2003) have presented major barriers to investigating the direct in vivo function(s) of DAZL in the male germline. Consequently, most previous DAZL studies have relied on re-constituted systems, including transfection of somatic cell lines (Maegawa et al., 2002; Xu et al., 2013), artificial tethering to in vitro synthesized RNAs injected into oocytes or zebrafish (Collier et al., 2005; Takeda et al., 2009), and in vitro-derived germ cells (Haston et al., 2009; Chen et al., 2014). These efforts have suggested diverse DAZL functions in different cell contexts, including roles in stress granule assembly, mRNA localization, stabilization, and translation (Fu et al., 2015). However, transfection assays have shown that DAZL can have opposite effects on the same reporter RNA in different somatic cell lines (Xu et al., 2013). It is not clear whether these discrepancies are due to cell context and/or non-physiological levels or recruitment of DAZL to RNA targets.
While the direct in vivo functions are not defined, in vitro binding assays and X-ray crystallography of the DAZL RNA recognition motif (RRM) identified GUU as a high-affinity binding site (Jenkins et al., 2011). However, the frequency of GUU across the transcriptome hampers bioinformatic predictions of functional DAZL-binding sites and RNA targets in vivo. Microarray analyses of RNAs that co-immunoprecipitate with DAZL have suggested potential targets (Reynolds et al., 2005, 2007; Chen et al., 2014); yet, few have been examined in Dazl KO mice, and these cannot account for the dramatic germ cell loss. Furthermore, different investigators have arrived at alternate conclusions about the role of DAZL as a translation repressor or activator based on immunofluorescence (IF) assays of wild-type (WT) and Dazl KO germ cells (Reynolds et al., 2005; Chen et al., 2014). Moreover, neither group explored whether the observed differences in protein abundance are associated with changes in mRNA levels.
These observations underscore the need to identify the direct in vivo RNA targets of DAZL in an unbiased and transcriptome-wide manner, as well as new strategies to both isolate limiting Dazl KO germ cells for transcriptome profiling and investigate how DAZL binds and regulates its RNA targets.
In this study, we used an integrative approach to elucidate the direct RNA targets and in vivo functions of DAZL in male germ cells. Multiple high-resolution, transcriptome-wide in vivo maps of DAZL-RNA interactions reveal DAZL binding to a vast set of mRNAs, predominantly through GUU sites in 3′ UTRs. Using transgenic mice with fluorescently labeled germ cells and fluorescence-activated cell sorting (FACS), we isolated germ cells from Dazl KO testes and WT controls and used RNA-sequencing (RNA-seq) to identify mRNAs that are sensitive to DAZL deletion. Integrating the RNA-seq and DAZL-RNA interaction datasets revealed that DAZL post-transcriptionally enhances the expression of a network of genes with essential roles in spermatogenesis and cell-cycle regulation. We also present multiple lines of evidence indicating that DAZL preferentially binds GUU sites in close proximity to polyA sequences and demonstrate that the polyA tail at the 3′ end of mRNAs has a critical role in DAZL-RNA binding. These data provide important insights into the mechanism of DAZL binding to its RNA targets, the molecular basis for postnatal germ cell loss caused by DAZL deletion, and reveal an mRNA regulatory program that is essential for postnatal germ cell survival.
RESULTS
DAZL Binds GUU-Rich Sequences across the Testis Transcriptome
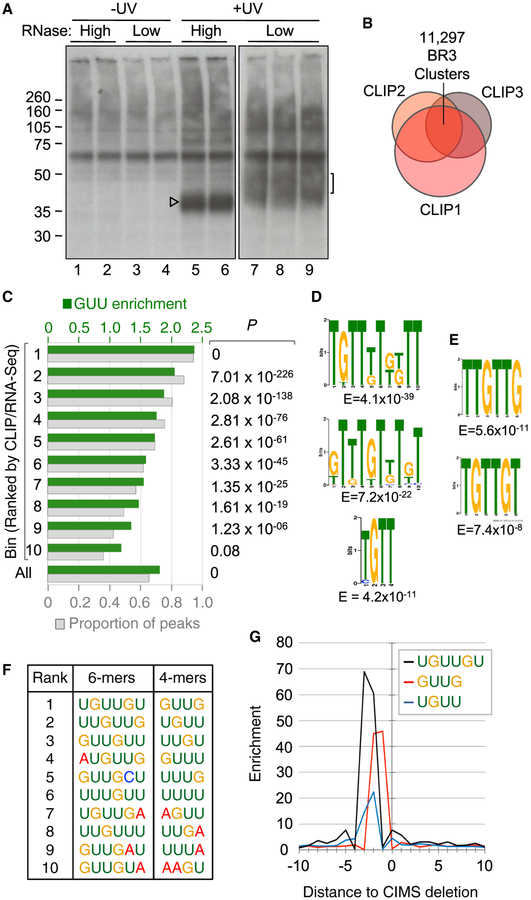
To comprehensively map DAZL-RNA interactions in vivo, high-throughput sequencing after in vivo cross-linking and immunoprecipitation (HITS-CLIP) libraries were generated from DAZL-RNA complexes purified from UV cross-linked adult mouse testes and sequenced using the Illumina platform (Figure 1A). The resulting CLIP reads from 3 biological replicates were filtered and mapped individually and were then intersected to reveal 11,297 genomic positions with overlapping CLIP reads in 3/3 libraries; these are hereafter designated as BR3 clusters (biologic reproducibility 3/3; Figure 1B). This interaction map reveals that DAZL directly and reproducibly binds distinct sites in >3,900 transcripts in adult testis (Data S1).
Figure 1. HITS-CLIP Identification of DAZL-RNA Contacts in the Adult Testis.
(A) Autoradiograph of radiolabeled cross-linked DAZL-RNA complexes purified from adult testes. Arrowhead indicates DAZL cross-linked to minimal RNA fragments. Bracket indicates RNA fragments of 35–50 nt excised for cDNA preparation.
(B) Venn diagram of overlapping CLIP reads from each replicate and identification of 11,297 genomic coordinates with overlapping CLIP reads from 3/3 biological replicates.
(C) Bins of CLIP peaks normalized to RNA-seq, with GUU enrichment (green) and proportion of peaks in each bin with GUU (gray), and P values from χ2 distribution of GUU enrichment at right.
(D) Top motifs present in bin 1, identified by MEME (top, middle) and DREME (bottom).
(E) Top motifs associated with CIMS sites identified by MEME.
(F) Top 10 6mers and 4mers associated with CIMS sites.
(G) Positioning of UGUUGU, GUUG, and UGUU motifs relative to CIMS deletion sites.
We next identified CLIP peaks within each cluster (Figure S1). Consistent with structural studies (Jenkins et al., 2011), these sites were enriched for GUU motifs compared to shuffled control sequences (1.8-fold; Figure 1C, All). Using RNA-seq data from age-matched testes, CLIP peaks were normalized to RNA levels and parsed into 10 bins based on CLIP:RNA-seq ratios. A clear correlation was evident between CLIP:RNA-seq read ratio, GUU enrichment, and the proportion of peaks in each bin that contained GUU (Figure 1C). Separately, de novo motif analysis using the MEME suite (Bailey et al., 2015) identified GTT-containing motifs as the most enriched sequence elements in genomic regions corresponding to peaks with the highest CLIP:RNA-seq ratios (Figure 1D).
DAZL binding sites were also examined using an independent read mapping and analysis pipeline that takes advantage of cross-link-induced mutation sites (CIMSs) that reflect positions of protein-RNA cross-linking in HITS-CLIP data (Zhang and Darnell, 2011). De novo motif analysis of the top 1,000 CIMS deletion sites (±10 nt) identified GUU-containing motifs as the most enriched sequence elements (Figure 1E). Examination of 6mers and 4mers associated with the top 1,000 CIMS sites ±20 nt relative to background sequences identified enrichment of GUU-containing sequences around deletions, particularly the GUUG motif, which was enriched 40- to 60-fold around CIMS deletion sites, with cross-linking occur ring at U residues within GUU triplets (Figures 1F and 1G). Thus, two independent mapping and bioinformatic workflows confirm that DAZL predominantly binds GUU-rich sequences in vivo.
DAZL Predominantly Binds GUU Sites in Close Proximity to mRNA PolyA Tails
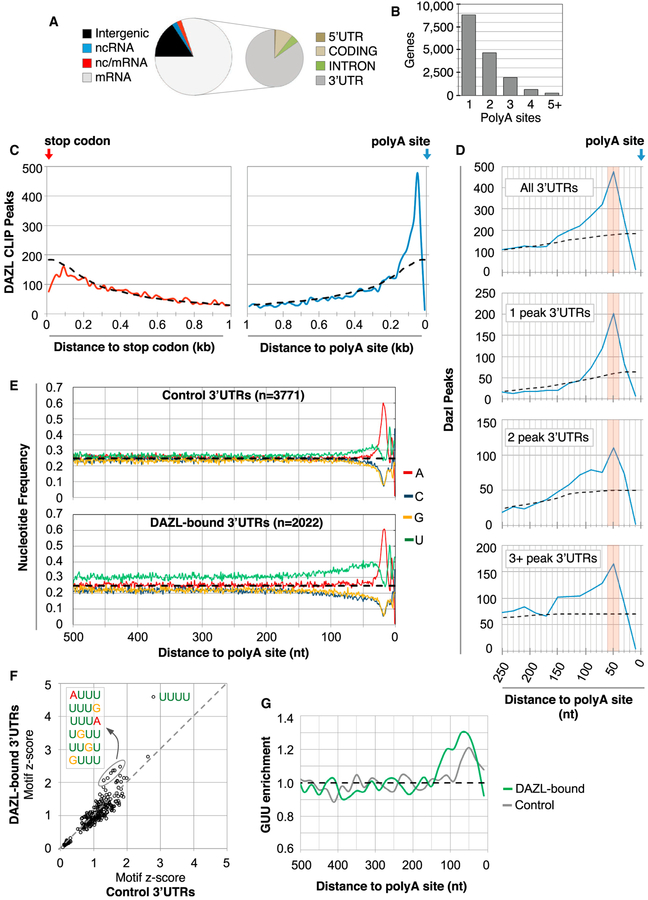
The majority of DAZL BR3 sites mapped to 3′ UTRs of protein-coding genes (Figure 2A; Table S1). To improve the annotation of DAZL-3′ UTR interactions, we used polyA-seq to generate a quantitative global map of polyA site utilization in adult testes (Figure S2A). Consistent with widespread transcription and alternative polyadenylation (APA) in the testis (Soumillon et al., 2013; Li et al., 2016), polyA-seq identified 28,032 polyA sites in RNAs from 16,431 genes (Figures 2B and S2B). Gene expression estimates from polyA-seq and RNA-seq showed a strong correlation (R = 0.83; Figure S2C), as did estimates of polyA site usage from polyA-seq and qRT-PCR (R = 0.72, 10 candidates examined; Figure S2D).
Figure 2. DAZL Predominantly Binds 3′ UTRs at PolyA-Proximal Sites.
(A) Distribution of BR3 CLIP sites in different genic regions. BR3 clusters mapped to regions with overlapping coding and non-coding RNAs were annotated as nc/mRNA.
(B) Number of genes with ≥1 polyA site, as determined by polyA-seq analysis.
(C) Metagene analysis of DAZL CLIP peaks in 20 nt bins relative to the stop codon (left, red line) or the polyA site (right, blue line). Dashed line represents the expected random distribution.
(D) Top corresponds to a higher magnification view of the right part of (C). The bottom three parts show CLIP peak distribution relative to the polyA site in 3′ UTRs with 1, 2, or more peaks in the entire 3′ UTR.
(E) Nucleotide frequency 500 nt upstream of polyA sites of control 3′ UTRs (no DAZL CLIP reads, top), and 3′ UTRs with DAZL-RNA interactions (bottom).
(F) Motif enrichment in 3′ UTRs with and without DAZL CLIP peaks (y and x axes, respectively), where values correspond to Z scores.
(G) GUU enrichment (observed/expected) in 3′ UTRs with and without Dazl CLIP peaks (green and gray lines, respectively).
Having defined testis-expressed 3′ UTRs, metagene analyses were performed to examine the distribution of 3′ UTR DAZL-RNA interactions relative to the upstream stop codon and downstream polyA site. Compared to the expected distribution if peaks were randomly distributed (Figure 2C, dashed line), no positional preference was observed when DAZL-3′ UTR interactions were measured relative to the stop codon (Figure 2C, compare red line to dashed line). In stark contrast, a prominent positional bias for DAZL-RNA contacts was observed relative to the polyA site, with strong enrichment within ~150 nt and the greatest number of interactions ~50 nt upstream of the polyA site (Figure 2C, compare blue line to dashed line). This positional preference is also evident on 3′ UTRs with multiple DAZL CLIP peaks, with broadening of DAZL binding in a 3′–5′ direction as the number of peaks per 3′ UTR increased (Figure 2D).
Further examination showed that 3′ UTRs with DAZL BR3 CLIP sites had a higher proportion of uridine residues compared to 3′ UTRs lacking DAZL CLIP reads (Figures 2E and S3A). Moreover, the most enriched motifs in DAZL-bound versus control 3′ UTRs were U rich, with 2 of the top 7 most-enriched 4mers containing GUU (Figure 2F). Similarly, 3 of the top 4 6mers (and 7 of the top 20) contained GUU (Figure S3B). Although both sets of 3′ UTRs had enrichment of GUU upstream of polyA sites, GUU enrichment was greater and extended over a broader region in DAZL-bound 3′ UTRs (Figure 2G).
Transcriptome-wide mapping of direct, biologically reproducible DAZL-RNA interactions in mouse testes indicates that DAZL binds U-rich 3′ UTRs of thousands of genes expressed in the mouse testis, with preferential binding to GUU-containing sequences upstream of polyA sites.
DAZL Directly Enhances Steady-State mRNA Levels of a Subset of In Vivo Targets
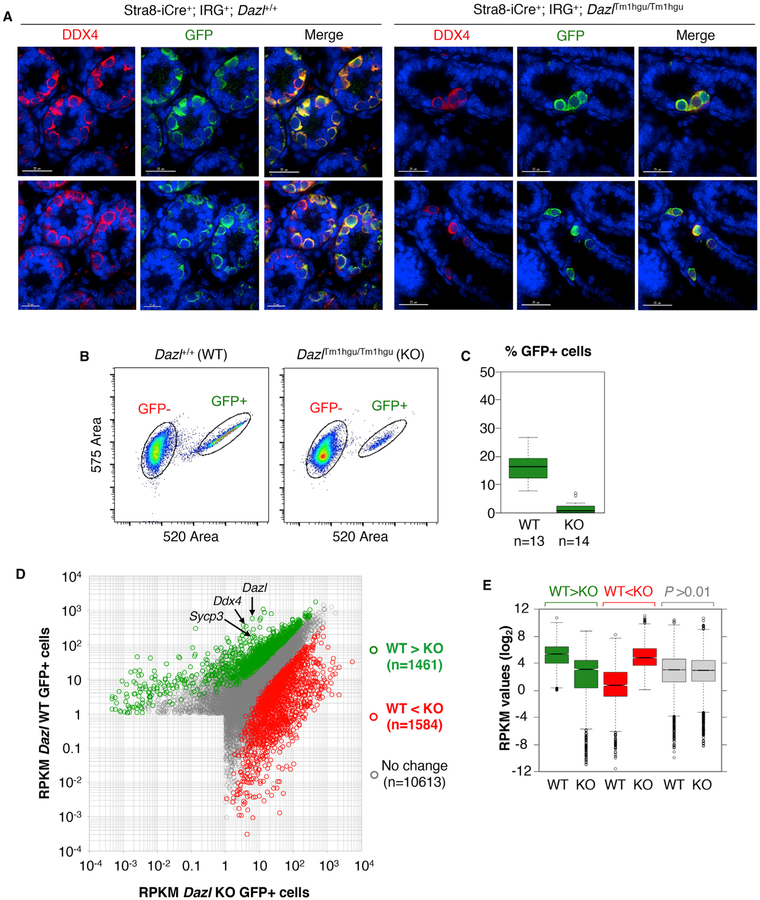
Significant barriers to understanding the in vivo functions of DAZL include the scarcity and variable number of germ cells in Dazl KO mice (Ruggiu et al., 1997; Lin and Page, 2005). To over-come these obstacles, we used Cre-lox to label germ cells with GFP followed by FACS to collect Dazl KO germ cells for RNA analyses. This approach (Zagore et al., 2015) uses the Stra8-iCre transgene that expresses Cre recombinase in postnatal germ cells (Sadate-Ngatchou et al., 2008) and the IRG transgene that expresses GFP following Cre-mediated recombination (De Gasperi et al., 2008). To generate mice with GFP+ DAZL-deficient postnatal germ cells (Figure 3A), we used mixed background breeders that are heterozygous for the null DazlTm1hgu allele (Ruggiu et al., 1997) (see Method Details). Testes of Stra8-iCre+; IRG+; Dazl2+ (WT) and Stra8-iCre+; IRG+; DazlTm1hgu/Tm1hgu (KO) mice were indistinguishable at postnatal day 0 (P0); however, germ cells declined steadily thereafter in the latter (data not shown). Cre expression from Stra8-iCre commences at ~P3; therefore, we selected animals at P6 for tissue collection and FACS. At this age, spermatogonia are the only germ cells present. Germ cell-restricted GFP expression in WT and KO mice was confirmed by IF microscopy (Figure 3A), and germ cell markers were significantly enriched in GFP+ cells (data not shown). As expected, GFP+ spermatogonia were significantly reduced in KO mice compared to littermate controls (Figures 3A–3C).
Figure 3. Genetic Labeling, Isolation, and Transcriptome Profiling of GFP+ WT and Dazl KO Germ Cells.
(A) IF of DDX4 and GFP in Stra8-iCre+; IRG+; Dazl2+ (Dazl WT) and Stra8-iCre+; IRG+; DazlTm1hgu/Tm1hgu (Dazl KO) testes at P6 (left and right, respectively). Sections from biological replicates are shown in the top and bottom rows. Scale bars for bottom WT panels represent 15 μM, all others represent 25 μM.
(B) Flow cytometry plots of GFP− and GFP+ cells from WT and KO mice.
(C) Percentage of GFP+ cells isolated by FACS from P6 WT and Dazl KO mice.
(D) Scatterplot of RPKM values for genes in WT and KO cells. Green and red dots denote genes with decreased and increased RNA levels in Dazl KO cells compared to WT controls.
(E) Boxplot showing the distribution of RPKM values for green, red, and gray genes indicated in (D).
To determine how the absence of DAZL affects global mRNA levels, RNA-seq analysis was performed on GFP+ cells collected by FACS. Overall, the number of expressed genes and the distribution of their reads per kilobase million (RPKM) values were comparable in WT and Dazl KO cells (11,739 and 12,269 expressed genes with RPKM >1, respectively). However, 1,462 transcripts had reduced RNA levels and 1,584 increased in Dazl KO cells (minimum 2-fold, adjusted p < 0.01; Figures 3D [green and red dots, respectively] and 3E). Notably, Ddx4 and Sycp3 were among the genes with reduced RNA in Dazl KO cells (86- and 26.5-fold, respectively; Figure 3D). Therefore, the reduced levels of DDX4 and SYCP3 proteins previously observed in Dazl KO germ cells by IF and attributed to reduced translation (Reynolds et al., 2005, 2007) are associated with significant reductions in their corresponding mRNAs.
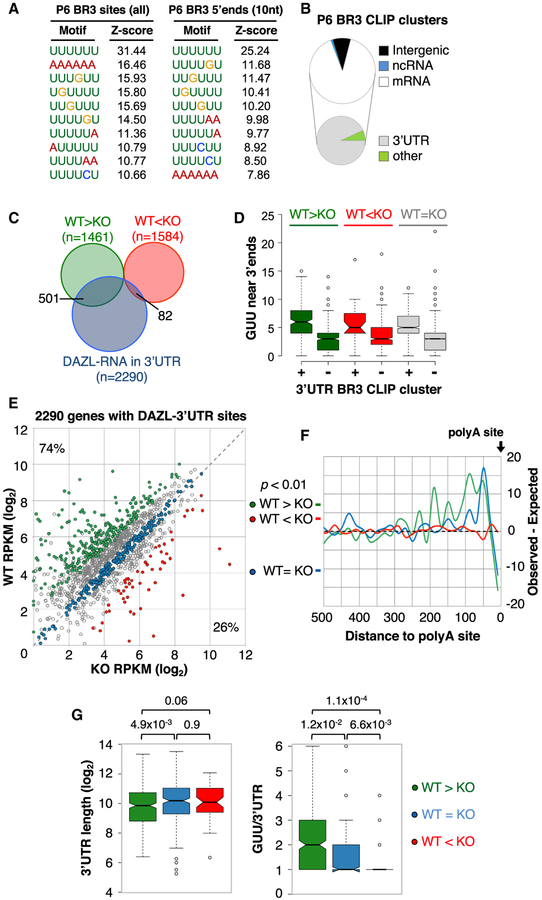
To investigate which RNA changes are primary or secondary consequences of DAZL loss, we mapped DAZL-RNA interactions in P6 testes using iCLIP (individual nucleotide resolution CLIP), which allows improved identification of direct cross-link sites over HITS-CLIP. As with DAZL BR3 sites in adult testes, P6 BR3 regions were enriched for GUU, with the majority in 3′ UTRs (Figures 4A and 4B). To accurately map positions of DAZL-3′ UTR contacts in spermatogonia, polyA-seq libraries were generated from GFP+ spermatogonia isolated by FACS from Stra8-iCre+; IRG+ testes. Of the 28,038 3′ UTRs defined by polyA-seq of whole adult testis (described above; Figure 2B), 16,502 were identified in the polyA-seq data from GFP+ spermatogonia, corresponding to 10,370 genes. Intersecting the P6 iCLIP and spermatogonia polyA-seq datasets showed that 84% of P6 DAZL BR3 sites (5,400/6,465) mapped to 3′ UTRs of 2,290 genes, indicating that DAZL also directly binds to a vast network of RNAs in spermatogonia via 3′ UTR interactions. Furthermore, metagene analysis showed that DAZL-RNA interactions in spermatogonia-expressed 3′ UTRs also had a polyA-proximal bias, with the greatest enrichment within 150 nt of the polyA site (Figure S4A). Thus, transcriptome-wide mapping of polyA sites and DAZL-RNA interactions confirm a polyA-proximal bias for DAZL-RNA binding to a broad set of mRNAs in both adult and juvenile testes.
Figure 4. DAZL Directly Enhances mRNA Levels through PolyA-Proximal Binding.
(A) Top 10 motifs (based on Z score ranking) for sequences of BR3 regions (left) or 10 nt windows centered at the 5′ end of each BR3 CLIP region.
(B) Distribution of 6,465 BR3 DAZL-RNA sites from iCLIP analysis of P6 testes.
(C) Venn diagram comparing genes with RNA level changes between WT and Dazl KO cells (≥2-fold, p < 0.01) and the set of 2,290 with DAZL-3′ UTR interactions.
(D) Examination of GUUs in the last 250 nt of 3′ UTRs with and without BR3 sites identified by iCLIP analysis of P6 testes.
(E) RPKM levels of 2,290 genes with DAZL-3′ UTR interactions in GFP+ WT and GFP+ KO cells. Green and red circles correspond to genes with RNA level differences between GFP+ WT and KO cells of ≥2-fold (adjusted p < 0.01). Blue dots correspond to genes with RPKM values that do not differ >20% between WT and KO cells.
(F) Metagene analysis of the distribution of BR3 DAZL-RNA sites in 3′ UTRs of DAZL-enhanced, DAZL-repressed, and DAZL-insensitive genes (green, red, and blue, respectively).
(G) Examination of 3′ UTR lengths (left) and frequency of GUU per 3′ UTR (right) for genes with increased RNA, unchanged RNA, or decreased RNA levels (green, blue, and red, respectively) in GFP+ Dazl KO cells compared to WT controls. P values (Wilcoxon rank sum test) for pairwise comparisons indicated above.
Of the 2,290 genes with DAZL-3′ UTR interactions in P6 testis, 583 also had differences in mRNA steady-state levels in KO compared to WT germ cells. A disproportionate number of these overlapping genes (85%) had decreased RNA levels in KO cells (501 downregulated and 82 upregulated genes, respectively) (Figures 4C and 4E). To investigate why 3′ UTR BR3 CLIP clusters are underrepresented on transcripts that increase in Dazl KO cells, GUU frequencies were examined in 3′ UTRs of different classes of genes with and without 3′ UTR BR3 CLIP clusters. Independent of whether genes had increased, decreased, or unchanged RNA levels in KO compared to WT germ cells, 3′ UTRs lacking BR3 sites had significantly fewer GUUs compared to 3′ UTRs with BR3 sites (Figure 4D). The paucity of GUUs in 3′ UTRs lacking BR3 CLIP clusters (in all gene categories) argues against ascertainment bias in the identification of RNAs directly bound by DAZL in vivo.
Metagene analyses revealed striking differences in the distribution of BR3 CLIP sites in the 3′ UTRs of genes with increased or decreased RNA levels in KO versus WT cells. BR3 sites in 3′ UTRs of genes repressed in a DAZL-dependent manner showed no positional preference (Figure 4F, red line). In contrast, DAZL binding sites in 3′ UTRs of genes enhanced by DAZL were enriched near the polyA tail, with the greatest number of interactions within 100 nt of the polyA site (Figure 4F, green line). Moreover, GUU motifs and DAZL BR3 interaction sites were significantly more abundant in DAZL-enhanced rather than DAZL-repressed genes, despite comparable 3′ UTR lengths (Figure 4G). These observations strongly suggest that the directly regulated mRNA targets of DAZL are those with reduced levels in KO germ cells, indicating that Dazl primarily functions as a positive post-transcriptional regulator of gene expression through polyA-proximal interactions.
We also examined DAZL-insensitive genes with DAZL-3′ UTR interactions whose RNA levels did not differ by >20% between WT and Dazl KO cells (Figure 4E, blue dots). Remarkably, DAZL-RNA interactions in these 3′ UTRs also displayed a strong polyA-proximal bias (Figure 4F, blue line). Furthermore, we identified several examples in which DAZL-3′ UTR binding patterns were indistinguishable between DAZL-insensitive and DAZL- enhanced 3′ UTRs (Figures S4B–S4D). These observations indicate that polyA-proximal binding alone is not sufficient for DAZL to enhance mRNA levels and suggest that DAZL acts in a 3′ UTR-specific manner. They also raise the possibility that a significant proportion of DAZL-3′ UTR interactions may not have regulatory function and may represent opportunistic interactions reflecting the general mechanism of DAZL recruitment and/or stabilization on its RNA targets (see below).
PolyA Tracts Are Enriched Near Positions of DAZL-RNA Interactions
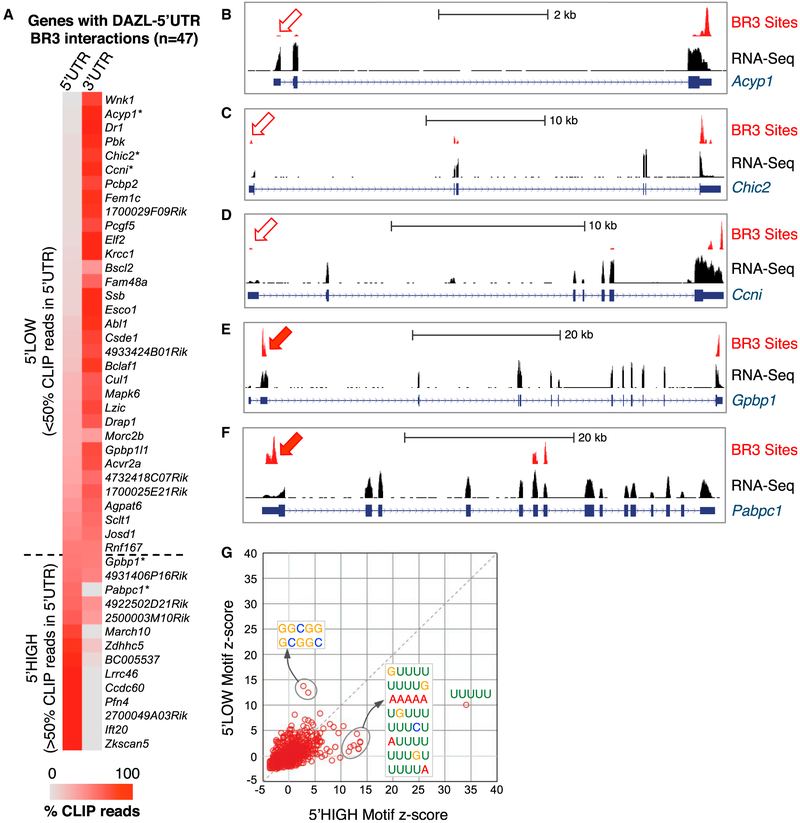
Multiple lines of evidence suggest a potential role for polyA sequences in facilitating DAZL-GUU interactions. This includes enrichment of DAZL-RNA contacts near polyA tails from metagene analyses integrating CLIP and polyA-seq datasets (Figures 2C and S4A). In addition, AAAAAA was among the most enriched 6mers when regions of overlapping iCLIP reads were examined in BR3 clusters, but significantly less enriched when motif analysis was performed on the 5′ ends of iCLIP BR3 sites where cross-link sites are enriched (König et al., 2010) (Figures 4A and 6B). These observations indicate that genomic-encoded polyA sequences are enriched near but not at direct sites of DAZL-RNA cross-linking. DAZL was previously shown to interact with PABPC1 (Collier et al., 2005), which binds both polyA tails and genomic-encoded A-rich RNA sequences, including autore-gulatory interactions in the 5′ UTR of its own mRNA (Hornstein et al., 1999). Pabpc1 was among a small set of 47 genes with DAZL BR3 CLIP sites in the 5′ UTR (56 sites in 5′ UTRs of 47 genes, compared to 6,397 sites in 3′ UTRs of 3,249 genes), prompting further examination of these outliers (Figure 5A; Data S1). For the majority (32/47 genes), the CLIP read number was greatest in the 3′ UTR and declined in a 3′–5′ manner (e.g., Acyp1, Chic2, and Ccni; Figures 5B–5D, 5′ low genes). Pabpc1 was 1 of only 15 genes for which >50% of the total CLIP reads were located in the 5′ UTR (Figures 5E and 5F, 5′ high genes). Marked differences were observed in the sequence features associated with these two sets of 5′ UTRs. Consistent with low CLIP coverage, 5′ UTRs of 5′ low genes were not enriched for GUU-containing sequences. In contrast, 5′ UTRs of 5′ high genes resembled DAZL-bound 3′ UTRs, for which the most enriched motifs included polyU and polyU tracts interrupted by a single G (Figure 5G). AAAAA was among the most enriched pentamers in 5′ UTRs of 5′ high genes, with a Z score equal to that of UGUUU (Figure 5G). Furthermore, polyA tracts (≥5 A’s) were present in 73% in 5′ high UTRs compared to 31% of 5′ low UTRs (Figure S5A). Thus, a common feature of biologically reproducible DAZL-RNA interactions across the transcriptome is the presence of local polyA sequence, either genomic encoded or added to mRNA via 3′ end processing.
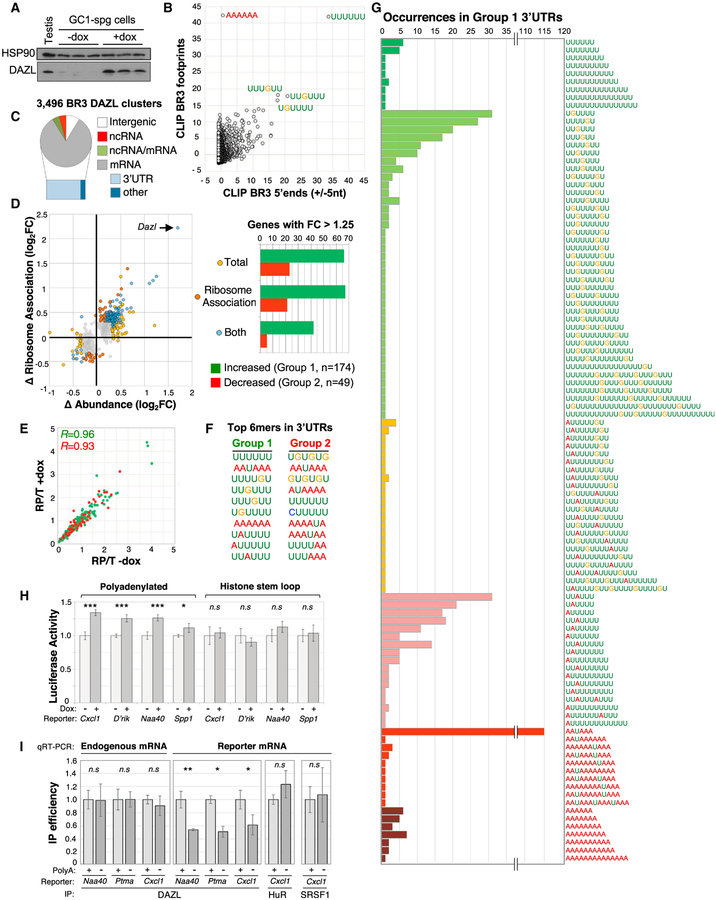
Figure 6. The PolyA Tail Has an Important Role in DAZL-RNA Binding and Regulation.
(A) Western blot showing dox-induced expression of DAZL in GC-1 spg cells used for RNA-seq analyses.
(B) Distribution of Z scores following motif-enrichment analysis of complete regions of BR3 positions bearing overlapping CLIP reads (y axis) or 10 nt regions centered at the 5′ end of BR3 sites (x axis).
(C) Distribution of BR3 CLIP sites across the genome.
(D) Left: fold change in mRNA ribosome association (y axis) and total mRNA level (x axis) for 894 genes following dox induction of DAZL expression. Right: bar chart of 223 genes with ≥1.25-fold change in mRNA association with ribosomes, change in total mRNA level, or both, following DAZL expression.
(E) Distribution of ribosome-protected to total mRNA (RP/T) ratios for group 1 and group 2 genes (green and red, respectively) before and after dox treatment.
(F) Top 10 most-enriched 6mers in last 250 nt of groups 1 and 2 3′ UTRs.
(G) Extended polyU tracts in group 1 3′ UTRs revealed by examining the distribution of the top 10 motifs (F) in each group 1 3′ UTR and merging any that overlap.
(H) Luciferase activity in untreated and dox-treated cells transfected with luciferase mRNA reporters bearing polyadenylated and non-polyadenylated 3′ UTRs of the indicated genes. D’Rik corresponds to reporters bearing the 3′ UTR of D030056L22Rik.
(I) Immunoprecipitation (IP) efficiency of endogenous D030056L22Rik mRNA and luciferase mRNA in dox-treated cells transfected with polyadenylated and non-polyadenylated versions of 3 different 3′ UTRs.
Values in (H) and (I) are averages and SDs from 3 to 5 replicate experiments.
*p < 0.05, **p < 0.01, ***p < 0.0001; n.s. denotes no statistically significant difference.
Figure 5. Rare DAZL-RNA Interactions in 5′ UTRs Are Associated with Enrichment of PolyA Tracts.
(A) Comparison of BR3 CLIP tag density in the 5′ and 3′ UTRs of genes with 5′ UTR BR3 CLIP sites. Color coding represents the percentage of BR3 tags in the gene that map to either the 5′ UTR or the 3′ UTR. Dashed line denotes cutoff between genes with <50% (above) or >50% (below) of the CLIP reads in the 5′ UTR. (B–F) Representative examples of 5′ low (B–D) and 5′ high (E and F) genes with CLIP read density in BR3 regions and RNA-seq read density shown.(G) Motif enrichment in 5′ UTRs with >50% or <50% of total BR3 CLIP reads in the 5′ UTR.
DAZL Post-transcriptionally Enhances Gene Expression in GC-1 spg Cells
Based on the data above, we hypothesized that local polyA sequences facilitate DAZL-GUU interactions, thus providing a potential explanation for the widespread binding of DAZL to GUU sites upstream of polyA tails. To explore this possibility, we first established a stable monoclonal cell line that allows doxycycline (dox)-inducible expression of DAZL. GC-1 spg cells were selected as the parental cell line, which are derived from spermatogonia (Hofmann et al., 1992) but express low levels of endogenous DAZL compared to mouse testis (Figure 6A). In these cells, dox treatment results in predominantly diffuse cytoplasmic localization of DAZL, similar to the pattern observed in WT germ cells (data not shown). As in the testis, iCLIP analysis in dox-treated GC-1 spg cells showed that DAZL cross-link sites were significantly enriched for GUU-containing sequences (Figures 6B and S5B), with 80.7% of BR3 sites that mapped to RefSeq mRNAs in 3′ UTRs (1,226 genes; Figure 6C). Furthermore, AAAAAA was overrepresented in entire BR3 regions with overlapping iCLIP reads (y axis), but not at 5′ ends (x axis) where direct sites of DAZL-RNA cross-linking are enriched (Figure 6B).
To determine how DAZL induction affects mRNA abundance and translation in GC-1 spg cells, RNA-seq datasets of total cytoplasmic RNA and ribosome-protected RNA fragments were generated from dox-treated and untreated cells. In total, 894 protein-coding genes had statistically significant differences in total cytoplasmic and/or ribosome-associated RNA levels between dox-treated and untreated cells (p < 0.05; Figure 6D, left). Gene expression changes quantified by qRT-PCR and RNA-seq had a strong positive correlation (R = 0.9, 13 targets examined; Figure S5C). The gene with the greatest mRNA differences was Dazl, corresponding to induction of the cDNA transgene, not the endogenous gene (evidenced by the lack of RNA-seq reads mapped to 5′ and 3′ UTRs of Dazl; data not shown). Excluding Dazl, we focused on a set of 223 genes with mRNA-level changes of ≥1.25-fold for further analysis. The majority had increased cytoplasmic- and/or ribosome-associated levels after dox treatment (174 dox-enhanced versus 49 dox-repressed genes) (Figure 6D, right). For both sets of genes, ribosome density was largely unchanged following dox treatment, indicating that mRNA changes in ribosome association following DAZL expression largely parallel changes in overall mRNA abundance (Figure 6E).
Comparing the 223 genes with dox-dependent RNA changes to those with DAZL-3′ UTR interactions identified a common set of 44 genes, all of which belonged to the group of genes with dox-induced increased mRNA levels (total and/or ribosome associated). Similarly, when the 223 genes were compared to the 3,907 with DAZL-3′ UTR interactions in adult testes, the majority of the overlapping genes (91.7%) were enhanced by DAZL (67/174 enhanced versus 6/49 repressed). Consistent with these biased distributions, GUU was enriched in 3′ UTRs of dox-enhanced genes but underrepresented in 3′ UTRs of dox-repressed genes(1.55-fold versus 0.85-fold, respectively). In addition, GUU-containing hexamers were among the most enriched motifs within 250 nt of the polyA site of dox-enhanced genes (Figure 6F). Closer examination of the distribution of the top 10 most-enriched hexamers in dox-enhanced 3′ UTRs showed that many overlap to form extended polyU tracts interrupted by a single A or, more commonly, a single G (Figure 6G). Thus, we conclude that genes with increased RNA levels following dox treatment are the direct targets of DAZL in GC-1 spg cells, and that DAZL post-transcriptionally enhances a subset of its RNA substrates in GC-1 spg cells via GUU interactions in 3′ UTRs, as in GFP+ spermatogonia.
DAZL Binding and Regulation of Its RNA Targets Require a PolyA Tail
To test the hypothesis that the 3′ polyA tail is important for DAZL binding and RNA regulation, we generated a series of reporter plasmids that express luciferase mRNAs bearing the 3′ UTR of DAZL target genes with either (1) endogenous sequences necessary for 3′ end cleavage and polyadenylation or (2) the stem loop structure from the 3′ end of histone mRNAs to generate nonpolyadenylated transcripts. Similar to effects on endogenous GC-1 spg genes, DAZL expression resulted in modest yet highly reproducible increases in luciferase activity in cells transfected with polyadenylated reporters bearing the Cxcl1, D030056L22Rik NAA40, and Spp1 3′ UTRs (Figures 6H [left] and S5D). In contrast, DAZL expression did not result in increased luciferase levels in cells transfected with non-polyadenylated mRNAs (Figure 6H, right). We next performed co-immunoprecipitation (coIP) assays to determine whether these differences in luciferase levels were associated with altered DAZL-3′ UTR binding. While DAZL binding to endogenous mRNAs was unaffected in cells transfected with any of the reporter plasmids, DAZL binding was markedly reduced to reporter mRNAs bearing the histone stem loop compared to polyA+ versions (Figure 6I). This was true for reporters bearing 3′ UTRs from DAZL-enhanced genes (Cxcl1, NAA40), as well as the Ptma 3′ UTR, which is bound by DAZL but insensitive to DAZL levels in testis and GC-1 spg cells (Figures 6I, S4C, S4D, and S5D). To determine whether the histone stem loop causes broad remodeling of RBP-3′ UTR interactions, we performed coIP experiments to test the binding of two RBPs previously shown to bind the Cxcl1 3′ UTR, SRSF1 and HUR (Herjan et al., 2018). No difference in SRSF1 or HUR binding was observed in polyadenylated and non-polyadenylated luciferase reporters bearing the Cxcl1 3′ UTR (Figure 6I). Examination of multiple CLIP datasets and reporter mRNAs indicate that DAZL interaction with GUU-containing RNAs is facilitated by local polyA sequences. These observations provide a potential rationale for the widespread binding of DAZL to polyA-proximal GUU sites that is observed in thousands of RNAs in the germ cell transcriptome.
DAZL Post-transcriptionally Controls a Network of Cell-Cycle Regulatory Genes
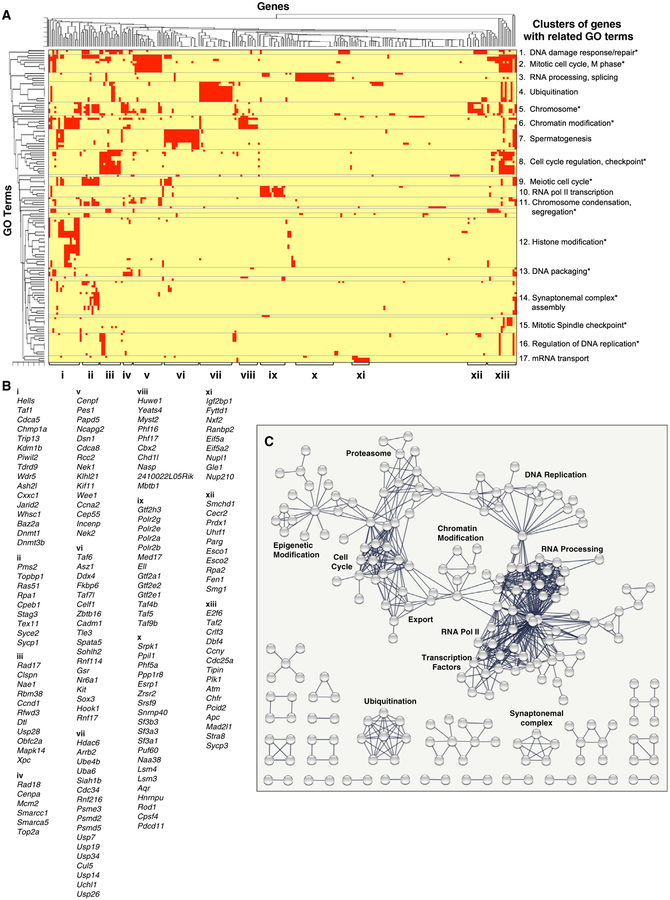
Collectively, the DAZL-RNA interaction maps, reporter mRNA assays, and RNA-seq analyses of isolated germ cells and dox-treated cells indicate that Dazl predominantly functions directly as a positive post-transcriptional regulator via 3′ UTR interactions. Therefore, to identify the potential primary causes of germ cell loss in Dazl KO mice, we more closely examined the 501 genes with DAZL-3′ UTR interactions and reduced mRNA levels in GFP+ Dazl KO cells. Hierarchical clustering revealed 17 groups of significantly enriched Gene Ontology (GO) terms, with the majority (12/17) associated with different aspects of cell-cycle regulation, including chromatin modification, condensation, or segregation; synaptonemal complex assembly; mitotic spindle checkpoint; DNA replication, repair, or packaging; and mitotic or meiotic cell-cycle regulation (Figure 7A, groups marked with asterisks). The remaining groups contained genes and GO terms associated with RNA processing, ubiquitination, spermatogenesis, transcription by RNA pol II, and mRNA transport. Among the DAZL-enhanced genes are several that are necessary for spermatogenesis (Figures 7A and 7B, groups i, vi, and xiii), many with essential roles in spermatogonial maintenance or proliferation, including Atm (Takubo et al., 2008), Dmrt1 (Zhang et al., 2016), Nxf2 (Pan et al., 2009), Sohlh2 (Hao et al., 2008), Sox3 (Raverot et al., 2005), Plzf/Zbtb16 (Costoya et al., 2004), and Taf4b (Lovasco et al., 2015).
Figure 7. DAZL Binds and Regulates a Broad Network of Essential Genes Encoding Interacting Proteins.
(A) Hierarchical clustering of enriched GO terms and genes associated with the set of 501 genes that are enhanced by DAZL and have DAZL-3′ UTR interactions. Subgroups of overlapping GO categories are indicated at left, while subgroups of genes are indicated with letters at bottom.
(B) List of all of the genes in each letter group indicated at the bottom of (A).
(C) Protein-protein interaction network for genes with DAZL-3′ UTR interactions and reduced RNA levels in Dazl KO cells compared to WT controls.
We also identified many examples in which genes encoding subunits of the same complex were among the 501 DAZL-enhanced genes (e.g., gene cluster ix in Figures 7A and 7B). To further assess physical interactions between the protein products of the 501 genes, we used the STRING database of protein-protein interactions (PPIs) (Szklarczyk et al., 2015). Focusing on high-confidence PPIs supported by experimental evidence or curated databases (minimum interaction score0.9), we observed a higher than expected number of the 501 genes encoding proteins that interact with one another (PPI enrichment p = 0). This includes a broad network of interacting proteins, including basal transcription factors, subunits of RNA pol II, RNA processing and export factors, chromatin-binding proteins, epigenetic regulators, and several components of the proteasome and ubiquitination machineries (Figure 7C).
Together, GO and PPI enrichment analyses in combination with DAZL CLIP, polyA-seq, and RNA-seq indicate that DAZL enhances postnatal germ cell survival via polyA-proximal interactions that promote high mRNA levels for a network of genes whose protein products physically interact, including several genes with essential roles in cell-cycle regulation and germ cell survival.
DISCUSSION
The necessity of the DAZ family of RBPs for germ cell survival is well established in multiple species. However, the direct targets, regulatory roles, and biological functions of these RBPs remained unclear. Our integrative analyses combining transgenic mice, FACS, and a panel of unbiased, transcriptome-wide profiling tools provide important insights into the molecular and biological functions of this important family of RBPs.
PolyA Tracts Facilitate DAZL-3′ UTR Interactions across the Germ Cell Transcriptome
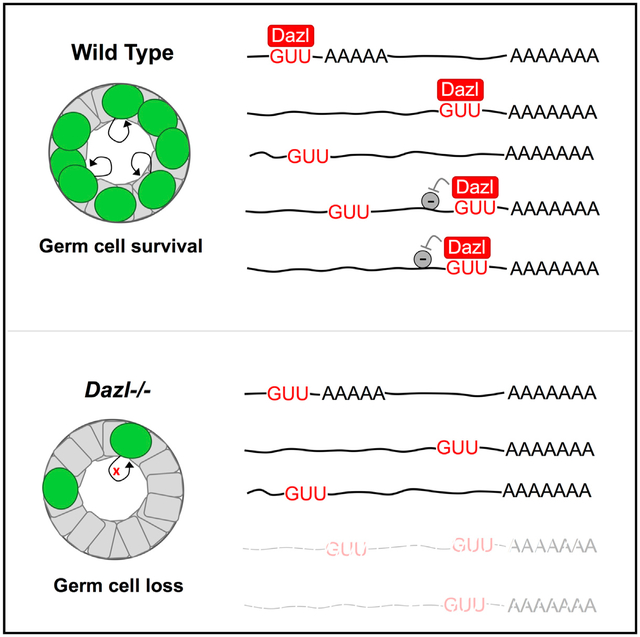
Transcriptome-wide DAZL-RNA interaction maps show that DAZL binds thousands of mRNAs expressed in adult and juvenile testes, with most of the interactions in 3′ UTRs. Given the widespread transcription and high RNA levels in male germ cells (Soumillon et al., 2013), it is not uncommon for a germ cell-ex-pressed RBP to reproducibly bind a vast set of RNAs (Hannigan et al., 2017). However, the DAZL CLIP maps are striking in that the binding sites have a strong positional preference, implying that additional sequence features specify which GUU sites are bound by DAZL. Our data suggest that widespread polyA-proximal DAZL-RNA interactions reflect a general mechanism of DAZL binding that depends on GUU and local polyA sequences in vivo. Supporting evidence includes the widespread preferential positioning of DAZL-RNA contacts upstream of polyA sites; overrepresentation of polyA tracts in the rare set of 5′ UTRs with high CLIP read levels (including the Pabpc1 5′ UTR); enrichment of genomic-encoded polyA tracts downstream of direct sites of DAZL-RNA cross-linking in two independent iCLIP datasets; and failure of DAZL to efficiently bind and enhance reporter mRNAs lacking a polyA tail. Based on these observations and previous evidence of RNA-independent DAZL-PABPC1 interactions (Collier et al., 2005), we propose that DAZL recruitment and/or stabilization on mRNA is mediated by local PABPC1-polyA interactions. Considering the prevalence of GUU throughout the transcriptome, a requirement for local PABPC1-polyA interactions in specifying sites of DAZL-RNA binding would increase the probability of DAZL loading on 3′ UTRs, where the majority of cytoplasmic RBPs exert their regulatory functions (Sweet and Licatalosi, 2014).
DAZL Enhances mRNA Levels of Specific Targets through polyA-Proximal Interactions
Regulation of mRNA translation and stability are intricately coupled processes (Bicknell and Ricci, 2017); therefore, it is not clear whether DAZL functions primarily to prevent mRNA decay or as a translational enhancer that indirectly affects mRNA levels. It is also possible that DAZL’s primary function differs in distinct mRNAs. Although translation rates of specific mRNAs in WT and Dazl KO germ cells were not measured, we found that changes in mRNA steady-state levels following DAZL expression in GC-1 spg cells generally paralleled changes in mRNA association with ribosomes.
Additional studies are necessary to determine why some mRNAs with polyA-proximal interactions have reduced RNA levels in Dazl KO cells, while others are insensitive to DAZL deletion. Notably, 3′ UTRs of the former had the highest number of GUU sites per 3′ UTR, despite modestly shorter lengths (Figure 4G). In addition, DAZL-RNA contacts on DAZL-insensitive genes were more concentrated near polyA sites, with reduced upstream 3′ UTR binding compared to contacts in DAZL-enhanced genes (Figure 4F). Thus, multiple DAZL-RNA contacts may be necessary for DAZL to post-transcriptionally enhance the expression of some genes. Examination of BR3 CLIP clusters with multiple distinct CLIP peaks showed that the peaks are often regularly spaced, with a median spacing of ~60 nt between adjacent peak regions (Figure S1F), perhaps indicating DAZL homodimerization as seen by in vitro and yeast two-hybrid analyses (Ruggiu and Cooke, 2000). Binding to a broader segment of the 3′ UTR in some mRNAs may displace or neutralize mRNA- specific cis- or trans-acting negative regulators. However, we also identified several examples in which the Dazl-RNA binding on 3′ UTRs of DAZL-enhanced and DAZL-insensitive genes were indistinguishable, suggesting that DAZL functions in a 3′ UTR-specific manner (Figure S4). These findings highlight the importance of combining RBP-RNA interaction maps with RNA profiling to distinguish functional from potentially opportunistic RBP-RNA interactions.
DAZL Controls a Network of Genes Essential for Germ Cell Survival
RNA-seq and CLIP showed a critical requirement for DAZL in maintaining the fidelity of the germ cell transcriptome through a combination of direct and indirect actions. While comparable numbers of genes had increased or decreased RNA levels in Dazl KO cells (1,584 and 1,461, respectively), they differed considerably with respect to enriched GO annotations and expression levels in WT cells. Genes with increased RNA in Dazl KO were significantly enriched for GO terms associated with the extracellular region and plasma membrane (Data S4). A total of 40% of the genes in this set had little to no expression in WT cells (<1 RPKM). The presence of epigenetic regulators among the list of DAZL-enhanced genes with DAZL-3′ UTR interactions (including Piwil2, Tdrd9, Dnmt1, and Dnmt3b; Figures 7A and 7B, group i) may partially explain this ectopic gene expression and the defects in erasure and re-establishment of DNA methylation that are observed in embryonic Dazl KO germ cells (Gill et al., 2011; Haston et al., 2009).
In stark contrast, genes with decreased RNA levels in Dazl KO cells were significantly enriched for GO terms associated with nuclear proteins involved in cell-cycle regulation, DNA repair, DNA methylation, transcription, RNA processing, and RNA transport (Figure S6; Data S4). Also enriched among DAZL-enhanced genes are GO terms associated with ubiquitin-protein ligase activity involved in cell-cycle control, a broad set of genes with the annotation “spermatogenesis,” and numerous genes essential for spermatogonial proliferation, differentiation, and/or survival (Figure S6, group G). Representative examples include Atm (Takubo et al., 2008), Ccnb1 (Tang et al., 2017), Dnd1 (Yamaji et al., 2017), Huwe1 (Bose et al., 2017), Kit (Blume-Jensen et al., 2000), Nxf2 (Pan et al., 2009), Plzf/Zbtb16 (Costoya et al., 2004), Sohlh2 (Hao et al., 2008), and Sox3 (Raverot et al., 2005) (see Data S4 for a more extensive list). These observations suggest that DAZL functions as a regulator of regulators that directly sustains mRNA levels of essential genes through RNA binding and indirectly by regulating mRNAs for transcription factors and epigenetic regulators that define which genes are transcribed. At this time, we cannot exclude the possibility that some of the RNA differences identified by RNA-seq reflect alterations in the relative levels of spermatogonial subtypes present in P6 WT and Dazl KO testes. However, pan-spermatogonial markers, as well as markers enriched in undifferentiated (Cdh1, Plzf/Zbtb16, and Gfra1) and differentiated spermatogonia (Kit and Stra8), had reduced RNA levels in Dazl KO cells.
Global measurements of protein and mRNA stability have indicated that genes with short mRNA and protein half-lives are significantly enriched for the GO terms “transcription,” “cell cycle,” and “chromatin modification” (Schwanhäusser et al., 2011), all of which are among the most-enriched GO terms associated with DAZL-enhanced genes with DAZL-3′ UTR interactions. Approximately 50% of genes with reduced mRNA levels in Dazl KO cells are among the top 20% of genes with the highest mRNA levels in WT cells (Figure 3E). Conversely, genes with elevated mRNAs following DAZL expression in GC-1 spg cells had very low levels of total and ribosome-associated mRNAs in the absence of dox, as well as significantly lower ribosome density (Figures S5E and S5F). Among these were Cxcl1 and Cxcl5, two well-studied genes that are subject to extensive negative post-transcriptional regulation (Herjan et al., 2013). Furthermore, pre-mRNA levels of DAZL-enhanced genes were unaltered by DAZL expression (Figure S5G), indicating that mRNA increases were due to regulation at the post-transcriptional level. These observations suggest that DAZL functions in the germline to increase steady-state levels of inherently unstable mRNAs to ensure high concentrations of regulatory factors important for spermatogonia proliferation.
In conclusion, our study provides insights into the molecular basis of germ cell loss in Dazl KO mice and demonstrates that germ cell survival depends on a DAZL-dependent mRNA regulatory program. Given the functional conservation between mouse DAZL, human DAZL, and DAZ (Vogel et al., 2002), our findings shed light on the molecular basis for azoospermia in 10%–15% of infertile men with Y chromosome microdeletions. The RNA targets of DAZL extend far beyond germ cell-specific genes and include many that encode core components of macromolecular complexes present in all proliferating cells. Therefore, our findings may also be relevant to other human diseases because Dazl is a susceptibility gene for human testicular cancer (Ruark et al., 2013) and is amplified or mutated in nearly 30% of breast cancer patient xenografts examined in a single study (Eirew et al., 2015). We propose a general model (Figure S7) whereby DAZL binds a vast set of mRNAs via polyA-proximal interactions facilitated by PABPC1-polyA binding and post-transcriptionally enhances the expression of a subset of mRNAs, namely a network of genes that are essential for cell-cycle regulation and mammalian germ cell maintenance. These observations provide insights into molecular mechanisms by which a single RBP is recruited to its RNA targets and coordinately controls a network of mRNAs to ensure germ cell survival.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Donny Licatalosi (ddl33@case.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J animals bearing the DazlTm1hgu allele were rederived at the Case Transgenic and Targeting Facility, and bred with animals bearing the Stra8-iCre or IRG transgenes. Mixed background (CD1xC57BL/6J) Stra8-iCre2+; DazlTm1hgu/+ males and IRG+; DazlTm1hgu/+ females were crossed to generate Dazl WT and KO offspring with GFP+ male germ cells. Day of birth was considered P0. For all procedures, animals were anesthetized by isoflourane inhalation and death confirmed by decapitation or cervical dislocation. HITS-CLIP, iCLIP, adult testis RNA-Seq, and adult testis Poly-Seq were performed using CD-1 animals purchased from Charles River labs. Spermatogonia PolyA-Seq libraries were generated from FACS-isolated cells from 8 week Stra8-iCre+; IRG+ C57BL/6J males as previously described (Zagore et al., 2015). All animal procedures were approved by the Institutional Animal Care and Use Committee at CWRU.
Tissue Culture Cells
GC-1 spg cells (an immortalized mouse spermatogonial cell line from ATCC) were cultured in DMEM, high glucose medium (ThermoFisher) supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (all from ThermoFisher) at 37°C, 5% CO2. GC-1 spg cells with inducible DAZL expression were maintained in DMEM high glucose medium (ThermoFisher) supplemented with 10% (v/v) Tet-system approved FBS (Clontech), 100 U/mL penicillin, 100 μg/mL streptomycin, 5 μg/mL blasticidin, and 300 μg/mL Zeocin (all from ThermoFisher) at 37°C, 5% CO2.
METHOD DETAILS
Spermatogonial cell isolation
Isolation of GFP+ cells was performed using dual fluorescence labeling followed by FACS as previously described (Zagore et al., 2015), except P6 cells were not stained with Hoechst and were collected using either BD Biosciences Aria or iCtye Reflection cytometry instrumentation. Briefly, p6 testes from Stra8-iCre+; IRG+; Dazl2+ and Stra8-iCre+; IRG+; DazlTm1hgu/Tm1hgu mice were decapsulated in cold 1X GBSS (Sigma). Enzyme dissociation of spermatogenic cells was performed using 3 mL GBSS (prewarmed to 33°C) containing 120 U/mL collagenase type I (Worthington Biochemical) and 10 μL 1 mg/mL DNase I (in 50% glycerol) in a 15 mL conical tube. Tubes were places horizontally at 33°C at 120 rpm for 15 minutes. Following incubation, tubules were allowed to settle for 1 minute at room temperature by placing tubes upright. The supernatant containing interstitial cells was removed using a wide-mouth pipette and the collagenase digestion was repeated. After the supernatant was removed a second time, the settled tubules were resuspended in 2.5 mL of prewarmed 1X GBSS containing 120 U/mL collagenase type I, 10 μL DNase I, and 50 μL of 50 mg/mL trypsin (Worthington Biochemical). Tubes were inverted gently to mix and placed horizontally for 15 minutes at 33°C, 120 rpm. A wide-mouth pipette was used for 3 minutes to disrupt the seminiferous tubules. Finally, 10 μL DNase I, and 30 μL of 50 mg/mL trypsin were added and the tubes were incubated again at 33°C for 15 minutes at 120 rpm. To inactivate trypsin, 400 μL prewarmed (33°C) filter sterilized FBS (ThermoFisher) was added followed by gentle mixing. Cells were pelleted by centrifugation for 5 minutes at 500 rcf. and resuspended in 1 mL prewarmed (33°C) 1X HBSS, supplemented with 10 μL DNase I. After sorting, cells were collected into tubes coated with 20% BSA, washed with 1X PBS and pelleted before snap freezing on dry ice.
Microscopy
P6 testes were decapsulated in cold HBSS, fixed overnight in 4% paraformaldehyde, and paraffin-embedded by the Histology Core Facility at CWRU. Slides containing 5 μM sections were de-paraffinized with 3 5-minute washes in xylene followed by a 5-minute rinse in 100% ethanol. Tissue was rehydrated with 5-minute incubations in 100%, 95%, 70% and 50% ethanol followed by tap water. Antigen retrieval was performed with citrate buffer, pH 6.0 (10mM Sodium Citrate, 0.05% Tween 20), at 95°C for 20 minutes. Slides were cooled with tap water for 10 minutes followed by a 5-minute wash in 1X PBS. Tissue was permeabilized for 10 minutes in 0.25% Triton X-100 in 1X PBS and rinsed in PBST (0.1% Tween 20 in 1X PBS). Slides were blocked in 1% BSA in PBST for 1 hour. Sections were incubated with 1:500 anti-GFP (Abcam) and 1:500 anti-DDX4 (Abcam) for 1 hour followed by 3 5-minute washes in PBST. An additional one hour incubation was performed using 1:100 anti-mouse Alexa 488 (ThermoFisher) and 1:100 anti-rabbit Cy3 (Jackson ImmunoResearch). Slides were washed 3 times in PBST before staining with 0.5 μg/mL DAPI for 5 minutes and rinsing in 1X PBS. Tissue was mounted on coverslips with Fluoromount G (Southern Biotech) and images captured using a Deltavision Deconvolution Microscope.
RNA-Seq
Adult testis RNA-Seq libraries (Tru-Seq, Illumina) were prepared from testes of two 8-week CD1 males. RNA-Seq libraries from WT and Dazl KO GFP+ cells purified by FACS were generated from two Stra8-iCre+; IRG+; Dazl2+ biological replicates (63,204 and 111,702 GFP+ cells) and three Stra8-iCre+; IRG+; DazlTm1hgu/Tm1hgu biological replicates (13,572, 13,430, and 30,088 GFP+ cells). RNA was extracted from samples via RNeasy Micro Kit (QIAGEN). Quality evaluation and quantification were performed on the Fragment Analyzer by Advanced Analytical. Total RNA was normalized to 250 picograms prior to oligo-dT capture and cDNA synthesis with the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara). The resulting cDNA was assessed on the Fragment Analyzer and quantified by the Life Technologies Qubit 3.0 fluorometer. Libraries were generated using the Illumina Nextera XT DNA Library Prep kit. High depth sequencing (50 million reads per sample) was performed with an Illumina HiSeq 2500 on a Rapid Run v2, 100 base pairs, Paired-End run. All RNA-Seq libraries were sequenced at CWRU Sequencing Core. Read mapping and gene expression quantification was performed using Olego and Quantas (Wu et al., 2013).
PolyA-Seq
Sequencing library construction
Adult testis PolyA-Seq libraries were generated from 8 week CD1 mice. Spermatogonia PolyA-Seq libraries were generated from 400ng of RNA from spermatogonia isolated by FACS from 8 week old Stra8-iCre+; IRG+ males as previously described (Zagore et al., 2015). PolyA+ RNA was selected by oligodT-hybridization on Dynabeads (ThermoFisher) and fragmented by alkaline hydrolysis. RNA fragments 50–100 nt long were gel purified from 10% PAGE/Urea gels, and used as template for reverse transcriptions with SuperScript III (Invitrogen). First strand cDNA was gel purified, then circularized with CircLigase (EpiCentre). Circularized DNA was used as the template for PCR. After cycle number optimization to obtain the minimal amount of PCR product detectable by SYBR Gold (ThermoFisher), the PCR product was gel purified and used as template for a second PCR with primers containing Illumina adaptor sequences. Primers used for library preparation are listed in Data S6. Adult and spermatogonia PolyA-Seq libraries were sequenced at CWRU and UC Riverside, respectively.
Read processing, filtering, and mapping
Reads consisting of two or more consecutive N’s or all poly(A) were filtered out, then remaining reads were processed to remove adaptor and poly(A) sequences. Remaining reads were mapped to mouse genome mm9 with GSNAP allowing 2 mismatches. Reads with identical genomic footprint and 4N’s were then collapsed into single reads as likely PCR duplicates. Mapped reads that were likely internal priming events rather than true poly(A) tails were discarded if they had 6 or more consecutive A’s or 7 or more A’s in the 10 nt window downstream of the read 3′ end (cleavage and polyadenylation site). We accepted only 3′ ends with 1 of 14 hexamers within 50 nt upstream of the 3′ end (as described in (Martin et al., 2012).
Clustering of reads into poly(A) sites
To focus on high confidence poly(A) sites, we accepted read 3′ ends that were ≥ 1 tag per million reads (TPM). Due to heterogeneity in cleavage site selection, reads that fell within 10 nt of each other were clustered into poly(A) sites, and a TPM was calculated for the entire cluster (polyA site region).
Identification of 3′UTRs
PolyA sites regions from adult PolyA-Seq were intersected with RefSeq genes. Intergenic sites within 10kb of an upstream stop codon were assigned to the upstream gene. For each polyA site region mapped to a protein coding gene, the closest upstream stop codon was identified and used to define 28,032 3′UTRs in adult testes (only polyA sites with at least 5% of the total PolyA-Seq reads in a gene were considered). To identify 3′UTRs expressed in spermatogonia, mapped reads from spermatogonia PolyA-Seq libraries were intersected with 100nt regions corresponding to the 28,032 adult testis polyA sites, identifying 16,502 sites with read counts greater than zero in both spermatogonia PolyA-Seq replicate libraries.
qRT-PCR validation
qRT-PCR was performed as previously described (Zagore et al., 2015). Sequences of all primers used in this study are available in Data S6.
DAZL HITS-CLIP from adult testes
Library construction
DAZL HITS-CLIP libraries were generated from testes from three biologic replicate 8-week old mice. Testes were detunicated in cold HBSS and seminiferous tubules UV-irradiated on ice. All steps were performed as previously described (Licatalosi et al., 2008; Licatalosi et al., 2012). Primers used for library preparation are listed in Data S6. Libraries were sequenced at the CWRU Sequencing Core and resulting reads mapped using Bowtie and Tophat (Trapnell et al., 2012) and filtered as previously described (Hannigan et al., 2017).
Identification of CLIP peaks
To identify peak regions in BR3 HITS-CLIP regions, the sum of all CLIP read lengths in a cluster was divided by the length of the cluster footprint to obtain an ‘expected’ density. Peaks are defined as regions of at least 10 nt long where the observed CLIP read density exceeded the expected density. 11,224 out of 11,297 had identifiable peaks (99.4% of BR3 clusters, 18364 peaks total).
Peak normalization and binning
For each BR3 peak, the sum of all observed-expected values per nt was calculated and divided by the average RNA-Seq read density per nt of the BR3 cluster. Only peaks within clusters with an average RNA-Seq read density per nt greater than 10 were considered for binning (13,106 peaks).
Genomic distribution of BR3 positions
To annotate BR3 sites, RefSeq coding regions, 5′UTRs, 3′UTRs, and introns were downloaded from the UCSC genome browser and intersected individually with BR3 coordinates.
Examination of genes with 5′UTR BR3 sites
For this analysis, only BR3 sites that mapped unambiguously to a single type of RefSeq gene fragment were examined (see Figure S2).
DAZL iCLIP from P6 testes and GC-1 spg cells
Library construction and analysis
Individual DAZL iCLIP libraries were generated from three P6 testes. Three 10 cm plates of GC-1 spg cells were induced with 10ng/mL doxycycline and harvested after 24 hours. The libraries were processed similar to HITS-CLIP libraries with the following changes. No 5′linker ligation step was performed, and the RT primer contained iSp18 spacers and phosphorylated 5′ end permitting circularization of first strand cDNA (after gel purification) to generate a PCR template without linearization (as described by (Ingolia et al., 2012). Primers used for library preparation are listed in Data S6. Libraries were sequenced at the CWRU Sequencing Core. Identical reads within each library were collapsed, and then mapped using Olego (Wu et al., 2013). Reads overlapping repetitive elements were discarded. After removing exon-spanning reads (0.06% of total), MultiIntersectBED (Quinlan and Hall, 2010) was used to identify genomic regions with overlapping CLIP reads from 3/3 libraries.
Genomic distribution of BR3 positions
BR3 regions were intersected with RefSeq genes and 3′UTRs of RefSeq protein coding genes.
Sequence analyses
Enriched motifs within CLIP regions and UTRs were performed using the EMBOSS tool Compseq. To generate z-scores, shuffled control sets were generated for each dataset analyzed using the EMBOSS tools Shuffleseq (10 shuffled versions of each sequence in each dataset). CIMS analysis was performed as previously described (Zhang and Darnell, 2011).
Distribution of DAZL-RNA contacts in 3′UTRs
Metagene analysis of DAZL-3′UTR interactions was performed on a subset of 3′UTRs defined by PolyA-Seq. 3′UTRs that overlapped with any intron sequence annotated in RefSeq were omitted. Only BR3 sites in genes with a single 3′UTR were analyzed. The resulting 5,284 BR3 HITS-CLIP peaks (adult testis) were examined in 2022 3′UTRs, and 3,321 BR3 iCLIP regions (P6 testis) were examined in 1,604 3′UTRs. To calculate distances, the midpoint of each BR3 site was measured relative to the upstream stop codon and the downstream polyA site. Positions of binding were then examined in 20nt windows relative to the stop codon or polyA site. For each 3′UTR, the expected distribution was determining by counting the number of BR3 sites in the 3′UTR and the number of 20nt bins in the 3′UTR, to determine the likelihood of any 20nt bin in a given 3′UTR containing a BR3 site. The control set of 3771 3′UTRs was identified by intersecting the 28032 3′UTRs from adult testis (identified by PolyA-Seq) with all adult testis CLIP reads, and selecting those with zero CLIP read coverage and no overlap with RefSeq introns.
Generation of inducible stable cell lines
The Dazl open reading frame was PCR amplified using Phusion polymerase (NEB) from WT CD1 mouse cDNA with the following primers: Dazl ORF F: (ACACTCGAGCCACCATGTCTGCCACAACTTCTGAGG), Dazl ORF R: ACAGGATCCTTAGCAGAGATGATCA GATTTAAGC). Dazl ORF was then cloned into pcDNA 4/TO/myc-His C response element (ThermoFisher). Stable monoclonal cell lines were generated from parental GC-1 spg cells using the T-Rex System (ThermoFisher) according to manufacturer’s instructions. Monoclonal cell populations were selected with blasticidin and Zeocin (ThermoFisher) antibiotics and tested for inducible DAZL expression by doxycycline induction and western blotting. Western blotting was performed as previously described in (Zagore et al., 2015) using anti-DAZL (Abcam) and anti-HSP90 (BD Bioscience) antibodies.
GC-1 spg Ribosome Profiling and RNA-Seq
Library construction and analysis
GC-1 spg cells were induced with 10 ng/mL doxycycline and harvested after 24 hours. Cells were immediately scraped into lysis buffer containing the translation elongation inhibitor emetine. (10 mM Tris-HCl(7.4), 5 mM MgCl2, 100 mM KCl, 1% Triton X-100,0.1 mg/mL emetine, 2 mM DTT, cOmplete Mini EDTA-free protease inhibitor). 50% of the lysate was treated with 0.2 U/uL RNaseI (ThermoFisher) for 30 minutes at room temperature. The remaining lysate (gradient input) was reserved for RNA-Seq library preparation. Ribosome protected footprints were run on a 10%–45% sucrose gradient and monosome associated RNA was purified using ethanol precipitation followed by miRNeasy Kit (QIAGEN). In parallel, RNA from the gradient inputs was purified using the RNeasy Mini Kit (QIAGEN), fragmented by partial alkaline hydrolysis and size selected. Purified RNA fragments from monosome fractions and gradient inputs were RQ1 DNase treated (Promega), rRNA depleted (RiboZero rRNA Removal kit – human/mouse/rat, Illumina) and processed concurrently. The libraries were prepared similar to iCLIP libraries except a preadenylated 3′ linker was used in place of the traditional 3′ RNA linker (Ingolia et al., 2012). Primers used for library preparation are listed in Data S6. Libraries were sequenced at the CWRU Sequencing Core. Identical reads within each library were collapsed and rRNA sequences removed. Read mapping, gene expression quantification, and differential gene expression analysis was performed using Olego and Quantas (Wu et al., 2013)
Luciferase reporter generation
DAZL target 3′UTRs and at least 100nt of downstream sequence were cloned into the pRL-TK vector (Promega), replacing the SV40 late poly(A) region. To generate non-polyadenylated reporter variants, the Hist2 histone stem loop (HSL) sequence was amplified from genomic CD1 mouse DNA using the following primers: Hist2 XbaI F (ACATCTAGAAAAAGGCTCTTTTCAGAGC), Hist2 BamHI R (TGTGGATCCTACCGTGACACAACTCTTTATC). After removing the SV40 late poly(A) region from pRL-TK, the HSL fragment was cloned downstream of the Renilla open reading frame. DAZL target 3′UTRs with disrupted poly A signals were inserted upstream of the HSL using blunt ended cloning. Oligonucleotides used for cloning are listed in Data S6.
Dual Luciferase Assays
GC-1 spg cells were induced with 1 ng/mL doxycycline. After 24 hours, pRL-TK 3′UTR reporters and pGL4.54[luc2/TK] (Promega) firefly luciferase control plasmids were transfected into GC-1 spg cells using Lipofectamine 2000 (Thermofisher). Media was replaced after 4–6 hours and cells were harvested after 24 hours. Dual luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer’s instructions. Reporter renilla luciferase levels were normalized to firefly luciferase activity. A two-tailed Student’s t test was used to analyze dual luciferase results and p value significance is defined as follows: p < 0.0001, ***; p < 0.01, **; p < 0.05, *.
Native RNA IP
Native RNA IPs (RIPs) were performed using conditions previously described (Gagliardi and Matarazzo, 2016) with the following antibodies: DAZL (Abcam), HUR (LifeTechnologies), and SRSF1 (Thermofisher). After IP, RNA was purified by phenol/CHCl3 extraction followed by ethanol precipitation. A rigorous Turbo DNase (ThermoFisher) treatment was used to remove residual plasmid DNA. qRT-PCR analysis was performed using Superscript III (ThermoFisher) and FastStart Universal SYBR Green Mastermix (ThermoFisher) to assess endogenous transcript or luciferase reporter bound to the RBP of interest. IP efficiency was calculated using 2^(ΔCT input-pellet). A two-tailed Student’s t test was used to analyze qPCR results and p value significance is defined as follows: p < 0.0001, ***; p < 0.01, **; p < 0.05, *.
GO Analysis
GO term analyses were performed with the Cytoscape application BiNGO (Saito et al., 2012; Cline et al., 2007) using a hypergeometric statistical test and Benjamini & Hochberg FDR correction (significance level of 0.05) to identify enriched terms after multiple testing correction. GO Slim settings were used to process the 656 set of genes (RPKM < 1 in WT), while GO Full was used for the 1462 and 1584 sets of genes (DAZL-enhanced and repressed, respectively). A set of 13,659 genes with RPKM > 1 in either WT or Dazl KO samples was used as the background gene set for enrichment. GO analysis of 501 Dazl-bound and enhanced genes was performed using GO Miner (Zeeberg et al., 2003).
Protein-Protein Interactions
The STRING database (Szklarczyk et al., 2015) was used to identify PPIs between protein products of the 501 DAZL-enhanced genes with DAZL-3′UTR interactions. Selected parameters were experiments and databases (for interaction sources), and highest confidence (0.900) (for minimum required interaction score).
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical tests performed in this research article are indicated in the figure legends along with sample size. P value significance is defined as follows: p < 0.0001, ***; p < 0.01, **; p < 0.05, *. Mean ± standard deviation is shown in all graphs. Analysis of sequencing data was performed as described in the following sections: ‘RNA-Seq Libraries’, ‘PolyA-Seq Libraries’, ‘DAZL HITS-CLIP from Adult Testis’, ‘DAZL iCLIP from P6 Testes and GC-1 spg Cells’, ‘GC-1 spg Ribosome Profiling and RNA-Seq’, ‘GO Analysis’, and ‘Protein-Protein Interactions’. Additional analysis of CLIP and RNA-Seq datasets was performed using tools from Galaxy (Afgan et al., 2018).
DATA AND SOFTWARE AVAILABILITY
The accession number for all deep sequencing datasets reported in this paper is GEO: GSE108997.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-DAZL | Abcam | Cat#ab34139; RRID: AB_731849 |
| Rabbit monoclonal anti-GFP | Abcam | Cat#ab183734; RRID: AB_2732027 |
| Mouse monoclonal anti-DDX4 | Abcam | Cat#ab27591; RRID: AB_11139638 |
| Goat anti-mouse IgG (H+L) Alexa Fluor 488 Conjugated | Thermo Fisher | Cat#A11017; RRID: AB_143160 |
| Cy3 Donkey polyclonal anti-rabbit | Jackson ImmunoResearch Labs | Cat#711-165-152; RRID: AB_2307443 |
| HuR monoclonal antibody | Thermo Fisher | Cat#39–0600; RRID:AB_2533394 |
| CUG-BP1 monoclonal antibody (3B1) | Thermo Fisher | Cat#MA1–16675; RRID: AB_2292334 |
| SRSF1 monoclonal antibody | Thermo Fisher | Cat#32–4500; RRID: AB_2533079 |
| Mouse monoclonal anti-Hsp90 | BD Bioscience | Cat#610418; RRID: AB_397798 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dynabeads Oligo(dT)25 | Thermo Fisher | Cat#61002 |
| Dynabeads Protein A for Immunoprecipitation | Thermo Fisher | Cat#10002D |
| SuperScript III Reverse Transcriptase | Thermo Fisher | Cat#18080044 |
| CircLigase ssDNA Ligase | Epicenter | Cat#CL4115K |
| SYBR Gold Nucleic Acid Gel Stain | Thermo Fisher | Cat#S11494 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#4693159001 |
| Phusion High-Fidelity DNA Polymerase | NEB | Cat#M0530S |
| FastStart Universal SYBR Green Master (Rox) | Sigma-Aldrich | Cat#4913850001 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher | Cat#11668019 |
| RQ1 RNase-Free DNase | Promega | Cat#M6101 |
| Proteinase K | Sigma-Aldrich | Cat#3115887001 |
| RNase I 100U/uL | ThermoFisher | Cat#AM2294 |
| Blasticidin | ThermoFisher | Cat#R21001 |
| Zeocin Selection Reagent | ThermoFisher | Cat#R25001 |
| DMEM, high glucose | ThermoFisher | Cat#11965084 |
| Fetal Bovine Serum (FBS) | ThermoFisher | Cat#10437028 |
| Penicillin/streptomycin | ThermoFisher | Cat#15140122 |
| Tet System Approved FBS | Clontech | Cat#631106 |
| Turbo DNA Free Kit | ThermoFisher | Cat#AM1907 |
| Critical Commercial Assays | ||
| RiboZero rRNA Removal kit - human/mouse/rat | Illumina | Cat#MRZH11124 |
| Dual Luciferase Reporter Assay System | Promega | Cat#E1910 |
| SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing | Takara | Cat#634890 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131–1024 |
| miRNeasy Kit | QIAGEN | Cat#217004 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| RNeasy Micro Kit | QIAGEN | Cat#74004 |
| Deposited Data | ||
| Raw and Analyzed Data | This paper | GSE108997 |
| Experimental Models: Cell Lines | ||
| Mouse: GC-1 spg cells (parental) | ATCC | Cat# CRL-2053, RRID: CVCL_8872 |
| GC-1 spg cells - inducible Dazl expression | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tg(Stra8-icre)1Reb/J | The Jackson Laboratory | RRID: IMSR_JAX:008208 |
| Mouse: B6;C3-Tg(CAG-DsRed,-EGFP)5Gae/J | The Jackson Laboratory | Stock#: 008605 |
| Mouse: Dazl Knockout - Dazl Tm1hgu | Marco Conti (UCSF) | Ruggiu et al., 1997 |
| Mouse: CD1-IGS | Charles River | Crl:CD1(ICR) |
| Oligonucleotides | ||
| Primers to clone Dazl ORF into pcDNA4/TO/myc-His C plasmid. Forward: ACACTCGAGCCACCATGTCTGCCACAACTTCTGAGG | This paper | N/A |
| Reverse: ACAGGATCCTTAGCAGAGATGATCAGATTTAAGC | ||
| Primers to generate histone stem loop to clone into pRTL-TK. Forward: ACATCTAGAAAAAGGCTCTTTTCAGAGC | This paper | N/A |
| Reverse: TGTGGATCCTACCGTGACACAACTCTTTATC | ||
| Primers for qRT-PCR, see Data S6 | This paper | N/A |
| Primers for HITS-CLIP, iCLIP, PolyA-Seq, Ribosome Profiling, and RNA-Seq library generation, see Data S6 | This paper | N/A |
| Primers for cloning, see Data S6 | This paper | N/A |
| Recombinant DNA | ||
| pRL-TK Vector | Promega | Cat#E2241 |
| pGL4.54[luc2/TK] Vector | Promega | Cat#E5061 |
| pcDNA 4/TO/myc-His C Mammalian Expression Vector | ThermoFisher | Cat#V1030–20 |
| pcDNA 6/TR | ThermoFisher | Cat#V1025–20 |
| Software and Algorithms | ||
| BiNGO Cytoscape Application | Saito et al., 2012; Cline et al., 2007 | http://apps.cytoscape.org/apps/bingo |
| String | Szklarczyk et al., 2015 | https://string-db.org/ |
| Olego | Wu et al., 2013 | https://zhanglab.c2b2.columbia.edu/index.php/Resources |
| Quantas | Wu et al., 2013 | https://zhanglab.c2b2.columbia.edu/index.php/Resources |
| GO Miner | Zeeberg et al., 2003 | https://discover.nci.nih.gov/gominer/index.jsp |
| MultiIntersectBED | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| Galaxy | Afgan et al., 2018 | https://usegalaxy.org |
| EMBOSS tools | Rice et al., 2000 | http://bioinfo.nhri.org.tw/cgi-bin/emboss/ |
| Other | ||
| Deltavision Deconvolution Microscope. | GE Healthcare | N/A |
Highlights.
Transcriptome-wide maps reveal broad DAZL binding to thousands of mRNAs near polyA tails
DAZL binding to GUU sites requires local polyA sequences
DAZL positively regulates a subset of its directly bound RNA targets
DAZL controls an mRNA regulatory program necessary for germ cell survival
ACKNOWLEDGMENTS
We are grateful to Marco Conti for providing the DazlTm1hgu/+ mice, Jo Ann Wise for comments on the manuscript, and the following Case Western Reserve University (CWRU) core facilities: Transgenic and Targeting Facility (mouse rederivation); Genomics (deep sequencing); Tissue Resources (embedding); Virology, Next Generation Sequencing, and Imaging Core (IF microscopy); Cytometry and Microscopy (FACS); and Applied Functional Genomics (low input RNA-seq libraries). This work was supported by funds from the NIH to L.L.Z. (T32 GM08056) and D.D.L. (R01 GM107331).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, and six data files and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.10.012.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, and Noble WS (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell AA, and Ricci EP (2017). When mRNA translation meets decay. Biochem. Soc. Trans 45, 339–351. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Jiang G, Hyman R, Lee KF, O’Gorman S, and Hunter T (2000). Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3′-kinase is essential for male fertility. Nat. Genet 24, 157–162. [DOI] [PubMed] [Google Scholar]
- Bose R, Sheng K, Moawad AR, Manku G, O’Flaherty C, Taketo T, Culty M, Fok KL, and Wing SS (2017). Ubiquitin Ligase Huwe1 Modulates Spermatogenesis by Regulating Spermatogonial Differentiation and Entry into Meiosis. Sci. Rep 7, 17759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Welling M, Bloch DB, Muñoz J, Mientjes E, Chen X, Tramp C, Wu J, Yabuuchi A, Chou YF, et al. (2014). DAZL limits pluripotency, differentiation, and apoptosis in developing primordial germ cells. Stem Cell Reports 3, 892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. (2007). Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, and Gray NK (2005). The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 24, 2656–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, and Pandolfi PP (2004). Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet 36, 653–659. [DOI] [PubMed] [Google Scholar]
- De Gasperi R, Rocher AB, Sosa MA, Wearne SL, Perez GM, Friedrich VLJ Jr., Hof PR, and Elder GA (2008). The IRG mouse: a two-color fluorescent reporter for assessing Cre-mediated recombination and imaging complex cellular relationships in situ. Genesis 46, 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. (2015). Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M, Urano J, and Reijo Pera RA (2005). Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics 85, 92–105. [DOI] [PubMed] [Google Scholar]
- Fu XF, Cheng SF, Wang LQ, Yin S, De Felici M, and Shen W (2015). DAZ Family Proteins, Key Players for Germ Cell Development. Int. J. Biol. Sci 11, 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi M, and Matarazzo MR (2016). RIP: RNA Immunoprecipitation. Methods Mol. Biol 1480, 73–86. [DOI] [PubMed] [Google Scholar]
- Gill ME, Hu YC, Lin Y, and Page DC (2011). Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc. Natl. Acad. Sci. USA 108, 7443–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan MM, Zagore LL, and Licatalosi DD (2017). Ptbp2 Controls an Alternative Splicing Network Required for Cell Communication during Spermatogenesis. Cell Rep. 19, 2598–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Yamamoto M, Richardson TE, Chapman KM, Denard BS, Hammer RE, Zhao GQ, and Hamra FK (2008). Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells 26, 1587–1597. [DOI] [PubMed] [Google Scholar]
- Haston KM, Tung JY, and Reijo Pera RA (2009). Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One 4, e5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herjan T, Yao P, Qian W, Li X, Liu C, Bulek K, Sun D, Yang WP, Zhu J, He A, et al. (2013). HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J. Immunol 191, 640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herjan T, Hong L, Bubenik J, Bulek K, Qian W, Liu C, Li X, Chen X, Yang H, Ouyang S, et al. (2018). IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat. Immunol 19, 354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann MC, Narisawa S, Hess RA, and Millán JL (1992). Immortalization of germ cells and somatic testicular cells using the SV40 large T antigen. Exp. Cell Res. 201, 417–435. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Harel H, Levy G, and Meyuhas O (1999). Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett. 457, 209–213. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins HT, Malkova B, and Edwards TA (2011). Kinked β-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc. Natl. Acad. Sci. USA 108, 18266–18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, and Ule J (2010). iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol 17, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Park JY, Zheng D, Hoque M, Yehia G, and Tian B (2016). Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol. 14, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD (2016). Roles of RNA-binding Proteins and Post-transcriptional Regulation in Driving Male Germ Cell Development in the Mouse. Adv. Exp. Med. Biol 907, 123–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. (2008). HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, and Darnell RB (2012). Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 26, 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, and Page DC (2005). Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev. Biol 288, 309–316. [DOI] [PubMed] [Google Scholar]
- Lovasco LA, Gustafson EA, Seymour KA, de Rooij DG, and Freiman RN (2015). TAF4b is required for mouse spermatogonial stem cell development. Stem Cells 33, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa S, Yamashita M, Yasuda K, and Inoue K (2002). Zebrafish DAZ-like protein controls translation via the sequence ‘GUUC.’. Genes Cells 7, 971–984. [DOI] [PubMed] [Google Scholar]
- Manku G, and Culty M (2015). Mammalian gonocyte and spermatogonia differentiation: recent advances and remaining challenges. Reproduction 149, R139–R157. [DOI] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, and Zavolan M (2012). Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 1, 753–763. [DOI] [PubMed] [Google Scholar]
- Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, Dorfman DM, and Pera RA (2003). Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. USA 100, 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Eckardt S, Leu NA, Buffone MG, Zhou J, Gerton GL, McLaughlin KJ, and Wang PJ (2009). Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev. Biol 330, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raverot G, Weiss J, Park SY, Hurley L, and Jameson JL (2005). Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev. Biol 283, 215–225. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. (1995). Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet 10, 383–393. [DOI] [PubMed] [Google Scholar]
- Reijo R, Alagappan RK, Patrizio P, and Page DC (1996). Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet 347, 1290–1293. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, Semple CA, Gray NK, and Cooke HJ (2005). Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum. Mol. Genet 14, 3899–3909. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Collier B, Bingham V, Gray NK, and Cooke HJ (2007). Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA 13, 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, and Bleasby A (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Ruark E, Seal S, McDonald H, Zhang F, Elliot A, Lau K, Perdeaux E, Rapley E, Eeles R, Peto J, et al. ; UK Testicular Cancer Collaboration (UKTCC) (2013). Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat. Genet 45, 686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, and Cooke HJ (2000). In vivo and in vitro analysis of homodimerisation activity of the mouse Dazl1 protein. Gene 252, 119–126. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, and Cooke HJ (1997). The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389, 73–77. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, and Braun RE (2008). Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 46, 738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y (2010). Function of Nanos2 in the male germ cell lineage in mice. Cell. Mol. Life Sci. 67, 3815–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, and Ideker T (2012). A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders PT, Turner JM, Ruggiu M, Taggart M, Burgoyne PS, Elliott D, and Cooke HJ (2003). Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction 126, 589–597. [DOI] [PubMed] [Google Scholar]
- Schrans-Stassen BH, Saunders PT, Cooke HJ, and de Rooij DG (2001). Nature of the spermatogenic arrest in Dazl −/− mice. Biol. Reprod 65, 771–776. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Scotti MM, and Swanson MS (2016). RNA mis-splicing in disease. Nat. Rev. Genet 17, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HW, and Wilkinson MF (2014). Transcriptional control of spermatogonial maintenance and differentiation. Semin. Cell Dev. Biol 30, 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A, et al. (2013). Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3, 2179–2190. [DOI] [PubMed] [Google Scholar]
- Sweet TJ, and Licatalosi DD (2014). 3′ end formation and regulation of eukaryotic mRNAs. Methods Mol. Biol 1125, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, and Inoue K (2009). DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One 4, e7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Ohmura M, Azuma M, Nagamatsu G, Yamada W, Arai F, Hirao A, and Suda T (2008). Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell 2, 170–182. [DOI] [PubMed] [Google Scholar]
- Tang JX, Li J, Cheng JM, Hu B, Sun TC, Li XY, Batool A, Wang ZP, Wang XX, Deng SL, et al. (2017). Requirement for CCNB1 in mouse spermatogenesis. Cell Death Dis. 8, e3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui S, Dai T, Warren ST, Salido EC, and Yen PH (2000). Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol. Reprod 62, 1655–1660. [DOI] [PubMed] [Google Scholar]
- Vangompel MJW, and Xu EY (2011). The roles of the DAZ family in spermatogenesis: more than just translation? Spermatogenesis 1, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Speed RM, Ross A, and Cooke HJ (2002). Partial rescue of the Dazl knockout mouse by the human DAZL gene. Mol. Hum. Reprod 8, 797–804. [DOI] [PubMed] [Google Scholar]
- Wu J, Anczuków O, Krainer AR, Zhang MQ, and Zhang C (2013). OLego: fast and sensitive mapping of spliced mRNA-seq reads using small seeds. Nucleic Acids Res. 41, 5149–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tan X, Lin Q, Schmidt B, Engel W, and Pantakani DV (2013). Mouse Dazl and its novel splice variant functions in translational repression of target mRNAs in embryonic stem cells. Biochim. Biophys. Acta 1829, 425–435. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Jishage M, Meyer C, Suryawanshi H, Der E, Yamaji M, Garzia A, Morozov P, Manickavel S, McFarland HL, et al. (2017). DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature 543, 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagore LL, Grabinski SE, Sweet TJ, Hannigan MM, Sramkoski RM, Li Q, and Licatalosi DD (2015). RNA Binding Protein Ptbp2 Is Essential for Male Germ Cell Development. Mol. Cell. Biol 35, 4030–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, et al. (2003). Go-Miner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, and Darnell RB (2011). Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol 29, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Oatley J, Bardwell VJ, and Zarkower D (2016). DMRT1 Is Required for Mouse Spermatogonial Stem Cell Maintenance and Replenishment. PLoS Genet. 12, e1006293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for all deep sequencing datasets reported in this paper is GEO: GSE108997.