Abstract
In the 80 years since their discovery the β-lactam antibiotics have progressed through structural generations, each in response to the progressive evolution of bacterial resistance mechanisms. The generational progression was driven by the ingenious, but largely empirical, manipulation of structure by medicinal chemists. Nonetheless, the true creative force in these efforts was Nature, and as the discovery of new β-lactams from Nature has atrophied while at the same time multi-resistant and opportunistic bacterial pathogens have burgeoned, the time for empirical drug discovery has passed. We concisely summarize recent developments with respect to bacterial resistance, the identity of the new β-lactams, and the emerging non-empirical strategies that will ensure that this incredible class of antibiotics has a future.
Introduction
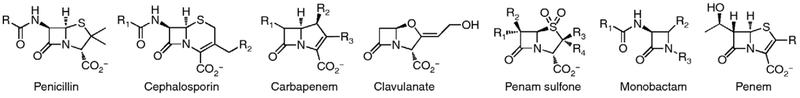
Penicillin was saving human life within fifteen years of Fleming’s providential contemplation of a curious Petri dish [1••,2••]. Over the next forty years a breadth of natural, synthetic, and semi-synthetic β-lactams encompassing the cephalosporin, carbapenem, clavulanate and penicillin sulfone, monobactam, and penem subclasses were discovered (Scheme 1). β-Lactams target the Penicillin-Binding Proteins (PBPs), enzymes charged with the biosynthesis and remodeling of the peptidoglycan structure of the bacterial cell wall [3–5,6•,7••]. They act as mimics of the d-Ala-d-Ala dipeptide in the peptidoglycan, and form an acyl-enzyme that is sterically blocked for further acyl transfer [8]. The β-lactams remain a therapeutic bulwark against bacterial infection [9••,10]. However, no new β-lactam subclass has been discovered in thirty years, research on the β-lactams has declined, and new β-lactam derivatives are a minority of new anti-infectives in clinical development [11•]. Several factors contribute to this circumstance: a diminished interest by pharma in all aspects of antibacterial discovery [12], a belief that the structure-activity interplay for the β-lactams has matured to such a level of complexity that the effort is no longer justified, and the concern that decades of profligate use of the β-lactams as antibiotics has selected such powerful resistance mechanisms that small promise for new β-lactams remains [13•]. Yet the inexorable emergence of multi-resistant bacterial pathogens, paired against the unparalleled efficacy and safety of the β-lactams, raises this question: can the β-lactams contribute to the pressing therapeutic need for new antibacterials? The properties of the β-lactams argue that they cannot possibly be ignored: the β-lactams target a uniquely prokaryotic structure – the bacterial cell wall – and their therapeutic safety derives from the absence of this target in eukaryotes. In this terse forum we forcefully summarize the recent discoveries that argue for future relevance of this most important antibacterial class.
Scheme 1.
The structures of the β-lactam family.
What has happened: β-Lactam-resistance in Gram-positive pathogens
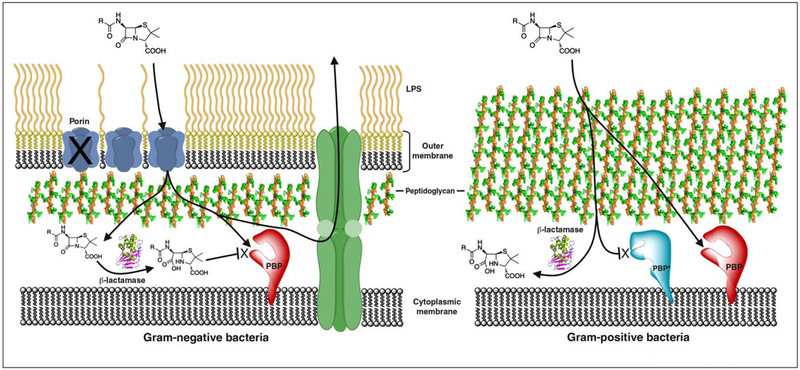
The susceptibility of Staphylococcus aureus to penicillins was lost quickly as a result of the acquisition of an enzyme capable of hydrolyzing the β-lactam ring of the penicillin, rendering the β-lactam ineffective (Figure 1). While the therapeutic advantage of the β-lactams was restored by new generations of penicillins (methicillin and oxacillin), methicillin-resistant S. aureus strain (MRSA) was identified shortly thereafter [14]. The high β-lactam resistance seen in MRSA arises from a different mechanism: acquisition of an accessory PBP (PBP 2a) that has lower reactivity toward β-lactams [15]. Intensive study of β-lactam-resistant PBPs has focused on the molecular basis for this resistance, to enable new β-lactam design [16–18]. Studies with peptidoglycan substrate mimetics, as allosteric effectors giving a more active conformation of PBP 2a, suggest value for allosteric effectors to synergize with existing β-lactams to render PBP 2a more β-lactam susceptible. Indeed, several new anti-MRSA cephalosporins by themselves trigger this conformational change [19]. In S. aureus both β-lactamase and PBP 2a expression are induced by β-lactam exposure [6•]. Likewise, inhibition of this signal transduction may also enable β-lactam synergy.
Figure 1.
A schematic representation of the entities involved in the antibacterial mechanism of the β-lactams, and the β-lactam resistance mechanisms exploited by Gram-negative and Gram-positive bacteria. Gram-negative bacteria (left panel) combine porin selection (or deletion) to minimize β-lactam access to the PBP, efflux pump expression to facilitate β-lactam removal, and β-lactamase expression in order to destroy the β-lactam. Gram-positive bacteria (right panel) acquire ‘replacement’ PBPs that are intrinsically less reactive toward β-lactams, often as a result of restricted access to the PBP active site, or revert to PBPs that remodel peptidoglycan biosynthesis in a way that evades the β-lactam mimicry of the –d-Ala-d-Ala peptide terminus otherwise used in the PBP-dependent cross-linking of the peptidoglycan.
PBP modification is also the primary basis for β-lactam resistance by the pneumococci and other α-hemolytic streptococci. For example, β-lactam challenge of Streptococcus pneumoniae induces an SOS response wherein mutation-prone transcription results in β-lactam resistance as a result of extensive mutation of three of its six PBPs [20]. High-level resistance occurs by mosaic gene formation of four of its PBPs. Enterococcus faecium exploits a different resistance mechanism. Following β-lactam exposure, a mutated PBP having minimal reactivity toward any β-lactam is expressed. Moreover, E. faecium (as do other Gram-positive bacteria) possesses an l,d-transpeptidase (Ltdfm) that catalyzes l,d-cross-linking of the peptidoglycan using l-Lys-d-Ala, rather than d-Ala-d-Ala, as the acyl donor. Production of the tetrapeptide substrate of Ltdfm is controlled by a two-component regulatory system and a metallo-d,d-carboxypeptidase [21]. This altered route is impervious to almost all β-lactams. Unexpectedly one β-lactam class, the carbapenems, acts as Ltdfm inhibitors [22]. Hence penicillin/cephalosporin and imipenem combination therapy may prove effective against resistant E. faecium. Non-classical 3 → 3 linkages generated by l,d-transpeptidases predominate in the peptidoglycan of nonreplicating Mycobacterium tuberculosis. Additionally, M. tuberculosis constitutively expresses a highly active β-lactamase. Recent recognition that the M. tuberculosis β-lactamase is inactivated by clavulanate, a classic β-lactamase inhibitor, suggests promise for dual β-lactam-clavulanate therapy against these mycobacteria [23]. Besides, loss of the LtdMt2 activity leads to loss of virulence and increased susceptibility to amoxicillin–clavulanate during the chronic phase of infection [24].
What has happened: β-Lactam-resistance in Gram-negative pathogens
Gram-negative bacteria combine porin selection, active efflux, and optimization of β-lactamase activity to acquire high-level β-lactam resistance (Figure 1). Modification of native PBPs also contributes to resistance in some species [25–27,28••]. Their β-lactamase activity is decisive. The serine-dependent β-lactamases use a catalytic serine nucleophile to form an acyl-enzyme intermediate, as an evolutionarily ancient adaptation of the PBP mechanism. The intensive therapeutic use of the β-lactams has resulted in the rapid diversification of the initially small β-lactamase family. Metallo-β-lactamases (MBLs) comprise a separate class that uses an active site metal for catalysis, and do not form a serine acyl-enzyme. The newest of these β-lactamases efficiently hydrolyze even the latest β-lactams.
Clavulanate and the penicillin sulfones (sulbactam and tazobactam) inhibit the serine-dependent β-lactamases [29,30]. Co-administration of a β-lactam antibiotic with one of these β-lactamase inhibitors has extended the therapeutic utility of early generation β-lactams [31]. Nonetheless, this combination therapy has further stratified serine-dependent β-lactamases such that some are now less efficiently inhibited. Reflecting their different mechanism, the metallo-β-lactamases are not inhibited by the serine β-lactamase inhibitors. Given the emerging role of the MBLs in β-lactam resistance, an effective MBL inhibitor is highly desirable. Notwithstanding recent progress, the prospect of a clinically useful MBL inhibitor is distant [30,32].
β-lactam compounds in late stages of development
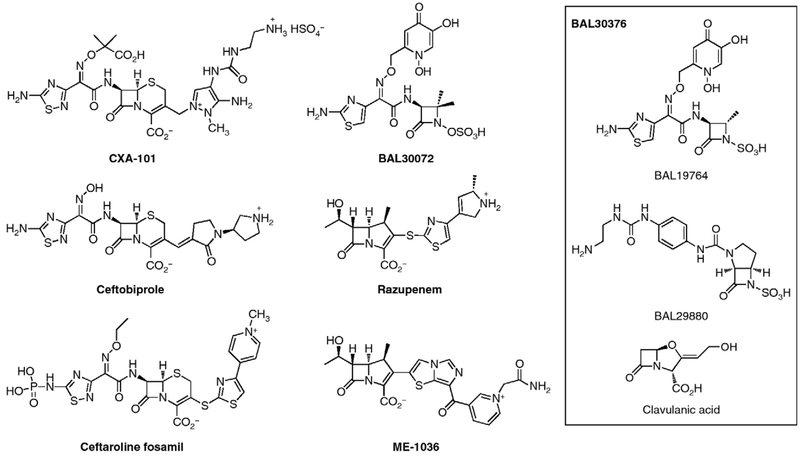
While there is no longer widespread interest in developing new β-lactams – a statement that equally pertains to other antibacterial classes – efforts toward new β-lactam structures have not been without reward. These efforts are directed toward the pressing needs of effective therapeutics against resistant Gram-positive pathogens such as MRSA, and against Gram-negative pathogens where the current cephalosporin and carbapenem structures have lost effectiveness. These efforts encompass four strategies. The first strategy is new cephalosporins that pair the N-(α-oxyimino)acyl sidechain of third-generation cephalosporins with structurally complex heterocycles at C-3 of the cephalosporin. The resulting cephalosporins (exemplified by CXA-101, ceftaroline and ceftobiprole) have exceptional activity against Gram-positives that additionally crosses over to some Gram-negatives. The second strategy is incorporation of heterocycles, structurally quite similar to those used for the cephalosporins, at C-2 of the carbapenems (exemplified by ME-1036 and razupenem) so as to impart broad-spectrum Gram-negative activity. The third strategy is new generation monobactams. Although the antibacterial spectrum of the monobactam subclass is limited to Gram-negative bacteria, monobactams are intrinsically stable to β-lactamase-catalyzed hydrolysis. The combination of a focused anti-bacterial spectrum and β-lactamase stability is advantageous given current opinion on antibiotic stewardship and evasion of the primary Gram-negative resistance mechanism, respectively. Last, the outstanding clinical success of combined β-lactam and β-lactamase inhibitor therapy is reflected by efforts toward more effective β-lactamase inhibitors. Progress toward each of these goals is summarized.
New generation cephalosporins
The C-7 N-(α-oxyimino)acyl sidechain of the third-generation cephalosporins improved β-lactamase stability and so enabled exceptional antibacterial efficacy even as the capabilities of the clinical β-lactamases evolved. As the cephalosporins now move to new generations, use of this N-acyl motif is now paired with heterocyclic substitution at C-3. All of these heterocycles show a positive charge at their terminus (as first seen in cefuroxime). In CXA-101 (Scheme 2) this positive charge is a primary amine; in ceftobiprole, a secondary amine; and in ceftaroline, a pyridinium. CXA-101 (currently in phase 2 clinical development) has exceptional Gram-negative activity and is especially active against P. aeruginosa [33]. CXA-101 has low propensity to induce resistance, and good stability against the AmpC β-lactamase. Ceftobiprole is the active component of the parenteral prodrug ceftobiprole medocaril [34]. The in vitro activity of ceftobiprole encompasses an unusually wide range of Gram-negative (including P. aeruginosa) and Gram-positive pathogens (including MRSA, methicillin-resistant Staphylococcus epidermidis, penicillin-resistant S. pneumoniae, and E. faecalis) [35]. Although ceftobiprole medocaril is approved in several countries, approval in the United States was denied owing to issues with the clinical data. The future of this antibiotic is uncertain. Ceftaroline, which is released upon in vivo hydrolysis of the water-soluble prodrug Ceftaroline fosamil, has potent activity against MRSA and S. pneumoniae owing to high-affinity for PBP 2a [19] and PBP 2x [36,37], respectively. The Gram-negative spectrum of ceftaroline is similar to that of other broad-spectrum cephalosporins. Ceftaroline is a substrate of the ESBL and AmpC β-lactamases. Accordingly, synergistic combinations of ceftaroline with β-lactamase inhibitors [38] and with aminoglycosides, are being explored [39].
Scheme 2.
Key β-lactam structures in current clinical development.
The molecular mechanism(s) for the superior activity of these cephalosporins against bacterial pathogens is uncertain. For Gram-negatives, a serendipitous combination of low affinity for drug export systems, low ability to induce β-lactamase expression [40], and high PBP affinity appears to operate. In the Gram-positive bacteria MRSA the catalytic activity of PBP 2a responds positively to the presence of the peptidoglycan substrate, opening the cleft, and allowing access to the PBP active site. This same opening is also facilitated by lower pH [41]. These new cephalosporins appear more adept at engaging the open cleft compared to preceding cephalosporin generations [19,42].
New generation carbapenems
Parenteral carbapenem therapy is used for serious Gram-negative infections. As a result of the acute angle between the conjoined β-lactam and dihydropyrrole rings forming the compact bicyclic core of the carbapenems, their unusual (in terms of structure and stereochemistry) 6α-2-hydroxyethyl substituent impairs the β-lactamase catalysis leading to long-lived acyl-enzymes. Nonetheless, new β-lactamase enzymes having clinically significant ‘carbapenemase’ activity now threaten this β-lactam class [32,43]. Incorporation of substitution into the carbapenem – to minimize their neurotoxicity, enable possible prodrug oral formulation, evade the carbapenemases, and extend carbapenem activity to Gram-positive pathogens – dominates the design of new carbapenems.
Imipenem was the first parenteral carbapenem. The ensuing generation of carbapenems (meropenem, ertapenem, and doripenem) now dominates clinical practice. The structural features evidenced by the newest carbapenems (biapenem, ME-1036, panipenem, razupenem, tebipenem, and tomopenem) strongly resemble those of new generation cephalosporins. Indeed, ME-1036 with its distal pyridinium at C-2 shows the same advantageous PBP access as does ceftaroline to PBP 2a of MRSA [19]. Likewise, razupenem (in phase 2 clinical trial) [44] has antimicrobial activity extending across a wide range of Gram-positive – including MRSA, vancomycin-intermediate S. aureus (VISA) and vancomycin-resistant Enterococcus faecium (VREF) – and Gram-negative bacteria, distinguishing it from previous carbapenems [45]. Its activity against P. aeruginosa is, however, mediocre as a result of diminished ingress via the outer membrane porins and efflux by the MexAB-OprM system, rather than as a result of reduced PBP affinity. Indeed, the notably high affinities of razupenem for PBP 2a of MRSA and PBP5 of ampicillin-resistant E. faecium implicate structural importance to its lipophilic C-2 thiazole for binding.
New generation monobactams
The clinical need for a Gram-negative β-lactam with exceptional β-lactamase stability is now far greater than at the time of the discovery of the monobactam subclass, some thirty years ago. The newest generation monobactams incorporate a siderophore substructure – that of an iron-binding chelate as is used by bacteria to secure iron from their environment – to facilitate uptake [46••]. Proper incorporation of the siderophore substructure into the β-lactam – to secure this objective while retaining Gram-negative activity – was a challenge. Nonetheless, these criteria appear fulfilled in the ‘siderophore’ monobactams BAL30072 and BAL30376. The dihydropyridone siderophore substituent of BAL30072 confers potent inhibitory activity against Acinetobacter, Burkholderia, and Pseudomonas spp., as well as many species of Enterobacteriaceae [47,48]. BAL30072 is highly resistant to hydrolysis by metallo-β-lactamases and is an inhibitor of class C β-lactamases. BAL30376 is a three-fold combination antibacterial, composed of a siderophore monobactam (analogous to aztreonam, with the dihydropyridone iron chelating group), a class-C β-lactamase inhibitor (BAL29880, a structurally unique monobactam that acts as a mechanism-based inhibitor) [49], and clavulanic acid (a mechanism-based inhibitor of other serine β-lactamases). This combination overcomes bacterial resistance by protection of the monobactam (which is inherently stable to metallo-β-lactamases) against the AmpC and ESBL enzymes, thus achieving activity against carbapenem-resistant strains of P. aeruginosa, among other multi-drug-resistant Gram-negatives. Against multi-resistant Enterobacteriaceae, BAL30376 overcame most AmpC- or ESBL-mediated resistance though less consistently than a carbapenem. However, BAL30376 was active against many metallo-β-lactamase producers.
New generation β-lactamase inhibitors
The concept of achieving antibacterial efficacy, without accelerating resistance development and without compromising safety, by synergism between a β-lactam and a β-lactamase inhibitor (inactivator) is proven by the decades of clinical success with the amoxicillin–clavulanate, ampicillin–sulbactam, and piperacillin–tazobactam combinations. With the forward evolution of β-lactamase capability resulting in the diminished effectiveness of clavulanate and the penam sulfones against these enzymes, there is renewed opportunity for β-lactamase inhibitor discovery. Specifically, recognizing the emergence of β-lactamases capable of carbapenem hydrolysis, future identification of an effective carbapenemase inhibitor has the potential to establish clinical longevity for existing carbapenems, as clavulanate and the penicillin sulfones have accomplished for the penicillins.
Several inhibitor classes – three of which are new – inactivate serine-dependent β-lactamases. While there are numerous structures that inhibit the metallo-β-tactamases in vitro, none has the potency, breadth of metallo-β-lactamase capability, and drug-like characteristics necessary for clinical use [32]. The new serine β-lactamase inhibitors are the 6-alkylidene-substituted penems and penicillin sulfones [50,51], the BAL29880 monobactam (in clinical development paired with a second PBP-specific monobactam, as cited above) [46••], and a bicyclic N-hydroxylactam-O-sulfate ester (NXL104, also in clinical development). NXL104 inhibits class A and class C (and some class D) serine-dependent β-lactamases, including those with extended spectrum β-lactam capability. The ceftazidime-NXL104 combination has potent activity against Enterobacteriaceae having the CTX-M ESBL. NXL104 also synergizes with the monobactam aztreonam against other Gram-negative pathogens, including those having carbapenemase activity [38,52] These collective data suggest promise for new β-lactamase inhibitor–β-lactam synergistic combinations.
The future of the β-lactams
The continuing evolution of resistance mechanisms threatens all current antibiotics. Until a strategy is in place that supports the discovery of new antibiotics and the cost of their clinical development, management of our antibiotic resources by reducing frivolous use, ensuring rigorous infection control and surveillance mechanisms, and selecting doses that disfavor emergence of resistance are necessary [27,53]. Accordingly, the future of the β-lactam antibiotics will involve identifying synergism between existing β-lactams and other antibiotic classes, while deferring use of these same β-lactams in favor of yet other antibiotics that have the potential to eliminate β-lactam-resistant bacteria [9••]. These strategies, coupled with the introduction of the new β-lactams now in late-stage clinical evaluation, offer promise with respect to management of Gram-positives and possibly – although less certainly – against the impending Gram-negatives. Where, however, will we find the β-lactams of the future? The combination of genetics, microbiology, and structural and mechanistic biochemistry will drive the expansion of the β-lactam structure. Manipulation of β-lactam biosynthetic gene clusters [54,55], augmented by epigenetic modulation of secondary metabolite expression [56], may uncover entirely new subclasses. The pathways used by bacteria to recognize the presence of antibiotics and mobilize their defense mechanisms are only now coming into focus. New targets [57] (or ‘targets’ to avoid) [26], new synergistic target pairings [57,58], and possible means to antagonize signal transduction [59] will be found. The global cooperation among industry, academia, government, medical and philanthropic leaders described by the ‘10 × 20 initiative’ of the Infectious Diseases Society of America [60••] is needed to ensure the perpetuity of antibiotics against future infectious diseases. The β-lactams will contribute to this quest.
Acknowledgements
The research activities of this laboratory are supported by the National Institutes of Health. Leticia I. Llarrull, Ph.D., is a Pew Latin American Fellow in the Biomedical Sciences, supported by The Pew Charitable Trusts. The opinions expressed are those of the authors and do not necessarily reflect the views of The Pew Charitable Trusts.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.••.Kong KF, Schneper L, Mathee K: β-Lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 2010, 118:1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]; A review that combines a concise history of the β-lactams combined with the current understanding of their molecular mechanisms.
- 2.••.Testero SA, Fisher JF, Mobashery S: β-Lactam antibiotics In Burger’s Medicinal Chemistry, Drug Discovery and Development, vol 7 Edited by Abraham DJ, Rotella DP. Wiley; 2010:257–402. [Google Scholar]; A comprehensive review of the medicinal chemistry of the β-lactams.
- 3.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P: The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 2008, 32:234–258. [DOI] [PubMed] [Google Scholar]
- 4.Schneider T, Sahl H-G: An oldie but a goodie–cell wall biosynthesis as antibiotic target pathway. Int J Med Microbiol 2010, 300:161–169. [DOI] [PubMed] [Google Scholar]
- 5.Kluge AF, Petter RC: Acylating drugs: redesigning natural covalent inhibitors. Curr Opin Chem Biol 2010, 14:421–427. [DOI] [PubMed] [Google Scholar]
- 6.•.Llarrull LI, Fisher JF, Mobashery S: Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new beta-lactams that meet the challenge. Antimicrob Agents Chemother 2009, 53:4051–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]; How fundamental biochemical study comparing the reactivity of β-lactam-susceptible to β-lactam-resistant S. aureus PBPs is relevant to β-lactam drug discovery for MRSA.
- 7.••.Kohanski MA, Dwyer DJ, Collins JJ: How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 2010, 8:423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]; How are bacteria ultimately killed by antibacterials? We know rather less about the answer to this basic question than we thought, and the perspectives given in this review have the potential to profoundly direct the experimental discovery of new targets and synergy with old targets.
- 8.Chen Y, Zhang W, Shi Q, Hesek D, Lee M, Mobashery S, Shoichet BK: Crystal structures of penicillin-binding protein 6 from Escherichia coli. J Am Chem Soc 2009, 131:14345–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.••.Shahid M, Sobia F, Singh A, Malik A, Khan HM, Jonas D, Hawkey PM: β-Lactams and β-lactamase-inhibitors in current- or potential-clinical practice: a comprehensive update. Crit Rev Microbiol 2009, 35:81–108. [DOI] [PubMed] [Google Scholar]; A thorough review of the clinical use of the β-lactams to control bacterial infection.
- 10.Peleg AY, Hooper DC: Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 2010, 362:1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.•.Devasahayam G, Scheld WM, Hoffman PS: Newer antibacterial drugs for a new century. Expert Opin Investig Drugs 2010, 19:215–234. [DOI] [PMC free article] [PubMed] [Google Scholar]; A summary of the entire scope of current antibacterial drug discovery.
- 12.Fischbach MA, Walsh CT: Antibiotics for emerging pathogens. Science 2009, 325:1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.•.Rice LB: The clinical consequences of antimicrobial resistance. Curr Opin Microbiol 2009, 12:476–481. [DOI] [PubMed] [Google Scholar]; The clinically important pathogens and the antibacterial options (and limitations) to control their infection.
- 14.Otto M: Looking toward basic science for potential drug discovery targets against community-associated MRSA. Med Res Rev 2010, 30:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antignac A, Tomasz A: Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob Agents Chemother 2009, 53:435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada M, Watanabe T, Baba N, Takeuchi Y, Ohsawa F, Gomi S: Crystal structures of biapenem and tebipenem complexed with penicillin-binding proteins 2X and 1A from Streptococcus pneumoniae. Antimicrob Agents Chemother 2008, 52:2053–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer P, Koch B, Zerfass I, Krauss J, van der Linden M, Frère JM, Contreras-Martel C, Hakenbeck R: Penicillin-binding protein 2x of Streptococcus pneumoniae: three new mutational pathways for remodelling an essential enzyme into a resistance determinant. J Mol Biol 2008, 376:1403–1416. [DOI] [PubMed] [Google Scholar]
- 18.Contreras-Martel C, Dahout-Gonzalez C, Martins Ados S, Kotnik M, Dessen A: PBP active site flexibility as the key mechanism for β-lactam resistance in pneumococci. J Mol Biol 2009, 387:899–909. [DOI] [PubMed] [Google Scholar]
- 19.Villegas-Estrada A, Lee M, Hesek D, Vakulenko SB, Mobashery S: Co-opting the cell wall in fighting methicillin-resistant Staphylococcus aureus: potent inhibition of PBP 2a by two anti-MRSA β-lactam antibiotics. J Am Chem Soc 2008, 130:9212–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fajardo A, Martinez JL: Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol 2008, 11:161–167. [DOI] [PubMed] [Google Scholar]
- 21.Sacco E, Hugonnet JE, Josseaume N, Cremniter J, Dubost L, Marie A, Patin D, Blanot D, Rice LB, Mainardi JL et al. : Activation of the l,d-transpeptidation peptidoglycan cross-linking pathway by a metallo-d,d-carboxypeptidase in Enterococcus faecium. Mol Microbiol 2010, 75:874–885. [DOI] [PubMed] [Google Scholar]
- 22.Mainardi JL, Hugonnet JE, Rusconi F, Fourgeaud M, Dubost L, Moumi AN, Delfosse V, Mayer C, Gutmann L, Rice LB et al. : Unexpected inhibition of peptidoglycan l,d-transpeptidase from Enterococcus faecium by the β-lactam imipenem. J Biol Chem 2007, 282:30414–30422. [DOI] [PubMed] [Google Scholar]
- 23.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CEr, Blanchard JS: Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science 2009, 323:1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G: The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 2010, 16:466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo TA, MacDonald U, Beanan JM, Olson R, MacDonald IJ, Sauberan SL, Luke NR, Schultz LW, Umland TC: Penicillin-binding protein 7/8 contributes to the survival of Acinetobacter baumannii in vitro and in vivo. J Infect Dis 2009, 199:513–521. [DOI] [PubMed] [Google Scholar]
- 26.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A: β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog 2009, 5:e1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishii K, Chiba N, Morozumi M, Hamano-Hasegawa K, Kurokawa I, Masaki J, Ubukata K: Diverse mutations in the ftsI gene in ampicillin-resistant Haemophilus influenzae isolates from pediatric patients with acute otitis media. J Infect Chemother 2010, 16:87–93. [DOI] [PubMed] [Google Scholar]
- 28.••.Zamorano L, Moya B, Juan C, Oliver A: Differential beta-lactam resistance response driven by ampD or dacB (PBP4) inactivation in genetically diverse Pseudomonas aeruginosa strains. J Antimicrob Chemother 2010, 65:1540–1542. [DOI] [PubMed] [Google Scholar]; Bacteria have sophisticated pathways to assess the presence of anti-bacterials and to mobilize defensive pathways. The biochemical entities within these pathways are surprisingly poorly known. Here – references [18,19] – a seemingly non-essential low Mr PBP of an important Gram-negative pathogen is identified as a key sensor initiating the defensive response to β-lactams.
- 29.Livermore DM, Hope R, Mushtaq S, Warner M: Orthodox and unorthodox clavulanate combinations against extended-spectrum β-lactamase producers. Clin Microbiol Infect 2008, 14(Suppl. 1):189–193. [DOI] [PubMed] [Google Scholar]
- 30.Drawz SM, Bonomo RA: Three decades of β-lactamase inhibitors. Clin Microbiol Rev 2010, 23:160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akova M: Sulbactam-containing β-lactamase inhibitor combinations. Clin Microbiol Infect 2008, 14(Suppl. 1):185–188. [DOI] [PubMed] [Google Scholar]
- 32.Oelschlaeger P, Ai N, Duprez KT, Welsh WJ, Toney JH: Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J Med Chem 2010, 53:3013–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moya B, Zamorano L, Juan C, Perez JL, Ge Y, Oliver A: Activity of a new cephalosporin, CXA-101 (FR264205), against beta-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob Agents Chemother 2010, 54:1213–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbour A, Schmidt S, Rand KH, Derendorf H: Ceftobiprole: a novel cephalosporin with activity against Gram-positive and Gram-negative pathogens, including methicillin-resistant Staphylococcus aureus (MRSA). Int J Antimicrob Agents 2009, 34:1–7. [DOI] [PubMed] [Google Scholar]
- 35.Henry X, Amoroso A, Coyette J, Joris B: Interaction of ceftobiprole with the low-affinity PBP 5 of Enterococcus faecium. Antimicrob Agents Chemother 2010, 54:953–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moisan H, Pruneau M, Malouin F: Binding of ceftaroline to penicillin-binding proteins of Staphylococcus aureus and Streptococcus pneumoniae. J Antimicrob Chemother 2010, 65:713–716. [DOI] [PubMed] [Google Scholar]
- 37.Kosowska-Shick K, McGhee PL, Appelbaum PC: Affinity of ceftaroline and other β-lactams for penicillin-binding proteins from Staphylococcus aureus and Streptococcus pneumoniae. Antimicrob Agents Chemother 2010, 54:1670–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mushtaq S, Warner M, Williams G, Critchley I, Livermore DM: Activity of chequerboard combinations of ceftaroline and NXL104 versus beta-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother 2010, 65:1428–1432. [DOI] [PubMed] [Google Scholar]
- 39.Vidaillac C, Leonard SN, Rybak MJ: In vitro evaluation of ceftaroline alone and in combination with tobramycin against hospital-acquired meticillin-resistant Staphylococcus aureus (HA-MRSA) isolates. Int J Antimicrob Agents 2010, 35:527–530. [DOI] [PubMed] [Google Scholar]
- 40.Mushtaq S, Livermore DM: AmpC induction by ceftaroline. J Antimicrob Chemother 2010, 65:586–588. [DOI] [PubMed] [Google Scholar]
- 41.Lemaire S, Fuda C, Van Bambeke F, Tulkens PM, Mobashery S: Restoration of susceptibility of methicillin-resistant Staphylococcus aureus to β-lactam antibiotics by acidic pH: role of penicillin-binding protein PBP 2a. J Biol Chem 2008, 283:12769–12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuda C, Hesek D, Lee M, Heilmayer W, Novak R, Vakulenko SB, Mobashery S: Mechanistic basis for the action of new cephalosporin antibiotics effective against methicillin- and vancomycin-resistant Staphylococcus aureus. J Biol Chem 2006, 281:10035–10041. [DOI] [PubMed] [Google Scholar]
- 43.Queenan AM, Bush K: Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 2007, 20:440–458 table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eguchi K, Kanazawa K, Eriguchi Y, Ueda Y: Pharmacodynamics of SMP-601 (PTZ601) against vancomycin-resistant Enterococcus faecium and methicillin-resistant Staphylococcus aureus in neutropenic murine thigh infection models. Antimicrob Agents Chemother 2009, 53:3391–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kihara R, Yanagihara K, Morinaga Y, Araki N, Nakamura S, Seki M, Izumikawa K, Kakeya H, Yamamoto Y, Tsukamoto K et al. : Potency of SMP-601, a novel carbapenem, in hematogenous murine bronchopneumonia caused by methicillin-resistant and vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 2008, 52:2163–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.••.Page MG, Heim J: New molecules from old classes: revisiting the development of β-lactams. IDrugs 2009, 12:561–565. [PubMed] [Google Scholar]; The newest β-lactam structures and why they were chosen for development.
- 47.Page MG, Dantier C, Desarbre E: In Vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant gram-negative bacilli. Antimicrob Agents Chemother 2010, 54:2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mushtaq S, Warner M, Livermore D: Activity of the siderophore monobactam BAL30072 against multiresistant non-fermenters. J Antimicrob Chemother 2010, 65:266–270. [DOI] [PubMed] [Google Scholar]
- 49.Endimiani A, Doi Y, Bethel CR, Taracila M, Adams-Haduch JM, O’Keefe A, Hujer AM, Paterson DL, Skalweit MJ, Page MG et al. : Enhancing resistance to cephalosporins in class C β-lactamases: impact of Gly214Glu in CMY-2. Biochemistry 2010, 49:1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Endimiani A, Bethel C, Choudhary Y, Bonomo RA: In vitro activity of penem-1 in combination with β-lactams against blaKPC-possessing Klebsiella pneumoniae isolates. Antimicrob Agents Chemother 2010, 54:1650–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drawz SM, Bethel CR, Doppalapudi VR, Sheri A, Pagadala SR, Hujer AM, Skalweit MJ, Anderson VE, Chen SG, Buynak JD et al. : Penicillin sulfone inhibitors of class D β-lactamases. Antimicrob Agents Chemother 2010, 54:1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stachyra T, Levasseur P, Pechereau MC, Girard AM, Claudon M, Miossec C, Black MT: In vitro activity of the β-lactamase inhibitor NXL104 against KPC-2 carbapenemase and Enterobacteriaceae expressing KPC carbapenemases. J Antimicrob Chemother 2009, 64:326–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hawkey PM: The growing burden of antimicrobial resistance. J Antimicrob Chemother 2008, 62(Suppl. 1):i1–i9. [DOI] [PubMed] [Google Scholar]
- 54.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H: Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci USA 2010, 107:2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin JF, Liras P: Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 2010, 13:263–273. [DOI] [PubMed] [Google Scholar]
- 56.Cichewicz RH: Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat Prod Rep 2010, 27:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bottcher T, Pitscheider M, Sieber SA: Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew Chem, Int Ed 2010, 49:2680–2698. [DOI] [PubMed] [Google Scholar]
- 58.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL: Inactivation of the glycoside hydrolase NagZ attenuates anti-Pseudomonal β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009, 53:2274–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S: Bacterial AmpD at the crossroads of peptidoglycan recycling and manifestation of antibiotic resistance. J Am Chem Soc 2009, 131:8742–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.••.Gilbert DN, Guidos RJ, Boucher HW, Talbot GH, Spellberg B, Edwards JE, Scheld WM, Bradley JS, Bartlett JG: The ‘10 × 20 initiative’: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis 2010, 50:1081–1083. [DOI] [PubMed] [Google Scholar]; The importance of immediate efforts in support of new antibacterial discovery, and what will be needed to ensure that these efforts happen.