Abstract
Background
The incorporation of novel biomarkers into therapy selection for patients with metastatic colorectal cancer (mcrc) has significantly improved outcomes. Optimal treatment planning now takes into account diverse characteristics of patients and their tumours to create personalized therapeutic plans.
Discussion
This review is split into two sections. In the first section, we review the prognostic and predictive significance of expanded RAS mutation testing, BRAF mutations, ERBB2 (her2) amplification, microsatellite instability (msi) and deficient mismatch repair (dmmr) protein, NTRK fusions, PIK3CA mutations, and met amplifications. The therapeutic implication of each of those biomarkers for personalizing therapies for each patient with mcrc is discussed. In the second section, we touch on testing methods and considerations of relevance to clinicians when they interpret companion diagnostics meant to guide therapy selection. The advantages and pitfalls of various methods are evaluated, and we also look at the potential of liquid biopsies and circulating tumour dna (ctdna) to change the landscape of therapeutic choice and biologic understanding of the disease.
Summary
Routine testing for extended RAS, BRAF, dmmr or high msi, and NTRK fusions is necessary to determine the best sequencing of chemotherapy and biologic agents for patients with mcrc. Although next-generation sequencing and ctdna are increasingly being adopted, other techniques such as immunohistochemistry retain their relevance in detection of her2 amplification, NTRK fusions, and dmmr.
Keywords: Biomarkers, colorectal cancer, metastatic, next-generation sequencing, ctdna, anti-egfr
INTRODUCTION
For 20 years, fluoropyrimidine doublet chemotherapy combined with either irinotecan (folfiri or capiri) or oxaliplatin (folfox or capox) has been the cornerstone of treatment for metastatic colorectal cancer (mcrc) in the first- and second-line settings1,2. Sequencing of those two regimens does not affect overall survival (os), and therefore, the choice of first-line therapy often depends on physician and patient preference, coupled with patient comorbidities (for example, pre-existing neuropathy)3. Although doublet chemotherapy is preferred because of its association with superior progression-free survival (pfs), sequential single-agent therapies have demonstrated similar os rates and might be appropriate for some patients who are frail or elderly4–6.
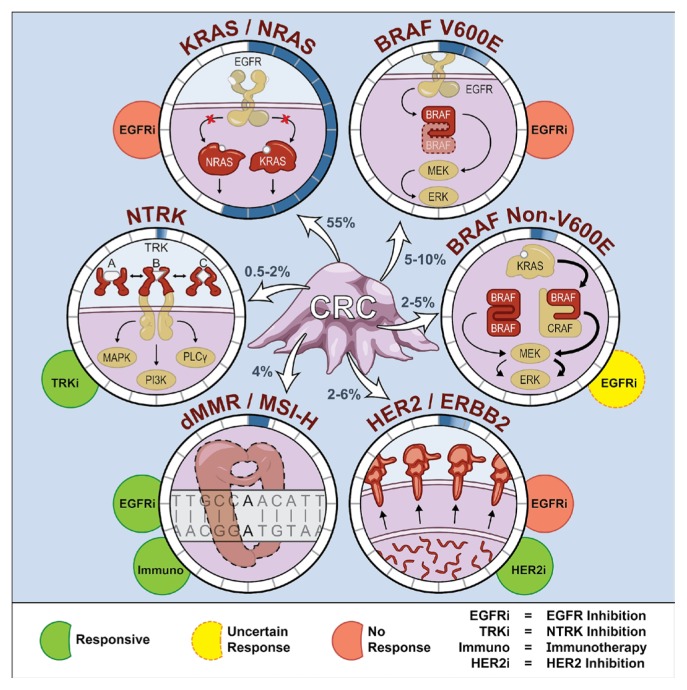
Since the introduction of targeted therapies, there has been an increased appreciation of the molecular stratification required for personalizing treatment. For example, antibodies against the epithelial growth factor receptor (“anti-egfr”) had limited activity in unselected patients, but were shown to have activity in KRAS wild-type cancers7,8. In the present review, we discuss clinically important alterations that drive treatment selection, including KRAS and NRAS (RAS) mutations, BRAF mutations, ERBB2 (her2) amplifications, deficient mismatch repair (dmmr) or high microsatellite instability (msi-h), NTRK fusions, PIK3CA mutations, and met amplification (Figure 1). In the second section, we review some practical and technical considerations to keep in mind when ordering biomarker tests, and we explore the relevance of next-generation sequencing (ngs) and circulating tumour dna (ctdna) or liquid biopsies.
FIGURE 1.
Current and emerging biomarkers used in personalizing treatment for patients with metastatic colorectal cancer (CRC). Prevalence of each biomarker in metastatic colorectal cancer is displayed with shading in the circle that surrounds the molecular alteration. Molecular alterations are not mutually exclusive and can co-occur. PLCγ = phospholipase C gamma; dMMR = deficient mismatch repair; MSI-H = high microsatellite instability.
MOLECULAR SUBTYPES IN mCRC
Expanded RAS Testing
KRAS codon 12 and 13 mutations were first identified as predictive biomarkers in third-line anti-egfr trials7,8. A subsequent retrospective analysis of the prime trial identified expanded mutations in KRAS and NRAS at codons 12, 13, 59, 61, 117, and 146 as predictive of the ineffectiveness of anti-egfr therapy9. International guidelines now mandate, as the standard of care, expanded RAS mutation testing before use of anti-egfr to identify the 55% of patients with mcrc for whom those agents will be ineffective9,10. There is even a suggestion of possible harm with the use of anti-egfr therapy in patients with RAS mutations11. Expanded RAS mutations are also a negative prognostic marker in the metastatic setting [median os (mos): 25 months vs. 32.1 months in wild-type disease; hazard ratio (hr): 1.52; 95% ci: 1.26 to 1.84; p < 0.001]12. Compared with KRAS, NRAS might be associated with shorter disease-free survival (33 months vs. 47 months; hr: 2.0; 95% ci: 1.3 to 2.8; p < 0.01) in early-stage disease and worse os in mcrc (hr: 1.83; 95% ci: 1.40 to 2.39; p < 0.001)13,14.
Effect of Primary Tumour Location (“Sidedness”) on Anti-EGFR Efficacy
Attention to the relevance of primary tumour location increased after a re-analysis of the Cancer and Leukemia Group B (calgb) 80405 trial showed that, in treatment-naïve patients with mcrc treated with either folfox or foliri (physician’s choice) and randomized to the addition of cetuximab or bevacizumab, no difference in mos was evident between the arms overall. However, survival differences were observed between patients with right- and left-sided tumours (mos: 19.4 months vs. 33.3 months; hr: 1.55; 95% ci: 1.32 to 1.82; p < 0.001), and the biologic associated with optimal results varied by side. Patients with left-sided tumours experienced improved outcomes with doublet chemotherapy plus cetuximab (mos: 36.0 vs. 31.4 months); those with right-sided tumours appeared to do better with a first-line doublet plus bevacizumab (mos: 24.2 months vs. 16.7 months; hr: 1.27; 95% ci: 0.98 to 1.63; p = 0.065)15. Those results were subsequently confirmed in numerous other first- and third-line trials that included anti-egfr therapy. Even when patients with BRAF mutations were excluded and adjustments were made in the right-sided tumour group for a higher proportion of female patients and patients with msi-h disease, primary tumour sidedness remained influential16–19.
Although those results were retrospectively identified, the reproducibility of the findings across studies has led to sidedness being accepted in many international guidelines as a predictive biomarker19,20. Although a doublet plus anti-egfr appears to be superior to bevacizumab for left-sided tumours, use of that combination in the first-line setting should be considered in balance with the added toxicity, particularly severe rash (~10%) and refractory hypomagnesemia (3%–7%). Those adverse reactions can have a significant effect on quality of life, but are potentially indicators for treatment efficacy21. In addition, compared with bevacizumab, anti-egfr therapy maintenance strategies are less well established22. For right-sided tumours, bevacizumab appears superior to anti-egfr when combined with doublet chemotherapy in the first-line setting15. In the third-line setting, patients with right-sided tumours might derive less benefit from anti-egfr therapy. However, in this patient population, effective treatment options are limited, and therefore, that inferior efficacy has less effect on treatment planning23.
Rechallenge with Anti-EGFR Therapy
After progression on an anti-egfr therapy, KRAS, NRAS, BRAF, and EGFR ectodomain mutations develop to drive signalling through the map kinase pathways despite egfr inhibition. The mutations provide a short-term selective advantage over other subclones, but are selected against with exponential decay after removal of anti-egfr; their half-life in ctdna is 4.4 months24. Retrospective studies have shown that, after intervening non–anti-egfr therapies, patients re-challenged with anti-egfr antibodies experience an overall response rate (orr) of 23%. In the same cohort, a wait of more than 2 half-lives from prior anti-egfr therapy was associated with an increase in the orr to 32%24. Similarly, in the prospective cricket trial, patients with evidence of persistent KRAS mutations experienced an inferior median pfs (mpfs) of 1.9 months compared with 4 months in patients with RAS wild-type disease re-challenged with anti-egfr (hr: 0.44; 95% ci: 0.18 to 0.98; p = 0.03)25. Based on ctdna, appropriate patients might be considered for re-challenge with anti-egfr therapy; however, further randomized prospective studies are needed.
BRAF
BRAF V600E Mutations
The serine–threonine braf kinase is found downstream from egfr in the mapk (mitogen-activated protein kinase) pathway. Hotspot mutations substituting glutamic acid for valine at codon 600 (V600E) result in kinase activity that is increased by a factor of 130 to 700 compared with that in wild-type BRAF26,27. BRAF V600E occurs in 5%–10% of cases of mcrc. It is nearly always mutually exclusive with RAS mutations and appears to predict a lack of, or reduced benefit from, anti-egfr therapy9,28,29. BRAF V600E mutations are also a strong negative prognostic marker independent of tumour sidedness (mos range: 10.5–13.5 months mutated vs. 28.3–30.6 months wild-type; hr: 2.01; 95% ci: 1.49 to 2.71; p < 0.001)9,11,12. Consideration of more aggressive firstline treatment with folfoxiri (fluorouracil–leucovorin–oxaliplatin–irinotecan) and bevacizumab is therefore advocated by some authors, given that folfoxiri has been associated with the greatest mos for a first-line regimen in patients with BRAF V600E mutations in tribe and a small Italian phase ii trial (mpfs: 7.5 months and 9.2 months respectively; mos: 19.0 months and 24.1 months respectively) 30,31. The tribe2 study compared upfront folfoxiri plus bevacizumab with a planned sequential switch of folfox plus bevacizumab to folfiri plus bevacizumab, confirming the superiority of a triplet in BRAF V600E–mutant mcrc (preliminary mos: 27.6 months vs. 22.6 months; p = 0.033)32.
In the second-line setting, single-agent BRAF inhibitors yielded disappointing results33. Combination approaches with irinotecan, cetuximab, and vemurafenib in the swog S1406 phase ii trial improved the mpfs (2.0 months vs. 4.3 months; hr: 0.48; 95% ci: 0.31 to 0.75; p = 0.001), but did not statistically improve os (5.9 months vs. 9.6 months, p = 0.19) in the context of 48% crossover from the control arm34. More recently, the beacon trial showed superior efficacy for the combination of encorafenib, binimetinib, and cetuximab compared with irinotecan or folfiri plus cetuximab (mos: 9.0 months vs. 5.4 months; hr: 0.52; 95% ci: 0.39 to 0.70; p < 0.0001) in the second or later lines of therapy35. Interestingly, the triplet did not appear more active than the combination of encorafenib and binimetinib (mos: 9.0 months vs. 8.4 months; hr: 0.79; 95% ci: 0.59 to 1.06). The magnitude of benefit seen in the the beacon trial resembled the estimated os of 9.1 months for dabrafenib–trametinib–panitumumab in another study36. Together, those trials support vertical inhibition of the mapk pathway as a therapeutic option for BRAF V600E–mutant mcrc.
Atypical BRAF Mutations
More than 200 non-V600E BRAF mutations or atypical BRAF mutations have been discovered, with a combined incidence ranging from 1.6% to 5.1%37–39. BRAF V600E mutations have been defined as class i mutations (active monomer); the atypical mutations are split into classes ii and iii. Class ii BRAF mutations are constitutively active dimers and have intermediate activity compared with their class i counterparts40. Class iii BRAF mutations result in a protein with a kinase domain having limited signal transduction activity, but it binds to craf and activates erk signalling in a ras-dependent manner41,42. Prognosis appears to be similar for patients with atypical BRAF mutations and those with wild-type BRAF mcrc43. However, the role of braf- and egfr-directed therapy is still being evaluated, although some studies suggest that those mutations might also show a reduced response to anti-egfr therapy44–47.
HER2 Amplification
Amplifications of her2 occur in 2%–6% of mcrc cases, but are enriched in KRAS/NRAS/BRAF wild-type mcrc, with a prevalence of approximately 13% in some series48–50. American Society of Clinical Oncology guidelines and the College of American Pathologists define her2 amplification as a her2:cep17 ratio of 2 or greater, or a copy number variant of 4 or greater51.
Amplifications of her2 appears to be a negative predictive marker for anti-egfr therapy, with mpfs being shorter regardless of line of therapy (2.8 months vs. 8.1–9.3 months depending on the study; hr: 7.05 to 10.66; p < 0.001)52–54. However, her2 amplification did not appear to impair the response to other non–anti-egfr therapies50. Dual-targeted anti-her2 therapy in heracles (trastuzumab–lapatinib) and MyPathway (pertuzumab–trastuzumab) both showed an orr of approximately 30%, with the pertuzumab–trastuzumab combination being associated with durable responses of 5.9 months in the second and subsequent lines of therapy when responses are often less than 5%49,55,56. Notably, anti-her2 therapies appear to be ineffective in patients with RAS mutations49. Those results have led to the prospective phase ii swog S1613 study comparing pertuzumab–trastuzumab with irinotecan–cetuximab in her2-amplified, RAS wild-type mcrc treated in the second or later line57. Additionally, investigation of neratinib–trastuzumab in a similar setting is ongoing (see NCT03457896 at https://ClinicalTrials.gov/).
Microsatellite Status
dMMR and MSI-H
Deficient mmr leads to nucleotide base insertion or deletion in dna regions with repetitive elements called “microsatellites,” resulting in a msi-h phenotype. The terms dmmr and msi-h are often used interchangeably, but dmmr refers to the missing proteins (mlh1, msh2, msh6, and pms2) that are usually detected by immunohistochemistry (ihc), and msi-h refers to the expanded microsatellites detected when 2 or more of 5 microsatellite loci are unstable in a polymerase chain reaction test58. Concordance of ihc staining for mmr protein loss with polymerase chain reaction measurement of msi-h is greater than 90%59. High msi can also arise from somatic hypermethylation of the MLH1 promotor, typically caused by aberrant methylation associated with the CpG island methylator phenotype, which might be associated with worse prognosis60. More recently, ngs panels used for detecting mutations have been modified to detect msi with high accuracy and represent an option for multiplexing biomarker tests61.
The prevalence of msi-h is 15%–20% in all crcs, but only 4% in mcrcs62. In mcrc, msi-h was previously associated with a worse prognosis, which could be driven partly by the fact that 20%–34.6% of msi-h crcs harbour BRAF V600E mutations63,64. Historically, the median os for mmr-intact mcrc was 17.9 months (95% ci: 16.2 months to 18.8 months) compared with 10.2 months (95% ci: 5.9 months to 19.8 months) for dmmr mcrc62. Recently, msi-h has shown importance in the metastatic setting because of its role as a tissue-agnostic biomarker for immunotherapy, regardless of germline or somatic origin65. Studies of monotherapy with checkpoint inhibitors have reported orrs of 31%–40%65,66. With combination nivolumab–ipilimumab, the orr increased to 55%, with 71% of patients being free from progression at 12 months, with only moderate increases in grade 3/4 toxicities (to 32% from 20%)64.
Microsatellite Stable mCRC and Immunotherapy
To date, immunotherapy has shown little activity in microsatellite-stable mcrc. Single-agent pembrolizumab and the combinations atezolizumab–cobimetinib (IMblaze 370) and 5-fluorouracil–atezolizumab–bevacizumab (modul) have all lacked activity in microsatellite-stable mcrc65,67,68. In contrast, the Canadian Cancer Trials Group co.26 trial showed that combination durvalumab–tremelimumab in microsatellite-stable mcrc was associated with a modest improvement in mos to 6.6 months from 4.1 months (unadjusted hr: 0.70; 90% ci: 0.53 to 0.92; p = 0.03)69.
In patients with non-hypermutated mcrc, PD-L1 has not been a useful biomarker to date66,67. In the Canadian Cancer Trials Group co.26 trial, retrospective analysis suggested that a high tumour mutational burden (≥28 mutations per megabase) from ctdna analysis might select patients (21% of the cohort) most likely to benefit from immunotherapy (hr: 0.34; 90% ci: 0.18 to 0.63; p = 0.004, with p = 0.07 for an interaction test of tumour mutational burden as a predictive biomarker)70. Comparatively, in calgb 80405, a tumour mutational burden of 8 or more mutations per megabase was a positive prognostic marker associated with improved mos (33.8 months vs. 28.1 months; hr: 0.73; 95% ci: 0.57 to 0.95; p = 0.020) in the firstline setting for patients receiving doublet chemotherapy and a biologic12,71. In several other studies, higher tumour mutational burden also appears to select patients with msi-h who are most likely to benefit from immunotherapy. However, the optimal threshold has yet to be defined, even in other cancer types (for example, lung cancer), where immunotherapy is more commonly used70,72,73.
NTRK Fusion
The NTRK genes encode 3 receptors—TrkA, TrkB, TrkC— recently identified as important partners in fusion events observed in multiple cancers that now have effective targeted therapies74. The prevalence of NTRK fusions is estimated to be 0.5%–2.0% in mcrc, but is enriched to 4% in msi-h mcrc22,74–76. NTRK1, 2, and 3 are normally involved in the development and function of the peripheral and central nervous system77–79. Targeted dna and rna panels, rna sequencing, florescence in situ hybridization (fish), and ihc are all options for detecting the fusions80,81. Larotrectinib and entrectinib are both U.S. Food and Drug Administration–approved Trk inhibitors with orrs of 75% and 57.4% respectively in more than 10 tumour types. Median duration of response to larotrectinib was not reached after 8.3 months of follow-up, with 55% of patients being free from progression at 1 year, and median duration of response to entrectinib was 10.4 months (95% ci: 7.1 months to not reached)82,83. The second-generation NTRK inhibitor LOXO-195 yielded an orr of 34% in 11 tumour types after patients had progressed on first-line Trk inhibitors and could represent a second-line option84.
PIK3CA Mutations
Activation of the epidermal growth factor receptor leads to signalling through both the mapk pathway and the pi3k pathway involved in cell survival and proliferation8. Intuitively, mutations in PIK3CA would result in resistance to anti-egfr agents, but mutations in exon 9 (68.4% of all PIK3CA mutations) have no effect on anti-egfr efficacy, and mutations in exon 20 (20.4% of PIK3CA mutations) have been associated with a lesser response to anti-egfr therapy only in select studies85. Exclusion of patients with PIK3CA mutations did not change the response to cetuximab in patients with KRAS/NRAS/BRAF wild-type tumours in calgb 8040512. Meta-analyses support the indeterminacy of an overall effect of PIK3CA mutation (regardless of exon), the interpretation of which is confounded by the fact that PIK3CA is often a co-mutation with others86,87. Currently, there is no clinical implication for PIK3CA mutations outside of a research context.
MET Alterations
The met receptor tyrosine kinase can be overexpressed, mutated, and amplified in mcrc, and its amplification is recognized as a potential mechanism of acquired resistance for mcrc treated with anti-egfr therapy88. However, multiple trials with various forms of met inhibition have been unsuccessful in mcrc89. Therefore, although genomic aberration in MET is commonly observed in mcrc, it remains in the research setting and has not been associated with innate resistance to anti-egfr therapy.
Consensus Molecular Subtyping
As defined by gene expression profiling, mcrc has been divided into 4 distinct consensus molecular subtypes (cmss). The 4 subtypes are cms1, with msi and immune activation (14%); cms2, with canonical crc alterations (37%); cms3, with metabolic dysregulation (13%); and cms4, with mesenchymal features (23%)90. Those subtypes reflect distinct biology and have been shown to be both prognostic for os and, in the calgb 80405 trial, predictive for benefit from cetuximab and bevacizumab91. Patients classified as cms1 appear to derive more benefit from bevacizumab, while those classified cms2 appear to derive more benefit from cetuximab (p for interaction: <0.001). Although many prospective trials using cms to stratify patients are planned, cms subtyping is not currently a standard of care and remains in the research environment.
UNDERSTANDING THE TESTS ORDERED
As the number of biomarkers of relevance discovered in mcrc increase, understanding the appropriate methods for identifying abnormalities becomes increasingly important to clinicians. In the second part of this review, we discuss the relevance of each technique in the era of precision oncology.
Conventional Laboratory Techniques
IHC
Widely available and low in cost, ihc is a method of detecting one or more specific antigens on a tissue by labelling them with an antibody against the whole protein or smaller epitopes. A secondary antibody is applied which is conjugated with either florescent or non-fluorescent markers (such as the brown 3,3′-diaminobenzidine stain commonly seen) that identify the antigen of interest under microscopy92. Immunohistochemistry remains an effective method for identifying dmmr and Lynch syndrome93. Similarly, the European Society for Medical Oncology recommends a 2-tier NTRK fusion detection algorithm with ihc population screening and confirmatory sequencing to reduce cost and handling time94. BRAF V600E mutation in melanoma can also be determined by ihc with high concordance. However, a high false-positive rate of 39% and a false-negative rate of 11% have been cited in the setting of mcrc95. Although other studies suggest better sensitivity and specificity, the conflicting evidence should caution against reliance on ihc alone for the detection of BRAF V600E mutations in mcrc96.
FISH
In the fish cytogenetic technique, florescent dna probes are hybridized to a sample, and the relative distribution of the probes determines the cytogenetic defect97. Increased numbers of fish probes can be used as confirmatory testing for her2 amplification when ihc shows intermediate staining98. Detection of fusions by fish (merging of two probes to give a different colour) is also a possibility, but can be labour-intensive if there are multiple potential fusion partners that are not known. In cases of fusions (such as NTRK, with its multiple fusion partners), targeted rna sequencing is therefore preferred to fish because of its high sensitivity, high throughput capability, and requirement for no prior knowledge of the fusion partners involved in the translocation80,81.
Sequencing Techniques
Sanger Sequencing
Sanger sequencing is a direct gene sequencing technique developed in 1977 to which all modern molecular testing assays are benchmarked. However, it has fallen out of favour because of newer technologies that allow for concurrent testing of multiple genes and greater sensitivity for low-allele-frequency mutations99. For example, single-gene companion diagnostic kits for KRAS use allele-specific polymerase chain reaction to increase assay sensitivity, allowing for detection of allele frequencies as low as 0.1%, compared with the 10% lower limit of detection with Sanger sequencing100. The enhanced sensitivity of newer testing assays results in the identification of up to 20% more patients with a RAS mutation, which has significant treatment implications101. The minimum detectable allele frequency is an important consideration. The crystal trial showed that RAS mutation frequency as low as 0.1% can predict the ineffectiveness of anti-egfr therapy102.
NGS
Next-generation sequencing refers to parallel sequencing reactions for multiple genes and multiple samples concurrently. An example of this technology uses fluorescence in added bases to detect the dna sequence. After dna purification and fragmentation, single-stranded dna is attached to the surface of a flow-cell channel. The single strands are amplified repeatedly to create dense clusters of template dna. Fluorescently labelled reversible terminator nucleotides, which can be detected by a camera, are added to the dna in subsequent cycles103. Other technologies use techniques such as pH changes when a nucleotide is added to provide sequence information.
NGS Sample Selection
When selecting which sample from a patient to use for sequencing, a few important considerations are necessary. In mcrc, high concordance for RAS and BRAF V600E mutation detection is observed between the primary tumour and metastases at various sites in the body104–106. Guidelines therefore suggest that testing a metastatic lesion is ideal; however, archival tissue can be used10. Fresh biopsies of recurrent tumours have the advantage of yielding higher-quality nucleic acids, particularly rna for transcriptomic analysis. Formalin-fixed, paraffin-embedded archival samples more than a few years old can introduce sequencing artefacts or errors107, but for most non-research biomarkers currently used, such archival samples are acceptable108.
Sequencing quality also depends on the ratio of tumour to normal tissue in a sample. If a particular nucleotide is sequenced 100 times and only 5% of the sample contains tumour cells, most of the measured dna will be from normal tissue, making cancer mutations hard to detect. Macrodissection by a pathologist to remove normal tissue under the microscope can help with that challenge. With such dissection, less normal dna remains to be sequenced, and so more of the sequencing reads are attributed to cancer dna.
Depth of Coverage
Another important metric in ngs is the concept of depth of coverage. “Depth of coverage” is defined as the number of times a particular nucleotide is sequenced; it is often denoted using a number that represents the average or median of the depth, followed by the letter x (that is, “400×”). With more depth, the variant detected is, importantly, more likely to be real rather than an artefact, and low-allele-frequency mutations are more likely to be detected, although other technical issues can affect sensitivity.
Panel Selection
Selecting the sequencing panel to use can be a challenge for clinicians and patients. At most academic institutions, individual companion diagnostic tests have been replaced by either a “home brew” ngs panel designed to meet local needs or a commercially available ngs panel such as FoundationOne CDx [324 genes (Hoffmann–La Roche, Mississauga, ON)] or Caris Molecular Intelligence [592 genes (Caris Life Sciences, Irving, TX, U.S.A.)]. For routine mcrc management, only a very targeted ngs panel is essential, but to balance the research interests of the institution and the ability to use one panel for multiple types of cancer, larger panels are often performed at a marginal increase in cost (for better economy of scale).
RNA Sequencing
Sequencing of rna involves extraction of rna, enrichment for messenger rna, and subsequent reverse transcription to create complementary dna that is then sequenced using ngs technologies just as dna would be. Other technologies such as array-based platforms are available for targeted assessment of gene expression. Apart from its previously mentioned role in fusion detection, rna sequencing can characterize the differences in gene expression that define the molecular subtypes of mcrc. Several iterations of those molecular subtypes have been presented; the cms version appears to be of greatest utility, but it remains in the research realm90. Sequencing of rna usually provides information about gene expression, but it can also be used to identify various types of mutations beyond fusions, such as single-nucleotide variants. However, because of sample degradation and artefacts, sequencing of rna can be more challenging to perform than conventional dna sequencing in archived formalin-fixed tissues.
Utility of Liquid Biopsy
When discussing liquid biopsies, the term “cell-free dna” refers to all dna found in the plasma, of which ctdna is the subset of tumour origin only. Typically, ctdna consists of short dna fragments approximately 166 bp long, with an estimated half-life of 16 minutes to 2.5 hours109. In treatment-naïve patients with mcrc, diagnostic molecular profiling of ctdna appears to have high concordance with tissue-based assays110. Although up to 15% of samples can be nondiagnostic because of a lack of detectable ctdna, the same problem can occur in tissue sequencing110,111. Detection of ctdna has also shown value in the risk-stratification of patients at risk of recurrence. In one study, the hr for stage ii patients with detectable ctdna after resection was 18 (95% ci: 7.9 to 40.0; p < 0.001)112. Amplifications can also be detected using ctdna. In the heracles trial, ctdna successfully detected amplifications in 46 of 48 patients with her2-amplified disease, and detection of higher copy numbers was associated with orr and pfs113. Some ctdna assays are also validated for detection of msi109.
SUMMARY
Increased appreciation of molecular subtypes beyond KRAS exon 2–mutant crc has refined the management of patients with mcrc. With the development of patient-friendly technologies such as ctdna, which allows for noninvasive molecular assessment, the integration of biomarkers can be expected to become more integral to every decision made in the clinic and will further improve outcomes for patients.
ACKNOWLEDGMENTS
The authors acknowledge Ryan Nini for assistance with graphics design.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare the following interests: JML has received fees as a consultant from Amgen, Taiho, Bayer, Novartis, and Ipsen, and has also received research support from Ipsen. MKCL has no conflicts of interest to disclose.
REFERENCES
- 1.de Gramont A, Figer A, Seymour M, et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol. 2000;18:2938–47. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- 2.Douillard JY, Cunningham D, Roth AD, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041–7. doi: 10.1016/S0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 3.Tournigand C, André T, Achille E, et al. folfiri followed by folfox6 or the reverse sequence in advanced colorectal cancer: a randomized gercor study. J Clin Oncol. 2004;22:229–37. doi: 10.1200/JCO.2004.05.113. [DOI] [PubMed] [Google Scholar]
- 4.Seymour MT, Maughan TS, Ledermann JA, et al. Different strategies of sequential and combination chemotherapy for patients with poor prognosis advanced colorectal cancer (mrc focus): a randomised controlled trial. Lancet. 2007;370:143–52. doi: 10.1016/S0140-6736(07)61087-3. [DOI] [PubMed] [Google Scholar]
- 5.Koopman M, Antonini NF, Douma J, et al. Sequential versus combination chemotherapy with capecitabine, irinotecan, and oxaliplatin in advanced colorectal cancer (cairo): a phase iii randomised controlled trial. Lancet. 2007;370:135–42. doi: 10.1016/S0140-6736(07)61086-1. [DOI] [PubMed] [Google Scholar]
- 6.Landre T, Uzzan B, Nicolas P, et al. Doublet chemotherapy vs. single-agent therapy with 5fu in elderly patients with metastatic colorectal cancer. a meta-analysis. Int J Colorectal Dis. 2015;30:1305–10. doi: 10.1007/s00384-015-2296-5. [DOI] [PubMed] [Google Scholar]
- 7.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 8.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–61. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, Siena S, Cassidy J, et al. Final results from prime: randomized phase iii study of panitumumab with folfox4 for first-line treatment of metastatic colorectal cancer. Ann Oncol. 2014;25:1346–55. doi: 10.1093/annonc/mdu141. [DOI] [PubMed] [Google Scholar]
- 10.Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35:1453–86. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 11.Douillard JY, Oliner KS, Siena S, et al. Panitumumab–folfox4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 12.Innocenti F, Ou FS, Qu X, et al. Mutational analysis of patients with colorectal cancer in calgb/swog 80405 identifies new roles of microsatellite instability and tumor mutational burden for patient outcome. J Clin Oncol. 2019;37:1217–27. doi: 10.1200/JCO.18.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cercek A, Braghiroli MI, Chou JF, et al. Clinical features and outcomes of patients with colorectal cancers harboring NRAS mutations. Clin Cancer Res. 2017;23:4753–60. doi: 10.1158/1078-0432.CCR-17-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Loree JM, Yu C, et al. Distinct impacts of KRAS, NRAS and BRAF mutations on survival of patients with metastatic colorectal cancer [abstract 3513] J Clin Oncol. 2018;36 [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2018.36.15_suppl.3513; cited 1 September 2019] [Google Scholar]
- 15.Venook AP, Niedzwiecki D, Innocenti F, et al. Impact of primary (1°) tumor location on overall survival (os) and progression-free survival (pfs) in patients (pts) with metastatic colorectal cancer (mcrc): analysis of calgb/swog 80405 (Alliance) [abstract 3504] J Clin Oncol. 2016;34 doi: 10.1200/JCO.2016.34.15_suppl.3504. [Available online at: https://ascopubs.org/doi/10.1200/JCO.2016.34.15_suppl.3504; cited 1 September 2019] [DOI] [Google Scholar]
- 16.Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and egfr directed antibodies in six randomized trials. Ann Oncol. 2017;28:1713–29. doi: 10.1093/annonc/mdx175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: a meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. doi: 10.1016/j.ejca.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Boeckx N, Koukakis R, Op de Beeck K, et al. Primary tumor sidedness has an impact on prognosis and treatment outcome in metastatic colorectal cancer: results from two randomized first-line panitumumab studies. Ann Oncol. 2017;28:1862–8. doi: 10.1093/annonc/mdx119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrahao ABK, Karim S, Colwell B, Berry S, Biagi J. The predictive effect of primary tumour location in the treatment of metastatic colorectal cancer: a Canadian consensus statement. Curr Oncol. 2017;24:390–400. doi: 10.3747/co.24.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Comprehensive Cancer Network (nccn) NCCN Clinical Practice Guidelines in Oncology: Colon Cancer. Ver. 2.2019. Fort Washington, PA: nccn; 2019. [Current version available online at: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf (free registration required); cited 1 September 2019. [Google Scholar]
- 21.Price T, Kim TW, Li J, et al. Final results and outcomes by prior bevacizumab exposure, skin toxicity, and hypomagnesaemia from aspecct: randomized phase 3 non-inferiority study of panitumumab versus cetuximab in chemorefractory wild-type KRAS exon 2 metastatic colorectal cancer. Eur J Cancer. 2016;68:51–9. doi: 10.1016/j.ejca.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Pietrantonio F, Di Nicolantonio F, Schrock AB, et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djx089. [DOI] [PubMed] [Google Scholar]
- 23.Brulé SY, Jonker DJ, Karapetis CS, et al. Location of colon cancer (right-sided versus left-sided) as a prognostic factor and a predictor of benefit from cetuximab in ncic co.17. Eur J Cancer. 2015;51:1405–14. doi: 10.1016/j.ejca.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 24.Parseghian CM, Loree JM, Morris VK, et al. Anti-egfr–resistant clones decay exponentially after progression: implications for anti-egfr re-challenge. Ann Oncol. 2018;30:243–9. doi: 10.1093/annonc/mdy509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cremolini C, Rossini D, Dell’Aquila E, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol. 2019;5:343–50. doi: 10.1001/jamaoncol.2018.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer KE, Pritchard CA. raf proteins and cancer: B-RAF is identified as a mutational target. Biochim Biophys Acta. 2003;1653:25–40. doi: 10.1016/s0304-419x(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 27.Roskoski R., Jr raf protein–serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399:313–17. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 28.Loree JM, Dowers A, Tu D, et al. Expanded RAS and BRAF V600 testing as predictive biomarkers for single agent cetuximab in the randomized phase iii co.17 trial [abstract 537] J Clin Oncol. 2019;37 doi: 10.1200/JCO.2019.37.4_suppl.537. [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.4_suppl.537; cited 1 September 2019] [DOI] [Google Scholar]
- 29.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loupakis F, Cremolini C, Salvatore L, et al. folfoxiri plus bevacizumab as first-line treatment in BRAF mutant metastatic colorectal cancer. Eur J Cancer. 2014;50:57–63. doi: 10.1016/j.ejca.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Cremolini C, Antoniotti C, Lonardi S, et al. Primary tumor sidedness and benefit from folfoxiri plus bevacizumab as initial therapy for metastatic colorectal cancer. Retrospective analysis of the tribe trial by gono. Ann Oncol. 2018;29:1528–34. doi: 10.1093/annonc/mdy424.021. [DOI] [PubMed] [Google Scholar]
- 32.Cremolini C, Antoniotti C, Lonardi S, et al. Updated results of tribe2, a phase iii, randomized strategy study by gono in the first- and second-line treatment of unresectable mcrc [abstract 3508] J Clin Oncol. 2019;37 doi: 10.1200/JCO.2019.37.15_suppl.3508. [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.15_suppl.3508; cited 1 September 2019] [DOI] [Google Scholar]
- 33.Kopetz S, Desai J, Chan E, et al. Phase ii pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol. 2015;33:4032–8. doi: 10.1200/JCO.2015.63.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopetz S, McDonough SL, Lenz HJ, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF- mutant metastatic colorectal cancer (swog S1406) [abstract 3503] J Clin Oncol. 2017;35 doi: 10.1200/JCO.20.01994. [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2017.35.15_suppl.3505; cited 1 September 2019] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopetz S, Grothey A, Van Cutsem E, et al. beacon crc: a randomized, 3-arm, phase 3 study of encorafenib and cetuximab with or without binimetinib vs. choice of either irinotecan or folfiri plus cetuximab in BRAF V600E–mutant metastatic colorectal cancer [abstract LBA-006] Ann Oncol. 2019;30 doi: 10.1093/annonc/mdz183.004. [Available online at: https://academic.oup.com/annonc/article/30/Supplement_4/mdz183.004/5526665; cited 1 September 2019] [DOI] [Google Scholar]
- 36.Corcoran RB, André T, Atreya CE, et al. Combined braf, egfr, and mek inhibition in patients with BRAF V600E–mutant colorectal cancer. Cancer Discov. 2018;8:428–43. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao Z, Torres NM, Tao A, et al. BRAF mutants evade erk-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015;28:370–83. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 39.Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33:125–36.e3. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the raf–erk signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 41.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead braf and oncogenic ras cooperate to drive tumor progression through craf. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant b-raf activate c-raf through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–9. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 43.Jones JC, Renfro LA, Al-Shamsi HO, et al. Non-V600 BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J Clin Oncol. 2017;35:2624–30. doi: 10.1200/JCO.2016.71.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated ras. Nature. 2017;548:234–8. doi: 10.1038/nature23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinozaki E, Yoshino T, Yamazaki K, et al. Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-egfr monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for Anti-EGFR Monoclonal Antibodies by Comprehensive Cancer Genomics (breac) study. Br J Cancer. 2017;117:1450–8. doi: 10.1038/bjc.2017.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dankner M. Targeted therapy for colorectal cancers with non-V600 BRAF mutations: perspectives for precision oncology. JCO Precision Oncol. 2018;2:1–12. doi: 10.1200/PO.18.00195. [Available online at: https://ascopubs.org/doi/10.1200/PO.18.00195; cited 1 September 2019] [DOI] [PubMed] [Google Scholar]
- 47.Johnson B, Loree JM, Jacome AA, et al. Atypical, non-V600 BRAF mutations as a potential mechanism of resistance to egfr inhibition in metastatic colorectal cancer. JCO Precision Oncol. 2019:1–10. doi: 10.1200/PO.19.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross JS, Fakih M, Ali SM, et al. Targeting her2 in colorectal cancer: the landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer. 2018;124:1358–73. doi: 10.1002/cncr.31125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meric-Bernstam F, Hurwitz H, Raghav KPS, et al. Pertuzumab plus trastuzumab for her2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019;20:518–30. doi: 10.1016/S1470-2045(18)30904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raghav K, Loree JM, Morris JS, et al. Validation of HER2 amplification as a predictive biomarker for anti–epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. JCO Precision Oncol. 2019:1–13. doi: 10.1200/PO.18.00226. [DOI] [PubMed] [Google Scholar]
- 51.Bartley AN, Washington MK, Ventura CB, et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American Society for Clinical Pathology, and American Society of Clinical Oncology. Am J Clin Pathol. 2016;146:647–69. doi: 10.1093/ajcp/aqw206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonesaka K, Zejnullahu K, Okamoto I, et al. Activation of erbb2 signaling causes resistance to the egfr-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies her2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–23. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 54.Martin V, Landi L, Molinari F, et al. HER2 gene copy number status may influence clinical efficacy to anti-egfr monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer. 2013;108:668–75. doi: 10.1038/bjc.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnold D, Prager GW, Quintela A, et al. Beyond second-line therapy in patients with metastatic colorectal cancer: a systematic review. Ann Oncol. 2018;29:835–56. doi: 10.1093/annonc/mdy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siena S, Sartore-Bianchi A, Trusolino L, et al. Final results of the heracles trial in her2- amplified colorectal cancer [abstract CT005] Cancer Res. 2017;77(suppl) [Available online at: https://cancerres.aacrjournals.org/content/77/13_Supplement/CT005; cited 1 September 2019] [Google Scholar]
- 57.Raghav KPS, McDonough SL, Tan BR, et al. A randomized phase ii study of trastuzumab and pertuzumab (tp) compared to cetuximab and irinotecan (cetiri) in advanced/metastatic colorectal cancer (mcrc) with her2 amplification: S1613 [abstract TPS3620] J Clin Oncol. 2018;36 [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2018.36.15_suppl.TPS3620; cited 1 September 2019] [Google Scholar]
- 58.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043–8. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 60.Advani SM, Advani P, DeSantis SM, et al. Clinical, pathological, and molecular characteristics of CpG island methylator phenotype in colorectal cancer: a systematic review and meta-analysis. Transl Oncol. 2018;11:1188–201. doi: 10.1016/j.tranon.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nowak JA, Yurgelun MB, Bruce JL, et al. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn. 2017;19:84–91. doi: 10.1016/j.jmoldx.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koopman M, Kortman GAM, Mekenkamp L, et al. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266–73. doi: 10.1038/sj.bjc.6604867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venderbosch S, Nagtegaal ID, Maughan TS, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the cairo, cairo2, coin, and focus studies. Clin Cancer Res. 2014;20:5322–30. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in dna mismatch repair–deficient/microsatellite instability–high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–9. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 65.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic dna mismatch repair–deficient or microsatellite instability–high colorectal cancer (Check-Mate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–91. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eng C, Kim TW, Bendell J, et al. on behalf of the IMblaze370 investigators. Atezolizumab with or without cobimetinib versus regorafenib in previously treated metastatic colorectal cancer (IMblaze370): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2019;20:849–61. doi: 10.1016/S1470-2045(19)30027-0. [DOI] [PubMed] [Google Scholar]
- 68.Grothey A, Tabernero J, Arnold D, et al. Fluoropyrimidine (fp) + bevacizumab (bev) + atezolizumab vs. fp/bev in BRAF wt metastatic colorectal cancer (mcrc): findings from cohort 2 of modul—a multicentre, randomized trial of biomarker-driven maintenance treatment following first-line induction therapy [abstract LBA19] Ann Oncol. 2018;29(suppl) doi: 10.1093/annonc/mdy424.020. [Available online at: https://academic.oup.com/annonc/article/29/suppl_8/mdy424.020/5141615; cited 1 September 2019] [DOI] [Google Scholar]
- 69.Chen EX, Jonker DJ, Kennecke HF, et al. cctg co.26 trial: a phase ii randomized study of durvalumab (d) plus tremelimumab (t) and best supportive care (bsc) versus bsc alone in patients (pts) with advanced refractory colorectal carcinoma (rcrc) [abstract 481] J Clin Oncol. 2019;37 [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.4_suppl.481; cited 1 September 2019] [Google Scholar]
- 70.Chen EX, Jonker DJ, Loree JM, et al. cctg co.26: updated analysis and impact of plasma-detected microsatellite stability (mss) and tumor mutation burden (tmb) in a phase ii trial of durvalumab (d) plus tremelimumab (t) and best supportive care (bsc) versus bsc alone in patients (pts) with refractory metastatic colorectal carcinoma (rmcrc) [abstract 3512] J Clin Oncol. 2019;37 [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.15_suppl.3512; cited 1 September 2019] [Google Scholar]
- 71.Innocenti F, Ou FS, Zemla T, et al. Somatic dna mutations, msi status, mutational load (ml): association with overall survival (os) in patients (pts) with metastatic colorectal cancer (mcrc) of calgb/swog 80405 (Alliance) [abstract 3504] J Clin Oncol. 2017;35 doi: 10.1200/JCO.2017.35.15_suppl.3504. [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2017.35.15_suppl.3504; cited 1 September 2019] [DOI] [Google Scholar]
- 72.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fabrizio DA, George TJ, Jr, Dunne RF, et al. Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J Gastrointest Oncol. 2018;9:610–17. doi: 10.21037/jgo.2018.05.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kheder ES, Hong DS. Emerging targeted therapy for tumors with NTRK fusion proteins. Clin Cancer Res. 2018;24:5807–14. doi: 10.1158/1078-0432.CCR-18-1156. [DOI] [PubMed] [Google Scholar]
- 75.Ardini E, Bosotti R, Borgia AL, et al. The TPM3–NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TrkA kinase inhibition. Mol Oncol. 2014;8:1495–507. doi: 10.1016/j.molonc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sartore-Bianchi A, Amatu A, Bonazzina E, et al. Pooled analysis of clinical outcome of patients with chemorefractory metastatic colorectal cancer treated within phase i/ii clinical studies based on individual biomarkers of susceptibility: a single-institution experience. Target Oncol. 2017;12:525–33. doi: 10.1007/s11523-017-0505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Arévalo JC, Wu SH. Neurotrophin signaling: many exciting surprises! Cell Mol Life Sci. 2006;63:1523–37. doi: 10.1007/s00018-006-6010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169:107–14. doi: 10.1016/S0304-3835(01)00530-4. [DOI] [PubMed] [Google Scholar]
- 79.Rubin JB, Segal RA. Growth, survival and migration: the Trk to cancer. Cancer Treat Res. 2003;115:1–18. doi: 10.1007/0-306-48158-8_1. [DOI] [PubMed] [Google Scholar]
- 80.Reeser JW, Martin D, Miya J, et al. Validation of a targeted rna sequencing assay for kinase fusion detection in solid tumors. J Mol Diagn. 2017;19:682–96. doi: 10.1016/j.jmoldx.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion–positive cancers in adults and children. N Engl J Med. 2018;378:731–9. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Demetri GD, Paz-Ares L, Farago AF, et al. Efficacy and safety of entrectinib in patients with NTRK fusion-positive (NTRK- fp) tumors: pooled analysis of startrk-2, startrk-1 and alka-372-001 [abstract LBA17] Ann Oncol. 2018;29 [Available online at: https://academic.oup.com/annonc/article/29/suppl_8/mdy424.017/5141583; cited 1 September 2019] [Google Scholar]
- 84.Hyman D, Kummar S, Farago AF, et al. Phase i and expanded access experience of LOXO-195 (BAY 2731954), a selective next-generation Trk inhibitor (Trki) [abstract CT127] Cancer Res. 2019;79 [Available online at: https://cancerres.aacrjournals.org/content/79/13_Supplement/CT127.short?rss=1; cited 1 September 2019] [Google Scholar]
- 85.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 86.Mei ZB, Duan CY, Li CB, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2016;27:1836–48. doi: 10.1093/annonc/mdw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-egfr monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 2011;23:1518–25. doi: 10.1093/annonc/mdr464. [DOI] [PubMed] [Google Scholar]
- 88.Bardelli A, Corso S, Bertotti A, et al. Amplification of the met receptor drives resistance to anti-egfr therapies in colorectal cancer. Cancer Discov. 2013;3:658–73. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raghav K, Bailey AM, Loree JM, et al. Untying the Gordian knot of targeting met in cancer. Cancer Treat Rev. 2018;66:95–103. doi: 10.1016/j.ctrv.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 90.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lenz HJ, Ou FS, Venook AP, et al. Impact of consensus molecular subtyping (cms) on overall survival (os) and progression free survival (pfs) in patients (pts) with metastatic colorectal cancer (mcrc): analysis of calgb/swog 80405 (Alliance) [abstract 3511] J Clin Oncol. 2017;35 [Available online at: https://ascopubs.org/doi/10.1200/JCO.2017.35.15_suppl.3511; cited 1 September 2019] [Google Scholar]
- 92.Oien KA. Pathologic evaluation of unknown primary cancer. Semin Oncol. 2009;36:8–37. doi: 10.1053/j.seminoncol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 93.Luchini C, Bibeau F, Ligtenberg MJL, et al. esmo recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review–based approach. Ann Oncol. 2019 doi: 10.1093/annonc/mdz116. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 94.Marchiò C, Scaltriti M, Ladanyi M, et al. esmo recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol. 2019 doi: 10.1093/annonc/mdz204. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 95.Estrella JS, Tetzlaff MT, Bassett RL, Jr, et al. Assessment of BRAF V600E status in colorectal carcinoma: tissue-specific discordances between immunohistochemistry and sequencing. Mol Cancer Ther. 2015;14:2887–95. doi: 10.1158/1535-7163.MCT-15-0615. [DOI] [PubMed] [Google Scholar]
- 96.Bledsoe JR, Kamionek M, Mino-Kenudson M. BRAF V600E immunohistochemistry is reliable in primary and metastatic colorectal carcinoma regardless of treatment status and shows high intratumoral homogeneity. Am J Surg Pathol. 2014;38:1418–28. doi: 10.1097/PAS.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genetics. 2005;6:782–92. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 98.Valtorta E, Martino C, Sartore-Bianchi A, et al. Assessment of a her2 scoring system for colorectal cancer: results from a validation study. Mod Pathol. 2015;28:1481–91. doi: 10.1038/modpathol.2015.98. [DOI] [PubMed] [Google Scholar]
- 99.Sanger F, Nicklen S, Coulson AR. dna sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Loree JM, Kopetz S, Raghav KPS. Current companion diagnostics in advanced colorectal cancer; getting a bigger and better piece of the pie. J Gastrointest Oncol. 2017;8:199–212. doi: 10.21037/jgo.2017.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Efrati E, Elkin H, Peerless Y, Sabo E, Ben-Izhak O, Hershkovitz D. lna-based pcr clamping enrichment assay for the identification of KRAS mutations. Cancer Biomark. 2010;8:89–94. doi: 10.3233/CBM-2011-0203. [DOI] [PubMed] [Google Scholar]
- 102.Van Cutsem E, Lenz HJ, Köhne CH, et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33:692–700. doi: 10.1200/JCO.2014.59.4812. [DOI] [PubMed] [Google Scholar]
- 103.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mariani P, Lae M, Degeorges A, et al. Concordant analysis of KRAS status in primary colon carcinoma and matched metastasis. Anticancer Res. 2010;30:4229–35. [PubMed] [Google Scholar]
- 105.Vignot S, Lefebvre C, Frampton GM, et al. Comparative analysis of primary tumour and matched metastases in colorectal cancer patients: evaluation of concordance between genomic and transcriptional profiles. Eur J Cancer. 2015;51:791–9. doi: 10.1016/j.ejca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 106.Miglio U, Mezzapelle R, Paganotti A, et al. Mutation analysis of KRAS in primary colorectal cancer and matched metastases by means of highly sensitivity molecular assay. Pathol Res Pract. 2013;209:233–6. doi: 10.1016/j.prp.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 107.Do H, Dobrovic A. Sequence artifacts in dna from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61:64–71. doi: 10.1373/clinchem.2014.223040. [DOI] [PubMed] [Google Scholar]
- 108.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating rna sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17:257–71. doi: 10.1038/nrg.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour dna. Nat Rev Cancer. 2017;17:223–38. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 110.Strickler JH, Loree JM, Ahronian LG, et al. Genomic landscape of cell-free dna in patients with colorectal cancer. Cancer Discov. 2018;8:164–73. doi: 10.1158/2159-8290.CD-17-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol. 2015;33:2753–62. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor dna analysis detects minimal residual disease and predicts recurrence in patients with stage ii colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Siravegna G, Sartore-Bianchi A, Nagy RJ, et al. Plasma her2/erbb2 copy number predicts response to her2-targeted therapy in metastatic colorectal cancer. Clin Cancer Res. 2019;25:3046–53. doi: 10.1158/1078-0432.CCR-18-3389. [DOI] [PubMed] [Google Scholar]