Abstract
The production of lactate under hypoxic conditions or by cancer cells was reported to promote the M2 polarization of tumor-associated macrophages. However, the exact effect of lactate on macrophages, particularly under hypoxic conditions, has remained largely elusive. In the present study, an in-depth bioinformatics analysis of previously published transcriptome data of macrophages was performed. A total of 6, 101 and 764 upregulated genes were identified in the lactate, hypoxia and hypoxia-lactate groups, respectively, whereas 4, 41 and 588 genes were downregulated in the same respective groups. Furthermore, differentially expressed genes (DEGs) of the hypoxia and hypoxia-lactate groups were significantly enriched in the hypoxia-inducible factor 1 (HIF-1) signaling pathway and the Hedgehog pathway. Upregulation of the mTOR and Hedgehog pathways in the hypoxia-lactate group was identified by gene set enrichment analysis. Furthermore, a set of HIF-1 pathway-associated genes was identified to be positively correlated with hypoxia using weighted gene co-expression network analysis. Lactate was indicated to inhibit the cell cycle in a hypoxia-independent manner. The DEGs of the hypoxia and hypoxia-lactate groups, including C-C motif chemokine receptor type 1 and 5, were enriched in the cytokine-cytokine receptor interaction pathway. In conclusion, under normoxic conditions, lactate exerted a weak effect on macrophages, while the combination of lactate and hypoxia markedly promoted the M2-polarization of macrophages via the HIF-1, Hedgehog and mTOR pathways. Lactate and hypoxia may also contribute to the formation of the spatial structure of tumor niches by inhibiting the proliferation of resident macrophages and by regulating the recruitment of peripheral macrophages.
Keywords: hypoxia, lactate, cancer, macrophage
Introduction
Reprogramming of the energy metabolism and evasion of immune surveillance are two emerging hallmarks of cancer (1). Cancer cells feature enhanced glucose uptake and utilize glycolysis as a major way of energy metabolism. The glycolytic pathway is active in cancer cells, even under normoxic conditions, and supports their rapid proliferation. This phenomenon is generally known as aerobic glycolysis or the Warburg effect (2,3). Due to the defective vasculature in tumor tissues, hypoxia is common among various types of cancer and is also an important trigger for glycolysis (4). Abnormal expression of oncogenes and mitochondrial DNA mutations may also contribute to the enhanced glycolysis in cancer cells (3). Previous studies have indicated that the Warburg effect may increase the production of ATP and provide more metabolic intermediates and precursors, which may be used for the biosynthesis of macromolecules. Furthermore, this process can enhance the tolerance of cancer cells to chemotherapies (5).
Recent studies have suggested that, in addition to the direct stimulation of the growth of cancer cells, glycolysis has an important role in the induction of immune tolerance of cancer. Carmona-Fontaine et al (6) reported that hypoxia and cancer-derived lactate, which is a by-product of glycolysis, may induce the expression of arginase 1 and macrophage mannose receptor 1 proteins, also known as M2-like markers, by tumor-associated macrophages (TAMs). TAMs are one of the most important components of the tumor microenvironment, accounting for 6–14% of total cells in the tumor tissue (7). A previous study by our group has also suggested that bladder cancer cells are able to re-program RAW264.7 cells into M2-like macrophages in microfluidic tumor microenvironment chips (8). However, the detailed effects of lactate on macrophages, particularly under hypoxic conditions, have remained largely elusive.
In the present study, an in-depth analysis of the transcriptome data of bone marrow-derived macrophages (BMDMs) stimulated by lactate and hypoxia, which were reported by Carmona-Fontaine et al (6), was performed. The algorithms, including gene enrichment analysis, gene set enrichment analysis (GSEA) and weighted gene co-expression network analysis (WGCNA), were jointly utilized in order to analyze the influence of lactate and hypoxia on macrophages, and to provide insight to the therapeutic applications of cancer cell metabolism in targeted therapies for cancer.
Materials and methods
Data accession and pre-processing
Raw mRNA read count data were retrieved from the GSE93702 dataset of the gene expression omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE93702 dataset contains the mRNA sequencing data of 4 groups of mouse BMDMs, including untreated BMDMs (control group) as well as BMDMs treated with 25 mM sodium lactate (lactate group), cultured in 1% oxygen (hypoxia group) or both (hypoxia-lactate group) in vitro for 24 h. Three duplicates were performed in each group (6). Rlog normalization was performed to obtain the expression matrix of the samples using the DESeq2 Package (version 1.14.1).
Screening for differentially expressed genes (DEGs) and enrichment analysis
Untreated BMDMs were used as controls and the DEGs of the lactate, hypoxia and lactate-hypoxia groups were separately calculated based on the Wald test and the generalized linear model using the DESeq2 package (9) of the R software. The threshold for DEGs was set at |log(fold change) |>log21.5, P-value <0.05 and Benjamini-Hochberg-adjusted P-value <0.1. Volcano plots and heat maps were used to display the DEGs of the three experimental groups vs. the control group. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.8), and P<0.05 was used as the threshold (10). The pathways, which exhibited significant alterations in at least two groups, were further displayed using the bubble plot. The volcano plot, heat map and bubble plot were drawn using the ggplot2 package (version 2.2.1) of the R software (11).
GSEA
GSEA was performed for each of the three groups separately using GSEA software (version 3.0), and the KEGG gene sets (version 6.2) were used as the annotation gene set (12). The cut-off values were set at P<0.05 and false discovery rate<0.25. Significantly enriched pathways were further displayed using the GSEA plot.
WGCNA
WGCNA was performed using the WCGNA package (version 1.63) of the R software. A step-by-step network construction and module detection approach were selected. The soft-threshold power was set at 10 according to the scale-free fit index and mean degree of connectivity. The method of dynamic tree cut from the package dynamic TreeCut was used for identifying modules. The correlation between the modules identified and the features of the culturing conditions in each group of cells was then determined. The modules with significant correlations with lactate or hypoxia treatment were further annotated using DAVID. The dendrogram, heat map and scatter plot were used to display the major results using the WGCNA package.
Results
Screening of DEGs and enrichment analysis
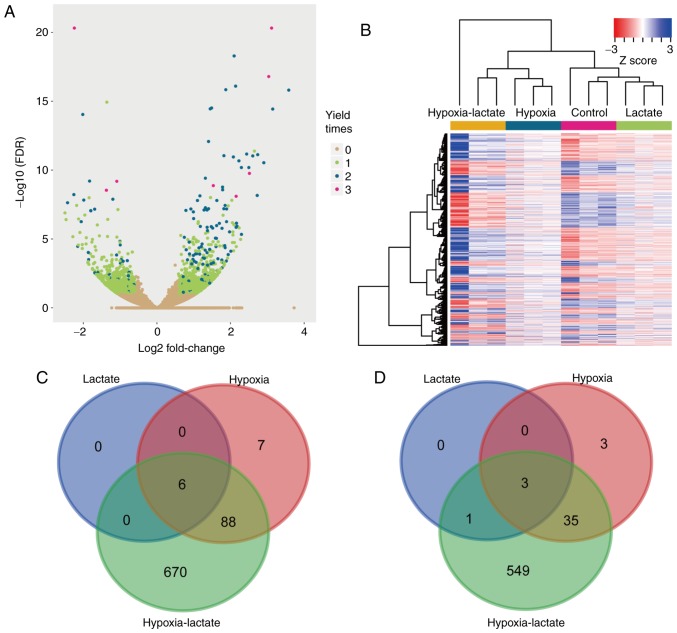
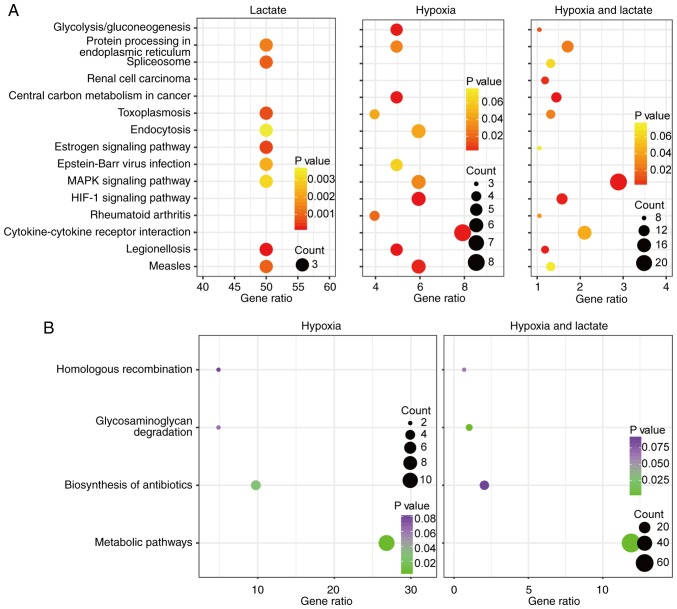
Compared with the untreated BMDM group, a total of 6, 101 and 764 upregulated genes were identified in the lactate, hypoxic and lactate-hypoxic group, respectively, while 4, 41 and 588 genes were downregulated in these three respective groups (Fig. 1A and B). Among them, heat shock protein family A member 1 like (HSPA1L), interferon-induced transmembrane protein 6, FXYD domain-containing ion transport regulator 2, HSPA1B, HSPA1A and metastasis-associated lung adenocarcinoma transcript 1 were upregulated in all three groups, while CD33, interleukin 12 receptor subunit β 2, cysteine- and glycine-rich protein 1 and transglutaminase 2 were downregulated in all three groups (Fig. 1C and D). Furthermore, KEGG pathway enrichment analyses were performed using DAVID software in order to assess the function of the DEGs identified. The upregulated genes of the lactate group were accumulated in the pathways legionellosis, antigen processing and presentation, estrogen signaling, toxoplasmosis and mitogen-activated protein kinase (MAPK) signaling, whereas the upregulated genes in the hypoxia group were accumulated in legionellosis, hypoxia-inducible factor-1 (HIF-1) signaling, central carbon metabolism in cancer, glycolysis/gluconeogenesis and cytokine-cytokine receptor interaction pathways. The focal adhesion, central carbon metabolism in cancer, MAPK signaling, legionellosis and HIF-1 signaling pathways were enriched by the upregulated genes of the hypoxia-lactate group. The downregulated genes in the hypoxia group were enriched in metabolic and biosynthesis of antibiotics pathways. Metabolic function, DNA replication, glycosaminoglycan degradation, mismatch repair and oxidative phosphorylation pathways were enriched by the downregulated genes of the hypoxia-lactate group. Details regarding the DEG-enriched pathways are presented in Table I. Significant pathways identified in at least two groups were further displayed using bubble plots to demonstrate any differences or commonalities between the three groups (Fig. 2A and B). The KEGG database of pathways is a collection of manually selected functionally-associated gene sets representing the knowledge on molecular interaction, reaction and association networks. There are various overlaps among pathways, as numerous genes have roles in several different pathways. In the present case, enrichment in legionellosis pathways does not necessarily mean that there is an on-going legionellosis infection in these cells, but it means that there may be common features with certain processes of legionellosis infection.
Figure 1.
Screening of DEGs. (A) Transcriptome data of the lactate, hypoxia and hypoxia-lactate group were compared with the data of the control group. The log2 (fold change) and -log10 (FDR) value of each gene in the hypoxia-lactate group are presented. Genes with no significant change in all three comparisons are displayed in yellow color and genes identified as DEGs in 1, 2 or 3 comparisons are indicated in green, blue and pink, respectively. (B) Expression of genes identified as DEGs in at least one comparison displayed in a heat map. The hypoxia-lactate group exhibited the most significant alteration in gene expression among the test groups. (C) Overlaps of upregulated genes of the hypoxia, lactate and hypoxia-lactate groups indicated in a Venn diagram. The hypoxia-lactate group had the highest number of DEGs. The expression of HSPA1L, interferon-induced transmembrane protein 6, FXYD domain-containing ion transport regulator 2, HSPA1B, HSPA1A and metastasis-associated lung adenocarcinoma transcript 1 was upregulated in all three groups, and most of the upregulated DEGs in the hypoxia group and lactate group were also DEGs in the hypoxia-lactate group. (D) Overlaps of downregulated genes in the hypoxia, lactate and hypoxia-lactate groups displayed in a Venn diagram. The expression of CD33, interleukin 12 receptor subunit β 2, cysteine- and glycine-rich protein 1 and transglutaminase 2 was downregulated in all three groups. DEG, differentially expressed gene; FDR, false discovery rate; HSPA1 L, heat shock protein family A member 1 like.
Table I.
Top 10 up- and down-regulated Kyoto Encyclopedia of Genes and Genomes pathways of differentially expressed genes.
| A, Lactate group | |
|---|---|
| Pathway | P-value |
| Upregulated | |
| Legionellosis | <0.001 |
| Antigen processing and presentation | <0.001 |
| Estrogen signaling pathway | <0.001 |
| Toxoplasmosis | 0.001 |
| Spliceosome | 0.001 |
| Measles | 0.001 |
| Protein processing in endoplasmic reticulum | 0.001 |
| Influenza A | 0.001 |
| Epstein-Barr virus infection | 0.002 |
| MAPK signaling pathway | 0.003 |
| B, Hypoxia group | |
| Pathway | P-value |
| Upregulated | |
| Glycolysis/gluconeogenesis | 0.001 |
| Cytokine-cytokine receptor interaction | 0.001 |
| Central carbon metabolism in cancer | 0.001 |
| HIF-1 signaling pathway | 0.001 |
| Measles | 0.002 |
| Legionellosis | 0.001 |
| Hematopoietic cell lineage | 0.021 |
| Biosynthesis of antibiotics | 0.016 |
| Rheumatoid arthritis | 0.019 |
| MAPK signaling pathway | 0.031 |
| Downregulated | |
| Metabolic pathways | 0.003 |
| Biosynthesis of antibiotics | 0.028 |
| C, Hypoxia-lactate group | |
| Pathway | P-value |
| Upregulated | |
| Focal adhesion | <0.001 |
| Transcriptional misregulation in cancer | <0.001 |
| Central carbon metabolism in cancer | <0.001 |
| Dorso-ventral axis formation | <0.001 |
| MAPK signaling pathway | 0.001 |
| Pathways in cancer | 0.001 |
| Regulation of actin cytoskeleton | 0.001 |
| Legionellosis | 0.001 |
| Proteoglycans in cancer | 0.002 |
| HIF-1 signaling pathway | 0.002 |
| Downregulated | |
| Metabolic pathways | <0.001 |
| DNA replication | <0.001 |
| Glycosaminoglycan degradation | <0.001 |
| Mismatch repair | 0.005 |
| Base excision repair | 0.005 |
| Oxidative phosphorylation | 0.006 |
| Nucleotide excision repair | 0.014 |
| Huntington's disease | 0.028 |
| Purine metabolism | 0.032 |
| Non-alcoholic fatty liver disease | 0.033 |
No downregulated pathway was obtained in the lactate group. HIF1, hypoxia-inducible factor-1; MAPK, mitogen-activated protein kinase.
Figure 2.
KEGG enrichment analysis of differentially expressed genes. Significantly enriched KEGG pathways yielded in at least two comparisons are displayed in bubble plots. The size of the dots represents the number of enriched genes; the X-axis coordinate represents the gene ratio and the color of the dots represents the P-value of enrichment analysis. The detailed enrichment score, P-value and false discovery rate value are provided in Table II. (A) Upregulated pathways. (B) Downregulated pathways. No downregulated pathways were obtained in the lactate group, which was therefore not included. HIF1, hypoxia-inducible factor-1; MAPK, mitogen-activated protein kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes.
GSEA analysis
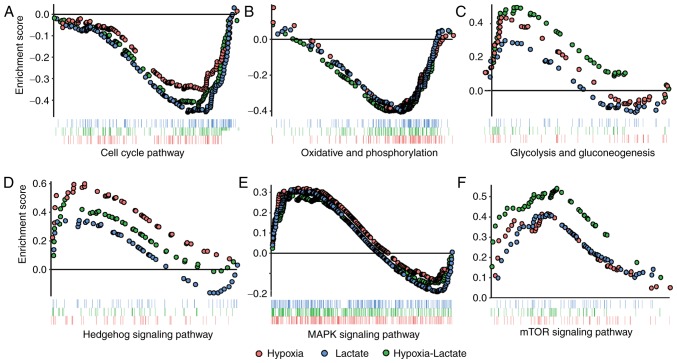
Subsequently, GSEA analysis was performed on the three groups in order to avoid the deviation caused by single-gene analysis. Downregulation of the cell cycle and oxidative phosphorylation pathways were identified in the lactate group, while enrichment of the Hedgehog signaling pathway and downregulation of oxidative phosphorylation was noted in the hypoxic group. Enhancement of the mTOR, Hedgehog and MAPK signaling pathways, as well as the glycolysis-gluconeogenesis pathway, were identified in the hypoxia-lactate group, whereas the cell cycle and oxidative phosphorylation pathways were downregulated in the hypoxia-lactate group (Fig. 3; Table II).
Figure 3.
GSEA for the lactate, hypoxia and hypoxia-lactate group. Pathways within the threshold of P<0.05 and false discovery rate <0.25 in at least one comparison were displayed using GSEA plots. The dots representing the genes in the hypoxia, lactate and hypoxia-lactate groups were colored in red, blue and green, respectively. (A) Cell cycle pathway. (B) Oxidative phosphorylation pathway. (C) Glycolysis and gluconeogenesis pathway. (D) Hedgehog pathway. (E) MAPK signaling pathway. (F) mTOR pathway. MAPK, mitogen-activated protein kinase; GSEA, gene set enrichment analysis.
Table II.
Significantly altered pathways in the gene set enrichment analysis.
| A, Lactate group | ||||
|---|---|---|---|---|
| Term | ES | NES | P-value | FDR |
| Cell cycle | −0.47 | −1.53 | <0.01 | 0.06 |
| Oxidative phosphorylation | −0.41 | −1.35 | 0.04 | 0.13 |
| B, Hypoxia group | ||||
| Term | ES | NES | P-value | FDR |
| Hedgehog signaling pathway | 0.60 | 1.66 | 0.01 | 0.04 |
| Oxidative phosphorylation | −0.40 | −1.39 | 0.02 | 0.14 |
| C, Hypoxia-lactate group | ||||
| Term | ES | NES | P-value | FDR |
| mTOR signaling pathway | 0.54 | 1.77 | 0.01 | 0.01 |
| Glycolysis gluconeogenesis | 0.49 | 1.59 | 0.01 | 0.04 |
| Hedgehog signaling pathway | 0.48 | 1.54 | 0.01 | 0.04 |
| Oxidative phosphorylation | −0.41 | −1.35 | 0.05 | 0.12 |
| Cell cycle | −0.44 | −1.41 | 0.02 | 0.15 |
| MAPK signaling pathway | 0.28 | 1.18 | 0.05 | 0.19 |
ES, enrichment score; NES, normalized ES; FDR, false discovery rate; MAPK, mitogen-activated protein kinase.
WGCNA analysis
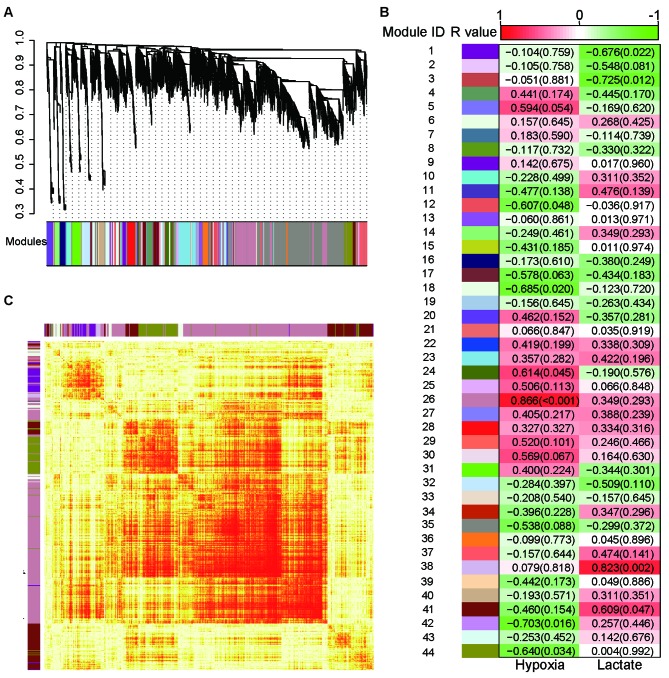
Furthermore, WGCNA analysis was performed to search for possible gene network modules with significant correlations to lactate treatment or hypoxic treatment. Using a step-by-step network construction and module detection approach, 44 gene modules were identified (Fig. 4A). Modules 38 and 41 were positively correlated with lactate treatment (r=0.823 and 0.609; P=0.002 and P=0.047, respectively), while modules 1 and 3 were negatively correlated with lactate treatment (r=−0.676 and −0.725; P=0.022 and 0.012, respectively). With regard to the oxygenation condition, modules 24 and 26 were positively correlated with hypoxia (r=0.614 and 0.866, P=0.045 and P<0.001, respectively), while modules 18, 12, 42 and 44 were negatively correlated with hypoxia (r=−0.685, −0.607, −0.703 and −0.640; P=0.020, 0.048, 0.016 and 0.034, respectively; Fig. 4B; Table III). Module 38 was mainly composed of enriched gene sets for the Rap1 signaling pathway, whereas module 41 was enriched in gene sets associated with base excision repair, pyruvate metabolism, hippo signaling and tight junction pathways. Module 1 was enriched in the focal adhesion pathway gene sets. Module 26 was enriched in the metabolic, central carbon metabolism in cancer, HIF-1 signaling pathway and glycolysis-gluconeogenesis gene sets. Module 24 was composed of enriched genes in dopaminergic synapse, adrenergic signaling in cardiomyocytes, neurotrophin signaling and sphingolipid signaling pathways. Module 12 was enriched in gene sets involved in the regulation of actin cytoskeleton, cell adhesion molecules, glycerophospholipid metabolism and Rap1 signaling pathways. Module 44 was enriched in gene sets involved in carbon metabolism, mTOR signaling and 5′-adenosine monophosphate-activated protein kinase signaling pathways. Finally, modules 18 and 42 were enriched in gene sets associated with protein processing of the endoplasmic reticulum and metabolic pathways, respectively (Table III). The topological overlap matrix among 1,000 randomly selected genes was presented using a heatmap (Fig. 4C).
Figure 4.
Weighted gene co-expression network analysis. (A) All genes were hierarchically clustered according to the topological overlap matrix and displayed using a dendrogram. (B) Module-feature associations. Each row corresponds to a module eigengene and the two columns correspond to hypoxia and lactate treatment. Each cell contains the R-value (corresponding correlation) and the P-value in parentheses. The cells are colored depending on the R-value. (C) The topological overlap matrix among 1,000 randomly selected genes in the analysis was displayed using a heatmap. Light colors represent low overlap and progressively darker red color represents higher overlap.
Table III.
Enrichment analysis of the modules significantly correlated with hypoxia or lactate treatment.
| A, Lactate | ||
|---|---|---|
| KEGG pathways | R-value | P-value |
| Module 41 | 0.61 | |
| Base excision repair | 0.016 | |
| Pyruvate metabolism | 0.023 | |
| Hippo signaling pathway | 0.027 | |
| Tight junction | 0.043 | |
| Module 38: | 0.82 | |
| Rap1 signaling pathway | 0.025 | |
| Module 1: | −0.68 | |
| Focal adhesion | 0.033 | |
| Module 3: | −0.73 | |
| Not applicable | >0.999 | |
| B, Hypoxia | ||
| KEGG pathways | R-value | P-value |
| Module 26 | 0.87 | |
| Renal cell carcinoma | <0.001 | |
| Pertussis | <0.001 | |
| Lysosome | <0.001 | |
| Metabolic pathways | <0.001 | |
| Amino sugar and nucleotide | <0.001 | |
| sugar metabolism | ||
| Central carbon metabolism in cancer | <0.001 | |
| Pathways in cancer | 0.001 | |
| Biosynthesis of antibiotics | 0.001 | |
| Focal adhesion | 0.001 | |
| HIF-1 signaling pathway | 0.001 | |
| Module 12 | −0.61 | |
| Regulation of actin cytoskeleton | 0.012 | |
| Cell adhesion molecules | 0.014 | |
| Morphine addiction | 0.022 | |
| Glycerophospholipid metabolism | 0.023 | |
| Rap1 signaling pathway | 0.029 | |
| Rheumatoid arthritis | 0.043 | |
| Regulation of lipolysis in adipocytes | 0.044 | |
| Hematopoietic cell lineage | 0.047 | |
| Module 42: Metabolic pathways | −0.70 | 0.008 |
| Module 44 | −0.64 | |
| Biosynthesis of antibiotics | <0.001 | |
| Metabolic pathways | <0.001 | |
| Colorectal cancer | 0.004 | |
| Progesterone-mediated oocyte maturation | 0.004 | |
| Carbon metabolism | 0.006 | |
| Steroid biosynthesis | 0.009 | |
| mTOR signaling pathway | 0.012 | |
| Central carbon metabolism in cancer | 0.017 | |
| Insulin signaling pathway | 0.017 | |
| Chronic myeloid leukemia | 0.027 | |
HIF1, hypoxia-inducible factor-1. The module names are consistent with those shown in Fig. 4B.
Discussion
Macrophages have an important role in the inflammatory response and tissue repair process. Macrophages are able to engulf pathogens, present antigens to T cells and secrete pro-inflammatory factors in order to activate and promote the immune response. In addition, they exhibit immune-modulatory functions by secreting anti-inflammatory factors, including IL-4, IL-10 and transforming growth factor-β (13). During the growth and progression of the tumor, macrophages are recruited to the tumor niche and contribute to the initiation of the anti-tumor immune response. However, cancer cells may also re-program macrophages into immune-modulatory macrophages, known as M2 TAMs. The functional status of TAMs is notably associated with the survival of cancer patients, while the M2-polarization of TAMs may be a promising target for cancer therapy (14,15).
Although previous studies have examined TAM reprogramming, the high heterogeneity noted in several types of solid tumor has led to a lack of consensus on the detailed mechanism of this process. However, solid tumors share certain characteristics, including malformation of tumor vessels and aerobic glycolysis, resulting in hypoxia and lactic acidosis, which are considered two hallmarks of the tumor microenvironment. Hypoxia has been indicated to induce the M2 polarization of TAMs via stabilization of HIF-1 (16). Previous studies have further suggested that melanoma, colon carcinoma and bladder transitional cell carcinoma cells may induce the M2-polarization of TAMs via lactate shuttling (8,17). Furthermore, previous studies that investigated inflammatory organ injury indicated that lactate was able to inhibit the pro-inflammatory function of lipopolysaccharide (LPS)-stimulated macrophages (18–20), and may therefore be a common regulator of TAM function in the tumor microenvironment.
Carmona-Fontaine et al (6) provided transcriptome data of BMDMs treated by hypoxia or lactate. The study simultaneously assessed the differences between the hypoxia+lactate group and all other samples, including the samples from the normoxic group, the normoxia+lactate group and the hypoxic group. The factors under investigation were lactate and hypoxia, and the heterogeneity between the three groups was taken into account. This may reduced the reliability of the results obtained of that study. In the present study, these transcriptome data were explored using DEG screening, KEGG enrichment, GSEA and WGCNA in order to enhance the current knowledge on the effect of hypoxia and lactate on TAMS.
Overall, the lactate treatment had a relatively mild effect on macrophages, with only two significantly altered GSEA pathways and 10 DEGs. In the hypoxic group, 142 DEGs and two GSEA pathways were identified. By contrast, the hypoxia+lactate treatment had a significant effect on the function of macrophages, with 1,352 DEGs and six significantly altered pathways. Furthermore, all DEGs in the lactate group exhibited the same trend in the hypoxia and hypoxia-lactate groups, whereas all altered pathways obtained in the lactate and hypoxia groups by GSEA analysis were also identified in the hypoxia-lactate group. Therefore, the production of lactate and the presence of hypoxia may work synergistically in the re-programming of macrophages.
The activation and regulation of the immune response are key functions of macrophages. In the present study, the HIF-1 and Hedgehog signaling pathways were enriched by the DEGs of the hypoxic and the hypoxia-lactate groups. Upregulation of the mTOR and the Hedgehog pathways in the hypoxia-lactate group was identified by GSEA. Furthermore, module 26, which was positively correlated with hypoxia, was enriched in the HIF-1 pathway gene sets. The HIF-1 pathway is considered the key pathway in the reprogramming of TAMs (8,17), and the mTOR and Hedgehog pathways were recently reported to contribute to the alternative activation of macrophages and to the inhibition of the secretion of pro-inflammatory factors (21,22). These results suggested that lactate and hypoxia promote the M2-like function of macrophages. However, the present results further demonstrated significant enrichment of the MAPK signaling pathway in the DEGs of all three groups. The upregulation of the DEGs of the MAPK pathway in the hypoxia-lactate group was also demonstrated by GSEA and was specifically identified in module 26. The MAPK pathway may increase the expression of pro-inflammatory factors, including IL1β and tumor necrosis factor-α (23). These data suggested that hypoxia and lactate may potentiate the M1-like pro-inflammatory function of macrophages. However, activation of MAPK by hypoxia and lactate may not be sufficient to activate an effective anti-tumor immune response. At present, there is insufficient data to support the alteration of the other inflammatory pathways and more importantly, due to several previous studies demonstrating that lactate was able to inhibit the pro-inflammatory response of LPS-stimulated macrophages (18,19).
The accumulation of immune cells was mostly noted in the well-perfused region, including the stroma and the invasive margin surrounding the tumor islets (24,25). The production of lactate by the tumor may decrease the pH and have an important role in the formation of this spatial structure (25). In the present study, GSEA demonstrated that lactate treatment downregulated the cell cycle pathway in a hypoxia-independent manner, even in a neutral pH environment. Thus, lactate may inhibit the in situ proliferation of macrophages under inflammatory conditions (26). The results further indicated that the DEGs of the hypoxia and hypoxia-lactate groups, including C-C motif chemokine receptor 1 (CCR1) and CCR5, were enriched in the cytokine-cytokine receptor interaction pathway. Module 26 was significantly associated with lactate levels and exhibited enrichment in the chemokine signaling pathway. These results suggested that hypoxia was further involved in the regulation of macrophage localization in the tumor microenvironment.
Several studies have indicated that hypoxia inhibits oxidative phosphorylation and promotes glycolysis (27,28). In the present study, the inhibition of oxidative phosphorylation and the activation of the glycolytic pathway were identified by the KEGG enrichment analysis, as well as GSEA and WGCNA, as specific biological processes that may be induced by hypoxia. Furthermore, GSEA demonstrated that lactate inhibits the oxidative phosphorylation pathway of macrophages, although it did not demonstrate any significant enhancement of the glycolysis pathway. The detailed energy metabolism status of macrophages under lactic acidosis conditions remains elusive and further studies are required to investigate this process.
In conclusion, lactate exhibited a weak effect on macrophages under normoxic conditions. However, the combination of lactate and hypoxia markedly promoted the M2-polarization of macrophages via the HIF-1, Hedgehog and mTOR pathways. Lactate and hypoxia may also contribute to the formation of the spatial structure of tumor niches by inhibiting the in situ proliferation of tissue-resident macrophages and by regulating the recruitment of peripheral macrophages.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the National Natural Science Foundation of China (grant no. 30901481) and the Major Research Program of Shandong Province (grant no. 2017GSF221016).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo) with the accession no. GSE93702.
Authors' contributions
WGCNA analysis was performed by XW, GG JHW and YX. GSEA was performed by BZ and YZ. YDS, TG and YJZ performed the DEG screening and KEGG enrichment analysis.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3108. [DOI] [PubMed] [Google Scholar]
- 4.Nurwidya F, Takahashi F, Minakata K, Murakami A, Takahashi K. From tumor hypoxia to cancer progression: The implications of hypoxia-inducible factor-1 expression in cancers. Anat Cell Biol. 2012;45:73–78. doi: 10.5115/acb.2012.45.2.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, Xavier JB. Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci USA. 2017;114:2934–2939. doi: 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eruslanov E, Neuberger M, Daurkin I, Perrin GQ, Algood C, Dahm P, Rosser C, Vieweg J, Gilbert SM, Kusmartsev S. Circulating and tumor-infiltrating myeloid cell subsets in patients with bladder cancer. Int J Cancer. 2012;130:1109–1119. doi: 10.1002/ijc.26123. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Wang D, Xu T, Liu P, Cao Y, Wang Y, Yang X, Xu X, Wang X, Niu H. Bladder cancer cells re-educate TAMs through lactate shuttling in the microfluidic cancer microenvironment. Oncotarget. 2015;6:39196–39210. doi: 10.18632/oncotarget.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33:111. doi: 10.1007/s12032-016-0829-6. [DOI] [PubMed] [Google Scholar]
- 11.Wickham H. ggplot2: Elegant graphics for data analysis. 2009 [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malyshev I, Malyshev Y. Current concept and update of the macrophage plasticity concept: Intracellular mechanisms of reprogramming and M3 macrophage ‘Switch’ phenotype. Biomed Res Int. 2015;2015:341308. doi: 10.1155/2015/341308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen SR, Schmid MC. Macrophages as key drivers of cancer progression and metastasis. Mediators Inflamm. 2017;2017:9624760. doi: 10.1155/2017/9624760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noman MZ, Hasmim M, Messai Y, Terry S, Kieda C, Janji B, Chouaib S. Hypoxia: A key player in antitumor immune response. A Review in the Theme: Cellular responses to hypoxia. Am J Physiol Cell Physiol. 2015;309:C569–C579. doi: 10.1152/ajpcell.00207.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoque R, Farooq A, Ghani A, Gorelick F, Mehal WZ. Lactate reduces liver and pancreatic injury in Toll-like receptor- and inflammasome-mediated inflammation via GPR81-mediated suppression of innate immunity. Gastroenterology. 2014;146:1763–1774. doi: 10.1053/j.gastro.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Errea A, Cayet D, Marchetti P, Tang C, Kluza J, Offermanns S, Sirard JC, Rumbo M. Lactate inhibits the Pro-inflammatory response and metabolic reprogramming in murine macrophages in a GPR81-independent manner. PLoS One. 2016;11:e0163694. doi: 10.1371/journal.pone.0163694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iraporda C, Romanin DE, Bengoa AA, Errea AJ, Cayet D, Foligné B, Sirard JC, Garrote GL, Abraham AG, Rumbo M. Local treatment with lactate prevents intestinal inflammation in the TNBS-induced colitis model. Front Immunol. 2016;7:651. doi: 10.3389/fimmu.2016.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira TA, Xie G, Choi SS, Syn WK, Voieta I, Lu J, Chan IS, Swiderska M, Amaral KB, Antunes CM, et al. Macrophage-derived Hedgehog ligands promotes fibrogenic and angiogenic responses in human schistosomiasis mansoni. Liver Int. 2013;33:149–161. doi: 10.1111/liv.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karnam A, Holla S, Balaji KN. Sonic hedgehog-responsive lipoxygenases and cyclooxygenase-2 modulate Dectin-1-induced inflammatory cytokines. Mol Immunol. 2015;68:280–289. doi: 10.1016/j.molimm.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Kim SC, Yu T, Yi YS, Rhee MH, Sung GH, Yoo BC, Cho JY. Functional roles of p38 mitogen-activated protein kinase in macrophage-mediated inflammatory responses. Mediators Inflamm. 2014;2014:352371. doi: 10.1155/2014/352371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmona-Fontaine C, Bucci V, Akkari L, Deforet M, Joyce JA, Xavier JB. Emergence of spatial structure in the tumor microenvironment due to the Warburg effect. Proc Natl Acad Sci USA. 2013;110:19402–19407. doi: 10.1073/pnas.1311939110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roiniotis J, Dinh H, Masendycz P, Turner A, Elsegood CL, Scholz GM, Hamilton JA. Hypoxia prolongs monocyte/macrophage survival and enhanced glycolysis is associated with their maturation under aerobic conditions. J Immunol. 2009;182:7974–7981. doi: 10.4049/jimmunol.0804216. [DOI] [PubMed] [Google Scholar]
- 28.Durnova GN, Kaplanskii AS, Portugalov VV. The effect of hypoxia on the function and metabolism of alveolar macrophages. Biull Eksp Biol Med. 1975;79:113–115. doi: 10.1007/BF00804233. (In Russian) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEO database (http://www.ncbi.nlm.nih.gov/geo) with the accession no. GSE93702.